Abstract
The genome-wide expression patterns of circular RNAs (circRNAs) are of increasing interest for their potential roles in normal cellular homeostasis, development, and disease. Thousands of circRNAs have been annotated from various species in recent years. Analysis of publically available or user-generated rRNA-depleted total RNA-seq data can be performed to uncover new circRNA expression trends. Here we provide a primer for profiling circRNAs from RNA-seq datasets. The description is tailored for the wet lab scientist with limited or no experience in analyzing RNA-seq data. We begin by describing how to access and interpret circRNA annotations. Next, we cover converting circRNA annotations into junction sequences that are used as scaffolds to align RNA-seq reads. Lastly, we visit quantifying circRNA expression trends from the alignment data.
Keywords: circRNA, Circular RNAs, Expression analysis, Ribo-depleted total RNA-seq
1. Introduction
Detection of circRNAs in RNA-seq data is limited to stranded, ribosomal RNA (rRNA/ribo)-depleted total RNA-seq, as opposed to library preparation proo9tocols requiring enrichment of polyadenylated RNAs. Total RNA-seq libraries are becoming increasingly common since they detect a variety of RNAs, including both mRNAs and non-polyadenylated RNAs. Over the past few years, thousands of circRNAs have been annotated in humans, mice, fish, insects, plants, and yeast from total RNA-seq data, using algorithms designed to detect “out-of-order” splicing [1–12]. These analysis pipelines include find_circ [2], circRNA_finder [8], CIRCexplorer [13], and CIRI [14], among o thers [15, 16]. Most of these algorithms and analysis pipelines can detect exonic, intronic, and intergenic circRNAs, whereas CIRCfinder is a pipeline designed to exclusively detect intronic circRNAs [17]. Despite thousands of circRNA annotations, novel circRNAs continue to be annotated, and the user should consider that specific tissues or cell types of interest might express novel circRNAs that necessitate the use of one of the previously mentioned circRNA detection algorithms.
It is generally required that the total RNA-seq datasets to be analyzed for circRNA expression trends are of very high depth, due to only circRNA junction spanning reads being used in the analysis. CircRNA junction spanning reads typically comprise less than 0.1% of the reads generated in a total RNA-seq experiment [5].
Here, we provide a primer for profiling circRNAs from RNAseq datasets using existing circRNA annotations. Table 1 provides a reference for reports and online databases containing circRNA annotations from various organisms. In this example, a circRNA annotation set is used to generate a circRNA junction scaffold Bowtie2 aligner index. Reads that align to the circRNA junction scaffold are summed using featureCounts and the number of reads that align to each circRNA junction is normalized to the read depth of the library. This normalization allows the direct comparison of circRNA expression from different conditions so that differentially expressed circRNAs can be identified.
Table 1.
circRNA annotations
| Annotations in reports | |||
|---|---|---|---|
| Organism | Genome annotation | Conditions | References |
| Mus musculus | mm9 | Brain; synaptoneurosomes; liver; heart; embryonic stem cell neuronal diff.; P19 cell neuronal diff.; primary neuron diff. | [2–5] |
| Homo sapiens | hg19 | Brain; SH-SY5Y cell neuronal diff.; HEK293 cells; CD34+ cells; CD19+ cells; neutrophils; Hs68 cells; DLD-1 cells; DKO-1 cells, DKs-8 cells | [1, 2, 4, 32, 33] |
| Drosophila melanogaster | dm3 | Heads; S2 cells | [3, 8] |
| Caenorhabditis elegans | ce6 | Various tissues and developmental stages | [2, 34] |
| Annotations in online databases | |||
|---|---|---|---|
| Database name | Web address | Description | References |
| circBase | www.circbase.org | circRNAs from various tissues, cell types, and developmental stages from human, mouse, fruit fly, C. elegans, and others | [35] |
| circRNAdb | http://reprod.njmu.edu.cn/circrnadb | Human circRNAs by cell type, proteincoding potential, gene symbol and PubMed ID | [36] |
| CircNet | http://circnet.mbc.nctu.edu.tw. | Human tissue-specific circRNA expression information and miRNA binding potential | [37] |
| Tissue-specific circRNA database | http://gb.whu.edu.cn/TSCD | Human and mouse circRNA from various tissues | [38] |
2. Materials
Computer running Linux or Mac OSX.
UNIX shell (e.g., bash).
- Installed open-access software:
- Bedtools [18]: github.com/arq5x/bedtools2/releases.
- Bowtie2 [19]: bowtie-bio.sourceforge.net/bowtie2/index.shtml.
- featureCounts [22]: subread.sourceforge.net.
rRNA-depleted total RNA-seq data in FASTQ format (user generated or downloaded from repository).
Genome sequence for species of interest in FASTA format.
circRNA annotations for species of interest in BED6 format.
Optional: SRAtools: github.com/ncbi/sra-tools.
3. Methods
3.1. Generate circRNA Junction Sequences
-
View contents of a circRNA annotation BED file. Many cir cRNA annotations are provided as a list of genomic coordinates or as Browser Extensible Data (BED) records that describe the genomic coordinates and other information (https://genome.ucsc.edu/FAQ/FAQformat#format1). The BED format includes a chromosome (chrom), a 0-based start (chromStart) position in base pair units, a stop position (chromStop), a feature name, a score, and strand information (Fig. 1a). The score can provide information about the expression level of a particular circRNA; however it is often assigned a “0” value. The strand assignment provides information about whether the circRNA is encoded from the sense or antisense DNA strand. Figure 1a illustrates a BED file (circRNA.bed) that contains annotations for two circRNAs (circ_1 and circ_2). The BED annotation is used to generate junction sequences circ_1 (Fig. 1b) and circ_2 (Fig. 1c). Both circ_1 and circ_2 are 20 nucleotides (nts) in length, and have the same genomic coordinates; however, circ_1 is encoded on the sense DNA strand and circ_2 is encoded on the antisense DNA strand to highlight how strand orientation impacts the directionality and sequence of circRNAs. CircRNAs encoded on the sense strand of DNA are assigned a “+” in the sixth “strand” field (Fig. 1a: circ_1), while those encoded on the antisense DNA strand are assigned a “−” (Fig. 1a: circ_2). Genes encoded on the antisense DNA strand run 3´ to 5´ in relation to the sense strand; therefore the chromStop field represents the “start” of the gene, and the chromStart field represents the “stop” of the gene. Because circ_2 is encoded on the antisense DNA strand, it is reverse complemented in relation to circ_1 (Fig. 1c). Note that 10 nt junction sequences for circ_1 and circ_2 join the last 5 nts with the first 5 nts of the circularizing exon and run in the 5´–3´ direction (Fig. 1b, c; junction sequence).
For this primer on circRNA expression profiling, we used the first four records in the mouse cortex annotation from Supplemental Table 2 in Gruner et al. (2016) [5], to make cortex_circRNA.bed. These four BED records will be used to generate circRNA junction sequences of 200 nt length in the manner illustrated in Fig. 1.# View contents of cortex_circRNA.bed $ more cortex_circRNA.bed chrX 58436422 58439349 CDR1as 0 + chr1 154691165 154691537 mm9_circ_000025 0 chr1 16430403 16453275 mm9_circ_000040 0 - chr18 51462263 51463167 mm9_circ_000042 0 +
- Split the annotations into circRNAs encoded from the sense “+” strand and circRNAs encoded from the antisense “−”strand. Write circRNAs encoded on the “+” strand to a file named “sense. cortex_circRNA.bed” and write circRNAs encoded on the “−” strand to a file named “antisense.cortex_circRNA.bed”.
$ awk ‘($6 == “+”)’ cortex_circRNA.bed > sense.cortex_circRNA.bed $ awk ‘($6 == “-”)’ cortex_circRNA.bed > antisense.cortex_circRNA.bed
-
Modify the BED start and stop coordinates to represent half the length of the desired junction scaffold sequence for the “left” and “right” sides of the circRNA junction (Fig. 1b, c). For this example, we generate a junction of 200 nts in length to accommodate RNA-seq reads of 125 nts in length [5]. Using these conditions, an end-to-end Bowtie2 alignment would at minimum require at least a 25-nt overlap of the circRNA junction. RNA-seq data generated from the Illumina platform will generally range in length from 50 to 150 nts. When using real RNA-seq data, consider the junction overlap, and choose a junction scaffold length that will accommodate the read length of your RNA-seq data.
The desired junction scaffold length for this example is 200 nts in length; therefore, the start and stop coordinates reported in the modified BED annotation for the “left” (Fig. 1b, c; blue arrows) and “right” (Fig. 1b, c; red arrows) sides of the junction will be 100 nts in length (half of the total junction scaffold length). The 100-nt “left” and “right” sequences will be joined to form the out-of-order circRNA junction scaffold sequence similar to the junctions depicted in Fig. 1b, c. For circRNAs encoded on the “+” strand, the “chromStop” position is used to generate the “left” side of the circRNA junction by subtracting 100 nts from the chromStop position, and then writing a new BED record to a file called “l_junc.sense.cortex_circRNA. bed” to represent the 100-nt left half of the junction. For the right side of the junction, 100 nts are added to the chromStart position, and the BED record is written to a file called “r_junc. sense.cortex_circRNA.bed” to represent the 100-nt right half of the junction. For circRNAs encoded on the “−” strand, the “chromStart” position is used to generate the “left” side of the circRNA junction by adding 100 nts to the chromStart position, and then writing the BED record to a file called “l_junc.antisense.cortex_circRNA.bed” to represent the 100-nt left half of the junction. For the right side of the junction, 100 nts are subtracted from the chromStop position, and the BED record is written to a file called “r_junc.antisense.cortex_circRNA.bed” to represent the 100-nt right side of the junction. There are some limitations with capturing circRNAs that are smaller than the desired circRNA junction scaffold length (see Note 1):$ awk ‘OFS=“\t” {$2=$3–100} {print $0}’ sense. cortex_circRNA.bed \ > l_junc.sense.cortex_circRNA.bed $ awk ‘OFS=“\t” {$3=$2+100} {print $0}’ sense. cortex_circRNA.bed \ > r_junc.sense.cortex_circRNA.bed $ awk ‘OFS=“\t” {$3=$2+100} {print $0}’ antisense.cortex_circRNA.bed \> l_junc.antisense.cortex_circRNA.bed $ awk ‘OFS=“\t” {$2=$3–100} {print $0}’ antisense.cortex_circRNA.bed \> r_junc.antisense.cortex_circRNA.bed -
Extract the “left” and “right” 100-nt sequences that will form the circRNA junction with the bedtools “getfasta” subcommand. Both a BED file (generated in the previous step) and a FASTA file containing the appropriate genome sequence are required to build the junction scaffold with the bedtools “getfasta” subcommand. Ensure that the annotations in the BED and genome FASTA file have the same chromosome name. iGenomes and Ensembl annotate genome FASTA chromosomes as a number only, whereas UCSC genome browser includes a “chr” prefix prior to the chromosome number (e.g., 1 vs. chr1). FASTA files of the genome or different genomes of interest organized by chromosome or contigs can be downloaded directly from UCSC Genome Browser: http://hgdownload.cse.ucsc.edu/downloads.html.
Ensembl: ftp://ftp.ensembl.org/pub/release-87/fasta.Illumina iGenomes: support.illumina.com/sequencing/sequencing_software/igenome.html. # Extract sequences for “left” and “right” sides of circRNA junctions for circRNAs encoded on “+” strand. $ bedtools getfasta -fi mm9.genome.fa -bed l_junc.sense.cortex_circRNA.bed \ -s -name -tab -fo l_junc.sense.cortex_circRNA.seq $ bedtools getfasta -fi mm9.genome.fa -bed r_junc.sense.cortex_circRNA.bed \-s -name -tab -fo r_junc.sense.cortex_circRNA.seq # Extract sequences for “left” and “right” sides of circRNA junctions for circRNAs encoded on the “-” strand. $ bedtools getfasta -fi mm9.genome.fa -bed l_junc.antisense.cortex_circRNA.bed \ -s -name -tab -fo l_junc.antisense.cortex_circRNA.seq $ bedtools getfasta -fi mm9.genome.fa -bed r_junc.antisense.cortex_circRNA.bed \ -s -name -tab -fo r_junc.antisense.cortex_circRNA.seq
- Combine the “left” and “right” sequences to form the 200-nt junction scaffold sequence in FASTA format. The FASTA output for the circRNA junctions is shown in Fig. 2.
$ paste l_junc.sense.cortex_circRNA.seq r_junc.sense.cortex_circRNA.seq \ | awk ‘{print “>“$1”\n”$2$4}’ > circRNA_junctions.fa # Append “-” strand circRNA junction sequence to “circRNA_junction.fa” $ paste l_junc.antisense.cortex_circRNA.seq r_junc.antisense.cortex_circRNA.seq \| awk ‘{print”>“$1”\n”$2$4}’ >> circRNA_junctions.fa
Fig. 1.
circRNA junction formation from BED annotations. (a) Representative BED file, circRNA.bed, a tabdelimited file that reports the chromosomal location and features of circRNAs. (b) Model of a 3-exon proteincoding gene with exon 2 encoding a circRNA. circ_1 maps to chr1 from nt 10 to 30. The downstream “left” end of the exon (at nt 30) joins with the upstream “right” end of the exon to form the circRNA junction. (c) circ_2 is encoded on the antisense DNA strand, but has the same start and stop coordinates as circ_1. The upstream “left” side of the exon joins with the downstream “right” side of the exon to form the circRNA junction. Because circ_2 is encoded on the antisense DNA strand, the sequence is reverse-complemented relative to circ_1. FASTA-formatted junction sequences (Subheading 3.1, step 4) are used to build the aligner index (Subheading 3.2, step 1) necessary for alignment of RNA-seq data to circRNA junctions (Subheading 3.2)
Fig. 2.
circRNA junction FASTA sequences. The resulting 200 nt circRNA junction sequences in FASTA format after joining the 100-nt “left” and “right” sequences (Subheading 3.1, step 5)
3.2. Align RNA-seq Reads to the circRNA Junction Scaffold
- Build the Bowtie2 aligner index using the circRNA junction scaffold. Six index files with the extension .bt2 are generated by bowtie2-build.
$ bowtie2-build circRNA_junctions.fa circRNA_junctions
-
Obtain RNA-seq datasets in FASTQ format if user-generated RNA-seq data is not available. Ensure that the library strategy used to generate the RNA-seq data is rRNA-depleted total RNA-seq. Publicly available RNA-seq datasets are available on ENCODE: www.encodeproject.org
NCBI Gene Expression Omnibus (GEO): http://www.ncbi.nlm.nih.gov/geo
NCBI Sequence Record Archive (SRA): http://www.ncbi.nlm.nih.gov/sra
SRAtools (https://github.com/ncbi/sra-tools) is needed to extract FASTQ data from SRA files downloaded from GEO and SRA. Obtain at least two RNA-seq datasets from different conditions to compare changes in circRNA expression. For the purposes of this example, we use the SRA prefetch tool to obtain total RNA/rRNA-depleted RNA-seq datasets deposited on the SRA for two conditions: one replicate each for 1-month-old (SRR4280863) mouse cortex and 22-month-old (SRR4280956) mouse cortex [5]. Once the SRA files have been downloaded, the SRA tool fastq-dump is used to extract the FASTQ files from the SRA files. Since these RNA-seq datasets are paired end, the read 1 data are contained in the “_1.fastq.gz” file and the read 2 data are contained in the “_2.fastq.gz” file. Both the read 1 and read 2 files will have the same number of records, and each record will have the same read ID (Fig. 3). The quality of the datasets can be assessed using FastQC (see Note 2):#use the prefetch tool from the SRA toolkit to download datasets from the SRA #The downloaded files will have a .sra extension $ prefetch SRR4280863 $ prefetch SRR4280956 #extract FASTQ data from .sra files and split into read1 and read2 using the SRA tool fastq-dump $ fastq-dump --split-files -F --gzip SRR4280863 $ fastq-dump --split-files -F --gzip SRR4280956 #rename datasets $ mv SRR4280863_1.fastq.gz 1m-cortex.R1.fastq.gz $ mv SRR4280863_2.fastq.gz 1m-cortex.R2.fastq.gz $ mv SRR4280956_1.fastq.gz 22m-cortex.R1.fastq.gz $ mv SRR4280956_2.fastq.gz 22m-cortex.R2.fastq.gz
- Align FASTQ datasets to the junction scaffold using Bowtie2. In this example, we use a minimum alignment score of −15 regardless of read length. Stringency of alignment output can be altered by various parameters in Bowtie2. These parameters are covered in the Bowtie2 manual (http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml). By default, Bowtie2 dumps its output to standard out (stdout). However, in this example, stdout is piped to samtools view to write a Binary Alignment Mapping (BAM) output file that includes only mapped reads sorted by the reference sequence and leftmost alignment position. BAM files are compressed binary versions of the Sequence Alignment Mapping (SAM) file. Checking the SAM/BAM alignment reports is critical to ensuring accurate alignment to the circRNA junctions (see Note 3). The circRNA alignment information provided in the SAM/BAM file is covered in Fig. 4 for the first aligned read reported in 1m-cortex.bam:
$ bowtie2 --score-min=C,−15,0 -x circRNA_junctions \ −1 1m-cortex.R1.fastq.gz −2 1m-cortex.R2.fastq.gz \ | samtools view -ShuF4 - \ | samtools sort - -m 8G 1m-cortex #repeat alignment for second condition
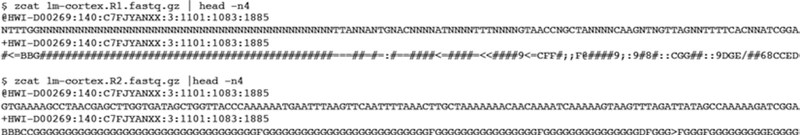
Fig. 3.
FASTQ records in read 1 and read 2 RNA-seq datasets. Each sequence record in the FASTQ file is composed of four lines. The first line always begins with an “@” character, followed by the read ID (in this example: HWI-D00269:140:C7FJYANXX:3:1101:1083:1885). The second line of FASTQ record is the read sequence, the third line is a “+” character with or without other read information, and the fourth line contains the sequencing qualities reported for each base. The RNA-seq FASTQ datasets for read 1 (1m-cortex.R1.fastq.gz) and read 2 (1m-cortex.R2.fastq.gz) contain the same number of records and the same read IDs
Fig. 4.
1m-cortex.bam output from Bowtie2 alignment for first mapped read. The meaning of fields in SAM/BAM output reports is described at samtools.github.io/hts-specs/. In 1m-cortex.bam, generated from the Bowtie2 alignment (Subheading 3.2, step 3), the first reported read is assigned a FLAG value of 163. This FLAG value indicates that the read is properly paired, is the second in the pair, and its mate is reverse complemented relative to the reference. The output also indicates that the read aligns to the CDR1as circRNA junction scaffold reference starting at nt 1 (field 4). The mapping quality (MAPQ) field value is 42, and the entire 125-nt (125M) RNA-seq read aligned to the CDR1as junction. An “=“ character is assigned to field 7 to indicate that the mate read aligns to the same reference sequence. Field 8 reports that the mate read aligns at position 29 of the reference sequence. The inferred fragment length of 153 nts from the read1 and read2 alignment is reported in field 9. The sequence and the quality scores for the read are reported in fields 10 and 11, and the alignment score is reported in field 12. A perfect alignment in Bowtie2 end-to-end mode will produce a score of 0, while various alignment penalties (e.g., mismatches, insertions, gaps) will report negative scores
3.3. Count Reads that Align to circRNA Junctions Using Featurecounts
- Generate a Gene Transfer Format (GTF) file using the cortex_ circRNA.bed file that contains the circRNA name information. The circRNA GTF output is shown in Fig. 5.
$ awk ‘(OFS=“\t”) \ {print $4, “Gruner_2016”, “junction”, “1”, “200”, “.”, “+”, “.”, “circID \”“$4”\”“} \ ‘ cortex_circRNA.bed > circRNA.gtf - Run featureCounts using the BAM files and the newly generated circRNA GTF file as input to count the reads that aligned to each circRNA junction. Multiple BAM files representing replicates and different conditions can be fed into feature-Counts simultaneously. The feature-Counts output is an easyto-interpret data frame that includes the read count for each circRNA for each condition (Fig. 6):
$ featureCounts -C -T 4 -t junction -g circID -a circRNA.gtf -o circRNA.counts.txt \ 1m-cortex.bam 22m-cortex.bam
Fig. 5.
circRNA GTF for featureCounts. The GTF generated in 3.3.1 provides (1) the circRNA name, (2) the source of data, (3) a feature, (4) the start position of the feature (1-based), (5) the end position, (6) the score (can be replaced with a “.”), (7) the strand, (8) the frame (can be replaced with a “.”), and (9) an attribute, which is a semicolon-separated list of tag-value pairs
Fig. 6.
FeatureCounts output. Output generated from featureCounts (Subheading 3.3, step 2) provides the count of reads for each circRNA feature. For example, 29 individual reads aligned to CDR1as circRNA junction in the 1m-cortex sample versus 45 individual reads in the 22m-cortex sample
3.4. Normalize circRNA Counts to Compare circRNA Expression Across Conditions
- Determine total library size for normalization using transcripts per million (TPM).TPM is one approach for library normalization [5]. For this normalization approach, total read number generated per library is required. For paired-end RNA-seq libraries, this includes the sum of raw FASTQ records from both the read 1 (.R1.fastq.gz) and read 2 (.R2.fastq.gz) RNAseq files. In this example, 1m-cortex.R1.fastq.gz includes 51,789,102 records, and 1m-cortex.R2.fastq.gz includes 51,789,102 records; thus, the total size of library is 103,678,204 reads. Because each FASTQ record is made up of four lines, the total number of lines in the RNA-seq FASTQ data file divided by four can be used to determine the total library size. In addition, the output log of Bowtie2 reports the total number of reads processed:
$ zcat 1m-cortex.R1.fastq.gz | wc -l 207156408 #Number of reads in Read1 FASTQ file: 207156408/4 = 51789102 reads $ zcat 1m-cortex.R2.fastq.gz | wc -l 207156408 #Number of reads in Read2 FASTQ file: 207156408/4 = 51789102 reads
- Normalize counts to TPM. Divide the number of reads that aligned to each circRNA by total library size. The result is multiplied by 1,000,000, and then divided by the scaling factor. The scaling factor is the length of the circRNA junction in kilobases (kb). The junction scaffold used for this example was 200 nts in length; thus the scaling factor is 0.2 kb:
$ awk ‘(NR > 2)’ circRNA.counts.txt \ | awk -v lib_size_1m=103678204 -v lib_size_22m=102442864 ‘OFS=“\t” \ {TPM_1m=((( $7/lib_size_1m)*10^6)/0.2)} \ {TPM_22m=((( $8/lib_size_22m)*10^6)/0.2)} \ {print $1, TPM_1m, TPM_22m}’ \ > circRNA_TPM.txt #view normalized data $ cat circRNA_TPM.txt CDR1as 1.39856 2.19635 mm9_circ_000025 0.530488 1.02496 mm9_circ_000040 0.241131 0.0488077 mm9_circ_000042 0.192905 0.780923 Quantify circRNA expression trends.Now that the circRNA counts are normalized, the different libraries/conditions can be directly compared. In our example, libraries of cortex RNA from 1-month-old mice have 1.4 TPM for CDR1as versus 2.2 TPM from 22-month-old mice. From here, fold change can be calculated to determine fluxes in circRNA expression (e.g., a 1.6-fold increase in CDR1as circRNA from 1 m to 22 m). The R base package is useful for performing statistical tests, and can be used in addition to the ggplot2 library for plotting expression changes in circRNAs [25, 26]. Expression trends of circRNAs observed with RNA-seq data should be validated using experimental methods such as RT-qPCR and Northern blot analysis. Additional chapters within this book detail these methodologies to study circRNAs (see Note 4).
4. Notes
A caveat of this method for the generation of junction is that it does not take into account the size of the circRNA. If a circRNA is derived from a single exon that is smaller than the size of the desired junction sequence length, the above script will grab flanking non-circRNA sequence, like introns or intergenic sequence. For circRNAs smaller than the desired junction, options include (1) generating a shorter junction sequence or (2) maintaining the desired length of the junction sequence by concatenating the circRNA sequence. A rough circRNA length can be determined by subtracting the chromStart position from the chromStop position in the BED record.
The quality of RNA-seq libraries can be assessed with fastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Among the quality measures of fastQC is sequence duplication, or repetitive reads due to PCR duplicates. Although the impact of PCR duplicates on RNA expression is debatable [27–29], PCR duplicates may impact circRNA profiling due to the low frequency of circRNA junction-specific reads. Picard MarkDuplicates (https://github.com/ broadinstitute/picard) or samtools rmdup can be used to remove PCR duplicates from alignment BAM files. FastQC also reports adapter contamination in the RNA-seq reads. Adapter contamination occurs if the cDNA insert is shorter than the desired read length, and the adapter sequence or a portion of the adapter sequence is included in the read output. Reads with considerable adapter sequence will not align to the circRNA junction scaffold in Bowtie2 end-to-end mode, so trimming adapter sequence can potentially enhance alignment to the circRNA junction scaffold. An option for trimming adapter sequence is Cutadapt [30]; however, adapter-trimmed sequences will result in variable read length, and filtering the alignments to require circRNA junction overlap will be necessary.
Spot check the junction sequences by loading circRNA BED annotations on IGV [23, 24] or UCSC genome browser [31] to ensure that the proper junction is accurately captured. The alignment BAM files can also be loaded onto IGV with the junction scaffold FASTA file (as the genome). A BAM index file (bam.bai) is required for each BAM file loaded onto IGV. To generate the index file, use samtools “index”. Visualizing the reads aligned onto the circRNA scaffold through IGV can help the user spot low mapping quality and other systemic errors.
As there are a huge number of predicted circRNAs, very few annotations have been validated by independent methods such as Sanger sequencing of circRNA RT-PCR products, RNase R treatment, or Northern blot techniques (see additional chapters in the book). It is important for the user to validate circRNA expression trends using independent molecular techniques.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences grant P20 GM103650 and National Institute on Aging grant R15 AG052931. We would also like to thank Matthew Bauer and David Knupp for critical review of this chapter.
References
- 1.Jeck WR, Sorrentino JA, Wang K et al. (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157. https://doi.org/10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Memczak S, Jens M, Elefsinioti A et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. https://doi.org/10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 3.Ashwal-Fluss R, Meyer M, Pamudurti NR et al. (2014) CircRNA biogenesis c ompetes with pre-mRNA splicing. Mol Cell 56:55–66. doi.org/10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 4.Rybak-Wolf A, Stottmeister C, Glazar P et al. (2014) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885. doi.org/10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 5.Gruner H, Cortés-López M, Cooper DA et al. (2016) CircRNA accumulation in the aging mouse brain. Sci Rep 6:38907 https://doi.org/10.1038/srep38907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang PL, Bao Y, Yee M-C et al. (2014) Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9:e90859 doi.org/10.1371/journal.pone.0090859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen Y, Guo X, Wang W (2017) Identification and characterization of circular RNAs in zebrafish. FEBS Lett 591:213–220. doi.org/10.1002/1873-3468.12500 [DOI] [PubMed] [Google Scholar]
- 8.Westholm JO, Miura P, Olson S et al. (2014) Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9:1966–1980. https://doi.org/10.1016/j.celrep.2014.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun X, Wang L, Ding J et al. (2016) Integrative analysis of Arabidopsis thaliana transcriptomics reveals intuitive splicing mechanism for circular RNA. FEBS Lett 590:3510–3516. https://doi.org/10.1002/1873-3468.12440 [DOI] [PubMed] [Google Scholar]
- 10.Ye C-Y, Chen L, Liu C et al. (2015) Widespread noncoding circular RNAs in plants. New Phytol 208:88–95. https://doi.org/10.1111/nph.13585 [DOI] [PubMed] [Google Scholar]
- 11.Nitsche A, Doose G, Tafer H et al. (2014) Atypical RNAs in the coelacanth transcriptome. J Exp Zool Part B Mol Dev Evol 322:342–351. https://doi.org/10.1002/jez.b.22542 [DOI] [PubMed] [Google Scholar]
- 12.Salzman J, Chen RE, Olsen MN et al. (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9:e1003777 doi.org/10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X-O, Wang H-B, Zhang Y et al. (2014) Complementary sequence-mediated exon circularization. Cell 159:134–147. doi.org/10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 14.Gao Y, Wang J, Zhao F (2015) CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 16:4 https://doi.org/10.1186/s13059-014-0571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szabo L, Morey R, Palpant NJ et al. (2015) Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16:126 https://doi.org/10.1186/s13059-015-0690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song X, Zhang N, Han P et al. (2016) Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 44:e87 https://doi.org/10.1093/nar/gkw075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Zhang XO, Chen T et al. (2013) Circular intronic long noncoding RNAs. Mol Cell 51:792–806. https://doi.org/10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 18.Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi.org/10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie2. Nat Methods 9:357–360. https://doi.org/10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. https://doi.org/10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao Y, Smyth GK, Shi W (2014) FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 23.Thorvaldsdóttir H, Robinson JT, Mesirov JP (2012) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. https://doi.org/10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson J, Thorvaldsdóttir H, Winckler W (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. https://doi.org/10.1038/nbt0111-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Core Team R (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org [Google Scholar]
- 26.Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer-Verlag, New York [Google Scholar]
- 27.Kivioja T, Vähärautio A, Karlsson K et al. (2012) Counting absolute numbers of molecules using unique molecular identifiers Nat Methods 9:72–74. https://doi.org/10.1038/nmeth.1778 [DOI] [PubMed] [Google Scholar]
- 28.Ebbert MTW, Wadsworth ME, Staley LA et al. (2016) Evaluating the necessity of PCR duplicate removal from next-generation sequencing data and a comparison of approaches. BMC Bioinformatics 17:239 https://doi.org/10.1186/s12859-016-1097-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parekh S, Ziegenhain C, Vieth B et al. (2016) The impact of amplification on differential expression analyses by RNA-seq. Sci Rep 6:25533 https://doi.org/10.1038/srep25533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. 10.14806/ej.17.1.200 [Google Scholar]
- 31.Kent WJ, Sugnet CW, Furey TS et al. (2002) the human genome browser at UCSC. Genome Res 12:996–1006. https://doi.org/10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salzman J, Gawad C, Wang PL et al. (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7:e30733 https://doi.org/10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou Y, Cha DJ, Franklin JL et al. (2016) Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep 6:37982 doi.org/10.1038/srep37982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanov A, Memczak S, Wyler E et al. (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10:170–177. https://doi.org/10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 35.Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20:1666–1670. https://doi.org/10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Han P, Zhou T et al. (2016) cir- RNAs in the human and mouse genomes. Brief cRNADb: a comprehensive database for Bioinform:1–9. https://doi.org/10.1093 human circular RNAs with protein-coding bib/bbw081 [Google Scholar]
- 37.Liu Y-C, Li J- R, Sun C-H et al. (2016) CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 44:D209–D215. https://doi.org/10.1093/nar/gkv940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia S, Feng J, Lei L et al. (2016) Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform:1–9. https://doi.org/10.1093/bib/bbw081 [DOI] [PubMed] [Google Scholar]