Abstract
Background
Identification of the Mycobacterium tuberculosis immunoproteome and antigens associated with serologic responses in adults has renewed interest in developing a serologic test for childhood tuberculosis (TB). We investigated IgG antibody responses against M. tuberculosis antigens in children with well-characterized TB.
Methods
We studied archived sera obtained from hospitalized children with suspected pulmonary TB, and classified as having confirmed TB (culture- confirmed), unlikely TB (clinical improvement without TB treatment), or unconfirmed TB (all others). A multiplexed bead-based assay for IgG antibodies against 119 M. tuberculosis antigens was developed, validated and used to test sera. The area under the curves (AUCs) of the empiric receiver–operator characteristic curves were generated as measures of predictive ability. A cross-validated generalized linear model was used to select the most predictive combinations of antigens.
Results
For the confirmed TB versus unlikely TB comparison, the maximal single antigen AUC was 0.63, corresponding to sensitivity 0.60 and specificity 0.60. Older (age: 60+ months old) children’s responses were better predictive of TB status than younger (age: 12–59 months old) children’s, with a maximal single antigen AUC of −0.76. For the confirmed TB versus unlikely TB groups, the most predictive combinations of antigens assigned TB risk probabilities of 0.33 and 0.33, respectively, when all ages were considered, and 0.57 (interquartile range: 0.48–0.64) and 0.35 (interquartile range: 0.32–0.40) when only older children were considered.
Conclusion
An antigen-based IgG test is unlikely to meet the performance characteristics required of a TB detection test applicable to all age groups.
Keywords: tuberculosis, pulmonary, diagnosis, serologic tests, biomarkers, antibodies
Childhood tuberculosis (TB) is estimated to account for approximately 10%–20% of the global TB disease burden.1 Most pediatric TB disease occurs in low- or middle-income countries where timely diagnosis may be especially challenging because of resource constraints, lack of capacity to perform microbiologic investigations and a focus on adult disease in TB programs.2 Microbiologic diagnosis remains the gold standard, but its widespread application is limited by the difficulties in obtaining good-quality respiratory specimens especially from very young children, cost and availability of laboratory facilities and expertise.3 Even in settings in which etiologic investigation is possible, up to 70% of children treated for pulmonary TB (PTB) are still diagnosed clinically using a combination of history, symptoms, tuberculin skin testing and chest radiography4,5 as microbiologic testing is often negative because of the paucibacillary nature of childhood TB.6,7 In children, detection of serum antibodies therefore represents an attractive diagnostic approach, focusing on host responses to Mycobacterium tuberculosis rather than attempting to detect bacteria or bacterial products. Furthermore, blood specimens can be obtained relatively easily.
Historically, serologic tests for TB have typically used 1 or 2 antigens or crude mixtures of components and products of mycobacteria. Recent meta-analyses of performance of commercially available and in-house tests for serodiagnosis of TB in adults and children8,9 concluded that test performance varied widely and that none of the tests performed well enough to be used routinely. The World Health Organization has issued a strong recommendation against the use of available serologic tests for TB diagnosis and has advocated for research aimed at developing improved assays.
The advent of proteomic approaches and the recognition of the heterogeneity of antibody responses have spawned interest in the identification of antibody signatures, that is, patterns of reactivity to a number of M. tuberculosis antigens, whose abundance and presence correlate with disease state.10–13 In previous work, high-density arrays of M. tuberculosis–recombinant protein antigens were used to identify sets of M. tuberculosis antigens whose seroreactivity could serve as a biomarker panel for TB diagnosis. This whole proteome approach combined with sera from adults with known TB status allowed for definition of the M. tuberculosis immunoproteome,13 comprised of 484 proteins that were recognized by serum from at least 1 adult patient with active TB disease. Furthermore, antibody reactivity against a subset of 13 proteins within the immunoproteome was associated with active TB in adults. The aim of our study was to identify antibody responses to M. tuberculosis protein antigens that are predictive of TB status in children 1–15 years of age. Considering that antibody responses vary by age, we also aimed to study the predictive power of these responses in the older versus younger children.
MATERIALS AND METHODS
Participants
Sera were obtained from children with suspected PTB hospitalized at Red Cross War Memorial Children’s Hospital in Cape Town, South Africa, and enrolled in a prospective study to evaluate new methods for diagnosis of PTB in children.6 Suspected PTB was defined based on cough or difficulty breathing, plus one or more of the following: household contact with an infectious TB source case within the preceding 3 months, loss of weight or failure to gain weight in the preceding 3 months, a positive (induration ≥10 mm) tuberculin skin test (TST) using purified protein derivative (PPD; 2TU, PPD RT23; Staten Serum Institute, Denmark, Copenhagen) and a chest radiograph suggestive of PTB. Children were excluded if they had received more than 72 hours of TB treatment or prophylaxis. All children had received Bacillus Calmette Guerin (BCG) vaccination at birth as provided in the National Immunisation Program. A history and physical examination were performed. Routine clinical investigations included chest radiography and TST, plus HIV testing in children whose HIV status was unknown. Two consecutive induced sputum specimens were obtained and submitted to the study laboratory for smear microscopy, Xpert MTB/RIF (Cepheid, Sunnyvale, CA) testing, and liquid culture. Standard TB therapy was initiated at the discretion of the treating doctor based on clinical, radiologic and microbiologic information.
Definition of TB Status
For study purposes, each child was classified using clinical and microbiologic criteria, according to the following definitions: “confirmed TB” (any culture or nucleic acid amplification test positive for M. tuberculosis); “unlikely TB,” that is, non-TB respiratory disease (culture-negative for M. tuberculosis, no TB treatment given and documented resolution or improvement of symptoms and signs at month 3 follow-up visit) or “unconfirmed TB” (all other children), consistent with the revised National Institutes of Health consensus definitions for diagnostic research.14 The unlikely TB group was further divided according to positive or negative TST result.
Blood Sample Collection, Processing and Storage
Blood specimens were collected at enrollment through venipuncture into a serum separator tube. Serum was separated (centrifuged at 1000g for 10 minutes at room temperature) within 2 hours of collection and immediately frozen in aliquots at −80°C.
Selection of Serum Specimens
Serum specimens were eligible for inclusion in this study if the child was 12–15 years old and HIV-negative at the time of serum collection, informed consent for use of stored samples had been provided, and there were complete data on microbiologic and clinical features. A sample size of 200 children was projected to provide adequate power to discriminate between TB groups. Consecutive sera were selected such that 40 (20%) were from children classified as confirmed TB, 80 (40%) were from children classified as unconfirmed TB, and 80 (40%) were from children classified as unlikely TB (39 TST-positive and 41 TST-negative). Specimens were transported on dry ice to the Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany, for serologic testing.
Ethics Approvals
Written informed consent for enrollment in the prospective study, including for use of stored samples, had been obtained from a parent or legal guardian, and assent had been obtained in children older than 7 years of age. Ethics committees of the Faculty of Health Sciences, University of Cape Town and Johns Hopkins University approved the study.
Antigen Selection
Antigens were selected from the whole proteome screen on the basis of receiver–operator characteristic curves from adult studies, random forest analysis and availability.13,15 We also studied additional antigens identified by collaborators as being of interest based on on-going vaccine work and/or their inclusion in other studies of antibody reactivity.16 There were 74 unique antigens, among which 21 were available from different producers, tested in the same multiplex. Thus, in total, the assay contained 119 antigens (including recombinant proteins and antigen cocktails, subsequently called “antigens”) (Table S1, Supplemental Digital Content 1, http://links.lww.com/INF/C768). The multiplex, bead-based Luminex assay was used to measure IgG antibodies. This assay was developed, validated and performed on study specimens as described in Ref.17 and the online data supplement (Supplemental Digital Content 1, http://links.lww.com/INF/C768). For each study sample, a single measurement in 1 dilution was performed.
Data Analysis and Sample Size
The aim of this analysis was to identify sets of antibody responses that were predictive of TB status. The area under the curve (AUC) and the best antibody response cut-off that maximized sensitivity and specificity for distinguishing between TB groups were generated for each of the antibody responses. This was done following the approach by Lopez-Raton et al18 implemented in the R package optimal.cutpoints. Pearson correlations plot of the antibody responses that had AUC >0.5 was generated. An optimal subset of antibody responses, among those that had AUC >0.5 derived from optimal.cutpoints, was selected using the cross-validated generalized linear model regularization path algorithm of Friedman et al.19 This uses the elastic net penalty for including predictors in the model, and it is implemented in the R package cv.glmnet.20 For each individual child, the model was used to provide an in-sample prediction of the probability of TB status and the box plots of these are presented. Further details of the implementation of cv.glmnet are provided in the online data supplement (Supplemental Digital Content 1, http://links.lww.com/INF/C768). These analyses were applied to determine predictive sets of antibody responses that discriminated between confirmed TB and unlikely TB groups. The age effect on the predictive power of antibody responses was investigated for the confirmed TB versus unlikely TB comparison, stratifying by 12–59 and ≥60 months of age. The empiric AUCs were also calculated to determine antibody responses that discriminated between the following pairs of TB groups: (1) confirmed TB versus unlikely TB/TST-positive; (2) confirmed TB versus unlikely TB/TST-negative and (3) confirmed TB versus unconfirmed TB. We estimated that a minimum of approximately 40 samples in the confirmed TB group was required to detect a significant reactivity difference between the confirmed TB group and the unlikely TB group assuming 60 peptides, a maximum of 0.05 false positives, a desired 0.35 reactivity difference, power of 85%, SD of 0.6 and a ratio of 2:1 for number of unlikely TB samples-to-confirmed TB samples.
RESULTS
Characteristics of study participants at enrollment are shown in Table 1. Median age was 41 months [interquartile range (IQR): 24–66 months), and 97 (49%) were male. The most prevalent symptoms and signs across groups were cough or other respiratory symptoms (69%–90%) and weight loss (63%–70%). There were no significant differences in the prevalence or duration of the assessed symptoms or signs among the 3 TB classification groups.
TABLE 1.
Characteristics of Study Participants at Enrollment (Time of Serum Collection)
| Characteristics | All, N = 200 |
Confirmed TB, N = 40 |
Unconfirmed TB, N = 80 |
Unlikely TB, N = 80 | |
|---|---|---|---|---|---|
|
| |||||
| TST-Negative, N = 41 |
TST-Positive, N = 39 |
||||
| Demographics | |||||
| Median age (months) (IQR) | 41 (24–66) | 50 (27–91) | 42 (26–66) | 22 (17–41) | 49 (31–61) |
| Age range (months) | 12–152 | 16–147 | 12–143 | 12–152 | 13–150 |
| Age 12–60 (months) | 136 (68) | 23 (58) | 52 (65) | 33 (80) | 28 (72) |
| Age ≥60 (months) | 64 (32) | 17(42) | 28(35) | 8 (20) | 11 (28) |
| Male, N (%) | 97 (49) | 21 (53) | 39 (49) | 21 (51) | 16 (41) |
| Household TB contact*, N (%) | 135 (69) | 24 (60) | 53 (69) | 22 (54) | 36 (92) |
| TB symptoms and signs | |||||
| Cough or other respiratory symptoms, N (%) | 160 (80) | 31 (78) | 61 (76) | 38 (93) | 30 (77) |
| Median cough duration (days) (IQR) | 14 (7–29) | 14 (7–14) | 14 (6–30) | 14 (3–30) | 14 (14–21) |
| Fever, N (%) | 105 (53) | 21 (53) | 49 (61) | 21 (51) | 14 (36) |
| Median fever duration (days) (IQR) | 7 (3–14) | 14 (7–14) | 5 (3–7) | 5 (3–14) | 11 (7–14) |
| Night sweats, N (%) | 116 (58) | 22 (55) | 51 (64) | 20 (49) | 23 (59) |
| Median night sweats duration (days) (IQR) | 14 (7–30) | 14 (7–28) | 14 (7–30) | 26 (7–60) | 14 (14–30) |
| Weight loss, N (%) | 128 (64) | 28 (70) | 50 (63) | 31 (76) | 19 (49) |
| Median days of weight loss (IQR) | 30 (14–31) | 30 (14–60) | 30 (14–31) | 30 (14–45) | 30 (18–31) |
| Results of diagnostic testing, N (%) | |||||
| TST positive† | 134 (68) | 28 (76) | 67 (84) | 0 (0) | 39 (100) |
| Culture-positive for Mycobacterium tuberculosis from respiratory sample | 40 (20) | 40 (100) | 0 (0) | 0 (0) | 0 (0) |
| Xpert MTB/RIF positive for M. tuberculosis from respiratory sample | 27 (14) | 23 (58) | 4 (5) | 0 (0) | 0 (0) |
| Chest radiograph suggestive of pulmonary TB‡ | 39 (40) | 12 (31) | 15 (23) | 4 (11) | 3 (8) |
N = 197 (3 recorded as not known).
N = 197 (TST not administered or not read for 3 participants).
N = 178.
Antibody Responses That Discriminate Between Confirmed TB Versus Unlikely TB
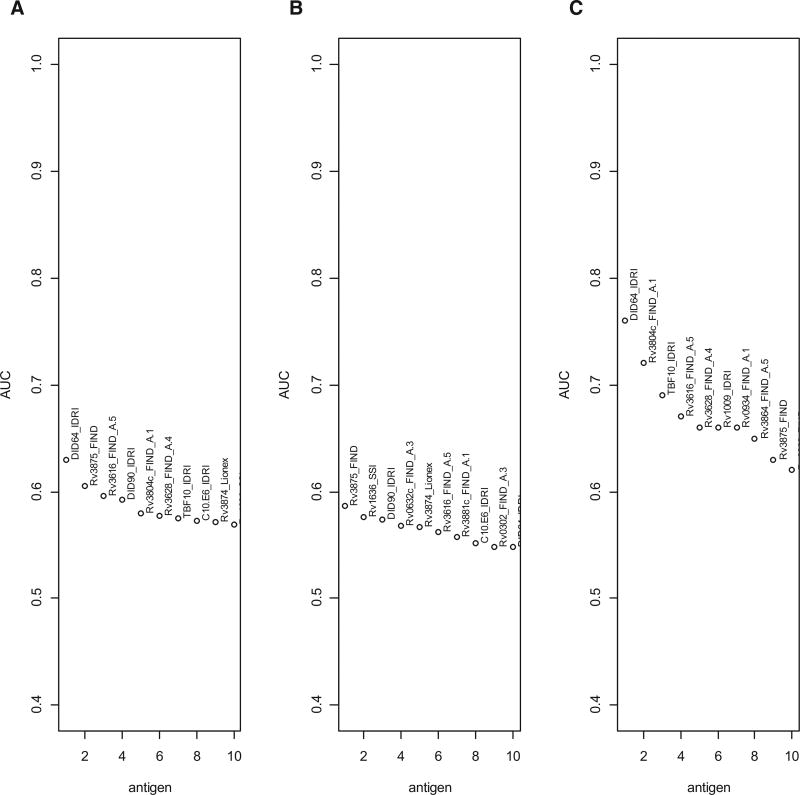
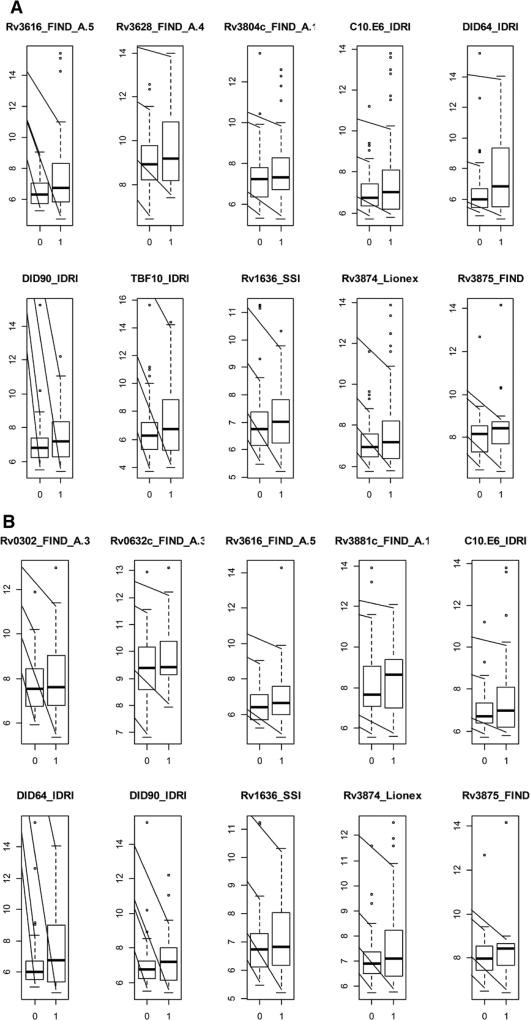
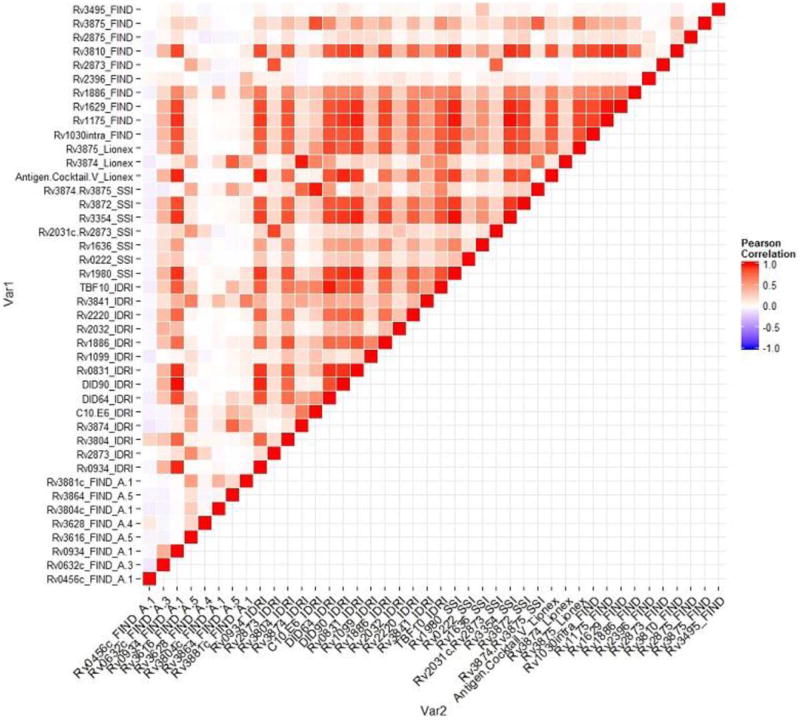
The AUCs and the antibody response cut-offs that optimized specificities and sensitivities for all antigens considered singly are shown in Table S2 (Supplemental Digital Content 1, http://links.lww.com/INF/C768). Forty-two of 119 antigens had AUCs between 0.5 and 0.63. The top 10 (of 119) of these antigens are shown in Figure 1A, and the log2-transformed antigen responses for those 10 antigens are shown in Figure 2A. The sensitivity was highest (60%) for DID64_IDRI, Rv3875_FIND and Rv2873_FIND, and specificity was highest for Rv3875_FIND (61%) and DID64_IDRI (60%) (Table S2, Supplemental Digital Content 1, http://links.lww.com/INF/C768). The heatmap of the Pearson correlations of the antibody responses with AUC >0.5 for the confirmed TB versus unlikely TB comparison (all ages) is shown in Figure 3.
FIGURE 1.
Plots of empiric AUCs for the 10 single antigens having the highest AUC for discrimination between children classified as confirmed TB versus children classified as unlikely TB for (A) all ages; (B) children 12–59 months old and (C) children 60 months old and older.
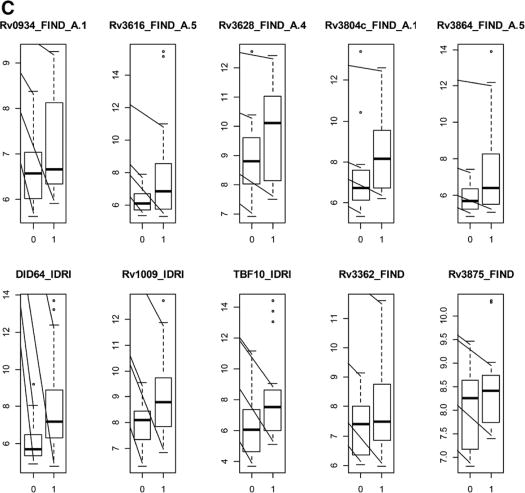
FIGURE 2.
Boxplots of the log2-transformed antibody responses for the 10 single antigens having the highest AUC for discrimination between children classified as unlikely TB (designated as 0 on the x axis) versus children classified as confirmed TB (designated as 1 on the x axis) for (A) all ages; (B) children 12–59 months old and (C) children 60 months old and older. The y axis represents log2-transformed antibody responses.
FIGURE 3.
Correlations plot of single antibody responses with AUC >0.5 for discrimination between all children classified as confirmed TB versus all children classified as unlikely TB.
For discrimination of confirmed TB versus unlikely TB, stratifying by age [12–59 months of age (n = 36) and ≥60 months of age (n = 84)] revealed better antigen prediction power in the older age group, with substantially higher AUCs (up to 0.76 for DID64_IDRI) for the older age group (Fig. 1C) than for the younger group (Fig. 1B). DID64_IDRI, Rv3875_FIND and Rv3616_FIND_A.5 were among the top 10 most predictive antigens common across all the stratifications antigens. To illustrate the distribution of these responses, Figure 2B and C show the log2-transformed antigen responses for the top 10 antigens for the younger group and the older group, respectively, for the comparison of confirmed TB versus unlikely TB. There was large variability and overlapping distributions between the 2 TB groups, but older children with confirmed TB had higher median values than older children in the unlikely TB group (Fig. 2C). The AUCs, optimal antibody response cut-points, as well as the sensitivities and specificities for all antigens considered singly, are shown in Table S3 (Supplemental Digital Content 1, http://links.lww.com/INF/C768) (children 60 months old or older) and Table S4 (Supplemental Digital Content 1, http://links.lww.com/INF/C768) (children 12–59 months old).
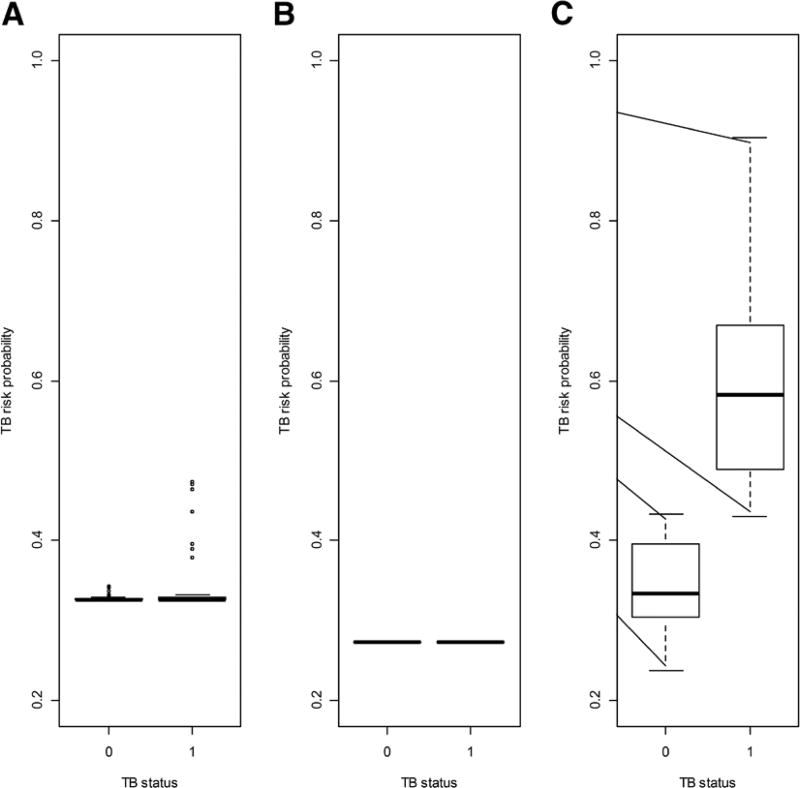
The antigens selected by the cross-validated generalized elastic.net algorithm are listed in Table 2, and the in-sample predictions of risk probabilities from the resultant model are shown in Figure 4 for all ages and by age strata. For all ages, 3 antigens were selected and the corresponding model assigned a similar median risk probability (0.33) to the confirmed TB and the unlikely TB groups. For the younger age group, all the antigens were considered by the algorithm to be noise and none of the antigens was selected, hence the resultant TB risk probability was equal to an average across both TB groups (0.27). For the older age group, the algorithm selected 24 antigens and the resultant model assigned a TB risk probability of 0.57 (IQR: 0.48–0.64) to the confirmed TB group versus 0.35 (IQR: 0.32–0.40) to the unlikely TB group.
TABLE 2.
Antigen Listings for the Optimal Antigen Combinations Selected by the Cross-validated Generalized Linear Model Regularization Path Algorithm for Discrimination Between Children Classified as Confirmed TB Versus Children Classified as Unlikely TB for All Ages, Children 12–59 Months Old and Children 60 Months Old and Older
| All Ages | Age 12–9 Months |
Age ≥60 Months |
|---|---|---|
| Rv3874_IDRI | None | Rv0456c_FIND_A.1 |
| C10.E6_IDRI | Rv0934_FIND_A.1 | |
| Rv3874.Rv3875_SSI | Rv3616_FIND_A.5 | |
| Rv3628_FIND_A.4 | ||
| Rv0934_IDRI | ||
| Rv2873_IDRI | ||
| C10.E6_IDRI | ||
| DID64_IDRI | ||
| Rv1099_IDRI | ||
| Rv1886_IDRI | ||
| Rv2032_IDRI | ||
| Rv2220_IDRI | ||
| TBF10_IDRI | ||
| Rv1636_SSI | ||
| Rv3872_SSI | ||
| Rv3874.Rv3875_SSI | ||
| Rv3874_Lionex | ||
| Rv3875_Lionex | ||
| Rv1886_FIND | ||
| Rv2396_FIND | ||
| Rv2873_FIND | ||
| Rv3810_FIND | ||
| Rv2875_FIND | ||
| Rv3875_FIND |
FIGURE 4.
Boxplots of risk probabilities and corresponding receiver–operator characteristic curves for optimal antigen combinations selected by the cross-validated generalized linear model regularization path algorithm for discrimination between children classified as confirmed TB versus children classified as unlikely TB for (A) all ages; (B) children 12–59 months old and (C) children 60 months old and older.
Antibody Responses That Discriminate Between Confirmed TB Versus Unlikely TB, by TST Status
The empiric AUC values and optimal cut-points for comparing the confirmed TB group (n = 40) to the group classified as unlikely TB/TST-negative (n = 41) are reported in Table S5 (Supplemental Digital Content 1, http://links.lww.com/INF/C768). All but 12 of the 119 antigens had AUC ≥0.5; Rv3875_FIND had the highest AUC (0.74), and sensitivity and specificity of 0.70 and 0.71, respectively. The empiric AUC values and optimal cut-points for comparing the confirmed TB group (n = 40) to the group classified as unlikely TB/TST-positive (n = 39) are reported in Table S6 (Supplemental Digital Content 1, http://links.lww.com/INF/C768). Only 9 of the 119 antigens had AUCs above 0.5; DID64_IDRI had the highest AUC (0.59), and sensitivity and specificity of 0.60 and 0.64, respectively.
Antibody Responses That Discriminate Between Confirmed TB Versus Unconfirmed TB
The empiric AUC values and optimal cut-points for comparing the confirmed TB group (n = 40) to the unconfirmed TB group (n = 39) are reported in Table S7 (Supplemental Digital Content 1, http://links.lww.com/INF/C768); Rv1886_FIND had the highest AUC (0.63), and sensitivity and specificity of 0.60 and 0.62, respectively.
DISCUSSION
To our knowledge, this is the first study assessing a well-defined multiplex panel of antigens for serodiagnosis of childhood TB. In this study of HIV-uninfected children, we found that those with active TB had serologic responses against a broad set of M. tuberculosis antigens. However, for the main comparison—children with confirmed TB versus children with unlikely TB—no single antigen or combination of antigens had sensitivity and specificity above about 60%. Findings were similar for the secondary but clinically important comparison of children with confirmed TB versus children with unconfirmed TB. To put our results into context, the World Health Organization target product profile for a TB detection test has a minimal requirement of specificity ≥98% and sensitivity ≥65%. The target product profile for a TB triage test (i.e., a sensitive but not necessarily highly specific rapid “rule-out” test intended to identify individuals who require additional TB testing) has minimum specificity and sensitivity requirements of ≥70% and ≥90%, respectively. Our results did not identify a single antigen or combination of antigens fulfilling these requirements and thereby holding promise for use as a detection or triage test suitable for application to children of all ages with TB signs/symptoms.
We found, however, that the capability of antibody responses to distinguish between TB groups was affected by age. Variation in antibody responses by age is initially due to maternal antibodies and subsequent maturation of B-cell responses during childhood; in TB, there also may be variations in antibody responses because of age-related differences in TB disease manifestations.9 We therefore stratified our main analysis by age, specifically 12–59 and 60+ months old, in line with the age categories used for notification of childhood TB. The best-performing single antigens achieved higher discriminatory power in older children than in younger children, and the generalized linear model selected a set of antigens that warrants further exploration in the older group. Thus, antibody responses may be of some use for TB diagnosis in older children, but it may be difficult to identify antigens that will deliver the same predictive power for all age groups or for younger children. Better TB diagnostic tests are of particular importance in young children in whom the confirmation of TB using conventional mycobacterial detection tests is especially challenging. The inability to demonstrate a discriminating pattern of antibody response in young children in this study is therefore disappointing.
Given that in TB endemic countries, where novel diagnostics are most urgently needed, a large proportion of individuals are sensitized to mycobacteria though repeated exposure, it is important to ascertain whether prior sensitization with M. tuberculosis interferes with test performance. We therefore stratified the main comparison by TST results and found that antibody responses were more discriminatory for the confirmed TB versus unlikely TB/TST-negative comparison than for the confirmed TB versus unlikely TB/TST-positive comparison. This may indicate that both humoral and cell-mediated immunity are affected by sensitization to mycobacteria, even in the absence of overt disease.
We were not able to identify antibody responses capable of distinguishing children with microbiologically confirmed TB from culture-negative children who were clinically diagnosed with TB (unconfirmed TB). This is a challenging area for pediatric TB diagnosis, since confirming TB among children who are clinically diagnosed, but for whom microbiologic investigations are negative, is currently impossible, and over-diagnosis with subsequent over-treatment frequently occurs.21 However, we cannot formally exclude the possibility that, in our study, children classified as unconfirmed TB truly had TB, a situation that could account for the overall similarity in antibody responses between these groups. Previous studies in adults have shown that antibody responses to M. tuberculosis antigens are highly heterogeneous among individuals.10,11 The current study has also found such heterogeneity, some of which was explained by the age of the children.
From a methodologic perspective, the first stage of our analytical process involved generating AUC from the empiric receiver–operator characteristics of the raw data and determining the optimal cut-offs for maximizing sensitivity and specificity. The second stage was aimed at finding combinations of antibody responses that improved predictive power. Because of the high correlations of some of the antibody responses, the cross-validated generalized elastic net regularization path algorithm (cv.glmnet) with a logit link was found to be superior to other available approaches for finding optimal combinations of correlated predictors. cv.glmnet was applied to a subset of those antibody responses that were at least weakly informative (AUC: >0.5), to reduce noise from noninformative antigens. The AUCs for each antigen were determined empirically and independently of other antibody responses, whereas the multivariable regression model fitted via cv.glmnet was a model-based optimization of the logit coefficients in the presence of confounding responses to other antigens. Thus, the 2 analytical approaches provide complementary information in the context of this exploratory study.22 The predicted link function from the cv.glmnet model is the optimal linear combination of selected antibody responses. This approach to estimation of risk probabilities to distinguish between TB groups is similar to the approach followed by Anderson et al23 who generated risk scores for HIV status using individual HIV RNA signature data in South African and Malawian children. In the current study, however, no validation samples were available on which to test the resultant model, and thus only the in-sample risk probabilities have been presented.
There are several additional limitations to our study. We studied only protein antigens, largely based on their identification in recent immunoproteome work,13 and we did not assess for antibody responses to other biochemical classes of antigens such as lipopolysaccharides. Serologic testing was restricted to IgG antibodies in our study. IgA and IgM responses to a number of M. tuberculosis antigens have been demonstrated in serum from TB patients and individuals with latent M. tuberculosis infection,8,24,25 and recent findings suggest that different antibody isotypes may differentially affect infection of cells by M. tuberculosis.26 Further, our experimental methods did not assess antibody glycosylation, the extent of which has been associated with activity in vitro.27 Finally, we cannot exclude the possibility of classification biases within the consensus case definitions. This is relevant to age as a confounder, since age is associated with diagnostic classification of children with suspect TB,28 and in our study, it is associated with serologic responses.
Positive antibody reactivity signals may have been confounded by cross-reactivity with Mycobacterium bovis BCG and/or non-TB mycobacteria. In South Africa, BCG vaccination is recommended at birth and coverage is estimated to be at least 90%.29 Comparative genomic studies have identified a number of genomic regions [regions of difference (RD)] that are present in M. tuberculosis complex but absent from M. bovis BCG and most non-TB mycobacteria.30–32 Our antigen set contained RD1 proteins (Rv3872, Rv3874, Rv3875, Rv3878, Rv3879c), RD2 proteins (Rv1980, Rv1980c, Rv1984), an RD3 protein (Rv1586c) and an RD4 protein (Rv0222). Interestingly, for discrimination of confirmed TB versus unlikely TB, the RD1 antigens Rv3874 and Rv3875 were among the most predictive when antigens were considered singly and also comprised the best-performing antigen combination as selected by the cross-validated generalized elastic net regularization path algorithm.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the contributions of the study clinical and laboratory staff at Red Cross Children Hospital, the National Health Laboratory Services diagnostic microbiology laboratory at Groote Schuur Hospital, the children and their caregivers. The authors thank the following for providing antigens used in this study: Peter Andersen at Statens Serum Institute (Copenhagen, Denmark); Mahavir Singh at Lionex GmbH (Braunschweig, Germany); Antigen Discovery Inc. (Irvine, CA) and Infectious Disease Research Institute (Seattle, WA).
This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number HHSN2722000900050C, “TB Clinical Diagnostics Research Consortium.” Additional support was provided by the National Institutes of Health, USA (K24AI104830 to S.E.D. and R01HD058971 to H.J.Z), the Foundation for Innovative New Diagnostics, the National Health Laboratory Services Research Trust, the Medical Research Council of South Africa and The Wellcome Trust (085251/B/08/Z).
M.D.P. and T.B. are employed by FIND (Geneva, Switzerland), a non-profit organization that collaborates with industry partners. Individuals and entities that provided antigens had no role in study design, implementation, analysis of results or decision to submit a manuscript for publication.
Footnotes
M.P.N., L.J.W., M.D.P., J.J.E., D.A., B.K., S.E.D. and H.J.Z. designed the study. M.P.N., L.J.W. and H.J.Z. enrolled children, performed clinical evaluations and collected serum during the previous prospective study conducted at Red Cross War Memorial Children’s Hospital. S.R., N.S.-M. and T.J. performed Luminex validation and assayed study specimens. B.A.S.N., N.J.A. and T.B. analyzed the data. B.A.S.N., S.R., N.S.-M., B.K., S.E.D. and H.J.Z. wrote the manuscript and all authors contributed. All authors reviewed and approved the final manuscript.
The other authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (www.pidj.com).
References
- 1.Dodd PJ, Gardiner E, Coghlan R, et al. Burden of childhood tuberculosis in 22 high-burden countries: a mathematical modelling study. Lancet Glob Health. 2014;2:e453–e459. doi: 10.1016/S2214-109X(14)70245-1. [DOI] [PubMed] [Google Scholar]
- 2.Dye C, Watt CJ, Bleed DM, et al. Evolution of tuberculosis control and prospects for reducing tuberculosis incidence, prevalence, and deaths globally. JAMA. 2005;293:2767–2775. doi: 10.1001/jama.293.22.2767. [DOI] [PubMed] [Google Scholar]
- 3.Moore HA, Apolles P, de Villiers PJ, et al. Sputum induction for microbiological diagnosis of childhood pulmonary tuberculosis in a community setting. Int J Tuberc Lung Dis. 2011;15:1185–1190. doi: 10.5588/ijtld.10.0681. i. [DOI] [PubMed] [Google Scholar]
- 4.Newton SM, Brent AJ, Anderson S, et al. Paediatric tuberculosis. Lancet Infect Dis. 2008;8:498–510. doi: 10.1016/S1473-3099(08)70182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zar HJ, Connell TG, Nicol M. Diagnosis of pulmonary tuberculosis in children: new advances. Expert Rev Anti Infect Ther. 2010;8:277–288. doi: 10.1586/eri.10.9. [DOI] [PubMed] [Google Scholar]
- 6.Zar HJ, Workman L, Isaacs W, et al. Rapid molecular diagnosis of pulmonary tuberculosis in children using nasopharyngeal specimens. Clin Infect Dis. 2012;55:1088–1095. doi: 10.1093/cid/cis598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicol MP, Allen V, Workman L, et al. Urine lipoarabinomannan testing for diagnosis of pulmonary tuberculosis in children: a prospective study. Lancet Glob Health. 2014;2:e278–e284. doi: 10.1016/S2214-109X(14)70195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Achkar JM, Ziegenbalg A. Antibody responses to mycobacterial antigens in children with tuberculosis: challenges and potential diagnostic value. Clin Vaccine Immunol. 2012;19:1898–1906. doi: 10.1128/CVI.00501-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steingart KR, Henry M, Laal S, et al. Commercial serological antibody detection tests for the diagnosis of pulmonary tuberculosis: a systematic review. PLoS Med. 2007;4:e202. doi: 10.1371/journal.pmed.0040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyashchenko K, Colangeli R, Houde M, et al. Heterogeneous antibody responses in tuberculosis. Infect Immun. 1998;66:3936–3940. doi: 10.1128/iai.66.8.3936-3940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Yang Y, Zhang J, et al. Humoral immune responses against the Mycobacterium tuberculosis 38-kilodalton, MTB48, and CFP-10/ESAT-6 antigens in tuberculosis. Clin Vaccine Immunol. 2010;17:372–375. doi: 10.1128/CVI.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steingart KR, Dendukuri N, Henry M, et al. Performance of purified antigens for serodiagnosis of pulmonary tuberculosis: a meta-analysis. Clin Vaccine Immunol. 2009;16:260–276. doi: 10.1128/CVI.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunnath-Velayudhan S, Salamon H, Wang HY, et al. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc Natl Acad Sci U S A. 2010;107:14703–14708. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham SM, Cuevas LE, Jean-Philippe P, et al. Clinical case definitions for classification of intrathoracic tuberculosis in children: an update. Clin Infect Dis. 2015;61(suppl 3):S179–S187. doi: 10.1093/cid/civ581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunnath-Velayudhan S, Davidow AL, Wang HY, et al. Proteome-scale antibody responses and outcome of Mycobacterium tuberculosis infection in non-human primates and in tuberculosis patients. J Infect Dis. 2012;206:697–705. doi: 10.1093/infdis/jis421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broger T, Basu Roy R, Filomena A, et al. Diagnostic performance of tuberculosis-specific IgG antibody profiles in patients with presumptive TB from two continents. Clin Infect Dis. 2017;64:947–955. doi: 10.1093/cid/cix023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Planatscher H, Rimmele S, Michel G, et al. Systematic reference sample generation for multiplexed serological assays. Sci Rep. 2013;3:3259. doi: 10.1038/srep03259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Raton M, Rodriguez-Alvarez MX, Cadarso-Suarez C, et al. OptimalCutpoints: an R package for selecting optimal cutpoints in diagnostic tests. J Stat Softw. 2014;61:1–36. [Google Scholar]
- 19.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman J, Hastie T, Simon N, Tibshirani R. Lasso and elastic-net regularized generalized linear models (R package 2017) Available at: http://cran.r-project.org.
- 21.Schumacher SG, van Smeden M, Dendukuri N, et al. Diagnostic test accuracy in childhood pulmonary tuberculosis: a Bayesian latent class analysis. Am J Epidemiol. 2016;184:690–700. doi: 10.1093/aje/kww094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vickers AJ, Cronin AM, Begg CB. One statistical test is sufficient for assessing new predictive markers. BMC Med Res Methodol. 2011;11:13. doi: 10.1186/1471-2288-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson ST, Kaforou M, Brent AJ, et al. ILULU Consortium; KIDS TB Study Group. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med. 2014;370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumann R, Kaempfer S, Chegou NN, et al. Serologic diagnosis of tuberculosis by combining Ig classes against selected mycobacterial targets. J Infect. 2014;69:581–589. doi: 10.1016/j.jinf.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Achkar JM, Chan J, Casadevall A. B cells and antibodies in the defense against Mycobacterium tuberculosis infection. Immunol Rev. 2015;264:167–181. doi: 10.1111/imr.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmermann N, Thormann V, Hu B, et al. Human isotype-dependent inhibitory antibody responses against Mycobacterium tuberculosis. EMBO Mol Med. 2016;8:1325–1339. doi: 10.15252/emmm.201606330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu LL, Chung AW, Rosebrock TR, et al. A functional role for antibodies in tuberculosis. Cell. 2016;167:433–443. e14. doi: 10.1016/j.cell.2016.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frigati L, Maskew M, Workman L, et al. Clinical predictors of culture-confirmed pulmonary tuberculosis in children in a high tuberculosis and HIV prevalence area. Pediatr Infect Dis J. 2015;34:e206–e210. doi: 10.1097/INF.0000000000000792. [DOI] [PubMed] [Google Scholar]
- 29.Zwerling A, Behr M, Verma A, et al. [Accessed January 25, 2017];The BCG World Atlas: A Database of Global BCG Vaccine Policies and Practices. 2011 doi: 10.1371/journal.pmed.1001012. Available at: http://www.bcgatlas.org. [DOI] [PMC free article] [PubMed]
- 30.Mahairas GG, Sabo PJ, Hickey MJ, et al. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philipp WJ, Nair S, Guglielmi G, et al. Physical mapping of Mycobacterium bovis BCG pasteur reveals differences from the genome map of Mycobacterium tuberculosis H37Rv and from M. bovis. Microbiology. 1996;142(pt 11):3135–3145. doi: 10.1099/13500872-142-11-3135. [DOI] [PubMed] [Google Scholar]
- 32.Behr MA, Wilson MA, Gill WP, et al. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.