Abstract
The well-being of all living organisms relies on the accurate duplication of their genomes. This is usually achieved by highly elaborate replicase complexes which ensure that this task is accomplished timely and efficiently. However, cells often must resort to the help of various additional “specialized” DNA polymerases that gain access to genomic DNA when replication fork progression is hindered. One such specialized polymerase family consists of the so-called “translesion synthesis” (TLS) polymerases; enzymes that have evolved to replicate damaged DNA. To fulfill their main cellular mission, TLS polymerases often must sacrifice precision when selecting nucleotide substrates. Low base-substitution fidelity is a well-documented inherent property of these enzymes. However, incorrect nucleotide substrates are not only those which do not comply with Watson-Crick base complementarity, but also those whose sugar moiety is incorrect. Does relaxed base-selectivity automatically mean that the TLS polymerases are unable to efficiently discriminate between ribonucleoside triphosphates and deoxyribonucleoside triphosphates that differ by only a single atom? Which strategies do TLS polymerases employ to select suitable nucleotide substrates? In this review, we will collate and summarize data accumulated over the past decade from biochemical and structural studies, which aim to answer these questions.
Keywords: DNA polymerase, translesion DNA synthesis, replicative bypass, ribonucleotide incorporation, steric gate, mutant polymerases
1. Introduction
In our recent review, “Translesion DNA polymerases in eukaryotes: what makes them tick?”(Vaisman and Woodgate, 2017), we summarized a substantial body of literature related to the biochemical and structural characterization of a specific group of enzymes known to be best adept at replicating imperfect DNA. Here, we intend to focus on a particular property of translesion synthesis (TLS) polymerases from all domains of life, i.e., their ability to discriminate between nucleotides with ribose and deoxyribose sugar rings. It goes without saying that efficient and accurate genome duplication relies on the capacity of DNA polymerases to select correct nucleotides during replication and repair DNA synthesis. For the incoming nucleotide substrate to be correct, its base not only has to be properly paired with the corresponding base on the DNA template, but it also has to be bonded with the correct furanose ring.
While the base substitution fidelity of DNA polymerases has been scrupulously and methodically measured, sugar selectivity of these enzymes (defined as the ratio of incorporation efficiencies of deoxyribonucleotide to ribonucleotide [dNTP/rNTP]), has only been studied sporadically. Even though early studies found that DNA polymerases do not strictly discriminate between deoxyribo- and ribo- substrates [reviewed in (Joyce, 1997)], and the presence of rNTPs in DNA has been detected in the genomes of some prokaryotes (Dalgaard, 2012), the importance of this subject was largely underappreciated, partly due to the fact that rNTPs and dNTPs have the same base-coding potential (Figure 1). Furthermore, a much larger number of rNTPs are transiently inserted during genome duplication as an integral part of de novo synthesis of small RNA stretches that are required to prime DNA replication on the lagging strand. Therefore, the seemingly insignificant number of ribonucleotides mistakenly incorporated into DNA by DNA polymerases appeared to be unworthy of in-depth analysis. However, subsequent studies revealed that due to the substantially (up to 1000-fold) greater intracellular concentration of rNTPs compared to dNTPs, ribonucleotides are embedded into DNA at much higher levels than previously assumed (Buckstein et al., 2008; Ferraro et al., 2010; Neuhard and Nygaard, 1987; Nick McElhinny et al., 2010c). For example, it was estimated that pol III holoenzyme, the primary replicase of the most widely studied prokaryotic model organism, Escherichia coli (E. coli), incorporates as many as 2,000 ribonucleoside monophosphates (rNMPs) during each round of its ~4 Mb genome duplication (Yao et al., 2013). Furthermore, more than 10,000 rNMPs are likely to be inserted within each replication cycle by the replicative polymerases α, δ, and ε into the 3-fold larger genome of Saccharomyces cerevisiae (S. cerevisiae), one of the most intensively studied eukaryotic model organisms (Nick McElhinny et al., 2010c). Last, but not least, quantitation of the number of rNMPs found in mammalian nuclear DNA implied that more than a million rNMPs are incorporated during genome duplication (Reijns et al., 2012). Even though cells seem able to tolerate such high levels of rNMPs sporadically inserted into DNA, their persistence in nuclear genomes can have detrimental consequences (Caldecott, 2014; Klein, 2017; Wallace and Williams, 2014; Williams and Kunkel, 2014; Williams et al., 2016). On the other hand, transient incorporation of rNMPs during DNA replication, or repair, has been shown to have important cellular functions (Dalgaard, 2012; Potenski and Klein, 2014). An eye-opening realization was that in replicating cells, rNMPs are the most abundant non-canonical nucleotides incorporated into DNA. Such observations fueled interest in the field and triggered a rapid expansion of research involving many laboratories that resulted in a large number of publications on the subject over the last decade. This work was recently summarized in several comprehensive reviews focusing on the mechanisms responsible for ribonucleotide incorporation and repair, as well as on the positive biological functions of ribonucleotides nested in DNA and on the potential danger when their prompt removal is compromised (Caldecott, 2014; Cerritelli and Crouch, 2016; Dalgaard, 2012; Jinks-Robertson and Klein, 2015; Klein, 2017; Potenski and Klein, 2014; Schroeder et al., 2015; Vaisman and Woodgate, 2015; Wallace and Williams, 2014; Williams and Kunkel, 2014; Williams et al., 2016).
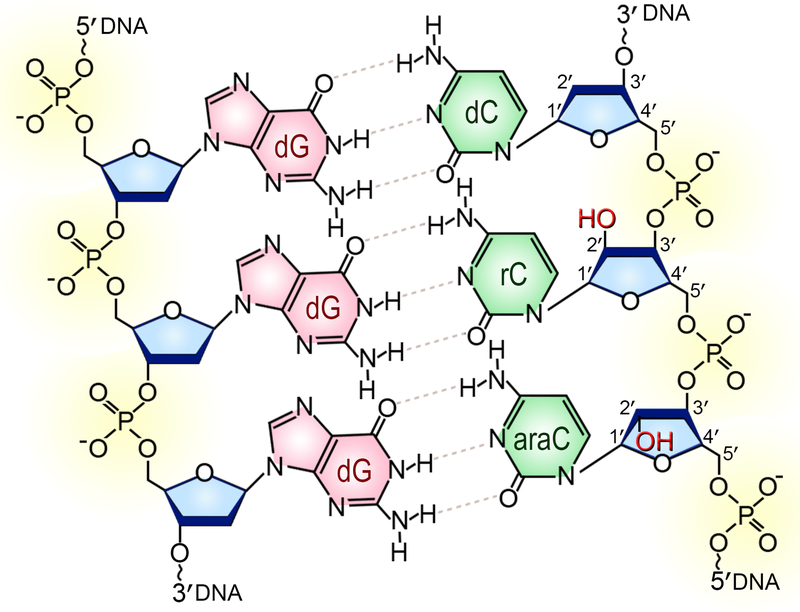
Figure 1. Chemical structure of double-stranded DNA formed by the covalently linked sugar rings (shown in blue), phosphate groups (yellow) and nitrogenous bases.
The segment shown on the diagram consists of three cytosine (green) / guanine (pink) Watson-Crick base pairs where two deoxycytidines (dC) are replaced with either cytidine (rC) or 1-β-D-arabinofuranosylcytosine (araC). The 2′-OH groups (highlighted in red) of the ribonucleotide and arabinofuranoside are on the opposite sides of the plane of the sugar. A color version of the figure is available online.
Our own interest in this subject was piqued by the remarkable efficiency of ribonucleotide incorporation that we detected while investigating the biochemical properties of the E. coli translesion polymerase, pol V (Vaisman et al., 2012a). In our follow-up studies, we also investigated the sugar selectivity of eukaryotic TLS polymerases, pols η and ι (Donigan et al., 2014; Donigan et al., 2015). These and other studies, related to the ability of polymerases implicated in TLS to discriminate between nucleotides with ribose and deoxyribose sugar rings is the subject matter of this review.
Defining which polymerases can be ranked as belonging to the TLS class is not a straightforward task. The capacity to traverse past damaged DNA sites has been demonstrated in vitro for virtually all DNA polymerases, including high fidelity replicases. However, the efficiency, fidelity, lesion specificity, and range of cognate substrates vary significantly between different enzymes. In this review, we not only consider so-called specialized TLS polymerases which, without doubt, play a central role in replicative bypass of DNA lesions, but also polymerases which are known to specialize in other cellular pathways, have narrow TLS activity and/or specificity, or are only recruited for TLS in specific cases.
Extensive in vivo and in vitro studies of TLS polymerases revealed that in addition to their ability to catalyze replication of damaged DNA, these enzymes are characterized by generally less accurate base selection than replicative DNA polymerases. This property is partially explained by the lack of intrinsic 3’→5’ exonucleolytic proofreading and by the relaxed physical constraints of the enzyme’s active site capable of accommodating bulky lesions and mismatched bases (McCulloch and Kunkel, 2008; Pata, 2010; Sale et al., 2012; Yang, 2005; Yang and Woodgate, 2007). Does this mean that TLS polymerases are also less discriminatory when it comes to ribose selection? To answer this question, we will assess the sugar selectivity of various TLS polymerases. Such characterization will not only include discrimination against rNMPs during incorporation, but also at the elongation step, even though extension of primers with a terminal rNMP is generally inhibited less efficiently than rNMP incorporation (Tables 1 & 2). We also will incorporate the available information about the ability of DNA polymerases to elongate RNA primers and DNA primers with a single rNMP at its 3’ end; to incorporate several rNMPs in a row; and to copy RNA templates and rNMP-containing DNA templates. Last but not least, we will analyze the molecular mechanisms underlying the capacity of TLS polymerases to distinguish between nucleotides differing only in the presence of a 2’ OH of the sugar ring (Figure 1) and very briefly discuss its implications for polymerase function.
Table 1.
Ribonucleotide discrimination factors for wild-type and mutant TLS DNA polymerases from B-, X- and Y-families.
| Family | Polymerase b | Residue c | Discrimination a | |||||
|---|---|---|---|---|---|---|---|---|
| Wild type polymerase | Steric Gate mutant | |||||||
| dNTP ×102 | rNTP×102 | araC | dNTP×102 | rNTP | araC | |||
| B d | pol ζ (Sc) f | Tyr-980 | 8 | 5-50 | ||||
| X e | pol β (Hs) g | Tyr-271 | 17 - 900 | 20 - 80 | 9 | 400 | 670 | 3.6 |
| pol λ (Hs) h | Tyr-505 | 10 - 800 | 30 - 500 | 2.4 - 18 | 12 - 520 | 360 - 700 | 8 | |
| pol μ (Hs) i | Gly-433 | j | 0.014 - 0.11 | 25 - 40 | ||||
| Y d | pol IV (Ec) j | Phe-13 | 17 - 440 | undetectable | 164 | |||
| DinB2 (Ms) k | Leu-14 | 13 - 750 | 1 - 3 | |||||
| Dbh (Sa) l | Phe-12 | 9 - 39 | 17 - 200 | 5 - 30 | 3.7 - 5 | |||
| Dpo4 (Ss) m | Tyr-12 | 12 - 29 | 50 - 200 | 3 - 30 | ||||
| pol η (Hs) n | Phe-18 | 0.1 - 3.8 | 4 - 34 | |||||
| pol ι (Hs) o | Tyr-39 | 0.006 - 0.67 | 11 - 67 | 0.025 - 7.4 | 1 - 2 | |||
| Rev1 (Hs) p | Phe-428 | 1.4 - 140 | 2.8 | 6 | ||||
Discrimination is defined as the ratio of catalytic efficiencies (correct nucleotide to nucleotide with wrong base or modified sugar) determined using steady-state or pre-steady-state kinetic assays. The identity of the template base/incoming nucleotide (N/dNTP or N/rNTP) and of the steric gate variant is indicated for each polymerase.
The species abbreviations are as follows: Ec, Escherichia coli; Ms, Mycobacterium smegmatis; Sc, Saccharomyces cerevisiae; Sa, Sulfolobus acidocaldarius; Ss, Sulfolobus solfataricus; Hs, Homo sapiens
Amino acid residues involved in the major mechanism of steric ribonucleotide exclusion are indicated.
The data for the B- and Y-family polymerases were obtained using recessed DNA substrates.
The data for the X-family polymerases were obtained using single-nucleotide gapped DNA substrates. On the recessed DNA substrates, discrimination against incorrect base by human pol μ ranges between 1.3 and 90 for all 12 base mispairs; against incorrect sugar it ranges between 0.5 and 11 for all four N/rNTP pairs (Roettger et al., 2004).
Sugar selectivity of the 4-subunit pol ζ was estimated based on the semi-quantitative assay using primer extension reactions containing each individual dNTP or rNTP at the concentrations estimated for unstressed yeast (Makarova et al., 2014). Base-substitution fidelity was determined using an M13 gap-filling assay (Zhong et al., 2006).
Wild type and Y271A - G/dTTP, G/rCTP, araC (Cavanaugh et al., 2010; Prakasha Gowda et al., 2010; Cavanaugh et al., 2011), T/rATP, G/rCTP (Nick McElhinny and Ramsden, 2003) and T/dNTP (Ahn et al., 1998).
Wild type pol λ - four N/rNTP, G/araC (Brown et al., 2010a; Garcia-Diaz et al., 2010; Gosavi et al., 2012) and 12 N/dNTP (Fiala et al., 2006); Y505G - A/rUTP and A/dGTP; Y505A - A/rUTP, A/dGTP, araC and T/dNTP (Brown et al., 2010a; Garcia-Diaz et al., 2010).
Wild type pol μ – four N/rNTP pairs (Nick McElhinny and Ramsden, 2003). Sugar selectivity of the G433Y mutant was estimated based on the gap-filling qualitative assay in the presence of each dNTP/rNTP mixture (Ruiz et al., 2003).
Wild type pol IV - G/rCTP (Jarosz et al., 2006) and three dNTPs opposite G (Kobayashi et al., 2002); F13V - G/rCTP (Jarosz et al., 2006).
Wild type DinB2 - four N/rNTP pairs (Ordonez and Shuman, 2014; Ordonez et al., 2014).
Wild type Dbh - four N/rNTP pairs, G/dTTP and T/dGTP; F12A - four N/rNTP pairs (DeLucia et al., 2003; DeLucia et al., 2006).
Wild type Dpo4 - four N/rNTP pairs (Sherrer et al., 2010) and 12 N/dNTP mispairs (Boudsocq et al., 2001); Y12A - four N/rNTP pairs (Sherrer et al., 2010).
Wild type pol η - G/rCTP, T/rATP, three dNTPs opposite G and T; Y39A - G/rCTP, G/dGTP, T/rATP, and T/dGTP (Mentegari et al., 2017; Su et al., 2016).
Wild type pol ι - G/rCTP, G/dGTP, T/rGTP, and T/dGTP; Y39A - G/rCTP, G/dGTP, T/rGTP, T/rATP, and T/dGTP (Donigan et al., 2014). Incorporation of rATP opposite the template T by wild type pol ι is undetectable.
Wild type Rev1 - G/rCTP, G/dNTPs and araC (Brown et al., 2010b).
Table 2.
Primer elongation on rNMP-containing templates and elongation of rNMP-containing primers by wild-type and mutant TLS DNA polymerases from B-, X- and Y-families.
| Family | Polymerase a | Wild type polymerase | Steric Gate mutant | |
|---|---|---|---|---|
| bypass b | elongation c | elongation c | ||
| A | pol θ (Hs) d | (0) e | ||
| B | pol ζ (Sc) f | 80-95% (1 - 4) | ||
| X g | pol β (Hs) h | 12.5% (1) | 90% (<8) | |
| pol λ (Hs) i | 70 - 300% (1) | |||
| pol μ (Hs) j | <1% (0) | 10 - 25% (1) | ||
| Y | pol IV (Ec) k | (0) | (15) | |
| DinB1 (Ms) l | (3) | (13) | ||
| DinB2 (Ms) m | 80 - 100 % (12) | (16) | (9) | |
| pol V (Ec) n | (>100) | (>1000) | ||
| Dbh (Sa) o | (0) | 90% (10) | ||
| Dpo4 (Ss) p | (10) | (20) | ||
| pol η (Hs) r | 95% | 100% (20) | ||
| pol ι (Hs) s | 100% (8) | 100% (15) | ||
| pol κ (Hs) t | 90% | 100% (3) | ||
The species abbreviations are as follows: Ec, Escherichia coli; Ms, Mycobacterium smegmatis; Sc, Saccharomyces cerevisiae; Sa, Sulfolobus acidocaldarius; Ss, Sulfolobus solfataricus; Hs, Homo sapiens.
Relative efficiency of TLS past rNMP. Number of efficiently bypassed consecutive rNMPs in the DNA template is shown in the brackets.
Relative efficiency of ribonucleotide elongation using either primers with single terminal rNMP or whole RNA primers. Number of sequentially incorporated rNMPs using either primers with terminal rNMP or whole DNA primers is shown in the brackets.
The data are from (Hogg et al., 2012; Kent et al., 2016).
Data obtained on primed double stranded DNA using Mg2+ (Lazzaro et al., 2012). Efficient extension of single-stranded DNA by rNTP incorporation especially in the presence of Mn2+ is described in the 2.1.1 section.
Data obtained using 2-subunit polymerase.
The data for the X-family polymerases were obtained using single-nucleotide gapped DNA substrates except those from (Bergoglio et al., 2003).
The data are from (Bergoglio et al., 2003; Cavanaugh et al., 2010; Cilli et al., 2015; Crespan et al., 2016; Nick McElhinny and Ramsden, 2003; Ramadan et al., 2003).
The data are from (Ramadan et al., 2003).
Whole RNA template was tested (Nick McElhinny and Ramsden, 2003).
The data are from (Jarosz et al., 2006).
The data are from (Ordonez and Shuman, 2014; Ordonez et al., 2014).
The data are from (Ordonez and Shuman, 2014; Ordonez et al., 2014).
The data are from (Vaisman et al., 2012a).
The data are from (DeLucia et al., 2003; DeLucia et al., 2006; Sherrer et al., 2010).
The data are from (Kirouac et al., 2011).
The data are from (Sassa et al., 2016).
The data are from (Donigan et al., 2014).
The data are from (Sassa et al., 2016; Su et al., 2017; Gali et al., 2017).
2. Sugar selectivity of TLS polymerases from different phylogenetic families
Based on primary amino acid sequence homology, DNA polymerases are divided into several discrete phylogenetic groups (Braithwaite and Ito, 1993; Ito and Braithwaite, 1991), most of which contain one or more members of the extended “TLS polymerase family”. Among the functionally-diverse A-family polymerases, are eukaryotic polymerases θ and ν that are involved in TLS and DNA repair pathways primarily related to processing the aberrant ends of strand breaks (Hogg et al., 2011; Rothwell and Waksman, 2005; Seki and Wood, 2008; Takata et al., 2006; Takata et al., 2010; Yamanaka et al., 2010). The B-family mainly comprised of high fidelity DNA polymerases involved in chromosomal replication, also contains bacterial (pol II) and eukaryotic (pol ζ); enzymes that play an important role in TLS (Fuchs and Fujii, 2007; Lange et al., 2011; Lawrence et al., 2000; Shcherbakova and Fijalkowska, 2006; Vaisman et al., 2012b). The X- and Y-families appear to consist of more functionally homogeneous polymerases which are all implicated in damaged DNA processing (Vaisman and Woodgate, 2017). Three eukaryotic X-family polymerases (pols β, λ, and μ), which are generally known as DNA repair enzymes, have all been shown to facilitate TLS (Vaisman and Woodgate, 2004; Waters et al., 2009). Y-family polymerases, which are conserved throughout three domains of life, include prokaryotic pol IV and pol V, archaeal Dbh and Dpo4, and eukaryotic pols η, ι, κ and Rev1, and are all classified as specialized TLS polymerases (Ohmori et al., 2001). Recently, the “cast” of TLS polymerases expanded by embracing a new member, PrimPol, a DNA-directed primase-polymerase belonging to the archaeo-eukaryotic primase superfamily (Bianchi et al., 2013; Garcίa-Gόmez et al., 2013; Mourόn et al., 2013). This enzyme is not only able to initiate DNA polymerization de novo and elongate existing DNA chains, but, remarkably, it can do it quite efficiently using damaged DNA substrates (Bianchi et al., 2013; Mourόn et al., 2013).
2.1. TLS DNA polymerases from A- and B- families
To date, four TLS polymerases from the A- and B- families have been identified; prokaryotic pol II and eukaryotic pols ζ, θ, and ν. While the B-family pol ζ was the first DNA polymerase characterised as specializing in TLS, A-family DNA pols θ and ν are among the newest additions to the broad superfamily of polymerases capable to copy imperfect DNA (Hogg et al., 2011; Seki and Wood, 2008; Takata et al., 2006; Takata et al., 2010; Yamanaka et al., 2010). However, thus far, only A-family TLS pol θ and B- family TLS pol ζ have been characterized with regard to their sugar selectivity.
2.1.1. DNA polymerase θ
Pol θ has the unusual ability to catalyze template-independent DNA synthesis using single-stranded DNA substrates in the presence of both dNTPs and rNTPs (Hogg et al., 2012; Kent et al., 2016). In reactions activated by Mg2+, pol θ transfers ribonucleotides to the 3’ termini of the single-stranded DNA much less efficiently than deoxyribonucleotides (Hogg et al., 2012). Pol θ extends DNA by two nucleotides in the presence of all four rNTPs and by one nucleotide in the presence of rGTP, but not in the presence of any other single ribonucleotide. Furthermore, with primed double-stranded DNA, rNTP insertion is barely detectable, even at high ribonucleotide concentrations and extended reaction times. In the presence of Mn2+ as a cofactor, pol θ is much more effective at transferring ribonucleotides onto single-stranded DNA termini. When elongation of single strand DNA is carried out in the presence of each nucleotide separately, pol θ is least efficient at incorporating uridines. In the presence of any other single rNTP, or all four ribonucleotides added simultaneously, it extends DNA by more than 50 nucleotides (Kent et al., 2016).
2.1.2. DNA polymerase ζ
The first bona fide TLS polymerase identified was the eukaryotic B-family pol ζ (Morrison et al., 1989; Nelson et al., 1996a) and to date, it is the only known eukaryotic B-family polymerase that plays a critical role in TLS. Pol ζ is a hetero-tetramer comprised of Rev3, Rev7, and two subunits of the replicative pol δ, Pol31 and Pol32 (Baranovskiy et al., 2012; Johnson et al., 2012; Lee et al., 2014; Makarova et al., 2012), whose primary responsibility appears to be the extension step of TLS after insertion opposite the lesion is accomplished by another TLS polymerase. Genetic and biochemical data indicate that S. cerevisiae pol ζ can efficiently copy DNA templates containing one to four consecutive rNMPs incorporated into DNA by replicative polymerases and is able to replicate RNA patches comprised of as many as 16 sequential rNMPs, although very ineffectively (Lazzaro et al., 2012). These studies strongly imply that pol ζ plays an important role in allowing cells to tolerate unrepaired ribonucleotides inserted into the genome by other polymerases.
The ability of pol ζ to tolerate a ribose sugar ring during ribonucleotide incorporation (Table 1) is much lower than its ability to copy embedded rNMPs in the template DNA, even when the 4-subunit polymerase complex is assayed in the presence of its cofactors, RPA (replication protein A), RFC (replication factor C) and PCNA (proliferating cell nuclear antigen) (Makarova et al., 2014). Despite the fact that the base substitution fidelity of pol ζ is substantially lower than that of replicative polymerases, the frequency of incorporation of ribonucleotides is only slightly higher for yeast pol ζ than for the high-fidelity enzymes (Table 1). Furthermore, under conditions which mimic cellular dNTP concentrations induced by DNA damage, pol ζ incorporates as little as one rNMP per 1,300 dNMPs insertions (Makarova et al., 2014). Based upon these observations, the incorporation of ribonucleotides is predicted to be an infrequent event when pol ζ is recruited to replicate damaged chromosomal DNA in yeast. It will be interesting to learn whether sugar discrimination of the recently purified human pol ζ4 (Lee et al., 2014) has characteristics similar to the yeast complex.
Another B-family DNA polymerase that can bypass lesions by itself, or in cooperation with other TLS polymerases, is prokaryotic pol II. In addition to its polymerase activity, it also possesses 3’−5’ exonuclease activity which not only sets it apart from the related eukaryotic pol ζ, but also from the majority of all other TLS polymerases. To date, the capacity of E. coli pol II to incorporate rNTPs and/or to extend primers with terminal rNMP, as well as its ability to proofread misinserted rNMPs has yet to be determined, but it is clearly a subject of great interest.
2.2. X- family of TLS DNA polymerases
The eukaryotic X-family DNA polymerase, pol β, is responsible for the majority of gap-filling during base excision repair (BER) (Sobol et al., 1996). However, pol β has also been implicated in TLS (Bassett et al., 2002; Vaisman et al., 2000). X-family polymerases, λ and μ have also been proposed to play a specific role in TLS (Belousova et al., 2010; Blanca et al., 2004; Krasikova et al., 2008; Maga et al., 2007; Shtygasheva et al., 2008; Zhang et al., 2002), even though their major cellular functions are thought to be related to other types of DNA transactions (such as BER, non-homologous end joining repair (NHEJ), and V(D)J recombination). The ability of X-family enzymes to distinguish between dNTPs and rNTPs has been studied extensively (Brown et al., 2010a; Cavanaugh et al., 2010; Cavanaugh et al., 2011; Gosavi et al., 2012; Nick McElhinny and Ramsden, 2003; Prakasha Gowda et al., 2010; Roettger et al., 2004). Most of these studies were performed using a single nucleotide-gapped DNA, the preferred substrate for pols β, λ and μ. Since all members of the X-family polymerases are characterized by a distinct set of properties, it is not surprising that each of them has adopted an individual approach to discriminate between ribonucleotide and deoxyribonucleotide incorporation.
2.2.1. DNA polymerase β
In contrast to primer extension reactions catalyzed by pol ζ, the efficiency of nucleotide incorporation by pol β opposite a template rNMP is ~8-fold lower relative to the incorporation opposite a dNMP [Table 2, (Cavanaugh et al., 2010)]. However, discrimination against rNTP incorporation is very similar for pol ζ and pol β.
Similar to high-fidelity DNA polymerases, the reduced efficiency of pol β to insert rNTPs is due to both weaker binding affinity and slower rate of incorporation (Cavanaugh et al., 2010; Cavanaugh et al., 2011; Gosavi et al., 2012; Prakasha Gowda et al., 2010). Even though ribonucleotide incorporation by pol β is significantly more efficient than incorporation of mismatched dNTPs, it is about 3–4 orders of magnitude less efficient than the correct dNTP incorporation (Table 1). Interestingly, DNA-dependent DNA polymerase β was even able to incorporate rNTP opposite a ribonucleotide templating residue, although with ~20-fold lower efficiency compared to rNTP insertion opposite the dNMP templating bases.
Inhibition of nucleotide incorporation by the 2’-OH on the furanose ring could be significantly reduced simply by inversion of this group configuration, as observed for a nucleotide with an arabinose sugar, in which the 2’-OH moiety is on the opposite side of the plane compared to the ribose of a normal rNTP (Figure 1). Thus, cytosine-1- β-d-arabinofuranoside (araC) is incorporated opposite template dG ~1000 times more efficiently than rCTP (Table 1) (Cavanaugh et al., 2010). However, when araC is present at the 3’- primer terminus, nucleotide insertion is inhibited, and primer extension is to a large extent halted (Cavanaugh et al., 2010). In contrast, the presence of an rNMP at the primer terminus has no effect on the efficiency and fidelity of primer elongation, suggesting that it does not cause sizable distortion of the DNA duplex. Pol β has also been shown to be able to catalyze incorporation of multiple sequential ribonucleotides (as many as 8 rNMPs) as well as to readily insert and elongate an oxidized ribonucleotide (Bergoglio et al., 2003; Cavanaugh et al., 2010; Cilli et al., 2015; Crespan et al., 2016). It is capable of error-prone insertion of rNTPs opposite damaged DNA and extension of the resulting (mis)pairs (Bergoglio et al., 2003; Cavanaugh et al., 2010; Cilli et al., 2015; Crespan et al., 2016). Interestingly, base substitution fidelity of pol β opposite 8-oxo-7,8-dihydroguanine (8-oxo-G) is much higher when the incorporated nucleotide has a ribose sugar ring (Crespan et al., 2016).
The data described above, appear to indicate that even though pol β has no problem extending primers after ribonucleotide incorporation, the exclusion of an incoming rNTP during the insertion step is quite efficient. However, these data were obtained using an in vitro approach without taking into account the intracellular imbalance of rNTPs over dNTPs. Re-evaluation of the biochemical data reveals that at concentrations close to physiological levels of competing rNTPs and dNTPs, pol β might insert as many as two rNTPs for every 100 dNTP incorporations (Cavanaugh et al., 2010). Furthermore, the rNTP/dNTP ratio in non-dividing cells is even higher, suggesting that in growth-arrested cells, accommodation of ribonucleotides and their oxidized derivatives by pol β is a very likely event. This can affect genome stability in non-dividing cells, since complex lesions formed by an oxidized base and a ribonucleotide can compromise base- and ribonucleotide excision repair (Cilli et al., 2015; Crespan et al., 2016). Consistent with such assessments is the hypothesis that an increased number of DNA breaks and enhanced chromosomal instability in cells overexpressing pol β is caused by frequent ribonucleotide incorporation (Bergoglio et al., 2003).
2.2.2. DNA polymerase λ
All of the major biochemical characteristics of pol λ related to its ability to distinguish between dNTP, rNTP and ara-NTP are similar to those described for pol β with which it shares 34% sequence identity [Table 1 and (Aoufouchi et al., 2000; Brown et al., 2010a; Garcia-Diaz et al., 2010; Gosavi et al., 2012)]. Accordingly, structures of the polymerase ternary complex with an incoming nucleotide were very similar for pols λ and β, both revealing energetically unstable binding of the rNTP (Cavanaugh et al., 2011). However, some differences in sugar discrimination by the two polymerases have also been detected. For example, compared to pol β, pol λ demonstrates somewhat stronger sugar selectivity and reduced base substitution fidelity upon ribonucleotide incorporation opposite 8-oxo-G (Crespan et al., 2016). Even though pol λ relatively effectively discriminates against rNTP incorporation, it nevertheless elongates RNA primers even more efficiently than DNA primers, which is not a characteristic of pol β [Table 2, (Ramadan et al., 2003)]. Until the recent discovery of eukaryotic PrimPols, such a property had only been described for pol α, a DNA polymerase responsible for the initiation of DNA replication from RNA primers.
2.2.3. DNA polymerase μ
While both pol β and pol λ are characterized by moderate sugar selectivity with a preference for dNTPs over rNTPs, the situation is very different for another X-family polymerase, pol μ (Table 1). Even when nucleotides are present at equal concentrations, pol μ inserts rNTPs with an efficiency that is only 1.4–11-fold lower than that of dNTPs (Nick McElhinny and Ramsden, 2003; Ruiz et al., 2003). Taking into account the intracellular rNTP/dNTP pool levels, it is likely that pol μ selects rNTPs even more often than dNTPs. Despite relaxed sugar selectivity during the nucleotide incorporation step, extension of a DNA primer with one ribonucleotide at its 3’ end is substantially (4 times) inhibited [Table 2, (Roettger et al., 2004; Ruiz et al., 2003)]. Likewise, elongation of RNA primers by pol μ although possible, is very inefficient (less than 10 % of DNA primer extension) (Nick McElhinny and Ramsden, 2003) and much less productive than when catalyzed by pol β or pol λ (Table 2).. Ribonucleotides inhibit polymerase activity of pol μ most dramatically when replication is carried out using RNA templates (Nick McElhinny and Ramsden, 2003). Such properties are likely very useful in promoting pol μ’s biological functions (Martin et al., 2013). Thus, it has been proposed that the ability to utilize rNTPs facilitates pol μ-catalyzed double-strand break repair (DSBR) outside S phase, when the intracellular concentration of dNTPs is low. It has also been shown that the fidelity of NHEJ reactions could be improved when pol μ uses rNTP as a substrate and the limited ability to extend primers with a terminal rNMP is important to ensure efficient NHEJ.
2.3. Y-family DNA polymerases
Although polymerases from different families can facilitate replication of damaged DNA, Y-family polymerases are, by far, the most proficient TLS enzymes. Phylogenetic analysis of the polymerases categorizes them into six branches: two branches containing prokaryotic UmuC homologs (found in gram-positive and gram-negative bacteria); the DinB-like enzymes with homologs in all three domains of life (bacteria, archaea and eukaryotes); and three branches consisting of eukaryotic Rad30A (pol η), Rad30B (pol ι) and Rev1 proteins (Ohmori et al., 2001). The structure and biochemical properties of prokaryotic (bacterial DinB paralogs and pol V), archaeal (Dbh and Dpo4), and eukaryotic (pols η, ι, κ, and Rev1) polymerases have been investigated in great detail by several groups with some of these studies focusing on the characterization of sugar recognition by these enzymes (Brown et al., 2010b; DeLucia et al., 2003; DeLucia et al., 2006; Donigan et al., 2014; Jarosz et al., 2006; Katafuchi et al., 2010; Ketkar et al., 2012; Kirouac et al., 2011; Kuban et al., 2012; McDonald et al., 2012; Niimi et al., 2009; Ordonez and Shuman, 2014; Ordonez et al., 2014; Sherrer et al., 2010; Shimizu et al., 2003; Vaisman et al., 2012a). Akin to their widely divergent capacities to promote lesion bypass, the ability of the Y-family polymerases to discriminate between rNTPs and dNTPs varies considerably.
2.3.1. Escherichia coli pol V
E. coli pol V is a heterotrimeric complex consisting of UmuD’2C (Tang et al., 1999). The polymerase has weak intrinsic catalytic activity (Reuven et al., 1999; Tang et al., 1999), which increases significantly in the presence of a trans-activating RecA nucleoprotein filament to form mutagenically active pol V Mut (consisting of UmuD′2C-RecA-ATP) (Jiang et al., 2009; Karata et al., 2012; Schlacher et al., 2006). When reconstituted in vitro, the complex displays optimal activity on an SSB-coated single-stranded circular DNA template in the presence of ATP and the β/γ (sliding-clamp/clamp-loader) complex (Karata et al., 2012). Under these reaction conditions, pol V exhibits a remarkable tolerance for the presence of a 2’-hydroxy group on the furanose moiety (Vaisman et al., 2012a). Consequently, 1 mM ATP (required in vitro for the γ–complex-driven assembly of the β-clamp around DNA) successfully competes with the dNTPs that are present at much lower concentrations. Even when levels of rNTPs and dNTPs are comparable, pol V Mut often chooses to use ribonucleotides as building blocks for DNA replication. In vitro, pol V Mut promptly incorporates all four rNTPs in an error-prone manner and catalyses efficient and processive RNA synthesis. Furthermore, RNA stretches synthesized by pol V are unusually long (Vaisman et al., 2012a), which is especially remarkable, since even mutant polymerases selected for the enhanced capacity to incorporate rNTPs are generally unable to incorporate more than several rNTPs sequentially (see Section 3 below).
2.3.2. DinB orthologs in bacteria, archaea, and eukarya
Unlike pol V, which is restricted to bacteria, DinB orthologs are found in all domains of life (prokaryotic pol IV, archaeal DinB and Dpo4, and eukaryotic pol κ) (Ohmori et al., 2001). Ribonucleotide discrimination by many members of this subfamily is as stringent as that of high fidelity polymerases. Thus, almost no insertion of rNTPs by E. coli pol IV, or human pol κ, was detected in the primer-extension reactions using an undamaged DNA template [Table 1, (Jarosz et al., 2006; Nevin et al., 2015; Niimi et al., 2009)]. Nevertheless, pol IV is able to incorporate rCTP opposite an N2-furfuryl-dG adduct, albeit with a 2000-fold lower efficiency than dCTP and ~140 times less efficiently than a mispaired dTTP or dATP (Jarosz et al., 2006). With regard to TLS of ribonucleotide-containing DNA, it has been shown that human pol κ is able to replicate past undamaged rNMP (Table 2) and relatively accurately bypasses 8-oxo-rG although with lower efficiency compared with 8-oxo-dG (Sassa et al., 2016). Furthermore, human pol κ can extend RNA primers by incorporating dNTPs even with an 8-oxo-dG in the DNA template strand (Su et al., 2017).
An intriguing discovery was made in the course of characterization of DinB homologs from gram-positive mycobacteria whose regulation and biology differ strikingly from their counterparts in other model microorganisms (Kana et al., 2010). Interestingly, in contrast to gram-negative E. coli, mycobacteria have more than one DinB homolog. Thus, Mycobacterium smegmatis has three DinB paralogs, DinB1, DinB2, and DinB3 (Ordonez et al., 2014). While DinB1 and DinB3 are similar to E. coli pol IV in that they discriminate strongly against ribonucleotides, DinB2 has vigorous RNA polymerase activity [Tables 1 & 2, (Ordonez and Shuman, 2014; Ordonez et al., 2014)]. Thus, DinB1 and DinB3 are able to incorporate no more than four, or one rNTP in a row respectively, and only when manganese was used as the divalent cation cofactor for catalysis. In contrast, DinB2 can promptly extend DNA primers by synthesizing significant stretches of consecutive rNMPs (at least 16). It is also able to utilize RNA as a template, incorporating not only dNTPs, but also rNTPs in the presence of Mn2+ (Ordonez and Shuman, 2014; Ordonez et al., 2014). The high rate of rNMP incorporation catalyzed by DinB2 is in the same order of magnitude as the rate of dNTP incorporation although the affinity for rNTPs is 26- to 78-fold lower than its affinity for dNTPs. DinB2 retains its low sugar selectivity during incorporation of modified nucleotides and TLS past some lesions (Ordonez and Shuman, 2014; Ordonez et al., 2014). Thus, DinB2 is highly base and sugar error-prone when using oxo-rGTP as a substrate for DNA synthesis, or when catalyzing TLS across an oxo-dG lesion in the template DNA strand. On the other hand, it is much more selective opposite an abasic site, especially in the presence of Mn2+ (Ordonez and Shuman, 2014; Ordonez et al., 2014). The properties of DinB2 therefore suggest that the polymerase might contribute to mycobacterial mutagenesis, especially under conditions of oxidative stress. On the other hand, owing to its relaxed sugar selectivity, DinB2 might be especially helpful for DNA repair during stationary phase or other quiescent phases, when dNTPs are scarce. Since rNMP patches, generated as a side effect of DinB2 synthetic activity, can be easily removed and replaced with dNMPs, they appear to be less dangerous for the cells than persistent strand breaks. Furthermore, even when RNA patches remain unrepaired during the onset of DNA replication, DinB2 itself can be recruited to overcome the potential obstacles by assisting the main replicase to copy problematic regions of genomic DNA.
Similar to their mammalian and most bacterial relatives, the archaeal Dbh and Dpo4 enzymes from Sulfolobus acidocaldarius and Sulfolobus solfataricus respectively, skillfully select dNTPs in preference to rNTPs. Even though they do incorporate ribonucleotides at detectable levels and Dpo4 is even able to insert several rNMPs sequentially, the efficiency of rNTP insertion by both Dpo4 and Dbh is about 3–4 orders of magnitude lower compared to dNTP incorporation [Tables 1 & 2, (DeLucia et al., 2003; DeLucia et al., 2006; Sherrer et al., 2010)]. Furthermore, in contrast to X-family polymerases, both Dbh and Dpo4 in general, discriminate against nucleotides with the wrong sugar better than against nucleotides with the wrong base (Table 1). Overall, these studies indicate that in contrast to pol V, but similar to high-fidelity polymerases, most DinB homologues are highly discriminatory against incoming ribonucleotides (DeLucia et al., 2003; DeLucia et al., 2006; Kirouac et al., 2011; Sherrer et al., 2010; Shimizu et al., 2003).
2.3.3. Polymerases η and ι
Eukaryotic cells have two RAD30 paralogs, pol η and pol ι. Interestingly, in the presence of rNTPs, the behavior of human pol η is closer to its human relative, pol ι, rather than to its S. cerevisiae pol η counterpart (Donigan et al., 2014; Donigan et al., 2015; Mentegari et al., 2017; Su et al., 2016).
In contrast to robust misinsertion of nucleotides with an incorrect base, incorporation of nucleotides with a wrong sugar is barely detectible in vitro for S. cerevisiae pol η, although the polymerase can efficiently extend RNA and rNMP-terminated primers using dNTPs and rNTPs (Donigan et al., 2014; Donigan et al., 2015; Gali et al., 2017). S. cerevisiae pol η is also able to insert rNTPs opposite damaged bases such as 8-oxo-G and T-T cyclobutane pyrimidine dimers in an error-free manner, but only while extending RNA, and not DNA primers. On the contrary, the human homolog is much less prone to distinguish the identity of the sugar moiety [Table 1. (Mentegari et al., 2017; Su et al., 2016)]. Human pol η (hpol η) not only efficiently synthesizes DNA/RNA and RNA fragments on undamaged DNA templates, but also does it much more accurately, compared to synthesis in the presence of dNTPs alone. Furthermore, hpol η displays a similar efficiency and accuracy of rNTP incorporation when replicating various damaged DNAs, such as 8-oxo-G, T-T cyclobutane pyrimidine dimers, 8-methyl-2’-deoxyguanosine, and cisplatin-GG intrastrand crosslink (Mentegari et al., 2017; Su et al., 2016). Based on these findings, it has been suggested that TLS by hpol η can contribute to the accumulation of rNMPs into genomic DNA, and particularly rCMPs paired with modified guanines. The impact of such specificity is especially significant, since human RNase H2 which is responsible for the initiation of ribonucleotide excision repair is least efficient while removing rCMPs and rGMPs and is often inhibited when the base paired with the rNMP is damaged (Mentegari et al., 2017).
Human pol η tolerates a ribose sugar not only during ribonucleotide incorporation opposite dNMP bases, but also during deoxyribonucleotide incorporation opposite undamaged and oxidized rNMPs embedded into DNA templates (Sassa et al., 2016). As a consequence, it catalyzes accurate and efficient TLS past rG and 8-oxo-rG. Furthermore, similar to pol κ, pol η exhibits much higher fidelity during bypass of 8-oxo-rG compared to bypass of 8-oxo-dG. These findings suggest that the ribose sugar affects the conformation of the 8-oxoguanine base itself, so as to promote the correct Watson-Crick pairing with cytosine in the polymerase active site (Sassa et al., 2016). The high efficiency and fidelity of pol η during TLS past 8-oxo-rG might play an important role protecting genomic integrity, since base- and ribonucleotide excision repair of oxidized guanine are suppressed by the ribose sugar moiety (Sassa et al., 2016).
Recent studies have demonstrated that similar to human pol κ, human pol η is able to replicate primed single-stranded DNA substrates in which one of the strands, either primer or template is replaced by RNA (Su et al., 2017). Pol η does so even in the presence of an equal amount of the DNA/DNA substrate by preferentially inserting dNMPs opposite undamaged bases, or opposite lesions such as 8-oxo-G or cyclobutane pyrimidine dimers. These findings, as well as similar studies with yeast enzyme (Gali et al., 2017), suggest that pol η might be involved in initiation of replication by substituting for pol α in extension of RNA primers especially when the DNA template is damaged. Furthermore, the RNA synthesis activity of yeast pol η suggests its involvement in lesion bypass during transcription (Gali et al., 2017), while reverse transcription activity of human pol η implicates this enzyme in DNA synthesis on an RNA template during double-strand break repair (Su et al., 2017).
While human pol η can incorporate ribonucleotides opposite an 8-oxo-G lesion, but not opposite an abasic site, the related human pol ι TLS polymerase readily inserts and extends rNTPs opposite both damaged templates (Donigan et al., 2014). Nevertheless, the efficiency of rNTP incorporation by human pol ι is more than 1000 times lower compared to incorporation of dNTPs (Table 1) and extension of ribonucleotide-terminated primers is efficient only in the presence of dNTPs (Table 2). Pol ι’s sugar selectivity appears to be template-dependent, since the probability of rNTP incorporation is higher opposite A and G than opposite T or C, when replicating an undamaged template and opposite an abasic site, than opposite 8-oxo-rG during TLS (Donigan et al., 2014). During ribonucleotide incorporation pol ι retains its preference for the insertion of guanine over adenine opposite an abasic site and opposite an undamaged T, although it inserts rGTP less efficiently than dATP [Table 1, (Donigan et al., 2014)].
2.3.4. Rev1
Unlike other Y-family DNA polymerases that incorporate all four dNTPs in a template-dependent fashion, Rev1 specializes in dCMP insertion and only scarcely utilizes the three other dNTPs (Nelson et al., 1996b). Human Rev1 discriminates between deoxy- and ribo-cytosine with moderate efficiency, incorporating dCTP 280-fold more frequently than rCTP [Table 1, (Brown et al., 2010b)]. This difference is caused mainly by the decrease in the rate of ribonucleotide incorporation, rather than nucleotide binding. As with other DNA polymerases, Rev1 readily incorporates araC, with only an ~2-fold reduced efficiency compared to dCTP [Table 1, (Brown et al., 2010b)].
2.4. Archaeo-eukaryotic primase polymerase
It is perhaps ironic that the most recent addition to the extended family of TLS polymerases is, probably, the oldest enzyme equipped to bypass DNA lesions (Bianchi et al., 2013; Garcίa-Gόmez et al., 2013; Rudd et al., 2013). DNA-directed primase-polymerase termed PrimPol is a versatile replicative enzyme belonging to the archaeo-eukaryotic primase superfamily of polymerases (Garcίa-Gόmez et al., 2013; Wan et al., 2013). Its unique set of properties has earned PrimPol a secure position within the intricate multi-protein network involved in DNA metabolic processes [summarized in several comprehensive reviews (Guilliam and Doherty, 2017; Rudd et al., 2014)]. PrimPol is well-suited to facilitate replication fork progression overcoming diverse obstacles through a variety of strategies. It can skip unreadable DNA sequences (Martinez-Jimenez et al., 2015), re-initiate DNA synthesis downstream of replisome-stalling lesions (Mourόn et al., 2013), as well as catalyze direct replication across damaged sites (Garcίa-Gόmez et al., 2013; Rudd et al., 2013). While PrimPol prefers to extend primers by inserting dNTPs, it is able to utilize both rNTPs and dNTPs during initiation of replication, as well as during chain elongation, at least when the reactions are activated by Mn2+, enzyme’s preferred metal cofactor. PrimPol can also quite efficiently catalyze ribonucleotide incorporation in the course of TLS, which at least in the case of 8-oxo-G bypass, leads to increased replication fidelity (Garcίa-Gόmez et al., 2013).
3. Molecular mechanisms of sugar discrimination by TLS polymerases
At first glance, rNTP incorporation seems harmless as long as the base of the incoming nucleotide forms a correct Watson-Crick pair with the template base. Furthermore, several recent publications suggest that errors in sugar selectivity made by DNA polymerases could actually be beneficial for the cells. For example, it has been suggested that rNMPs incorporated during chromosomal replication might serve as physiological markers of the nascent DNA in eukaryotes and gram-positive bacteria (Ghodgaonkar et al., 2013; Yao et al., 2013). In this case, the presence of ribonucleotides plays a role similar to the hemi-methylation in gram-negative bacteria by directing the mismatch repair proteins to correct replication errors in the newly synthesized DNA strands and thus preserve genomic integrity. We have recently shown that ribonucleotides might also play a protective role in E. coli by triggering repair pathways specifically directed at their removal, but simultaneously correcting deoxyribo- and ribo-nucleotide base mismatches (McDonald et al., 2012; Vaisman et al., 2013). Another example of when rNMP incorporation might be beneficial for the cell, is double strand break repair (DSBR) through non-homologous end joining (NHEJ), which appears to be more efficient during pol μ-guided formation of a ribonucleotide-containing DNA substrate (Nick McElhinny and Ramsden, 2003).
On the other hand, an excessive number of ribonucleotides embedded into genomic DNA can potentially threaten the cell’s well-being, if left unrepaired. Notably, the presence of a reactive 2’-hydroxyl on the ribose ring makes the DNA strand more susceptible to spontaneous and enzymatic hydrolytic cleavage, thus reducing its overall chemical stability and increasing genome instability. Substitution of dNMPs with rNMPs also distorts the structure of the double helix and as consequence, inhibits nucleosome formation, disrupts the ability of DNA-binding proteins to recognize DNA, and interferes with various DNA processing pathways. For example, even an isolated rNMP located within a DNA template can slow down the progression of a replication fork, and when a single rNMP is present at the 3’-end of the primer, the resulting distortion of the 3’-OH group can potentially terminate primer elongation [Table 2, (Yao et al., 2013)]. Furthermore, a DNA double helix with multiple sequential rNMPs undergoes conformational changes from the standard B-form to A-form, which explains the inability of the majority of DNA polymerases to extend RNA primers and to incorporate several rNMPs in a row. The accumulation of clustered ribonucleotides also leads to replication fork stalling (Clausen et al., 2013; Nick McElhinny et al., 2010a; Nick McElhinny et al., 2010b; Watt et al., 2011), and the failure to process ribonucleotides properly has been shown to cause replication stress and genome instability in yeast (Nick McElhinny et al., 2010b). Removal of random ribonucleotides from the mammalian genome is essential for mouse embryonic development (Reijns et al., 2012) and the inability to repair ribonucleotide lesions has been linked to Aicardi-Goutières syndrome, a chronic inflammatory disorder in humans (Rabe, 2013). Taking into consideration that cellular rNTP levels are higher than the corresponding dNTPs, it becomes obvious that prevention of ribonucleotide accumulation in double-stranded DNA is an important task, which is achieved by a living cell using a variety of strategies.
The first line of defense against incorporation of ribonucleotides into genomic DNA comes from DNA polymerases by themselves. At a first glance, it might seem that because of the relaxed constrains of their active site and inability to proofread errantly inserted rNMPs due to the lack of the 3’−5’ exonucleolytic activity, low fidelity TLS polymerases in general should be less likely to prevent binding of an incoming rNTP than a high fidelity replicative DNA polymerase. In reality, discrimination factors are not directly related to the base-substitution fidelity of the enzyme and vary greatly not only within the same polymerase family, but also between paralogs in the same organism and between orthologs of the same gene in different organisms. As discussed above, among TLS polymerases there are enzymes that are naturally adept to accommodate nucleotides with an extra OH group at the 2’ position of the sugar ring and those that are remarkably strict in selecting a nucleotide with the appropriate sugar (Table 1).
3.1. The steric exclusion mechanism for protecting DNA from ribonucleotide invasion
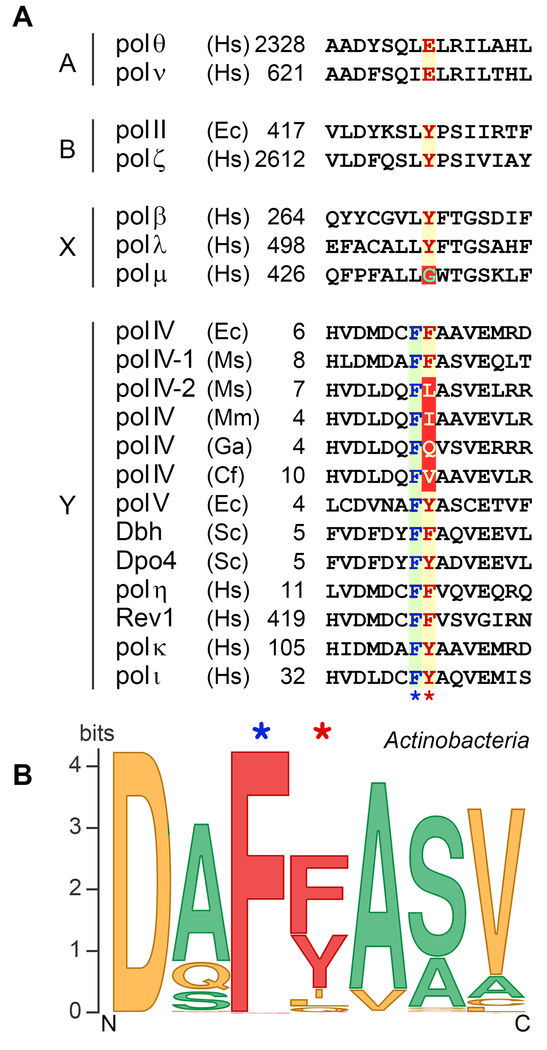
Even though the ability to discriminate between deoxyribo- and ribo- substrates varies greatly in different DNA polymerases, most of them favor dNTP incorporation with a high degree of selectivity. Various kinetic, site-directed mutagenesis and crystallographic studies have suggested that the responsibility to monitor the 2’ position on the sugar ring of the incoming nucleotide is confined to a specific amino acid within the nucleotide binding pocket in the polymerase active site [reviewed in (Joyce, 1997)] that clashes with the 2’-OH of the incoming rNTP, thus physically blocking a nucleotide with the wrong sugar from being aligned for incorporation into DNA. Based on these studies, the structural model of suitable sugar selection was borne (Joyce, 1997). According to this model, the backbone nitrogen of the so-called “steric gate” residue forms the hydrogen bond with the OH group at the 3’ position on the sugar ring of the incoming nucleotide, while the side chain of the same residue is positioned such that it occupies the space which otherwise could accommodate a hydroxyl group at the C2′ position of rNTP. The residue that acts as the steric barrier is highly conserved within each polymerase family. In the A-family it is glutamic acid, in the C-family it is histidine, and in the B- and Y- family polymerases it almost always is tyrosine or phenylalanine (Figure 2A). For polymerases from the X-family, the model of steric exclusion of ribonucleotides is somewhat different. In order to prevent rNTP accommodation, these polymerases rely on a protein backbone segment, rather than on a large side chain of a specific amino acid (described below). As any other protective system, the steric exclusion defence mechanism is not one hundred percent “fool-proof”. In the next sections, we will discuss how stringent the steric barrier is in Y- and X-family TLS polymerases and how easily can it be breached.
Figure 2. Multiple sequence alignments of the amino acid region involved in sugar discrimination in TLS DNA polymerases.
(A) Sequence alignments for TLS polymerases from A-, B, X, and Y families. The highly conserved single amino acid residue within each sequence responsible for the steric exclusion of ribonucleotides is shown in red letters over a yellow background and is indicated by the red star below the alignment. The unusual steric gate Gly in pol μ is indicated in green over a red background. The unconventional steric gate residues [I, L, Q and V] found in some DinB polymerases from Actinobacteria are shown in yellow letters over a red background. Almost invariant amino acid [F] juxtaposed to the steric gate residue of Y-family polymerases is shown in blue letters over a green background and is indicated by the blue star below the alignment. The numbers indicate the amino acid position of the first residue shown and are relative to the N-terminus for each polymerase. The species abbreviations are as follows: Ec, Escherichia coli; Ms, Mycobacterium smegmatis; Mm, Microbacterium mangrovi; GA, Gordonia araii; Cf, Cellulomonas fimi; Sa, Sulfolobus acidocaldarius; Ss, Sulfolobus solfataricus; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens. (B) Steric gate polymorphism among actinobacterial DinB homologs. A sequence logo for the fragment of DNA polymerase IV from Actinobacteria consisting of steric gate residue and three flanking residues on each side was created using Weblogo program (Crooks et al., 2004; Schneider and Stephens, 1990). The original sequence set consisted of 3759 actinobacteria sequences from Uniprot depository (including computationally translated sequences from TrEMBL database), predicted by InterPro signature IPR022880 as belonging to DNA polymerase IV family (retrieved on September 3, 2017, http://www.ebi.ac.uk/interpro/entry/IPR022880) (Finn et al., 2017). Sequences exceeding the threshold of 90% identity were removed, resulting in the smaller set of 1550 proteins. Multiple sequence alignment was performed using Kalign program (Lassmann et al., 2009). Sequences poorly aligned or with the gaps in the steric gate region were removed, which left 1347 proteins that were used for the sequence logo construction. Amino acids are color-coded according to their molecular weight (MW). Steric gate position in the wild type DNA polymerases are most often occupied by bulky residues (shown in red). Amino acids with lower MW identified in DinB homologs from Actinobacteria are shown in yellow. Amino acids containing the smallest side chains (shown in green) are often used to replace the steric gate residues in recombinant mutant polymerases. We thank William Taft and Iosif Vaisman (George Mason University) for creating the Dpo4 alignments and logo. A color version of the figure is available online.
3.2. The “steric gate” in Y-family DNA polymerases
Although the steric gate in Y-family polymerases is the key barrier preventing rNMPs from infesting genomic DNA, it is not the only sugar recognition determinant, as clearly illustrated by the dramatic differences in sugar selectivity of human and yeast pols η despite the fact that the steric gate in both orthologs is phenylalanine (Phe35 and Phe18 in S.cerevisiae and human pol η, respectively). What causes human and yeast enzymes to behave so differently is presently unknown and would be interesting to elucidate.
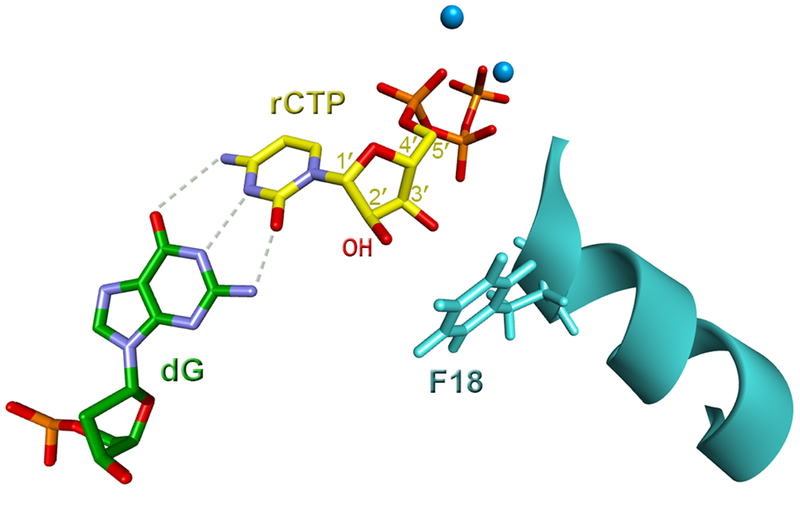
An important contribution into understanding the mechanism regulating the ability of hpol η to incorporate ribonucleotides has been made using the crystal structure of the human DNA polymerase with an incoming ribonucleotide (rC) opposite both an undamaged DNA base (dG) (Figure 3) and an 8-oxodG lesion (Mentegari et al., 2017; Su et al., 2016). It has been shown that pol η scaffolds the incoming rNTP to pair with the complementary template with a significant propeller twist. As a result of the somewhat different orientation of the incoming rNTP relative to dNTP in the polymerase active site, a steric clash between the 2’-OH group of the ribose and the steric gate of pol η is reduced even when the DNA strand is damaged. However, the distance between the primer terminus and the incoming rNTP increases, thus elevating the energy barrier. Consequently, even though human pol η readily inserts rNTPs, the catalytic efficiency of this reaction is still substantially lower compared with the efficiency of dNTP incorporation (Table 1) (Su et al., 2016).
Figure 3. Insertion of an incoming rCTP (shown in yellow) opposite template dG (shown in green) by human pol η (light blue).
The F18 steric gate residue of hpol η, the 2’ OH group of the sugar ring in rCTP, and Ca2+ ions (blue) are indicated. (PDB ID 5EWE) (Su et al., 2016). For details see section 3.2. A color version of the figure is available online.
Knowledge of the position and the identity of a specific residue that shapes sugar selectivity of DNA polymerases lies at the basis of another highly informative approach used to study the cause and cost of ribonucleotide incorporation. Indeed, significant insights have been gained through the investigation of the consequences of the replacement of the steric gate residues. It should be noted that here, we will mainly summarize existing information about the effects of such substitution on the biochemical properties of the polymerases while its biological outcome, although extremely valuable and useful, is beyond the scope of this review.
Multiple studies using different DNA polymerases demonstrated that by simply replacing the steric gate residue with an amino acid containing a smaller side chain, ribonucleotide discrimination can be reduced by 2–3 orders of magnitude (Table 1). This is hardly surprising, since such replacement clears a space in the polymerase binding-pocket which can be used to accommodate the 2’-OH group of the ribose [Figure 3, (Kirouac et al., 2011; Su et al., 2016)]. However, compromised sugar selectivity in most mutant polymerases from different families is not only caused by the increased efficiency of rNTP incorporation, but also by the decreased efficiency of dNTP incorporation, as well as a diminished ability to extend primers containing the 3’-terminal rNMP (Cavanaugh et al., 2011; DeLucia et al., 2003; Donigan et al., 2014; Donigan et al., 2015; Katafuchi et al., 2010; Kirouac and Ling, 2009; Kirouac et al., 2011; Niimi et al., 2009; Sherrer et al., 2010), possibly due to the loss of stacking interactions between the steric gate residue and the sugar ring of the incoming nucleotide (Kirouac et al., 2011; Nevin et al., 2015; Su et al., 2016). Therefore, the bulky side chain of the steric gate is not only important for the effective exclusion of ribonucleotides, but also for the efficient incorporation of correct deoxynucleotides. One exception to this rule is the F13V substitution at the steric-gate of E. coli pol IV which results in the increased efficiency of the correct dNTP incorporation (Jarosz et al., 2006). Interestingly, while the Phe to Val change is sufficient to remove the steric barrier blocking rNTP incorporation into undamaged DNA (Table 1), the efficiency of rCTP incorporation opposite a N2-furfuryl-dG adduct is the same for the wild type and F13V mutant pol IV (Jarosz et al., 2006).
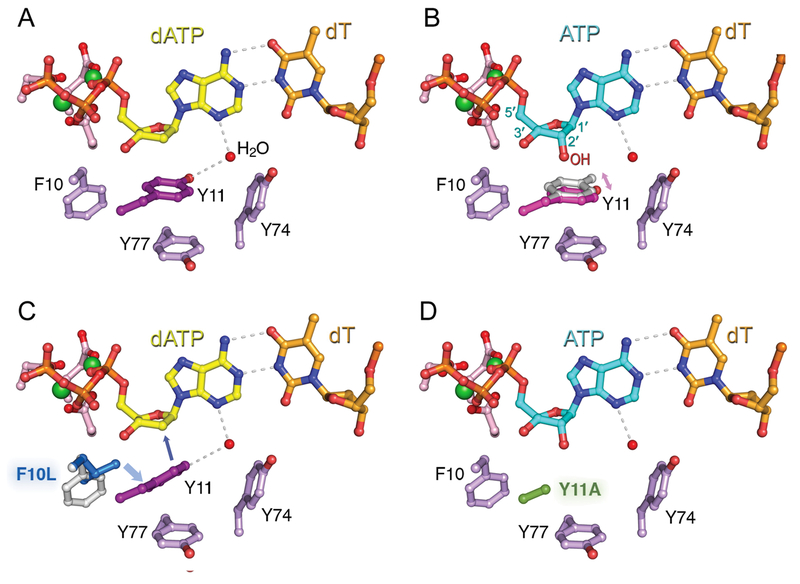
Similar levels of ribonucleotide incorporation opposite damaged sites by wild-type and a steric gate mutant was also observed for E. coli pol V using DNA templates with a T-T cyclobutane pyrimidine dimer (Kuban et al., 2012). However, the steric gate substitution reduces the already poor sugar discrimination of pol V on undamaged DNA (Figure 4D) such that the Y11A variant becomes a de facto DNA-dependent RNA polymerase capable of synthesizing long RNA stretches with the efficiency, processivity, and base substitution fidelity comparable to that of DNA synthesis (Vaisman et al., 2012a). Even when presented with a mixture of ribonucleotides and deoxyribonucleotides, Y11A often prefers to incorporate rNTPs. Interestingly, when Tyr11 was replaced with phenylalanine, another residue conserved as a steric gate in Y-family polymerases, the sugar discrimination of the Y11F mutant actually improved, suggesting that phenylalanine might sense a ribose moiety better than tyrosine, at least in UmuC (Vaisman et al., 2012a).
Figure 4. Structural models of ribonucleotide exclusion by wild type and steric gate variants of E. coli DNA pol V.
Models of wild type and mutant UmuC inserting dATP (shown in yellow) or rATP (blue) opposite a template T (orange) are based on the structure of human DNA pol η (PDB 3MR3). Comparison of the structures of wild type UmuC incorporating dATP (A) and rATP (B) provides an explanation of the unusually low sugar selectivity of pol V. The steric gate residue, Y11, of the wild type enzyme is characterized by increased flexibility. Rotation of the Y11A benzene ring (alternate positions shown in pink and grey) allows for the accommodation of the hydroxyl group at the 2’ position of the nucleoside (indicated in red). (C) Substitution of the highly conserved F10 residue (shown in grey) with leucine (blue) results in dramatic increase in sugar selectivity because the side chain of leucine presses on the benzene ring of Y11 positioning it closer to the C2 position of the sugar moiety of the incoming nucleotide thereby favoring deoxyribonucleotide (dATP) selection and preventing ribonucleotide incorporation. (D) Substitution of the steric gate Y11 with alanine (shown in green) containing smaller side chain creates a void in the active site of UmuC thus relaxing the minor groove alignment for correct Watson-Crick base pair and facilitating accommodation of the 2’-OH group of the incoming nucleotide (shown for ATP). Adapted from (Vaisman et al., 2012a), with permission. A color version of the figure is available online.
The effects of a steric gate alanine substitution was also tested for the S.cerevisiae pol η which is in contrast to its human homolog and E.coli pol V, is highly skillful when choosing nucleotides with the correct sugar (Donigan et al., 2014; Donigan et al., 2015). The resulting F35A variant of yeast pol η acquired the ability to incorporate rNMPs with a high degree of base selectivity on both undamaged DNA and a T-T cyclobutane pyrimidine dimer-containing DNA template. Nevertheless, it was still less prone to accommodate rNMPs than the wild-type human pol η (Donigan et al., 2014; Donigan et al., 2015; Mentegari et al., 2017; Su et al., 2016). The mutant enzyme also acquired the capacity to generate tracks of up to six consecutive rNMPs, but at the cost of a reduction in its overall catalytic activity (Donigan et al., 2014; Donigan et al., 2015).
Very interesting data were obtained in the course of the characterization of a steric gate mutant of human pol ι. Unexpectedly, a Y39A steric gate substitution not only resulted in a dramatic reduction in Mg2+-dependent DNA polymerization activity, but quite remarkably, rNMP insertion also became much less efficient (Donigan et al., 2014). However, when Mg2+ was replaced with Mn2+ as the metal ion for catalysis, the mutant pol ι exhibited a marked increase in rNTP incorporation and extension using both undamaged and damaged DNA substrates [Table 1, (Donigan et al., 2014)]. Strikingly, the mutant polymerase also lost the unique and characteristic pol ι base-selection infidelity. This unexpected change in behavior was explained by molecular modeling which revealed that the Y39A substitution reduced the normally error-prone constraints on the active site in the steric gate mutant, so as to support canonical Watson-Crick base pairing and an increase in base selectivity (Donigan et al., 2014). An improved fidelity was also described for the Y112A and F35A steric gate variants of human pol κ (Katafuchi et al., 2010) and yeast pol η (Donigan et al., 2014; Donigan et al., 2015), respectively. However, the correlation between the reduced size of the side chain of the amino acid located at the steric gate and an increase in polymerase accuracy is not universal in TLS polymerases, since there is no increase in the fidelity of dNTP incorporation in the archaeal Dpo4 or Dbh, or E. coli pol V steric gate mutants (DeLucia et al., 2003; Katafuchi et al., 2010; Kirouac and Ling, 2009; Sherrer et al., 2010; Shimizu et al., 2003; Vaisman et al., 2012a).
While the Mg2+-dependent reduction of rNMP incorporation in the pol ι steric gate mutant was unexpected, the same effect caused by the steric gate replacement in M. smegmatisa DinB2 raised no eyebrows. One look at the primary amino acid sequence alignment (Figure 2) and everything becomes clear. As mentioned above, tyrosine and phenylalanine, both of which are equipped with bulky aromatic side chains, alternate in order to fulfill the function of a steric gate preventing incorporation of rNTPs into DNA by the majority of Y- family polymerases. The intriguing exception from this rule was recently found in some bacterial DinB homologs such as M. smegmatis and M. tuberculosis DinB2 which has a leucine residue at the steric gate position [Figure 2, (Kana et al., 2010; Ordonez et al., 2014)]. The leucine amino acid with its small side chain can not serve as a very effective protector against ribonucleotide incorporation. Indeed, sugar discrimination of M. smegmatis DinB2 is among the lowest found in TLS DNA polymerases (Ordonez et al., 2014), while other mycobacterial DinB homologs with their conventional steric gates (Phe in DinB1 and Tyr in DinB3), display a strong preference for dNTP incorporation (Ordonez et al., 2014). Therefore, it comes as no surprise that replacement of leucine 14 by phenylalanine “closes” the loose steric gate of M. smegmatis DinB2 and suppresses its ribonucleotide incorporation activity (Ordonez et al., 2014). And vice versa, the F23L variant of M. smegmatis DinB1 acquires vigorous rNTP insertion activity. Interestingly, the F23L change has no obvious effect on the DNA polymerase activity of DinB1, while the L14F change in DinB2 diminishes DNA synthesis by insertion of dNTPs, although to a lesser degree than it reduces efficiency of rNTP incorporation. With regard to the replacement of the steric gate residue by alanine, the L14A mutation had very little effect on the ability of M. smegmatis DinB2 to perform DNA synthesis with either dNTPs or rNTPs, whereas F23A replacement in DinB1, without affecting dNTP incorporation, significantly improved RNA synthesis although to a lesser extent compared to the F23L change (Ordonez et al., 2014).
In addition to having a key function in selection of nucleotide with appropriate sugar ring and facilitating the correct deoxynucleotide incorporation, the steric gate residue has also been proposed to play an important role in enabling efficient TLS by some DNA polymerases, as seen in the bypass of N2-dG lesions by various DinB orthologs. For example, E. coli pol IV and S. acidocaldarius Dbh in which the steric gate residues (F13 and F12, are replaced by Val or Ala, respectively), lost their ability to extend primers past the N2-furfuryl-dG and N2-B[a]P-dG adducts (Jarosz et al., 2006). The TLS capacity of the Y112A and Y112V steric gate variants of human pol κ is also compromised when dealing with lesions such as thymine glycol, or methylated bases (Niimi et al., 2009). However, the effect of the steric gate identity on the TLS efficiency is not predetermined, even for the same DNA polymerase, but it is rather lesion-specific. Thus, bypass of 8-oxo-guanine does not depend on the status of the steric gate of pol κ, and moreover, opposite the BPDE-N2-dG adduct, the Y112A mutant incorporates dCTP even more efficiently than the wild-type pol κ. Similar or even higher TLS efficiency in the steric gate mutants compared to the wild type enzymes has been also shown for other Y-family polymerases (pol V, pol η and pol ι (Donigan et al., 2014; Donigan et al., 2015; Kuban et al., 2012).
3.3. The “steric fence” in X-family DNA polymerases
Structural analysis of X-family polymerases suggested that the putative steric gate position is occupied by tyrosine in pol β (Tyr271) and pol λ (Tyr505 in human and Tyr 503 in mouse orthologs) and by a more flexible glycine (Gly433) residue in pol μ (Figure 2A), which correlates well with the sugar selectivity of these polymerases (Table 1). Accordingly, substitution of the Gly433 residue of pol μ with the Tyr causes dramatic 25- to 40-fold increase in the sugar selectivity (Brown et al., 2010a; Ruiz et al., 2003). However, the reverse replacement of Tyr with Gly or Ala diminishes the sugar selectivity of human pol λ (Y505G and Y505A) and human pol β (Y271A) only moderately (4–12 times) such that ribonucleotide discrimination of mutant pols β and λ is still 40–250 times lower than that of wild-type pol μ [Table 1. (Brown et al., 2010a; Cavanaugh et al., 2010; Ruiz et al., 2003)]. Furthermore, the decrease in sugar selectivity caused by the Y271A substitution is not that much different from the decrease resulting from the replacement of the tyrosine with the very similar phenylalanine in the Y271F pol β variant (~3-fold) (Brown et al., 2010a; Cavanaugh et al., 2011). The modest reduction in nucleotide selectivity of the pol β and pol λ steric gate variants compared to changes observed for other DNA polymerases with equivalent mutations is explained in part by a minuscule decrease in the efficiency of the Watson-Crick dNTP incorporation (Cavanaugh et al., 2011). These data are consistent with a proposed structural model, according to which, X-family polymerases unlike polymerases from other families, do not rely solely on the side chain of a steric gate in order to prevent incorporation of rNTPs into genomic DNA (Pelletier et al., 1994). Instead, the primary responsibility for maintaining high sugar selectivity in these polymerases lies with the protein backbone segment near the C-terminus of an α-helix (Brown et al., 2010a; Cavanaugh et al., 2011). According to this model, the backbone carbonyl (Y271 in pol β and Y505 in pol λ) behaves more like a steric “fence” than a “gate” and the backbone segment (Y271–G274 in pol β and Y505-G508 in pol λ) rather than the backbone of a single residue, determines the sugar selectivity of these enzymes (Brown et al., 2010a; Pelletier et al., 1994). In addition, an interaction between the primer terminus and the side-chain of Y271 in pol β, is important for shaping the active site geometry, and plays a supportive role in deterring ribonucleotide insertion (Cavanaugh et al., 2011).
Similar to pols β and λ who need more than the backbone of a single residue to ensure effective ribonucleotide exclusion, the reason for the inability of the X-family pol μ to distinguish between rNTPs and dNTPs (Table 1) is more complex than the small size of the side chain of the Gly433 residue. Molecular dynamics simulations (Li and Schlick, 2013) have shown that compared to pols β and λ, pol μ has much more flexible active site. These findings provide an additional explanation for the unique ability of pol μ to accommodate ribonucleotides in the binding cleft and to catalyze their insertion. Further insights were gained through recent structural studies of the catalytic domain of pol μ, in complex with a single-strand break gapped DNA and incoming ribonucleotides (Moon et al., 2017). This study suggests that binding and incorporation of a correctly paired rNTP occurs without distortion of active site geometry which, surprisingly, does not differ much from the active sites of pols β or λ. Furthermore, the structural and biochemical data obtained using wild-type and various mutants of pol μ agree with previous assessments that similar to pols β or λ, sugar selectivity of pol μ cannot be simply attributed to a single steric gate residue. However, the uncovered synergistic interactions with multiple active site residues, including a residue positioned too far to directly sense the presence of a 2′-OH of rNTP have not been detected in pols β and λ, X-family polymerases with better sugar selectivity.
It should be noted that the lack of sugar discrimination was detected for pol μ only when the polymerase’s preferred substrate, i.e., a short-gapped DNA (Pryor et al., 2015), was used in the in vitro reconstituted reactions. On primed single-stranded templates, the sugar selectivity of pol μ is as high as of pols β and λ, leading to the suggestion that pol μ utilizes different exclusion mechanisms on gapped and non-gapped DNA substrates (Roettger et al., 2004). Further structural studies are clearly necessary to test this hypothesis and to gain a better understanding of the molecular mechanism of the steric fence ribonucleotide exclusion.
3.4. Other factors affecting ribonucleotide discrimination
Despite a dramatic reduction in sugar selectivity caused by the replacement of a steric gate residue by an amino acid with a smaller side chain, most mutant polymerases still generally prefer dNTPs over rNTPs (Table 1). These findings imply that besides the active site architecture and the basic mechanism of sugar selection, other factors affect furanose recognition. Indeed, it has been shown that ribonucleotide discrimination can be modulated by amino acids other than the actual steric gate, thereby serving as additional barriers assisting in prevention of rNTP incorporation by DNA polymerases [reviewed in (Brown and Suo, 2011)].
Analysis of the crystal structure of human pol η with incoming rNTP revealed that the active site of the polymerase contains a residue which appears to provide a second line of defense against the accommodation of nucleotides with an incorrect sugar in the minor groove of the enzyme (Su et al., 2016). This hypothesis was based on the ability of the side chain of Tyr-92 to form a stacking interaction with the benzene ring of Phe-18 thus stabilizing the position of the side chain of the steric gate residue. Indeed, a Y92A substitution in hpol η relaxed the discrimination against ribonucleotides by more than an order of magnitude. A similar model based on the stacking interaction of the steric gate with the tyrosine residue acting as the so-called “second guard” has also been proposed for other Y family DNA polymerases including pol ι (Tyr-102), human pol κ (Tyr-174) and its S. solfataricus homolog Dpo4 (Tyr-81).
Several studies suggested that sugar tolerance can be modulated by residues directly flanking the steric gate [(Brown and Suo, 2011; Nick McElhinny et al., 2010b) and references therein]. For example, biochemical characterization of E. coli pol V variants revealed that when the highly conserved F10 residue that is juxtaposed to the Y11 steric gate, was replaced by leucine (Figures 2 and 4), incorporation of nucleotides with the wrong sugar and/or wrong base was severely inhibited (Vaisman et al., 2012a). Even after prolonged incubation, F10L was only able to elongate primers by no more than four rNTPs before synthesis stopped. Under the same reaction conditions, wild-type pol V catalyzed synthesis of an RNA strand of at least several hundred nucleotides in length. These data suggest that Leu10 dramatically strengthens the capacity of the designated steric gate, Y11, to protect the E. coli genome from incorporation of errant rNMPs, while also improving the overall fidelity of the enzyme. In support of these findings, structural modeling of UmuC revealed that the branched side-chain of L10 presses on the benzene ring of Y11 forcing its side chain closer to the C2′ position of the incoming nucleotide thereby firmly closing the “steric gate” [Figure 4C, (Vaisman et al., 2012a)]. As a result, the F10L mutant is characterized by a reduction in TLS activity in addition to enhanced base substitution fidelity and an improved ability to sense the 2′-hydroxy group of incoming nucleotides.
The ability of the residues neighboring the steric gate to influence sugar selectivity has been a very useful tool to study the consequences and processing of ribonucleotides incorporated into genomic DNA. For example, the ability to modulate rNTP exclusion through the substitution of an amino acid adjacent to the steric gate has been successfully utilized with replicative polymerases (yeast pols ε and δ (Nick McElhinny et al., 2010b; Lujan et al., 2013) and E. coli pol III (our unpublished observations) where substitution of a steric gate residue is lethal. Interestingly, replacement of methionine adjacent to the steric gate tyrosine in yeast DNA polymerase ε by leucine or glycine had opposite effects, with leucine decreasing and glycine increasing rNMP incorporation (Nick McElhinny et al., 2010b).
In contrast to E. coli pol V and yeast pol ε, mutation of F272, an amino acid with an aromatic side chain immediately adjacent to the steric gate of pol β (Y271), had very little effect on sugar selectivity, despite the fact that this residue is very close to the ribose ring of the incoming nucleotide (Cavanaugh et al., 2011). Although replacing the F272 by alanine substantially reduced the efficiency of the correct nucleotide insertion compared with wild type enzyme, ribonucleotide discrimination of the mutant polymerase increased slightly. Therefore, the contribution of amino acid residues in the vicinity of the steric gate into sugar selectivity appears to be specific for the particular DNA polymerase.
4. Concluding Remarks
Significant progress in understanding the mechanisms regulating ribonucleotide incorporation by various TLS DNA polymerases has been made during the past two decades. However, we still have much to learn, considering that for a number of DNA polymerases, the biochemical characterization of sugar selectivity is still lacking. It is hard to overestimate the importance of emerging structure-function studies which will help us not only to learn how each individual polymerase selects the appropriate substrate from a nucleotide pool and to uncover common mechanisms governing the behavior of individual polymerases, but also to better understand how such properties are used by each enzyme to fulfill its biological functions.
As with any other field of research, the more we learn, the more questions arise, many of which can open new fascinating avenues of future research. Why the steric gate position in the recently discovered DinB2 paralog found in some species of the actinobacterial genera Mycobacterium, is occupied by an ineffective Leu residue instead of the canonical Phe or Tyr found in all other Y family polymerases, is just an example of such questions. Finding an answer to this question can potentially open a new chapter in the studies of sugar discrimination by DNA-dependent DNA polymerases. Interestingly, multiple sequence alignments uncovered other uncommon steric gate residues (isoleucines, glutamines, and valines) in DinB-like polymerases from various species of Actinobacteria (Ordonez and Shuman, 2014; Ordonez et al., 2014) (Figure 2). Today, no significant naturally-occurring steric gate polymorphism has been identified in other type of organisms raising the question of how unique is this phenomenon?
Footnotes
Disclosure of interest:
Funding for this review was provided by the National Institute of Child Health and Human Development/ National Institutes of Health Intramural Research Program.
References
- Ahn J, Kraynov VS, Zhong X et al. (1998). DNA polymerase β: effects of gapped DNA substrates on dNTP specificity, fidelity, processivity and conformational changes. Biochem J 331:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoufouchi S, Flatter E, Dahan A et al. (2000). Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res 28:3684–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Lada AG, Siebler HM et al. (2012). DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J Biol Chem 287:17281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett E, Vaisman A, Tropea KA et al. (2002). Frameshifts and deletions during in vitro translesion synthesis past Pt-DNA adducts by DNA polymerases β and η. DNA Repair 1:1003–16. [DOI] [PubMed] [Google Scholar]
- Belousova EA, Maga G, Fan Y et al. (2010). DNA polymerases β and λ bypass thymine glycol in gapped DNA structures. Biochemistry 49:4695–704. [DOI] [PubMed] [Google Scholar]
- Bergoglio V, Ferrari E, Hubscher U et al. (2003). DNA polymerase β can incorporate ribonucleotides during DNA synthesis of undamaged and CPD-damaged DNA. J Mol Biol 331:1017–23. [DOI] [PubMed] [Google Scholar]
- Bianchi J, Rudd SG, Jozwiakowski SK et al. (2013). PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell 52:566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanca G, Villani G, Shevelev I et al. (2004). Human DNA polymerases λ and β show different efficiencies of translesion DNA synthesis past abasic sites and alternative mechanisms for frameshift generation. Biochemistry 43:11605–15. [DOI] [PubMed] [Google Scholar]
- Boudsocq F, Iwai S, Hanaoka F, Woodgate R. (2001). Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DNA polymerase with lesion-bypass properties akin to eukaryotic polη. Nucleic Acids Res 29:4607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braithwaite DK, Ito J. (1993). Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res 21:787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Fiala KA, Fowler JD et al. (2010a). A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. J Mol Biol 395:282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Fowler JD, Suo Z. (2010b). Kinetic basis of nucleotide selection employed by a protein template-dependent DNA polymerase. Biochemistry 49:5504–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Suo Z. (2011). Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry 50:1135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckstein MH, He J, Rubin H. (2008). Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol 190:718–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW. (2014). Molecular biology. Ribose--an internal threat to DNA. Science 343:260–1. [DOI] [PubMed] [Google Scholar]
- Cavanaugh NA, Beard WA, Wilson SH. (2010). DNA polymerase β ribonucleotide discrimination: insertion, misinsertion, extension, and coding. J Biol Chem 285:24457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh NA, Beard WA, Batra VK et al. (2011). Molecular insights into DNA polymerase deterrents for ribonucleotide insertion. J Biol Chem 286:31650–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. (2016). The balancing act of ribonucleotides in DNA. Trends Biochem Sci 41:434–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilli P, Minoprio A, Bossa C et al. (2015). Formation and repair of mismatches containing ribonucleotides and oxidized bases at repeated DNA sequences. J Biol Chem 290:26259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen AR, Murray MS, Passer AR et al. (2013). Structure-function analysis of ribonucleotide bypass by B-family DNA replicases. Proc Natl Acad Sci U S A 110:16802–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespan E, Furrer A, Rosinger M et al. (2016). Impact of ribonucleotide incorporation by DNA polymerases β and λ on oxidative base excision repair. Nat Commun 7:10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. (2004). WebLogo: a sequence logo generator. Genome Res 14:1188–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard JZ. (2012). Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet 28:592–7. [DOI] [PubMed] [Google Scholar]
- DeLucia AM, Grindley ND, Joyce CM. (2003). An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res 31:4129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucia AM, Chaudhuri S, Potapova O et al. (2006). The properties of steric gate mutants reveal different constraints within the active sites of Y-family and A-family DNA polymerases. J Biol Chem 281:27286–91. [DOI] [PubMed] [Google Scholar]
- Donigan KA, McLenigan MP, Yang W et al. (2014). The steric gate of human DNA polymerase ι regulates ribonucleotide incorporation and deoxyribonucleotide fidelity. J Biol Chem 289:9136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donigan KA, Cerritelli SM, McDonald JP et al. (2015). Unlocking the steric gate of DNA polymerase η leads to increased genomic instability in Saccharomyces cerevisiae. DNA Repair 35:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro P, Franzolin E, Pontarin G et al. (2010). Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res 38:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala KA, Duym WW, Zhang J, Suo Z. (2006). Up-regulation of the fidelity of human DNA polymerase λ by its non-enzymatic proline-rich domain. J Biol Chem 281:19038–44. [DOI] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC et al. (2017). InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res 45:D190-D99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RP, Fujii S. (2007). Translesion synthesis in Escherichia coli: lessons from the NarI mutation hot spot. DNA Repair (Amst) 6:1032–41. [DOI] [PubMed] [Google Scholar]
- Gali VK, Balint E, Serbyn N et al. (2017). Translesion synthesis DNA polymerase η exhibits a specific RNA extension activity and a transcription-associated function. Sci Rep 7:13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz M, Murray MS, Kunkel TA, Chou KM. (2010). Interaction between DNA polymerase λ and anticancer nucleoside analogs. J Biol Chem 285:16874–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcίa-Gόmez S, Reyes A, Martίnez-Jimenez MI et al. (2013). PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell 52:541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodgaonkar MM, Lazzaro F, Olivera-Pimentel M et al. (2013). Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell 50:323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosavi RA, Moon AF, Kunkel TA et al. (2012). The catalytic cycle for ribonucleotide incorporation by human DNA pol λ. Nucleic Acids Res 40:7518–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliam TA, Doherty AJ. (2017). PrimPol-prime time to reprime. Genes (Basel) 8:E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Seki M, Wood RD et al. (2011). Lesion bypass activity of DNA polymerase θ (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol 405:642–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Sauer-Eriksson AE, Johansson E. (2012). Promiscuous DNA synthesis by human DNA polymerase θ. Nucleic Acids Res 40:2611–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Braithwaite DK. (1991). Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res 19:4045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Godoy VG, Delaney JC et al. (2006). A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439:225–8. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Karata K, Woodgate R et al. (2009). The active form of DNA polymerase V is UmuD’2C-RecA-ATP. Nature 460:359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S, Klein HL. (2015). Ribonucleotides in DNA: hidden in plain sight. Nat Struct Mol Biol 22:176–8. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S. (2012). Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc Natl Acad Sci U S A 109:12455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce CM. (1997). Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci U S A 94:1619–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana BD, Abrahams GL, Sung N et al. (2010). Role of the DinB homologs Rv1537 and Rv3056 in Mycobacterium tuberculosis. J Bacteriol 192:2220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karata K, Vaisman A, Goodman MF, Woodgate R. (2012). Simple and efficient purification of Escherichia coli DNA polymerase V: cofactor requirements for optimal activity and processivity in vitro. DNA Repair 11:431–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katafuchi A, Sassa A, Niimi N et al. (2010). Critical amino acids in human DNA polymerases η and κ involved in erroneous incorporation of oxidized nucleotides. Nucleic Acids Res 38:859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent T, Mateos-Gomez PA, Sfeir A, Pomerantz RT. (2016). Polymerase θ is a robust terminal transferase that oscillates between three different mechanisms during end-joining. Elife 5:e13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketkar A, Zafar MK, Banerjee S et al. (2012). Differential furanose selection in the active sites of archaeal DNA polymerases probed by fixed-conformation nucleotide analogues. Biochemistry 51:9234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac KN, Ling H. (2009). Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase ι. EMBO J 28:1644–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac KN, Suo Z, Ling H. (2011). Structural mechanism of ribonucleotide discrimination by a Y-family DNA polymerase. J Mol Biol 407:382–90. [DOI] [PubMed] [Google Scholar]
- Klein HL. (2017). Genome instabilities arising from ribonucleotides in DNA. DNA Repair, 10.1016/j.dnarep.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Valentine MR, Pham P et al. (2002). Fidelity of Escherichia coli DNA polymerase IV: preferential generation of small deletion mutations By dNTP-stabilized misalignment. J Biol Chem 277:34198–207. [DOI] [PubMed] [Google Scholar]
- Krasikova YS, Belousova EA, Lebedeva NA et al. (2008). Interaction between DNA polymerase λ and RPA during translesion synthesis. Biochemistry (Mosc) 73:1042–6. [DOI] [PubMed] [Google Scholar]
- Kuban W, Vaisman A, McDonald JP et al. (2012). Escherichia coli UmuC active site mutants: effects on translesion DNA synthesis, mutagenesis and cell survival. DNA Repair 11:726–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Takata K, Wood RD. (2011). DNA polymerases and cancer. Nat Rev Cancer 11:96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Frings O, Sonnhammer EL. (2009). Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res 37:858–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW, Gibbs PEM, Murante RS et al. (2000). Roles of DNA polymerase ζ and Rev1 protein in eukaryotic mutagenesis and translesion replication. Cold Spring Harb Symp Quant Biol 65:61–69. [DOI] [PubMed] [Google Scholar]
- Lazzaro F, Novarina D, Amara F et al. (2012). RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell 45:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Gregory MT, Yang W. (2014). Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc Natl Acad Sci U S A 111:2954–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Schlick T. (2013). “Gate-keeper” residues and active-site rearrangements in DNA polymerase μ help discriminate non-cognate nucleotides. PLoS Computational Biology 9:e1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Kunkel TA. (2016). DNA Polymerases Divide the Labor of Genome Replication. Trends Cell Biol 26:640–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Clausen AR et al. (2013). Evidence that ribonucleotides are signals for mismatch repair of leading strand replication errors. Mol Cell 50:437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Villani G, Crespan E et al. (2007). 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature 447:606–8. [DOI] [PubMed] [Google Scholar]
- Makarova AV, Stodola JL, Burgers PM. (2012). A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res 40:11618–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, McElhinny SA, Watts BE et al. (2014). Ribonucleotide incorporation by yeast DNA polymerase ζ. DNA Repair (Amst) 18:63–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MJ, Garcia-Ortiz MV, Esteban V, Blanco L. (2013). Ribonucleotides and manganese ions improve non-homologous end joining by human Polμ. Nucleic Acids Res 41:2428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Jimenez MI, Garcia-Gomez S, Bebenek K et al. (2015). Alternative solutions and new scenarios for translesion DNA synthesis by human PrimPol. DNA Repair (Amst) 29:127–38. [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA. (2008). The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res 18:148–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Vaisman A, Kuban W et al. (2012). Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by pol V. PLoS Genet 8:e1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentegari E, Crespan E, Bavagnoli L et al. (2017). Ribonucleotide incorporation by human DNA polymeraseη impacts translesion synthesis and RNase H2 activity. Nucleic Acids Res 45:2600–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AF, Pryor JM, Ramsden DA et al. (2017). Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu. Nucleic Acids Res 45:9138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Christensen RB, Alley J et al. (1989). REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol 171:5659–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourόn S, Rodriguez-Acebes S, Martίnez-Jimenez MI et al. (2013). Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol 20:1383–9. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. (1996a). Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272:1646–49. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. (1996b). Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382:729–31. [DOI] [PubMed] [Google Scholar]
- Neuhard J, Nygaard P. (1987). Purines and pyrimidines In Ingraham JL, Low KB, Neidhardt FC, Magasanik B, Schaechter M, Umbarger HE, eds. EcoSal:Escherichia coli and Salmonella, Cellular and Molecular Biology. Washington D.C., American Society for Microbiology, 445–73 [Google Scholar]
- Nevin P, Engen JR, Beuning PJ. (2015). Steric gate residues of Y-family DNA polymerases DinB and pol κ are crucial for dNTP-induced conformational change. DNA Repair 29:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Ramsden DA. (2003). Polymerase μ is a DNA-directed DNA/RNA polymerase. Mol Cell Biol 23:2309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kissling GE, Kunkel TA. (2010a). Differential correction of lagging-strand replication errors made by DNA polymerases α and δ. Proc Natl Acad Sci U S A 107:21070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kumar D, Clark AB et al. (2010b). Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol 6:774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D et al. (2010c). Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A 107:4949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi N, Sassa A, Katafuchi A et al. (2009). The steric gate amino acid tyrosine 112 is required for efficient mismatched-primer extension by human DNA polymerase κ. Biochemistry 48:4239–46. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RPP et al. (2001). The Y-family of DNA polymerases. Mol Cell 8:7–8. [DOI] [PubMed] [Google Scholar]
- Ordonez H, Shuman S. (2014). Mycobacterium smegmatis DinB2 misincorporates deoxyribonucleotides and ribonucleotides during templated synthesis and lesion bypass. Nucleic Acids Res 42:12722–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez H, Uson ML, Shuman S. (2014). Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Res 42:11056–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pata JD. (2010). Structural diversity of the Y-family DNA polymerases. Biochim Biophys Acta 1804:1124–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier H, Sawaya MR, Kumar A et al. (1994). Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science 264:1891–903. [PubMed] [Google Scholar]
- Potenski CJ, Klein HL. (2014). How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res 42:10226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakasha Gowda AS, Polizzi JM, Eckert KA, Spratt TE. (2010). Incorporation of gemcitabine and cytarabine into DNA by DNA polymerase β and ligase III/XRCC1. Biochemistry 49:4833–40. [DOI] [PubMed] [Google Scholar]
- Pryor JM, Waters CA, Aza A et al. (2015). Essential role for polymerase specialization in cellular nonhomologous end joining. Proc Natl Acad Sci U S A 112:E4537-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe B (2013). Aicardi-Goutieres syndrome: clues from the RNase H2 knock-out mouse. J Mol Med 91:1235–40. [DOI] [PubMed] [Google Scholar]
- Ramadan K, Maga G, Shevelev IV et al. (2003). Human DNA polymerase λ possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J Mol Biol 328:63–72. [DOI] [PubMed] [Google Scholar]
- Reijns MA, Rabe B, Rigby RE et al. (2012). Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149:1008–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. (1999). The mutagenesis protein UmuC is a DNA polymerase activated by UmuD’, RecA, and SSB and Is specialized for translesion replication. J Biol Chem 274:31763–66. [DOI] [PubMed] [Google Scholar]
- Roettger MP, Fiala KA, Sompalli S et al. (2004). Pre-steady-state kinetic studies of the fidelity of human DNA polymerase μ. Biochemistry 43:13827–38. [DOI] [PubMed] [Google Scholar]
- Rothwell PJ, Waksman G. (2005). Structure and mechanism of DNA polymerases. Advances in Protein Chemistry 71:401–40. [DOI] [PubMed] [Google Scholar]
- Rudd SG, Glover L, Jozwiakowski SK et al. (2013). PPL2 translesion polymerase is essential for the completion of chromosomal DNA replication in the African trypanosome. Mol Cell 52:554–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd SG, Bianchi J, Doherty AJ. (2014). PrimPol-A new polymerase on the block. Mol Cell Oncol 1:e960754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JF, Juarez R, Garcia-Diaz M et al. (2003). Lack of sugar discrimination by human Pol μ requires a single glycine residue. Nucleic Acids Res 31:4441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. (2012). Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 13:141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa A, Caglayan M, Rodriguez Y et al. (2016). Impact of ribonucleotide backbone on translesion synthesis and repair of 7,8-dihydro-8-oxoguanine. J Biol Chem 291:24314–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Cox MM, Woodgate R, Goodman MF. (2006). RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature 442:883–7. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18:6097–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JW, Randall JR, Matthews LA, Simmons LA. (2015). Ribonucleotides in bacterial DNA. Crit Rev Biochem Mol Biol 50:181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Wood RD. (2008). DNA polymerase θ (POLQ) can extend from mismatches and from bases opposite a (6–4) photoproduct. DNA Repair (Amst) 7:119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Fijalkowska IJ. (2006). Translesion synthesis DNA polymerases and control of genome stability. Front Biosci 11:2496–517. [DOI] [PubMed] [Google Scholar]
- Sherrer SM, Beyer DC, Xia CX et al. (2010). Kinetic basis of sugar selection by a Y-family DNA polymerase from Sulfolobus solfataricus P2. Biochemistry 49:10179–86. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Gruz P, Kamiya H et al. (2003). Erroneous incorporation of oxidized DNA precursors by Y-family DNA polymerases. EMBO Rep 4:269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtygasheva AA, Belousova EA, Rechkunova NI et al. (2008). DNA polymerases β and λ as potential participants of TLS during genomic DNA replication on the lagging strand. Biochemistry (Mosc) 73:1207–13. [DOI] [PubMed] [Google Scholar]
- Sobol RW, Horton JK, Kuhn R et al. (1996). Requirement of mammalian DNA polymerase-β in base-excision repair. Nature 379:183–86. [DOI] [PubMed] [Google Scholar]
- Su Y, Egli M, Guengerich FP. (2016). Mechanism of ribonucleotide Incorporation by human DNA polymerase η. J Biol Chem 291:3747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Egli M, Guengerich FP. (2017). Human DNA polymerase η accommodates RNA for strand extension. J Biol Chem 292:18044–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K, Shimizu T, Iwai S, Wood RD. (2006). Human DNA polymerase ν (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem 281:23445–55. [DOI] [PubMed] [Google Scholar]
- Takata K, Arana ME, Seki M et al. (2010). Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic Acids Res 38:3233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG et al. (1999). UmuD’2C is an error-prone DNA polymerase, Escherichia coli, DNA pol V. Proc Natl Acad Sci U S A 96:8919–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Masutani C, Hanaoka F, Chaney SG. (2000). Efficient translesion replication past Oxaliplatin and Cisplatin GpG adducts by human DNA polymerase η. Biochemistry 39:4575–80. [DOI] [PubMed] [Google Scholar]
- Vaisman A, Woodgate R. (2004). Translesion DNA polymerases, eukaryotic In Lennarz WJ, Lane MD, eds. Encyclopedia of Biological Chemistry. Oxford, Academic Press, Elsevier Science, 247–50 [Google Scholar]
- Vaisman A, Kuban W, McDonald JP et al. (2012a). Critical amino acids in Escherichia coli responsible for sugar discrimination and base-substitution fidelity. Nucleic Acids Res 40:6144–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, McDonald JP, Woodgate R. (2012b). Translesion DNA Synthesis In Böck A, Curtiss R, Kaper JB, Karp PD, Neidhardt FC, Slauch JM, Squires CL, eds. EcoSal: Escherichia coli and Salmonella: cellular and Mol Microbiol. Washington, DC, American Society for Microbiology [Google Scholar]
- Vaisman A, McDonald JP, Huston D et al. (2013). Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair. PLoS Genet 9:e1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Woodgate R. (2015). Redundancy in ribonucleotide excision repair: Competition, compensation, and cooperation. DNA Repair (Amst) 29:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Woodgate R. (2017). Translesion DNA polymerases in eukaryotes: what makes them tick? Crit Rev Biochem Mol Biol 52:274–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BD, Williams RS. (2014). Ribonucleotide triggered DNA damage and RNA-DNA damage responses. RNA Biol 11:1340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Lou J, Xia Y et al. (2013). hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep 14:1104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME et al. (2009). Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 73:134–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt DL, Johansson E, Burgers PM, Kunkel TA. (2011). Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst) 10:897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Kunkel TA. (2014). Ribonucleotides in DNA: origins, repair and consequences. DNA Repair 19:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Lujan SA, Kunkel TA. (2016). Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat Rev Mol Cell Biol 17:350–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Minko IG, Takata K et al. (2010). Novel enzymatic function of DNA polymerase ν in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links. Chem Res Toxicol 23:689–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W (2005). Portraits of a Y-family DNA polymerase. FEBS Lett 579:868–72. [DOI] [PubMed] [Google Scholar]
- Yang W, Woodgate R. (2007). What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci U S A 104:15591–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao NY, Schroeder JW, Yurieva O et al. (2013). Cost of rNTP/dNTP pool imbalance at the replication fork. Proc Natl Acad Sci U S A 110:12942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Guo D et al. (2002). Lesion bypass activities of human DNA polymerase μ. J Biol Chem 277:44582–7. [DOI] [PubMed] [Google Scholar]
- Zhong X, Garg P, Stith CM et al. (2006). The fidelity of DNA synthesis by yeast DNA polymerase ζ alone and with accessory proteins. Nucleic Acids Res 34:4731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]