Abstract
Tropical reefs are shifting from coral to macroalgal dominance, with macroalgae suppressing coral recovery, potentially via effects on coral microbiomes. Understanding how macroalgae affect corals and their microbiomes requires comparing algae- versus coral-dominated reefs without confounding aspects of time and geography. We compared survival, settlement, and post-settlement survival of larvae, as well as the microbiomes of larvae and adults, of the Pacific coral Pocillopora damicornis between an Marine Protected Area (MPA) dominated by corals versus an adjacent fished area dominated by macroalgae. Microbiome composition in adult coral, larval coral, and seawater did not differ between the MPA and fished area. However, microbiomes of adult coral were more variable in the fished area and Vibrionaceae bacteria, including strains most closely related to the pathogen Vibrio shilonii, were significantly enriched, but rare, in adult and larval coral from the fished area. Larvae from the macroalgae-dominated area exhibited higher pre-settlement mortality and reduced settlement compared to those from the coral-dominated area. Juveniles planted into a coral-dominated area survived better than those placed into a fished area dominated by macroalgae. Differential survival depended on whether macroalgae were immediately adjacent to juvenile coral rather than on traits of the areas per se. Contrary to our expectations, coral microbiomes were relatively uniform at the community level despite dramatic differences in macroalgal cover between the MPA (~2% cover) and fished (~90%) area. Reducing macroalgae may elicit declines in rare but potentially harmful microbes in coral and their larvae, as well as positive intergenerational effects on offspring survival.
Keywords: coral microbiome, coral larvae, coral-algae interactions, marine protected areas, Pocillopora damicornis
Introduction
Coral reefs support great biodiversity and provide critical ecosystem services (Cesar et al. 2003). They buffer coastal populations from storms, provide a primary source of protein for many island and coastal nations (Dalzell et al. 1996, Cesar et al. 2003), and generate billions of dollars annually in tourism-related income (Cesar et al. 2003). However, coral reefs are in rapid global decline, with coral cover decreasing by 80% in the Caribbean since the 1970’s (Gardner et al. 2003, Jackson et al. 2014) and by >50% in the Pacific since the 1980’s (Bruno & Selig 2007, De’ath et al. 2012). Threats to reefs include overfishing, pollution, disease (Bellwood et al. 2004), thermal stress, and ocean acidification (Hoegh-Guldberg et al. 2007, Hughes et al. 2017). These stressors may directly harm corals, but may also indirectly lower coral health by allowing proliferation of competitive macroalgae (Hughes et al. 2010). Contact with algae has been linked to bleaching, disease, and tissue death in adult corals (Nugues et al. 2004, Rasher and Hay 2010), potentially harming coral through diverse mechanisms, including allelopathy, oxygen depletion, and destabilization of coral-associated microbial communities (microbiomes) (Rasher & Hay 2010, Barott & Rohwer 2012, Zaneveld et al. 2016, Morrow et al. 2017). Furthermore, macroalgae can directly inhibit both settlement and survivorship of coral larvae (Kuffner et al. 2006, Hughes et al. 2007, Dixson et al. 2014, Webster et al. 2015), often in a species-specific manner (Vermeij et al. 2009). Macroalgae may also disrupt microbiomes of coral larvae, but to our knowledge effects of macroalgae- versus coral-dominance of reefs on larval microbiomes or pre-settlement survival has not been investigated. Identifying the mechanisms and consequences of coral-algae interactions is vital for understanding coral resilience under changing ocean conditions, as well as for creating effective conservation strategies.
Coral microbiomes may play important roles in coral acclimation to variable ocean environments (Rosenberg et al. 2007, Krediet et al. 2013, Peixoto et al. 2017). For example, corals that maintain or acquire thermotolerant strains of the symbiotic alga Symbiodinium have a lower risk of bleaching and mortality in response to fluctuating water temperatures (Pettay et al. 2015), and disrupting coral microbiomes with antibiotics can increase tissue loss in response to temperature stress (Gilbert et al. 2012). It is thus worrisome that microbial dysbiosis (i.e., a shift to higher abundances of harmful microbes or lower abundances of beneficial microbes) is becoming more common on degraded reefs (Dinsdale et al. 2008, Dinsdale & Rohwer 2011) and may render corals more susceptible to bleaching and mortality (Ritchie 2006, Harvell et al. 2007, Rosenberg et al. 2007).
Coral-macroalgae interactions on degraded reefs may drive dysbiosis, shifting the coral microbiome to an alternative state via mechanisms such as the production of algal allelochemicals (Morrow et al. 2012, Morrow et al. 2017), release of dissolved organic matter (Dinsdale & Rohwer 2011, Barott & Rohwer 2012, Haas et al. 2016), or transfer of harmful bacteria to corals interacting with algae (Nugues et al. 2004, Sweet et al. 2013, Pratte et al. 2017). Alternatively, changes in coral microbiomes in response to increasing algal cover could be a mechanism by which corals cope with algal competition or other biotic and abiotic stressors (Rosenberg et al. 2007).
Comparisons of adjacent reef areas that vary in algal cover due to protection status provide unique opportunities to explore coral-algae-microbiome interactions in situ without the confounding effects of contrasts across large spatial or temporal scales. Marine Protected Areas (MPAs) that prohibit fishing are valuable conservation tools for maintaining or restoring reef health. Corals in “no take” MPAs benefit from enhanced herbivore grazing that removes competing seaweeds (Mumby et al. 2007, Rasher et al. 2013) or via reduced fishing-associated damage to corals that increases coral susceptibility to disease (Lamb et al. 2015, 2016). Healthy MPA corals may serve as a source of coral larvae to “rescue” degraded areas beyond reserve boundaries (Almany et al. 2009, McCook et al. 2010, Selig & Bruno 2010), but this rescue will depend upon survival of exported larvae during dispersal and on post-settlement survival if the larvae recruit to degraded reefs. Furthermore, ecosystem processes within MPAs, such as predation or herbivory, might also aid in conservation of microbiota required for coral health (Krediet et al. 2013), development (Vermeij et al. 2009, Tran & Hadfield 2011, Sneed et al. 2014), and ecosystem function (Ainsworth et al. 2010). By comparing islands that span ~2,000 km in the Pacific, reefs from populated islands were found to differ in reef fish biomass, abundances of fleshy algae, and benthic reef water microbiomes compared to reefs on unpopulated islands, suggesting that human use alters reef microbiomes (Dinsdale et al. 2008, Sandin et al. 2008, Kelly et al. 2014). Haas et al. (2016) also found positive correlations between fleshy algal cover and microbial abundance and community composition in benthic water across 60 reef sites spanning three ocean systems, while Zaneveld et al. (2016) found that herbivore exclusion plots had higher algal abundances and more variable coral microbiomes compared to plots with herbivores. However, to our knowledge, no studies have compared coral microbiomes in MPAs versus fished areas or investigated how microbiome composition may relate to survival of larvae produced from these areas. Such comparisons would help determine the extent to which coral microbiomes change across reefs dominated by corals versus macroalgae when not confounded by time or large distances.
We evaluated the effects of differing macroalgal abundance (resulting from reef protection status) on coral microbiomes using reef areas separated by only 100 – 500 meters. We conducted experiments in long-term (>10 yr) MPAs and adjacent fished areas (two MPAs and two fished areas – one pair of sites for pre-settlement experiments and one pair of sites for post-settlement experiments) along the southwest coast of Viti Levu, Fiji. Corals within the fished areas experience 5 to 15-fold more frequent and 23 to 67-fold more extensive algal contact (measured by proportion of colony perimeter in contact with macroalgae) than those in adjacent MPAs (Bonaldo & Hay 2014), allowing us to investigate how chronic interactions with macroalgae affect microbiomes of adult coral and their offspring under natural conditions and how this relates to juvenile coral survivorship. Specifically, we asked whether: 1) coral and seawater microbiome composition differed between a coral-dominated MPA and an adjacent fished area, 2) potentially harmful microbial taxa were less abundant in coral from the MPA compared to the fished area, 3) larvae from the MPA experience higher survivorship prior to settlement compared to larvae from the fished area, 4) post-settlement juvenile coral experience higher survivorship in an MPA compared to a fished area, and 5) higher juvenile survivorship depends on settlement on substrate free of macroalgae.
Methods
Study sites and focal coral
We focused on the coral Pocillopora damicornis because it occurs commonly in both MPA and fished areas and produces brooded larvae that could be obtained easily. Our study sites were shallow back-reef lagoons of 1–3 m water depth within two, small (0.5 – 0.8 km2), locally managed MPAs and their adjacent fished areas at Vatu-o-lalai (18°12.26′ S, 177°41.26′ E) and Votua villages (18°13.08′ S, 177°42.59′ E) along the southwest coast of Viti Levu, Fiji. The MPA was established in 2002 at Vatu-o-lalai and in 2003 at Votua. These sites are approximately three kilometers apart and the MPA and fished area at each site experience similar physical regimes as judged by algal and coral growth rates when relieved of biotic pressures (Rasher et al. 2012, Dell et al. 2016, Clements et al. in press). All sites experience comparable flushing of reef water, with oceanic water flowing over the reef crest at high tide and washing out through deep channels at low tide. The MPAs have high coral cover (~57%) and low macroalgal cover (≤ 2%) on hard substrates; the fished areas have low coral cover (4–16%) and high macroalgal cover (50–90%) on hard substrates (Rasher et al. 2013). Consequently, coral contact with macroalgae is 5–15 times more frequent and 23–67 times more extensive in the fished areas than in the MPAs (Bonaldo & Hay 2014). MPAs also have 2–3 times higher diversity and 7–17 times higher biomass of herbivorous fishes than fished areas (Rasher et al. 2013).
Coral collection and maintenance of coral larvae
Between 29 October and 6 November 2014 (1–10 days before the full moon), portions from individual P. damicornis colonies were collected from the MPA and adjacent fished area at Votua village (12 colonies per area, collected with permissions from the Korolevu-i-Wai District Environment Committee). Collection locations for MPA versus fished area coral were separated by ~100 to 500 m. Each coral was placed in a separate bucket with approximately 19 liters of water from the respective collection site and monitored at dusk for larval release. Four colonies from the MPA and four from the fished area released larvae at dusk on the day they were collected. To characterize the microbiome of larvae from the MPA and fished area, we collected 10 larvae per colony upon release. Each larva was rinsed 3 times in 0.22 μm filter-sterilized (Corning disposable vacuum filter/storage systems 0.22 μm cellulose acetate 45 mm filter, ThermoFisher Scientific, Waltham, MA) seawater (FSW), preserved separately in RNAlater (ThermoFisher Scientific, Waltham, MA), and stored at −20° C. We simultaneously collected four clippings from each adult coral colony that released larvae and preserved these in the same manner.
Of the eight colonies used for microbiome analysis, four colonies from the MPA and three from the fished area produced sufficient numbers of larvae (≥ 100 per colony) for use in subsequent larval survival and settlement experiments (see text below and Figure S1 for a diagram of the experimental design). These larvae were pooled by area (MPA or fished area) and maintained in 600 mL polystyrene plastic containers filled with 400 mL of unfiltered water collected from a deep channel on the back reef that is open to the outer reef. Larvae of P. damicornis are packed with Symbiodinium and can remain viable for 100 days in the lab with water changes every 2–3 days (Richmond 1987, Isomura & Nishihira 2001). We changed water daily until the start of all experiments (which were all run simultaneously). Larval age at the start of experiments ranged from 7–16 days due to larval release occurring on different days. Any inactive larvae that failed to exhibit swimming behavior after three gentle pipette aspirations in the plastic dish were not used in any experiments. All larvae were transferred with sterile wide bore pipette tips (Axygen 1000 μL universal pipette tips: wide bore, ThermoFisher Scientific, Waltham, MA).
DNA extractions and amplicon sequencing of the 16S gene
Sequencing of the 16S rRNA gene was used to compare microbiome composition between MPA and fished area coral and seawater. DNA was extracted from coral larvae and adults using the PowerSoil DNA extraction kit and from water samples (polyethersulfone filters) using the PowerWater DNA extraction kit (both kits from MoBio Laboratories, QIAGEN, Carlsbad, CA). To account for intra-colony variation, DNA from five larvae and four clippings of adult coral branches were extracted individually per colony. For the larval survival experiment (see below), DNA from two larvae per dish was extracted individually (with one exception when only one larva was alive at the end of the experiment). Additionally, for each sample, we centrifuged the residual RNAlater solution (10,000 rpm, 10 min) to collect any dissociated cells, re-suspended the resulting pellet in solution C1 (MoBio Laboratories, QIAGEN), and added these cells to the power bead tube. PCR reactions were performed in triplicate with dual-barcoded primers (F515 and R806) targeting the V4 region of the 16S rRNA gene, following standard protocols described in Kozich et al. (2013). PCR reactions included 45 μL of Platinum PCR SuperMix (Life Technologies, Thermo Scientific, Waltham, MA), 3 μL of template DNA (of 100 μL total DNA elution volume), and 1 μL each of forward and reverse primer. The thermal cycling protocol was as follows: initial denaturation at 94°C (3 min), followed by 35 cycles of denaturation at 94°C (45 sec), primer annealing at 50°C (45 sec) primer extension at 72°C (90 sec), and a final extension at 72°C (10 min). Amplicons were cleaned and DNA concentrations were normalized using SequalPrep plates (ThermoFisher Scientific, Waltham, MA). Amplicons were then pooled at equimolar concentrations and sequenced on Illumina’s MiSeq platform using a 500 cycle kit (250 × 250 nt paired end reads) spiked with 10% PhiX to increase nucleotide diversity. Raw sequence reads can be found under NCBI bioproject number PRJNA382809.
Amplicon data analyses
We used Trim Galore! (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to demultiplex, trim (100 bp cutoff length), and filter low-quality reads (Phred score cutoff 25), and FLASH (Magoč & Salzberg 2011) to merge paired-end reads (read length 250 bp, fragment length 300, fragment standard deviation 30). QIIME (Caporaso et al. 2010) was used to assess community composition based on merged reads. Briefly, chimeric sequences were identified and removed in QIIME using USEARCH (Edgar 2010). Amplicons were clustered into Operational Taxonomic Units (OTUs) at 97% similarity using the UCLUST algorithm (Edgar 2010) in open-reference OTU picking. The Greengenes database (McDonald et al. 2012, Werner et al. 2012) was used to assign taxonomy to OTUs. Chloroplast-affiliated OTUs were removed from downstream analyses. A total of 1,066,315 sequences (from 6,012,330 originally) remained after quality filtering and removal of chimeras and chloroplast sequences. The number of sequences per sample ranged from 235 to 20,812 for the initial collection of coral larvae and adults, 970 to 46,941 for coral larvae maintained in MPA or fished area water, and 31,816 to 59,734 for water samples. To avoid confounding sequencing depth with biological or environmental variables, as discussed recently in Weiss et al. (2017), diversity analyses were performed using a uniform sequence count identified as the highest count permitted without losing any replicates for a given experiment: 1,650 for coral larvae, coral adults, and water and 1,175 for larvae maintained in MPA or fished area water, and water samples. OTU abundances from each non-independent subsample were collapsed on the mean for a given independent replicate to avoid pseudoreplication (where coral colonies are spatially segregated and confounded within factor: area of origin) using the QIIME script collapse_samples.py. All of the following analyses were performed on the mean OTU abundance for each replicate.
Primer E (Clarke 1993) was used to perform principal coordinates analysis (PCO) on Bray-Curtis dissimilarity matrices from OTU tables based on 97% similarity clusters of 16S rRNA gene sequences. Statistical significance of a priori groupings were tested with PERMANOVA and PERMDISPERSION within Primer E version 7.
Two-factor ANOVA (factor 1: area of origin, factor 2: life stage), implemented via the aov function within the lm package in RStudio version 3.0, was used to test for differences in the relative abundances of microbial taxa among adult and larval coral using proportion data. When groups did not meet the parametric assumption of homoscedasticity, we applied a permutation ANOVA, via the aovp function within the lm package of RStudio 3.0, on proportion data. We first included only those taxa (Family level) contributing to 2% or greater relative abundance within at least one sample group, using data from coral larvae and adults from the MPA and fished area, collected when larvae were initially released. Taxa contributing < 2% were pooled to generate ‘Low Abundance Bacteria’ and ‘Low Abundance Archaea’ datasets that were also tested by two-factor permutation ANOVA (factor 1: area of origin, factor 2: life stage). A Bonferonni correction was implemented to account for multiple comparisons (critical p-value p < 0.004). Upon detecting large differences between adult and larval coral microbial taxa, we chose to also conduct an additional evaluation of the effects of area of origin on relative abundances of taxa contributing to 2% or greater, ‘Low Abundance Bacteria’ pooled, and ‘Low Abundance Archaea’ pooled with a one-factor (area of origin) ANOVA or permutation ANOVA (if data were not homoscedastic) on proportion data for adult coral and larval coral separately. This additional testing increases our risk of a type one statistical error (discussed in the context of our findings in the results section below) but allowed us to examine our samples for any microbial taxa that may differ by area of origin within each coral life stage, while reducing the number of factors and contrasts involved in the analyses.
We also tested for indicator OTUs of coral from the MPA and fished area, analyzing adult coral and larval coral separately with multi-level pattern analysis within the indicspecies package in RStudio 3.0.
Two-factor ANOVA was also used as above to test for differences in the relative abundances of potential pathogens among sample groups. OTU tables (species level) were screened for bacterial groups that have been described as coral pathogens, both those verified with Koch’s postulates and those that have not been verified (see the following reviews for described coral pathogens: Harvell et al. 2007, Rosenberg et al. 2007, Rosenberg & Kushmaro 2011). We detected bacterial OTUs (97% similarity clusters) most closely related to Vibrio shilonii, a bacterium previously shown to cause disease in Oculina patagonica (Kushmaro et al. 2001) and closely related to bacterial strains that cause bacterial bleaching (Ben-Haim et al. 2003b, Harvell et al. 2007) and white syndromes (Sussman et al. 2008) in P. damicornis and other coral species. These were the only OTUs closely affiliated with a known coral pathogen, with the exception of an OTU most closely related to Serratia marcescens (posited coral pathogen of coral species in the Caribbean, Harvell et al. 2007), which was present in only 1 adult sample at 0.06% relative abundance. We therefore tested for differences in the abundances of OTUs identified as belonging to the Vibrionaceae family, and to V. shilonii specifically (based upon Greengenes classification) in adults and larvae (factor 1: area of origin, factor 2: life stage).
Larval survivorship in MPA or fished area water
To test for the effect of water from the MPA versus fished area on larval survivorship before settlement, MPA and fished area larvae were aliquoted in a full factorial design into 600 mL polystyrene dishes with 400 mL unfiltered water collected ~1–2 meters above the benthos daily from the MPA or fished area and used immediately in water changes. There were 10 replicate dishes per level of each factor (factor 1: larval area of origin; factor 2: water area of origin), dishes were randomly interspersed, and each replicate dish held 10 larvae. To maintain similar conditions between experiments and to reduce the influence of ‘home reef’ (i.e., enhanced larval preferences for or survival on or within substrates or water from the site where the parent coral was collected) effects on larval responses (survival or settlement), we collected water for both experiments from the MPA and fished area of Vatu-o-lalai village, approximately 3 km from where adults that released these larvae were collected at Votua village. Water was collected approximately 1–2 meters above the benthos and changed daily for the first five days of the experiment. A 250 mL aliquot of this freshly collected water was filtered through a 0.22 μm polyethersulfone filter each day, and the filter was preserved in RNALater for microbiome analysis (i.e., non-filtered water was used to hold the larvae, but the filter was used to assess the water’s microbiome). No settlement substrate was added during the experiment, and larvae avoid settling on the polystyrene surfaces of the dishes (KB Ritchie, personal communication). We recorded metamorphosis (on the dish or in the water column, which was rare) daily for six days and assessed survivorship at the end of the six-day experiment. Larvae were considered alive if they exhibited swimming behavior after three gentle pipette aspirations within the dish. Larvae alive at the end of the experiment were collected (n = 10 independent samples per level of each factor in our design, i.e., dishes considered independent, not individual larvae from within dishes), rinsed three times in filter-sterilized seawater, and preserved in RNAlater individually for microbiome analysis. Following DNA extraction and sequencing of the 16S gene, MPA and fished area larvae maintained in MPA or fished area water were screened for potential coral pathogens. OTUs identified as V. shilonii were the only hypothesized coral pathogens detected in these samples. We tested for differences in the abundance of taxa identified as V. shilonii and Vibrionaceae with a two-factor ANOVA via the aov function within the lm package of RStudio 3.0 (factor 1: larval area of origin, factor 2: water area of origin). Lastly, we tested for differences in the abundance V. shilonii and Vibrionaceae in water samples by a one-factor ANOVA (factor: water area of origin). Primer E (Clarke 1993) was used to perform principal coordinates analysis (PCO) on Bray-Curtis dissimilarity matrices from OTU tables based on 97% similarity clusters of 16S rRNA gene sequences. Statistical significance of a priori groupings were tested with PERMANOVA within Primer E version 7.
Settlement behavior and post-settlement survivorship of MPA and fished area larvae offered MPA and fished area substrates
To test for the effects of MPA vs. fished area substrates on larval settlement and survivorship, we set up a full-factorial experiment with MPA and fished area larvae offered coral rubble from either the MPA or the fished area. To prevent confounding home reef with MPA vs. fished area effects, we collected rubble pieces from the MPA and fished area at Vatu-o-lalai village (approximately 3 km from sites where adult colonies were collected) and used these in settlement assays with larvae from the MPA and fished area at Votua village. Rubble pieces were similar in size and collected from haphazard locations throughout the MPA and fished area. All rubble collected from the MPA was naturally free of macroalgal fouling, whereas rubble collected from the fished area was either fouled with some macroalgae (characteristic of the benthos in the fished area - Rasher et al. 2013, Bonaldo & Hay 2014) or free of fouling. All three types of substrate were fouled with comparable amounts of CCA and short (< 0.5 cm) turf. Crustose coralline algae (CCA) may stimulate settlement of coral larvae; therefore, we also quantified CCA cover on rubble from each location. Photos of rubble collected from the MPA and fished area were analyzed with Coral Point Count Software (Nova Southeastern University, Kohler & Gill 2006). CCA abundances between the three types of rubble collected (MPA rubble without macroalgae, fished area rubble without macroalgae, and fished area rubble with macroalgae) were tested with a one-factor ANOVA in JMP Pro 13 software (SAS Institute Inc.). Rubble fouled with macroalgae had short algal fronds ~0.5–4 cm in height. These pieces of rubble were used to test for the mean effect of naturally occurring multi-species assemblages of macroalgae on larval settlement and post-settlement survival. Water for these experiments was collected from the MPA and fished area at Vatu-o-lalai simultaneously with the rubble and then daily thereafter for use in the larval settlement experiments described below.
For the first settlement experiment, larvae from MPA and fished area adults were separately aliquoted to 600mL polystyrene plastic dishes (10 larvae per dish; n = 20 dishes per level of each factor, dishes randomly interspersed) and offered only MPA substrate (without macroalgae) with 400 mL of unfiltered water from the MPA or only fished area substrate (with macroalgae) with 400 mL of unfiltered water from the fished area, with daily water changes. All water used in experiments was collected from ~1–2 meters above the benthos. These two substrates were chosen for the first experiment because they are typical of the MPA vs. fished area site differences (Rasher et al. 2013, Bonaldo & Hay 2014). Within each replicate, larvae could either settle on the added substrate or remain in the water column. Settlement was recorded at 24 and 48 h. After 48 h, all non-settled larvae were removed and the settled coral were held on their substrate in the lab. The effect of settlement substrate on post-settlement survival was assessed on day four following the 48 h settling period; surviving juveniles were then out-planted to the reef.
Juveniles on MPA rubble were out-planted to the MPA and juveniles on fished area rubble were out-planted to the fished area. To reduce the possibility of home reef effects confounding MPA vs. fished area effects, juvenile coral were out-planted to MPA and fished area sites at Vatu-o-lalai village approximately 3km from Votua, where the fragments of adult coral colonies had initially been collected. Zip-ties were used to attach the rubble to u-nails driven into the reef bottom, with each rubble piece containing 4–9 juveniles at the time of out-planting. For each replicate, similarly sized pieces of control rubble (without any juvenile coral) were attached to the benthos in the same manner as above to test for natural coral recruitment to rubble (MPA rubble without macroalgae in the MPA and fished area rubble with macroalgae in the fished area) that might be confused with, and falsely increase survivorship rates of, our out-planted juveniles. Survivorship of out-planted juveniles and natural recruitment to control pieces of rubble were recorded after four and twenty-six days on the reef (when experimental coral were eight and thirty days post-settlement). Recruitment to control rubble was low in each area (0–0.1 recruit/replicate), and average recruitment to control rubble was deducted from the appropriate treatment before calculating the proportion of surviving juveniles at each time-point. Across all treatment combinations, nine replicates were lost due to rubble becoming unattached from the benthos over 26 days on the reef.
Differences between survivorship of juvenile coral on MPA substrate planted in the MPA vs. fished area substrate planted in the fished area could be due to differences in macroalgal abundance on the settlement substrate or due to other unrecognized physical or biotic differences between the MPA and fished area sites (hereafter referred to as ‘site’ effects). To test for a site effect vs. the effect of macroalgae on the settlement rubble, we performed a second experiment to test for settlement of MPA larvae (too few larvae remained from fished area adults to conduct this experiment with those larvae) on similarly sized rubble from either i) the MPA (without macroalgae), ii) the fished area but without macroalgae, or iii) the fished area but with macroalgae (n = 14 – 15 for each treatment). Experimental procedures were the same as in the settlement experiment. Briefly, larvae from MPA adults were aliquoted to 600 mL polystyrene plastic dishes with 400 mL of unfiltered water and substrate from either the MPA or fished area (10 larvae per dish, dishes randomly interspersed). Water changes were performed daily with freshly collected unfiltered water from the MPA or fished area collected ~1–2 meters above the benthos. Settlement was assessed at 24 and 48 hours. Survivorship of newly-settled-juvenile coral was assessed four days after the initial 48-hour settlement experiment. Juvenile coral that had settled on these substrates were then out-planted into the field (MPA rubble to the MPA site and fished area rubble to the fished area site) using the procedures described above. Again, natural recruitment to MPA or fished area control rubble at both sites was low (0.0–0.07 recruit/replicate) and was deducted before calculating the proportion of surviving juvenile coral. Four replicates planted in the MPA became detached and were lost by day 26. No replicates from the fished area were lost.
Statistical analyses of larval behavior and recruit survivorship
JMP Pro 12 (SAS Institute Inc.) was used to analyze larval metamorphosis, larval settlement, and larval and juvenile survivorship. Larval metamorphosis and survivorship were analyzed by a two-factor ANOVA on proportion data. Settlement was analyzed with repeated measures ANOVA on square root transformed proportion data. Juvenile coral survivorship was analyzed with repeated measures ANOVA on proportion data. All data were homoscedastic; when needed, square root transformations were performed to improve normality.
Results
Coral and water microbiomes from coral-dominated Marine Protected Areas (MPAs) and macroalgae-dominated fished areas
Microbiome community composition of adult coral, larvae, and water did not differ as a result of collection site (MPA or fished area), but did differ between sample types (Figure 1A PERMANOVA sample type p = 0.001, area of origin p = 0.426, sample type * area of origin p = 0.803). Water and larval microbiomes were more diverse than adult microbiomes (Figure 1B, number of OTUs: sample type p < 0.001, area of origin p = 0.764, sample type * area of origin p = 0.909; Figure 1C, Shannon diversity index: sample type p < 0.001, area of origin p = 0.539, sample type * area of origin p = 0.282), despite under-sampling water microbial communities at a rarefaction depth of 1,650 (see rarefaction curve in supplementary Figure S2). Findings were similar if water samples were excluded from the analyses; microbial community composition was dictated by life stage (adult or larvae), not by the area (MPA vs. fished area) (Figure S3B, PERMANOVA life stage p = 0.001, area of origin p = 0.338, life stage * area of origin p = 0.584). Findings were also similar whether based on the mean for each independent replicate colony or all subsamples from each independent colony (Figure S3A and S3B). However, differences detected with PERMANOVA are partially due to dispersion differences among groups, with microbiome composition exhibiting lower dispersion among MPA adults compared to dispersion among fished area adults, or to larvae from either area (Figure S3B PERMDISP area of origin p = 0.743, life stage p = 0.002, area of origin * life stage p = 0.013, see Table S1 for pairwise comparisons). We also tested for area of origin effects using adults alone and larvae alone and did not detect effects in either analysis (adults p = 0.246; larvae p = 0.588, Monte Carlo PERMANOVA, Figure S3C–D).
Figure 1.
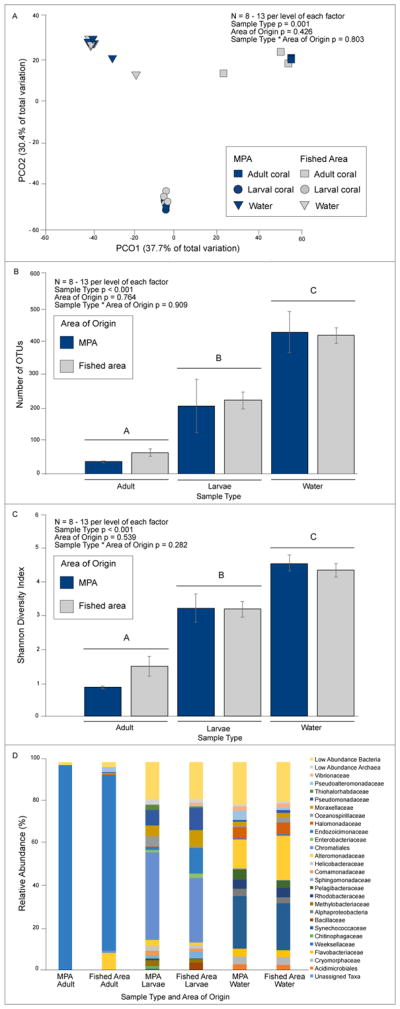
A) PCO and PERMANOVA analysis of Bray Curtis dissimilarity matrix of coral larvae, coral adults, and water microbiomes from the MPA and fished area. B) Diversity of OTUs in coral and water samples. C) Shannon Diversity Index for coral and water samples. OTUs and Shannon Diversity were analyzed by a two-factor ANOVA with Tukey post-hoc analysis. D) Taxonomic groups that contribute to 2% or greater of the microbial community composition of coral adults, coral larvae, and water are depicted at the level of family with the exception of Chromatiales (order). Low abundance taxa, contributing less than 2% of community composition were pooled to generate ‘Low Abundance Bacteria’ and ‘Low Abundance Archaea’ groups. For analyses in A–D above, each coral larva, and coral adult data point represents the mean community composition for a single replicate.
We also compared each common taxonomic group that comprised ≥ 2% relative abundance and the pooled group of uncommon bacterial and archaeal taxa (< 2% relative abundance) between MPA and fished area sites (Figure 1D, Table S2A–B, and S3A–B). None of these taxonomic groups differed significantly between MPA and fished area sites and this was true whether adults and larvae were tested together (2-factor ANOVA) or separately (1-factor ANOVA), despite biasing our analyses toward a higher probability of a false positive through multiple statistical tests on these data sets. In contrast, certain taxonomic groups differed notably in relative abundance between adults and larvae. Endozoicimonaceae were enriched 13-fold in adults compared to larvae (~ 90% vs ~ 7%; two-factor ANOVA source area p = 0.722, life stage p < 0.001, source area * life stage p = 0.113), whereas larvae contained 58–243 fold more Chromatiales (p < 0.001), Methylobacteriaceae (p = 0.001), Sphingomonadaceae (p < 0.001), Pseudomonadaceae (p = 0.003), and Helicobacteraceae (p = 0.002) (Table S2A and S2B). Larvae were also enriched 8-fold in low abundance bacteria (p = 0.001) and 90-fold in low abundance archaea (p = 0.002) compared to adult coral (Table S2A and S2B).
Indicator OTU analysis on coral from the MPA and fished area did not find any OTU that was enriched in MPA or fished area adults. However, two OTUs are indicative of fished area larvae. An OTU classified as Ruminococcus gnavus within the family Lachnospiraceae, and an unclassified OTU within the family Lachnospiraceae were found to have high specificity (100% and 91%, respectively, of the reads for each OTU were found in fished area larvae) and high fidelity (each of these OTUs were found in 100% of fished area larvae).
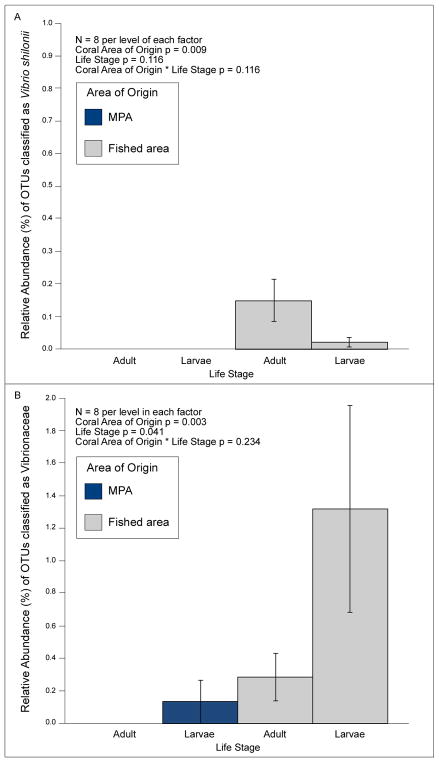
OTUs classified as Vibrio shilonii (at 97% clustering; see discussion below regarding limitations of 16S-based classification of microbial species) were the only potential coral pathogens in more than one of our 16 coral replicates. We did not detect V. shilonii in any our MPA coral at a sampling depth of 1,650; however, it occurred at low (< 1%), but significantly higher, relative abundances in our fished area coral (Figure 2A, Table S4, two-factor ANOVA coral area of origin p = 0.009, life stage p = 0.116, coral area of origin * life stage p = 0.116). We also detected higher abundances of taxa within the Vibrionaceae family in fished area versus MPA coral, especially in larvae (Figure 2B, Table S4, two-factor ANOVA coral area of origin p = 0.003, life stage p = 0.041, coral area of origin * life stage p = 0.234). Vibrionaceae were not detected on MPA adults but were detected at low (mean ± SE; 0.13 ± 0.13%) abundances on their larvae. We detected low abundances of V. shilonii in both MPA (0.32 ± 0.12%) and fished area (0.53 ± 0.14%) water, with these values not differing significantly (Table S4, p = 0.269, n = 5). The abundances of Vibrionaceae also did not differ significantly between MPA and fished area water (Table S4, 2.08 ± 0.66% vs. 0.93 ± 0.39%, respectively, p = 0.898, n = 5).
Figure 2.
Abundances and two-factor ANOVA analyses of Vibrio shilonii (A) and taxa within the family Vibrionaceae (B) in corals (adults and larvae) from the MPA and fished area.
Larval microbiomes, survivorship, settlement, and post-settlement survival
Despite the similarity of coral microbiomes between the coral-dominated MPA and macroalgae-dominated fished sites, when held in the lab for six days in MPA or fished area water, survivorship of MPA larvae was 94% regardless of water source while survivorship of fished area larvae was significantly lower – only 26%, when in fished area water and 66% when in MPA water (Figure 3A; two-factor ANOVA, Tukey HSD post-hoc analysis; larval area of origin p < 0.001, water area of origin p = 0.008, larval area of origin * water area of origin p = 0.008). Larval metamorphosis (in the water column or on the plastic dish) did not bias these results, as < 1% of individuals underwent metamorphosis in any treatment, and this percentage did not differ significantly among treatments (two-factor ANOVA larval area of origin p = 0.332, water area of origin p = 0.332, larval area of origin * water area of origin p = 0.332).
Figure 3.
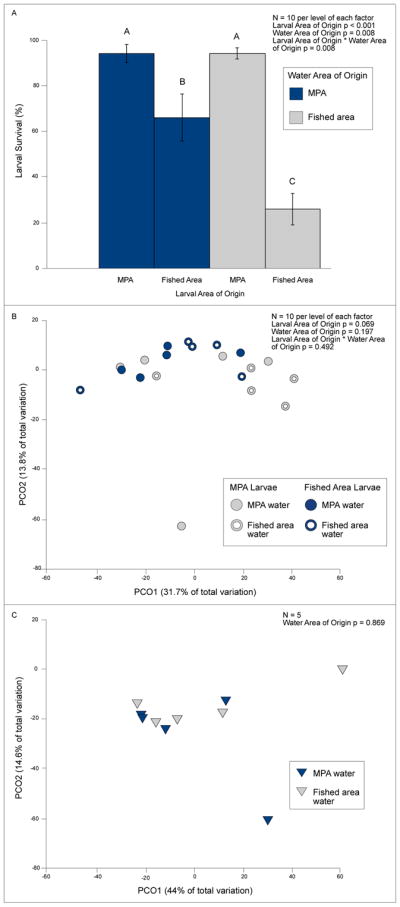
A) Survival and two-factor ANOVA analysis of larvae (mean ± SE) from the MPA and fished area maintained in MPA or fished area water for six days (n = 10 per level of each factor). Letters above bars indicate significant groupings by Tukey HSD post-hoc analysis. B) PCO and PERMANOVA analysis of Bray Curtis dissimilarity matrix of microbiomes from surviving larvae from the MPA or fished area maintained in MPA or fished area water for six days. C) PCO and PERMANOVA analysis of Bray Curtis dissimilarity matrix of water microbiomes from the MPA and fished area, used to maintain larvae in the lab from the MPA or fished area for six days.
When comparing the microbiomes of larvae that survived the 6-day experiment, we did not detect differences among larvae from the fished area or the MPA when maintained in water from the fished area or from the MPA (Figure 3B: PERMANOVA larval area of origin p = 0.069, water area of origin p = 0.197, larval area of origin * water area of origin p = 0.492), nor between the MPA and fished area water in which the larvae were held (Figure 3C; p = 0.869). The only suggested coral pathogens found on larvae in this experiment were OTUs classified as Vibrio shilonii. The mean relative abundance (± SE) of V. shilonii on MPA larvae was 0.0 ± 0.0% for larvae held in MPA water and 0.01 ± 0.01% for larvae in fished area water (Table S4). V. shilonii abundance on fished area larvae was 4.16 ± 4.14% for larvae in MPA water and 0.28 ± 0.17% for larvae in fished area water (Table S4). These abundances did not differ significantly among treatments (two-factor ANOVA larval area of origin p = 0.177, water area of origin p = 0.551, larval area of origin * water area of origin p = 0.495). We detected low abundances of V. shilonii in both MPA (0.32 ± 0.12%) and fished area (0.53 ± 0.14%) water (Table S4), with these values not differing (p = 0.269, n = 5). Furthermore, the abundance of Vibrionaceae as a group did not differ significantly between MPA and fished area water (Table S4, 2.08 ± 0.66% vs. 0.93 ± 0.39%, respectively, p = 0.898, n = 5).
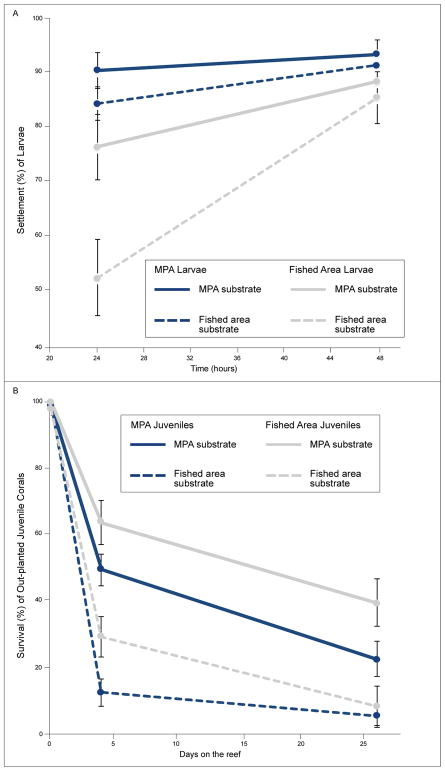
When larvae were offered rubble from either the MPA or fished area as settlement substratum in a no-choice experiment (i.e., larvae are given the option to settle on the type of rubble provided or remain in the water column), MPA larvae settled more rapidly than larvae from the fished area (Figure 4A, Table 1A larval area of origin * time interaction p < 0.001). For both MPA and fished area larvae, settlement was more rapid in response to MPA than to fished area substrate (Figure 4A, Table 1A, substrate type * time p = 0.010). For MPA larvae, 84–90% had settled by 24 h, whereas 52–76% of fished area larvae settled in this time period. After 48 h of isolation with a particular substrate type, 85–93% of all larvae had settled regardless of larval origin or substrate type.
Figure 4.
A) Settlement (mean ± SE) of MPA and fished area larvae on rubble from the MPA without macroalgae and from the fished area with macroalgae at 24 and 48 h (n = 20 per level of each factor; absolute percentages provided). See Table 1A for statistical analyses of repeated measures ANOVA on square root transformed proportion data. B) Survival (mean ± SE) of newly settled MPA and fished area juvenile corals on MPA versus fished area substrates that were out-planted to their corresponding reef (MPA rubble planted in the MPA and fished area rubble planted in the fished area) when corals were four and 26 days old (n = 13 – 18 per level of each factor due to loss of zip-tied rubble on the reef over time). See Table 1B for statistical analyses of repeated measures ANOVA on proportion data.
Table 1.
Repeated measures ANOVA on A) square root transformed settlement of larvae (originating from the MPA or fished area) on substrate from the MPA (no macroalgae) or fished area with macroalgae and B) survival of recently settled juvenile corals over 26 days on the reef. Juveniles that settled on MPA substrate were out-planted to the MPA and juveniles that settled on the fished area substrate were out-planted to the fished area.
| A | |||
|---|---|---|---|
| Source | DF | F Ratio | P |
| Larval Area of Origin | 1 | 15.75 | < 0.001 |
| Substrate Type | 1 | 5.95 | 0.020 |
| Time | 1 | 31.62 | < 0.001 |
| Larval Area of Origin*Substrate Type | 1 | 2.10 | 0.156 |
| Larval Area of Origin*Time | 1 | 14.26 | < 0.001 |
| Substrate Type*Time | 1 | 7.40 | 0.010 |
| Larval Area of Origin*Substrate Type*Time | 1 | 3.72 | 0.062 |
| B | |||
|---|---|---|---|
| Source | DF | F Ratio | P |
| Larval Area of Origin | 1 | 8.16 | 0.007 |
| Substrate Out-plant Treatment | 1 | 46.39 | < 0.001 |
| Time | 2 | 446.69 | < 0.001 |
| Larval Area of Origin*Substrate Out-plant Treatment | 1 | 0.77 | 0.387 |
| Larval Area of Origin*Time | 2 | 4.67 | 0.013 |
| Substrate Out-plant Treatment*Time | 2 | 22.53 | < 0.001 |
| Larval Area of Origin*Substrate Out-plant Treatment*Time | 2 | 1.15 | 0.322 |
When recently-settled juvenile coral were out-planted to the sites from which their settlement substrates had been collected (i.e., MPA substrate to the MPA, fished area substrate to the fished area), survival was higher in the MPA than in the fished area regardless of larval area of origin (Figure 4B, Table 1B, substrate out-plant treatment * time p < 0.001). Survival on fished area substrate out-planted to the fished area was 12–29% by day four and 5–8% by day 26. In contrast, survival on MPA substrate out-planted to the MPA was 49–64% on day four and 22–39% on day 26. Surprisingly, given lower survivorship of fished area larvae pre-settlement (Figure 3A), larvae from fished area adults survived better as newly-settled juveniles when out-planted to the field than did those from MPA adults (Figure 4B, Table 1B, larval area of origin p = 0.007; larval area of origin * time p = 0.013). Greater post-settlement survivorship of fished area larvae was not due to selective pre-settlement mortality of less hardy individuals among the fished area larvae. Mortality of larvae during the initial settlement experiment (48 h) was ≤ 4% and did not differ among treatments (larval area of origin p = 0.336, substrate type p = 0.747, larval area of origin * substrate type p = 0.747).
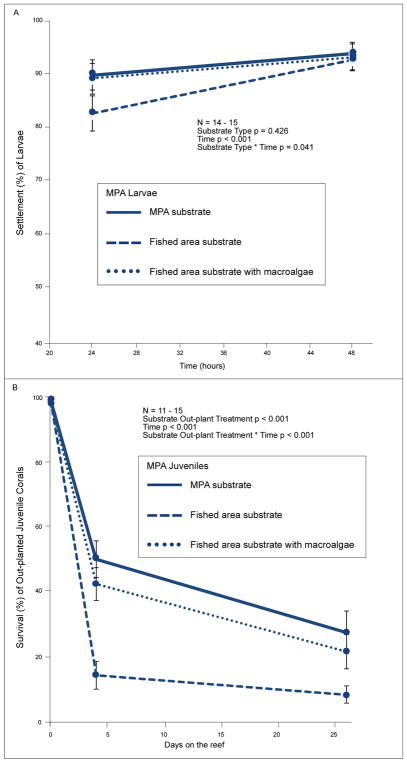
Lower survivorship of juvenile coral (regardless of larval area of origin) planted in the fished area versus the MPA could be due to larger-scale site differences, or smaller-scale differences of the substrate used (i.e., differences in abundances of macroalgae or crustose coralline algae on rubble). CCA cover did not contribute to differences between MPA and fished area rubble. CCA cover was high and did not differ among treatment groups (ANOVA p = 0.308, 66% ± 0.12 SE, 79% ± 0.04, 84% ± 0.03 on fished area rubble with macroalgae, fished area rubble without macroalgae, and MPA rubble, respectively). To assess the potentially confounding factors of site differences and macroalgal presence on rubble, we conducted a second, no-choice settlement experiment that ran simultaneously, but used only MPA larvae (due to insufficient larvae produced by fished area adults). In this experiment, larvae settled more rapidly (by 24 h) on rubble without macroalgae than rubble with macroalgae, even if both types of rubble originated from the fished area; this difference disappeared by 48 h (Figure 5A; substrate type * time p = 0.041). Mortality of larvae during the 48 h settlement experiment was low (<3%) and did not differ among treatments (substrate type p = 0.841). When these juveniles were out-planted back to their respective field sites (MPA rubble to the MPA and fished area rubble to the fished area), survival of juveniles differed between treatment types (Figure 5B, substrate out-plant treatment p < 0.001), with the lowest survival occurring for juveniles on substrate fouled with macroalgae within the fished area. Coral on rubble not fouled by macroalgae survived similarly well whether placed in the MPA or fished area (Figure 5B, 43–51% survival on day four and 22–28% on day twenty-six). In contrast, those on macroalgae- fouled rubble in the fished area experienced only 15% survival to day four and 9% to day 26 (Figure 5B). On day 4 and 26 after out-planting, survival of juvenile coral in the fished area was ~190% and ~150% higher, respectively, if on fished area rubble without macroalgae than on fished area rubble fouled with macroalgae. In contrast, survivorship of juvenile coral on non-macroalgal fouled rubble was only ~20% to 30% higher when out-planted to the MPA compared to the fished area on day 4 and 26, respectively.
Figure 5.
A) Percent settlement (mean ± SE) of MPA larvae on rubble from the MPA without macroalgae, the fished area without macroalgae, and the fished area with macroalgae at 24 and 48 hours (n = 15 with the exception of one lost replicate due to sloughing of macroalgae during the settlement experiment; absolute percentages provided). Repeated measures ANOVA was performed on square root transformed proportion data. B) Percent survival (mean ± SE) of newly settled juvenile corals on rubble from the MPA without macroalgae, the fished area without macroalgae, and the fished area with macroalgae that were out-planted to their corresponding reef (MPA rubble planted in the MPA and fished area rubble planted in the fished area) when the juvenile corals were four days old. Repeated measures ANOVA was performed on proportion data (n = 11 – 15 due to loss of replicates planted on the reef over time).
Discussion
Our experiments within an MPA dominated by corals and an adjacent fished area dominated by macroalgae allowed an assessment of how microbiomes of the coral Pocillopora damicornis are shaped by chronically (up to 12 years) higher macroalgal abundances and whether these habitat differences are correlated with changes in larval behavior or survivorship. By sampling coral from coral- versus macroalgae-dominated reefs that are only ~100–500 meters apart, we were able to examine microbiomes on degraded and healthy reefs that are not confounded in time or by large spatial scales. These study areas differ dramatically in the extent (23–67 fold greater) and frequency (5–15 fold greater) of coral-macroalgae contact (Bonaldo & Hay 2014), and these differences have persisted for the 7+ years we have worked on these reefs (M.E. Hay, personal observation).
Responses of coral microbial communities
Despite the large difference in algal cover (primarily brown seaweeds [Sargassum, Turbinaria, Dictyota], and a lesser abundance of red and green seaweeds [Galaxaura, Amphiroa, Liagoria, and Halimeda], Rasher et al. 2013) between the fished area and MPA, the microbiome composition of adult and larval P. damicornis did not differ between the coral-dominated MPA and the macroalgae-dominated fished area. This result contrasts with evidence suggesting that macroalgae alter the physiochemical environment, the microbial load, and community composition in surrounding seawater (Wild et al. 2010, Haas et al. 2011, Nelson et al. 2013), and the microbiome of associated corals (Wild et al. 2010, Haas et al. 2011, Morrow et al. 2012, 2013, 2017, Thurber et al. 2012, Nelson et al. 2013). Specifically, algae are predicted to affect corals through DOM release that promotes microbial growth in surrounding seawater, declines in local oxygen concentrations, and enrichment of copiotrophic and pathogenic microbes that may overwhelm the native coral microbiota (Dinsdale & Rohwer 2011, Barott & Rohwer 2012). Algae may also release allelochemicals that alter coral microbial communities on contact (Morrow et al. 2011, 2012, 2017), or act as vectors for pathogenic microbes (Nugues et al. 2004, Sweet et al. 2013). All of these mechanisms may operate on relatively small spatial scales, exerting strongest effects in zones of direct algae-coral contact (Barott et al. 2009, 2011, Pratte et al. 2017). Here, our sampling did not assess microbiome variation relative to algal contact sites. Nonetheless, the similarity of microbiomes (both coral and water) from sites with ~ 2% cover of macroalgae versus ~90% cover of high-biomass macroalgae (Rasher et al. 2013) suggests that enhanced algal coverage at the reef scale does not systemically alter the microbiome of P. damicornis. Lack of differences in water microbiomes from our coral- and algae-dominated reefs may result from sampling water ~1–2 meters above the benthos, where reef water is readily exchanged with oceanic water flowing over the reef crest. The positioning of small protected areas within the larger background of fished areas may also facilitate dispersal and mixture of microbes at scales of hundreds of meters, helping to homogenize both coral and seawater microbiomes across degraded algae-dominated and protected coral-dominated reefs. However, if this is the case, it is not suppressing corals in the MPAs, where coral cover on hard substrates is nearly 60% (Rasher et al. 2013). Additionally, corals in the fished area grow as well as those in the MPA when macroalgae within 50 cm of the coral colonies are removed (Clements et al. in press). This suggests minimal effects of macroalgal DOM on growth of corals at the scale of ≥ 50 cm.
It is also possible that our results are specific to P. damicornis (but preliminary data for other corals from these sites suggest that this is not the case; D. Beatty unpublished data). Previous work has indicated that coral-algae interactions and their outcomes are often species-specific, with effects of macroalgae on coral microbiomes varying from undetectable to strong (Morrow et al. 2012, 2013, Thurber et al. 2012). Here, in contrast to numerous non-Pocilloporid coral taxa, which decline in abundance at macroalgae-dominated sites, the abundance of Pocilloporid coral does not differ significantly between the MPA and fished areas at Votua village (Bonaldo & Hay 2014). This persistence may be due in part to the ability of Pocilloporids to maintain a stable microbiome in spite of drastic differences in benthic cover. In support of this hypothesis, we found similarly high relative abundances (>80%) of Endozoicimonaceae bacteria in adult P. damicornis coral from both healthy and degraded reefs. Recent evidence suggests that these bacteria are functionally important members of the healthy coral holobiont in multiple coral species (Meyer et al. 2014, Lee et al. 2015, Ding et al. 2016, Neave et al. 2016), including P. damicornis (Bayer et al. 2013). We also detected similar abundances of Endozoicimonacaeae (1–12%) on larvae from both the MPA and fished area, adding further support for the hypothesized importance of these bacteria in P. damicornis persistence. Adult corals were not maintained in filter-sterilized seawater (FSW) before larval release; therefore, we do not know if Endozoicimonaceae were rapidly acquired from the environment or vertically transferred to brooded larvae. However, larvae were rinsed in FSW three times before preservation, so it is unlikely that the presence of Endozoicimonaceae represents contamination from seawater because this group’s abundance was <0.5% in our seawater samples.
While we did not detect a significant community-level shift in coral microbiomes between our macroalgae-dominated and coral-dominated sites, we did detect differences in the abundances, although rare, of Vibrionaceae, with this bacterial family being significantly enriched in both adults and larvae from the macroalgae-dominated reef compared to those from the coral-dominated MPA. The enriched bacteria included OTUs classified, as Vibrio shilonii, a demonstrated coral pathogen (Kushmaro et al. 2001). Caution should be taken when interpreting ecological function and pathogenicity when using the 16S rRNA gene for classification because bacterial strains identified as the same species by this method can vary in genome size and functional gene content, including genes involved in pathogenicity (discussed in Franzosa et al. 2015, Land et al. 2015). However, it is interesting to note that the 16S rRNA gene of V. shilonii shares 96.6% similarity with that of a P. damicornis pathogen (Vibrio coralliilyticus) that causes coral bleaching (Ben-Haim et al. 2003a) and with other Vibrio spp. that cause white syndromes in many Indo-Pacific coral species (Sussman et al. 2008). The overall abundance of taxa falling within the Vibrionaceae family was low (≤ 2%) and comparable to abundances (0–3%) found in healthy corals (Lee et al. 2017, Morrow et al. 2017, Tout et al. 2015), even in the macroalgae-dominated area, suggesting that the sampled P. damicornis were not in a ‘diseased’ state. Nevertheless, the differences in Vibrionaceae abundance may indicate that coral in the protected area are more resistant to colonization by potentially harmful bacteria, consistent with a recent investigation by Lamb et al. (2016) that found lower abundances of coral disease in no-take reserves. While our findings provide evidence of proportionally lower abundances of Vibrionaceae on coral in a no-take protected area compared to an adjacent fished reef, more work is needed to confirm a pathogenic role for the detected bacteria and the reproducibility of findings in other coral species and protected areas. Indeed, Vibrio species are also found in healthy corals (Chimetto et al. 2008, Raina et al. 2009) and may function as coral mutualists by providing fixed nitrogen (Ceh et al. 2013). We also found R. gnavus as an indicator species of fished area larvae. R. gnavus is an anaerobic gut microbe that has been implicated in human disease and is capable of breaking down mucins (Crost et al. 2013). Its impact on adult coral or their larvae is unknown.
We also found that microbiomes of adult coral from the macroalgae-dominated fished area were more variable in community composition than those from the coral-dominated MPA. This is consistent with Zaneveld et al. (2016) who found that corals in experimental plots where macroalgal cover increased due to the absence of fish grazing exhibited greater microbial beta diversity. Thus, microbiome variance may be an early indicator of coral stress, but further investigations are needed to test this hypothesis. We also found that while adult MPA coral were less variable in their microbiome composition compared to their larvae, levels of inter-individual microbiome variability did not differ between adult and larval coral from the macroalgae-dominated fished area. Taken together, these patterns suggest that P. damicornis adults from the MPA have more constrained microbial communities than their adult counterparts from the fished area and than juveniles from both areas, adding support to the notion that coral-algae interactions may increase the variance (Thurber et al. 2012, Zaneveld et al. 2016) of coral microbiomes. However, greater inter-individual variability in coral microbiomes could indicate either 1) the loss of regulatory mechanisms within the coral holobiont, thereby predisposing corals to microbial dysbiosis (Krediet et al. 2013, Thompson et al. 2015), or 2) the holobiont’s adaptive response to counter local biotic or abiotic stressors (Rosenberg 2007). To better understand our microbial community data in the context of coral fitness and health, we concurrently investigated how more frequent and chronic algal interactions (in the macroalgae-dominated fished area) affected larval behavior, and larval and juvenile survivorship.
Effects of parentage, habitat, and substrate on larval survival
Based on prior evidence showing that larvae of P. damicornis are packed with photosynthate providing Symbiodinium and that larvae can settle in under two hours or remain viable in the plankton for 100 days (Richmond 1987, Isomura & Nishihira 2001), we expected high larval survivorship over the short duration of our larval survival experiment. In contrast, we found rapid mortality within some treatments. During six days of exposure to MPA or fished area water without a choice of appropriate settlement substrates, larvae from MPA adults experienced only 6% mortality regardless of water source, while larvae from fished area adults experienced significantly higher, 74% and 34%, mortality in both fished area and MPA water, respectively (Figure 3A). Thus, larvae produced by adults in the fished area appear less robust than those produced by adults in the MPA. We failed to detect differences in the relative abundance of potentially pathogenic bacterial OTUs classified as Vibrio shilonii on coral larvae that experienced higher mortality. It is possible that Vibrio shilonii OTUs could have been at greater abundance on, and differentially impacted survivorship of, fished area larvae but that we failed to detect differences in bacterial relative abundances because we analyzed only the less infected, or most resistant, larvae living at the end of the six-day experiment. Microbiomes of dead larvae were not analyzed due to rapid shifts in microbial communities following mortality.
Given that we were unable to document significant differences in potential pathogens or microbial community composition between MPA and fished area larvae or between MPA and fished area water (Fig. 3B & 3C), it may be that differential mortality is due to differential larval provisioning by adults rather than microbial effects. Dense macroalgae, which is typical of the fished area, commonly suppress coral recruitment, growth, and survivorship (Hughes et al. 2007, Burkepile and Hay 2008, Thurber et al. 2012, Zaneveld et al. 2016). However, to our knowledge, this is the first documentation of negative intergenerational effects of algal dominance on coral.
Experimental studies indicate that many species of macroalgae deter coral larval settlement (Kuffner et al. 2006, Vermeij et al. 2009, Diaz-Pulido et al. 2010, Dixson et al. 2014). However, as reefs globally continue to degrade, larvae may not be able to avoid settlement near macroalgae. We found that both MPA and fished area larvae settled more rapidly on MPA substrate free of macroalgae than on fished area substrate fouled with macroalgae. However, by the end of the 48 h experimental period, almost all larvae had settled, regardless of substrate type. When newly-settled juvenile coral were out-planted to the sites from where their substrates originated, juvenile survivorship was ~5 times greater in the MPA than the fished area (Fig 4B day 30), confirming a strong positive effect of the no-take MPA on juvenile coral survival.
However, lower survivorship of juveniles in the fished area could have been due to macroalgae on the substrate onto which they settled, other differences between the MPA and fished area (site differences), or both. We therefore investigated the relative impact on juvenile survival of site and of macroalgal presence on the settlement substrate. Survivorship of juveniles in the fished area on day 4 and 26 was ~190% and 150% higher, respectively, if on fished area rubble without macroalgae than on fished area rubble fouled with macroalgae (Figure 5B day 4 & 26). In contrast, the increase in survival due to site was modest (Figure 5B day 4 & 26, 20–30% higher). Thus, nearby macroalgae on the same piece of rubble–not general traits of the macroalgae-dominated area (i.e., site effects)–were largely responsible for reductions in juvenile survivorship in the fished area.
Although pre-settlement larvae from MPA adults experienced greater survival than larvae from fished area adults, this relationship was reversed for post-settlement survivorship in the field. This pattern occurred regardless of settlement substrate type (MPA or fished area origin) or the site into which the coral were out-planted. This was not due to selective mortality of less hardy fished area larvae during the initial 48 h settlement experiment; in that period, mortality was low (≤ 4%) and did not differ between treatments. It is possible that degraded reefs have selected for hardier post-settlement populations of P. damicornis, but this hypothesis is difficult to reconcile with the lower survival of fished area larvae during the pre-settlement period. If degraded reefs have selected for hardier coral, then these populations may become increasingly valuable as global change and other anthropogenic stressors continue to impact reefs.
Conclusion
The composition of P. damicornis microbial communities did not differ significantly between the MPA and fished area despite drastic differences in benthic cover between these sites and substantial differences in larval survivorship. However, adults within the coral-rich MPA exhibited lower variability in their microbial community composition than those from the macroalgae-dominated fished area. Additionally, larval and adult P. damicornis from the MPA had significantly lower abundances of Vibrionaceae and OTUs classified as the coral pathogen Vibrio shilonii. Taken together, our findings indicate that coral within a coral-dominated MPA with abundant and diverse herbivore populations and low abundances of macroalgae experience greater larval survivorship, reduced variability in their adult microbial community composition, and reduced abundances of rare but potentially harmful bacteria. However, overall microbial community composition remained relatively uniform despite reef protection status and a 45-fold difference in macroalgal cover (~2% vs 90%) between these sites. Reproductive adults were only collected from one MPA and one fished area (following permitting guidelines). Further studies will be needed to understand how frequency of coral-algae interactions in natural reef environments affects coral microbiomes and coral fitness in other species of reef-building corals and if findings are reproducible among other coral- and algae-dominated areas. At present, our study suggests that investigating macroalgal impacts on coral health via alterations of their microbiomes may require understanding the importance of subtle microbiome alterations such as changes in rare taxa of potential pathogens or changes in variability of coral microbial communities rather than drastic differences in microbial community composition.
Supplementary Material
Acknowledgments
We thank the Fijian government and the Korolevu-i-wai district elders for collection and research permissions, G. O. Longo for statistical guidance, K. Ritchie for comments on the manuscript and guidance regarding larval collection and maintenance, and V. Bonito for communicating research objectives to the Korolevu-i-wai Environment Committee. Financial support came from the National Institutes of Health (2 U19 TW007401-10), the National Science Foundation (OCE 0929119), the Simons Foundation (346253), and the Teasley Endowment to the Georgia Institute of Technology. Credit for images in supplemental figure S1: “Coral larvae” and “Sargassum spp.” to Tracey Saxby, “Cauliflower coral” and “Coralline algae” to Joanna Woerner, Integration and Application Network, University of Maryland Center for Environmental Science (ian.umces.edu/imagelibrary/).
References
- Ainsworth TD, Thurber RV, Gates RD. The future of coral reefs: a microbial perspective. Trends Ecol Evol. 2010;25:233–240. doi: 10.1016/j.tree.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Almany GR, Connolly SR, Heath DD, Hogan JD, Jones GP, McCook LJ, Mills M, Pressey RL, Williamson DH. Connectivity, biodiversity conservation and the design of marine reserve networks for coral reefs. Coral Reefs. 2009;28:339–351. [Google Scholar]
- Barott K, Smith J, Dinsdale E, Hatay M, Sandin S, Rohwer F. Hyperspectral and physiological analyses of coral-algal interactions. PLoS ONE. 2009;4:e8043. doi: 10.1371/journal.pone.0008043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barott KL, Rodriguez-Mueller B, Youle M, Marhaver KL, Vermeij MJA, Smith JE, Rohwer FL. Microbial to reef scale interactions between the reef-building coral Montastraea annularis and benthic algae. Proc R Soc B. 2011;279:1655–1664. doi: 10.1098/rspb.2011.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barott KL, Rohwer FL. Unseen players shape benthic competition on coral reefs. Trends Microbiol. 2012;20:621–628. doi: 10.1016/j.tim.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Bayer T, Neave MJ, Alsheikh-Hussain A, Aranda M, Yum LK, Mincer T, Hughen K, Apprill A, Voolstra CR. The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol. 2013;79:4759–4762. doi: 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellwood DR, Hughes TP, Folke C, Nystrom M. Confronting the coral reef crisis. Nature. 2004;429:827–833. doi: 10.1038/nature02691. [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y, Thompson F, Thompson C, Cnockaert M, Hoste B, Swings J, Rosenberg E. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol. 2003a;53:309–315. doi: 10.1099/ijs.0.02402-0. [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y, Zicherman-Keren M, Rosenberg E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl Environ Microbiol. 2003b;69:4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldo RM, Hay ME. Seaweed-coral interactions: variance in seaweed allelopathy, coral susceptibility, and potential effects on coral resilience. PLoS ONE. 2014;9:e85786. doi: 10.1371/journal.pone.0085786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno JF, Selig ER. Regional decline of coral cover in the Indo-Pacific: timing, extent, and subregional comparisons. PLoS ONE. 2007;2:e711. doi: 10.1371/journal.pone.0000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkepile DE, Hay ME. Herbivore species richness and feeding complementarity affect community structure and function on a coral reef. Proc Natl Acad Sci USA. 2008;105:16201–16206. doi: 10.1073/pnas.0801946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceh J, Kilburn MR, Cliff JB, Raina JB, van Keulen M, Bourne DG. Nutrient cycling in early coral life stages: Pocillopora damicornis larvae provide their algal symbiont (Symbiodinium) with nitrogen acquired from bacterial associates. Ecol Evol. 2013;3:2393–2400. [Google Scholar]
- Cesar H, Burke L, Pet-Soede L. The economics of worldwide coral reef degradation. Cesar Environmental Economics Consulting (CEEC) Technical Report 2003 [Google Scholar]
- Chimetto LA, Brocchi M, Thompson CC, Martins RC, Ramos HR, Thompson FL. Vibrios dominate as culturable nitrogen-fixing bacteria of the Brazilian coral Mussismilia hispida. Syst Appl Microbiol. 2008;31:312–319. doi: 10.1016/j.syapm.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Clarke KR. Nonparametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- Clements CS, Rasher DB, Hoey AS, Bonito VJ, Hay ME. Spatial and temporal limits of coral-macroalgal competition: negative impacts on corals depend on macroalgal density, proximity, and duration of contact. Mar Ecol Prog Ser. doi: 10.3354/meps12410. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS ONE. 2013;8:e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalzell P, Adams T, Polunin N. Coastal fisheries in the Pacific Islands. Oceanograph Lit Rev. 1997;5:516. [Google Scholar]
- De’ath G, Fabricius KE, Sweatman H, Puotinen M. The 27–year decline of coral cover on the Great Barrier Reef and its causes. Proc Natl Acad Sci. 2012;109:17995–17999. doi: 10.1073/pnas.1208909109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell CLA, Longo GO, Hay ME. Positive feedbacks enhance macroalgal resilience on degraded coral reefs. PLoS ONE. 2016;11:e0155049. doi: 10.1371/journal.pone.0155049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Pulido G, Harii S, McCook L, Hoegh-Guldberg O. The impact of benthic algae on the settlement of a reef-building coral. Coral Reefs. 2010;29:203–208. [Google Scholar]
- Ding JY, Shiu JH, Chen WM, Chiang YR, Tang SL. Genomic insight into the host-endosymbiont relationship of Endozoicomonas montiporae CL-33T with its coral host. Front Microbiol. 2016;7:251. doi: 10.3389/fmicb.2016.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, Hatay M, Hall D, Brown E, Haynes M, Krause L, Sala E, Sandin SA, Vega Thurber R, Willis B, Azam F, Knowlton N, Rohwer F. Microbial ecology of four coral atolls in the Northern Line Islands. PLoS ONE. 2008;3:e1584. doi: 10.1371/journal.pone.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Rohwer F. Coral Reefs: An Ecosystem in Transition. Springer; Netherlands: 2011. Fish or germs? Microbial dynamics associated with changing trophic structures on coral reefs; pp. 231–240. [Google Scholar]
- Dixson DL, Abrego D, Hay ME. Chemically-mediated behavior of recruiting corals and fishes: a tipping point that may limit reef recovery. Science. 2014;345:892–897. doi: 10.1126/science.1255057. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, Morgan XC, Huttenhower C. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nature Rev Microbiol. 2015;13:360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TA, Côté IM, Gill JA, Grant A, Watkinson AR. Long-term region-wide declines in Caribbean corals. Science. 2003;301:958–960. doi: 10.1126/science.1086050. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Hill R, Doblin MA, Ralph PJ. Microbial consortia increase thermal tolerance of corals. Mar Biol. 2012;159:1763–1771. [Google Scholar]
- Haas AF, Nelson CE, Kelly LW, Carlson CA, Rohwer F, Leichter JJ, Wyatt A, Smith JE. Effects of coral reef benthic primary producers on dissolved organic carbon and microbial activity. PLoS ONE. 2011;6:e27973. doi: 10.1371/journal.pone.0027973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AF, Fairoz MF, Kelly LW, Nelson CE, Dinsdale EA, Edwards RA, Giles S, Hatay M, Hisakawa N, Knowles B, Wei Lim Y, Maughan H, Pantos O, Roach TNF, Sanchez SE, Silveira CB, Sandin S, Smith JE, Rohwer F. Global microbialization of coral reefs. Nat Microbiol. 2016;1:16042. doi: 10.1038/nmicrobiol.2016.42. [DOI] [PubMed] [Google Scholar]
- Harvell D, Jordán-Dahlgren E, Merkel S, Rosenberg E, Raymundo L, Smith G, Weil E, Willis B. Coral disease, environmental drivers, and the balance between coral and microbial associates. Oceanography. 2007;20:172–195. [Google Scholar]
- Hoegh-Guldberg O, Mumby P, Hooten A, Steneck R, Greenfield P, Gomez E, Harvell CD, Sale PF, Edwards AJ, Caldeira K, Knowlton N, Eakin CM, Iglesias-Prieto R, Muthiga N, Bradbury RH, Dubai A, Hatziolos ME. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318:1737–1742. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- Hughes TP, Kerry JT, Álvarez-Noriega M, Álvarez-Romero JG, Anderson KD, Baird AH, Babcock RC, Beger M, Bellwood DR, Berkelmans R. Global warming and recurrent mass bleaching of corals. Nature. 2017;543:373–377. doi: 10.1038/nature21707. [DOI] [PubMed] [Google Scholar]
- Hughes TP, Graham NA, Jackson JB, Mumby PJ, Steneck RS. Rising to the challenge of sustaining coral reef resilience. Trends Ecol Evol. 2010;25:633–642. doi: 10.1016/j.tree.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Hughes TP, Rodrigues MJ, Bellwood DR, Ceccarelli D, Hoegh-Guldberg O, McCook L, Moltschaniwskyj N, Pratchett MS, Steneck RS, Willis B. Phase shifts, herbivory, and the resilience of coral reefs to climate change. Curr Biol. 2007;17:360–365. doi: 10.1016/j.cub.2006.12.049. [DOI] [PubMed] [Google Scholar]
- Isomura N, Nishihira M. Size variation of planulae and its effect on the lifetime of planulae in three pocilloporid corals. Coral Reefs. 2001;20:309–315. [Google Scholar]
- Jackson JBC, Donovan MK, Cramer KL, Lam VV, editors. Status and Trends of Caribbean Coral Reefs: 1970–2012. Global Coral Reef Monitoring Network, IUCN; Gland, Switzerland: 2014. [Google Scholar]
- Kelly LW, Williams GJ, Barott KL, Carlson CA, Dinsdale EA, Edwards RA, Haas AF, Haynes M, Lim YW, McDole T, Nelson CE, Sala E, Sandin SA, Smith JE, Vermeij MJA, Youle M, Rohwer F. Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proc Natl Acad Sci USA. 2014;111:10227–10232. doi: 10.1073/pnas.1403319111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler KE, Gill SM. Coral point count with Excel extensions (CPCe): A Visual Basic program for the determination of coral and substrate coverage using random point count methodology. Comput and Geosci. 2006;32:1259–1269. [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krediet CJ, Ritchie KB, Paul VJ, Teplitski M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc R Soc B. 2013;280:20122328. doi: 10.1098/rspb.2012.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffner IB, Walters LJ, Becerro MA, Paul VJ, Ritson-Williams R, Beach KS. Inhibition of coral recruitment by macroalgae and cyanobacteria. Mar Ecol Prog Ser. 2006;323:107–117. [Google Scholar]
- Kushmaro A, Banin E, Loya Y, Stackebrandt E, Rosenberg E. Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int J Syst Evol Microbiol. 2001;51:1383–1388. doi: 10.1099/00207713-51-4-1383. [DOI] [PubMed] [Google Scholar]
- Lamb JB, Wenger AS, Devlin MJ, Ceccarelli DM, Williamson DH, Willis BL. Reserves as tools for alleviating impacts of marine disease. Phil Trans R Soc B. 2016;371:20150210. doi: 10.1098/rstb.2015.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JB, Williamson DH, Russ GR, Willis BL. Protected areas mitigate diseases of reef-building corals by reducing damage from fishing. Ecology. 2015;96:2555–2567. doi: 10.1890/14-1952.1. [DOI] [PubMed] [Google Scholar]
- Land M, Hauser L, Se-Ran J, Nookaew I, Leuze MR, Ahn T, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics. 2015;15:141–161. doi: 10.1007/s10142-015-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee STM, Davy SK, Tang SL, Fan TY, Kench PS. Successive shifts in the microbial community of the surface mucus layer and tissues of the coral Acropora muricata under thermal stress. FEMS Microbiol Ecol. 2015;91:fiv142. doi: 10.1093/femsec/fiv142. [DOI] [PubMed] [Google Scholar]
- Lee STM, Davy SK, Tang SL, Fan TY, Kench PS. Water flow buffers shifts in bacterial community structure in heat-stressed Acropora muricata. Sci Rep. 2017;7 doi: 10.1038/srep43600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCook LJ, Ayling T, Cappo M, Choat JH, Evans RD, De Freitas DM, Heupel M, Hughes TP, Jones GP, Mapstone B, Marsh H, Mills M, Molloy FJ, Pitcher CR, Pressey RL, Russ GR, Sutton S, Sweatman H, Tobin R, Wachenfeld DR, Williamson DH. Adaptive management of the Great Barrier Reef: A globally significant demonstration of the benefits of networks of marine reserves. Proc Natl Acad Sci USA. 2010;107:18278–18285. doi: 10.1073/pnas.0909335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JL, Paul VJ, Teplitski M. Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLoS ONE. 2014;9:e100316. doi: 10.1371/journal.pone.0100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow K, Paul VJ, Liles M, Chadwick N. Allelochemicals produced by Caribbean macroalgae and cyanobacteria have species-specific effects on reef coral microorganisms. Coral Reefs. 2011;30:309–320. [Google Scholar]
- Morrow KM, Bromhall K, Motti CA, Munn CB, Bourne DG. Allelochemicals produced by brown macroalgae of the Lobophora genus are active against coral larvae and associated bacteria, supporting pathogenic shifts to Vibrio dominance. Appl Environ Microbiol. 2017;83:e02391–02316. doi: 10.1128/AEM.02391-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow KM, Liles MR, Paul VJ, Moss AG, Chadwick NE. Bacterial shifts associated with coral-macroalgal competition in the Caribbean Sea. Mar Ecol Prog Ser. 2013;488:103–117. [Google Scholar]
- Morrow KM, Ritson-Williams R, Ross C, Liles MR, Paul VJ. Macroalgal extracts induce bacterial assemblage shifts and sublethal tissue stress in Caribbean corals. PLoS ONE. 2012;7:e44859. doi: 10.1371/journal.pone.0044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumby PJ, Harborne AR, Williams J, Kappel CV, Brumbaugh DR, Micheli F, Holmes E, Dahlgren CP, Paris CB, Blackwell PG. Trophic cascade facilitates coral recruitment in a marine reserve. Proc Natl Acad Sci USA. 2007;104:8362–8367. doi: 10.1073/pnas.0702602104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave MJ, Rachmawati R, Xun L, Michell CT, Bourne DG, Apprill A, Voolstra CR. Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 2016;11:186–200. doi: 10.1038/ismej.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Goldberg SJ, Kelly LW, Haas AF, Smith JE, Rohwer F, Carlson CA. Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J. 2013;7:962–979. doi: 10.1038/ismej.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugues MM, Smith GW, Hooidonk RJ, Seabra MI, Bak RPM. Algal contact as a trigger for coral disease. Ecol Lett. 2004;7:919–923. [Google Scholar]
- Pettay DT, Wham DC, Smith RT, Iglesias-Prieto R, LaJeunesse TC. Microbial invasion of the Caribbean by an Indo-Pacific coral zooxanthella. Proc Natl Acad Sci USA. 2015;112:7513–7518. doi: 10.1073/pnas.1502283112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto RS, Rosado PM, de Assis Leite DC, Rosado AS, Bourne DG. Beneficial microorganisms for coral health and resilience. Front Microbiol. 2017;8:341. doi: 10.3389/fmicb.2017.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratte ZA, Longo GO, Burns AS, Hay ME, Stewart FS. Contact with turf algae alters the coral microbiome: contact versus systemic impacts. Coral Reefs. 2017 https://doi.org/10.1007/s00338-017-1615-4
- Raina JB, Tapiolas D, Willis BL, Bourne DG. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol. 2009;75:3492–3501. doi: 10.1128/AEM.02567-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasher DB, Engel S, Bonito V, Fraser GJ, Montoya JP, Hay ME. Effects of herbivory, nutrients, and reef protection on algal proliferation and coral growth on a tropical reef. Oecol. 2012;169:187–198. doi: 10.1007/s00442-011-2174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasher DB, Hay ME. Chemically rich seaweeds poison corals when not controlled by herbivores. Proc Natl Acad Sci USA. 2010;107:9683–9688. doi: 10.1073/pnas.0912095107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasher DB, Hoey AS, Hay ME. Consumer diversity interacts with prey defenses to drive ecosystem function. Ecology. 2013;94:1347–1358. doi: 10.1890/12-0389.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond R. Energetics, competency, and long-distance dispersal of planula larvae of the coral Pocillopora damicornis. Mar Biol. 1987;93:527–533. [Google Scholar]
- Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser. 2006;322:1–14. [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Kushmaro A. Coral Reefs: An Ecosystem in Transition. Springer; Netherlands: 2011. Microbial diseases of corals: pathology and ecology; pp. 451–464. [Google Scholar]
- Sandin SA, Smith JE, DeMartini EE, Dinsdale EA, Donner SD, Friedlander AM, Konotchick T, Malay M, Maragos JE, Obura D. Baselines and degradation of coral reefs in the northern Line Islands. PLoS ONE. 2008;3:e1548. doi: 10.1371/journal.pone.0001548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selig ER, Bruno JF. A global analysis of the effectiveness of Marine Protected Areas in preventing coral loss. PLoS ONE. 2010;5:e9278. doi: 10.1371/journal.pone.0009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneed JM, Sharp KH, Ritchie KB, Paul VJ. The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Phil Trans R Soc B. 2014;281:20133086. doi: 10.1098/rspb.2013.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M, Willis BL, Victor S, Bourne DG. Coral pathogens identified for white syndrome (WS) epizootics in the Indo-Pacific. PLoS ONE. 2008;3:e2393. doi: 10.1371/journal.pone.0002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet MJ, Bythell JC, Nugues MM. Algae as reservoirs for coral pathogens. PLoS ONE. 2013;8:e69717. doi: 10.1371/journal.pone.0069717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Rivera HE, Closek CJ, Medina M. Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Front Cell Infect Microbiol. 2015;4:176. doi: 10.3389/fcimb.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber RV, Burkepile DE, Correa AM, Thurber AR, Shantz AA, Welsh R, Pritchard C, Rosales S. Macroalgae decrease growth and alter microbial community structure of the reef-building coral, Porites astreoides. PLoS ONE. 2012;7:e44246. doi: 10.1371/journal.pone.0044246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tout J, Siboni N, Messer LF, Garren M, Stocker R, Webster NS, Ralph PJ, Seymour JR. Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran C, Hadfield MG. Larvae of Pocillopora damicornis (Anthozoa) settle and metamorphose in response to surface-biofilm bacteria. Mar Ecol Prog Ser. 2011;433:85–96. [Google Scholar]
- Vermeij M, Smith J, Smith C, Thurber RV, Sandin S. Survival and settlement success of coral planulae: independent and synergistic effects of macroalgae and microbes. Oecol. 2009;159:325–336. doi: 10.1007/s00442-008-1223-7. [DOI] [PubMed] [Google Scholar]
- Webster FJ, Babcock RC, Van Keulen M, Loneragan NR. Macroalgae inhibits larval settlement and increases recruit mortality at Ningaloo Reef, Western Australia. PLoS ONE. 2015;10:e0124162. doi: 10.1371/journal.pone.0124162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vazquez-Baeza Y, Birmingham A. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, Angenent T, Knight R, Ley RE. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2012;6:94–103. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C, Niggl W, Naumann MS, Haas AF. Organic matter release by Red Sea coral reef organisms—potential effects on microbial activity and in situ O2 availability. Mar Ecol Prog Ser. 2010;411:61–71. [Google Scholar]
- Zaneveld JR, Burkepile DE, Shantz AA, Pritchard CE, McMinds R, Payet JP, Welsh R, Correa AMS, Lemoine NP, Rosales S, Fuchs C, Maynard JA, Thurber RV. Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat Commun. 2016;7:11833. doi: 10.1038/ncomms11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.