Abstract
The use of blood biomarkers for stroke has been long considered an excellent method to determine the occurrence, timing, subtype, and severity of stroke. In this study, venous blood was obtained from ischemic stroke patients after stroke onset and compared with age and sex-matched controls. We used a multiplex panel of 37 inflammatory molecules, analyzed using Luminex MagPix technology, to identify the changes in plasma proteins after ischemic stroke. We identified eight key molecules that were altered within the blood of stroke patients as compared to controls. Plasma levels of interleukin 6 signal transducer (sIL-6Rβ/gp130), matrix metalloproteinase-2 (MMP-2), osteopontin, sTNF-R1 and sTNF-R2 were significantly higher in stroke patients compared to controls. Interferon-β, interleukin-28, and thymic stromal lymphopoietin (TSLP) were decreased in plasma from stroke patients. No other immunological markers were significantly different between patient groups. When stroke patients were treated with tissue plasminogen activator (t-PA), plasma levels of interferon-α2 significantly increased while interleukin-2 and pentraxin-3 decreased. The discriminatory power of the molecules was evaluated by receiver operating characteristic (ROC) analysis. According to ROC analysis, the best markers for distinguishing stroke occurrence were MMP-2 (AUC=0.76, sensitivity 62.5%, specificity 88.5%), sTNF-R2 (AUC=0.75, sensitivity 83.3%, specificity 65.3%) and TSLP (AUC=0.81, sensitivity 66.7%, specificity 96.2%). Multivariate logistic regression, used to evaluate the combination of proteins, identified a biomarker panel with high specificity and sensitivity (AUC=0.96, sensitivity 87.5%, specificity 96.2%). These results indicate a novel set of blood biomarkers that could be used in a panel to identify stroke patients and their responsiveness to therapeutic intervention.
Keywords: blood biomarker panels, ischemic stroke, inflammation, luminex, plasma
1. Introduction
Stroke is a debilitating neurological condition with limited treatment options [1]. Imaging techniques (e.g., computed tomography (CT) and Magnetic resonance imaging (MRI)) are useful in detecting the occurrence, type and severity of strokes. However, there is a need to develop rapid, accessible, affordable, and easy to use diagnostic tools to identify and treat stroke symptoms in conjunction with imaging techniques. The use of blood biomarkers for stroke has been long considered an excellent method to determine the occurrence, timing, subtype, and severity of strokes [2–4]. Blood biomarkers can also be used to determine the efficacy of existing and novel stroke treatment strategies. Panels of multiple biomarkers have been shown to predict stroke diagnosis with high sensitivity and specificity [5].
The existing body of stroke research suggests inflammation is an important part of the pathophysiology of cerebral ischemia [6]. Many cytokines, including interleukin-1β (IL-1β), interleukin-6 (IL-6), interleukin-10 (IL-10) and tumor necrosis factor-α (TNF-α) have been studied extensively within the context of stroke [2, 7–10]. This concentration on general components of the immune response makes it difficult to identify unique immunological properties for stroke. Our increasing understanding of the complexities of the immune system advances the utility of probing multiple immunological factors in stroke patients seeking unique and useful biomarker signatures or panels to aid in stroke diagnosis, guide patient treatments, and track prognosis or recovery.
Previous published studies showed promising results in human patients when using genomic tools to screen for novel biomarker signatures of large vessel stroke [11–13]. However, the use of RNA for biomarker analysis may not be clinically practical in a limited time frame. For this study, we collected blood plasma from stroke patients and age-matched controls to examine the levels of multiple immunological proteins in plasma samples. We used a panel of 37 immunological molecules and examined their levels in plasma taken from patients ~24 hours after confirmed stroke events. We identified eight potential stroke biomarkers, several of which are novel (not previously associated with stroke). These biomarkers were used to develop a multi-protein panel with high specificity and sensitivity for detecting stroke occurrence. We also identified potential markers for patients who received tissue plasminogen activator (t-PA) treatment following stroke.
2. Results
2.1. Study Population Demographics
Plasma samples used in this study were obtained from peripheral blood drawn from consenting patients admitted to the Grady Memorial Hospital Emergency Room presenting stroke symptoms and age-matched participants from an out-patient clinic. The average time between stroke and blood draw was 22.9 ± 4.5 hours (mean ± SE). Demographic data for the 24 stroke patients and 26 control patients analyzed in this study are shown in Table 1. Patient age ranged from 39 to 100; control samples were selected to be age-matched, resulting in a similar spread of ages. (i.e., stroke patients spanned the entire 39 to 100 year range with median age of 58 and an interquartile range from 51-74 years; control participants ranged from 40 to 85 years with a median of 61 and interquartile range from 51-76). The gender breakdown of both groups was also similar, with the 24 stroke patients consisting of 15 males and 9 females and the 26 controls including 16 males and 10 females. Overall, the subjects in this study displayed no significant differences in age or sex distribution between stroke and control groups. We were able to obtain additional demographic information on all of the controls and 17 of the 24 stroke patients, including risk factors, stroke subtypes and thrombolytic treatment.
Table 1:
Demographic information about patients
| Demographic Factor | Stroke (n=24) | Control (n = 26) |
|---|---|---|
| Age | 58 (51-74) | 61 (51-76) |
| Female Gender | 9 (36%) | 10 (38.46%) |
| Prior stroke or TIA1 | 0 | 0 |
| Hypertension1 | 9 (53%) | 26 (100%) |
| Coronary artery disease1 | 3 (18%) | 0 |
| Congestive heart failure1 | 4 (24%) | 0 |
| Diabetes mellitus1 | 4 (24%) | 1 (4%) |
| Atrial fibrillation1 | 1 (6%) | - |
| MCA stroke1 | 17 (100%) | - |
| Atherogenic stroke1 | 12 (71%) | - |
| Cardioembolic stroke1 | 5 (6%) | - |
| Thrombolytic therapy1 | 11 (65%) | - |
All samples were from patients who were self-determined as African-American. Age shown as median (quartile 1 – quartile 3).
Data from all control and 17 of the 24 stroke patients.
2.2. Immunological Profiling of Plasma in Total Study Population
Of the 37 inflammatory proteins probed using the multiplex ELISA assay, 19 were detected in all samples, resulting in 26 control and 24 stroke measurements. Levels of four proteins (IL-27, IL-32, IL-34, and LIGHT/TNFSF14) were below detection limit levels in all samples and were eliminated from further analysis. The remaining 14 protein biomarkers yielded at least one stroke and one control measurement, with the exception of Pentraxin-3 which was detected only in a sub-set of stroke samples and no control samples. Protein concentrations of all 37 proteins with individual n numbers available for analysis are shown in Table 2.
Table 2:
Immunological Profile
| Biomarker | Stroke (pg/ml) [n=24] | Control (pg/ml) [n = 26] | p-value |
|---|---|---|---|
| APRIL/TNFSF13 | 108387 (70178-155143) | 81108 (58714-108851) | 0.093 |
| BAFF/TNFSF13B | 4096 (2989-5934) | 3473 (2840-3980) | 0.10 |
| Chitinase 3-like 1 | 10588 (8248-13716) | 8736 (5466-12061) | 0.20 |
| Gp130/sIL-6Rβ | 29933 (18999-39481) | 22285 (18367-29580) | 0.046 |
| IFN-α2* | 6.868 (3.495-9.48) [17] | 6.705 (5.22-11.15) | 0.52 |
| IFN-β | 25.4 (20.45-33.59) | 29.52 (25.27-38.36) | 0.033 |
| IFN-γ | 8.389 (4.336-11.27) | 9.36 (8.42-12.18) | 0.066 |
| IL-2* | 1.14 (0.79-1.39) [13] | 0.79 (0.41-1.64) [19] | 0.86 |
| IL-8* | 10.04 (6.377-13.95) [22] | 10.36 (8.11-14.38) | 0.51 |
| IL-10* | 0.2895 (0.24-1.09) [4] | 0.128 (0.128-0.128) [1] | n/a |
| IL-11 | 0.9975 (0.7171-1.184) | 0.81 (0.645-1.07) | 0.22 |
| IL-12 (p40) | 16.22 (13.44-17.45) | 16.87 (14.46-19.89) | 0.58 |
| IL-12 (p70)* | 0.17 (0.06369-0.2575) [6] | 0.24 (0.065-0.44) [7] | 0.53 |
| IL-19* | 3.105 (0.89-11.79) [4] | 4.9 (1.05-23.86) [3] | 0.69 |
| IL-20* | 18.82 (10.54-23.38) [21] | 19.48 (15.96-27.52) | 0.12 |
| IL-22* | 2.42 (2.42-2.42) [1] | 31.37 (1.44-444.4) [5] | n/a |
| IL-26 | 3.319 (2.259-4.232) | 3.385 (2.52-3.92) | 0.92 |
| IL-27 (p28)† | none | none | n/a |
| IL-28A/IFN-λ2 | 14.01 (9.525-17.56) | 18.12 (15.31-22.79) | 0.0059 |
| IL-29/IFN-λ1* | 3.35 (2.715-12.57) [9] | 10.25 (6.33-12.22) [10] | 0.32 |
| IL-32† | none | none | n/a |
| IL-34† | none | none | n/a |
| IL-35* | 20.87 (16.4-28.93) [15] | 27 (16.4-39.81) [23] | 0.31 |
| LIGHT/TNFSF14† | none | none | n/a |
| MMP-1* | 614.8 (169.8-1044) [14] | 721.2 (217.24-843.43) [15] | 0.96 |
| MMP-2 | 4268 (2129-5787) | 2033 (1233-3535) | 0.0011 |
| MMP-3* | 3872 (2370-5782) [22] | 3944(2276.08-6348.75) | 0.87 |
| Osteocalcin | 560.6 (383.3-838.9) | 575.7 (413-685.4) | 0.90 |
| Osteopontin (OPN) | 6931(2768-10827) | 4316(2037-5867) | 0.020 |
| Pentraxin-3* | 6.342 (1.426-73.43) [9] | none | n/a |
| sCD163 | 623.4 (473-771.4) | 530.8 (394.39-759.43) | 0.44 |
| SCD30/TNFRSF8 | 218.5 (142-271.7) | 168.8 (137.22-277.63) | 0.37 |
| sIL-6Rα* | 6.868 (3.495-9.48) [17] | 6.705 (5.22-11.15) | 0.59 |
| sTNF-R1 | 1055 (783.7-1230) | 807.3 (629.43-970.58) | 0.033 |
| STNF-R2 | 7688 (5866-11642) | 4422 (2913.77-7074.6) | 0.0019 |
| TSLP | 11.14 (9.401-15.75) | 17.2 (13.61-20.27) | 0.0001 |
| TWEAK/TNFSF12 | 129.4 (106.9-166.3) | 119.3 (96.95-154.92) | 0.41 |
Median values and interquartile ranges (IQR) shown
only expressed in a subset of samples, n # shown italicized in square brackets
Biomarker is below detection level in stroke and control samples
Significant p-vales bold
Statistical analysis indicated that eight of the 33 detectable proteins in our samples were significantly different between stroke and control samples (Figure 1 and Table 2). The eight biomarkers were IFN-β, IL-28, sIL-6Rβ/gp130, MMP-2, osteopontin, sTNF-R1, sTNF-R2 and TSLP. Plasma levels of sIL-6Rβ/gp130, MMP-2, osteopontin, sTNF-R1 and sTNF-R2 were significantly higher in stroke patients compared to controls, while IFN-β, IL-28, and TSLP were significantly lower in plasma from stroke patients. None of the other immunological markers significantly differed among groups.
Fig 1: Significantly different immunological profiles of stroke and control patients.
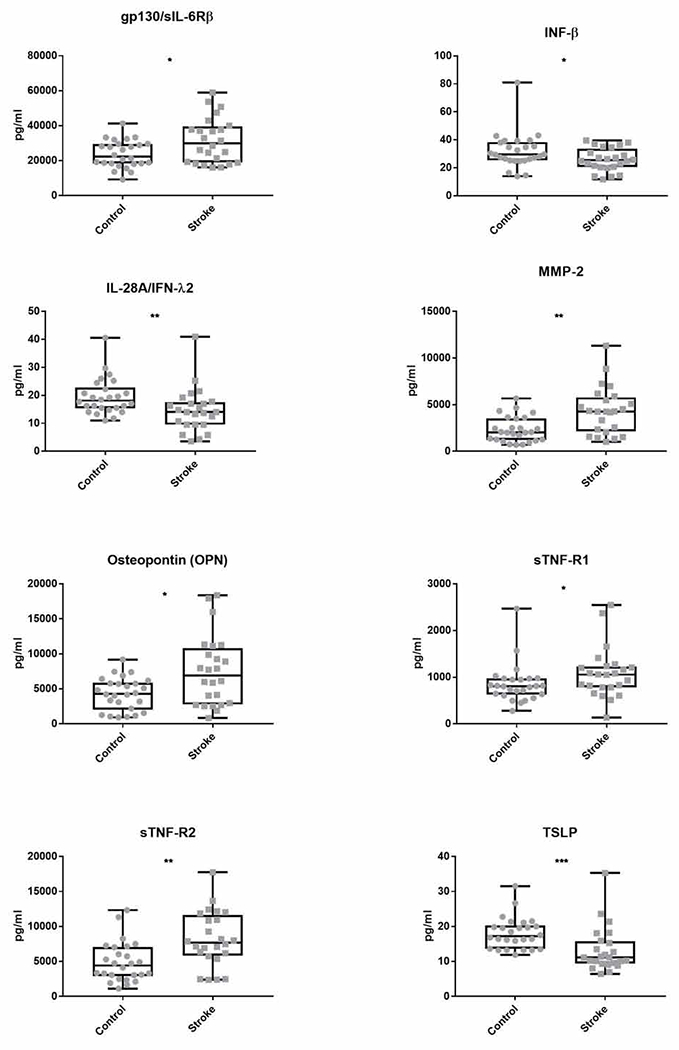
Levels of 37 immunological proteins in the plasma of stroke patients and age-matched controls were determined using a multiplexed ELISA. Recorded fluorescence intensities were converted to concentrations using standard curves for each protein. Plasma concentrations, in picograms per milliliter (pg/ml), of the eight proteins that were significantly different between stroke and control populations are shown here as box and whisker plots. All concentration values are shown. Whiskers extend from minimum to maximum concentration. Box encloses the range between the first and third quartiles with horizontal line at median value. P-values calculated using post-hoc pairwise non-parametric Mann-Whitney U-test. *p≤.05 **p≤.01 ***p≤.001
2.3. Effects of rt-PA Treatment on Immunological Profile
Thirteen of the 24 stroke patients in this study received recombinant tissue plasminogen activator (rt-PA) treatment after arriving at the hospital. To investigate the possible effects of this treatment on blood immunological profiles, we compared protein levels between rt-PA treated and untreated stroke patient samples. Statistical analysis identified IFN-α2, IL-2, and Pentraxin-3 as significantly different between rt-PA treated and non-treated groups (Table 3). Pentraxin-3 was notable because in the majority of samples (9) the levels were below the detection limit of our assay. However, all of the detectable pentraxin-3 samples were those of stoke patients, which might suggest elevated pentraxin-3 in stroke. Further evaluation of these nine samples showed that pentraxin-3 levels were an order of magnitude higher in those patients that did not receive rt-PA treatment compared to those that did receive treatment. While the sample size is small, our statistical analysis shows that the difference in pentraxin-3 levels between treatment and untreated patients is statistically significant. None of these factors duplicated those found to be significantly different between stroke and control samples, suggesting that the eight biomarkers previously identified could be of use in all stages of stroke diagnosis and monitoring, regardless of patient treatment status.
Table 3:
Immunological profile of stroke patients who did and did not receive rt-PA treatment
| Analyte | No rt-PA (pg/ml) [n=9] | rt- PA (pg/ml) [n=13] | p-value |
|---|---|---|---|
| APRIL/TNFSF13 | 119079 (90590-208421) | 93130 (66009-148935) | 0.21 |
| BAFF/TNFSF13B | 4558 (3471-6308) | 4551 (2912 – 5913) | 0.30 |
| Chitinase 3-like 1 | 13597 (9719 – 1445) | 9770 (7778 – 11844) | 0.21 |
| gp130/sIL-6Rβ | 36834 (16714 – 38900) | 26096 (20324 – 42258) | 0.90 |
| IFN-α2 | 8.93 (7.78 – 11.09) [7] | 5.53 (3.14 – 7.169) [10] | 0.031 |
| IFN-β | 26.25 (22.53 – 37.18) | 23.13 (20.23 – 31.13) | 0.29 |
| IFN-γ | 10.88 (5.765 – 13.18) | 8.357 (5.458 – 9.67) | 0.23 |
| IL-2 | 1.14 (0.97 – 1.8) [7] | 0.79 (0.3163 – 1.14) [6] | 0.047 |
| IL-8 | 13.66 (6.968 – 20.02) | 9.41 (7.432 – 13.66) [11] | 0.49 |
| IL-10 | 0.25 (0.23 – 0.27) [2] | 0.8295 (0.309 – 1.35) [2] | 0.33 |
| IL-11 | 1.13 (0.9595 – 1.625) | 0.9621 (0.7554 – 1.12) | 0.13 |
| IL-12(p40) | 17.45 (14.47 – 22.11) | 15 (13.49 – 17.21) | 0.23 |
| IL-12(p70) | 0.17 (0.085 – 0.4125) | 0.1225 (0.06492 – 0.18) | 0.93 |
| IL-19 | 3.105 (0.89 - 11.79) [4] | none | n/a |
| IL-20 | 25.27 (12.61 – 31.27) [8] | 16.13 (10.54 – 20.14) [11] | 0.10 |
| IL-22 | n/a | n/a | n/a |
| IL-26 | 3.586 (2.259 – 4.815) | 3.317 (2.195 – 3.86) | 0.64 |
| IL-27(p28) | n/a | ||
| IL-28A/IFN-λ2 | 16.25 (11.62 – 23.35) | 14.01 (9.525 – 17) | 0.20 |
| IL-29/IFN-λ1 | 8.995 (2.558 – 15.69) [6] | 3.35 (2.08 – 3.35) [3] | 0.25 |
| IL-32 | n/a | n/a | n/a |
| IL-34 | n/a | n/a | n/a |
| IL-35 | 25.95 (19.24 – 30.35) [8] | 17.79 (4.881 – 20.87) [7] | 0.067 |
| LIGHT/TNFSF14 | n/a | n/a | n/a |
| MMP-1 | 771.2 (484.7 – 1138) [5] | 304.5 (169.8 – 917.5) [8] | 0.12 |
| MMP-2 | 4334 (4216 – 5317) | 2571 (1548 – 6242) | 0.56 |
| MMP-3 | 3872 (2830 – 6253) [8] | 3889 (1692 – 5352) [12] | 0.58 |
| Osteocalcin | 572.7 (411.1 – 829.1) | 459.9 (349.6 – 1187) | 0.60 |
| Osteopontin (OPN) | 7737 (2730 – 10029) | 6125 (3421 – 10572) | 0.74 |
| Pentraxin-3 | 73.43 (26.82 – 406.5) [4] | 1.426 (0.2529 – 5.232) [4] | 0.029 |
| SCD30/TNFRSF8 | 229.1 (163.5 – 359.7) | 221.1 (160.8 – 275.8) | >0.9999 |
| sCD163 | 613.3 (487.1 – 935.1) | 653 (482.8 – 768.9) | 0.90 |
| sIL-6Rα | 4951 (3480 – 5986) | 3987 (3351 – 4899) | 0.19 |
| sTNF-R1 | 1202 (823.5 – 1535) | 931.6 (718.7 – 1184) | 0.36 |
| STNF-R2 | 7878 (4431 – 12026) | 8002 (6558 – 11497) | >0.9999 |
| TSLP | 12.72 (9.209 – 19.8) | 10.26 (9.849 – 12.46) | 0.36 |
| TWEAK/TNFSF12 | 133.8 (110 – 164.6) | 127.8 (99 – 160.1) | 0.44 |
Median values and interquartile ranges (IQR) shown; all values in pg/ml
only expressed in a subset of samples, n # shown italicized in square brackets
Biomarker is below detection level in stroke and control samples
Significant p-vales bold
2.4. Univariate Analysis of Significant Immunological Profile for Stroke
ROC curves were generated for the eight biomarkers identified as having significantly different concentrations between stroke and control samples (Figure 2). Table 4 shows AUC, sensitivity, specificity and accuracy calculated at the optimal cut-off value for each of the eight proteins. Sensitivity and specificity of the test are inversely related and change depending upon the cutoff level chosen to assign positive and negative testing results. Table 4 lists the sensitivity and specificity proportions for the “optimized” cut-off point, determined by the highest combined sensitivity and (1-specificity) values. Four of the eight identified proteins (Figure 2), IL-28, MMP-2, sTNF-R2 and TSLP were determined acceptable discriminators (AUC ≥ 0.7) between stroke and control conditions when used individually and only TSLP qualified as a “good” discriminator (AUC ≥ 0.8). While decreased IL-28A/IFN-λ2 level is specific for stroke, the sensitivity of this measure is relatively low compared to other proteins assessed. Increased sTNF-R2 and MMP-2 and decreased TSLP levels were predictive of stroke with a high degree of sensitivity, specificity and accuracy.
Figure 2: ROC curves of Individual Immunological factors.
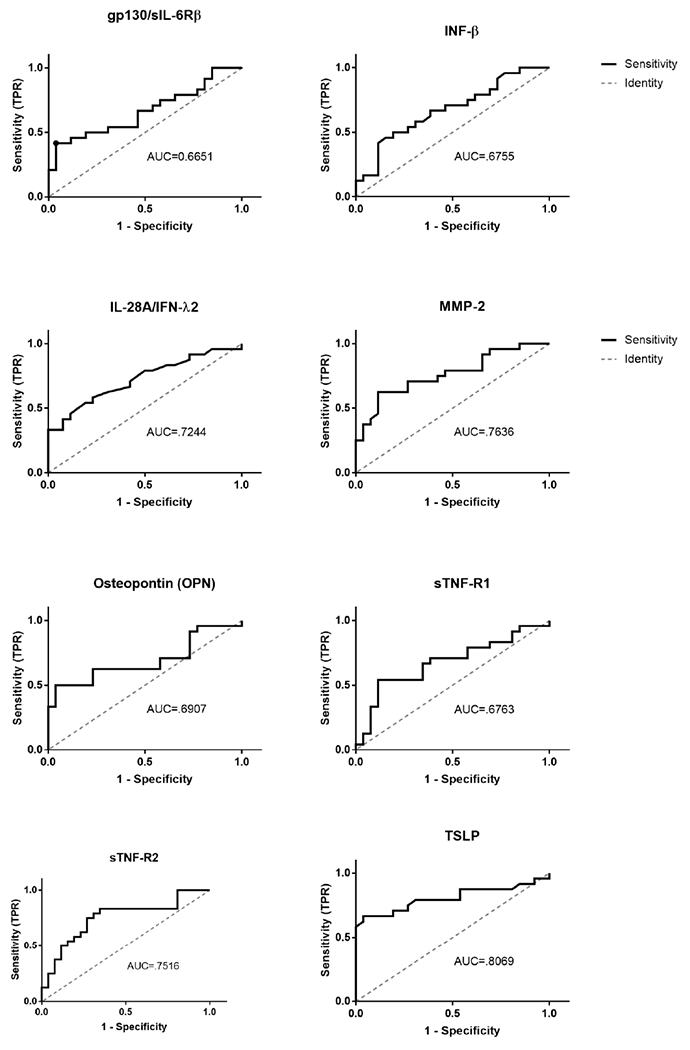
ROC curve analysis was based on 24 stroke patients and 26 control participants. Optimal cut-offs were defined as the point where the sum of sensitivity and specificity were maximized. Utility in discriminating stroke and control samples is indicated by area under the curve. Classification performance: AUC ≥ 0.7 = acceptable, AUC ≥ 0.8 = good, AUC ≥ 0.9 = excellent.
Table 4:
ROC analysis of significant analytes
| Biomarker | Cut-off | AUC (95% CI) | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|---|
| Gp130/sIL-6Rb | > 35095 | .6651 (.5114-.8187) | .4167 | .9615 | .7 |
| IFN-β | < 25.07 | .6755 (.5259 - .825) | .5 | .8077 | .66 |
| IL-28A/IFN-λ2 | < 15.13 | .7244 (.5815 - .8672) | .5833 | .7692 | .66 |
| MMP-2 | > 4169 | .7636 (.6301 - .8972) | .625 | .8846 | .76 |
| Osteopontin (OPN) | > 7599 | .6907 (.5365 - .8449) | .5 | .9615 | .74 |
| sTNF-R1 | > 1040 | .6763 (.5227 - .8299) | .5417 | .8846 | .72 |
| STNF-R2 | > 5335 | .7516 (.6119 - .8913) | .8333 | .6538 | .76 |
| TSLP | < 12.98 | .8069 (.6743 - .9404) | .6667 | .9615 | .82 |
Cut-off values (pg/mL), AUC (95% CI), sensitivity, specificity and accuracy of each analyte different between stroke and control patients. Cut-off values were chosen as the point of the curve with the highest sum of specificity and sensitivity.
2.5. Multivariate Analysis of Biomarker Panels
In addition to using biomarkers individually to discriminate between stroke patients and healthy controls, we also examined whether multiple biomarkers could be combined into diagnostic panels. We divided our eight biomarkers into two panels and evaluated each for its ability to discriminate between stroke and healthy samples. For the first panel, we combined the top univariate performers (Figure 2), IL-28A/IFN-λ2, MMP-2, sTNF-R2, and TSLP, using a logistic regression analysis to predict the outcome (i.e., “disease” or “healthy”). Combining these four molecules, each of which achieved only “acceptable” or “good” discrimination between groups assessed individually, we were able to achieve an AUC of .955, which is considered excellent, with sensitivity and specificity of .875 and .962 respectively (Table 5).
Table 5:
ROC for Biomarker Panel
| Panel | AUC | Sensitivity | Specificity |
|---|---|---|---|
| Gp130/sIL-6Rb, IFN-β, Osteopontin (OPN), sTNF-R1, | .926 | .870 | .962 |
| IL-28A/IFN-λ2, MMP-2, sTNF-R2, TSLP | .955 | .875 | .962 |
In a second panel we combined the least successful univariate performers, Gp130/sIL-6Rb, IFN-β, Osteopontin (OPN), and sTNF-R1, none of which could be used individually to successfully discriminate between stroke and healthy samples. The resulting panel showed greatly improved diagnostic value, with an AUC of .926, sensitivity of .870 and specificity of .962.
3. Discussion
The goals of this study were to identify candidate stroke biomarkers in the blood of patients and develop a panel of biomarkers to detect stroke with high specificity and sensitivity. We identified eight immunologically relevant proteins present in significantly different amounts in the plasma of stroke patients compared to age-matched controls. These eight molecules are diverse in their form and function, including chemokines (IL-28A/IFN-g2, IFN-β, TSLP), metalloproteases (MMP-2), transmembrane receptor domains (sTNF-R1, sTNF-R2, gp130/sIL-6RB) and extracellular structural proteins (OPN). The diversity of molecules suggests the complexity of the systemic immunological environment that is triggered as a result of ischemic stroke. Of these eight molecules, to our knowledge only sTNF-R1 and MMP-2 had previously been identified as biomarkers of stroke [14, 15].
Among the biomarkers that we identified as being significantly altered in the blood of stroke patients were factors that play important roles in both the innate and adaptive arms of the immune response. Gp130/sIL-6RB, IFN-β, IL-28A/IFN-g2 and MMP-2 are involved in innate immune cascades [16, 17]. On the other hand, IL-28A/IFN-g2 plays an active role in directing adaptive immune responses as a co-stimulatory factor in T cell activation, inhibiting Th2 responses [18]. In addition, many of these biomarkers are involved in multiple, sometimes overlapping and intersecting, molecular signaling cascades. Specifically, IL-28A/IFN-g2 and TSLP regulate the transcription factor STAT5, which is critical for immune cell development [19]. OPN (also known as secreted phosphoprotein 1, or SPP1) regulates MMP2. Therefore this regulatory pathway might be a significant feature of the stroke-induced immune response [14]. Finally, sTNF-R1 has been identified as biomarker associated with mortality not only in stroke, but also sepsis [15, 20], indicating that this molecule may be indicative of immune dysregulation leading to negative outcomes or complications.
ROC curves are commonly used to measure the predictive power of medical tests [21]. Specificity is a measure of the proportion of patients negative for the condition who accurately test as negative. Therefore, high specificity is an ideal property for tests designed to “rule in” patients as having some condition. Sensitivity is a measure of the proportion of patients positive for the condition who accurately test as positive, so high sensitivity indicates high negative predictive power which would be ideal for tests to “rule out” patients as having a particular condition. While many individual inflammatory molecules have been shown to be regulated following stroke, none have shown sufficient specificity to be used clinically as diagnostic tools for stroke. [2, 5]. Here, we identified eight molecules as statistically different between stroke patients and controls, but only four proteins (IL-28, MMP-2, sTNF-R2 and TSLP) were determined acceptable to discriminate between stroke and control conditions when used individually based on our AUC criteria. While our analysis showed that Gp130, IFN-β, OPN and sTNF-R1 levels were significantly different between stroke and control samples, these proteins alone could not accurately determine stroke occurrence.
The levels of some biomarkers are tightly controlled, but clinical practice has long established a wide variation between individuals as normal. Potential biomarkers can be present in the plasma of normal individuals over a wide range of concentrations. While the average levels of an individual biomarker among individual stroke patients are different than in controls, there may be overlap in the distribution of protein levels. Therefore, rather than selecting rigid cutoffs for single biomarkers, it can be beneficial to look at levels of many biomarkers and establish clusters of relative elevation or depression that may be more widely useful as indicators of disease. Following this reasoning, we designed diagnostic tests that include combining the eight single tests into two panels of biomarkers. We used a simple logistic model as proof of concept that looking at the relative ways proteins cluster into high and low expression can be more predictive of disease than simple univariate tests with rigid cut-off levels. For the first panel, we combined the four biomarkers determined acceptable or good (i.e., IL-28A/IFN-λ2, MMP-2, sTNF-R2 and TSLP) using a logistic regression analysis to predict stroke or control samples. Using this four biomarker panel, we were able to achieve an AUC of .955, which is considered excellent, with high sensitivity and specificity. In a second panel, we chose the four biomarkers that were not shown to individually discriminate between stroke and control samples based on AUC criteria (i.e., Gp130/sIL-6Rb, IFN-β, OPN and sTNF-R1). This four marker panel showed an excellent AUC of .926 with high sensitivity and specificity, indicating the improved potential of biomarker panels compared to tests of individual proteins. Overall, both panels performed as well or better on all measures than any individual biomarker.
In addition to identifying biomarkers that differ in stroke patients as compared to controls, we further divided the stroke patients into those that received rt-PA treatment and those that did not receive this treatment. We did not find that any of the previously discussed stroke biomarkers were significantly affected by the rt-PA treatment. However, found that rt-PA treatment resulted in significant differences between three of the analytes in patients that received treatment compared to those that did not, including IFN-α2, IL-2, and pentraxin-3. To our knowledge IFN-α2 has not been explored as a biomarker for stroke, but it has been identified as a prognostic indicator in other conditions including viral infections with HIV, herpes, and hepatitis, autoimmune conditions such as lupus, and certain cancers. IL-2 is a pro-inflammatory cytokine that has been examined in a variety of inflammatory contexts, including stroke. One study suggested IL-2 as a useful prognostic indicator in acute ischemic strokes, as IL-2 levels that decreased over the week following stroke was predictive of survival [22]. Pentraxin-3 has been indicated as a stroke biomarker, but reports in the literature are mixed with some studies concluding that elevated pentraxin-3 is positively correlated to stroke occurrence [23] or mortality [24] while others report no such relation [25].
We recognize that there are limitations to this study. While we were able to collect some demographic data on all patients (including stroke diagnosis, age, gender and t-PA administration), we were only able to retrieve data related to co-morbidity on a subpopulation of patients. Although limited by the small sample size and the missing demographic data from a few of our patients, the findings have shown that analyzing the levels of immunological factors in blood collected systemically can be an effective method for identifying patients who have recently had ischemic strokes. The next step will be to continue this study with a larger, better characterized sample population. We will also extend our findings to determine if the blood markers correlate well with stroke interventions (e.g. t-PA and endovascular thrombectomy) and treatment effect on clinical endpoints such as infarct volume and neurological outcomes.
The underlying concept of this study posits that evaluating the systemic immunological profile of patients can be an effective way to non-invasively diagnose disease, even for conditions such as stroke in which damage is isolated mainly in one region of the body. This study serves as proof of concept that there is a unique and identifiable systemic immunological signature for ischemic stroke, and that it can be used to diagnose patients from simple blood tests.
4. Experimental Procedures
4.1.
The study was carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving human subjects. All procedures were approved by the Institutional Review Boards at Morehouse School of Medicine and Grady Memorial Hospital. Written informed consent was received from all participants prior to inclusion in the study. Patient samples were assigned random numerical identifiers prior to analysis and were a subset of those collected at Grady Memorial Hospital, described in a previously published study [26]. In this study, we analyzed plasma samples from 24 patients of varied age, gender, and ethnicity admitted to Grady Memorial Hospital with confirmed strokes and 26 healthy controls from an out-patient clinic matched for hypertensive profiles without history of stroke. All stroke patients included in this study experienced ischemic strokes. For all participants (patients and controls), ~8 ml of whole blood was collected into vacutainer plasma tubes and the resulting plasma was stored at −80°C until use. Blood draw from stoke patients was performed the morning following admission to the hospital. Average time between stroke and blood draw was 22.9 ± 4.5 hours (mean ± SE). Final clinical stroke diagnosis was verified by review of the medical records by a panel of neurologists at Grady Memorial Hospital.
4.2. Immunological/Inflammatory Profiling
Prior to experiment, plasma samples were allowed to thaw at room temperature. All plasma samples were analyzed by multiplex immunoassay based Luminex MAGPIX technology (Bio-Rad, Hercules CA, USA) using a multiplex kit (kits and reagents were purchased from Bio-Rad, Hercules CA, USA). We used the Bio-Plex Pro™ Human Inflammation Panel 1, which tests for a panel of 37 biomarkers of inflammation, including members of the tumor necrosis factor (TNF) superfamily of proteins, interferon (IFN) family of proteins, Treg cytokines, and matrix metalloproteinases (MMPs). The full panel included antibodies to detect the following immunologically relevant proteins: TNF superfamily 13/a proliferation-inducing ligand (TNFSF13/APRIL), TNF superfamily 13b/B-cell activating factor (TNFSF13B/BAFF), TNF superfamily member 8 (TNFRSF8/sCD30), soluble cluster of differentiation 163 (sCD163), Chitinase-3-like 1, interleukin 6 signal transducer (sIL-6Rβ/gp130), IFN alpha-2 (IFN-α2), IFN beta (IFN-β), IFN gamma (IFN-γ), interleukin-2 (IL-2), soluble interleukin-6 receptor (sIL-6Rα), interleukin-8 (IL-8), interleukin-10 (IL-10), interleukin-11 (IL-11), interleukin-12 p40 (IL-12 p40), interleukin-12 p70 (IL-12 p70), interleukin-19 (IL-19), interleukin-20 (IL-20), interleukin-22 (IL-22), interleukin-26 (IL-26), interleukin-27 (IL-27 p28), interferon lambda 2 (IFN-λ2/IL-28A), interferon lambda 1 (IFN-λ1/IL-29), interleukin-32 (IL-32), interleukin-34 (IL-34), interleukin-35 (IL-35), TNF superfamily 14 (TNFSF14/LIGHT), matrix metallopeptidase 1 (MMP-1), matrix metallopeptidase 2 (MMP-2), matrix metallopeptidase 3 (MMP-3), osteocalcin, osteopontin (OPN), pentraxin-3, soluble TNF receptor superfamily member 1A (sTNF-R1), soluble TNF receptor superfamily member IB (sTNF-R2), thymic stromal lymphopoietin (TSLP), TNF superfamily 12/ TNF-related weak inducer of apoptosis (TNFSF12/TWEAK).
The multiplex assay was performed following the manufacturer’s instructions: standards were prepared by serial dilution and all plasma samples were diluted 1:1 with provided sample diluent. Fifty μl of microbeads conjugated to capture antibodies were loaded into 96 well plates, to which 50 μl of standards, controls, and experimental samples were added and incubated for 1 hour at room temperature with shaking. Following incubation, unreacted protein was washed away and biotinylated detection antibodies were allowed to incubate for 30 minutes with shaking. Excess antibody was washed away and streptavidin, conjugated to the fluorescent reporter tag phycoerythrin (PE), was incubated for 10 minutes. Unreacted florescent reporter was washed and the beads were suspended in 125 μl of assay buffer in preparation for fluorescence measurements to be read thorough the MAGPIX system. The ELISA plates were read using a MAGPIX system running Bio-Plex Manager MP software (Bio-Rad, Hercules, USA).
4.3. Statistical Analysis
Statistical analysis was carried out using GraphPad Prism version 7.01 (San Diego, CA, USA). Median values and interquartile ranges (IQR) for continuous variables or percentages for categorical variables were calculated. The non-parametric Mann-Whitney test was used to determine p values between groups, with a probability of < 0.05 considered statistically significant.
4.31. Univariate Single Biomarker Analysis
The discriminatory power of statistically significant immunological proteins was determined via receiver operating characteristic (ROC) analysis. ROC curves were built and area under the curve (AUC) and 95% confidence interval (CI) were calculated using GraphPad Prism software. Area under the curve (AUC) is a measure of the ability of the test to accurately discriminate between two states: in this case, subjects with stroke versus subjects without stroke. Cut-off values were chosen to maximize the sum of sensitivity and specificity. AUC above 0.70 was considered acceptable discrimination and AUC above 0.80 was considered good [27].
Measures of the accuracy of diagnostic tests are based upon how the test classifies subjects into the true positive (TP), true negative (TN), false positive (FP), and false negative (FN) groups.
Specifically,
Sensitivity = TP/(TP+FN), or the proportion of true cases correctly identified. This can also be called the true positive rate (TPR)
Specificity = TN/ (TN+FP), or the proportion of true negative. The compliment of this measurement, 1-Specificity, is therefore the proportion of false positives correctly identified, also called the false positive rate (FPR)
The ROC curve plots TPR versus FPR.
Accuracy = (TN + TP)/(TN+TP+FN+FP), or the proportion of correctly identified cases
Positive predictive value (PPV) = TP/ (TP+FP), or the proportion of correctly identified positive cases.
Negative predictive value (NPV) = TN/(TN+FN), or the proportion of correctly identified negative cases.
Accuracy, NPV, and PPV are dependent on incidence of disease.
4.3.2. Multivariate Biomarker Panel Analysis
Using R statistical software and the pROC and Epi packages, ROC curves were constructed and sensitivity and specificity calculated for these biomarker panels. Multivariate logistic regression was used to evaluate combinations of biomarkers in panels so that the ROC curve was based on model based probability of the form:
Where β values are coefficients determined from the regression, x values are protein concentrations, and index i represents the biomarker rank included in the model.
Highlights:
We used a multiplex panel of 37 molecules to identify the changes in human plasma proteins after stroke
We identified eight key molecules that were altered within the blood of stroke patients as compared to controls.
Levels of sIL-6Rβ/gp130, MMP-2, osteopontin, sTNF-R1 and sTNF-R2 were higher in stroke patients compared to controls.
Multivariate logistic regression identified a stroke biomarker panel with high specificity and sensitivity
Acknowledgements
We thank Dr. Shaokui Ge for his advice on biostatistical methods. This project was supported in part by NIH grants U54 NS060659; U54 RR026137, G12RR003034 and S21MD000101.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heron M Deaths: Leading Causes for 2014. Natl Vital Stat Rep. 2016;65(5):1–96. PMID: 27376998. [PubMed] [Google Scholar]
- 2.Foerch C, Montaner J, Furie KL, Ning MM, Lo EH. Invited Article: Searching for oracles? Blood biomarkers in acute stroke. Neurology. 2009;73(5):393–9. PMID: 19652144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen MB, Chacon MR, Sattin JA, Levine RL, Vemuganti R. Potential biomarkers for the diagnosis of stroke. Expert Rev Cardiovasc Ther. 2009;7(4):389–93. PMID: 19379063. [DOI] [PubMed] [Google Scholar]
- 4.Laskowitz DT, Kasner SE, Saver J, Remmel KS, Jauch EC. Clinical usefulness of a biomarker-based diagnostic test for acute stroke: the Biomarker Rapid Assessment in Ischemic Injury (BRAIN) study. Stroke. 2009;40(1):77–85. Epub 2008/10/25. doi: 10.1161/STROKEAHA.108.516377 PMID: 18948614. [DOI] [PubMed] [Google Scholar]
- 5.Jickling GC, Sharp FR. Biomarker panels in ischemic stroke. Stroke. 2015;46(3):915–20. doi: 10.1161/STROKEAHA.114.005604 PMID: 25657186; PubMed Central PMCID: PMCPMC4342265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iadecola C, Anrather J. The immunology of stroke: from mechanisms to translation. NatMed. 2011;17(7):796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JS, Yoon SS, Kim YH, Ryu JS. Serial measurement of interleukin-6, transforming growth factor-beta, and S-100 protein in patients with acute stroke. Stroke. 1996;27(9):1553–7. PMID: 8784129. [DOI] [PubMed] [Google Scholar]
- 8.Dziedzic T, Bartus S, Klimkowicz A, Motyl M, Slowik A, Szczudlik A. Intracerebral hemorrhage triggers interleukin-6 and interleukin-10 release in blood. Stroke. 2002;33(9):2334–5. PMID: 12215608. [DOI] [PubMed] [Google Scholar]
- 9.Ormstad H, Aass HC, Amthor KF, Lund-Sorensen N, Sandvik L. Serum cytokine and glucose levels as predictors of poststroke fatigue in acute ischemic stroke patients. J Neurol. 2011;258(4):670–6. doi: 10.1007/s00415-011-5962-8 PMID: 21365457; PubMed Central PMCID: PMCPMC3065647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng L, Wang Y, Liu J, Wang L, Weng S, Chen K, et al. Pro-inflammatory cytokine network in peripheral inflammation response to cerebral ischemia. Neurosci Lett. 2013;548:4–9. doi: 10.1016/j.neulet.2013.04.037 PMID: 23643982. [DOI] [PubMed] [Google Scholar]
- 11.Barr TL, Conley Y, Ding J, Dillman A, Warach S, Singleton A, et al. Genomic biomarkers and cellular pathways of ischemic stroke by RNA gene expression profiling. Neurology. 75 United States2010. p. 1009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stamova B, Xu H, Jickling G, Bushnell C, Tian Y, Ander BP, et al. Gene expression profiling of blood for the prediction of ischemic stroke. Stroke. 2010;41(10):2171–7. doi: 10.1161/STROKEAHA.110.588335 PMID: 20798371; PubMed Central PMCID: PMCPMC2987675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jickling GC, Xu H, Stamova B, Ander BP, Zhan X, Tian Y, et al. Signatures of cardioembolic and large-vessel ischemic stroke. Ann Neurol. 2010;68(5):681–92. doi: 10.1002/ana.22187 PMID: 21031583; PubMed Central PMCID: PMCPMC2967466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang R, Pan X, Huang Z, Weber GF, Zhang G. Osteopontin enhances the expression and activity of MMP-2 via the SDF-1/CXCR4 axis in hepatocellular carcinoma cell lines. PLoS One. 2011;6(8):e23831 Epub 2011/08/31. doi: 10.1371/journal.pone.0023831 PMID: 21909361; PubMed Central PMCID: PMCPMC3166084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greisenegger S, Segal HC, Burgess AI, Poole DL, Mehta Z, Rothwell PM. Biomarkers and mortality after transient ischemic attack and minor ischemic stroke: population-based study. Stroke. 2015;46(3):659–66. Epub 2015/02/03. doi: 10.1161/STROKEAHA.114.007624 PMID: 25649803; PubMed Central PMCID: PMCPMC4820048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng YW, Li H, Yu JP, Zhao H, Wang SE, Ren XB. Interferon-λs: special immunomodulatory agents and potential therapeutic targets. J Innate Immun. 2013;5(3):209–18. Epub 2012/11/30. doi: 10.1159/000345365 PMID: 23207147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H, Koh G. Lipopolysaccharide activates matrix metalloproteinase-2 in endothelial cells through an NF-kappaB-dependent pathway. Biochem Biophys Res Commun. 2000;269(2):401–5. doi: 10.1006/bbrc.2000.2308 PMID: 10708565. [DOI] [PubMed] [Google Scholar]
- 18.Khalaj AJ, Hasselmann J, Augello C, Moore S, Tiwari-Woodruff SK. Nudging Oligodendrocyte Intrinsic Signaling to Remyelinate and Repair: Estrogen Receptor Ligand Effects. The Journal of Steroid Biochemistry and Molecular Biology. doi: http://dx.doi.org/10.1016/j.jsbmb.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell BD, Kitajima M, Larson RP, Stoklasek TA, Dang K, Sakamoto K, et al. The transcription factor STAT5 is critical in dendritic cells for the development of TH2 but not TH1 responses. Nat Immunol. 2013;14(4):364–71. Epub 2013/02/24. doi: 10.1038/ni.2541 PMID: 23435120; PubMed Central PMCID: PMCPMC4161284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikacenic C, Hahn WO, Price BL, Harju-Baker S, Katz R, Kain KC, et al. Biomarkers of Endothelial Activation Are Associated with Poor Outcome in Critical Illness. PLoS One. 2015;10(10):e0141251 Epub 2015/10/22. doi: 10.1371/journal.pone.0141251 PMID: 26492036; PubMed Central PMCID: PMCPMC4619633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Florkowski CM. Sensitivity, specificity, receiver-operating characteristic (ROC) curves and likelihood ratios: communicating the performance of diagnostic tests. Clin Biochem Rev. 2008;29 Suppl 1:S83–7. PMID: 18852864; PubMed Central PMCID: PMCPMC2556590. [PMC free article] [PubMed] [Google Scholar]
- 22.Nayak AR, Kashyap RS, Purohit HJ, Kabra D, Taori GM, Daginawala HF. Evaluation of the inflammatory response in sera from acute ischemic stroke patients by measurement of IL-2 and IL-10. Inflamm Res. 2009;58(10):687–91. Epub 2009/04/02. doi: 10.1007/s00011-009-0036-4 PMID: 19340396. [DOI] [PubMed] [Google Scholar]
- 23.Sezer S, Uçar F, Ulusoy EK, Erdogan S, Bilen S, Züngün C, et al. Serum amyloid A, fetuin-A, and pentraxin-3 levels in patients with ischemic stroke: novel prognostic biomarkers? Turk J Med Sci. 2014;44(1):16–23. PMID: 25558553. [DOI] [PubMed] [Google Scholar]
- 24.Ryu WS, Kim CK, Kim BJ, Kim C, Lee SH, Yoon BW. Pentraxin 3: a novel and independent prognostic marker in ischemic stroke. Atherosclerosis. 2012;220(2):581–6. Epub 2011/12/01. doi: 10.1016/j.atherosclerosis.2011.11.036 PMID: 22178425. [DOI] [PubMed] [Google Scholar]
- 25.Ceylan M, Yalcin A, Bayraktutan OF, Atis O, Acar E. Serum pentraxin-3 levels in acute stroke: No association with stroke prognosis. Atherosclerosis. 2015;243(2):616–20. Epub 2015/11/11. doi: 10.1016/j.atherosclerosis.2015.10.089 PMID: 26546709. [DOI] [PubMed] [Google Scholar]
- 26.Meller R, Pearson AN, Hardy JJ, Hall CL, McGuire D, Frankel MR, et al. Blood transcriptome changes after stroke in an African American population. Ann Clin Transl Neurol. 2016;3(2):70–81. doi: 10.1002/acn3.272 PMID: 26900583; PubMed Central PMCID: PMCPMC4748310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarchielli P, Nardi K, Chiasserini D, Eusebi P, Tantucci M, Di Piero V, et al. Immunological profile of silent brain infarction and lacunar stroke. PloS one. 2013;8(7):e68428. doi: 10.1371/journal.pone.0068428 PMID: 23874624; PubMed Central PMCID: PMCPMC3706426. [DOI] [PMC free article] [PubMed] [Google Scholar]


