Abstract
Genetic analyses have linked MicroRNA-137 (MIR137) to neuropsychiatric disorders, including schizophrenia and autism spectrum disorder (ASD). MiR-137 plays important roles in neurogenesis and neuronal maturation, but the impact of miR-137 loss of function in vivo remains unclear. Here we show the complete loss of miR-137 in the mouse germline (gKO) or nervous system (cKO) leads to postnatal lethality, while heterozygous gKO and cKO mice remain viable. Partial loss of miR-137 in heterozygous cKO mice results in dysregulated synaptic plasticity, repetitive behavior, and impaired learning and social behavior. Transcriptomic and proteomic analyses revealed that the miR-137 mRNA target, Phosphodiesterase 10a (Pde10a), is elevated in heterozygous KO mice. Treatment with the PDE10A inhibitor papaverine or knockdown of Pde10a ameliorates the deficits observed in the heterozygous cKO mice. Collectively, our results suggest that MIR137 plays essential roles in postnatal neurodevelopment, and that dysregulation of miR-137 potentially contributes to neuropsychiatric disorders in humans.
Introduction
MicroRNAs (miRNAs) are a class of endogenous and non-coding single-stranded ~22-nucleotide RNAs, many of which are evolutionarily conserved. Based on sequence complementarities, miRNAs target mRNAs and regulate posttranscriptional gene expression1,2. With a high degree of spatial and temporal specificity, miRNAs regulate neurogenesis, synaptic plasticity, and circadian rhythm3–6. Thus, miRNAs could orchestrate neuronal development and plasticity by modulating complex gene networks. The perturbation of miRNA expression could potentially contribute to the etiology of human diseases, including neuropsychiatric disorders.
Previous meta-analyses combining genome-wide association studies (GWAS) from 17 separate studies identified a locus on chromosome 1p21.3 to be highly associated with schizophrenia7–10. MIR137 resides in this locus, which encodes microRNA-137 (miR-137). Intriguingly four other loci achieving genome-wide significance identified in the same studies contain the genes predicted to be regulated by miR-137. Moreover, MIR137 is also associated with autism spectrum disorders (ASD); such as in a large-scale copy number variation (CNV) analyses of ASD patients, the risk pathogenic CNV overlap with the loci containing MIR137 gene7,10,11. Besides schizophrenia and ASD, MIR137 has been linked to bipolar disorder as well12. These findings together suggest that miR-137 contributes to the pathogenesis of neuropsychiatric disorders. However, whether the dysregulation of miR-137 leads directly to the phenotypes associated with neuropsychiatric disorders remains to be determined.
Previously published works have demonstrated the critical roles of miR-137 in neurogenesis, neuronal maturation, and dendritic morphogenesis during neurodevelopment13,14. More recently, using induced human neurons, the overexpression of miR-137 impairs synaptic plasticity and learning and memory in the hippocampus15. The minor allele associated with schizophrenia can increase MIR137 expression, suggesting that miR-137 gain of function might be associated with this disease 8,10,12,16. However, little is known about the in vivo impact of the loss of miR-137, which is critical given that the microdeletions of MIR137 are linked to ASD in multiple reports11,17–20.
In this study, we generated a miR-137 conditional allele to investigate the impact of miR-137 loss of function in vivo. We show that miR-137 is critical for postnatal mouse development. Partial loss of miR-137 leads to the dysregulation of synaptic plasticity, altered repetitive or stereotyped behaviors, and impaired sociability/social novelty preference. Combining large-scale proteomic analyses with gene expression profiling identified the mRNAs that miR-137 post-transcriptionally regulates. By overlapping with the known genes linked to neuropsychiatric disorders, we found a significant enrichment of ASD-associated genes among miR-137 targets. One of the miR-137 mRNA targets, Phosphodiesterase 10a (PDE10A), is significantly upregulated in vivo upon partial loss of miR-137. A specific PDE10A inhibitor, papaverine, can ameliorate the deficits associated with the partial loss of miR-137. Additional shRNA-mediated knockdown of Pde10a results in similar rescue effects, supporting that the increased expression of Pde10a is responsible for the observed abnormalities. These results together highlight the critical roles of MIR137 in postnatal neurodevelopment and suggest that the dysregulation of miR-137 contributes to neuropsychiatric disorders in humans.
Results
Loss of miR-137 in germline and nervous system leads to postnatal lethality
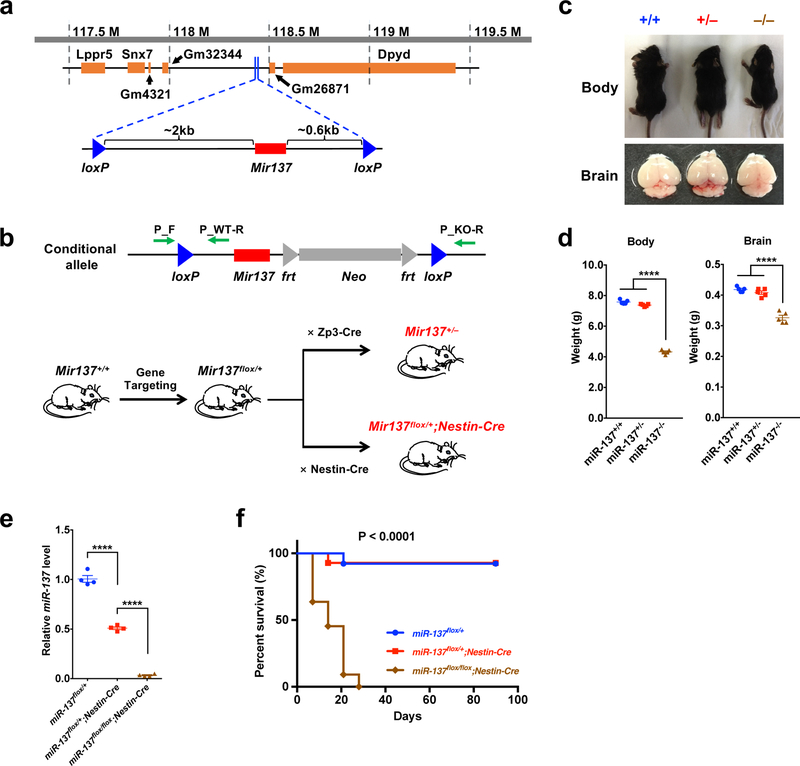
To generate miR-137 knockout mice, we designed a targeting vector to disrupt the Mir137 gene via homologous recombination in mouse embryonic stem cells, where two loxP sites were inserted upstream (~2 kb) and downstream (~0.6 kb) of the Mir137 gene, and derived mice carried the floxed allele of miR-137 (Fig. 1a). By crossing with either Zp3-Cre or Nestin-Cre line, we deleted Mir137 in the germline or nervous system and generated the heterozygous miR-137 global knockout (gKO) and conditional knockout (cKO) mice (Fig. 1b). MiR-137 wild-type (miR-137+/+ or miR-137flox/+), heterozygous (miR-137+/– or miR-137flox/+;Nestin-Cre) and homozygous (miR-137–/– or miR-137flox/flox;Nestin-Cre) mice were used in our following analyses. We found significantly reduced levels of mature miR-137 in the brain upon the loss of Mir137 (Supplementary Fig. 1a,b and Supplementary Table 1). The deletion of Mir-137 did not affect the expression of an adjacent gene, dihydropyrimidine dehydrogenase (Dpyd) (Supplementary Fig. 1c). The complete loss of miR-137 (gKO) resulted in postnatal lethality in miR-137–/– mice, which survived up to postnatal day 21 (P21), while the partial loss of miR-137 did not affect the survival rate of miR-137+/– mice (Supplementary Fig. 1d). Comparing to miR-137+/+ littermates, miR-137+/– mice showed few differences in body and brain weight; while the body and brain weights were significantly decreased in miR-137–/– mice (Fig. 1c,d).
Figure 1: Loss of miR-137 leads to postnatal lethality.
a, Generation of the miR-137 conditional allele. A targeting vector was designed to disrupt the Mir137 gene via homologous recombination in mouse embryonic stem cells, where two loxP sites were inserted upstream (~2 kb) and downstream (~0.6 kb) of the Mir137 gene (the image was modified from the UCSC Genome Browser).
b, Schematic of the crosses to generate miR-137 knockout mice. Green arrows indicate the primer sets that designed for PCR genotyping. By crossing with either Zp3-Cre or Nestin-Cre line, we could specifically delete Mir137 in the germline or central and peripheral nervous system and generate the heterozygous global knockout (miR-137+/–) and conditional knockout (miR-137flox/+;Nestin-Cre) mice.
c–d, Gross appearance of miR-137 gKO mice. Representative wild-type (miR-137+/+), heterozygous (miR-137+/–) and homozygous (miR-137–/–) of miR-137 knockout mice and brain tissues (c). At postnatal day 16 (P16), the body and brain weight of miR-137–/– mice are much smaller than miR-137+/+ and miR-137+/– mice (n = 5 mice per group) (d). Data represent means ± s.e.m; Body: F2,12 = 1123, P < 0.0001; Brain: F2,12 = 67.05, P < 0.0001; One-way ANOVA with Bonferroni post hoc test; n.s., nonsignificant; ****, P < 0.0001.
e, Verification of decreased expression of mature miR-137 in miR-137flox/+;Nestin-Cre and miR-137flox/flox;Nestin-Cre mice in the hippocampus using independent real-time PCR (n = 4 mice per group). Data represent means ± s.e.m; F2,9 = 505.6, P < 0.0001; One-way ANOVA with Bonferroni post hoc test; ****, P < 0.0001.
f, Kaplan-Meier graph shows survival curves of the miR-137flox/+ (n = 13 mice), miR-137flox/+;Nestin-Cre (n = 14 mice) and miR-137flox/flox;Nestin-Cre (n = 11 mice) mice groups. Mice in the homozygous (miR-137flox/flox;Nestin-Cre) group exhibited significantly decreased survival (dot) relative to the other groups (Two-Tailed log-rank tests; P < 0.0001). All 11 tested miR-137flox/flox;Nestin-Cre mice died by postnatal day 28.
To examine whether the loss of miR-137 resulted in impaired embryonic and early postnatal development, we performed timed mating using heterozygous mice. We collected embryos at embryonic day 15.5 (E15.5) and found few morphological differences among miR-137+/+, miR-137+/– and miR-137–/– mice (Supplementary Fig. 2a). At the early postnatal stage (P0), miR-137+/+ and miR-137–/– mice were similar in terms of size and weight (Supplementary Fig. 2b), but the differences were usually observed as early as P7 (data not shown), suggesting a postnatal effect of miR-137 loss-of-function. At P14 the major organs (heart, liver, lung, and kidney) of miR-137–/– mice are smaller than those of their miR-137+/+ and miR-137+/– littermates (Supplementary Fig. 2c). Major organ sections stained with H&E (haematoxylin and eosin) showed no apparent morphological differences (Supplementary Fig. 2d).
In miR-137 cKO mice, miR-137 was deleted specifically in the nervous system to more accurately reflect the neuronal phenotype; we found the mature miR-137 expression in brain significantly reduced in miR-137flox/+;Nestin-Cre and miR-137flox/flox;Nestin-Cre mice compared to the littermate control (Fig. 1e). Also, we observed similar postnatal lethality in homozygous miR-137 cKO (miR-137flox/flox;Nestin-Cre) mice during postnatal development (Fig. 1f), strongly suggesting that the phenotypes in miR-137 gKO mice are from the loss of miR-137 in the nervous system. Therefore, we mainly focused on using miR-137 cKO mice to understand the in vivo roles of miR-137 in the brain.
Loss of miR-137 results in the dysregulation of synaptic plasticity
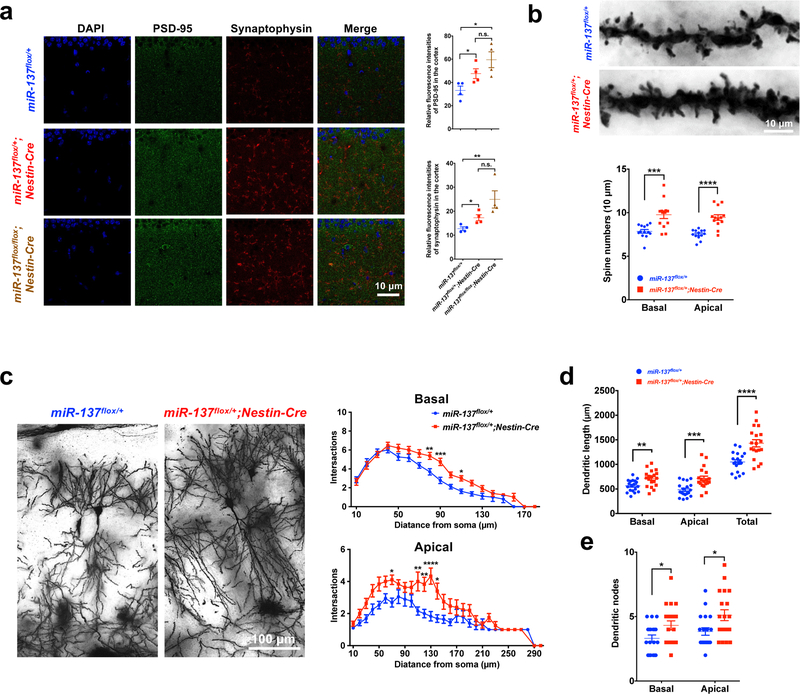
PSD-95 and Synaptophysin immunohistochemistry staining was performed using hippocampal or cortical tissues from miR-137 cKO mice at the age of P18 to determine whether the loss of miR-137 influences synaptic growth in vivo. We observed in both hippocampal and cortical tissues, the relative fluorescence intensities and protein levels of PSD-95 and Synaptophysin increased in miR-137flox/+;Nestin-Cre and miR-137flox/flox;Nestin-Cre mice when compared to their miR-137flox/+ siblings, indicating the overgrowth of synapse upon loss of miR-137 (Fig. 2a and Supplementary Fig. 3a–c). Also, we performed the Caspase-3 staining in brain sections of miR-137 cKO mice at different developmental stages (P0, P18, and P60), but did not see any significant change in Caspase-3 expression between miR-137flox/+ and miR-137flox/+;Nestin-Cre mice, indicating that partial loss of miR-137 has limited impact on neuronal viability/apoptosis (Supplementary Fig. 3d). To further examine the spine structure in vivo, we performed Golgi staining in miR-137flox/+ and miR-137flox/+;Nestin-Cre mice and analyzed their dendritic spines in the hippocampal CA1 region. The spine densities of basal and apical spines were significantly increased in the miR-137flox/+;Nestin-Cre mice compared to the littermate control, indicating the loss of miR-137 results in synaptic pruning deficits (Fig. 2b). Also, compared with the neurons of miR-137flox/+ mice, the neurons in miR-137flox/+;Nestin-Cre mice exhibited increased dendritic complexity and enhanced dendritic growth (Fig. 2c–e). Consistently, we also observed a similar increase of PSD-95 and Synaptophysin in miR-137 gKO mice (Supplementary Fig. 4a,b). These results together suggest that miR-137 plays essential roles in regulating synaptic plasticity.
Figure 2: Loss of miR-137 in the nervous system leads to synaptic overgrowth and impaired dendritic growth in vivo.
a, Immunohistochemistry (IHC) staining of PSD-95 and Synaptophysin in miR-137 cKO mice hippocampal tissues. PSD-95 and Synaptophysin IHC staining were carried out on 40-μm thick floating sections containing hippocampal tissue from four pairs of littermate cKO mice, i.e., miR-137flox/+, miR-137flox/+;Nestin-Cre and miR-137flox/flox;Nestin-Cre mice, at the age of postnatal day 18 (P18) (n = 4 mice per group). Relative fluorescence intensity of PSD-95 (F2,9 = 9.494, P = 0.0061) and Synaptophysin (F2,9 = 12.73, P = 0.0024) elevated upon the loss of miR-137 in the hippocampus. Image analyses and quantification were performed using ImageJ software. Data represent means ± s.e.m; One-way ANOVA with Bonferroni post hoc test; n.s., nonsignificant; *, P < 0.05; **, P < 0.01.
b–e, Golgi staining reveals dendritic growth in the hippocampal CA1 region in miR-137flox/+ and miR-137flox/+;Nestin-Cre mice (n = 13 slices from 4 mice per group). (b)The spine densities of basal (t = 3.895, P = 0.0007) and apical (t = 5.406, P < 0.0001) spines were significantly increased in the miR-137flox/+;Nestin-Cre mice compared to the littermate control (Data represent means ± s.e.m. Unpaired Two-Tailed t-test; ***, P < 0.001; ****, P < 0.0001). (c) Sholl analysis showing, compared with neurons in miR-137flox/+ mice, the CA1 pyramidal neurons in miR-137flox/+;Nestin-Cre mice exhibited increased dendritic complexity (Data represent means ± s.e.m; Basal spines: t80 = 3.952, P = 0.0016; t90 = 4.534, P = 0.0001; t110 = 3.047, P = 0.0437. Apical spines: t70 = 3.210, P = 0.0388; t110 = 4.105, P = 0.0013; t120 = 4.004, P = 0.0019; t130 = 5.242, P < 0.0001; t140 = 3.493, P = 0.0143. Two-way ANOVA with Bonferroni post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; distance points with P < 0.05 were not marked in the figure). Partial loss of miR-137 significantly increased dendritic length (tBasal = 3.275, P = 0.0023; tApical = 3.905, P = 0.0003; tTotal = 4.422, P < 0.0001) (d) and dendritic nodes (tBasal = 2.118, P = 0.0377; tApical = 2.675, P = 0.0092) (e) in miR-137flox/+;Nestin-Cre neurons (Data represent means ± s.e.m. Unpaired Two-Tailed t-test; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
Partial loss of miR-137 leads to the deficits in learning and memory
Given the link of miR-137 to neuropsychiatric disorders, we then determined whether the loss of miR-137 led to any behavioral abnormalities relevant to neuropsychiatric disorders. We subjected miR-137 cKO/gKO mice to a battery of behavioral tests. Due to the postnatal lethality of the complete loss of miR-137, we focused on 2- to 3-month-old wildtype and heterozygotic miR-137 cKO/gKO mice.
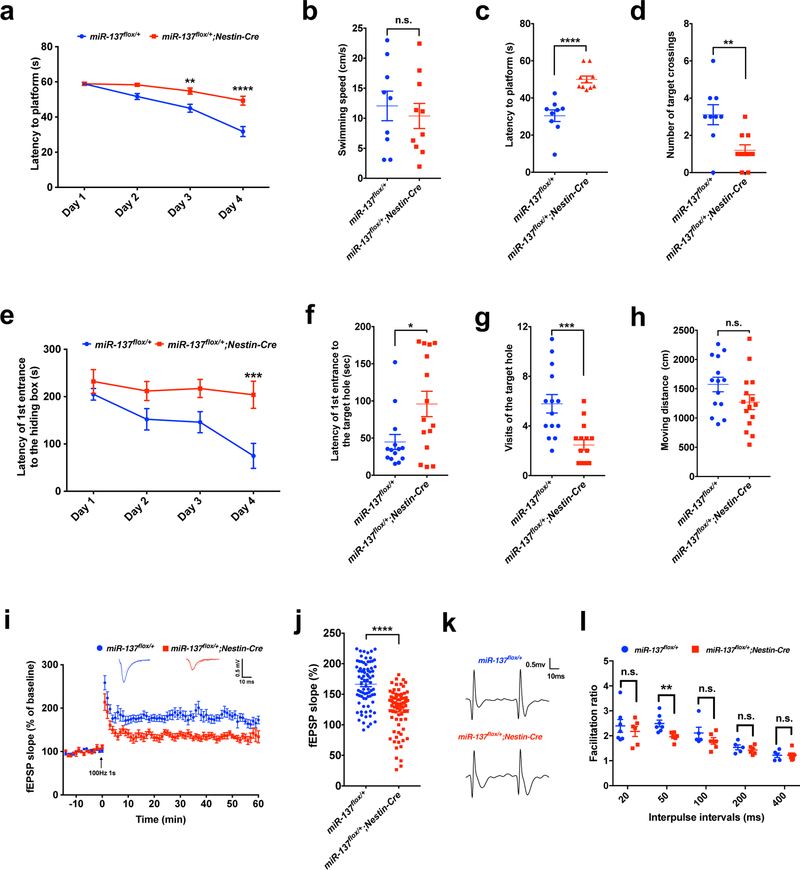
Previous reports indicate that overexpression of miR-137 in the dentate gyrus impairs hippocampus-dependent learning15. We therefore asked whether the loss of miR-137 would also alter learning and memory in our miR-137 knockout mice. First, we performed the Morris water maze test (Fig. 3a–d). During the training phase of four trials per day for four successive days, although both miR-137flox/+ and miR-137flox/+;Nestin-Cre mice showed improved latency to the platform, miR-137flox/+;Nestin-Cre mice exhibited a significant delay in finding the platform (Fig. 3a). There was no significant difference in swimming speeds between the two groups of mice (Fig. 3b). In the subsequent probe test phase, miR-137flox/+;Nestin-Cre mice exhibited significantly higher latency to targets and fewer target crossings (Fig. 3c,d). Consistently we observed similar changes in miR-137 gKO mice (Supplementary Fig. 5a–d). To further assess the impaired spatial learning and memory deficit in miR-137 cKO and gKO mice, we performed Barnes maze test. Mice received training in the Barnes maze for four consecutive days, consisting of four trials per day to find the hiding box. We found that both miR-137flox/+;Nestin-Cre and miR-137+/– mice failed to diminish the latency of the first entry to the hiding box (Fig. 3e and Supplementary Fig. 5e). Probe trials on Day 5 demonstrated that both miR-137flox/+;Nestin-Cre and miR-137+/– mice had spatial memory deficits as they visited the target hole less often and spent less time in target quadrant than miR-137flox/+ mice and miR-137+/+ (Fig. 3f,g and Supplementary Fig. 5f,g). The swimming speeds were not significantly different between the two groups (Fig. 3h and Supplementary Fig. 5h).
Figure 3: Partial loss of miR-137 leads to the learning and memory deficits.
a–d, MiR-137flox/+;Nestin-Cre mice exhibited learning and memory deficits in the Morris water maze test. During the training phase, in which four trials were conducted per day for four successive days, both miR-137flox/+ (n = 9 mice) and miR-137flox/+;Nestin-Cre (n = 10 mice) mice showed improved latency to locate the platform, but miR-137flox/+;Nestin-Cre mice exhibited a significant delay to diminish the latency to locate the platform. (Data represent means ± s.e.m; tDay1 = 0.026, P > 0.9999; tDay2 = 2.565, P = 0.0501; t Day3 = 3.766, P = 0.0014; tDay4 = 6.761, P < 0.0001; Two-way ANOVA with Bonferroni post hoc test. **, P < 0.01; ****, P < 0.0001) (a). In probe trials on Day 5, there was no significant difference in swimming speeds between the two groups of mice (t = 0.524, P = 0.6073) (b). Compare to miR-137flox/+, miR-137flox/+;Nestin-Cre mice exhibited a significantly longer latency to locate the platform (t = 5.530, P < 0.0001) (c), but less number of target crossings (t = 3.213, P = 0.0051) (d). Data shown are means ± s.e.m. Unpaired Two-Tailed t-test; **, P < 0.01; ****, P < 0.0001.
e–h, The miR-137 cKO mice were subject to the Barnes maze test. During the training phase, in which four trials were conducted per day for four successive days, the miR-137flox/+;Nestin-Cre mice (n = 15 mice) failed to diminish the latency of first entrance to the hiding box, indicating that loss of miR-137 results in impaired spatial memory (e). (Data represent means ± s.e.m; tDay1 = 0.842, P > 0.9999; tDay2 = 1.864, P = 0.2603; t Day3 = 2.220, P = 0.1141; tDay4 = 4.025, P = 0.0004; Two-way ANOVA with Bonferroni post hoc test. ***, P < 0.001. Probe trials on Day 5 demonstrated that miR-137flox/+;Nestin-Cre mice also had spatial memory deficits as they spent less time (t = 2.537, P = 0.0173) in the target quadrant (f) and visited the target hole less often (t = 3.978, P = 0.0005) (g) than miR-137flox/+ mice (n = 14 mice). The swimming speeds showed no significant difference between two groups (t = 1.713, P = 0.0981) (h). Data shown are means ± s.e.m. Unpaired Two-Tailed t-test; n.s., nonsignificant; *, P < 0.05; ***, P < 0.001.
i–j, Partial loss of miR-137 resulted in impaired long-term potentiation (LTP) in miR-137flox/+;Nestin-Cre mice. A typical experiment showing the time course of CA1 LTP for a single recording. fEPSP traces before (gray) and after (blue or red) are shown in the inset above. Pooled data showing the time course of LTP from all recordings made from miR-137flox/+ or miR-137flox/+;Nestin-Cre mice (i). Average LTP amplitude measured at 55–60 min post-induction (j). n = 8 slices from 4 mice per group; 10 fEPSP slope (%) values were collected from 1 slice. Data represent means ± s.e.m; t = 7.552, P < 0.0001; Unpaired Two-Tailed t-test; ****, P < 0.0001.
k-l, Paired-pulse facilitation (PPF). Representative recording of the paired-pulse ratio at the interpulse interval of 50 ms from the slices prepared from miR-137flox/+ (n = 7 slices from 4 mice) and miR-137flox/+;Nestin-Cre (n = 6 slices from 4 mice) mice (k). PPF studies across different interpulse intervals (20 ms, 50 ms, 100 ms, 200 ms and 400 ms) revealed a significant difference in the paired-pulse ratio at 50 ms intervals (l). Data represent means ± s.e.m; t20ms = 0.660, P = 0.5226; t50ms = 3.389, P = 0.0060; t100ms = 1.387, P = 0.2301; t200ms = 0.905, P = 0.3893; t 400ms = 0.025, P = 0.9809. Unpaired Two-Tailed t-test; n.s., nonsignificant; **, P < 0.01.
With these observed hippocampus-dependent learning deficits, we further investigated the effect of miR-137 deficiency in neuronal plasticity. We examined long-term potentiation (LTP) in acute hippocampal slices prepared from miR-137flox/+ and miR-137flox/+;Nestin-Cre mice. Compared to miR-137flox/+, LTP was attenuated in miR-137flox/+;Nestin-Cre mice (Fig. 3i). The amplitude of LTP 55–60 min after induction was significantly attenuated in miR-137flox/+;Nestin-Cre mice compared with miR-137flox/+ mice (Fig. 3j). Also, we examined paired-pulse facilitation (PPF) across different interpulse intervals, including 20, 50, 100, 200, and 400 ms. Compared to miR-137flox/+ mice, we found a significant reduction of the paired-pulse ratio in miR-137flox/+;Nestin-Cre mice at the 50 ms interval (Fig. 3k,l). The electrophysiological results indicate that miR-137 plays vital roles in maintaining the normal synaptic plasticity. Together, these data suggest that the loss of miR-137 results in the deficits of learning and memory. Prior studies found that the overexpression of miR-137 could result in impaired learning and memory as well, indicating that miR-137-mediated regulation could be dosage-dependent.
Partial loss of miR-137 leads to altered repetitive/stereotyped behaviors and impaired sociability and social novelty preference
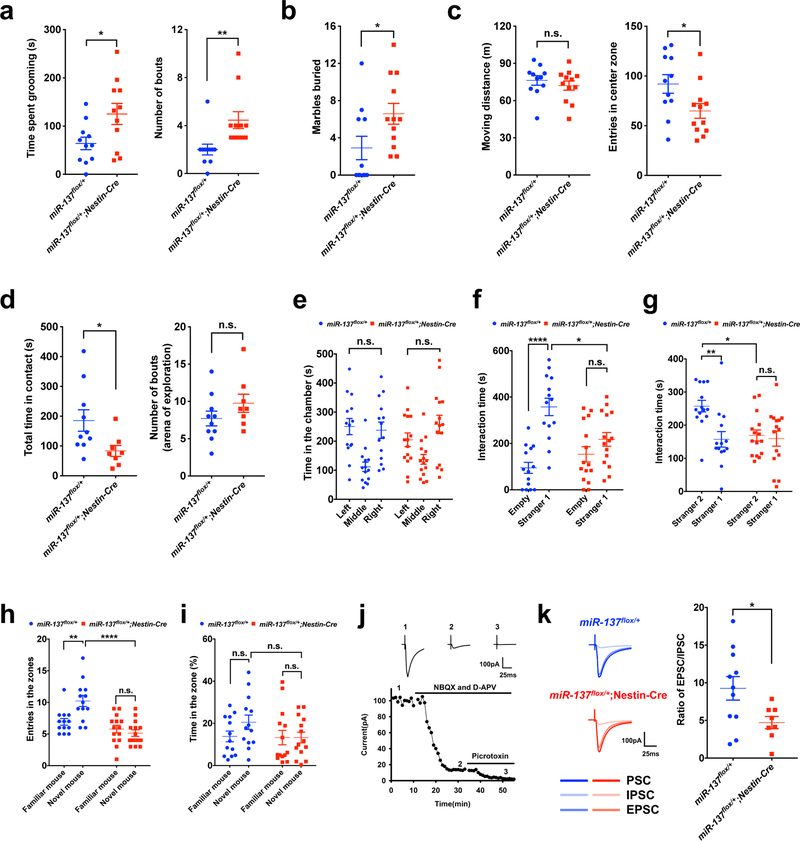
Impaired social functioning is a characteristic phenotype associated with schizophrenia, ASD, and deficits in social memory are also characteristic of some forms of intellectual disability (ID)21,22. The genomic deletion of MIR137 in patients usually accompany with ASD phenotypes; e.g., 12 individuals carrying a microdeletion in chromosome 1p21.3, containing MIR137 gene, are reported with a 92% incidence of ASD10–13. Therefore, we decided to examine the changes in social and repetitive behaviors, which are the core features of ASD. To assess repetitive/stereotyped behaviors, we recorded self-grooming bouts and performed the marble-burying test. We found that miR-137flox/+;Nestin-Cre mice exhibited significantly more bouts and time spent in self-grooming (Fig. 4a), and buried more marbles than miR-137flox/+ littermates (Fig. 4b). In the open field test, there was slight difference in the total distance moved during the 30-minutes period (Fig. 4c), suggesting similar locomotor activities between miR-137flox/+ and miR-137flox/+;Nestin-Cre mice. On the other hand, the miR-137flox/+;Nestin-Cre mice entered the center zone significantly fewer times than miR-137flox/+ mice (Fig. 4c), indicating elevated anxiety-like behavior. To characterize the social behavior of the miR-137flox/+;Nestin-Cre mice, we performed the social interaction test. We found that, compared to miR-137flox/+ littermates, miR-137flox/+;Nestin-Cre mice displayed similar arena exploration levels, but underwent excessive self-grooming in less time when in contact with unfamiliar mice (Fig. 4d and Supplementary Fig. 6a). These data together suggest that partial loss of miR-137 leads to a specific social deficit and unusual repetitive behavior in miR-137flox/+;Nestin-Cre mice.
Figure 4: Partial loss of miR-137 causes impaired social behaviors in mice.
a, Self-grooming test showing an increased total time spent grooming (t = 2.410, P = 0.0257) and number of bouts (t = 2.939, P = 0.0081) in miR-137flox/+;Nestin-Cre mice than in miR-137flox/+ mice (n = 11 mice per group). Data represent means ± s.e.m; Unpaired Two-Tailed t-test; n.s., nonsignificant; *, P < 0.05, **, P < 0.01.
b, In the marble-burying test, compared with miR-137flox/+ mice (n = 11 mice), miR-137flox/+;Nestin-Cre mice (n = 12 mice) showed a significant number of marble burying bouts. Data represent means ± s.e.m; t = 2.195, P = 0.0396; Unpaired Two-Tailed t-test; *, P < 0.05.
c, In the open field test, miR-137flox/+;Nestin-Cre mice (n = 12 mice) traveled similar distances (t = 0.7779, P = 0.4453) in 30-minutes duration and exhibited significantly less number of entries in the center zone (t = 2.292, P = 0.0323) when compared with control miR-137flox/+ mice (n = 11 mice). Data represent means ± s.e.m; Unpaired Two-Tailed t-test; n.s., nonsignificant; *, P < 0.05.
d, Social interactions tests are showing total time in contact and number of arena exploration. When comparing with miR-137flox/+ mice (n = 10 mice), miR-137flox/+;Nestin-Cre mice (n = 8 mice) displayed significantly less time in contact with unfamiliar mice (t = 2.326, P = 0.0335) and showed a similar arena exploration level (t = 1.320, P = 0.2055). Data represent means ± s.e.m; Unpaired Two-Tailed t-test; n.s., nonsignificant; *, P < 0.05.
e–g, MiR-137flox/+;Nestin-Cre mice exhibited impaired social behavior in the three-chamber test. Both miR-137flox/+ (n =14 mice) and miR-137flox/+;Nestin-Cre (n = 15 mice) mice had no preference for either left chamber or right chamber during the habituation phase (miR-137flox/+ mice: tleft vs. right = 0.3572, P > 0.9999; miR-137flox/+;Nestin-Cre mice: tleft vs. right = 1.545, P = 0.3785) (e), but miR-137flox/+;Nestin-Cre mice lacked a social preference for a mouse over the empty cage (miR-137flox/+ mice: tEmpty vs. Stranger1 = 5.827, P < 0.0001; miR-137flox/+;Nestin-Cre mice: tEmpty vs. Stranger1 = 1.476, P = 0.8747; miR-137flox/+ vs. miR-137flox/+;Nestin-Cre: tStranger1 = 3.159, P = 0.0155) (f). Moreover, unlike miR-137flox/+ control mice, miR-137flox/+;Nestin-Cre mice displayed impaired social novelty recognition by demonstrating no preference for a novel mouse over a familiar mouse (miR-137flox/+ mice: tStranger2 vs. Stranger1 = 3.438, P = 0.0068; miR-137flox/+;Nestin-Cre mice: tStranger2 vs. Stranger1 = 0.3359, P > 0.9999; miR-137flox/+ vs. miR-137flox/+;Nestin-Cre: tStranger2 = 3.039, P = 0.0188) (g). Data represent means ± s.e.m; Two-way ANOVA with Bonferroni post hoc test. n.s., nonsignificant; **, P < 0.01; ****, P < 0.0001.
h-i, In the social discrimination test, miR-137flox/+;Nestin-Cre mice exhibited impaired social discrimination ability. When comparing with miR-137flox/+ mice (n = 13 mice), miR-137flox/+;Nestin-Cre mice (n = 14 mice) exhibited significantly fewer entries number (miR-137flox/+ mice: tNovel vs. Familiar = 3.581, P = 0.0046; miR-137flox/+;Nestin-Cre mice: tNovel vs. Familiar = 0.722, P > 0.9999; miR-137flox/+ vs. miR-137flox/+;Nestin-Cre: tNovel = 5.609, P < 0.0001) (h) and relatively lower % time spent in novel mouse zone (miR-137flox/+ mice: tNovel vs. Familiar = 1.538, P = 0.7822; miR-137flox/+;Nestin-Cre mice: tNovel vs. Familiar = 0.011, P > 0.9999; miR-137flox/+ vs. miR-137flox/+;Nestin-Cre: tNovel = 1.689, P = 0.5846) (i). Data represent means ± s.e.m; Two-way ANOVA with Bonferroni post hoc test. n.s., nonsignificant; **, P < 0.01; ****, P < 0.0001.
j–k, Partial loss of miR-137 resulted in imbalanced excitation/inhibition (E/I) ratio and impaired social behavior in miR-137 knockout mice. A representative whole-cell recording experiment (repeated independently three times with similar results) and samples trace at indicated time points showing the time course of evoked synaptic currents in the absence or presence of various inhibitors to determine the E/I ratio. Excitatory and inhibitory postsynaptic currents (EPSCs and IPSCs) were pharmacologically isolated by using respective inhibitors specific to glutamate or GABA receptors. E/I ratio by sequentially recording evoked synaptic responses, firstly in the absence of any inhibitors to obtain total synaptic currents (i.e., EPSC+IPSC), secondly in the presence of NBQX/APV to obtain IPSC, and finally in the presence of NBQX/APV/Picrotoxin to verify the IPSC component. Sample traces are showing various components of synaptic currents (j). Summary data shows a decreased E/I ratio in miR-137flox/+; Nestin-Cre (n = 8 slices from 4 mice) compared to miR-137flox/+ control mice (n = 11 slices from 6 mice) (k). t = 2.311, P = 0.034; Unpaired Two-Tailed t-test. *, P < 0.05.
We next evaluated the sociability and social novelty of miR-137 knockout mice by using the three-chamber test. In miR-137 cKO mice, both miR-137flox/+ and miR-137flox/+;Nestin-Cre mice showed no preference for either left chamber or right chamber during the habituation phase (Fig. 4e and Supplementary Fig. 6b), but miR-137flox/+;Nestin-Cre mice showed a limited social preference for a mouse over an object (Fig. 4f). Unlike miR-137flox/+ control mice, miR-137flox/+;Nestin-Cre mice demonstrated no preference for a strange mouse over a familiar mouse (Fig. 4g). We also confirmed these abnormal social behaviors in miR-137 gKO mice (Supplementary Fig. 6c–e).
To further assess the impaired social behavior associated with the partial loss of miR-137, we then used another modified protocol of the social discrimination test to probe the mice for their social discrimination ability23,24. We found miR-137+/– and miR-137flox/+;Nestin-Cre mice had significantly fewer entries into the novel mouse zone and spent significantly less time with the novel mouse compared to their littermate controls (Fig. 4h,i and Supplementary Fig. 6f,g).
The alteration of the balance of excitation and inhibition (E/I ratio) is associated with, and thought to cause, a wide range of neurological and mental disorders, including autism and schizophrenia25–27. In support of this, we found that upon the partial loss of miR-137, the E/I ratio significantly decreased in miR-137flox/+;Nestin-Cre mice compared to their miR-137flox/+ littermates (Fig. 4j,k). Other reports indicate the overexpression of miR-137 has minimal impact on social behaviors15; in contrast, our observations show that the partial loss of miR-137 (both in germline and CNS) leads to abnormal sociability, social novelty, and recognition in mice, supporting the idea that the partial loss of miR-137 leads to the phenotypes associated with ASD.
Genome-wide identification of in vivo mRNA targets of miR-137
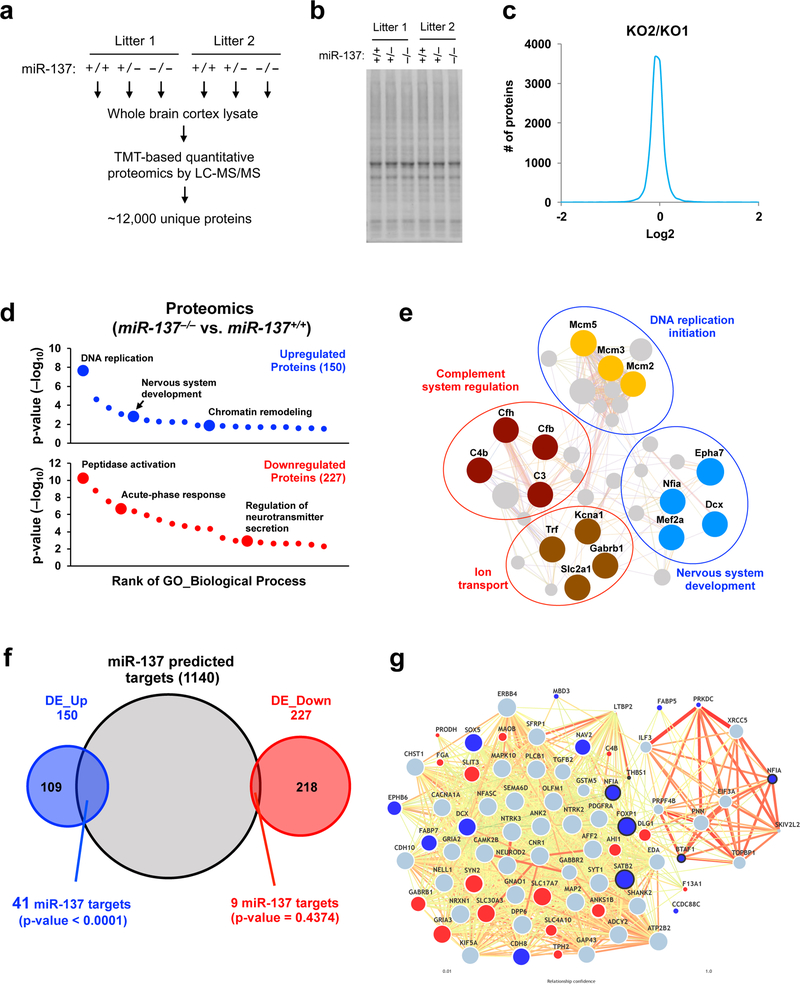
Given the role of miRNAs in posttranscriptional gene regulation, we conducted TMT (Tandem Mass Tag)-based quantitative proteomics analysis using total protein from miR-137+/+, miR-137+/– and miR-137–/– mice (P12, two biological replicates from two littermates) brain cortical tissues (Fig. 5a). Replicates possessed consistent TMT protein quantification results (Fig. 5b,c and Supplementary Fig. 7a), and the relative abundance of selective proteins derived from the TMT-based MS quantification was selectively validated by Western blotting (Supplementary Fig. 7b).
Figure 5: Systematic identification of in vivo mRNA targets of miR-137 by integrating proteomic, transcriptomic and bioinformatic analyses.
a, Workflow of the TMT (Tandem Mass Tag)-based quantitative proteomic analysis. Six brain cortical samples from two litters (3 genotypes per little) were used as described in Method section. There were total ~1,800, 000 MS2 scans, among which ~140, 000 unique peptides and ~12,000 unique proteins were identified and quantified.
b, SDS-PAGE and Coomassie staining of the whole brain cortical tissues homogenized in the lysis buffer before protein digestion for the proteomics analysis.
c, The consistency of protein TMT quantification result was found between the two replicates. As a representative, miR-137–/– samples in litter 1 (KO1) and litter 2 (KO2) mice showed high correlation regarding protein TMT quantification intensity.
d, GO enrichment analysis of differentially expressed (DE) proteins (miR-137–/– / miR-137+/+) identified in the proteomic analysis (n = 150 for upregulated DE genes; n = 227 for downregulated DE genes). The DE genes were enriched in the biological process of “DNA replication,” “Nervous system development” and “Peptidase activity regulation.” The full result of the analysis is presented in Supplementary Fig. 8b.
(e) Interactome of the differentially expressed (DE) genes upon the loss of miR-137. Predicted protein-protein interaction network, which was generated with represented upregulated genes (blue circle) and downregulated genes (red circle), indicates the impact of the miR-137 loss of function in various biological processes. Colored dots indicate the genes are differentially expressed in proteomics analysis.
f, The overlap between miR-137 predicted targets and upregulated proteins. Proteomic analysis confidently identified 417 differentially expressed (DE) protein (377 genes) in miR-137–/– mice compared to miR-137+/+ mice. By overlapping with miR-137 predicted targets (1140 transcripts with conserved sites), we found that miR-137 predicted targets significantly overlapped with upregulated DE genes (DE_Up, 41 out of 150), but not down-regulated genes (DE_Down). Pearson’s chi-squared test calculated the p-value.
g, MiR-137-mediated regulation in ASD candidate genes interactome. Predicted ASD-gene network exploring brain-specific interactions between ASD candidate genes were generated, including 15 upregulated and 16 downregulated DE proteins. Upregulated miR-137 predicted targets might potentially influence the adjacent ASD candidate gene’s expression in mRNA and protein levels, such as significantly downregulated GRIA3 and GABRB1 were previously reported as associated with ASD. The dots marked with bold lines indicate the ASD candidate genes targeted by miR-137 as shown in Supplementary Fig. 9.
Using a widely used algorithm, Sylamer28, we checked the correlation between miRNA seed match enrichment in 3’ UTRs and protein log2 fold changes (Supplementary Fig. 8a). We found the upregulated proteins in miR-137–/– cortexes are specifically and significantly enriched for miR-137 binding sites (the top two enriched 7-nt sequences are predicted to be targeted by miR-137). When comparing miR-137–/– / miR-137+/+, miR-137+/– / miR-137+/+, and miR-137–/– / miR-137+/–, we identified 417, 94 and 76 differentially expressed (DE) proteins respectively (Supplementary Table 2). The GO analyses showed that the upregulated proteins (miR-137–/– / miR-137+/+) were significantly involved in DNA replication, nervous system development, and chromatin remodeling while the downregulated proteins were enriched in the genes that play important roles in peptidase activity regulation, acute-phase response, and regulation of neurotransmitter secretion (Fig. 5d, Supplementary Fig. 8b and Supplementary Table 2). The interactome analysis using the selected DE genes revealed a complex gene regulatory network, suggesting that these DE genes might be associated with each other and result in dysfunction upon loss of miR-137 (Fig. 5e). Notably, several significantly downregulated genes are involved in ion transport and complement system regulation, which are reported to play essential roles in autism and schizophrenia29–32. These data together suggest that the loss of miR-137 leads to the dysregulation of the genes involved in selective biological processes. These genes could be co-regulated by miR-137 to influence postnatal neurodevelopment.
To identify the direct mRNA targets of miR-137, we overlapped the predicted miR-137 targets with those DE proteins and identified 41 upregulated, and 9 downregulated miR-137 predicted targets (Fig. 5f). We found that those upregulated DE genes significantly associated with miR-137 predicted targets (Pearson’s chi-square test, p-value < 0.0001). Among the 41 upregulated miR-137 predicted targets, the mRNA levels of 37 genes (90%) as determined by RNA-seq (Supplementary Table 3) changed little, suggesting that miR-137 regulates its direct targets in a posttranscriptional manner.
We then explored whether miR-137 targets the known candidate genes of neuropsychiatric disorders. To do this, we overlapped the DE proteins with ASD, schizophrenia, and ID candidate genes (Supplementary Fig. 9 and Supplementary Table 4). Notably, we found that the upregulated ASD candidate genes were significantly associated with miR-137 predicted targets, suggesting that miR-137 may play essential roles in ASD by directly regulating the genes involved in ASD. Given the potential impact of miR-137 in ASD, we generated a predicted ASD-gene network to explore brain-specific interactions between ASD candidate genes, including the 15 upregulated and 16 downregulated DE proteins (Fig. 5g and Supplementary Fig. 9). In the predicted network, we found the upregulated miR-137-predicted targets potentially influence the adjacent ASD candidate gene expression in protein levels. For example, downregulated GRIA3 (Glutamate receptor subunit 3) and GABRB1 (Gamma-aminobutyric acid receptor subunit beta-1) have been reported in association with ASD33,34, and our results also showed significantly downregulation of GRIA3 and GABRB1 in miR-137–/– mice brain, which may partially explain the altered repetitive and social behaviors in mice upon loss of miR-137.
By overlapping with previously reported PSD/PSD95 core complex proteins35, we found a significant fraction of PSD/PSD95 core complex proteins were downregulated in miR-137–/– mice (Supplementary Fig. 10a). Many of these downregulated PSD/PSD95 core complex proteins are associated with ASD and schizophrenia (Supplementary Fig. 10b). Also, we found a sizeable number of PSD/PSD95 core complex genes could potentially be bound by miR-137 (Supplementary Fig. 10c). These results together suggest that the loss of miR-137 leads to the dysregulation of PSD/PSD95 core complex proteins.
miR-137 regulates its mRNA targets post-transcriptionally
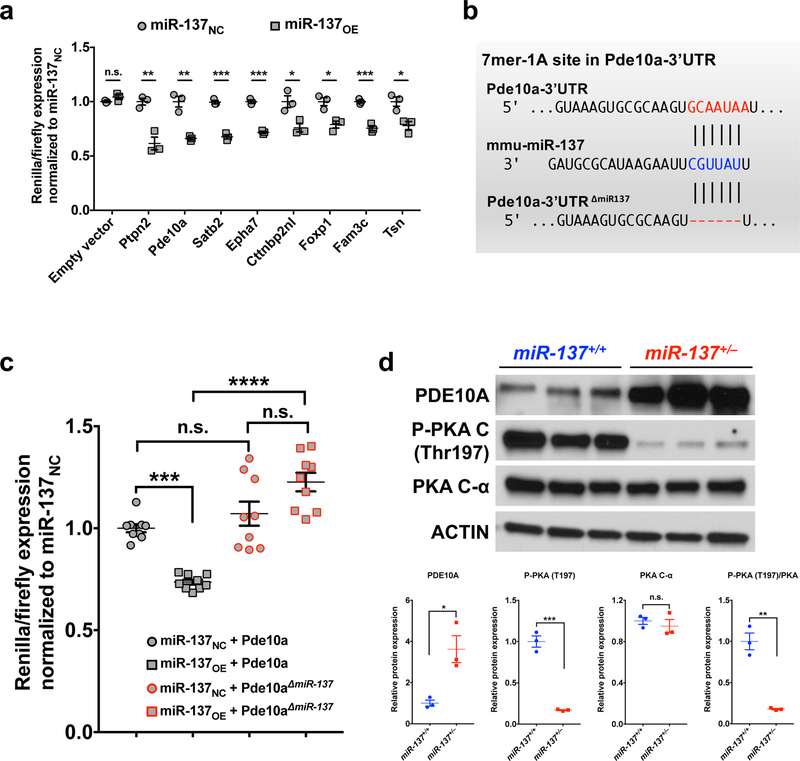
To investigate the miR-137-mediated regulation, we focused on the most 5 upregulated miR-137 targets, including Fam3c (Family with sequence similarity 3, member C), Pde10a (Phosphodiesterase 10a), Epha7 (Ephrin type-A receptor 7), Foxp1 (Forkhead box protein P1) and Shroom2 (Shroom family member 2), as well as 5 miR-137 targets that associate with neurological function/disorders, including Ptpn2 (Protein Tyrosine Phosphatase, Non-Receptor Type 2), Satb2 (Special AT-rich sequence-binding protein 2), Trim67 (Tripartite motif containing 67), Cttnbp2nl (Cortactin Binding Protein 2 N-terminal like), and Tsn (Translin) (Supplementary Fig. 11a). Using a dual luciferase reporter assay with the 3’-UTR of these genes, we found that 8 out of the 10 miR-137 predicted targets could be regulated by miR-137 in vitro (Fig. 6a). The decreased luciferase level was regulated by the overexpressed miR-137 rather than altered luciferase mRNA level (Supplementary Fig. 11b and Supplementary Table 1). To identify the critical genes regulated by miR-137, we examined the protein levels of the most changed targets identified by the dual luciferase assay (i.e., Ptpn2, Pde10a and Stab2), and found Pde10a showed a significant increase (Supplementary Fig. 11c), suggesting that the expression of Pde10a is sensitive to miR-137 expression.
Figure 6: PDE10A is a key mRNA target of miR-137.
a, Primary screen of miR-137 predicted targets in HEK293FT cells. 3’UTR-dependent luciferase assays were performed using both sh-control (miR-137NC), and sh-miR-137 (miR-137OE) for each of 10 miR-137 predicted targets. For each 3’UTR, luciferase expression was normalized (hRluc/hluc) to the miR-137NC control treatment. 8 out of 10 predicted targets were significantly regulated by miR-137 in vitro. n = 3 independent experiments; Data represent means ± s.e.m; tEmpty vector = 1.951, P = 0.1227; tPtpn2 = 5.854, P = 0.0042; tPde10a = 6.736, P = 0.0025; tSatb2 = 12.79, P = 0.0002; tEpha7 = 15.58, P = 0.0009; tCttnbp2nl = 3.631, P = 0.0221; tFoxp1 = 4.305, P = 0.0126; tFam3c = 9.047, P = 0.0008; tTsn = 3.890, P = 0.0177; Unpaired Two-Tailed t-test; n.s., nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
b–c, MiR-137 regulates the expression of Pde10a through the predicted binding site in the 3’UTR of Pde10a. (b) The miR-137 7mer-1A target site in the Pde10a-3’UTR as predicted by TargetScan was removed. (c) Co-transfection experiment was performed for the four plasmids, including “miR-137NC (miR-137 negative control)”, “miR-137OE (miR-137 overexpression)”, “Pde10a-3’UTR” and “Pde10a-3’UTR∆miR−137” constructs in HEK293FT cells (combination as indicated in the figure). All the other three samples were normalized to “miR-137NC + Pde10a-3’UTR”. There is no significant difference when directly comparing Pde10a-3’UTR and Pde10a-3’UTR∆miR−137 constructs. Pde10a-3’UTR-dependent expression of a luciferase reporter gene was suppressed by miR-137 overexpression. MiR-137-mediated suppression of luciferase was specific, as deletion of the miR-137 target site in the Pde10a-3’UTR (Pde10a-3’UTR∆miR−137) abolished repression by miR-137 overexpression. n = 9 independent experiments. Data represent means ± s.e.m; miR-137NC + Pde10a vs. miR-137OE + Pde10a: t = 4.511, P = 0.0009; miR-137NC + Pde10a vs. miR-137 NC + Pde10a∆miR−137: t = 1.22, P > 0.9999; miR-137OE + Pde10a vs. miR-137OE + Pde10a∆miR−137: t = 8.373, P < 0.0001; miR-137 NC + Pde10a∆miR−137 vs. miR-137OE + Pde10a∆miR−137: t = 2.642, P = 0.0856; Two-way ANOVA with Bonferroni post hoc test; n.s., nonsignificant; ***, P < 0.001; ****, P < 0.0001.
d, Partial loss of miR-137 resulted in a significant increase in PDE10A (relative to ACTIN), which associated with the reduced phosphorylation of PKA (P-PKA / PKA-Cα) without changing total PKA levels (n = 3 mice). Full-length blots are presented in Supplementary Fig. 15. Data represent means ± s.e.m; tPDE10A = 3.92, P = 0.0172; tp-PKA = 12.12, P = 0.0003; tPKA-Cα = 0.691, P = 0.5274; tP-PKA/PKA-Cα = 8.059, P = 0.0013; Unpaired Two-Tailed t-test. n.s., nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further confirm that miR-137 regulates the expression of Pde10a through the predicted binding site in the 3’UTR of Pde10a, we mutated Pde10a-3’UTR by deleting the miR-137 target site (Pde10a-3’UTRΔmiR−137) (Fig. 6b). We found that miR-137 overexpression led to significant gene downregulation in the presence of original psi-CHECK2-Pde10a-3’UTR but had a limited effect with psi-CHECK2-Pde10a-3’UTRΔmiR−137, validating that Pde10a is a specific target of miR-137 (Fig. 6c). We also assessed whether the Pde10a-3’UTR is, in fact, a target of the endogenous miR-137. To do so, we used mouse Neuro-2a cells, which contain high miR-137 expression levels (Supplementary Fig. 12a), to perform miR-137 loss-of-function studies. We found that endogenous miR-137 could significantly suppress the gene expression in the presence of original psi-CHECK2-Pde10a-3’UTR, but not with psi-CHECK2-Pde10a-3’UTRΔmiR−137 (Supplementary Fig. 12b). Also, Pde10a-3’UTR-dependent expression of a luciferase reporter gene was increased with miR-137 inhibition, suggesting that Pde10a is a specific target of endogenous miR-137.
Previous studies showed that PDE10A is enriched in brain and degrades cAMP (Cyclic adenosine monophosphate) and cGMP (Cyclic guanosine monophosphate), two critical signaling molecules that act as “second messengers” to trigger physiological changes36, and linked to psychiatric disorders37. We found that the increased Pde10a in miR-137+/– mice was associated with the reduced phosphorylation of cAMP-dependent protein kinase (P-PKA), which is directly regulated by the cAMP status in the cAMP/PKA signaling pathway, without changing total PKA protein levels (Fig. 6d).
Inhibition of PDE10A ameliorates the abnormalities associated with the partial loss of miR-137
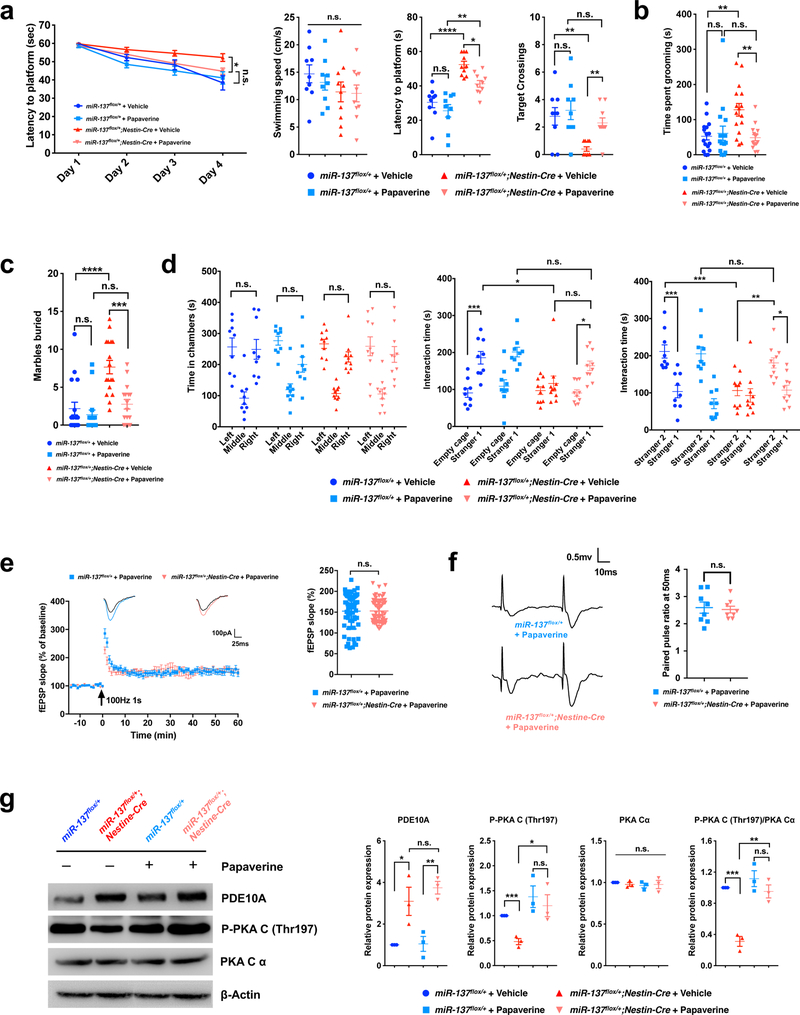
Inhibition of PDE10A has been shown to ameliorate both dopaminergic and glutamatergic dysfunction, which made PDE10A an attractive possible therapeutic38. Given the significant increase of Pde10a protein level upon the partial loss of miR-137, we thus reasoned that a PDE10A inhibitor could potentially be used to inhibit the increase of PDE10A activity and might ameliorate the impaired behaviors associated with the partial loss of miR-137. To test this, we applied a widely used PDE10A-specific inhibitor, papaverine38, in the behavioral assays discussed above. We found that, to a certain extent, papaverine could rescue the impaired spatial memory in the Morris water maze and Barnes maze test (Fig. 7a and Supplementary Fig. 13a). In the two repetitive behavioral assays, we found that in miR-137flox/+;Nestin-Cre mice, papaverine treatment resulted in significantly less time spent grooming (Fig. 7b) and fewer numbers of marbles buried (Fig. 7c), like those in miR-137flox/+ mice. We found that papaverine rescues the impaired social behaviors associated with the partial loss of miR-137 (Fig. 7d and Supplementary Fig. 13b). With papaverine treatment, although miR-137flox/+ and miR-137flox/+;Nestin-Cre mice showed no significant difference in preference to entering the left or right chamber during the habituation phase, miR-137flox/+;Nestin-Cre mice exhibited significantly more interaction time with introduced and novel mice. In the open field test, papaverine did not significantly change the locomotor activity and number of entries in the center zone (Supplementary Fig. 13c).
Figure 7: Inhibition of PDE10A ameliorates the abnormal behaviors associated with the partial loss of miR-137.
a, In the Mrris water maze test, the mean escape latency for the trained mice decreased over the course of the 4 learning days in all groups, but papaverine significantly improved the latency to locate the platform in miR-137flox/+;Nestin-Cre mice than in miR-137flox/+ mice (Day4: miR-137flox/+;Nestin-Cre + Vehicle vs. miR-137flox/+;Nestin-Cre + Papaverine: t = 2.831, P = 0.0321; miR-137flox/+ + Papaverine vs. miR-137flox/+;Nestin-Cre + Papaverine: t = 1.36, P > 0.9999; Data represent means ± s.e.m; Two-way ANOVA with Bonferroni post hoc test. n.s., nonsignificant; *, P < 0.05). In the spatial probe test performed on day 5, although papaverine did not significantly change the swimming speed (F3,34 = 1.053, P = 0.8466), it ameliorated the impaired latency to the platform (F3,34 = 19.93, P < 0.0001) and increased the number of target crossings (F3,34 = 6.532, P = 0.0013) in miR-137flox/+;Nestin-Cre mice. Data represent means ± s.e.m; miR-137flox/++Vehicle: n = 9 mice; miR-137flox/++Papaverine: n = 9 mice; miR-137flox/+;Nestin-Cre+Vehicle: n = 10 mice; miR-137flox/+;Nestin-Cre+Papaverine: n = 10 mice. One-way ANOVA with Bonferroni post hoc test. n.s., nonsignificant; *, P < 0.05; **, P < 0.01 ****, P < 0.0001.
b, In self-grooming test, papaverine resulted in significantly less time spent grooming in miR-137flox/+;Nestin-Cre mice (F3,59 = 5.96, P = 0.0013). Data represent means ± s.e.m; miR-137flox/++Vehicle: n = 16 mice; miR-137flox/++Papaverine: n = 16 mice; miR-137flox/+;Nestin-Cre+Vehicle: n = 16 mice; miR-137flox/+;Nestin-Cre+Papaverine: n = 15 mice. One-way ANOVA with Bonferroni post hoc test. n.s., nonsignificant; **, P < 0.01.
c, In the marble-burying test, papaverine ameliorated the impaired repetitive behaviors in miR-137flox/+;Nestin-Cre mice demonstrating by the improved number of marbles buried to the same level in miR-137flox/+ mice (F3,62 = 14.01, P < 0.0001). Data represent means ± s.e.m; miR-137flox/++Vehicle: n = 17 mice; miR-137flox/++Papaverine: n = 16 mice; miR-137flox/+;Nestin-Cre+Vehicle: n = 17 mice; miR-137flox/+;Nestin-Cre+Papaverine: n = 16 mice. One-way ANOVA with Bonferroni post hoc test. n.s., nonsignificant; ***, P < 0.001; ****, P < 0.0001.
d, In the three-chamber test, papaverine had no significant effect on the preference to the left or right chamber of miR-137flox/+ and miR-137flox/+;Nestin-Cre mice during the habituation phase. Left vs. right: miR-137flox/++Vehicle mice (n = 9 mice), t = 0.2517, P > 0.9999; miR-137flox/++Papaverine mice (n = 9 mice), t = 2.417, P > 0.9999; miR-137flox/+;Nestin-Cre+Vehicle mice (n = 10 mice), t = 1.397, P > 0.9999; miR-137flox/+;Nestin-Cre+Papaverine mice (n = 10 mice), t = 0.8727, P > 0.9999. In the subsequent task probing phase, the application of papaverine ameliorated the impaired sociability and social novelty in miR-137flox/+;Nestin-Cre mice, as indicated by the significantly increased interacting time with a mouse versus empty cage (miR-137flox/+;Nestin-Cre+Papaverine mice: tEmpty cage vs. Stranger1 = 3.705, P = 0.0119) or with novel mice (Stranger 2) versus familiar mice (Stranger 1) (miR-137flox/+;Nestin-Cre+Papaverine mice: tStranger2 vs. Stranger1 = 3.356, P = 0.0364). Data represent means ± s.e.m; Two-way ANOVA with Bonferroni post hoc test. n.s., nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
e, Papaverine rescued the impaired long-term potentiation (LTP) in miR-137flox/+;Nestin-Cre mice. Left panel, A typical experiment showing the time course of CA1 LTP for a single recording. fEPSP traces before and after are shown in the inset above. Pooled data are showing the time course of LTP from all recordings made from miR-137flox/+ or miR-137flox/+;Nestin-Cre mice. Right panel, Average LTP amplitude measured at 55–60 min post-induction. n = 6 slices from 4 mice per group; 10 fEPSP slope (%) values were collected from 1 slice. Data represent means ± s.e.m; t = 0.078, P = 0.9379; Unpaired Two-Tailed t-test; n.s., nonsignificant.
f, Increased paired-pulse facilitation (PPF) in papaverine-treated miR-137flox/+;Nestin-Cre mice. PPF studies across different interpulse intervals (20 ms, 50 ms, 100 ms, 200 ms and 400 ms) revealed that papaverine could restore the decreased paired-pulse ratio in miR-137flox/+;Nestin-Cre mice. Left panel, Representative recording of the paired-pulse ratio at the interpulse interval of 50 ms from the slices prepared from papaverine-treated miR-137flox/+ and miR-137flox/+;Nestin-Cre mice. Left panel, Paired-pulse ratio at an interval of 50 ms measured for up to 40 events for each recording in papaverine-treated miR-137flox/+ and miR-137flox/+;Nestin-Cre mice. n = 8 slice from 4 mice per group; Data represent means ± s.e.m; t = 0.2954, P = 0.7720; Unpaired Two-Tailed t-test; n.s., nonsignificant.
g, Partial loss of miR-137 resulted in a significant increase in PDE10A (relative to β-Actin), which associated with the reduced phosphorylation of PKA (P-PKA / PKA-Cα) without changing total PKA levels. After injecting papaverine, the reduced phosphorylation of PKA in miR-137flox/+;Nestin-Cre mice has resorted to the same level in miR-137flox/+ mice (n = 3 mice). Full-length blots are presented in Supplementary Fig. 15. Data represent means ± s.e.m; One-way ANOVA with Bonferroni post hoc test. Pde10a: F3,8 = 11.66, P = 0.0027; P-PKA: F3,8 = 6.085, P = 0.0184; PKA-Cα: F3,8 = 0.2374, P = 0.8679; P-PKA/PKA-Cα: F3,8 = 24.66, P = 0.0002. n.s., nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Also, papaverine could rescue the impaired LTP in miR-137flox/+;Nestin-Cre mice (Fig. 7e). The treatment of papaverine could lead to a significant increase of paired-pulse ratio at the interpulse interval of 50 ms in miR-137flox/+;Nestin-Cre mice (Fig. 7f). Consistent with the observation in gKO mice, we also found the partial loss of miR-137 in miR-137flox/+;Nestin-Cre resulted in a significant increase in PDE10A (compared to β-Actin), which is associated with reduced phosphorylation of PKA without changing total PKA levels (Fig. 7g). Papaverine reduced P-PKA in miR-137flox/+;Nestin-Cre mice to its normal level in miR-137flox/+ mice (Fig. 7g).
To examine whether papaverine could rescue the impaired non-behavioral phenotype in vitro, we directly treated the primary hippocampal neurons isolated from P0 littermate of miR-137flox/+ and miR-137flox/+;Nestin-Cre mice treated with papaverine. We found that papaverine could significantly reduce the dendritic complexity, dendritic length, and nodes of miR-137flox/+;Nestin-Cre neurons (Supplementary Fig. 13d–g). These data together suggest that PDE10A is a key mRNA target of miR-137 and potentially serves as a therapeutic target for the phenotypes associated with the loss of MIR137.
Knockdown of Pde10a rescues the deficits associated with the partial loss of miR-137
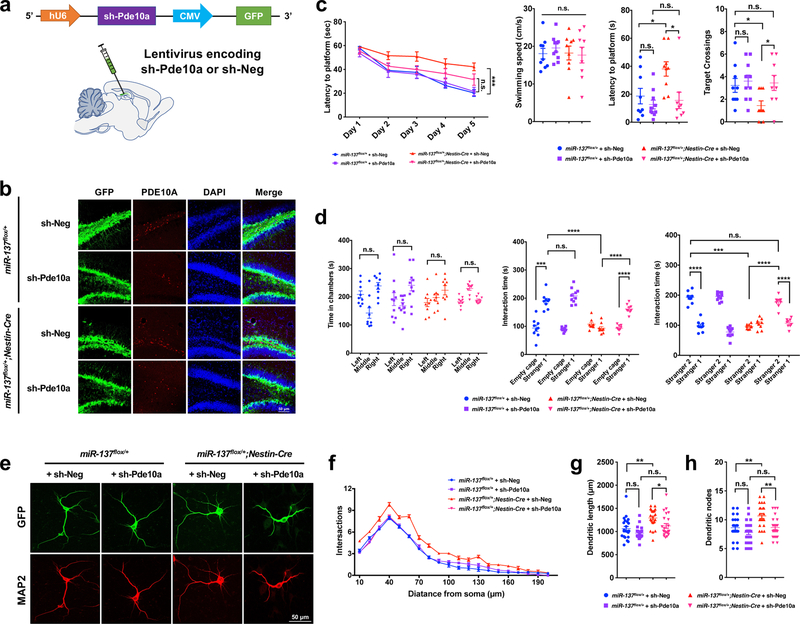
To strengthen the link between miR-137 and Pde10a, we generated the lentiviruses encoding shRNA targeting Pde10a (sh-Pde10a) and injected lenti-sh-Pde10a in adult miR-137flox/+ and miR-137flox/+;Nestin-Cre mice and assessed the impact of Pde10a knockdown on the impaired phenotypes associated with the partial loss of miR-137 (Fig. 8a–8b and Supplementary Fig. 14a).
Figure 8: Knockdown of Pde10a ameliorates the abnormal behaviors associated with the partial loss of miR-137.
a, Schemati illustration of the sh-Pde10a lentivirus constructs. Lentivirus encoding shRNA targeting Pde10a (sh-Pde10a) or negative control (sh-Neg) were injected into adult miR-137flox/+ and miR-137flox/+;Nestin-Cre mice 3 weeks before the behavioral assays.
b, The ICC staining by using PDE10A antibody revealed a high knockdown efficiency of sh-Pde10a. The experiment was repeated independently three time with similar results.
c, In the Morris water maze test, the mean escape latency for the trained mice decreased over the course of the 5 learning days in all groups, and sh-Pde10a resulted in the improved latency to locate the platform in miR-137flox/+;Nestin-Cre mice (Day5: miR-137flox/+ + sh-Pde10a vs. miR-137flox/+;Nestin-Cre + sh-Pde10a: t = 2.738, P = 0.2172; miR-137flox/+ + sh-Neg vs. miR-137flox/+;Nestin-Cre + sh-Neg: t = 5.873, P = 0.0003; Data represent means ± s.e.m; Two-way ANOVA with Bonferroni post hoc test. n.s., nonsignificant; ***, P < 0.001). In the spatial probe test performed on day 6, although sh-Pde10a did not change the swimming speed (F3,33 = 0.2683, P = 0.8478), it significantly ameliorated the impaired latency to the platform (F3,33 = 5.377, P = 0.0040) and increased the number of target crossings (F3,33 = 3.073, P = 0.0411) in miR-137flox/+;Nestin-Cre mice. Data represent means ± s.e.m; miR-137flox/++sh-Neg: n = 9 mice; miR-137flox/++sh-Pde10a: n = 10 mice; miR-137flox/+;Nestin-Cre+sh-Neg: n = 9 mice; miR-137flox/+;Nestin-Cre+sh-Pde10a: n = 9 mice. One-way ANOVA with Bonferroni post hoc test. n.s., nonsignificant; *, P < 0.05.
d, In the three-chamber test, sh-Pde10a had no significant effect on the preference to the left or right chamber of miR-137flox/+ and miR-137flox/+;Nestin-Cre mice during the habituation phase. Left vs. right: miR-137flox/++sh-Neg mice (n = 9 mice), t = 1.446, P > 0.9999; miR-137flox/++sh-Pde10a mice (n = 10 mice), t = 2.464, P > 0.9999; miR-137flox/+;Nestin-Cre+sh-Neg mice (n = 9 mice), t = 2.079, P > 0.9999; miR-137flox/+;Nestin-Cre+sh-Pde10a mice (n = 9 mice), t = 0.2697, P > 0.9999. In the subsequent task probing phase, the application of sh-Pde10a ameliorated the impaired sociability and social novelty in miR-137flox/+;Nestin-Cre mice, as indicated by the significantly increased interacting time with a mouse versus empty cage (miR-137flox/+;Nestin-Cre+sh-Pde10a mice: tEmpty cage vs. Stranger1 = 6.742, P < 0.0001) or with novel mice (Stranger 2) versus familiar mice (Stranger 1) (miR-137flox/+;Nestin-Cre+sh-Pde10a mice: tStranger2 vs. Stranger1 = 9.225, P < 0.0001). Data represent means ± s.e.m; Two-way ANOVA with Bonferroni post hoc test. n.s., nonsignificant; ***, P < 0.001; ****, P < 0.0001.
e-h, sh-Pde10a could rescue the impaired neuronal phenotype in vitro. (e) Primary hippocampal neurons were isolated from P0 littermate of from miR-137flox/+ and miR-137flox/+;Nestin-Cre mice (n = 3 mice). After infecting lentivirus encoding sh-Pde10a, we confirmed its efficiency with GFP and immunocytochemistry staining of MAP2 were performed at DIV 7. The experiment was repeated independently three time with similar results. (f) sh-Pde10a reduced dendritic complexity in miR-137flox/+;Nestin-Cre neurons compared with controls, as determined by Sholl analysis (Data represent means ± s.e.m; Interaction: F57,1340 = 3.286, P < 0.0001; Distance: F19,1340 = 795.4, P < 0.0001; Treatment: F3,1340 = 177.5, P < 0.0001. Two-way ANOVA with Bonferroni post hoc test.). sh-Pde10a significantly reduced the dendritic length (miR-137flox/+;Nestin-Cre mice: tsh-Neg vs. sh-Pde10a = 2.822, P = 0.0354) (g) and the number of dendritic nodes (miR-137flox/+;Nestin-Cre mice: tsh-Neg vs. sh-Pde10a = 3.485, P = 0.0046) (h) in miR-137flox/+; Nestin-Cre neurons. n = 23 slice from 3 mice per group. Data represent means ± s.e.m; Two-way ANOVA with Bonferroni post hoc test. n.s., n onsignificant; *, P < 0.05; **, P < 0.01.
In the Morris water maze test, the mean escape latency for the trained mice decreased over the course of the 5 learning days in all groups, and sh-Pde10a resulted in the improved time in miR-137flox/+;Nestin-Cre mice took to find the platform. In the probe trials, although sh-Pde10a did not change the swimming speed, it significantly improved the impaired latency to the platform and increased the number of target crossings in miR-137flox/+;Nestin-Cre mice (Fig. 8c). In the 3-chamber test, we found Pde10a knockdown significantly improved the impaired sociability and social novelty in miR-137flox/+;Nestin-Cre mice, without influencing their choice of the left or right chamber during the habituation phase (Fig. 8d). In the self-grooming test, although not statistically significant, sh-Pde10a resulted in less time spent grooming in miR-137flox/+;Nestin-Cre mice in self-grooming test (Supplementary Fig. 14b). In the marble-burying test, sh-Pde10a ameliorated the impaired repetitive behaviors in miR-137flox/+;Nestin-Cre mice to the same level of miR-137flox/+ mice (Supplementary Fig. 14c). In the open field test, sh-Pde10a did not change the moving distance among the groups, but significantly ameliorated the anxiety-like behavior in miR-137flox/+;Nestin-Cre mice (Supplementary Fig. 14d).
Our earlier data indeed showed that papaverine resulted in the impaired dendritic growth of both miR-137flox/+ and miR-137flox/+;Nestin-Cre neurons, which may occur due to the toxicity of papaverine solution (Supplementary Fig. 13e–g). To confirm that Pde10a plays a role in dendritic growth, we infected primary cultured miR-137flox/+ and miR-137flox/+;Nestin-Cre neurons with sh-Pde10a lentivirus (Fig. 8e). We found that knockdown of Pde10a rescued the impaired dendritic complexity and growth in miR-137flox/+;Nestin-Cre neurons, but not in miR-137flox/+ neurons (Fig. 8f–h). Also, we observed that knockdown of Pde10a could ameliorate the increased spine numbers of PSD-95 and Synaptophysin in miR-137flox/+;Nestin-Cre neurons to the similar level in miR-137flox/+ neurons (Supplementary Fig. 14e). Together, these data provide the direct evidence that increased expression of Pde10a is responsible for the observed abnormalities.
Discussion
Genome-wide association studies (GWAS) have identified multiple loci that are likely involved in neuropsychiatric disorders, including schizophrenia, ASD, bipolar disorder, and depression7,10,11,39,40. Among them, MIR137 was found significantly associated with schizophrenia and ASD. Also, in 5 independent studies, 12 individuals carrying a microdeletion in chromosome 1p21.3 reported with a 92% incidence of ASD and 100% incidence of intellectual disability (ID)11,17–20. The minimal overlapping region of their deletions on chromosome 1p21.3 only includes the MIR137 gene, indicating that miR-137 mediates the core clinical features of ASD in patients carrying 1p21.3 deletion. Furthermore, gene expression analyses found that the overexpression of MIR137 could be associated with schizophrenia15, while the loss of MIR137 is likely associated with ASD11. Here we present the first characterization of miR-137 knockout mice at molecular, cellular, electrophysiological, and behavioral levels. Our results suggest that miR-137 plays important roles in postnatal neurodevelopment and provides the direct evidence to support that the loss of miR-137 potentially causes ASD.
We observed that the loss of miR-137 significantly influenced the expression of many known ASD genes. Particularly the upregulated ASD genes were significantly enriched with miR-137 predicted targets, such as Foxp1, Nfia, Satb2, and Tsn. For these genes, we only observed the changes at the protein level, but not at RNA level. Given the critical roles of these genes in neurodevelopment, our results suggest important roles of miR-137 in posttranscriptional gene regulation in ASD. In a previously published study focusing on MIR137 gain-of-function, Siegert et al. found the mRNAs and proteins of Cplx1, Nsf, Syn3, and Syt1 were consistently downregulated upon overexpression of miR-137 in vitro and in vivo15. In our RNA-seq and proteomics analyses, however, we did not observe significant changes of these genes, suggesting that it is possible that the overexpression of miR-137 could target additional mRNAs compared to endogenous miRNA. Moreover, both our and their results15 suggest that either the miR-137 loss-of-function or gain-of-function could result in impaired learning and memory, indicating that the miR-137-mediated regulation during neurodevelopment could be dosage-dependent.
PDE10A is one of the miR-137 mRNA targets that is previously linked to neuropsychiatric disorders37. The inhibition of PDE10A can ameliorate both dopaminergic and glutamatergic dysfunction, suggesting its attractive therapeutic potential in treating neuropsychiatric disorders38. Papaverine is a widely used selective inhibitor of PDE10A. Administration of papaverine produced an increase in striatal tissue levels of cGMP and an increase in extracellular cAMP. These effects of papaverine on biochemical and behavioral assays were absent in PDE10A knockout mice, indicating specific inhibitory effects of papaverine on PDE10A activity41. Given that the PDE10A protein level is significantly elevated in the heterozygous miR-137 KO mice, we conducted several social deficit rescue experiments, and our results demonstrate that papaverine could indeed restore the impaired repetitive behaviors and sociability/social novelty in miR-137flox/+;Nestin-Cre mice. Also, we found that papaverine can partially ameliorate learning and memory deficits as well as repair neuronal morphological alterations associated with the partial loss of miR-137. These significant rescue effects suggest that PDE10A inhibition efficiently improves those behavioral abnormalities found in miR-137flox/+;Nestin-Cre mice. An earlier clinical trial using a selective PDE10A inhibitor for the treatment of schizophrenia failed42. Rather, our results presented here suggest that PDE10A inhibitors should be targeted to a subgroup of neuropsychiatric disorders associated with the partial loss of MIR137. Notably, given that miR-137 has highly pleiotropic functions, it may be difficult to treat patients using only single-drug therapies aimed at specific pathways.
In summary, we show here that the partial loss of miR-137 leads to the molecular, cellular, electrophysiological, and behavioral abnormalities associated with ASD. Given the known genetic association between MIR137 and neuropsychiatric disorders, our data provide the first direct evidence that the dysregulation of miR-137 directly contributes to the molecular pathogenesis of neuropsychiatric disorders.
Methods
Ethics statement
All the animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Institute of Zoology Chinese Academy of Sciences and Emory University.
Mice
All mice used were from the 129S6/SvEvTac genetic background. We designed a targeting vector to disrupt the Mir-137 gene via homologous recombination in mouse embryonic stem cells, where two loxP sites were inserted upstream (~2 kb) and downstream (~0.6 kb) of the Mir137 gene. By crossing with either Zp3-Cre or Nestin-Cre line, we were able to specifically delete Mir137 in the germline or central and peripheral nervous system and generate the heterozygous global knockout (gKO) and conditional knockout (cKO) mice. We then crossed heterozygous mice to generate miR-137 wild-type (miR-137+/+ or miR-137flox/+), heterozygous (miR-137+/– or miR-137flox/+;Nestin-Cre), and homozygous (miR-137–/– or miR-137flox/flox;Nestin-Cre) mice. Genotyping occurred using tail DNA, and the specific primers are shown in Supplementary Table 1. Kaplan-Meier analysis was performed, and survival curves were generated using Prism 7 software (GraphPad).
Timed mating was performed by crossing heterozygous × heterozygous. The embryos were collected at embryonic day 15.5 (E15.5). Postnatal mice at postnatal day 0 (P0) were collected for morphology analysis. Mice at P14 were used for histological analysis. Major organ tissues, such as heart, liver, lung, and kidney from miR-137 gKO mice were dissected immediately after the animals were euthanized, and then fixed in 4% (mass/vol) paraformaldehyde (PFA) at 4°C overnight, stored in 70% (vol/vol) ethanol, and embedded in paraffin. Paraffin blocks were sectioned at 5-μm. After deparaffinization, the sections were stained according to hematoxylin and eosin (H&E) stained procedure. H&E staining was performed on tissues from at least three mice for each genotype.
Immunocytochemistry (ICC)
ICC staining occurred as described previously13. In brief, cells were first preblocked using DPBS containing 5% normal goat serum (Vector Laboratories) and 0.1% Triton X-100 for 30 minutes, followed by overnight incubation with primary antibodies: mouse microtubule-associated protein 2 (MAP2, 1:400; MAB3418; Millipore), mouse postsynaptic density protein 95 (PSD-95; 1:500; ab2723; Abcam), or mouse synaptophysin (1:300; ab8049; Abcam). After washing with DPBS, cells were incubated with secondary antibodies that included goat anti-mouse Alexa Fluor 568 (1:500; A11031; Invitrogen) or goat anti-mouse Alexa Fluor 488 (1:500; A11001; Invitrogen), followed by counterstaining with DAPI (D9542; Sigma-Aldrich). Coverslips were then mounted on slides with Mowiol mounting medium.
Immunohistochemistry (IHC)
Four pairs of littermates of P18 cKO mice were anesthetized with 2.5% Avertin (200 mg/kg body weight) and transcardially perfused with cold PBS followed by 4% paraformaldehyde in phosphate buffer. The brains were removed and postfixed in 4% paraformaldehyde overnight, and then soaked overnight in 30% sucrose in PBS. Finally, the brains were sectioned into 40-μm thickness using a Leica Sliding Microtome (SM2010 R). IHC staining of PSD-95 (1:500; ab2723; Abcam) and synaptophysin (1:300; ab8049; Abcam) was carried out on 1-in-3 40-μm thick floating sections containing hippocampus and cortex from 4 pairs of littermates. The IHC staining was also performed in P18 gKO miR-137+/+ and miR-137–/–mice, and 8-week-old miR-137+/+ and miR-137+/– mice.
Golgi staining
Golgi staining was performed as described previously 43. In brief, brains from 4 pairs of independent littermate miR-137 cKO mice were harvested whole and rinsed with double distilled water. Brains were then incubated in a 1:1 mixture of FD Solution A:B for 2 weeks at room temperature in the dark. Brains were then transferred into FD Solution C and stored in the dark at room temperature for 48 hours. Coronal sections (200μm) were cut with a Leica CM1950 cryostat and mounted on 3% gelatin-coated slides. Staining procedures were followed according to the manufacturer’s protocol (FD NeuroTechnologies), and slides were dehydrated in ethanol and mounted with Permount medium. Dendrites were traced, and their lengths were measured using the Simple Neurite Tracer plugin of Fiji. Spines were counted on secondary and tertiary branches of apical and basal dendrites in the CA1 hippocampal region individually.
Image analysis
Z-stack images (at 0.3-μm intervals) were obtained on a ZEISS 710 confocal laser-scanning microscope with an X63 oil-immersion lens at an electronic zoom factor of 1 or 2. Image analyses and quantification were performed using ImageJ software.
Behavioral assays
Mice were housed in groups of 3–5 animals on a 12:12H light: dark cycle. For behavioral tests, mice were moved to the testing room 24 hours before behavioral testing for acclimation. All experimental areas were cleaned with 70% ethanol before the tests and between subjects. All experiments occurred during the light phase. Data were collected blindly using the software Smart V3.0.03 (Panlab, Barcelona, Spain). Only 2- to 3-month-old male mice obtained from at least two independent cohorts were used for behavioral testing.
Morris Water maze test.
A circular water tank (diameter 120 cm) was filled with water, and the water was made opaque with nontoxic white paint. A round platform (diameter 13 cm) was hidden 1 cm beneath the surface of the water at the center of a given quadrant of the water tank. Mice received training in the Morris water maze for 4 successive days, and each session consisted of 4 trials. For each trial, the mouse was released from the wall of the tank and allowed to search, find, and stand on the platform for 20 seconds within the 60-second trial period. For each training session, the starting quadrant and sequence of the 4 quadrants from where the mouse was released into the water tank were randomly chosen, such that it was different among the separate sessions for each animal and was different for individual animals. A probe test was conducted 24 hours after completion of the training. During the probe test, the platform was removed from the pool, and the task performances were recorded for 60 seconds. Mice in the water pool were recorded using a video camera, and the task performances, including swimming tracks, speed, and time spent in each quadrant were recorded for analysis.
Barnes Maze Task.
The Barnes maze platform is circular, 122 cm in diameter, and has 20 evenly spaced holes (5 cm diameter, 2 cm away from the edge), with only one hole leading to a removable hiding box that is situated directly below the escape hole on the maze top. Testing begins on the first day by habituating the mice in the hiding box for one minute. Following this, four consecutive days of acquisition trials with four trials per day in which mice were allowed 5 min per trial to find the hiding box. Mice that did not find the hiding box by the end of the 5 min were gently guided to the escape hole by the investigator. Mice were returned to their home cage for 30 minutes between trials. A 60-sec probe trial (no hiding box) was performed the day following the last day of acquisition. Animal movements in the maze were digitally recorded.
Self-grooming test.
Mice were placed in a new Plexiglas cage with fresh bedding without nesting or cardboard material. Self-grooming behavior was recorded for 10 min. Cumulative time spent grooming, and the number of bouts was scored for each mouse.
Marble-burying task.
Mice were individually placed in Plexiglas cages containing 5-cm-deep fresh bedding, followed by gently overlaying 20 black glass marbles (15 mm diameter) equidistant in a 4 × 5 arrangement. Testing was conducted for 30 min. After the test period, unburied marbles were counted. Marbles were considered buried if they were at least one half covered with bedding.
Open-field test.
The open field was made of plywood (painted white) sized 72 cm length × 72 cm width × 36 cm height. A central square (18 × 18 cm) was drawn in the middle of the open field and was used to measure exploration. Mice were placed in one corner of the open field apparatus and could explore the apparatus for 30 minutes while a video camera recorded their movements.
Social interaction test.
Social interaction assay was performed as described previously 44. Briefly, an adult miR-137flox/+ or miR-137flox/+;Nesting-Cre male mouse was placed with an unfamiliar juvenile male C57BL/6J mouse in a clean neutral Plexiglas cage (325 × 210 × 180 mm). The 10-minute session of social behaviors (nose-to-nose sniffing, nose-to-anogenital sniffing, self-grooming, following, crawling over or under the partner’s body) of target mice were recorded and analyzed by an observer who was uninformed of the genotypes.
Three-chamber social test.
Sociability testing occurred in a rectangular, three-chambered box as previously described45. Each chamber was 20 × 40 × 22 cm, and the dividing walls were made from clear Plexiglas, with small square openings (5 × 5 cm) allowing access into each chamber. The test animal was placed in the middle chamber and left to freely explore the test arena with an empty cage in each side chamber for the 10 min habituation period. Then, the mouse was led to the middle chamber, and the openings in the apparatus were closed. An unfamiliar C57BL/6J male (Stranger 1) was then placed in a wire cage in one of the side chambers together with an empty wire cage in the other side chamber. The doors between chambers were then removed, and the test animal freely explored all three chambers over a 10 min session. Following this, a novel stranger C57BL/6J mouse (Stranger 2) was placed in the previously empty wire cage, and the subject mouse was allowed to explore for 10 min. Animal movements were digitally recorded.
Social discrimination test.
Two weeks before testing, subject animals and stimulus animals (female C57BL/6 mice aged 3–4 months) were housed alone in individual clean cages in the testing room. The social discrimination test was performed according to published protocols with minor modifications23,24. Testing took place in a novel, clean box (72 cm length × 72 cm width × 36 cm height) during the light phase. The subject mouse was habituated in the testing box for 5 minutes. After habituation, one stimulus female was introduced into one of two transparent bottles having holes throughout (9 cm in diameter, 12 cm high), and interactions between the mice were videotaped for 5 minutes. The experimental animal was then returned to its home cage. Thirty minutes later another unknown female was placed in the other bottle in the testing box; the experimental animal was placed back into the testing box, facing to both the familiar and the novel female at the same distances. Interactions of the subject mouse with the familiar and the novel female were also videotaped for 5 minutes. The amount of time spent investigating each stimulus female and the entries into each stimulus female zone were recorded and analyzed.
Drug treatment in the animals.
The PDE10A inhibitor, papaverine, was purchased from Sigma-Aldrich (Cat# P3510; Papaverine hydrochloride; powder form). On test mornings, mice received papaverine (30 mg/kg, dissolved in ddH2O with 2% Tween 80 and 0.5% hydroxyethylcellulose) or vehicle (2% Tween 80 and 0.5% hydroxyethylcellulose in ddH2O) intraperitoneal injection. Behavioral testing occurred 30 minutes after papaverine treatment.
Drug treatment in the primary hippocampal neuron.
Primary neurons were prepared as described previously 46. Primary hippocampal neurons were isolated from P0 newborn littermates of miR-137flox/+ and miR-137flox/+;Nesting-Cre mice and cultured on poly-D-lysine-coated glass coverslips with a density of 5 × 104 cells per well in a 24-well plate. Cultures were exposed to a medium containing papaverine (1 × 10–5 M) on day 4 and throughout the incubation period.
Electrophysiology
Long-term potentiation (LTP).
Four-week-old miR-137flox/+;Nesting-Cre mice and their wild-type littermates were used for recording hippocampal LTP. Hippocampal slices were prepared as previously described 47. The brain, including the two hippocampi, was removed into an ice-cold artificial cerebrospinal fluid (ACSF) containing the following components (in mM): 125 NaCl, 3.25 KCl, 1.5 MgCl2, 1.25 NaH2PO4, 25 NaHCO3, 10 glucose, 2.5 CaCl2, saturated with 95% O2/5% CO2. Next, the brain was positioned on the stage of a vibratome slicer (Campden instruments, 7000smz) and cut to obtain 400μm thick transverse hippocampal slices. These slices were kept continuously oxygenated for at least 1 h before use. All the experiments were carried out at room temperature 22–25 °C. Field excitatory postsynaptic potentials (fEPSPs) were recorded in the CA1 region of the hippocampus. A bipolar concentric stimulating electrode (FHC Inc., Bowdoin, ME) was placed in the Schaffer collaterals to deliver test and conditioning stimuli. A glass recording electrode (3–4MΩ) filled with ACSF were positioned in stratum radiatum of CA1. fEPSPs in CA1 were induced by test stimuli at 0.033 Hz with an intensity that elicited a fEPSP amplitude of 40–50% of maximum. Test responses were recorded for 30–40 min before beginning the experiment, to ensure the stability of the response. After at least 15 min of stable baseline recording, LTP was induced by 100 Hz tetanic stimulation (1 train of 100 Hz for 1s) at baseline stimulation intensity. The fEPSP signals were digitized using the Digidata 1440A interface board. The data were sampled at 10 kHz and filtered at 2 kHz. Traces were obtained by pClamp 10.6 and analyzed using the Clampfit 10.6 (Axon Instruments, Foster City, CA).
Paired-pulse facilitation (PPF).
4-week-old miR-137flox/+;Nesting-Cre mice and their wild-type littermates were used for the PPF experiment. The hippocampal slices were treated as described above. Extracellular stimuli were administered on the border of areas CA3 and CA1 along the Schaffer-collaterals using a bipolar concentric stimulating electrode (FHC Inc., Bowdoin, ME). fEPSPs were recorded in stratum radiatum with an ACSF-filled glass recording electrode (1–3MΩ). All test stimuli were 100μs in duration and 1/2–2/3 maximal stimulation strength. fEPSPs in CA1 were induced by test stimuli at 0.05 Hz with different interstimulus intervals (ISIs) of 20, 50, 100, 200, and 400 ms. Traces were obtained by pClamp 10.6 and analyzed using the Clampfit 10.6 (Axon Instruments, Foster City, CA).
The balance of excitation and inhibition (E/I ratio).
The procedures for the preparation and recording of hippocampal slices were described previously48. Briefly, the brain of 4-week-old miR-137f/+;Nesting-Cre mice and their wild-type littermates were quickly dissected and transferred to ice-cold ACSF saturated with 95% O2/5% CO2 and sliced to 360 μm sagittal slices. The slices were stored at 32°C for at least 2 h before a single slice was transferred to the recording chamber. ACSF contained (in mM): 120 NaCl, 3.0 KCl, 1.0 NaH2PO4, 26 NaHCO3, 11 D-glucose, 2.0 CaCl2, and 1.2 MgSO4. The stimulation electrode was placed near the stratum pyramidal layer of the CA1 area to execute 0.1Hz stimulation to obtain evoked synaptic responses. Whole-cell recordings occurred under the voltage clamp mode with a holding potential of –70 mV except in those to construct I/V curves of synaptic currents. Series resistance was monitored by a –3 mV step throughout the entire experiment of whole cell access, and if it fluctuated more than 20%, the data were excluded from the analysis. For the E/I ratio experiments, slices were first perfused by ACSF to record total postsynaptic current (PSC) for 10 min, then 10 μM NBQX/50 μM APV was added to specifically record inhibitory postsynaptic current (IPSC). Then, 100 μM picrotoxin was added to verify the inhibitory response. The E/I ratio was calculated as (PSC-IPSC)/IPSC.
RNA isolation, quantitative RT-PCR, and RNA-seq
Mouse brain samples were homogenized in Trizol (Invitrogen) and processed according to the manufacturer’s instructions. RNA was then reverse-transcribed using a SuperScript® III First-Strand Synthesis System (Invitrogen). qPCR quantified cDNA via TaqMan assay (Invitrogen). Each reaction was run in triplicate and analyzed following the ∆∆Ct method using glyceraldehyde-3-phosphate dehydrogenase (Gapdh) as a normalization control. Mature miR-137 was quantified as described in our previous work 13. Luciferase mRNA was examined followed by the Universal SYBR Green Quantitative PCR Protocol. All primer information is listed in Supplementary Table 1.
Four μg of total RNA (from P12 mice cortical tissues) was used for RNA-seq library construction following instructions of the Illumina mRNA sample prep kit (Cat# RS-100–0801). In brief, the poly-A–containing mRNA was purified using poly-T oligo-attached magnetic beads. The mRNA was then fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into the first-strand cDNA using reverse transcriptase and random primers. Second-strand cDNA synthesis followed using DNA Polymerase I and RNaseH. These cDNA fragments underwent an end-repair process, the addition of a single “A” base, and then ligation of the adapters. These products were gel purified and enriched with PCR to create the final cDNA libraries. The library constructs were run through the bioanalyzer to verify size and concentration before sequencing on the Illumina HiSeq2000 machines. In total, four biological replicates were carried out for each condition. Short read alignment, transcript expression estimation, and differential analysis were carried out using the Tophat2 (v2.1.1) and Cufflinks (v2.2.1) pipelines. The short reads were aligned against a reference gene database (Ensembl: Mus musculus, NCBIM37.62) without attempting to assemble novel transcripts. Both bias detection (–frag-bias-correct) and multi-read correction (–multi-read-correct) options were used to obtain a more accurate estimate of transcript abundance. A gene was considered differentially expressed with a q-value < 0.05.
Quantitative analysis of proteome by multiplexed TMT-LC/LC-MS/MS
Protein extraction, digestion, labeling, and pooling.
Quantitative proteomics analysis was performed by the TMT strategy as described previously49. In brief, the P12 mouse cortical tissues were dissected and immediately homogenized in buffer (8M urea, 50 mM Hepes, pH8.5) on ice within 1 min. After quantification by BCA assay (Thermo Fisher Scientific) and protein integrity evaluation by SDS-PAGE, 200 μg protein aliquots from each sample were adjusted to 4 μg/μl by digestion buffer supplemented with 10% acetonitrile and 4 mM Dithiothreitol. The digestion was performed in the presence of 2 μg Lys-C (Wako, 1:100 w/w) at room temperature for 2 h, and then diluted 4 times by 50 mM HEPES, pH 8.5, without urea before 4 μg trypsin (Promega, 1:50 w/w) was added. After that, digestion continued for 4 more hours at room temperature before quenching with 1% trifluoroacetic acid. The following peptide desalting, TMT labeling, and efficiency testing procedures were performed according to our previous report49.
Offline peptide fractionation by basic pH reverse phase liquid chromatography.
The equally mixed TMT-labeled peptides from all samples were desalted and then fractionated on a C18 column (XBridge, 3.5 μm particle size, 4.6 mm × 25 cm, Waters) at a flow rate of 0.4 ml/min. 15% to 55% gradient of buffer B (95% acetonitrile, 10 mM Ammonium formate, pH 8) created by buffer A (buffer B omitting acetonitrile) for 60 minutes with adjusted time intervals to achieve requal amount of peptides in each fraction. About 60 fractions were finally collected.
Long gradient acidic pH reverse phase liquid chromatography coupled with tandem MS.
Each fractionated peptide sample 5% formic acid was run individually and sequentially without concatenation. The reverse phase column (100 µm I.D., 60 cm long) was packed by 1.9 µm C18 resins (Dr. Maisch GmbH, Germany) and was heated by 65°C50. Peptides were eluted by buffer B (65% acetonitrile, 0.2% formic acid) with 25%~30% gradient change in 2 h at a flow rate of 0.15 µl/min in the presence of 5% DMSO. The specific starting and ending buffer B concentrations were determined by the LC chromatography of the previous run of the sample fraction next to the current sample to be analyzed. The mass spectrometry analyzer was Q Exactive hybrid quadrupole-Orbitrap (Thermo Fisher Scientific), which was operated in data-dependent mode with a survey MS scan (60,000 resolution, 2 × 105 AGC target and 50 ms maximal ion time) followed by MS/MS scans [60,000 resolution, 1 × 105 AGC target, 150 ms maximal ion time, HCD, 38% collision energy (stepped: 5%), 1.2 m/z isolation window and 0.25 m/z offset, and 45 s dynamic exclusion].
Peptide identification and quantification by JUMP software.
The MS data analysis was performed by our JUMP software (version 1.0.55) with improved sensitivity and specificity51. The acquired MS/MS raw files were first converted into the mzXML format and then searched by JUMP against the integrated target database from Uniprot (mouse, 52,490 protein entries) with a decoy database (reversed sequences of all proteins in the target database)52. Specific parameters included full trypticity, two maximal miscleavages, 10 ppm mass tolerance for both precursor ion and product ions, static mass addition (+229.16293) on lysine and N-terminus for TMT-tag, and the assignment of a, b, and y ions. Crude peptide-spectrum matches were first analyzed to determine the mass accuracy and shift; then filtered using 4 standard deviations (~2 ppm) according to the results of doubly charged peptides with Jscore of ≥ 30.0. The cleaned matches were grouped by precursor ion charge state and further filtered by Jscore and dJn values. The cutoff values for these values were adjusted until a protein false discovery rate (FDR) lower than 1% was achieved. If one peptide was matched to multiple proteins, the peptide was represented by the protein with the highest peptide-spectrum matches (PSM) according to the rule of parsimony. The peptide and protein quantification steps included: (i) extract TMT reporter ion intensities from each PSM; (ii) correct the distorted raw intensities due to isotopic distribution of each TMT tag (e.g., TMT126 also generates 7.9% and 0.3% signals onto TMT127 and TMT128 in addition to 91.8% on its own.); (iii) exclude very low intensity PSMs (e.g. minimum intensity of 1,000 and median intensity of 5,000); (iv) normalize the sample loading bias with the trimmed median intensity of all PSMs; (v) calculate the mean-centered intensities across samples (e.g. relative intensities between each sample and the mean), using all PSMs with removal of outliers detected by Dixon’s Q-test or generalized extreme Studentized deviate test; (vi) average all corresponding PSMs of a protein; (vii) finally calculate protein absolute intensities by multiplying the intensity average by the grand-mean of three most highly abundant PSMs53.
Identification of differentially expressed (DE) protein.
In total, 11,729 unique proteins at 1% FDR (0.1% unique peptide FDR and 0.05% PSM FDR). We showed the confidence intervals of the identified peptides in three aspects: i) Matching scores: the score distribution of accepted PSMs is vastly different from that of decoy PSMs; ii) Mass accuracy: almost all PSMs have precursor mass error within ± 4 ppm; and iii) De novo amino acid tags: almost all (99.9%) of accepted PSMs are supported by tags, with the median tag length of 6 AAs. These results strongly support the reliability of the 11,729 identified proteins. Besides the above stringent criteria, we further required a minimum of 9 PSMs for each protein, resulting in only 7,810 proteins for subsequent TMT-based quantification. No decoy protein passed the threshold of 9 PSMs, resulting in 0 FDR for this set of 7,810 proteins. However, we recognized that the FDR estimation might not be accurate and further calculated the standard deviation of the FDR using Target-decoy method54. The actual FDR falls between 0 and 0.07% within a 99% confidence interval. For each pairwise comparison (e.g., KO vs. WT), we required a consistent trend of significant changes in both experiments and defined differentially expressed (DE) proteins with the following criteria: (i) absolute z-score ≥1.96 (that corresponds to a P-value < 0.05) in both experiments; (ii) the same trend in both experiments: the signs of z-scores should be consistent. This procedure resulted in 417, 94, and 76 DE proteins, when comparing KO vs. WT, Het vs. WT, and KO vs. Het, respectively.
Integrated bioinformatics analyses
Gene ontology (GO) analysis was performed by Database for Annotation, Visualization and Integrated Discovery (DAVID v6.8)55. The WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) was used for additional GO analysis56. GeneMANIA57 and Pathway Commons58 were used to predict the function and interaction of the differentially expressed genes. Predicted ASD-associated gene interaction networks were performed by genome-wide predictions of autism-associated genes (http://asd.princeton.edu/)). For Sylamer analysis28, 7,810 identified proteins were ranked from upregulated to downregulated in miR-137–/– cortex versus miR-137+/+ cortex. Sylamer software (https://www.ebi.ac.uk/research/enright/software/sylamer) was used to assess 3’UTR miRNA seed match enrichment p-values across the ranked protein list. A FASTA file containing 3’UTRs of protein-encoding genes was obtained and exported from Sylarray software (available at https://www.ebi.ac.uk/research/enright/software).
Lists of miR-137 predicted targets and schizophrenia-, ASD- and ID-associated genes
Conserved miR-137 predicted targets were obtained from TargetScanMouse Release 7.1 (http://www.targetscan.org/mmu_71/). Schizophrenia-associated genes were obtained from the Schizophrenia Gene Resource (http://bioinfo.mc.vanderbilt.edu/SZGR/)) and Gene List Automatically Derived For You (GLAD4U)59. ASD-associated genes were obtained from SFARI (https://gene.sfari.org/autdb/HG_Home.do). ID-associated genes were obtained from the Intellectual Disability Project (http://gfuncpathdb.ucdenver.edu/iddrc/home.php). A detailed gene list is in Supplementary Table 4. Galaxy online tools were used to overlap different gene lists60.
Lentiviral constructs
The U6-shRNA lentiviral construct was used to insert Pde10a shRNA sequence driven by the U6 promoter and GFP reporter gene driven by CMV promoter61,62. PCR-based generation of Pde10a driven by a U6 promoter was done using a PCR Shagging approach as previously described63. To knockdown the expression of Pde10a, three Pde10a shRNA sequences were designed by using Invitrogen website. Lentiviruses encoding for Pde10a shRNA sequences were produced by Lipofectamine 2000-mediated co-transfection of 293T cells with pREV, pVSVg, pMDL, and the pLenti-U6-GFP plasmid. The filtered and concentrated virus was applied to a primary neuron, and the knockdown efficiency of lentivirus determined at 72 h after virus infection. Lentivirus grating in vivo was performed as described previously61.
Western blotting
Protein samples were separated on SDS-PAGE gels and then transferred to polyvinylidene fluoride membranes (Millipore). Membranes were processed according to the ECL Western Blotting Protocol (GE Healthcare). Anti-PDE10A (SAB2700582; Sigma-Aldrich), anti-SATB2 (ab34735; Abcam), anti-PTPN2 (MABS1753; Millipore), anti-DNMT3A (3598S; CST), anti-PKA C-α (4782S; CST), anti-Phospho-PKA C (Thr197) (5661S; Cell Signaling Technology), and anti-PSD-95 (MAB1598; Millipore) were used as primary anntibodies at a 1:1,000 dilution. HRP-labeled secondary antibodies were obtained from Cell Signaling Technology (7074S & 7076S) and were used at a dilution of 1:5,000. The antibodies against GAPDH (AM4300; Thermo Fisher) or Actin (A5060; Sigma-Aldrich) were used for loading controls. All Western blot quantifications were performed using ImageJ software. Full Western blotting images are summarized in Supplementary Fig. 15.
Dual luciferase assays
3’UTR cloning and dual luciferase assays were performed as previously described13. In brief, 3’UTR sequences of candidate mRNAs of miR-137 predicted targets were PCR amplified directly from the first-strand cDNA generated from 5 µg of TRIZOL-isolated total RNA using oligo-dT SuperScript III reverse transcription, according to the manufacturer’s instructions (Invitrogen). All primers were designed incorporating XhoI and NotI restriction sites and 3–4 bp of the other random sequence to aid in restricting digestion (Supplementary Table 1). XhoI- and NotI-digested PCR products were cloned into XhoI- and NotI-digested psiCHECK-2 dual luciferase vector (Promega).
As a primary screen of candidate miR-137 targets, 293 FT cells (24-well plate, grown overnight before transfection) were transfected with U6-sh-miR-137 cloned into a pCR2.1 TOPO vector (sh-miR-137) and psiCHECK-2–3’UTR using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. As a control, a psiCHECK-2 plasmid with no 3’ UTR and U6-neg-shRNA (sh-Control) were co-transfected with U6-miR-137-shRNA TOPO or psiCHECK-2–3’UTR, respectively. All transfections used a total of 1 µg of plasmid DNA. The ratio of luciferase-3’UTR/shRNA plasmid was 1:16 for all experiments. Luciferase expression was detected using the Dual-Luciferase Reporter 1000 System (Promega) according to the manufacturer’s instructions. 48 h after transfection, hRLuc activity was normalized to hLuc+ activity to account for variation in transfection efficiencies, and miR-137-mediated knockdown of hRLuc activity was calculated as the ratio of normalized hRLuc activity in the U6-miR-137-shRNA treatments to normalized hRLuc activity in the U6-neg-shRNA treatments. All luciferase readings were taken from either three individual wells for each psiCHECK-2–3’UTR construct and control construct tested.
The miR-137 target site in the Pde10a-3’UTR was deleted using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies) to delete 7 bases (GCAAUAA) from the miR-137 seed site in the Pde10a-3’UTR luciferase reporter (Pde10a-3’UTR∆miR−137) (primers were designed with the QuikChange Primer Design online tool provided by Agilent Technologies and shown in Supplementary Table 1). Target site deletion was verified by Sanger sequencing. We performed the transfection experiment for the four plasmids, including “sh-control (miR-137 negative control, miR-137NC)”, “sh-miR-137 (miR-137 overexpression, miR-137OE)”, “Pde10a-3’UTR” and “Pde10a-3’UTR∆miR−137” constructs in HEK293FT cells. All transfections used a total of 1 μg of plasmid DNA, and the ratio of luciferase-3’UTR/shRNA plasmid was 1:16 for all experiments. We have normalized all other three samples to “miR-137NC + Pde10a-3’UTR”.
To assess whether the Pde10a-3’UTR is, in fact, a target of the endogenous miR-137, we performed the above-mentioned luciferase reporter assay in mouse Neuron-2a cells. To maximum the miR-137 inhibition effect, we transfected either mirVana miR-137 inhibitor (ID: MH10513, 10uM) or Negative Control #1 to Neuron-2a cells on Day 1 by Lipo RNAiMAX. On Day 2, we performed the transfection experiment for the two plasmids (4ng per well in 24-well plate), including “Pde10a-3’UTR” and “Pde10a-3’UTR∆miR−137” constructs, in Neuron-2a cells. On Day 3, the cells were collected to examine luciferase expression. All the other three samples were normalized to “Negative Control + Pde10a-3’UTR.”
Data collection
Littermates were housed in the same cages. For behavioral assays and electrophysiological experiments, each cage contains mixed genotypes, and the people who in charge of data collection did not know the genotype information during the experiments. Z-stack images (at 0.3-μm intervals) were obtained on a ZEISS 710 confocal laser-scanning microscope with an X63 oil-immersion lens at an electronic zoom factor of 1 or 2. Image analyses and quantification were performed using ImageJ software (1.49v). Behavioral assays: Data were collected blindly using the software Smart V3.0.03 (Panlab, Barcelona, Spain). Electrophysiology: The fEPSP signals were digitized using the Digidata 1440A interface board. Traces were obtained by pClamp 10.6 and analyzed using the Clampfit 10.6 (Axon Instruments, Foster City, CA).
Statistical analyses
All statistical analyses were performed using Prism 7.0 (GraphPad Software). Data sets were analyzed for significance using either unpaired Student’s two-tailed t-test or ANOVA measures with multiple comparison post-hoc tests; all data presented as mean ± s.e.m. Pearson’s Chi-squared tests with Yates’ continuity correction were performed in R software (http://www.r-project.org/). The sample size and description of the sample collection are reported in each figure legend. Quantification of immunostaining was performed blindly. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications (Ref. 45). Data distribution was assumed to be normal, but this was not formally tested.
Supplementary Material
Acknowledgements
We would like to thank S. Warren and D. Cook for critical reading of the manuscript. This work was supported by the National Key R&D Program of China (2018YFA0108001 to Z.T.), Natural Science Foundation of China (31590831 and 91640204 to DC, 81571212 to Z.T.), Strategic Priority Research Program of Chinese Academy of Science (Grant XDB 19000000), National Institutes of Health (NS051630, NS079625, and MH102690 to P.J., AG047928 to J.P.), and Simons Foundation Autism Research Initiative (239320 to P.J.). Hundred Talents Program of Chinese Academy of Science to Z.T.. P.J. is NARSAD Independent Investigator Award-Suzanne and John Golden Investigator.
Footnotes
Accession codes
Genome-wide datasets are deposited at GEO under accession number GSE79661 (RNA-seq) and at the ProteomeXchange database under accession number PXD003874 (proteomics).
Competing interests
The authors declare no competing interests.
Reporting Summary
Additional information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Genome-wide datasets are deposited at GEO under accession number GSE79661 (RNA-seq) and at the ProteomeXchange database under accession number PXD003874 (proteomics).
References
- 1.Pasquinelli AE MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nature reviews. Genetics 13, 271–282 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Ha M & Kim VN Regulation of microRNA biogenesis. Nature reviews. Molecular cell biology 15, 509–524 (2014). [DOI] [PubMed] [Google Scholar]
- 3.He L & Hannon GJ MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. Genetics 5, 522–531 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Li X & Jin P Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci 11, 329–338 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Issler O & Chen A Determining the role of microRNAs in psychiatric disorders. Nature reviews. Neuroscience 16, 201–212 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Schratt G microRNAs at the synapse. Nature Reviews Neuroscience 10, 842–849 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Cross-Disorder Group of the Psychiatric Genomics, C., et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ripke S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 45, 1150–1159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schizophrenia Psychiatric Genome-Wide Association Study, C. Genome-wide association study identifies five new schizophrenia loci. Nature genetics 43, 969–976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinto D, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. American journal of human genetics 94, 677–694 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan J, et al. A rare functional noncoding variant at the GWAS-implicated MIR137/MIR2682 locus might confer risk to schizophrenia and bipolar disorder. American journal of human genetics 95, 744–753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szulwach KE, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol 189, 127–141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smrt RD, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28, 1060–1070 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegert S, et al. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nature neuroscience (2015). [DOI] [PMC free article] [PubMed]
- 16.Guan F, et al. MIR137 gene and target gene CACNA1C of miR-137 contribute to schizophrenia susceptibility in Han Chinese. Schizophrenia research 152, 97–104 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Carter MT, et al. Hemizygous deletions on chromosome 1p21.3 involving the DPYD gene in individuals with autism spectrum disorder. Clinical genetics 80, 435–443 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Willemsen MH, et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. Journal of medical genetics 48, 810–818 (2011). [DOI] [PubMed] [Google Scholar]
- 19.D’Angelo CS, Moller Dos Santos MF, Alonso LG & Koiffmann CP Two New Cases of 1p21.3 Deletions and an Unbalanced Translocation t(8;12) among Individuals with Syndromic Obesity. Mol Syndromol 6, 63–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tucci A, Ciaccio C, Scuvera G, Esposito S & Milani D MIR137 is the key gene mediator of the syndromic obesity phenotype of patients with 1p21.3 microdeletions. Mol Cytogenet 9, 80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinkham AE, Hopfinger JB, Pelphrey KA, Piven J & Penn DL Neural bases for impaired social cognition in schizophrenia and autism spectrum disorders. Schizophrenia research 99, 164–175 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dykens EM Psychopathology in children with intellectual disability. Journal of child psychology and psychiatry, and allied disciplines 41, 407–417 (2000). [PubMed] [Google Scholar]
- 23.Macbeth AH, Edds JS & Young WS 3rd. Housing conditions and stimulus females: a robust social discrimination task for studying male rodent social recognition. Nature protocols 4, 1574–1581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleschevnikov AM, et al. Deficits in cognition and synaptic plasticity in a mouse model of Down syndrome ameliorated by GABAB receptor antagonists. The Journal of neuroscience : the official journal of the Society for Neuroscience 32, 9217–9227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marin O Interneuron dysfunction in psychiatric disorders. Nature reviews. Neuroscience 13, 107–120 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477, 171–178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eichler SA & Meier JC E-I balance and human diseases - from molecules to networking. Front Mol Neurosci 1, 2 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dongen S, Abreu-Goodger C & Enright AJ Detecting microRNA binding and siRNA off-target effects from expression data. Nature methods 5, 1023–1025 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren RP, et al. Increased Frequency of the Null Allele at the Complement C4b Locus in Autism. Clinical and experimental immunology 83, 438–440 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odell D, et al. Confirmation of the association of the C4B null allelle in autism. Hum Immunol 66, 140–145 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Sekar A, et al. Schizophrenia risk from complex variation of complement component 4. Nature (2016). [DOI] [PMC free article] [PubMed]
- 32.Fatemi SH, Reutiman TJ, Folsom TD & Thuras PD GABA(A) receptor downregulation in brains of subjects with autism. J Autism Dev Disord 39, 223–230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fatemi SH, Folsom TD, Reutiman TJ & Thuras PD Expression of GABA(B) receptors is altered in brains of subjects with autism. Cerebellum 8, 64–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramanathan S, et al. A case of autism with an interstitial deletion on 4q leading to hemizygosity for genes encoding for glutamine and glycine neurotransmitter receptor sub-units (AMPA 2, GLRA3, GLRB) and neuropeptide receptors NPY1R, NPY5R. BMC medical genetics 5, 10 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez E, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Molecular systems biology 5, 269 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beavo JA & Brunton LL Cyclic nucleotide research -- still expanding after half a century. Nature reviews. Molecular cell biology 3, 710–718 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 149, 525–537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grauer SM, et al. Phosphodiesterase 10A inhibitor activity in preclinical models of the positive, cognitive, and negative symptoms of schizophrenia. The Journal of pharmacology and experimental therapeutics 331, 574–590 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Wellcome Trust Case Control, C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Major Depressive Disorder Working Group of the Psychiatric, G.C., et al. A mega-analysis of genome-wide association studies for major depressive disorder. Molecular psychiatry 18, 497–511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siuciak JA, et al. Inhibition of the striatum-enriched phosphodiesterase PDE10A: a novel approach to the treatment of psychosis. Neuropharmacology 51, 386–396 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Wilson LS & Brandon NJ Emerging biology of PDE10A. Current pharmaceutical design 21, 378–388 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Tang GB, et al. The Histone H3K27 Demethylase UTX Regulates Synaptic Plasticity and Cognitive Behaviors in Mice. Frontiers in Molecular Neuroscience 10(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silverman JL, Yang M, Lord C & Crawley JN Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci 11, 490–502 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472, 437–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vicario-Abejon C Long-term culture of hippocampal neurons. Curr Protoc Neurosci Chapter 3, Unit 3 2 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Terashima A, et al. An essential role for PICK1 in NMDA receptor-dependent bidirectional synaptic plasticity. Neuron 57, 872–882 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia S, et al. p21-activated kinase 1 restricts tonic endocannabinoid signaling in the hippocampus. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pagala VR, et al. Quantitative protein analysis by mass spectrometry. Methods in molecular biology 1278, 281–305 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Wang H, et al. Systematic optimization of long gradient chromatography mass spectrometry for deep analysis of brain proteome. Journal of proteome research 14, 829–838 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, et al. JUMP: a tag-based database search tool for peptide identification with high sensitivity and accuracy. Molecular & cellular proteomics : MCP 13, 3663–3673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng J, Elias JE, Thoreen CC, Licklider LJ & Gygi SP Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2, 43–50 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Mertz J, et al. Sequential Elution Interactome Analysis of the Mind Bomb 1 Ubiquitin Ligase Reveals a Novel Role in Dendritic Spine Outgrowth. Molecular & cellular proteomics : MCP 14, 1898–1910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elias JE & Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature methods 4, 207–214 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Huang DW, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Wang J, Duncan D, Shi Z & Zhang B WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic acids research 41, W77–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic acids research 38, W214–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic acids research 39, D685–690 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jourquin J, Duncan D, Shi Z & Zhang B GLAD4U: deriving and prioritizing gene lists from PubMed literature. BMC Genomics 13 Suppl 8, S20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goecks J, Nekrutenko A, Taylor J & Galaxy T Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome biology 11, R86 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lie DC, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature 437, 1370–1375 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Liu C, et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 6, 433–444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu PP, et al. MiR-203 Interplays with Polycomb Repressive Complexes to Regulate the Proliferation of Neural Stem/Progenitor Cells. Stem cell reports 9, 190–202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.