Abstract
Reporter genes play important roles in transgenic research. LacZ is a widely used reporter gene that encodes Escherichia coli β-galactosidase, an enzyme that is well known for its ability to hydrolyze X-gal into a blue product. It is unknown whether transgenic LacZ has any adverse effects. R26R reporter mice, containing a LacZ reporter gene, were generated to monitor the in vivo recombination activity of various transgenic Cre recombinase via X-gal staining. P0-Cre is expressed in neural crest-derived cells, which give rise to the majority of the craniofacial bones. Herein, we report that 12% of the R26R reporter mice harboring P0-Cre had unexpected mid-facial developmental defects manifested by the asymmetrical growth of some facial bones, thus resulting in tilted mid-facial structure, shorter skull length, and malocclusion. Histological examination showed a disorganization of the frontomaxillary suture, which may at least partly explain the morphological defect in affected transgenic mice. Our data calls for the consideration of the potential in vivo adverse effects caused by transgenic β-galactosidase.
Keywords: R26R, LacZ, β-galactosidase, reporter gene, P0-Cre, Wntl-Cre, transgenic mouse
Introduction
Transgenic techniques have been widely used in scientific research. Many reporter mouse models have been generated to determine the expression pattern of endogenous genes and transgenes, to trace cell lineages, and to map cell fate (Abe and Fujimori 2013). The Gt(ROSA)26Sor (Rosa26) locus is one of the most widely used harbors for transgene insertion because of its ubiquitous expression (Casola 2010; Chen et al. 2011). R26R is a Cre indicator mouse line in which the endogenous Rosa26 locus was engineered with a targeting construct containing a LacZ gene downstream of a floxed STOP sequence (Soriano 1999). Following Cre-mediated excision of this floxed sequence, β-galactosidase (β-gal) is expressed, which can be detected by X-gal staining (Araki et al. 1995). Green Fluorescent Protein (GFP) is another commonly used reporter. It is widely recognized that GFP has cytotoxicity both in vitro and in vivo, which can potentially confound the data interpretation (Ansari et al. 2016). It has been shown that LacZ overexpression is toxic to cortical neurons in culture (Detrait et al. 2002). However, it is unknown whether transgenic LacZ has adverse effects on cellular homeostasis in vivo.
P0-Cre is driven by the promoter of myelin protein zero (P0) gene. P0-Cre transgenic mice can be used to recombine the floxed alleles in Schwann cells (Feltri et al. 1999) and neural crest-derived progenitors (Yamauchi et al. 1999). The latter is a population of pluripotent cells which can migrate and differentiate into multiple cell types such as osteoblasts and chondrocytes, which form the majority of the craniofacial bones (Minoux and Rijli 2010). We previously reported that deletion of Tscl by P0-Cre increases craniofacial bone mass (Fang et al. 2015a; Fang et al. 2015b). In an effort to use R26R reporter mice to characterize the recombination activity of P0-Cre in craniofacial tissues, we surprisingly found that some R26R reporter mice harboring P0-Cre had unexpected mid-facial developmental defects in association with the disorganization of fibrous sutural structures connecting affected facial bones.
Materials and methods
Mice
The P0-Cre, Wnt1-Cre and R26R transgenic mice were described previously (Danielian et al. 1998; Giovannini et al. 2000; Soriano 1999). Both P0-Cre and R26R reporter mice were maintained on the C57BL/6 background. Heterozygous P0-Cre male mice (P0-Cre/+) were mated with heterozygous R26R reporter female mice (R26R/+) to generate P0-Cre/+;R26R/+ mice. P0-Cre/+;R26R/R26R and R26R/R26R mice were generated by mating P0-Cre/+;R26R/+ mice with R26R/+ mice. P0-Cre/+;R26R/+ and P0-Cre/+;R26R/R26R mice were designated as P0-Cre;R26R mice. R26R/+ and R26R/R26R mice were designated as R26R mice (Control). Heterozygous Wnt1-Cre male mice (Wnt1-Cre/+) were mated with homozygous R26R reporter female mice (R26R/R26R) to generate Wnt1-Cre/+;R26R/+ (Wnt1-Cre;R26R) and R26R/+ (Control) mice. Experimental mice were housed under a standardized condition at the University of Michigan School of Dentistry and all handling protocols were approved by IACUC at University of Michigan. 2-month-old mice were euthanized by CO2 asphyxia and 7-day-old mice were euthanized by decapitation. To detect the engineered Rosa26 locus and Cre transgene, PCR was performed using tail DNA as described previously (He et al. 2017; Liu et al. 2004; Soriano 1999).
X-gal staining
Postnatal 7-day-old mouse skulls were dissected in cold PBS to separate the calvaria and the cranial base. The skulls were fixed in 2% PFA and 1% glutaraldehyde co-fixative for 15min at room temperature and were then rinsed in PBS. The skulls were stained in X-gal staining solution (1mg/ml X-gal, 5mM Kferri/Kferro, 2mM MgCl2, 0.02% NP-40, 0.01% sodium deoxycholate dissolved in PBS) at 37°C overnight. The skulls were then rinsed in PBS the next morning and pictured using stereomicroscope (Leica M165 FC).
Skull morphological measurement
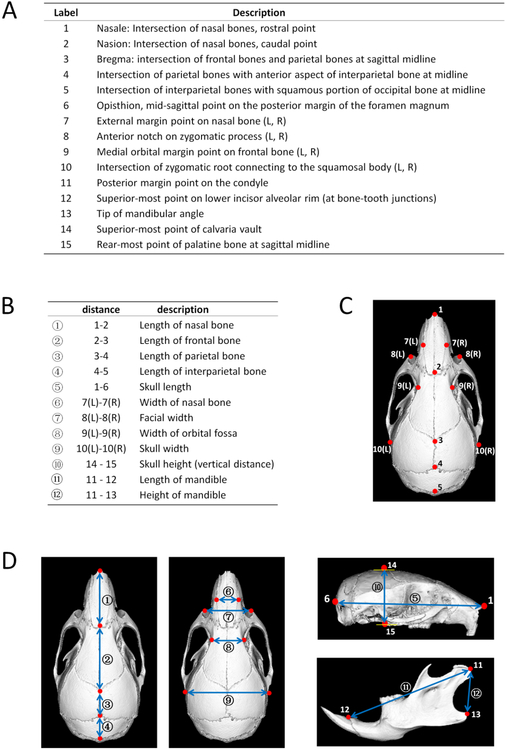
Skulls were measured by using Micro-CT (eXplore Locus SP, GE Healthcare Pre-Clinical Imaging, London, ON, Canada) and digital caliper (Fowler 6"/150 mm Ultra-Cal V, Sylvac SA) as previously described (Wei et al. 2017). Micro-CT images were reoriented and reconstructed to 3D through Microview (version 2.2). A total of 19 landmarks (11 single, 4 paired, see Fig 1A, 1C) and 12 linear measurements (5 anterior-posterior [A-P], 4 transversal, 1 vertical, 2 mandible items, see Fig 1B, 1D) were chosen to analyze the morphology of the mouse skulls. All parameters were measured by the same operator twice and the average values were used for calculations.
Fig 1.
Landmarks and linear measurement parameters of mouse skulls. There were a total of 19 (11 single, 4 paired) skeletal landmarks (A, C, D) and 12 (anterior-posterior①-呤, transversal⑥-⑨, vertical⑩, mandibular⑪-⑫) linear measurements (B, D).
Histological examination
2-month-old mouse calvariae were decalcified in 14% EDTA at 4°C for 2 weeks and embedded in Tissue-Plus O.C.T. Compound (Fisher Healthcare) in a coronal direction. 10 μm tissue sections were obtained using cryostat (Leica CM1950) for Hematoxylin and Eosin (H&E) staining.
Statistical analysis
Student’s t-test was employed for the comparison between two groups and one-way ANOVA with subsequent post-hoc analysis was used for multiple-group comparisons by GraphPad Prism 7.0 (GraphPad, San Diego, CA, USA). Statistical significance was defined asp < 0.05.
Results and Discussion
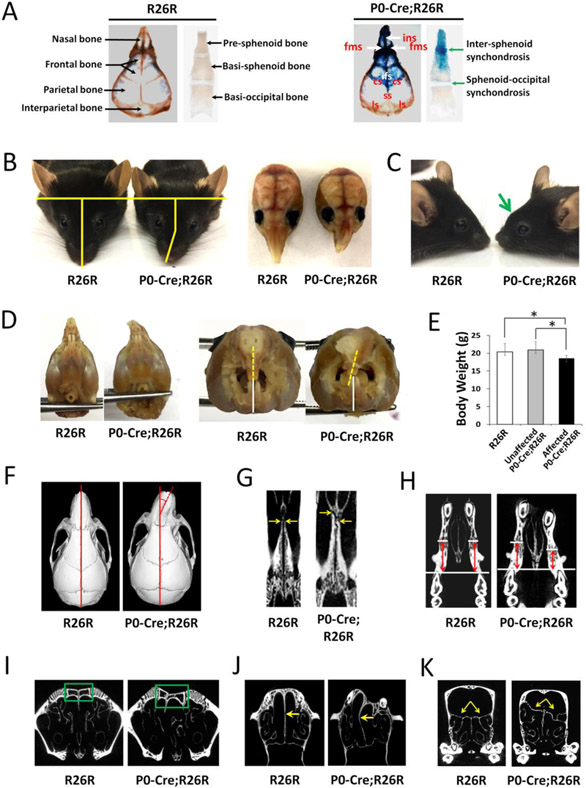
To characterize the recombination activity of P0-Cre in craniofacial tissues, we performed X-gal staining on P0-Cre;R26R mice. Our data showed that P0-Cre had recombination activity in the neural crest-derived nasal bones, frontal bones, and adjacent sutures connecting these bones in the calvaria, as well as the pre-sphenoid bone, rostral part of basi-sphenoid bone, and intersphenoid synchondrosis between them (Fig 2A). In addition, X-gal staining was also positive in facial bones including the ethmoid bone, palatine bones, maxilla, and mandible that are derived from neural crest (data not shown). In contrast, X-gal staining was negative in the mesoderm-derived parietal bones, interparietal bones, basi-occipital bones, and sphenoid-occipital synchondrosis (Fig 2A). Interestingly, we found 11.6% (5/43) of the P0-Cre;R26R mice showed mid-facial morphological defects, which were absent from all control mice (n=45). In affected mice, the nasal bone was tilted to one side (Fig 2B) and formed an abnormal protrusion at the junction between the nasal bone and the frontal bone on lateral view (Fig 2C). The nasal bone could tilt toward either side (two mice toward left side and three mice toward right side in this study). The upper incisors tilted to one side while the lower incisors were not affected and thus caused malocclusion (Fig 2D). The malocclusion could lead to compromised masticatory function, which may explain a 11.3% body weight decrease in affected P0-Cre;R26R mice compared to unaffected P0-Cre;R26R mice (Fig 2E). Analysis on Micro-CT three-dimensional image showed that there was a 14.6°±5.5° tilt of nasal bone to the anterior-posterior axis of skull (Fig 2F). Furthermore, there was asymmetric growth in other bilateral facial structures, such as the palatine bone (Fig 2G), maxilla (Fig 2H), nasal sinus (Fig 2I), perpendicular plate, (Fig 2J) and ethmoid bone (Fig 2K). The craniofacial morphology of unaffected P0-Cre;R26R mice showed no difference compared to control mice. In this study, the five affected P0-Cre;R26R mice were from three different litters that were generated from two different breeding units. Among them, two male mice were P0-Cre;R26R/R26R and three female mice were P0-Cre;R26R/+, which may indicate that the craniofacial defect was independent of LacZ gene dose and gender.
Fig 2.
P0-Cre;R26R mice had mid-facial developmental defects. (A) X-gal staining of 7-day-old P0-Cre;R26R and R26R mice. The arrows and labels indicate respective bones, sutures, and synchondroses, ins = intemasal suture; fms = frontomaxillary suture; ifs = interfrontal suture; cs = coronal suture; ss = sagittal suture; Is = lambdoidal suture. Affected 2-month-old P0-Cre;R26R mice showed (B) nasal tilting, (C) protruding at the junction between the nasal bone and the frontal bone at lateral view (arrow), and (E) incisor malocclusion (dashed line indicates midline of upper incisors, solid line indicates midline of lower incisors). (E) Body weight of affected 2-month-old P0-Cre;R26R mice (n=5) were significantly decreased compared to unaffected P0-Cre;R26R mice (n=38) and R26R mice (n=45), *p<0.05. Micro-CT images showed a tilted nasal bone (F), asymmetric growth of the palatine bone (G), maxilla (H), nasal sinus (I), perpendicular plate (J), and ethmoid bone (K) in affected P0-Cre;R26R mice. The dashed line in F indicates the angle between the axis of the nasal bone and that of skull; The arrows in G indicates the rostral point of the palatine bone in a horizontal section; the lines and arrows in H indicate the distance between the mesial point of the first molar to the premaxilla-maxilla suture in a horizontal section; the box in I indicates the nasal bone and sinus in a coronal section; the arrows in J indicate the perpendicular plate in a horizontal section; the arrows in K indicate the superior border of the ethmoid bone in a coronal section.
To further determine the effects of the mid-facial defects of affected P0-Cre;R26R mice on the craniofacial morphology, we performed linear measurements of mouse skulls in three dimensions. Our data showed a compromised mouse skull growth in the anterior-posterior dimension of affected P0-Cre;R26R mice. There was a significant decrease in nasal bone length (−17.5%), frontal bone length (−9.9%), and skull length (−10.1%) in affected P0-Cre;R26R mice compared to control mice, and a 16.4%, 9.0%, and 9.5% decrease in those parameters compared to unaffected P0-Cre;R26R mice (Table 1摠摡⑤). There was no significant difference among the three groups in the transverse or vertical dimensions of both the skull and the mandible. No difference was detected in all dimensions of craniofacial growth between control and unaffected P0-Cre;R26R mice.
Table 1.
Linear measurements of 2-month-old mouse skulls.
| Dimension | Parameter | R26R (mm) |
Unaffected P0Cre;R26R (mm) |
Affected P0Cre;R26R (mm) |
|---|---|---|---|---|
| A-P | ① | 7.55 ± 0.28 | 7.45 ± 0.07 | 6.23 ± 0.17 *# |
| ② | 7.39 ± 0.14 | 7.32 ± 0.20 | 6.66 ± 0.14 *# | |
| ③ | 3.75 ± 0.13 | 3.70 ± 0.15 | 3.74 ± 0.07 | |
| ④ | 3.37 ± 0.07 | 3.40 ± 0.11 | 3.35 ± 0.04 | |
| ⑤ | 22.41 ± 0.24 | 22.26 ± 0.22 | 20.15 ± 0.36 *# | |
| Transversal | ⑥ | 2.52 ± 0.07 | 2.52 ± 0.03 | 2.52 ± 0.07 |
| ⑦ | 6.08 ± 0.08 | 6.05 ± 0.11 | 6.23 ± 0.18 | |
| ⑧ | 4.15 ± 0.04 | 4.11 ± 0.02 | 4.11 ± 0.03 | |
| ⑨ | 10.37 ± 0.14 | 10.40 ± 0.13 | 10.45 ± 0.10 | |
| Vertical | ⑩ | 7.47 ± 0.15 | 7.48 ± 0.15 | 7.38 ± 0.11 |
| Mandible | ⑪ | 11.52 ± 0.22 | 11.59 ± 0.14 | 11.37 ± 0.23 |
| ⑫ | 3.69 ± 0.09 | 3.64 ± 0.12 | 3.68 ± 0.16 |
Affected P0-Cre;R26R mice had a significant decrease in the length of the nasal bone (①), frontal bone, (②) and skull (⑤) compared to unaffected P0-Cre;R26R mice and R26R mice, n=5 for each group. No difference was detected in all parameters between unaffected P0-Cre;R26R mice and R26R mice.
p<0.05 compared to R26R mice;
p<0.05 compared to unaffected P0-Cre;R26R mice.
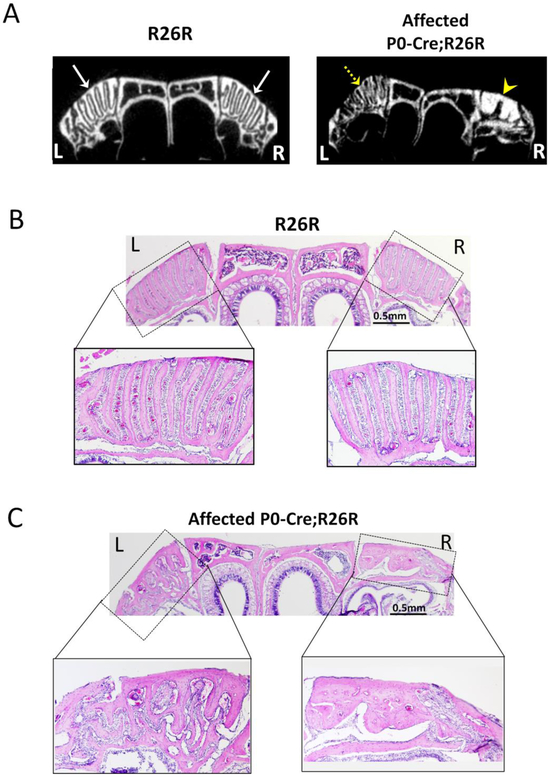
Since most craniofacial bones are formed via intramembranous ossification, the growth and expansion of these bones considerably rely on fibrous sutures between adjacent bones. Recent literature reported that Glil positive cells in the sutures form a niche of osteogenic stem cells contributing to craniofacial bone formation (Zhao et al. 2015). In human craniosynostosis, premature fusion or dysfunction of sutures depresses the bone growth of the affected side with no effect on or compensatory overgrowth in the contralateral bone, which can consequently lead to the asymmetrical growth of craniofacial structures (Morriss-Kay and Wilkie 2005; Senarath-Yapa et al. 2012). To determine whether there was an alteration in sutures in affected P0-Cre;R26R mice, we examined the frontomaxillary suture, which is the intersection of the nasal bone, premaxilla, and frontal bone (Fig 3). Both Micro-CT (Fig 3A) and histological examination (Fig 3B, 3C) showed that the frontomaxillary sutures were well-arranged in control mice. However, in affected P0-Cre;R26R mice, the frontomaxillary suture was irregular on both sides. Ectopic bone formation was observed in some areas, which was more evident on the side that the nasal bone tilted toward. The disorganization of the sutural area may subsequently disturb the growth of adjacent bones and result in mid-facial defects in affected mice.
Fig 3.
Structural disorganization of the frontomaxillary suture in affected 2-month-old P0-Cre;R26R mice. (A) Micro-CT images of the nasal bone and frontomaxillary suture in a coronal section. Arrows point to the well-organized suture at both sides in control mice; dashed arrow points to the disorganized sutural structure and arrow head point to the ectopic bone formation in the suture area in affected P0-Cre;R26R mice. (B-C) Histological images of the nasal bone and frontomaxillary suture in a coronal section in control (B) and affected P0-Cre;R26R mice (C). L indicates left side and R indicates right side.
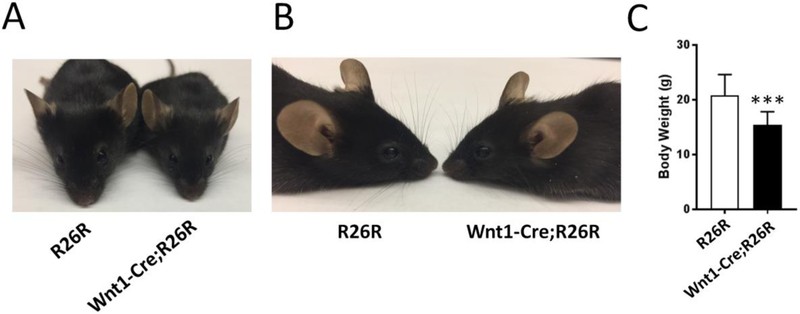
To determine whether the observed craniofacial abnormality in P0-Cre;R26R mice was caused by the P0-Cre transgene itself, we analyzed the P0-Cre/+ mice (n=22) and did not find any abnormality. In addition, we did not find the nasal bone tilting in more than 100 other transgenic mice harboring P0-Cre including the conditional knockout mice of Tscl (Fang et al. 2015a), Fip200 (unpublished), and Fak (unpublished). Thus, we conclude that the craniofacial abnormality in affected P0-Cre;R26R mice is caused by the expression of the LacZ gene in P0-Cre targeted cells. Furthermore, to determine whether the observed craniofacial defects were specific to P0-Cre;R26R mice, we analyzed Wnt1-Cre;R26R mice. Wnt1-Cre is another transgenic Cre which also targets neural crest-derived cells (Danielian et al. 1998; Jiang et al. 2002). However, there are significant differences in the Cre distribution patterns between P0-Cre and Wnt1-Cre (Chen et al. 2017). Similar to control mice (n=19), Wnt1-Cre;R26R (n=20) mice showed no abnormality in craniofacial morphology (Fig 4A, 4B), which indicated that the mid-facial defect in P0-Cre;R26R mice might be due to the expression of the LacZ gene in neural crest-derived cells that are specifically targeted by P0-Cre but not Wnt1-Cre. Notably, Wnt1-Cre;R26R mice had lower body weight compared to control mice (Fig 4C) but unaffected P0-Cre;R26R mice had no body weight decrease (Fig 2E). This further indicated the difference between Wnt1-Cre and P0-Cre. It is known that Cre-recombinase itself has toxicity and could have adverse effects on multiple organs (Janbandhu et al. 2014; Lexow et al. 2013; Wang et al. 2015). Although our data showed that P0-Cre recombinase alone did not cause any obvious defects, the mid-facial defects in P0-Cre;R26R mice could be theoretically due to the combinatory effect of both Cre and LacZ.
Fig 4.
Wnt1-Cre;R26R mice had no mid-facial developmental defects. Front (A) and lateral (B) view of representative 2-month-old R26R and Wnt1-Cre;R26R mice. (C) Body weight of Wnt1-Cre;R26R mice (n=14) were significantly decreased compared to R26R mice (n=7), ***p<0.001.
In our study, only 12% P0-Cre;R26R mice had mid-facial defects. By X-gal staining, we observed that the P0-Cre mice had very variable degrees of Cre expression patterns in the craniofacial region. Only small percentage of P0-Cre;R26R mice showed extensive X-gal staining pattern at one-week-old (data not shown). Thus, we suspect that the affected P0-Cre;R26R mice had more P0-Cre positive cells, and thus had more LacZ-expressing cells. Alternatively, there may be an underlying genetic predisposition in the affected mice.
Conclusions
We have identified a mid-facial developmental defect caused by the expression of the widely used LacZ reporter gene. Although the underlying mechanism of the effect of LacZ expression in P0-Cre targeted tissue on craniofacial morphology is unknown, our data clearly demonstrated an in vivo abnormality caused by transgenic LacZ, which calls for attention to the potential adverse effects caused by this widely used transgenic reporter gene. When the LacZ reporter gene is used, data should be interpreted with careful consideration of the potential adverse effect caused by exogenous β-galactosidase.
Acknowledgements
We thank Neil Thomas for help in editing the manuscript. We also thank Drs. Marco Giovannini, Andrew McMahon, and Phillipe Soriano for genetically modified mouse lines. This work was supported by NIH (AR062030 to FL) and NSFC (81470764 to MH). Micro-CT work was partly supported by the P30 Core Center award to the University of Michigan from NIAMS (AR 69620).
Footnotes
Conflict of interest
None of the authors have any conflicts of interest to disclose.
References
- Abe T, Fujimori T (2013) Reporter mouse lines for fluorescence imaging Dev Growth Differ 55:390–405 doi: 10.1111/dgd.12062 [DOI] [PubMed] [Google Scholar]
- Ansari AM et al. (2016) Cellular GFP Toxicity and Immunogenicity: Potential Confounders in in Vivo Cell Tracking Experiments Stem Cell Rev 12:553–559 doi: 10.1007/s12015-016-9670-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Araki M, Miyazaki J, Vassalli P (1995) Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase Proc Natl Acad Sci U S A 92:160–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola S (2010) Mouse models for miRNA expression: the ROSA26 locus Methods Mol Biol 667:145–163 doi:10.1007/978-1-60761_811-9_10 [DOI] [PubMed] [Google Scholar]
- Chen CM, Krohn J, Bhattacharya S, Davies B (2011) A comparison of exogenous promoter activity at the ROSA26 locus using a PhiiC31 integrase mediated cassette exchange approach in mouse ES cells PLoS One 6:e23376 doi:10.1371/journal.pone.0023376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G et al. (2017) Specific and spatial labeling of P0-Cre versus Wnt1-Cre in cranial neural crest in early mouse embryos Genesis 55 doi:10.1002/dvg.23034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP (1998) Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase Curr Biol 8:1323–1326 [DOI] [PubMed] [Google Scholar]
- Detrait ER, Bowers WJ, Halterman MW, Giuliano RE, Bennice L, Federoff HJ, Richfield EK (2002) Reporter gene transfer induces apoptosis in primary cortical neurons Mol Ther 5:723–730 doi:10.1006/mthe.2002.0609 [DOI] [PubMed] [Google Scholar]
- Fang F, Sun S, Wang L, Guan JL, Giovannini M, Zhu Y, Liu F (2015a) Neural Crest-Specific TSC1 Deletion in Mice Leads to Sclerotic Craniofacial Bone Lesion J Bone Miner Res 30:1195–1205 doi:10.1002/jbmr.2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Wei X, Hu M, Liu F (2015b) A mouse model of craniofacial bone lesion of tuberous sclerosis complex Musculoskelet Regen 1 doi:10.14800/mr.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltri ML, D’Antonio M, Previtali S, Fasolini M, Messing A, Wrabetz L (1999) P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells Ann N Y Acad Sci 883:116–123 [PubMed] [Google Scholar]
- Giovannini M et al. (2000) Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2 Genes Dev 14:1617–1630 [PMC free article] [PubMed] [Google Scholar]
- He Y, Sun X, Wang L, Mishina Y, Guan JL, Liu F (2017) Male germline recombination of a conditional allele by the widely used Dermo1-cre (Twist2-cre) transgene Genesis 55 doi:10.1002/dvg.23048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbandhu VC, Moik D, Fassler R (2014) Cre recombinase induces DNA damage and tetraploidy in the absence of loxP sites Cell Cycle 13:462–470 doi:10.4161/cc.27271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM (2002) Tissue origins and interactions in the mammalian skull vault Dev Biol 241:106–116 doi:10.1006/dbio.2001.0487 [DOI] [PubMed] [Google Scholar]
- Lexow J, Poggioli T, Sarathchandra P, Santini MP, Rosenthal N (2013) Cardiac fibrosis in mice expressing an inducible myocardial-specific Cre driver Dis Model Mech 6:1470–1476 doi:10.1242/dmm.010470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M, Kream BE (2004) Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues Int J Dev Biol 48:645–653 doi:10.1387/ijdb.041816f1 [DOI] [PubMed] [Google Scholar]
- Minoux M, Rijli FM (2010) Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development Development 137:2605–2621 doi:10.1242/dev.040048 [DOI] [PubMed] [Google Scholar]
- Morriss-Kay GM, Wilkie AO (2005) Growth of the normal skull vault and its alteration in craniosynostosis: insights from human genetics and experimental studies J Anat 207:637–653 doi:10.1111/j.1469-7580.2005.00475.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senarath-Yapa K, Chung MT, McArdle A, Wong VW, Quarto N, Longaker MT, Wan DC (2012) Craniosynostosis: molecular pathways and future pharmacologic therapy Organogenesis 8:103–113 doi:10.4161/org.23307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain Nat Genet 21:70–71 doi:10.1038/5007 [DOI] [PubMed] [Google Scholar]
- Wang L, Mishina Y, Liu F (2015) Osterix-Cre transgene causes craniofacial bone development defect Calcif Tissue Int 96:129–137 doi:10.1007/s00223-014-9945-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Thomas N, Hatch NE, Hu M, Liu F (2017) Postnatal Craniofacial Skeletal Development of Female C57BL/6NCrl Mice Front Physiol 8:697 doi:10.3389/fphys.2017.00697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y et al. (1999) A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice Dev Biol 212:191–203 doi:10.1006/dbio.1999.9323 [DOI] [PubMed] [Google Scholar]
- Zhao H, Feng J, Ho TV, Grimes W, Urata M, Chai Y (2015) The suture provides a niche for mesenchymal stem cells of craniofacial bones Nat Cell Biol 17:386–396 doi:10.1038/ncb3139 [DOI] [PMC free article] [PubMed] [Google Scholar]