Abstract
Objective
Rheumatoid arthritis (RA) is characterized by anti-citrullinated protein antibodies (ACPAs), nevertheless, the origin, specificity, and functional properties of ACPAs remain poorly understood. To characterize the evolution of ACPAs, we sequenced the plasmablast antibody repertoire at serial timepoints in subjects with established RA.
Methods
Blood samples were obtained at up to four timepoints from eight anti-CCP+ individuals with established RA. We single-cell sorted CD19+CD3−IgD−CD14−CD20−CD27+CD38++ plasmablasts and co-stained with citrullinated-peptide tetramers to identify ACPA-expressing plasmablasts. Cell-specific oligonucleotide barcodes were utilized followed by large-scale sequencing and bioinformatic analysis to obtain error-corrected, paired, heavy and light chain antibody gene sequences for each B cell.
Results
Bioinformatic analysis revealed 170 persistent plasmablast lineages of which 19% included multiple isotypes. Among IgG- and IgA-expressing plasmablasts, we observed significantly more IgA-expressing persistent lineages compared to IgG (P < 0.01). We identified shared CDR3 sequence motifs across subjects. A subset of lineages comprised of later-timepoint derived members with divergent somatic hypermutations encoded antibodies that bind an expanded set of citrullinated antigens. Further, these recombinant, differentially mutated plasmablast antibodies formed immune complexes that stimulated higher levels of macrophage TNF-α production compared to antibodies representing earlier-timepoint, less-mutated lineage members.
Conclusions
Our findings demonstrate that established RA is characterized by a persistent IgA ACPA response that exhibits ongoing affinity maturation. This observation suggests the presence of a persistent mucosal antigen that continually promotes the production of IgA plasmablasts, and their affinity maturation and epitope spreading to generate ACPAs that bind additional citrullinated antigens and more potently stimulate macrophage TNF-α production.
Rheumatoid arthritis (RA) is characterized by the generation of autoantibodies including rheumatoid factor and anti-citrullinated protein antibodies (ACPAs) [1]. Here, we sequenced the plasmablast antibody repertoire over serial timepoints in established RA, to gain insights into the evolution of ACPAs and further define the role of ACPAs in promoting the pathogenesis of RA.
ACPAs target citrullinated epitopes arising from post-translational modifications of arginine to citrulline by peptidyl arginine deiminase (PAD) [2]. Epitope spreading of ACPAs precedes the onset of clinical arthritis [3]. Previous studies demonstrated that recombinant murine ACPAs can increase the severity of arthritis in mice [4,5]. B cell depletion using rituximab provides clinical benefit in seropositive RA [6], and its efficacy is associated with mild reductions in circulating ACPA levels [7]. A mucosal drive has been postulated in RA [8]. Cigarette smoking represents a risk factor for ACPA+ RA [9], and ACPAs are detectable in sputum and/or serum from early RA subjects and individuals at risk for developing RA [10].
ACPAs may contribute to the pathogenesis of RA by stimulating immune effector cells including macrophages [11], which produce TNF-α and other cytokines [12]. Macrophages can be activated by pro-inflammatory cytokines, immune complexes (ICs), and toll-like receptor (TLR) agonists [13]. Studies from our lab and others have shown that ICs composed of RA blood-derived ACPAs and citrullinated proteins stimulate blood-derived [14] and synovial fluid-derived [15] macrophages to produce TNF-α. These ACPA ICs stimulate macrophages via FcγRII [15–18] and TLR4 [16,17]. Nevertheless, the role and mechanisms by which affinity maturation and the evolving ACPA repertoire contribute to the development and persistence of RA remain unclear.
Here, we utilized our cell-barcoding antibody repertoire sequencing method [19] to investigate the evolution of the ACPA B cell response in RA. Previous studies utilizing this approach focused on the blood IgG repertoire in established RA [19] or the IgA and IgG repertoire of pre-RA/early RA subjects [20], at a single timepoint. In this study, we sequenced blood plasmablasts, which have been shown to produce ACPAs [19,21], at up to four timepoints from eight anti-CCP+ individuals with established RA. Bioinformatic analysis revealed plasmablast lineages persisting across serial timepoints and shared HC and LC CDR3 motifs across subjects. Using antigen microarrays and ELISAs, we demonstrated that, compared to earlier-timepoint plasmablast lineage members, a subset of later-timepoint, differentially mutated lineage members encode antibodies that target additional citrullinated antigens. In vitro macrophage stimulation assays revealed that these affinity-matured, differentially mutated, later-timepoint, recombinant plasmablast antibodies generate ICs that more potently stimulate macrophages to produce TNF-α, which could promote the pathogenesis of RA.
Materials and Methods
Human samples
After obtaining written informed consent under protocols approved by the Stanford University Institutional Review Board, blood samples were collected in heparin tubes at serial timepoints ≥2 months apart from eight anti-CCP+ subjects with RA (Table 1) recruited at VA Palo Alto Healthcare System. RA subjects met American College of Rheumatology 1987 and 2010 criteria [22,23]. Monocyte-derived macrophages were generated from blood obtained from Stanford Blood Center. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation with Ficoll-Paque™ PLUS (GE Healthcare Life Sciences).
Table 1.
Demographics of subjects studied
| Subject | Age at First Time Point | Gendera | Number of Tender + Swollen Jointsb | |||
|---|---|---|---|---|---|---|
|
| ||||||
| T1 | T2 | T3 | T4 | |||
| Subject 1 | 63 | F | 18 | 14 (4 mo) | 7 (10 mo) | |
| Subject 2 | 61 | M | 5 | 2 (3 mo) | 3 (6 mo) | |
| Subject 3 | 60 | M | 4 | 10 (3 mo) | 22 (8 mo) | 18 (10 mo) |
| Subject 4 | 67 | M | 0 | 0 (4 mo) | ||
| Subject 5 | 76 | M | 0 | 10 (7 mo) | ||
| Subject 6 | 66 | M | 0 | 4 (12 mo) | ||
| Subject 7 | 83 | M | 10 | 1 (11 mo) | 12 (13 mo) | |
| Subject 8 | 74 | M | 5 | 4 (10 mo) | ||
Gender demographics at the VA Hospital, where samples were obtained, are predominantly male
Number of tender and swollen joints was added at each time point and months (mo) from initial sampling are indicated in parentheses
Single-cell sorting of plasmablasts
CD19+CD3−IgD−CD14−CD20−CD27+CD38++ plasmablasts were single-cell sorted (BD FACSAria) as described [19,24] (Supplementary Methods). PBMC-staining used fluorophore-conjugated antibodies against CD19 (HIB19; Biolegend), CD3 (UCHT1; BD), IgD (IA6-2; BD), CD14 (MφP9; BD), CD20 (L27; BD), CD27 (CLB-27/1; LifeTechnologies), CD38 (HB7; BD), IgA (IS11-8E10; Miltenyi), and IgM (MHM-88; Biolegend). PBMCs were co-stained with pooled citrullinated-peptide tetramers comprised of 14 citrullinated peptides (Supplementary Table 1) to identify ACPA-producing plasmablasts (Supplementary Methods).
Cell barcode-enabled sequencing of the plasmablast antibody repertoire
Sequencing of immunoglobulin genes from individual plasmablasts was performed using cell barcodes as described [19,20] with minor modifications (Supplementary Methods). Well-specific oligonucleotide barcodes (TruGrade Oligos; IDT) were added to cDNA by template switching using Maxima Reverse Transcriptase (ThermoFisher). HC and LC genes were amplified from pooled cDNA using gene-specific PCR primers, and paired-end sequencing (2×330) performed using Illumina MiSeq. Samples underwent two [20] or three PCR rounds (Supplementary Table 2).
Bioinformatic analysis of immunoglobulin sequences
Sequence data was processed, HC VDJ and LC VJ aligned using ImMunoGeneTics (IMGT) HighV-Quest [25], and phylogenetic trees generated as described [19,20,24] (Supplementary Methods). IMGT-based analysis of V-region somatic hypermutations (SHM) was utilized to compare mutation levels across subjects and timepoints. Clonal families and persistent lineages were assigned based on shared, IMGT-based assignments of HC and LC VJ genes and 60% identity within HC and LC CDR3 regions (Levenshtein distance [26]), similar to others with different identity thresholds [27].
To evaluate HC and LC CDR3 sequence motifs across patients, cluster analysis was performed using CD-HIT [28,29]. Persistent lineage-derived HC and LC CDR3 sequences were linked by a 4-X spacer, clustered using a 70% identity cutoff, and results visualized with igraph in R [30]. For clusters containing >1 subject, linked HC and LC CDR3 sequences were used to generate sequence logos (WebLogo [31]). Plasmablast sequences were aligned to germline VJ genes by ClustalX2 [32], input to IgTree [33], and lineage trees visualized in Graphviz 2.38 [34]. Insertions and D gene segments were included in this mutation count from germline. Geneious 7.0.3 [35] alignments identified amino acid differences between lineage members.
Monoclonal antibody expression
A subset of subject-derived plasmablast antibody sequences representing clonal families/lineages and/or antibodies binding to the citrullinated-peptide tetramers and negative control antibodies (Supplementary Methods) were recombinantly produced in the Robinson lab or by LakePharma (San Carlos, CA). For consistency in characterization assays, all antibodies were expressed on the human IgG1 Fc domain. In-house production was done as described using the Expi293 Expression System (ThermoFisher) with Expi293F cells [36] (Supplementary Methods).
Characterizing antibody binding specificities
The QUANTA Lite® CCP3.1 IgG/IgA kit (Inova) was used to test CCP-binding activity of plasma and recombinant antibodies using the manufacturer’s protocol. For recombinant antibodies, an activity cutoff of three standard deviations above negative control antibodies was used. Recombinant antibodies were further screened for binding to citrullinated and native peptides/proteins by planar and bead-based antigen arrays as described [3,37]. Planar (50 μg/mL) and bead-based arrays (25 μg/mL) evaluated antibody binding to ~350 or 40 proteins/peptides, respectively. Using fluorophore-conjugated anti-human IgG secondary antibodies to detect binding, median fluorescent intensities were determined from quadruplicate print spots (planar array) or >50 beads (bead array).
ELISAs were performed using peptides and full-length proteins citrullinated in vitro with PAD from rabbit skeletal muscle (Sigma) or recombinant PAD4 (Supplementary Methods). ELISA plates were coated with full-length proteins (1–5 μg/mL) and peptides (10–15 μg/mL) diluted in bicarbonate/carbonate buffer (pH 9.5) (Supplementary Methods), and HRP-based detection with the 1-Step Ultra TMB-ELISA Substrate (ThermoFisher) was used. Fold change from an activity cutoff (three standard deviations above the average activity of negative controls) was calculated.
Macrophage stimulation assays
Macrophages were isolated and cultured from plated PBMCs by adhesion with 30 ng/mL of human macrophage stimulating factor (hMCSF, Peprotech) as described [16,17] (Supplementary Methods). This approach is commonly used for culturing macrophages and has been proposed as a reference standard [13]. To form plate-bound ICs, plates were coated with 50 μL of protein (20 μg/mL), washed with PBS, blocked with 1% low-endotoxin BSA in PBS (150 μL), and 50 μL of recombinant, subject-derived antibodies (50 μg/mL) was added. After further washing, differentiated macrophages were added (50,000 cells/well) in media (5% FBS, without hMCSF). To block FcγRII and/or TLR4, cells were pre-incubated (37°C) for one hour with anti-CD32 (Stemcell Technologies, clone IV.3) and/or InSolution™ TLR4 Inhibitor, TAK-242 (EMD Millipore). Lipopolysaccharide (50 ng/mL; LPS, Sigma-Aldrich) was used as a positive control. After 24-hours (37°C), supernatants were harvested and TNF-α levels measured by ELISA (Peprotech).
Statistical analysis
Statistical analysis was performed in Prism 7 (GraphPad). One-way or two-way ANOVA followed by Tukey’s test was used for multiple comparisons, and P < 0.05 was considered statistically significant. The relationship between tender and swollen joints and average normalized clonal family size was determined by Spearman correlation.
Results
Profiling the evolution of the blood plasmablast antibody repertoire across serial timepoints in RA
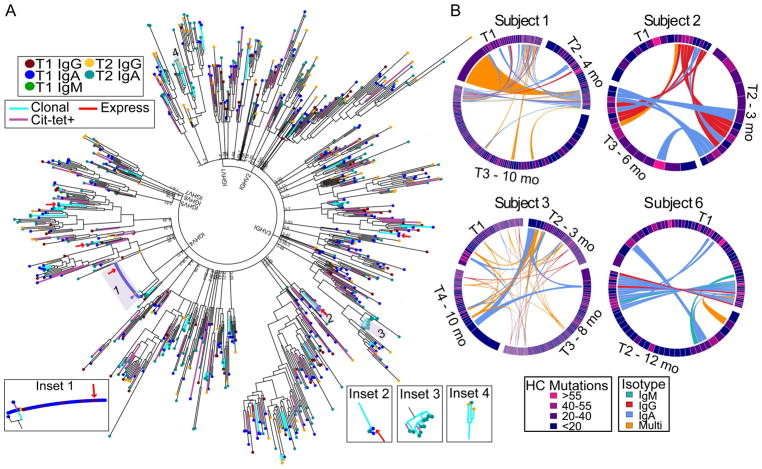
We used our cell barcode-enabled antibody repertoire sequencing method [19] to sequence the blood CD19+CD3−IgD−CD14−CD20−CD27+CD38++ plasmablast antibody repertoires from up to four timepoints from anti-CCP+ individuals with established RA (Table 1). Bioinformatic analyses assigned sequencing reads and obtained error-corrected consensus sequences of the HC and LC variable regions of each plasmablast. These datasets were used to construct phylogenetic trees to provide an overview of the plasmablast antibody repertoire, including identification of clonally expanded families/lineages and antibodies only observed once (singletons) (Figure 1A). This representative tree depicts trends observed in multiple subjects, and these observations are further highlighted in the inset images. We identified clonal lineages with representatives in multiple timepoints (Insets 1 and 2), hereby identified as persistent lineages. Some clonal expansions consisted of plasmablasts expressing antibodies of a single isotype (Insets 2 and 3) while others included multiple isotypes (Insets 1 and 4). By combining bioinformatic analysis of the sequence data with flow-based staining data of citrullinated-peptide tetramers, we identified candidate ACPA-expressing clonal expansions (Insets 1–3) and singletons (Figure 1A). A combination of affinity and cell-surface receptor expression may lead to differential binding among clonal family members and false negatives among the sorted plasmablasts. Conversely, the increased avidity of the citrullinated-peptide tetramers may lead to some nonspecific (false positives) staining during our flow-based analysis. Thus, the flow-based tetramer staining represents a screening tool for ACPAs.
Figure 1.
Plasmablast clonal lineages persist and evolve over time in anti-CCP+ RA. A, Representative phylogenetic tree captures the relationships among HC and LC sequences from blood plasmablasts obtained at two timepoints from an anti-CCP+ RA individual. Each leaf (colored by isotype and timepoint (T)) represents the concatenated, error-corrected, IMGT-aligned, consensus HC and LC sequence for a single CD19+CD3−IgD−CD14−CD20−CD27+CD38++ plasmablast, and the tree is anchored by HC V gene assignment. Branch colors indicate clonally expanded sequences (cyan) and sequences derived from plasmablasts that bound citrullinated-peptide tetramers (pink), including singletons and those belonging to clonal expansions. Sequences selected for recombinant expression are indicated (red arrows/lines). The inset images highlight representative clonal families/lineages encoding antibodies comprised of ≥1 isotypes or present at ≥1 timepoints. B, Representative chord diagrams depict plasmablast clonal families and persistent lineages observed at serial timepoints (T) taken months (mo) after the initial timepoint (T1). Persistent lineages include singleton observations at any one timepoint. Sector width and color indicate size and average number of mutations, respectively, for the given family/lineage at each timepoint. Connections between related lineages at different timepoints are colored by isotype. Lineages shared by ≥3 timepoints for Subject 3 are shaded darker to highlight these persistent lineages.
Our bioinformatic analysis identified clonally expanded plasmablast lineages, including families observed at a single timepoint and lineages persisting across multiple timepoints (Figure 1B, Supplementary Figure 1). Across the eight subjects, we identified 170 persistent lineages comprised of single and/or multiple members at different timepoints, and these chord plots highlight several important trends. We observed persistent lineages in all RA subjects examined. For Subject 3, select lineages persisted across four timepoints, spanning almost one year. Most lineages consistently expressed one isotype while 33 (19%) included multiple isotypes – orange-colored connecting chords across sectors (Figure 1B, Supplementary Figure 1). Although some sequences were identical across timepoints, certain plasmablast lineages exhibited changes (e.g. increases or decreases) in the average number of HC V gene SHMs between timepoints – changing sector colors.
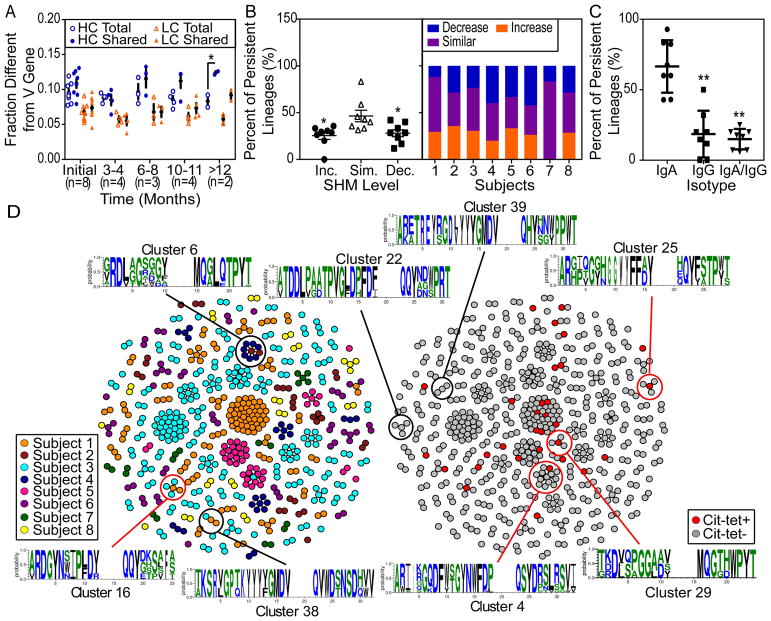
Further bioinformatic analysis of SHM levels showed that plasmablast sequences belonging to lineages shared across multiple timepoints possess a higher SHM level within the HC V gene, compared to the total sequenced plasmablast population. The number of mutations from the aligned germline sequence was normalized by the length of the V region identified. We observed a statistically significant increase in the SHM level in the persistent lineage-derived HC sequences compared to the total population at ≥12 months from T1 (Figure 2A, n = 2). Without sequence length normalization, we observed a mean of 29.6 mutations in the HC V gene among the T1-derived sequences that belong to persistent lineages, across each subject’s average mutation level (Supplementary Figure 2A). We observed similar trends when focusing on the full-length sequences, with a mean of 30.6 mutations in the HC V gene among the seven subjects with full-length T1-derived persistent lineage sequences (Supplementary Figure 2B). Overall, we identified 46 (27%) persistent lineages demonstrating a high level of SHM with a mean of ≥40 nucleotide mutations in the HC V gene compared to germline in ≥1 timepoint.
Figure 2.
A subset of the persistent lineages increase in SHMs over time, and persistent lineages predominantly express the IgA isotype. A, Sequences from eight anti-CCP+ RA subjects were analyzed for differences in SHM levels among all consensus, IMGT-aligned sequences (Total) compared to the persistent lineage-derived subset (Shared). * = P < 0.05 by two-way ANOVA, Tukey correction. B, Persistent lineages were evaluated for SHM levels and categorized according to average SHM level over time: increasing (Inc.), maintaining similar levels (Sim.), or decreasing (Dec.). * = P < 0.05 by one-way ANOVA, Tukey correction. C, For IgA/IgG-expressing plasmablasts, the percent of persistent lineages consistently expressing IgA was compared to that utilizing IgG or both IgG and IgA. ** = P < 0.01 by one-way ANOVA, Tukey correction. In panels A–C, dots represent individuals and bars indicate mean ± SEM. D, Persistent lineage-derived sequences were evaluated for similarities in CDR3 regions, and clusters visualized by igraph. Each dot represents the sequence from a single cell. Dots are colored by subject (left) or by citrullinated-peptide tetramer staining (right). Clusters with ≥1 member binding to citrullinated-peptide tetramers are circled in red. Motifs of clusters comprised of ≥1 subject were visualized (WebLogo3.5.0).
Focusing on the persistent lineages, we determined the average fold change in normalized SHMs across different timepoints and identified subsets of lineages that were increasing (>1.1-fold change), decreasing (<0.9-fold change) or staying the same (Supplementary Figure 2C–E). Although most lineages were changing (i.e, increasing or decreasing) in SHM levels over time, evaluating each subset separately showed significantly more lineages maintaining similar SHM levels (P <0.05; Figure 2B). Across the individual subjects, we observed that different subjects exhibited varying percentages of lineages belonging to each category. Subjects 4 and 6 had the highest percentage of lineages with decreasing SHMs, and Subject 4 also had the second lowest percentage of lineages with increasing SHMs. Compared to other subjects, Subject 4 also exhibited a lower mutation level in the HC and LC V genes among persistent lineage-derived sequences. Interestingly, Subject 4 showed low levels of disease with zero tender/swollen joints at T1 and four months later (Table 1).
We characterized isotype usage among IgA and IgG-expressing plasmablasts. We observed a significantly higher number of IgA plasmablasts captured at each timepoint for each subject (P < 0.01; Supplementary Figure 2F). Further, our analysis of persistent lineages revealed that a significantly higher percentage of persistent lineages expressed antibodies of the IgA isotype compared to IgG (P < 0.01; Figure 2C). Although no correlation was found with mutation levels, we observed an inverse correlation between the number of tender and swollen joints and normalized average size of clonal families (Supplementary Figure 2G). Together, our observations suggest persistent antigen stimulation of the plasmablast response, and these persistent lineages predominantly express IgA. Among all of the persistent lineages identified, we observe a subset that continue to increase in average SHM levels over months in individuals with established anti-CCP+ RA.
Analysis of persistent lineages across patients provides evidence of convergence
To characterize amino acid CDR3 sequence motifs among persistent lineages identified in different subjects, we performed cluster analysis on the paired amino acid HC and LC CDR3 sequences from individual plasmablasts using CD-HIT [28,29]. This analysis revealed multiple clusters containing shared sequence motifs across subjects (70% identity) within the identified persistent lineages (Figure 2D). Sequences of two highly related clusters were grouped for motif generation (Cluster 6, Figure 2D). Within the clusters, we identified sequences produced by plasmablasts that bound the citrullinated-peptide tetramers (Figure 2D). Thus, CD-HIT cluster analysis demonstrated that individuals with established RA generate persistent B cell lineages that exhibit shared amino acid motifs across individuals, some of which express ACPA.
Identification of the antigen targets of recombinant antibodies derived from RA plasmablast lineages
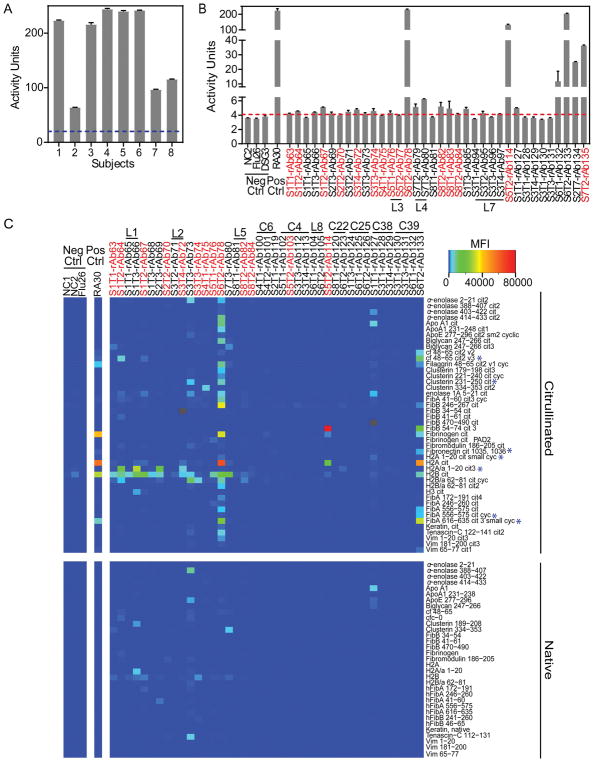
We selected 71 plasmablast antibody sequences for recombinant expression (Supplementary Table 3) as representatives of (i) clonal families/persistent lineages unique to individuals, (ii) lineages possessing HC and/or LC CDR3 amino acid sequence motifs shared across subjects, and (iii) singleton antibodies derived from plasmablasts that bound citrullinated-peptide tetramers. We used negative control antibodies reactive to influenza [38], desmoglein 3 [39], or derived from ongoing projects investigating non-RA diseases. All antibodies were expressed on the human IgG1 Fc domain. Subject plasma (Figure 3A), and all subject-derived recombinant antibodies (Figure 3B; Supplementary Figure 3A) were assayed for citrullinated peptide binding by CCP3.1 ELISA. We used RA antigen planar (Figure 3C) and bead-based (Supplementary Figure 3B) arrays to further characterize binding specificities. These analyses identified clonal family/lineage recombinant antibodies that bound citrullinated epitopes in fibrinogen, histones, clusterin, or α-enolase. Importantly, certain later-timepoint-derived clonal lineage members (e.g. rAb66 and rAb82) bound an expanded set of antigen targets compared to antibodies derived from earlier timepoints (Figure 3C).
Figure 3.
Recombinant antibodies representing clonal lineages bind citrullinated antigens. The CCP3.1 ELISA was used to measure anti-CCP antibody binding A, in RA subject plasma and B, among the expressed recombinant antibodies. Using the manufacturer’s protocol, activity units and plasma activity cutoff (blue dotted line) were determined. For recombinant antibodies, the activity cutoff (red dotted line) was set to three standard deviations above the average binding activity of negative control antibodies. Data presented as mean ± SEM from duplicates. C, Recombinant antibodies were evaluated for epitope specificity using an RA antigen microarray that includes ~350 citrullinated and native proteins/peptides. Microarrays were probed with recombinant antibodies representing clonal families/lineages and singletons derived from plasmablasts binding to citrullinated-peptide tetramers. The presented heatmap depicts median fluorescent intensities (MFI) from quadruplicate print spots. Gray indicates an antibody-antigen combination for which data could not be obtained. Each antibody is labeled according to the subject (S) and timepoint (T) from which it is derived. The recombinant antibodies derived from a plasmablast that stained for the citrullinated-peptide tetramers during sorting are marked in red. Further, similar peptide sequences to those used to generate the tetramers are marked with a blue asterisk.
Affinity maturation drives epitope spreading
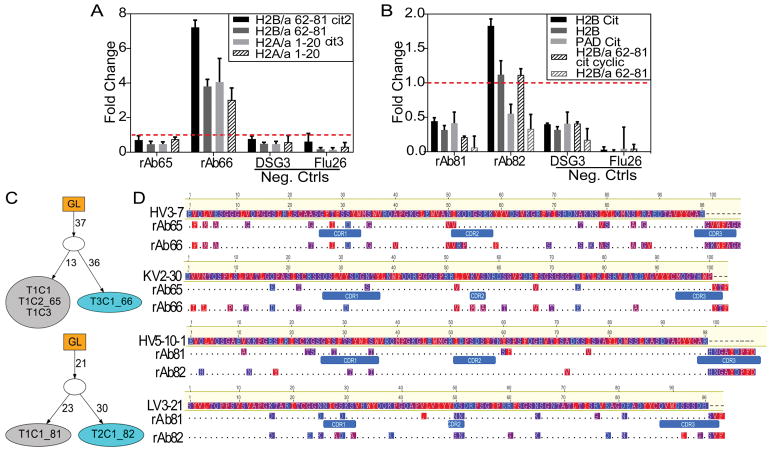
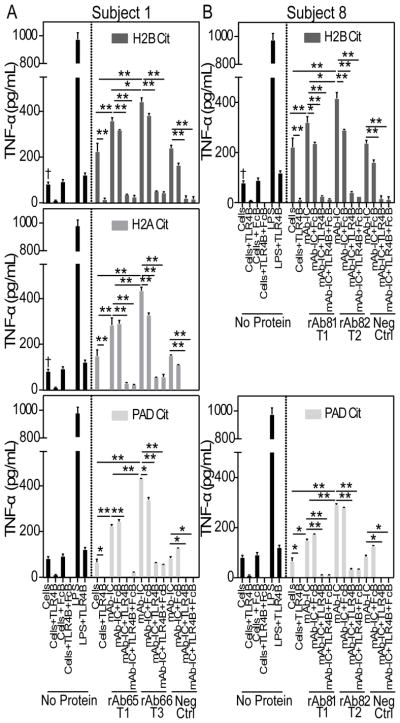
In-depth characterization of persistent lineages from different subjects revealed mutation patterns suggesting that affinity maturation drives epitope spreading. ELISA analysis of antibodies from a Subject 1-derived lineage confirmed that the later-timepoint-derived antibody, rAb66, bound specific epitopes of H2B and H2A, while the earlier member, rAb65, did not bind these epitopes (Figure 4A). We also observed substantial binding differences for rAb65 and rAb66 to full-length, in vitro citrullinated H2A, H2B and PAD, which can auto-citrullinate [40] (Supplementary Figure 4A). Similarly, for a Subject 8-derived lineage, the later-timepoint, more-mutated rAb82 bound full-length, citrullinated H2B and an H2B-derived epitope while rAb81 did not bind (Figure 4B). IgTree-based [33] analysis revealed a bifurcation from a common parent with 37 and 21 shared alterations for rAb65/rAb66 and rAb81/rAb82, respectively, followed by continued, divergent evolution (Figure 4C). Direct alignment with germline HC and LC V sequences demonstrated substantial differences between the lineage members in regions outside of CDR3 (e.g. CDR1, CDR2, and framework; Figure 4D).
Figure 4.
Alternatively mutated plasmablast recombinant antibodies exhibit differential binding to citrullinated antigens. Binding activity of recombinant antibodies representing persistent lineages from A, Subject 1 and B, Subject 8 to full-length proteins and specific epitopes was assessed by ELISA. The fold change (dotted line) above an activity cutoff of three standard deviations above the negative control antibodies was calculated. Data presented as the mean ± SEM from duplicates. C, These persistent lineage members were subjected to IgTree analysis to characterize the degree of relatedness between members and the distinct SHMs accumulated in each B cell-encoded antibody over the course of the affinity maturation process. Connectors between nodes of family trees indicate the number of mutations accumulated for each member compared to the germline HC and LC VJ gene sequences. Cyan-colored nodes represent lineage members binding to additional epitopes by ELISA. D, Nucleotide-based, full-length HC and LC antibody sequences were converted to amino acid sequences and aligned to germline sequences (Geneious). Segments corresponding to different CDRs are indicated. The observed differences from germline of these evolving ACPAs are highlighted and colored by hydrophobicity with red indicating more hydrophobic residues.
Persistent lineage-derived antibodies (rAb71, rAb72) identified in Subject 3 with differential reactivity by planar antigen array exhibited similar binding reactivity to H2A, H2B, and an H2A-derived peptide by ELISA (Supplementary Figure 4B). Evaluating antibodies with similar CDR3 sequence motifs, ELISA analysis confirmed that two clusters (C38 and C39) contained at least one antibody that bound citrullinated antigens (Supplementary Figures 4C and D). The later-timepoint member from Subject 6 (rAb133) exhibited differential binding activity compared to the earlier-timepoint member. Using antigen microarray and ELISA analyses, we confirmed that certain later-timepoint, persistent plasmablast lineage members encoded antibodies that differentially bound citrullinated antigens. Further, antibodies present at later timepoints possessed increased levels of and/or divergent SHMs. Together, these findings suggest that affinity maturation over time drives epitope spreading of the ACPA response.
Increased stimulation of macrophage TNF-α production by affinity-matured recombinant antibodies
To examine the ability of representative, persistent plasmablast lineage-derived, recombinant antibodies to stimulate macrophage TNF-α production, we generated plate-bound ICs of these antibodies with citrullinated H2A, H2B, or PAD. Human monocyte-derived macrophages produced significantly higher levels of TNF-α in response to ICs generated with recombinant ACPAs representative of persistent lineages from two different subjects compared to citrullinated antigen alone (P < 0.001; Figures 5A and B). Importantly, the negative control antibody did not induce significantly higher levels of macrophage TNF-α production compared to citrullinated antigen alone (Figures 5A and B). Additionally, citrullinated H2A and H2B significantly increased TNF-α production compared to cells alone (P < 0.05; Figure 5A and B); however, plate-bound PAD, which included all in vitro citrullination reagents but lacked H2A or H2B, resulted in a similar level of TNF-α production compared to cells alone.
Figure 5.
RA subject-derived recombinant antibody:citrullinated-antigen immune complexes stimulate macrophages to produce TNF-α. Plate-bound immune complexes were formed by coating wells with a citrullinated antigen (H2B, H2A or PAD) followed by incubation with recombinant antibodies representing persistent lineages from A, Subject 1 or B, Subject 8. Monocyte-derived macrophages were added to each well, and after 24 hrs, supernatants harvested and TNF-α levels measured using ELISA. A portion of macrophages were pre-incubated with the TLR4 inhibitor TAK-242 (TLR4B) and FcγRII inhibitor anti-CD32 clone IV.3 (FcRB) prior to addition to the plate. LPS was used as a positive control, and cells alone with buffers as negative controls. The same controls are displayed in multiple panels for comparison. TNF-α levels were compared between antibodies observed at different timepoints within the same lineage, and differences in levels produced in the presence of inhibitors were compared to IC alone. ** = P < 0.001; * = P < 0.05 by two-way ANOVA, Tukey correction. Cells incubated with citrullinated proteins without IC were also compared to cells alone. † = P < 0.05 by one-way ANOVA, Tukey correction. Representative data presented as mean ± SEM, with similar trends observed in two independent experiments.
To investigate the role of FcγRII and TLR4, we pre-incubated cells with blocking reagents prior to incubation with immobilized ICs. Blocking FcγRII reduced the levels of TNF-α produced compared to IC alone, with statistically significant reductions observed for rAb81 and rAb82 complexed with citrullinated H2B (P < 0.05; Figure 5B) and rAb66 complexed with citrullinated H2A or PAD (Figure 5A). Further, the TLR4 inhibitor significantly reduced TNF-α levels stimulated by citrullinated antigens alone or in complex with recombinant antibody (P < 0.001; Figures 5A and B). As shown in previous publications [16,17], TLR4 blockade exhibited a dominant effect in inhibiting citrullinated antigen-IC stimulation of macrophages to produce TNF-α. No differences were observed between TLR4 inhibitor alone or in combination with blocking FcγRII.
We further evaluated the effect of accumulated SHMs on IC-based macrophage stimulation. Recombinant antibodies observed at later timepoints and possessing higher levels of SHMs (i.e. rAb66 and rAb82) generated ICs that stimulated macrophages to produce significantly more TNF-α compared to less-mutated antibodies derived from the same lineage at earlier timepoints (P < 0.05; Figure 5A and B). Thus, individual recombinant ACPA form ICs that stimulate macrophage TNF-α production. These results reveal that affinity maturation resulted in ACPAs that formed ICs that more potently stimulated macrophage TNF-α production through FcγRII and TLR4.
Discussion
Here, we characterized the evolving ACPA plasmablast response in individuals with established RA by integrating antibody sequence information with specificity and functional activity of the encoded antibodies. We identified clonal families and persistent lineages expressing ACPA using citrullinated-peptide tetramers, and representative recombinant antibodies were characterized for their binding specificity and functional properties. Our analysis revealed that divergently mutated, later-timepoint plasmablast antibodies exhibited differential binding specificity and increased potency to stimulate macrophages to produce TNF-α, compared to less-mutated, earlier-timepoint lineage members.
Our findings suggest that persistent antigen stimulation of B cell lineages in individuals with RA results in continued affinity maturation and high SHM levels. Persistent lineages shared IMGT-based assignments of HC and LC VJ genes and 60% identity within HC and LC CDR3 (Levenshtein distance [26]). Persistent lineage-derived sequences exhibited higher levels of HC V gene SHM compared to the total sequenced plasmablast population. Across the eight subjects, we observed a mean of 29.6 mutations within the HC V gene of persistent lineages at T1 and 27% of persistent lineages averaging ≥40 mutations in the HC V gene in ≥1 timepoint. These mutation levels are substantially higher compared to HC V genes of H1N1-induced B cells (mean ~19 mutations per patient) from five subjects [41] or IgG-expressing memory B cells from three healthy donors (mean 18 ± 8.1 mutations) [42]. This high SHM level is likely generated through germinal center (GC)-mediated affinity maturation and lineage expansion during which B cells undergo iterations of division and mutation in the GC dark zone (DZ) and selection in the light zone (LZ), with up to six divisions observed in the DZ [43]. At a mutation rate of ~10−3 per base pair per cell division, it is estimated that there will be ≥1 mutation across HC and LC V genes per every two divisions [44]. Thus, we estimate that these highly mutated plasmablast lineages present in established RA resulted from >80 cell divisions and repeated rounds of division/selection in GCs. At later timepoints, multiple persistent lineages exhibited increased and differential mutations among lineage members, while other lineages exhibited fewer mutations at later timepoints. Since we only sampled the repertoire at set timepoints, these observations may arise through (i) recruitment of a common progenitor that undergoes successive rounds of division, mutation, and selection in the GC, or (ii) stimulation of distinct branches of the lineage tree at different timepoints.
A significantly higher percentage of the persistent plasmablast lineages express IgA compared to IgG. Since IgA represents the dominant isotype in mucosally-driven immune responses, this suggests that a persistent mucosal antigen stimulates the ongoing IgA plasmablast response in established RA. This is consistent with previous studies demonstrating an elevated percentage of IgA plasmablasts in anti-CCP+ individuals at risk for RA [20], association of periodontal disease with RA [45], and the presence of ACPA in sputum of individuals with early RA [10]. In summary, our findings demonstrate a continued, ongoing IgA plasmablast response in established RA, suggesting that a persistent mucosal antigen stimulus plays a key role in mediating the persistence of RA.
Our results support a mechanism by which evolving B cell lineages produce ACPAs that contribute to the pathogenesis of RA by stimulating macrophage TNF-α production. Previous studies from our lab demonstrated that citrullinated fibrinogen or H2B ICs generated using pooled IgG from RA plasma stimulate macrophages to produce TNF-α [16,17]. Building upon these previous observations, we expressed the recombinant antibodies on the IgG1 backbone as a research tool and demonstrated that individual, RA subject-derived ACPAs formed ICs capable of stimulating macrophage TNF-α production. Further, persistent lineage antibodies observed at later timepoints that possess increased levels of SHM encode antibodies that (i) exhibit differential binding to citrullinated antigens and (ii) more potently stimulate macrophages to produce TNF-α. We note that the antibodies derived from later timepoints do not fully share all SHMs as that observed at the earlier timepoint; however, our IgTree analysis showed that plasmablasts derived at different timepoints share a common parent cell. Thus, differential, continued affinity maturation from this common parent cell results in the generation of antibodies that exhibit epitope spreading and more potently promote inflammation in RA.
Data from this study provides evidence of a common process by which individuals with RA generate persistent B cell lineages with shared CDR3 sequence motifs. Previous studies provided evidence of convergent evolution of the antibody response, such as in response to Dengue virus infection [46] and antibodies against the CD4 binding site of HIV-1 [47]. For RA, we demonstrate that certain persistent B cell lineages share CDR3 amino acid sequence motifs across individuals. In two of these shared clusters, we confirmed that a representative member bound citrullinated antigens by ELISA, suggesting that these motifs contain key CDR3 residues that mediate citrullinated-epitope binding.
There are several limitations to this study. First, we focused on characterizing the ongoing immune response through sequencing the plasmablast repertoire. We did not sequence memory B cells, which might contain common B cell progenitors. Second, we performed single-cell sequencing with limited depth. Future studies could utilize a droplet-based method to sequence a larger number of B cells. Third, recombinant antibody binding specificity was analyzed using an antigen microarray containing ~350 distinct proteins/peptides, and it is likely that certain antigen targets are not included on this array. Fourth, to isolate ICs for macrophage stimulation, we formed plate-bound ICs; however, this approach may lead to structural constraints, potentially reducing the capacity of these ICs to stimulate macrophage TNF-α production compared to in vivo IC formation. Finally, we used 14 different citrullinated-peptide tetramers to identify ACPA-producing plasmablasts; however, future studies could more thoroughly characterize ACPA-expressing B cells by including additional citrullinated peptides and/or proteins.
In summary, sequencing blood plasmablasts over serial timepoints revealed that ACPA-encoding B cell lineages persist and evolve, exhibiting extensive, progressive and differential SHM. Intriguingly, these persistent lineages predominantly express IgA, suggesting a continued mucosal drive in established RA, an observation that may provide important insight into the mechanisms underlying the persistence of RA. Continued affinity maturation results in epitope spreading such that B cell lineage members observed at later timepoints express ACPA with expanded citrullinated antigen reactivities. Importantly, differentially evolved ACPAs observed at later timepoints formed ICs that more potently stimulated macrophage TNF-α production, highlighting the potential for an increased pathogenic role of these affinity-matured ACPAs. The level of TNF-α produced by IC stimulation of macrophages is reduced by TLR4 blockade, demonstrating the potential of TLR4 as a therapeutic target in RA, as suggested by previous studies [16,17,48,49]. Together, our findings demonstrate that affinity maturation plays a critical role in epitope spreading and generation of ACPAs that more potently drive macrophage TNF-α production in RA.
Supplementary Material
Acknowledgments
Funding: Funding was provided by NIH NIAMS R01 AR063676, NIH NIAID U19 AI11049103, and NIH NIAID U01 AI101981 to WHR. SEE received support from NIH NIAID 5T32 AI07290-30.
We thank the Stanford Functional Genomics Facility (SFGF) and Stanford Shared FACS Facility (SSFF) for their assistance. The antibody repertoire sequencing data has been deposited in the NCBI Sequence Read Archive (SRA), accession SRP150122.
Footnotes
Conflict of interest: William Robinson is a Founder, member of the Board of Directors, consultant for, and owner of equity in Atreca, Inc. All other authors declare no conflicts.
Author Contributions
All authors assisted with drafting or revising the manuscript, and all authors approved the final version to be published. Dr. Robinson had full access to all of the data in this study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design: Elliott, Robinson
Acquisition of data: Elliott, Kongpachith, Lingampalli, Adamska, Cannon
Analysis and interpretation of data: Elliott, Mao, Blum, Robinson
References
- 1.Helmick CG, Felson DT, Lawrence RC, Gabriel S, Hirsch R, Kwoh CK, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States: Part I. Arthritis Rheum. 2008;58:15–25. doi: 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- 2.Wang S, Wang Y. Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis. Biochim Biophys Acta. 2013;1829:1126–1135. doi: 10.1016/j.bbagrm.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sokolove J, Bromberg R, Deane KD, Lahey LJ, Derber LA, Chandra PE, et al. Autoantibody epitope spreading in the pre-clinical phase predicts progression to Rheumatoid Arthritis. PLOS One. 2012;7:e35296. doi: 10.1371/journal.pone.0035296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhn KA, Kulik L, Tomooka B, Braschler KJ, Arend WP, Robinson WH, et al. Antibodies against citrullinated proteins enhance tissue injury in experimental autoimmune arthritis. J Clin Invest. 2006;116:961–973. doi: 10.1172/JCI25422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uysal H, Bockermann R, Nandakumar KS, Sehnert B, Bajtner E, Engström A, et al. Structure and pathogenicity of antibodies specific for citrullinated collagen type II in experimental arthritis. J Exp Med. 2009;206:449–462. doi: 10.1084/jem.20081862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isaacs JD, Cohen SB, Emery P, Tak PP, Wang J, Lei G, et al. Effect of baseline rheumatoid factor and anticitrullinated peptide antibody serotype on rituximab clinical response: a meta-analysis. Ann Rheum Dis. 2013;72:329–336. doi: 10.1136/annrheumdis-2011-201117. [DOI] [PubMed] [Google Scholar]
- 7.Cambridge G, Leandro MJ, Lahey LJ, Fairhead T, Robinson WH, Sokolove J. B cell depletion with rituximab in patients with rheumatoid arthritis: Multiplex bead array reveals the kinetics of IgG and IgA antibodies to citrullinated antigens. J Autoimmun. 2016;70:22–30. doi: 10.1016/j.jaut.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Demoruelle MK, Deane KD, Holers VM. When and Where Does Inflammation Begin in Rheumatoid Arthritis? Curr Opin Rheumatol. 2014;26:64–71. doi: 10.1097/BOR.0000000000000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klareskog L, Stolt P, Lundberg K, Källberg H, Bengtsson C, Grunewald J, et al. A new model for an etiology of rheumatoid arthritis: Smoking may trigger HLA–DR (shared epitope)–restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006;54:38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 10.Willis VC, Demoruelle MK, Derber LA, Chartier-Logan CJ, Parish MC, Pedraza IF, et al. Sputum Autoantibodies in Patients With Established Rheumatoid Arthritis and Subjects at Risk of Future Clinically Apparent Disease. Arthritis Rheum. 2013;65:2545–2554. doi: 10.1002/art.38066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.England BR, Thiele GM, Mikuls TR. Anticitrullinated protein antibodies: origin and role in the pathogenesis of rheumatoid arthritis. Curr Opin Rheumatol. 2017;29:57–64. doi: 10.1097/BOR.0000000000000356. [DOI] [PubMed] [Google Scholar]
- 12.Kinne RW, Bräuer R, Stuhlmüller B, Palombo-Kinne E, Burmester G-R. Macrophages in rheumatoid arthritis. Arthritis Res Ther. 2000;2:189. doi: 10.1186/ar86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clavel C, Ceccato L, Anquetil F, Serre G, Sebbag M. Among human macrophages polarised to different phenotypes, the M-CSF-oriented cells present the highest pro-inflammatory response to the rheumatoid arthritis-specific immune complexes containing ACPA. Ann Rheum Dis. 2016;75:2184–2191. doi: 10.1136/annrheumdis-2015-208887. [DOI] [PubMed] [Google Scholar]
- 15.Laurent L, Clavel C, Lemaire O, Anquetil F, Cornillet M, Zabraniecki L, et al. Fcγ receptor profile of monocytes and macrophages from rheumatoid arthritis patients and their response to immune complexes formed with autoantibodies to citrullinated proteins. Ann Rheum Dis. 2011;70:1052–1059. doi: 10.1136/ard.2010.142091. [DOI] [PubMed] [Google Scholar]
- 16.Sokolove J, Zhao X, Chandra PE, Robinson WH. Immune Complexes Containing Citrullinated Fibrinogen Co-Stimulate Macrophages via Toll-like receptor 4 and Fcγ receptor. Arthritis Rheum. 2011;63:53–62. doi: 10.1002/art.30081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohn DH, Rhodes C, Onuma K, Zhao X, Sharpe O, Gazitt T, et al. Local Joint Inflammation and Histone Citrullination in a Murine Model of the Transition From Preclinical Autoimmunity to Inflammatory Arthritis. Arthritis Rheumatol. 2015;67:2877–2887. doi: 10.1002/art.39283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clavel C, Nogueira L, Laurent L, Iobagiu C, Vincent C, Sebbag M, et al. Induction of macrophage secretion of tumor necrosis factor α through Fcγ receptor IIa engagement by rheumatoid arthritis–specific autoantibodies to citrullinated proteins complexed with fibrinogen. Arthritis Rheum. 2008;58:678–688. doi: 10.1002/art.23284. [DOI] [PubMed] [Google Scholar]
- 19.Tan Y-C, Kongpachith S, Blum LK, Ju C-H, Lahey LJ, Lu DR, et al. Barcode-enabled sequencing of plasmablast antibody repertoires in rheumatoid arthritis. Arthritis Rheumatol Hoboken NJ. 2014;66:2706–2715. doi: 10.1002/art.38754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinslow JD, Blum LK, Deane KD, Demoruelle MK, Okamoto Y, Parish MC, et al. Elevated IgA Plasmablast Levels in Subjects at Risk of Developing Rheumatoid Arthritis. Arthritis Rheumatol. 2016;68:2372–2383. doi: 10.1002/art.39771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerkman PF, Rombouts Y, Voort EIH, van der Trouw LA, Huizinga TWJ, Toes RE, et al. Circulating plasmablasts/plasmacells as a source of anticitrullinated protein antibodies in patients with rheumatoid arthritis. Ann Rheum Dis. 2013;72:1259–1263. doi: 10.1136/annrheumdis-2012-202893. [DOI] [PubMed] [Google Scholar]
- 22.Arnett FC, Edworthy SM, Bloch DA, Mcshane DJ, Fries JF, Cooper NS, et al. The american rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 23.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 24.Lu DR, Tan Y-C, Kongpachith S, Cai X, Stein EA, Lindstrom TM, et al. Identification of functional anti-staphylococcus aureus antibodies by sequencing patient plasmablast antibody repertoires. Clin Immunol. 2014 doi: 10.1016/j.clim.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alamyar E, Giudicelli V, Li S, Duroux P, Lefranc M-P. IMGT/HighV-QUEST: the IMGT® web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 2012;8:26. [Google Scholar]
- 26.Levenshtein VI. Binary Codes Capable of Correcting Deletions, Insertions and Reversals. Sov Phys Dokl. 1966;10:707–710. [Google Scholar]
- 27.Gupta NT, Heiden V, AJ, Uduman M, Gadala-Maria D, Yaari G, et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinforma Oxf Engl. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 29.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinforma Oxf Engl. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal. 2006:1695. Complex Systems. [Google Scholar]
- 31.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A Sequence Logo Generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2. 0. Bioinforma Oxf Engl. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 33.Barak M, Zuckerman NS, Edelman H, Unger R, Mehr R. IgTree: creating Immunoglobulin variable region gene lineage trees. J Immunol Methods. 2008;338:67–74. doi: 10.1016/j.jim.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Gansner ER, North SC. An open graph visualization system and its applications to software engineering. Softw Pract Exp. 2000;30:1203–1233. [Google Scholar]
- 35.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinforma Oxf Engl. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair N, Feng N, Blum LK, Sanyal M, Ding S, Jiang B, et al. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Sci Transl Med. 2017;9:eaam5434. doi: 10.1126/scitranslmed.aam5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hueber W, Kidd BA, Tomooka BH, Lee BJ, Bruce B, Fries JF, et al. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 2005;52:2645–2655. doi: 10.1002/art.21269. [DOI] [PubMed] [Google Scholar]
- 38.Tan Y-C, Blum LK, Kongpachith S, Ju C-H, Cai X, Lindstrom TM, et al. High-throughput sequencing for natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin Immunol. 2014;151:55–65. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeh S-W, Cavacini LA, Bhol KC, Lin M-S, Kumar M, Duval M, et al. Pathogenic human monoclonal antibody against desmoglein 3. Clin Immunol. 2006;120:68–75. doi: 10.1016/j.clim.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Andrade F, Darrah E, Gucek M, Cole RN, Rosen A, Zhu X. Autocitrullination of human peptidyl arginine deiminase type 4 regulates protein citrullination during cell activation. Arthritis Rheum. 2010;62:1630–1640. doi: 10.1002/art.27439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wrammert J, Koutsonanos D, Li G-M, Edupuganti S, Sui J, Morrissey M, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal center by regulated proliferation and hypermutation. Nature. 2014;509:637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kindt TJ, Goldsby RA, Osborne BA, Kuby J. Kuby Immunology. Macmillan; 2007. [Google Scholar]
- 45.Potikuri D, Dannana KC, Kanchinadam S, Agrawal S, Kancharla A, Rajasekhar L, et al. Periodontal disease is significantly higher in non-smoking treatment-naive rheumatoid arthritis patients: results from a case-control study. Ann Rheum Dis. 2012;71:1541–1544. doi: 10.1136/annrheumdis-2011-200380. [DOI] [PubMed] [Google Scholar]
- 46.Parameswaran P, Liu Y, Roskin KM, Jackson KK, Dixit VP, Lee J-Y, et al. Convergent antibody signatures in human dengue. Cell Host Microbe. 2013;13:691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatterer E, Shang L, Simonet P, Herren S, Daubeuf B, Teixeira S, et al. A specific anti-citrullinated protein antibody profile identifies a group of rheumatoid arthritis patients with a toll-like receptor 4-mediated disease. Arthritis Res Ther. 2016:18. doi: 10.1186/s13075-016-1128-5. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5053084/ [DOI] [PMC free article] [PubMed]
- 49.Davis MLR, LeVan TD, Yu F, Sayles H, Sokolove J, Robinson W, et al. Associations of toll-like receptor (TLR)-4 single nucleotide polymorphisms and rheumatoid arthritis disease progression: An observational cohort study. Int Immunopharmacol. 2015;24:346–352. doi: 10.1016/j.intimp.2014.12.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.