Abstract
Objective
Lung cancer is classified as a single entity comprised of multiple histological subtypes. But how similar are these subtypes on a genetic level? This commentary aims to address this question through a concise overview of germline and somatic differences between small-cell lung cancer (SCLC), lung adenocarcinoma (LUAD), and lung squamous cell carcinoma (LUSC).
Methods
We reveal the weak overlap found between these three lung cancer subtypes using published data from one of the largest germline genetic studies of lung cancer to date and somatic mutation data from Catalogue Of Somatic Mutations In Cancer (COSMIC).
Results
These data indicate these three subtypes share very little with each other at the genetic level. At the germline single-nucleotide polymorphism (SNP) level, only 24 independent SNPs from two chromosomes were shared across all three subtypes. We also demonstrate that only 30 unique cancer-specific mutations overlap the three subtypes from COSMIC and that this is fewer than overlapping mutations chosen at random. Finally, we show that only 3 somatic mutational signatures are shared between these three subtypes.
Conclusion
This commentary highlights that these three lung cancer subtypes may be distinct diseases at the genetic level. In the era of precision medicine, we feel that these genomic differences will be of utmost importance in the choice of lung cancer therapy in the future.
Keywords: Lung Cancer, Mutation, Genomics, SNPs, Small Cell Lung Cancer, Non-Small-Cell Lung Cancer
Introduction
Lung cancer is the leading cause of cancer mortality in the world [1]. Histological classifications separate lung cancer into several subtypes, but accumulating evidence [2–4] suggests that outside of their location in the lung, these subtypes of lung cancer share little in common and may in fact be distinct diseases at the genetic level. As the genetic evidence described below suggests, lung cancer subtypes can be characterized by germline and somatic genetic features that may represent unique biology and may also respond differently to therapies.
Lung cancer is first classified into two main types by its characteristic cellular size: non-small cell lung cancer (NSCLC) and small-cell lung cancer (SCLC). NSCLC comprises 85% of lung cancer, and two of its major histologic subtypes, adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC), comprise 40% and 30% of new lung cancer cases, respectively [5]. SCLC comprises approximately 15% of new lung cancer cases and can be further divided into several rare histologic subtypes but is often studied as a single entity. While these common classifications are based upon histology, the recent influx of genetic data has allowed for deeper characterization. These genetic data have been identified from both the germline and somatic genomes of lung cancer patients.
Functional differences between subtypes
We recently performed a study to identify the regulatory common genetic variants (SNPs) associated with each of these lung cancer subtypes [6]. Our approach identified regulatory SNPs within expression quantitative trait loci (eQTLs) and enhancers and their target genes which were common to all lung cancer subtypes using regulatory data from The Functional Annotation Of the Mammalian genome (FANTOM) project, the Genotype-Tissue Expression (GTEx) project and two additional studies [7, 8]. Strikingly, we found that although more than 150 target genes were associated with each subtype, there were only five genes (from one genomic region) that overlapped between LUAD, LUSC, and SCLC. We also found that this weak overlap extended to biological pathways and the regulatory mechanisms in each subtype.
Differences in germline genetic risk for three lung cancer subtypes
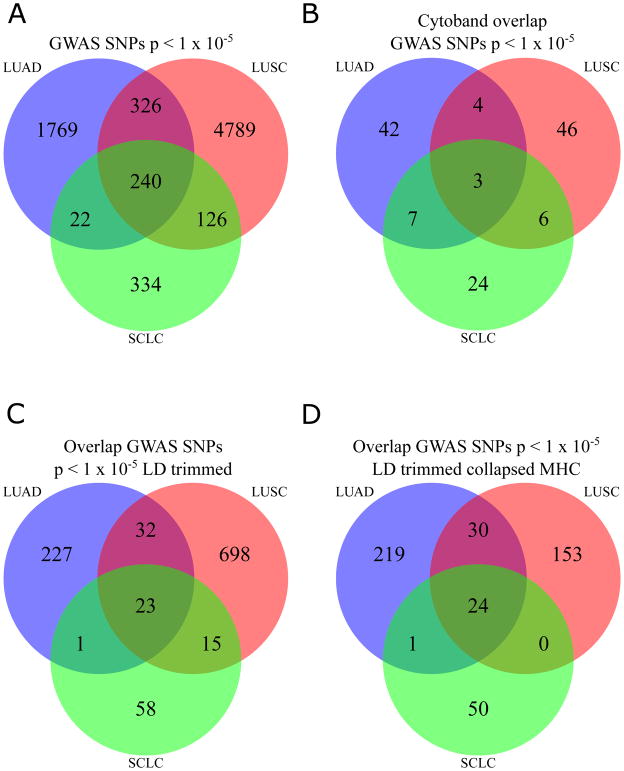
In confirmation of our results, a genome-wide association study (GWAS) recently published by McKay et al. [9] examining these three lung cancer subtypes in a European population came to similar conclusions. Importantly, the authors reported distinct genetic variants associated with each subtype at a statistical threshold of p < 5 × 10−8. To determine the overlap between the genetic variants across the lung cancer subtypes at a less stringent statistical threshold, we selected all SNPs with p-value < 1 × 10−5 from McKay et al. [9] and identified the overlap between subtypes at the SNP level. Interestingly, only 240 variants from two chromosomes (chr. 6 and chr. 15) representing three cytobands (6p22.1, 6p21.32, 15q25.1) were associated with the three most frequent lung cancer histologic groups, i.e. LUAD, LUSC, and SCLC (Fig. 1A,1B). These SNPs may represent shared genetic signals, thus we trimmed these SNPs based on linkage disequilibrium (LD) from the 1000 Genomes Phase III European Population and discovered that only 23 unlinked SNPs were common across the three lung cancer subtypes as illustrated in Figure. 1C. However, all of these overlapping SNPs were located on chromosome 15 and no overlap was observed on chromosome 6. We hypothesized that this lack of overlap is due to variants in the major histocompatibility complex (MHC) on chromosome 6 and the complex LD patterns in this region. To address these complex LD patterns, we collapsed all SNPs in the MCH region into one block for each subtype and again identified the overlap for LUAD, LUSC, and SCLC. After the MHC collapse, we found a total of 24 SNPs that overlap from chromosome 15 including the one block of the MHC region (Fig. 1D). This phenomenon of little overlap between lung cancer subtypes has also been observed in prior studies [10, 11] and highlights the distinct differences in the genetic predisposition to these cancer subtypes of the same tissue. The identification of SNPs unique to a specific lung cancer subtype is not only in European populations, but also extends to other populations as well. For example, a recent GWAS for lung cancer in African Americans found two significant SNPs near CHRNA5 and TERT where one of the SNPs, rs2853677, near the TERT gene was only significant in LUAD but not in LUSC [12]. These two loci were previously reported to be associated with lung cancer in other population types [10, 11]. In another study, Hu et al. [13] performed a GWAS for lung cancer in a Han Chinese population and found six significant SNPs (representing two novel loci 13q12.2 and 22q12.2) in four loci. For these six SNPs, two (rs2736100 and rs448809) were significantly different per subtype using LUAD, LUSC, and SCLC. This observation of difference in genetic susceptibility to lung cancer by subtypes also extends to never smokers of Asian descent. Lan et al. [14] performed a GWAS of never smoking women of Asian descent and found six loci with statistical association (three of them were new at the time of publication). They discovered that rs9387478 and rs2395185 were only associated with LUAD while rs7086803 showed a stronger effect in LUSC than LUAD. They discussed that more sample sizes would be needed for confirmation of this result. This genetic evidence also exists for somatic mutations characterizing each subtype.
Figure 1. Weak overlap between germline genetic variants in three lung cancer subtypes.
Panel A shows the overlap between single nucleotide polymorphisms (SNPs) at p < 1 × 10−5 from McKay et al. Panel B shows the overlap between subtypes by the cytobands represented by the SNPs in Panel A. The full set of SNPs trimmed by linkage disequilibrium (LD) (r2 > 0.8, 1000 Genomes Phase III) reveals few genomic regions shared by all three lung cancer subtypes in Panel C. To account for the strong LD in the MHC region, we represent all SNPs in MHC as one genomic loci in the LD trim and show the final overlapping independent genomic regions in Panel D. Of note, the 24 independent regions represented in Panel D are from two chromosomes: chromosome 6 and chromosome 15.
Somatic mutational differences between three lung cancer subtypes
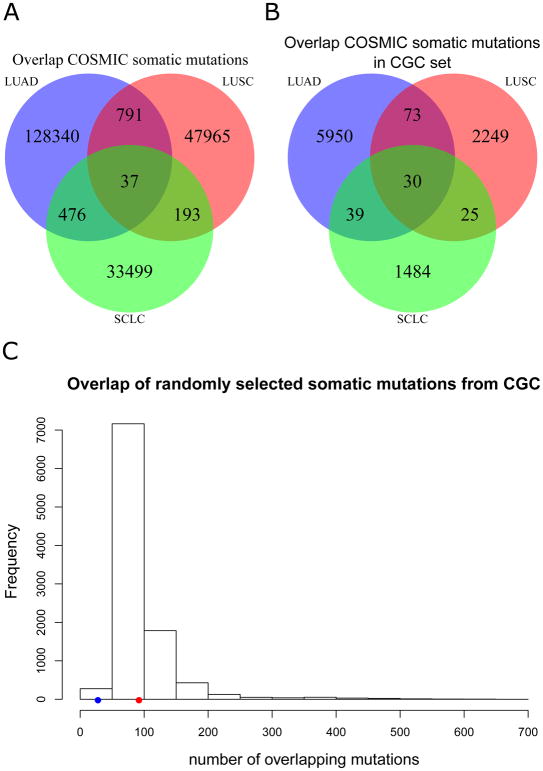
There are two major types of somatic mutations in cancer cells: driver mutations and passenger mutations. Driver mutations usually confer selective uncontrolled growth to the cells allowing them to survive, while passenger mutations are usually benign mutations that have been acquired during the life of the cell [15]. Many driver mutations occur in the same genomic location [16], so we identified the exact locations of somatic mutations in genes reported in the Catalogue Of Somatic Mutations In Cancer (COSMIC) database [17], the largest somatic mutation repository to date, to confirm whether this weak overlap between LUAD, LUSC, and SCLC germline variants expands to the somatic genome. We obtained all COSMIC somatic mutations from genome-wide screens, that do not target any specific genes, and removed mutations identified in cell lines, non-primary tumors, and silent mutations. Interestingly, only 37 identical mutations were shared between all three lung cancer subtypes out of more than 30,000 mutations in each subtype (Fig. 2A). These 37 mutations represent 10 different genes, and the majority of the mutations (> 70%) are in the well-known tumor suppressor gene TP53 that is mutated in many cancer types [17]. However, these genes that contain the exact same mutations may still be passenger genes that do not lead to cancer. To identify relevant cancer-related driver genes that are in each subtype, we additionally filtered the mutations to include genes identified in the Cancer Gene Census (CGC), a list of genes causally implicated in cancer [17]. After filtering for CGC genes, we found only 30 unique mutations that overlap LUAD, LUSC, and SCLC and these mutations were all found within three genes: TP53, BRAF, and PIK3CA (Fig. 2B). To determine if this overlap was more than would be observed by chance, we randomly selected samples from COSMIC based on matched numbers of samples for each subtype. These randomly chosen samples also matched the mutation load of each subtype. After randomly choosing these mutations, we discovered on average that 94 mutations were shared between these randomly chosen samples by chance alone. (Fig. 2C). This finding suggests that the 30 shared mutations in LUAD, LUSC, and SCLC may not be due to any shared biology and are the result of random chance. These findings agree with a recent review published by Herbst et al. [18] earlier this year, which provides a great overview of somatic alterations in key biological pathways and their mutation frequencies in LUAD and LUSC. In confirmation of the COSMIC data, the authors show that TP53, BRAF, and PIK3CA have evidence for mutation in LUAD and LUSC; albeit at different frequencies (authors did not report statistics for SCLC). Additionally, they report that many of the genes (e.g. ATM and FGFR1) are only uniquely mutated in LUAD or LUSC in confirmation of the genetic differences between lung cancer subtypes at the somatic level. For more details, see Box 1 from Herbst et al [18]. This somatic mutation evidence supports the germline variant evidence of different disease processes for LUAD, LUSC, and SCLC. We then used somatic mutational signatures to provide insight into biological processes for each disease type. Somatic mutational signatures are unique patterns of mutations and can be used to gain an understanding of the mutational processes as well as environmental exposures of a cancer sample [19]. COSMIC investigators have identified 30 different mutational signatures across 40 cancer types using over 10,000 exomes and ~ 1,000 genomes [17]. According to these 30 well-characterized mutational signatures in COSMIC, LUAD, LUSC, and SCLC share only three mutational signatures in common. Two of the three signatures (signature 1 and signature 5) have been found in each of the 40 cancer types analyzed by COSMIC and the third (signature 4) is indicative of cigarette smoking due to its high number of C → A transversions that implicate prior exposure to tobacco smoke [19].
Figure 2. Weak overlap between somatic mutations in three lung cancer subtypes.
In panel A, we show the number of identical somatic mutations shared amongst three lung cancer subtypes. Panel B shows the overlap between identical somatic mutations that are only found in the Cancer Gene Census (CGC) list. We show in panel C that randomly choosing CGC somatic mutations from all somatic mutations in COSMIC after normalizing by mutation load reveals more sharing on average (red dot, n = 94) than observed (blue dot, n = 30) between three lung cancer subtypes.
Functional pathways between subtypes
In addition to the weak overlap observed between the variants at the germline and somatic genomes, there is also weak overlap between subtypes based upon biological pathways that are enriched with these genes. For example, we found in our study [6] that only two KEGG pathways (metabolic pathways and proteasome pathways) overlapped between LUAD, LUSC, and SCLC at the germline level. Most of the biological pathways enriched with germline associated genes were unique to each subtype. At the somatic level, there also exists a weak overlap of biological pathways enriched with genes associated with the lung cancer subtypes. For example, Wang et al. [20] examined differentially expressed genes (DEGs) between LUAD and LUSC and assessed biological pathways enriched with these genes. They found many biological differences between these two subtypes based upon DEGs. Their analysis also revealed several GO terms and KEGG pathways unique to each subtype in confirmation of the trends at the germline level. Although Wang et al. did not study SCLC, a similar study was performed by Ni et al. [21], published in 2018, that found KEGG pathways and GO terms enriched with DEGs for SCLC versus normal controls. Their functional results shared little with either LUAD or LUSC and there were no KEGG pathways that overlapped between SCLC and LUAD or LUSC from these studies.
Together, these results suggest that although weak overlap occurs in genetic features at the somatic level, the overlap consists of factors relevant in most cancer types and is non-specific to lung cancer. This is especially true for the overlapping genes and mutational signatures that are likely to be found in most cancer types irrelevant of tissue type. This evidence is further supported by reports that show these subtypes are more similar to cell-like tumors in other organs. For example, LUSC is more similar to other squamous like tumors such as head and neck squamous cell carcinoma [2, 3] than to LUAD.
Additional differences between subtypes
In addition to the genetic differences between subtypes, there are also epidemiologic and clinical differences. For example, most never smokers who develop lung cancer develop LUAD. LUAD also has a higher prevalence among women [22]. Conversely, SCLC and LUSC are heavily influenced by smoking and LUSC is more commonly found in males than females. Additionally, LUAD is found peripherally in the lungs and is the least aggressive tumor type in comparison to the more aggressive centrally located LUSC and SCLC [22].
Summary and conclusions
Together, this genetic evidence suggests that these three lung cancer subtypes are distinct from each other. Their germline and somatic profiles vastly differ as well as their potential environmental causes, physiological properties, and clinical features. Current treatments are expanding into therapies that target specific genetic mutations rather than treating by specific histological subtype [23, 24]. In the era of precision medicine, we believe that the increased use of genomics will be necessary to further treat each subtype as a distinct disease and to identify the most appropriate therapeutic target with clinical significance.
Acknowledgments
We thank members of the Zhao lab for helpful comments on this work. This work was partially supported by National Institutes of Health grants (R01LM012806 to Z.Z. and K07CA172294 to M.A.).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MD, Niu B, McLellan MD, Uzunangelov V, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pietanza MC, Ladanyi M. Bringing the genomic landscape of small-cell lung cancer into focus. Nat Genet. 2012;44:1074–1075. doi: 10.1038/ng.2415. [DOI] [PubMed] [Google Scholar]
- 5.Bender E. Epidemiology: The dominant malignancy. Nature. 2014;513:S2–3. doi: 10.1038/513S2a. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien TD, Jia P, Caporaso NE, Landi MT, Zhao Z. Weak sharing of genetic association signals in three lung cancer subtypes: evidence at the SNP, gene, regulation, and pathway levels. Genome Med. 2018;10:16. doi: 10.1186/s13073-018-0522-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao K, Bosse Y, Nickle DC, Pare PD, Postma DS, Laviolette M, Sandford A, Hackett TL, Daley D, Hogg JC, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He B, Chen C, Teng L, Tan K. Global view of enhancer-promoter interactome in human cells. Proc Natl Acad Sci U S A. 2014;111:E2191–2199. doi: 10.1073/pnas.1320308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, Caporaso NE, Johansson M, Xiao X, Li Y, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet. 2017;49:1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeboller H, Risch A, McKay JD, Wang Y, Dai J, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980–4995. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanetti KA, Wang Z, Aldrich M, Amos CI, Blot WJ, Bowman ED, Burdette L, Cai Q, Caporaso N, Chung CC, et al. Genome-wide association study confirms lung cancer susceptibility loci on chromosomes 5p15 and 15q25 in an African-American population. Lung Cancer. 2016;98:33–42. doi: 10.1016/j.lungcan.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12. 2 in Han Chinese. Nat Genet. 2011;43:792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 14.Lan Q, Hsiung CA, Matsuo K, Hong YC, Seow A, Wang Z, Hosgood HD, 3rd, Chen K, Wang JC, Chatterjee N, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet. 2012;44:1330–1335. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 19.Jia P, Pao W, Zhao Z. Patterns and processes of somatic mutations in nine major cancers. BMC Med Genomics. 2014;7:11. doi: 10.1186/1755-8794-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Zhang L, Tian P, Tian S. Identification of differentially-expressed genes between early-stage adenocarcinoma and squamous cell carcinoma lung cancer using meta-analysis methods. Oncol Lett. 2017;13:3314–3322. doi: 10.3892/ol.2017.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni Z, Wang X, Zhang T, Li L, Li J. Comprehensive analysis of differential expression profiles reveals potential biomarkers associated with the cell cycle and regulated by p53 in human small cell lung cancer. Exp Ther Med. 2018;15:3273–3282. doi: 10.3892/etm.2018.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, editors. Pathology and Genetics of Tumors of the Lung, Pleura, Thymus and Heart. Lyon: IARC Press; 2004. World Health Organization Classification of Tumors. [Google Scholar]
- 23.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res. 2015;4:36–54. doi: 10.3978/j.issn.2218-6751.2014.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]