Abstract
Homotypic interactions of viral capsid proteins are common, driving viral capsid self-formation. By taking advantage of such interactions of norovirus shell (S) domain that naturally builds the interior shells of norovirus capsids, we have developed a technology to produce 60-valent, icosahedral S60 nanoparticles through the E. coli system. This has been achieved by several modifications to the S domain, including an R69A mutation to destruct an exposed proteinase cleavage site and triple cysteine mutations (V57C/Q58C/S136C) to establish inter-S domain disulfide bonds for enhanced inter-S domain interactions. The polyvalent S60 nanoparticle with 60 exposed S domain C-termini offers an ideal platform for antigen presentation, leading to enhanced immunogenicity to the surface-displayed antigens for vaccine development. This was proven by constructing a chimeric S60 nanoparticle displaying 60 rotavirus (RV) VP8* proteins, the major RV neutralizing antigen. These S60-VP8* particles are easily produced and elicited high IgG response in mice toward the displayed VP8* antigens. The mouse antisera after immunization with the S60-VP8* particles exhibited high blockades against RV VP8* binding to its glycan ligands and high neutralizing activities against RV infection in culture cells. The three-dimensional structures of the S60 and S60-VP8* particles were studied. Furthermore, the S60 nanoparticle can display other antigens, supporting the notion that the S60 nanoparticle is a multifunctional vaccine platform. Finally, the intermolecular disulfide bond approach may be used to stabilize other viral-like particles to display foreign antigens for vaccine development.
Keywords: norovirus S particle, protein nanoparticle, disulfide bond, subunit vaccine, vaccine platform, norovirus, rotavirus
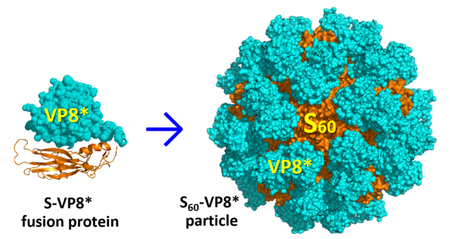
Graphical Abstract
The generation of biomaterials through bioengineering is a rapidly-growing area of modern medicine. Examples include the various polyvalent protein nanoparticles and complexes that have been constructed by taking advantage of the self-formation feature of viral capsid proteins.1–3 Viral capsid proteins are responsible for many basic functions necessary for viral life cycles, such as viral attachment and entry, and thus can elicit neutralizing antibodies against viral infection after immunization to humans and animals. Consequently, viral capsid proteins are promising vaccine targets against viral infection. Indeed, various capsid protein nanoparticles and complexes have been developed and used as non-replicating subunit vaccines to combat against different infectious diseases.1–3 Unlike traditional live-attenuated and inactivated virus vaccines that need cultivation of infectious virions and are associated with certain safety concerns, the non-replicating subunit vaccines derived from bioengineered viral capsid proteins do not involve in an infectious agent and, therefore, are safer and have lower manufacturing costs than the traditional vaccines. Thus, subunit vaccines represent a next generation of innovative vaccine strategy.
Noroviruses (NoVs) are members the Norovirus genus in the family Caliciviridae, causing epidemic acute gastroenteritis in humans with significant morbidity and mortality.4 Structurally, a NoV virion is encapsulated by a protein capsid that is composed of a single major structural protein, the capsid protein or viral protein 1 (VP1). The crystal structures of NoV capsids revealed that NoV VP1 contains two principle domains, the N-terminal shell (S) and the C-terminal protruding (P) domains, linked by a short hinge.5 The S domain builds the interior, icosahedral shell supporting the basic scaffold of a NoV virion, while the P domain constitutes the dimeric protrusions6–9 to stabilize NoV capsid and recognize cell surface glycans as NoV host attachment factors or receptors.10–13
In vitro expression of full-length NoV VP1 via a eukaryotic system resulted in self-formation of 180-valent virus-like particles (VLPs) that are structurally and antigenically similar to the authentic viral capsids,5, 14 while production of the P domain via the E. coli system formed P dimers that are structurally indistinguishable from those of NoV capsids.6–10, 15–18 In addition, generation of modified NoV P domains assembled into different higher order particles or complexes, including the 12-valent small P particles,19 the 24-valent P particles,20, 21 and the 36-valent P complexes.22
Unlike the P domain, NoV S domain is less-well studied. However, “thin layer” S particles, produced by the expression of the S domain via the baculovirus/insect cell system,10, 23 and likely equivalent to the 180-valent inner shells of NoV capsids, have been reported. In the present study, we developed a 60-valent S particle, referred as S60 nanoparticle, via the simple prokaryotic E. coli system and applied it as a multifunctional vaccine platform for antigen presentation for subunit vaccine development against rotavirus (RV) and other pathogens.
RVs cause severe gastroenteritis primarily in infants and young children, leading to ~200,000 deaths, 2.3 million hospitalizations, and 24 million outpatient visits among children < 5 years of age globally each year.24–26 The two current RV vaccines, RotaTeq (Merck) and Rotarix (GlaxoSmithKline, GSK), are effective in developed countries.27, 28 However, they have not shown satisfactory efficacies in most developing countries29–31 in Africa and Asia, where the majority of RV infection, morbidity, and mortality occurs and, thus, where RV vaccines are needed most. Our recent studies suggested that the low RV vaccine efficacy in the developing countries could be due to mismatched P types of the vaccines with the changing predominant RV P types in the middle- and low-income nations.32, 33 In addition, both current live attenuated vaccines remain costly and the replications of vaccine RVs in intestine after oral administration may be the cause of the increased risk of intussusception in vaccinated children.34–36 Thus, next generations of RV vaccines that can overcome the aforementioned limitations of the two current RV live vaccine are needed.
RV P types are determined by viral protein 4 (VP4) that constitutes the spike proteins of a RV virion. Structurally each spike protein contains two major parts, the stalk formed by VP5* and the distal head built by VP8*.37 The VP8* is responsible for interaction with RV host attachment factor or receptors that are a group of cell surface glycans, including histo-blood group antigens (HBGAs).32, 38–40 Previous studies have shown that VP8* antigens elicit neutralizing antibodies that inhibited RV infection and replication in culture cells and protected immunized mice from RV infection.41, 42 The VP8* antigen is, therefore, an important vaccine target against RVs.41–44
However, many defined neutralizing antigens, including RV VP8*, face a common problem of low immunogenicity for subunit vaccine development, due to their small sizes with low valences. This problem can be solved via fusion or conjugation of the antigens to a large, polyvalent protein platform for enhanced immunogenicity. In this study, we provided solid evidence of significantly enhanced immunogenicity of the RV VP8* antigens when displayed by the NoV S60 nanoparticle vaccine platform. Our data indicated that the S60-VP8* particle was easy to produce, stable, and highly immunogenic toward the displayed RV VP8* antigens and, thus, is likely a promising subunit vaccine against RV infection.
RESULTS
Low particle formation efficiency of native NoV S domains.
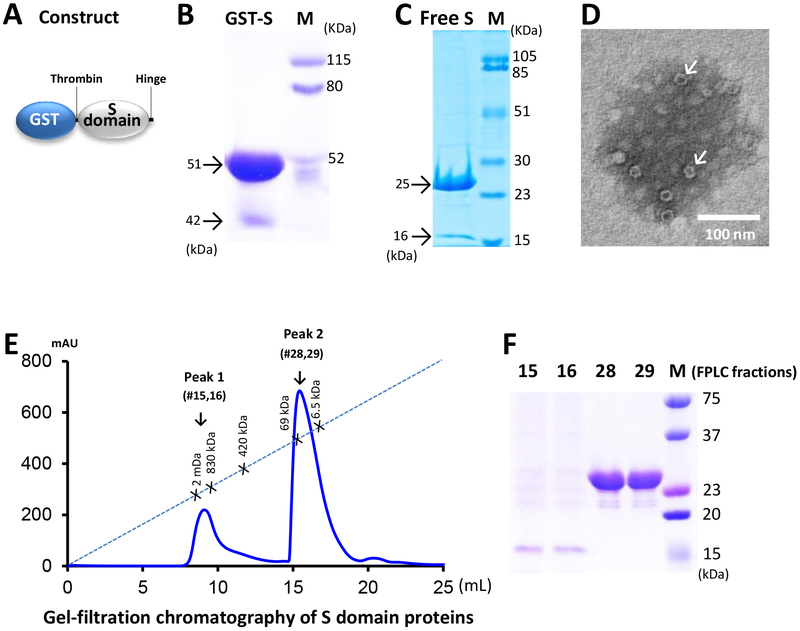
Our study started with the production of the native S domain with the hinge of a GII.4 NoV (VA387) via E. coli using the GST-Fusion System through vector pGEX-4T-1, resulting in GST-S domain fusion proteins with a molecular weight (MW) of ~51 kDa (Figure 1, A and B). Free S domain proteins at ~25 kDa (Figure 1C) without GST were obtained through thrombin cleavage, while the GST remained bound to the sepharose beads. EM examination of the S protein revealed few ring-shaped structures in ~20 nm in diameter (Figure 1D), likely corresponding to assembled S particles or complexes. Gel-filtration chromatography of the S domain proteins revealed two broad peaks (Figure 1E). SDS PAGE (Figure 1F) followed by a Western analysis (data not shown) using NoV VLP hyperimmune serum14 and N-terminally sequencing (Figure 2, see below), confirmed that both peaks were formed by the S proteins. Peak 1 with high MWs containing apparently the self-assembled S particles or complexes was much smaller than peak 2 that mostly contained S domain monomers (~25 kDa) and/or dimers (~50 kDa), confirming the EM observation that only minor portion of the S proteins formed the S particles.
Figure 1.
Native norovirus (NoV) S domain proteins assembled into particles or complexes at low efficiency. (A) Schematic diagram of the expression construct of the GST-S domain fusion protein, showing positions of the thrombin cleavage site and the hinge. (B and C) SDS-PAGE analysis of the GST-S fusion protein (GST-S, ~51 kDa) (B) and the free S protein (~25 kDa) (C). (D) An EM micrograph of the S proteins showing few assembled S particles (arrows). (E) Elution curve of a gel-filtration chromatography of the S protein via a size-exclusion column (Superdex 200). The gel-filtration column was calibrated by the Gel Filtration Calibration Kit and the purified NoV P particles,20, 21 small P particles,19 and P dimers.10 The elution positions of the blue Dextran 2000 (~2000 kDa, void), P particles (~830 kDa), small P particles (~420 kDa), P dimers (~69 kDa), and aprotinin (~6.5 kDa) are indicated. (F) SDS-PAGE analysis of the proteins from the two elution peaks, peak 1 (fraction #15 and 16) and peak 2 (fraction #28 and 29). In all SDS PAGE, Lane M is pre-stained protein markers with indicated molecular weights. Minor S protein bands at ~42, and ~16 kDa were indicated in (B), (C) and (F), respectively.
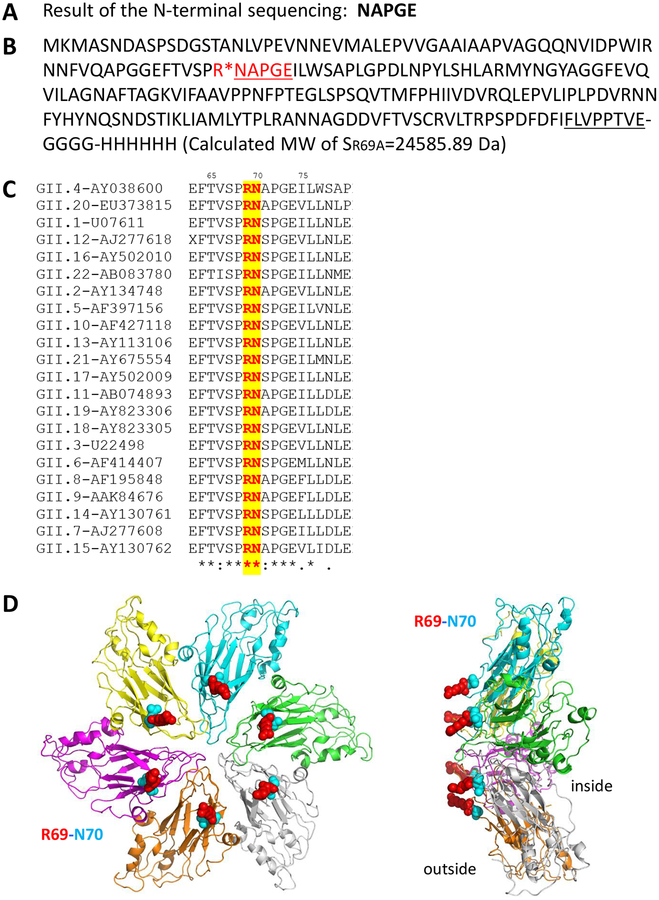
Figure 2.
Identification of the exposed protease site in the S domain. (A) N-terminal sequencing of the protease cleaved S protein resulted in penta-residue sequences, NAPGE. (B) The S domain sequences showing the NAPGE sequences (underlined red fonts) and indicating the protease cutting site (red star symbol). The C-terminal hinge (underlined), the four-residue linker (GGGG), and the end fused Hisx6 peptide are indicated. The calculated molecular weight of this recombinant S domain protein is also indicated. (C) Sequence alignment among representations of all GII norovirus genotypes indicated that the protease site is highly conserved (red fonts with yellow highlight). (D) Inspection of a partial GII NoV shell structure (Wen Jiang, unpublished data) at the 3-fold axis shows the exposed proteinase sites formed by R69 (red)-N70 (cyan). The six S domains are shown in cartoon representation in different colors, while the proteinase sites are shown in sphere representations. Left panel: top view; right panel: side view.
We also observed a minor protein band with lower MW in the GST-S fusion proteins (Figure 1B, 42 kDa) and the free S proteins (Figure 1C, 16 kDa), respectively, which should be proteinase-cleaved forms of the S proteins as these minor protein bands reacted with the NoV VLP-specific antibody (data not shown) and showed S domain sequences in N-terminal peptide sequencing (Figure 2, see below). We further noted that the S domain proteins that assembled into S particles or complexes were mostly digested into the smaller S domain proteins (Figure 1 E and F, peak 1, fractions #15 and #16). By contrast, the unassembled S proteins remained intact (Figure 1 E and F, peak 2, fraction #28 and #29), suggesting that the assembled S particles or complexes were sensitive to a proteinase, while the unassembled S proteins were not. Because peak 1 represents only a minor portion (<25%) of the total S proteins, we further concluded that the native NoV S domain proteins assembled into particles with low efficiency.
Identification of the exposed protease cleavage site in the S protein.
The above findings prompted us to identify the protease cleaved site. This was achieved by N-terminally sequencing of the two cleaved S protein bands at ~16 kDa (Figure 1, C and F), resulted in the same five-residue sequences of NAPGE (Figure 2A). These penta-residues matched the S domain sequences from N70 to E74, indicating that the cleavage site is between R69 and N70 (Figure 2B), which is a trypsin/clostripaina recognition site. Genetic analysis of NoV VP1 sequences showed that this protease site is highly conserved among all GII NoVs (Figure 2C). Structural analysis of a GII NoV shell structure at the three-fold axis (Wen Jiang, unpublished data) indicated that this protease site is exposed on the shell surface of a NoV capsid (Figure 2D).
Destruction of the protease site for high S particle formation efficiency.
Based on the above data, we introduced an R69A mutation to destruct the proteinase cleavage site, resulting in the SR69A protein. In addition, we used a C-terminally linked Hisx6 peptide to replace the N-terminal GST tag to avoid the thrombin cleavage step to simplify purification procedure (Figure 3A). We also inserted a short linker (GGGG) between the hinge and the Hisx6 peptide for flexibility to the Hisx6 peptide to prove the concept of antigen presentation of the S particles.
Figure 3.
Production and characterization of the SR69A proteins and the S60 nanoparticles. (A) Schematic diagram of the expression construct of the SR69A protein showing the hinge, a linker (GGGG), and the Hisx6 peptide (an orange ball labeled as H). Its complete sequences are shown in Figure 2B. (B) SDS-PAGE analysis of the SR69A proteins (~25 kDa). Lanes 1 to 5 were elution fractions from the TALON CellThru Resin. Lane M represents pre-stained protein markers with indicated molecular weights. (C) An EM micrograph of the SR69A protein showing self-assembled S60 nanoparticles in uniform sizes. (D and E) Analysis of the SR69A proteins by gel-filtration chromatography (D), followed by an SDS PAGE analysis of the elution peaks (E). (D) Elution curve of gel-filtration chromatography of the SR69A proteins via a size-exclusion column (Superdex 200, 10/300 GL). The gel-filtration column was calibrated as done in Figure 1E. The elution positions of the blue Dextran 2000 (~2000 kDa), P particle (~830 kDa), P dimer (~69 kDa), and aprotinin (~6.5 kDa) are indicated by (x) labeled as 1, 2, 3, and 4, respectively. (E) SDS-PAGE analysis of the SR69A proteins from the three major peaks in the gel-filtration (D), in which lane C is the control SR69A proteins before being loaded to the size-exclusion column; lane M is the pre-stained protein markers with indicated molecular weights; lanes 8 and 9 were from fractions #8 and 9 of peak 1, lane 16 was from fraction #16 of peak 2; while lane 19 was from fraction #19 of peak 3. (F) Electrospray ionization mass spectrometry (ESI-MS) analysis of the SR69A proteins. ESI mass spectrum acquired in positive ion mode for aqueous ammonium acetate solutions (500 mM, pH 6.8 and 25 °C) of 80 μM SR69A protein (based on monomer). Both the SR69A domain monomers (25.047 kDa) and dimers (50.095 kDa) were detected. A broad feature centered at m/z ~15,500 was observed. Although the mass resolution was insufficient to establish the charge states, the MW of these ions is estimated, based on reported m/z of large protein complexes,59 to be approximately 1.47 MDa, corresponding to the MWs of the 60 valent S60 nanoparticles.
The SR69A protein (~25 kDa) was produced well in the E. coli system and could be purified by the Hisx6-binding TALON CellThru resin at extremely high yield (~40 mg/liter bacterial culture) and high stability (Figure 3B). EM observation indicated many ring/hexagon/pentagon-shapes structures in uniform sizes, representing the self-assembled S particles in diameters of ~22 nm (Figure 3C). Gel-filtration revealed one major and two minor peaks (Figure 3, D and E) that should represent the S particles (>1 MDa), S dimers (~50 kDa), and S monomers (~25 kDa), respectively, based on their MWs, which were supported by EM observations and electrospray ionization mass spectrometry (ESI-MS) analysis (below). SDS-PAGE confirmed the three peaks being formed by the S protein (Figure 3, B, D, and E). SDS-PAGE also often revealed minor bands at ~50 kDa (Figure 3, B and E) that reacted with NoV VLP-specific antibody (data not shown), indicating the un-denatured S domain dimers after SDS and reduction treatment during the SDS-PAGE analysis. This was particularly obvious in the S particles factions (peak 1: fractions #8 and #9) of the gel-filtration chromatography (Figure 3, D and E) compared with the dimer (peak 2: #16) and monomer (peak 3: #19) fractions. These data indicated that vast majority of the SR69A protein assembled into uniform S particles.
Self-assembly of the 60-valent SR69A nanoparticles.
Based on the calculated MW of the recombinant SR69A protein (24585.89 Daltons, Figure 2B), ESI-MS analysis revealed three forms of the SR69A proteins, each representing S monomers (25.047 kDa), S dimers (50.095 kDa), and S particles (~1.47 MDa) (Figure 3F). Accordingly, the self-assembled SR68A particles were 60-valent, designated as S60 nanoparticles. The fact that no further complex signals larger than ~1.47 MDa were found indicated that the 180-valent native NoV S particle did not occur, consistent with the uniform particle sizes observed by EM (Figure 3C). These S60 nanoparticles were further confirmed by the structural reconstruction of the S60-VP8* chimeric particle by cryoEM (Figure 7, see below).
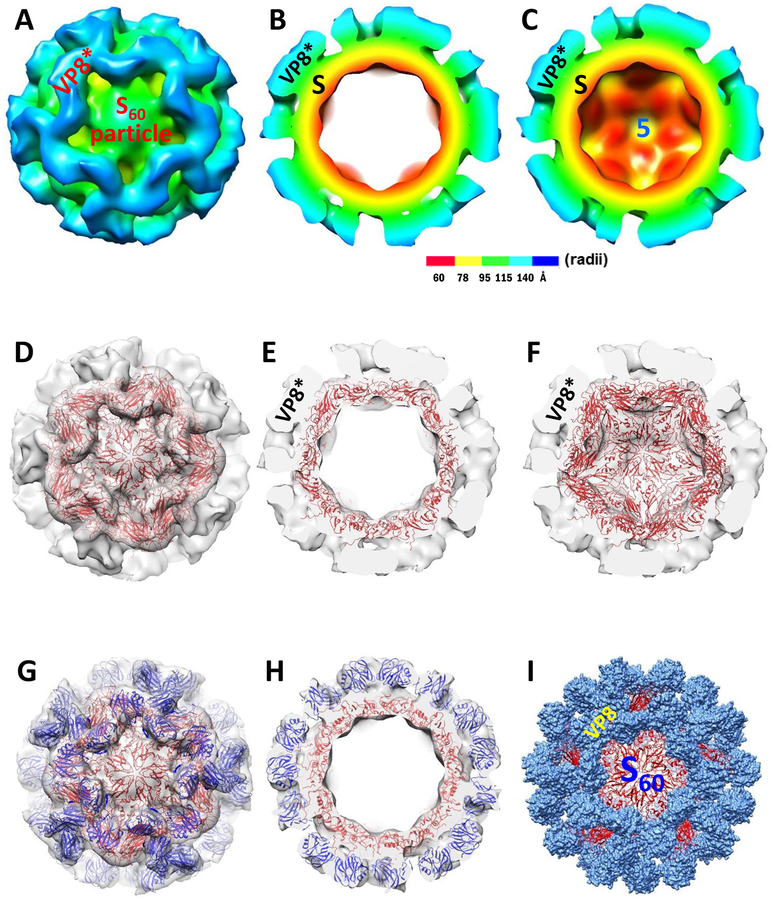
Figure 7.
Structure of the S60-VP8* particle. (A to C) The three-dimensional structures of the S60-VP8* particles were reconstructed by cryoEM technology. (A) Surface structure of the S60-VP8* particle at the five-fold axis. (B and C) The cut structures of the middle slice (B) and the second half (C) of the S60-VP8* particle showing the external and internal structures. The interior S60 nanoparticle (S) and the protruding VP8* antigens are indicated. The color schemes-based radii are shown. “5” indicates the five-fold axis. (D to F) Fitting of the 60 valent FCV shell structure (red, cartoon representation) into the cryoEM density map (transparent grey) of the inner S60 nanoparticle region of the S60-VP8* particle. The fittings results are shown by three transparent cut views, showing the first half (D), the middle slice (E), and the second half (F) of the S60-VP8* particle viewing from the front five axis. (G and H) Fitting of 60 VP8* crystal structures (PDB code: 2DWR) of a P[8] RV into the protruding regions of the S60-VP8* particle cryoEM density maps. The fitted FCV inner shell crystal structures in the S60 nanoparticle region of the S60-VP8* particle are shown in cartoon representation (red), while the fitted VP8* crystal structures in the protruding regions are shown in blue cartoon representation. (I) An S60-VP8* particle model based on the above fitting outcomes. The interior S60 nanoparticle is shown in red cartoon representation, while 60 protruding VP8* antigens are indicated in dot representation in light blue color.
Structural modeling of the S60 nanoparticle.
Using the crystal structure of the 60-valent feline calicivirus (FCV) VLP (PDB #: 4PB6)45 as a template, we built structure model of the S60 nanoparticles to understand their structure features (Figure 4, B to D). This revealed somewhat pentagon (Figure 4C) and hexagon (Figure 4D) at the five- and two-fold axis, respectively. These pentagonal and hexagonal S60 nanoparticles can be easily recognized in the EM micrographs (Figure 4A and Figure 3C). In addition, this S60 particle model fits well into the S60 particle region of the cryoEM density map of the S60-VP8* particle (Figure 7, D to F, see below), supporting the high structural similarity between the 60-valent FCV shell and the NoV S60 particle. As expected, 60 C-terminal hinges are exposed on the surface of each S60 nanoparticles (Figure 4, B to D), providing excellent fusion sites for foreign antigens to be displayed by the S60 nanoparticle.
Figure 4.
Structural modeling of the S60 nanoparticles based on the crystal structure (PDB #: 4PB6) of the 60 valent feline calicivirus (FCV) VLPs. (A) An EM micrograph showing the S60 nanoparticles. (B to D) The structures of the SR69A protein monomer (orange) in cartoon representation (B) and the S60 nanoparticles at five- (C) and two-fold (D) axis, respectively, in surface representation. The exposed C-terminal hinges (surface representation) are shown in green. (E to G) The structures of the SR69A protein monomer (orange) in cartoon representation with a C-terminally fused linker (GGGG, magenta) and a Hisx6 peptide (sky-blue) in dot representation (E) and the resulting S60 nanoparticles at five- (F) and two-fold (G) axis, respectively, in surface representation. The exposed C-terminal hinges, linkers, and Hisx6 peptides are shown in dot representations.
We also modeled the S60 particle with C-terminal linkers (GGGG) and Hisx6 peptides to mimic antigen presentation by the S60 nanoparticle (Figure 4, E to G). 60 Hisx6 peptides were displayed on the surface of each S60 particle, supporting the fact the S60 nanoparticles with the C-terminally linked Hisx6 peptides were purified efficiently by the Hisx6-binding resin (Figure 3B). Therefore, it is plausible to anticipate that various antigens from other pathogens can also be displayed by the S60 nanoparticles through fusing to the exposed C-terminus of the S protein.
The S60-nanoparticle as a potent platform for antigen presentation.
The RV surface spike protein VP8* was studied to test the feasibility of the S60 nanoparticle as a platform for antigen presentation for enhanced immunogenicity. S60-VP8* chimeric particles with the RV VP8* being displayed on the surface were made by fusion of the RV VP8* protein to the C-terminus of the SR69A protein via a four-histidine (HHHH) linker (Figure 5A). A Hisx6 peptide was added to the C-terminus of the VP8* protein for purification purpose. High yields (>30 mg/liter bacterial culture) of the chimeric protein (~45 kDa) were obtained (Figure 5B). Gel-filtration analysis of the SR69A-VP8* proteins revealed three typical peaks, most likely representing the SR69A-VP8* particles (peak 1), dimers (peak 2), and monomer (peak 3), respectively, based on their MWs (Figure 5C). Retention comparison of the three peaks indicated that about a half of the SR69A-VP8* proteins self-assembled into particles (peak 1 vs. peak 2 and peak 3).
Figure 5.
Characterization of the S60-VP8* chimeric nanoparticles. (A) Schematic diagram of the SR69A-VP8* chimeric protein. The VP8* antigen (green) of rotavirus was fused to the hinge via a linker (HHHH). A Hisx6 peptide (orange) was fused to the C-terminus of the VP8* antigen. (B) SDS-PAGE analysis of the SR69A-VP8* protein (~45 kDa). (C) Gel-filtration chromatography of the SR69A-VP8* protein through the size-exclusion column (Superdex 200, 10/300 GL). The column was calibrated as done in Figure 1E. The elution positions of the blue Dextran 2000 (~2000 kDa), P particle (~830 kDa), P dimer (~69 kDa), and aprotinin (~6.5 kDa) are indicated by (x) labeled as 1, 2, 3, and 4, respectively. (D) EM micrographs of the S60-VP8* particles from peak 1 of the gel-filtration (C). (E) Electrospray ionization mass spectrometry (ESI-MS) analysis of the SR69A-VP8* proteins. ESI mass spectrum acquired in negative ion mode for aqueous ammonium acetate solutions (500 mM, pH 6.8 and 25 °C) of 80 μM SR69A-VP8* protein (based on monomer). The SR69A-VP8* monomers (44.966 kDa) and a degraded product (19.981 kDa) were detected. A broad feature centered at m/z ~23,700 was observed. Although the mass resolution was insufficient to establish the charge states, the MW of these ions is estimated based on reported m/z of large protein complexes59 to be approximately 3.4 MDa, corresponding to the MWs of the 60 valent SR69A-VP8* nanoparticles.
EM examination of the proteins from peak 1 revealed many SR69A-VP8* particles (referred as S60-VP8* particles) in uniform sizes with recognizable protrusions likely to be the surface displayed VP8* proteins (Figure 5D), differing from the surface smooth S60 particles (Figures 3C, and 4A). ESI-MS analysis of the peak 1 proteins revealed anticipated 60-valent S60-VP8* particles with MWs of ~3.4 MDa (Figure 5E). Again, no further signal of 180-valent particles was observed, consistent with the uniform sizes of the S60-VP8* particles on the micrographs of the same proteins (Figure 5D). However, the ESI-MS analysis did reveal monomers (44.966 kDa) of the SR69A-VP8* protein and trace amount of degraded product at 19.981 kDa, indicating that the S60-VP8* particles could disassemble into monomers.
Further improvement of the S60-nanoparticle platform.
The presence of the dimer and monomer peaks in the S60- and S60-VP8* particle productions (Figures 3D and5D, peaks 2 and 3) indicated room for improvement by seeking factors to increase inter-S domain interactions. Inspection of a GII.4 shell structure (Wen Jiang, unpublished data) indicated that V57 and Q58 of an S domain are sterically close to M140’ and S136’ of the neighboring S domain, respectively, with distances of 5.7 to 5.9 Å (Figure 6. A and B). This suggested that the two pair residues are ideal locations to introduce inter-S domain disulfide bonds to further enhance S60-VP8* particle formation. To test this hypothesis, a double mutant (S69A/57C/140C-VP8) was made by changing V57 and M140 of the SR69A-VP8* protein into cysteines (Figure 6C) and this led to significantly increased yields of the mutant proteins (>50 mg/liter bacterial culture, Figure 6D). Gel-filtration analysis indicated that majority (~70%) of the S69A/57C/140C-VP8* proteins assembled into the S60-VP8* particles (Figure 6E) that was confirmed by EM observations (data not shown). It was noted that peak 3 representing the S monomers was completely gone, supporting the increased inter-S domain interactions after the disulfide bond introduction.
Figure 6.
Further stabilization of the S60-VP8* particles by introducing inter-S domain disulfide bonds. (A and B) Structural analysis of a GII.4 shell structure. (A) Partial shell structure of a GII.4 NoV (Wen Jiang, unpublished data) at three-fold axis revealed that V57 and Q58 of an S domain are sterically close to M140’ and S136’ of the neighboring S domain, respectively. The six S domains are shown in cartoon representation in grey, while the mentioned four amino acids are shown in sphere representation in different colors. (B) A close-up of the steric relationship among V57 (red)/Q58 (cyan) of one S domain and S136’ (green)/M140’ (orange) of the neighboring S domain with distances of 5.7 to 5.9 Å. (C to E) Characterization of the SR69A/V57C/M140C-VP8* proteins. (C) The expression construct of the SR69A/V57C/M140C-VP8* protein. (D) SDS PAGE analysis of the SR69A/V57C/M140C-VP8* protein. Lanes 1, 2, 3, and 4 are four eluted protein fractions from the affinity column. 15 μl of each fraction were loaded in each lane. M, prestained protein markers with indicated MWs. (E) The elution curve of a gel-filtration chromatography of the SR69A/V57C/M140C-VP8* proteins through the size-exclusion column (Superdex 200, 10/300 GL). The gel-filtration column was calibrated as done in Figure 1E. The elution positions of the blue Dextran 2000 (~2000 kDa), P particle (~830 kDa), P dimer (~69 kDa), and aprotinin (~6.5 kDa) are indicated by (x) labeled as 1, 2, 3, and 4, respectively. (F to J) Characterization of the SR69A/V57C/Q58C/S136C-VP8* proteins. (F) The expression construct of the SR69A/V57C/Q58C/S136C-VP8* protein. (G) SDS PAGE analysis of the SR69A/V57C/Q58C/S136C-VP8* proteins. Lanes 1, 2, and 3 are three eluted protein fractions from the affinity column. 10 μl of each fraction were loaded to each lane. M, prestained protein markers with indicated MWs. (H) Gel-filtration analysis of the SR69A/V57C/Q58C/S136C-VP8* proteins through the size-exclusion column (Superdex 200, 10/300 GL). The gel-filtration column was calibrated as done in Figure 1E. The elution positions of four proteins with different MWs are indicated as (E). (I) EM micrograph of the S60-VP8* particles from peak 1 of the gel filtration (H). (J) SDS PAGE analysis of the proteins from peak 1 (fraction # 7 to 10), peak 2 (faction # 21), and peak 3 (fraction # 23). Lane C is control protein before loading to the column. M, prestained protein markers with indicated MWs.
To further improve the S60-VP8* particle formation efficiency, V57, Q58, and S136 of the SR69A-VP8* protein were simultaneously mutated into cysteines (Figure 6F), the S69A/57C/58C/136C-VP8* proteins can be produced at high yields to >40 mg/liter bacterial culture (Figure 6G). Gel-filtration analysis (Figure 6, H and J) demonstrated that vast majority (>90%) of the S69A/57C/58C/136C-VP8* proteins self-assembled into the S60-VP8* particles that was confirmed by EM observations (Figure 6I). Noteworthy, both peaks 2 and 3 representing the S-VP8* dimers and monomers, respectively, disappeared (Figure 6, H and J), supporting the notion that the S60-VP8* particles formation efficiency increased dramatically, as a result of the improved inter-S domain interactions via disulfide bonds. We also performed quadruple cysteine mutations to all V57, Q58, S136, and M140 of the SR69A-VP8* protein; the outcomes in protein yields and S60-VP8* particle formation efficiency (data not shown) were similar to those of S69A/57C/58C/136C-VP8* proteins (Figure 6, F to J), indicating that triple cysteine mutations were good enough to produce highly stable S60-VP8* particles.
Structure reconstruction of the S60-VP8* nanoparticles.
We reconstructed the three-dimensional (3-D) structure of the S60-VP8* particle by cryoEM technology (see Materials and methods) to a resolution of 14 Å, exhibiting a T=1 symmetry containing 60 S-VP8* proteins (Figure 7). The structure of the S60-VP8* nanoparticle (Figure 7A) indicated that the VP8* antigens are displayed on the surface of the S60-VP8* particle, forming the protrusions extending from the interior S60 nanoparticle. The cut structures of the middle slice (Figure 7B) and the second half (Figure 7C) of the S60-VP8* particle showed the structures of the exterior VP8* antigens (blue, cyan, and partial green) and the interior S60 nanoparticle (red, yellow and partial green). The five-fold axis of the icosahedral S60 nanoparticle can be recognized (Figure 7C). The diameter of the S60-VP8* chimeric particle is ~28 nm.
To validate the cryoEM structures, the crystal structure of the 60-valent FCV shell (PDB #: 4PB6) was fitted into the cryoEM density map of the S60-nanoparticle portion of the S60-VP8* particle. Transparent cryoEM density maps with the fitted FCV shell structure of the first half (Figure 7D), the middle slice (Figure 7E), and the second half (Figure 7F) of the S60-VP8* particle demonstrated an excellent fit between the FCV 60-valent shell structure and the NoV S60 nanoparticle region of the S60-VP8* particle, validating the 60-valent, icosahedral structure of our S60 nanoparticle (Figure 4) and the S60-VP8* particle.
We then fitted 60 copies of the VP8* crystal structure (PDB code: 2DWR) of the P[8] RV Wa strain into the protruding regions of the S60-VP8* particle cryoEM density maps (Figure 7, G and H). Transparent cryoEM density maps of the first half (Figure 7G) and the middle slice (Figure 7H) of the S60-VP8* particle with the fitted VP8* crystal structures indicated excellent fit between the protruding regions of the S60-VP8* particle and the 60 VP8* structures, further proving the structures and orientations of the VP8* antigens on the surface of the S60 nanoparticle. Based on these fittings, a crystal structure model of the S60-VP8* particle was made (Figure 7I).
The S60-VP8* particles reacted with RV VP8*- and NoV VLP-specific antibodies.
The S60-VP8* particles were loaded on a CsCl density gradient. After ultracentrifugation a protein band was seen in the middle of the gradient (data not shown). The S60-VP8* particles in the fractionated gradient were detected by antibodies specific to P[8] RV VP8* and GII.4 NoV VLPs, respectively. In both cases, a defined peak of the S60-VP8* particles was detected by EIA as a single peak at the middle of the gradient (Figure 8), corresponding to a density of 1.311 mg/cm3. These data also indicated that both VP8* and S domain portions of the S60-VP8* particles are accessible by corresponding antibodies, respectively.
Figure 8.
The S60-VP8* particles reacted with RV VP8*- and NoV VLP-specific antibodies. The S60-VP8* particles were loaded on a CsCl density gradient. After ultracentrifugation the S60-VP8* particles in the fractionated gradient were detected by P[8] RV VP8*-specific antibodies (A) and hyperimmune sera against GII.4 NoV VLPs (B), respectively. In both cases, a defined peak of the S60-VP8* particles was detected at the middle of the gradient.
The S60 nanoparticle-displayed VP8*s retain ligand-binding function.
Similar to our previous study40 that showed VP8* proteins of P[8] RVs bound Leb-positive saliva, we showed here that the S60-VP8* particles bound the Leb-antigen-positive saliva samples, but not those that were negative for Leb antigens (Figure 9). These data indicated that the S60 particle-displayed VP8* antigens are in correct folding with the original ligand-binding function, validating the S60-VP8* particle as a RV vaccine candidate.
Figure 9.
The S60 nanoparticle-displayed RV VP8*s retain glycan ligand-binding function. Saliva-based binding assay indicated that the S60-VP8* particles bound the Leb antigen-positive saliva samples, but not those Leb antigen-negative ones. The S60 nanoparticles without VP8* did not bind any of these saliva samples. Both S60-VP8* and S60 nanoparticles were used at 25 μg/mL in the saliva binding assays.
Improved immunogenicity toward the S60 nanoparticle-displayed VP8* antigens.
The S60-VP8* particles were immunized to mice (N=6) and the VP8*-specific immune responses were measured using the free VP8* antigens as controls for comparisons. After three immunizations, the VP8*-specific IgG response after immunization with the S60-VP8* particles was 11.6 folds higher than that induced by the free VP8* antigens (P=0.0004) (Figure 10A), although 10 μg of the S60-VP8* particles that were used for immunization contained less VP8* molecules than those of 10 μg of the used free VP8* antigens. As negative controls the S60 nanoparticles did not elicit VP8*-specific IgG response. These data indicated that the S60 nanoparticle can significantly improve the immunogenicity of the displayed RV VP8* antigens. No abnormality in appearance and behavior of the immunized mice were observed, suggesting that the S60 nanoparticle platform and the S60-VP8* particles are not toxic to mice.
Figure 10.
The S60-VP8* particle enhanced immunogenicity toward the displayed RV VP8* antigens. Same dose/dosage of the S60-VP8* particles, free VP8* antigens, and S60 nanoparticles without VP8* were immunized to mice (N=6), respectively, followed by measurements of the VP8*-specific IgG responses (A), 50% blocking titers (BT50) against RV VP8*-glycan ligand interaction (B), and neutralization activity against RV infection/replication in culture cells (C) of the resulting mouse antisera. (A) VP8*-specific IgG responses/titers elicited by the S60-VP8* particles, free VP8* antigens, and the S60 nanoparticles, respectively. (B) BT50 against RV VP8*-ligand interactions by the mouse sera after vaccination with the same three immunogens, respectively. (C) Neutralizing activity against RV infection/replication in culture cells by mouse sera after immunization with the same three immunogens, respectively. In all these experiments mouse sera after immunization with diluent (PBS) are used as negative controls. The statistical differences between data groups are shown by star symbols (* P < 0.05, ** P < 0.01, *** P<0.001).
The S60-VP8* particle-elicited antisera enhanced blockade against RV VP8*-ligand binding.
Binding of VP8*s to RV host ligands or receptors is a key step in RV infection38. Accordingly, an in vitro blocking assay against the binding of RV VP8* proteins to their HBGA ligands has been developed as a surrogate RV neutralization assay.42 We performed such blocking assays using the previously developed P-VP8* particles42 (referred as PP-VP8*) and Leb-positive saliva samples as the RV ligands.40 We found that the mouse antisera after immunization with the S60-VP8* particles exhibited 22.8 folds higher 50% blocking titer (BT50) than that of the antisera after immunization with the free VP8* antigens (P=0.0003) (Figure 10B), further supporting the notion that the S60 nanoparticle significantly improved the immunogenicity of the displayed RV VP8* antigens. As a negative control, mouse sera after immunization with the S60 nanoparticles without VP8* antigens did not reveal such blockades.
The S60-VP8* particle-immunized mouse antisera exhibited enhanced neutralization against RV replication.
We also performed conventional cell culture-based neutralization assays to determine the neutralizing activity of the mouse antisera after vaccination with the S60-VP8* particles against infection/replication of the cell-culture adapted RV Wa strain (P[8]). In consistence with their BT50s (see above), the mouse antisera after immunization with the S60-VP8* particles exhibited significantly higher neutralizing activities at three different serum dilutions (1:75, 1:150, and 1:300) than those of the antisera after immunization with the free VP8* antigens (P=0.0003, P=0.0001, and P=0.0016, respectively) (Figure 10C). The mouse antisera after immunization with the S60 nanoparticles without VP8* did not reveal such neutralization activity. These data further supported the notions that the S60 nanoparticles is a capable vaccine platform for significantly increased immunogenicity of the displayed RV VP8* antigens and that the S60-VP8* particle is a promising vaccine candidate against RV infection.
The S60 nanoparticle as a multifunctional vaccine platform.
In addition to the RV VP8* antigen, we have been able to fuse several other antigens to the S60 nanoparticle to the same exposed S domain C-terminus via the same linker. These included 1) the HA1 antigen or receptor-binding domain (RBD) (223 amino acids) of the hemagglutinin (HA) of an H7N9 influenza A virus; 2) the TSR antigen (67 amino acids) of the circumsporozoite surface protein (CSP) of a malaria parasite Plasmodium falciparum; 3) the protruding domain antigen (187 amino acids) of a hepatitis E virus; 4) a longer version of the RV VP8* antigen (231 amino acids); and 5) the VP8* antigen (159 amino acids) of the murine RV (mRV) EDIM strain (Table 1). Particle formations of these fusion proteins have been shown by gel-filtration and/or EM (Table 1). In addition, we have shown that the S60 nanoparticle-displayed HA1 and mRV VP8* antigens elicited significantly higher HA1- and mRV VP8*-specific antibody titers, respectively, than those elicited by the free HA1 or mRV VP8* antigens (Table 1, detailed data will be published in separate manuscripts), a scenario similar to that of the S60-nanparticle-displayed RV VP8* antigen (Figure 10A). These data support the notion that the rationally developed S60 nanoparticle may serves as a multifunctional platform for vaccine development.
Table 1.
A list of antigens that have been displayed by the S60 nanoparticles.
| Epitope/Antigen | Size (residue) | Yield (mg/liter bacteria culture) | S60-antigen particle formation | Significant immune enhancement in mice6 |
|---|---|---|---|---|
| RV VP8* antigen | 159 | ~40 | yes | Yes |
| HA1 antigen1 | 223 | ~10 | yes | Yes |
| TSR/CSP antigen2 | 67 | ~10 | yes | ND |
| Full RV VP8* antigen3 | 231 | ~20 | yes | ND |
| Murine RV VP8* antigen4 | 159 | ~5 | yes | Yes |
| HEV protruding domain antigen5 | 187 | ~10 | yes | ND |
HA1 antigen containing the receptor binding site is the head portion of the hemagglutinin (HA) of H7N9 influenza A virus.
TSR/CSP antigen is the C-terminal portion of the major surface protein of a circumsporozoite (CSP) that play a key role in host cell invasion of a malaria parasite plasmodium falciparum.
Full RV VP8* antigen is the full-length VP8* domain of the spike protein of a human P[8] rotavirus.
Murine RV VP8* antigen is the core portion of the VP8* protein constituting the head of the spike protein of a murine rotavirus EDIM strain.
HEV protruding domain antigen is a part of the protruding domain of a hepatitis E virus capsid.
Immune enhancements of the S60 nanoparticle-displayed antigens were measured in mice using free monomeric antigens as control for comparisons. ND, not determined.
DISCUSSION
In this study we have developed a technology to produce uniform 60-valent NoV S60 nanoparticles with high efficiency via the simple bacterial expression system. This was achieved by taking advantage of the homotypic interactions of NoV VP1 S domain that naturally builds the interior shells of NoV capsids, as well as several modifications to stabilize the S domain proteins and enhance the inter-S domain interactions, respectively. Specifically, we introduced an R69A mutation to destruct the exposed protease cleavage sites on the surface of the native shell that otherwise leads to easy degradation of the S proteins. In addition, we introduced triple (V57C/Q58C/S136’C) or quadruple (V57C/Q58C/S136’C/M140’C) cysteine mutations to establish inter-S domain disulfide bonds between two pairs of sterically close residues (V57/M140’ and Q58/S136’, Figure 6) that belong two neighboring S domains. This led to significantly enhanced stability and yields of the self-assembled S60 nanoparticles produced by the simple E. coli system.
The self-assembled, polyvalent S60 nanoparticle with 60 flexibly exposed S domain C-termini is an ideal vaccine platform for antigen presentation and immunogenicity enhancement. This has been demonstrated by constructing a chimeric S60 nanoparticle displaying 60 RV VP8* proteins, the major RV neutralizing antigen. The S60-VP8* particles can be easily produced with high stability. They elicited significantly higher IgG response in mice toward the displayed VP8* antigens than that induced by the free VP8* proteins. The mouse antisera after vaccination with the S60-VP8* particles exhibited significantly stronger blockade against RV VP8* binding to its glycan ligands and significantly higher neutralizing activities against RV replication in culture cells than those of sera after immunization with the free VP8* antigens. While protective efficacy of the S60-VP8* particle vaccine is being determined using a murine RV challenge model in our lab, the presented data in this report strongly supported the notion that the S60-VP8* particle is a promising vaccine candidate against RV infection and thus the S60 nanoparticle is an excellent platform for antigen display for vaccine development.
The enhanced immune responses to an antigen that is displayed by a polyvalent platform over that elicited by its free monovalent counterpart is a widely accepted concept (reviewed in2, 3, 46, 47). This can be explained by the polyvalent B- and T-cell epitopes of the antigens on the polyvalent platform to induce stronger humoral and cellular immune responses, respectively, in animals and humans compared with those elicited by the monovalent epitopes of the free antigen.46, 47 Thus, the polyvalent platform is likely to function as adjuvants to increase the immunogenicity of the displayed antigens. The observed high immune responses of our S60-VP8* particles toward the displayed VP8* antigens in mice may follow the same principle.
Native NoV capsids contain 180 VP1s, while in vitro expression of NoV VP1 via a eukaryotic system often resulted in a mixture of 180- and 60-valent VLPs and the two VLP formats were exchangeable via artificial denature and renature treatments.48 Although it has not yet been thoroughly studied, previous expression of truncated S domains via the baculovirus/insect cell system appeared to self-assemble 180-valent S particles.10, 23 However, uniform 60-valent NoV VLPs or S particles via an expression system has never been produced before. Importantly, our S60-nanoparticles maintained the native conformation with authentic antigenicity, thus our NoV S60 nanoparticle technology represents a significant bioengineering advancement. We wish to highlight that uniform complexity and size of vaccine particles are important factors in quality control of vaccine products, as variations of complexity and size will result in variations in immunization outcomes of the vaccines.
One important feature of our technology was to rationally introduce inter-molecular disulfide bonds to stabilize the S60-nanoparticles. This approach can also be used to stabilize other viral protein particles or complexes. During our previous construction of NoV P particles, we found that an addition of a cysteine-containing peptide to the end of NoV P domain promoted and stabilized P particle formation via inter-P dimer disulfide bonds.19–22 In this current study, the S60 nanoparticles self-assembled efficiently (Figure 3D), but the formation efficiency of the original version of the S60-VP8* particles was compromised (Figure 5C), due to an addition of the monomeric VP8* antigen. Noteworthy, the self-formation efficiency of the S60-VP8* particles was significantly enhanced by introducing inter-S domain disulfide bonds. This was achieved by two basic steps. First, we analyzed the shell structure of a GII.4 NoV (Wen Jiang, unpublished data) to identify two pairs of sterically close (5.7 to 5.9 Å) residues (V57/M140’ and Q58/S136’) that belong to two adjacent S domains (Figure 6, A and B), respectively. Then two to four of these residues were mutated to cysteines simultaneously in different combinations: 1) V57C/M140C, 2) V57C/Q58C/M140C, 3) V57C/Q58C/S136C, and 4) V57C/Q58C/S136C/S140C, followed by production and measurement of the self-formation efficiency of the resulted S60-VP8* particles. In addition to the two tested pairs of the residues, we assume that introduction of disulfide bonds to other pairs of amino acids that belong to adjacent S domains but are sterically close will also stabilize the S60-nanoparticles. We further hypothesize that this method of inter-molecular disulfide bonds will also help to stabilize other viral protein particles or complexes.
Among these mutations, the S60-VP8* particles with the triple cysteine mutations (V57C/Q58C/S136C) exhibited the highest particle formation efficiency with >95% the S-VP8* proteins self-assembling into the S60-VP8* particles (Figure 6, F to J). The dimer and monomer forms of the mutated S-VP8* proteins were completely gone (compared Figure 6H with Figure 5C and Figure 6E). We also noted that the S60-VP8* particles with quadruple cysteine mutations (V57C/Q58C/S136C/M140C) exhibited similar high efficiency of S60-VP8* particle formation (data not shown) as the ones with the triple cysteine mutations. However, the detailed structural bases or mechanisms behind these different outcomes among various cysteine mutation combinations remain elusive. These results and our previous studies on the P particles19–22 strongly suggested that introduction of inter-molecular disulfide bonds may be utilized as a general approach to promote and stabilize a viral protein particle or complex formation. According to these data, the S60-VP8* particles with the R69A and V57C/Q58C/S136C mutations was used to perform downstream experiments, while the modified S domain with the same mutations was and will be used to produce stable S60 nanoparticles as a platform to display other antigens.
The S60- and S60-VP8* nanoparticles in this study were purified via a small Hisx6 peptide that was linked to the exposed C-terminus of the S domain or the S-VP8* protein. The purified nanoparticles were found to be stable for at least two months at 4 or −20 °C. Our data showed that the GST tag is not a good choice for the S60- and S60-VP8* particle production due to its large size (220 residues) that disturbs the S60 nanoparticle formation, and thus needs to be removed by an extra thrombin cleavage step, complicating the purification procedure. We also tested a tag-free purification method and found that the both S60 and S60-VP8* nanoparticles can be selectively precipitated by ammonium sulfate and then resolved in phosphate buffered saline (PBS) and other buffers (data not shown). We further noted that the S60-VP8* particles formed a defined single peak after CsCl density gradient (Figure 8) or sucrose gradient ultracentrifugation (data not shown). Finally, we discovered that the S60 and the S60-VP8* particles were eluted as a single peak through the gel-filtration size exclusion column (Figures 3, 5, 6) and in an anion exchange chromatography (data not shown). These data collectively indicated that the S60 and the S60-VP8* particles, and most likely other S60-antigen chimeric particles can be purified through a tag-free approach.
The 60 freely exposed C-termini are a key feature facilitating the S60 nanoparticle to be a useful vaccine platform. Foreign antigens or epitopes can simply be fused to the end of the S domain via a flexible linker through recombinant DNA technology. This study showed clearly that the Hisx6 peptide and the RV VP8* antigen can be presented well by the S60 particle, as shown by the structurally stable S60-Hisx6 and the S60-VP8* particles, as well as by their excellent binding to the TALON CellThru Resin (Hisx6) and the Leb ligands (RV VP8*). In addition, the fact that several other tested antigens can be presented by the S60 nanoparticles supports the notion that the S60 nanoparticles may serve as a multifunctional vaccine platform.
The modeling of the S60 nanoparticle and the S60-Hisx6 using the crystal structure of the 60 valent FCV VLP and the reconstruction of the 3-D structures of the S60-VP8* particle via cryoEM technology provide insights into the structural basis of how the S60 nanoparticle displays the Hsix6 peptides and the RV VP8* antigens. The excellent fit between the S60 nanoparticle model and the S60 nanoparticle region, as well as between the 60 VP8* antigen crystal structures and the protruding regions of the S60-VP8* particle cryoEM density map validated the reconstructed cryoEM structure of the S60-VP8* particle. These elucidated structures also shed light on the structural relationship between the S60 nanoparticle and their displayed antigens, which will help designing future presentations of other foreign antigens by the S60 nanoparticle. Finally, these structural studies also confirmed the 60 valent T=1 icosahedral symmetry of the S60 nanoparticle and the S60-VP8* particle.
CONCLUSIONS
We have developed a self-assembled, polyvalent protein nanoparticle featured with easy production, high stability, and high immunogenicity, serving as an ideal platform for antigen display. As a proof of concept, a chimeric S60 nanoparticle displaying 60 copies of RV neutralizing VP8* antigens has been constructed. Our data indicated that the highly immunogenic S60-VP8* particle is likely a promising vaccine candidate against RV infection and that the S60 nanoparticle is a multifunctional platform to enhance immunogenicity of various antigens for vaccine development against different pathogens. In addition, the intermolecular disulfide bond method can be used to stabilize other VLPs and similar protein particles as platforms for antigen displays.
METHODS
Plasmid constructs.
1) Expression construct of glutathione-s-transferase (GST)-tagged S domain protein. The S domain with the hinge-encoding sequences of a GII.4 NoV strain VA387 (GenBank AC#: AY038600.3; residue 1 to 221) were inserted into the multiple cloning sites of the pGEX-4T-1 vector (GST Gene Fusion System, GE Healthcare Life Sciences) via the BamH1/Sal I sites. The resulting S domain protein had an N-terminal GST with a thrombin cleavage site in between. 2) Plasmid construct for the Hisx6-fused SR69A domain expression. The same NoV S domain-hinge-encoding sequences with an R69A mutation were inserted into the multiple cloning sites of the pET-24b vector (Novagen) via the BamH1/Not I sites. The resulting S domain protein had a C-terminally fused Hisx6 peptide with a four-glycine linker (GGGG) in between. 3) DNA construct for SR69A-VP8* chimeric protein expression. A DNA fragment containing RV VP8*-encoding sequences of a P[8] human RV strain BM14113, equivalent to the amino acid sequences from 64 to 231 of the VP8* of WA strain (G1P[8], GenBank AC#: VPXRWA), was fused to the C-terminal end of the SR69A domain-hinge with a four-histidine linker (HHHH) in between. RV strain BM14113 was isolated directly from a RV positive stool sample40. A Hisx6-peptide was added to the C-terminus of the VP8* antigen for purification purpose. 4) Expression constructs of the SR69A/V57C/M140C-VP8*, the SR69A/V57C/Q58C/S136C-VP8*, and SR69A/V57C/Q58C/S136C/M140C-VP8* chimeric proteins. These DNA constructs were made by introductions of other two (V57C/M140C), three (V57C/Q58C/S136C), or four (V57C/Q58C/S136C/M140C) mutations to the expression construct of the SR69A-VP8* chimeric proteins through site-directed mutagenesis. In addition, plasmid DNA constructs for other SR69A/V57C/Q58C/S136C-based chimeric particles displaying antigens of various pathogens, including a) the VP8* antigen of the murine RV EDIM (epizootic diarrhea of infant mice) strain;49 b) the full-length RV VP8* antigen of human RV (strain BM14113); c) the surface TSR antigen of the circumsporozoite protein (CZP) (GenBank AC#:CAB38998, residues 309–375) of Plasmodium falciparum parasite 3D7 strain;50 d) the H7 HA1 antigen (the receptor binding domain) of influenza A virus; and e) the P domain antigens of hepatitis E viruses,51–53 were constructed using the construct of SR69A/V57C/Q58C/S136C-VP8* chimeric proteins as the starting construct, in which the RV VP8*-encoding sequences were replaced with those encoding the corresponding antigens.
Production and purification of recombinant proteins.
The recombinant GST- and Hisx6-fused proteins were expressed in E. coli (BL21, DE3) as described previously.10, 42, 53, 54 The resulting recombinant proteins were purified using Sepharose 4 Fast Flow purification resin (GE Healthcare Life Sciences) for GST tagged and the TALON CellThru Resin (ClonTech) for the Hisx6-peptide fused proteins according to the instructions of the manufacturers. The GST was removed from the target proteins by thrombin (GE Healthcare Life Sciences) cleavage, while the GST-fusion proteins still bound to the purification resin.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and protein quantitation.
Purified proteins were analyzed by SDS-PAGE using 10% or 12% separating gels. Proteins were quantitated by SDS-PAGE using serially diluted bovine serum albumin (BSA, Bio-Rad) as standards on same gels.41
Gel filtration chromatography.
This was performed to analyze the size distributions of proteins and protein complexes as described previously10, 54, 55 through an AKTA Fast Performance Liquid Chromatography System (AKTA Pure 25L, GE Healthcare Life Sciences) using a size exclusion column (Superdex 200, 10/300 GL, GE Healthcare Life Sciences). The column was calibrated by gel-filtration calibration kits (GE Healthcare Life Sciences) and purified NoV P particles (~830 kDa),55 small P particles (~420 kDa),19 and P dimers (~69 kDa)10 as described previously.52, 54 The proteins of the elution peaks were analyzed by SDS-PAGE.
Cesium chloride (CsCl) density gradient ultracentrifugation.
0.5 mL of resin-purified S60-VP8* particles were mixed with 11 mL CsCl solution with a density of 1.300 g/mL and then packed in a 12-ml centrifuge tube. After centrifugation at 288,000 g for 45 h in the Optima L-90K ultracentrifuge (Beckman Coulter) using a SW41Ti rotor, the gradient was fractionated by bottom puncture into 23 fractions with ~0.5 mL each. CsCl densities of fractions were determined based on the refractive index. The S60-VP8* particles in factions were analyzed by ELISA, in which individual fractions were diluted 20 folds in PBS and coated on microtiter plates. The coated proteins were detected by NoV VLP- and RV VP8*-specific antibodies.
Electron microscopy.
Protein samples were prepared for electron microscopy (EM) inspection for nanoparticle formation using 1% ammonium molybdate as the staining solution.21 Specimens were observed under an EM10 C2 microscope (Zeiss, Germany) at 80 kV at a magnification of 10,000× to 40,000×.
Electrospray ionization mass spectrometry (ESI-MS).
ESI-MS measurements were carried out in either positive or negative ion mode using a Synapt G2S quadrupole-ion mobility separation-time of flight (Q-IMS-TOF) mass spectrometer (Waters, Manchester, UK) equipped with a nanoflow ESI (nanoESI) source. Each sample solution was prepared in 500 mM aqueous ammonium acetate buffer (pH 6.8, 25°C) and loaded into a nanoESI tip, which was produced from a borosilicate capillary (1.0 mm o.d., 0.68 mm i.d.) pulled to ~5 μm using a P-1000 micropipette puller (Sutter Instruments, Novato, CA). To perform ESI, a platinum wire was inserted into the nanoESI tip and a voltage of +1.1 kV (positive ion mode) or −0.9 kV (negative ion mode) was applied. A source temperature of 60 °C was used. The Cone, Trap and Transfer voltages were 100 V, 5 V and 2 V, respectively, and the Trap gas flow rate was 6.0 mL· min−1; all other parameters were set at their default values. Data acquisition and processing were performed using Waters MassLynx software (version 4.1).
N-terminal amino acid sequencing.
The SDS-PAGE gel slice containing the cleaved S domain protein was cut out and N-terminal amino acid sequencing was conducted by 494 Procise Protein Sequencer/140C Analyzer (Applied Biosystem, Inc.) at the Protein Facility of Iowa State University (http://www.protein.iastate.edu/).
Immunization of mice.
BALB/c mice (Harlan-Sprague-Dawley, Indianapolis, IN) at 3–4 weeks of age were divided into four groups (N = 6) for immunizations with following immunogens (10 μg/dose): 1) S60-VP8* chimeric particles; 2) free VP8* protein; 3) S60 nanoparticle without VP8* antigen; and 4) diluent (PBS, pH7.4) in approximately the same volume. Immunization was performed intramuscularly three times with Inject Alum adjuvant (Thermo Scientific, 50 μl/mouse) at two-week intervals. Immunized mice were observed for any abnormality in appearance and behavior as possible toxicity indications of the immunogens. Bloods were collected two weeks after the third immunization and sera were prepared from blood samples via a standard protocol.
Enzyme immunoassay (EIA).
EIA was performed to measure the antibody titers of mouse sera after immunization with different immunogens, as described previously.41 Gel-filtration-purified free VP8* antigen at 1 μg/mL was coated on 96-well microtiter plates and incubated with serially diluted mouse sera.42 Bound VP8*-specific antibodies were detected by goat-anti-mouse secondary antibody-HRP conjugates (MP Biomedicals, Inc). Antibody titers were defined by the end-point dilutions with a cutoff signal intensity of OD450=0.1.
HBGA binding assay.
Saliva-based HBGA binding assays were performed to measure the glycan binding function of the S60 nanoparticle-displayed VP8*s to their HBGA ligands.40 Briefly, boiled saliva samples (1:1000 diluted) that are Leb positive or Leb negative were coated on 96-well microtiter plates and incubated with various S60-VP8* particles or S60 nanoparticles without RV VP8* at indicated concentrations. The bound proteins were measured by guinea pig anti-VP8* antiserum (for S60-VP8* particles) or guinea pig hyperimmune serum against GII.4 NoV VLPs (for S60 nanoparticles), followed by an incubation of HRP-conjugated goat anti-guinea pig IgG (ICN Pharmaceuticals).
Serum blocking titers against RV VP8*-HBGA ligand attachment.
This was performed as a surrogate neutralization assay as described previously.42 Boiled and diluted (1:1000) human saliva samples with positive Lewis b (Leb) antigens, the ligands of P[8] RV,40, 56 were coated on microtiter plates. The P particle-displayed RV VP8* (PP-VP8*)41 at 625 ng/mL was pre-incubated with the post-immune sera after immunization with various immunogens (S60-VP8* particles, free VP8* antigens, S60 nanoparticles, and PBS) at different dilutions before the PP-VP8* was added to the coated saliva samples. The 50% blocking titer (BT50) was defined as the lowest serum dilution causing at least 50% reduction against the binding of PP-VP8* particles to HBGAs/saliva samples compared with the unblocked positive control.
RV neutralization assay:
This was performed as described previously.54 Briefly, MA104 cells were cultivated in 6-well plates and tissue culture-adapted RV Wa strain (G1P[8]) at a titer of ~50 PFU/well was used as the inoculum. Trypsin-treated Wa RVs were incubated with mouse sera after immunization with indicated immunogens (see above) for one hour and then were added to the cells. The cells were overlaid with media including trypsin (Invitrogen) and 0.8% agarose. After a four-day incubation, the plaques were stained and counted. The neutralization (%) of the sera was calculated by the reduction in plaque numbers in the wells treated with antisera relative to the number in untreated control wells.
Structural modeling of the S60 nanoparticle.
The structures of the S60 nanoparticles with or without Hisx6 peptide were modeled using the crystal structure (PDB #: 4PB6) of the 60-valent feline calicivirus (FCV) VLPs45 as template using software PyMOL Molecular Graphics System, version 1.8.2.0 (Schrodinger, LLC). All crystal structure-based images were made by this software.
Three-dimensional structural reconstruction of the S60-VP8* chimeric nanoparticles by cryoEM.
This was performed using a similar cryo-EM approach as described in our previous studies.19, 20, 41 Briefly, aliquots (3 to 4 μL) of gel-filtration-purified S60-VP8* chimeric particles were flash frozen onto 0.001% poly-L-Lysine coated graphene oxide grids that were then loaded into the microscope. Low-electron-dose images (~20 e/Å2) were recorded on a Gatan 4k CCD using a CM200 cryomicroscope at a nominal magnification of x50,000 with a calibrated pixel size of 2.31 Å. The defocus value for each micrograph was determined with Gctf-v1.06.57 1899 particles were selected with Gautomatch v0.53 (http://www.mrc-lmb.cam.ac.uk/kzhang/). After 2D classification with Relion/2.0,58 1234 particles were left and subjected to cryoSPARC for initial reconstruction and homogeneous refinement. Analysis of cryo-EM models, including fitting of the S60 nanoparticle model (see above) and the crystal structure of P[8] RV VP8* (2DWR) were performed using UCSF Chimera software (version 1.12; http://www.rbvi.ucsf.edu/chimera).
Statistical analysis.
Statistical differences among data sets were calculated by software GraphPad Prism 6 (GraphPad Software, Inc) using t test. P-values were set at 0.05 (P < 0.05) for significant difference, 0.01 (P<0.01) for highly significant difference, and 0.001 (P<0.001) for extremely significant difference.
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (23a) of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the Cincinnati Children’s Hospital Research Foundation (Animal Welfare Assurance no. A3108–01).
ACKNOWLEDGMENTS
The research in this article was supported by the National Institute of Health, the National Institute of Allergy and Infectious Diseases (R21 AI092434–01A1 and 1R21AI097936–01A1 to M.T./Jarek Meller, R56AI114831 and R01 AI089634 to X. J.), as well as Cincinnati Children’s Hospital Medical Center (Innovation Fund to M.T.) and the Center for Clinical and Translational Science and Training (CCTST) of the University of Cincinnati (T1 Pilot Grant to M.T.).
REFERENCES
- 1.Tan M; Jiang X Norovirus P Particle: A Subviral Nanoparticle for Vaccine Development against Norovirus, Rotavirus and Influenza Virus. Nanomedicine (Lond) 2012, 7, 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan M; Jiang X Subviral Particle as Vaccine and Vaccine Platform. Curr. Opin. Virol 2014, 6, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan M; Jiang X Recent Advancements in Combination Subunit Vaccine Development. Hum. Vaccin. Immunother 2017, 13, 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel MM; Widdowson MA; Glass RI; Akazawa K; Vinje J; Parashar UD Systematic Literature Review of Role of Noroviruses in Sporadic Gastroenteritis. Emerg Infect Dis 2008, 14, 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prasad BV; Hardy ME; Dokland T; Bella J; Rossmann MG; Estes MK X-Ray Crystallographic Structure of the Norwalk Virus Capsid. Science 1999, 286, 287–290. [DOI] [PubMed] [Google Scholar]
- 6.Bu W; Mamedova A; Tan M; Xia M; Jiang X; Hegde RS Structural Basis for the Receptor Binding Specificity of Norwalk Virus. J virol 2008, 82, 5340–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao S; Lou Z; Tan M; Chen Y; Liu Y; Zhang Z; Zhang XC; Jiang X; Li X; Rao Z Structural Basis for the Recognition of Blood Group Trisaccharides by Norovirus. J virol 2007, 81, 5949–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y; Tan M; Xia M; Hao N; Zhang XC; Huang P; Jiang X; Li X; Rao Z Crystallography of a Lewis-Binding Norovirus, Elucidation of Strain-Specificity to the Polymorphic Human Histo-Blood Group Antigens. PLoS Pathog 2011, 7, e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi JM; Hutson AM; Estes MK; Prasad BV Atomic Resolution Structural Characterization of Recognition of Histo-Blood Group Antigens by Norwalk Virus. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 9175–9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan M; Hegde RS; Jiang X The P Domain of Norovirus Capsid Protein Forms Dimer and Binds to Histo-Blood Group Antigen Receptors. J virol 2004, 78, 6233–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan M; Jiang X Norovirus Gastroenteritis, Carbohydrate Receptors, and Animal Models. PLoS Pathog 2010, 6, e1000983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan M; Jiang X Norovirus-Host Interaction: Multi-Selections by Human Histo-Blood Group Antigens. Trends Microbiol 2011, 19, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan M; Jiang X Histo-Blood Group Antigens: A Common Niche for Norovirus and Rotavirus. Expert Rev. Mol. Med 2014, 16, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P; Farkas T; Zhong W; Tan M; Thornton S; Morrow AL; Jiang X Norovirus and Histo-Blood Group Antigens: Demonstration of a Wide Spectrum of Strain Specificities and Classification of Two Major Binding Groups among Multiple Binding Patterns. J virol 2005, 79, 6714–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W; Chen Y; Jiang X; Xia M; Yang Y; Tan M; Li X; Rao Z A Unique Human Norovirus Lineage with a Distinct Hbga Binding Interface. PLoS Pathog 2015, 11, e1005025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shanker S; Choi JM; Sankaran B; Atmar RL; Estes MK; Prasad BV Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus Gii.4 Epidemic Variant: Implications for Epochal Evolution. J virol 2011, 85, 8635–8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shanker S; Czako R; Sankaran B; Atmar RL; Estes MK; Prasad BV Structural Analysis of Determinants of Histo-Blood Group Antigen Binding Specificity in Genogroup I Noroviruses. J virol 2014, 88, 6168–6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansman GS; Biertumpfel C; Georgiev I; McLellan JS; Chen L; Zhou T; Katayama K; Kwong PD Crystal Structures of Gii.10 and Gii.12 Norovirus Protruding Domains in Complex with Histo-Blood Group Antigens Reveal Details for a Potential Site of Vulnerability. J virol 2011, 85, 6687–6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan M; Fang PA; Xia M; Chachiyo T; Jiang W; Jiang X Terminal Modifications of Norovirus P Domain Resulted in a New Type of Subviral Particles, the Small P Particles. Virology 2011, 410, 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan M; Fang P; Chachiyo T; Xia M; Huang P; Fang Z; Jiang W; Jiang X Noroviral P Particle: Structure, Function and Applications in Virus-Host Interaction. Virology 2008, 382, 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan M; Jiang X The P Domain of Norovirus Capsid Protein Forms a Subviral Particle That Binds to Histo-Blood Group Antigen Receptors. J virol 2005, 79, 14017–14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bereszczak JZ; Barbu IM; Tan M; Xia M; Jiang X; van Duijn E; Heck AJ Structure, Stability and Dynamics of Norovirus P Domain Derived Protein Complexes Studied by Native Mass Spectrometry. J. Struct. Biol 2012, 177, 273–282. [DOI] [PubMed] [Google Scholar]
- 23.Bertolotti-Ciarlet A; White LJ; Chen R; Prasad BV; Estes MK Structural Requirements for the Assembly of Norwalk Virus-Like Particles. J virol 2002, 76, 4044–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tate JE; Burton AH; Boschi-Pinto C; Steele AD; Duque J; Parashar UD; Network, W. H.-c. G. R. S. 2008 Estimate of Worldwide Rotavirus-Associated Mortality in Children Younger Than 5 Years before the Introduction of Universal Rotavirus Vaccination Programmes: A Systematic Review and Meta-Analysis. Lancet Infect Dis 2012, 12, 136–141. [DOI] [PubMed] [Google Scholar]
- 25.Parashar UD; Gibson CJ; Bresse JS; Glass RI Rotavirus and Severe Childhood Diarrhea. Emerg Infect Dis 2006, 12, 304–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker CL; Rudan I; Liu L; Nair H; Theodoratou E; Bhutta ZA; O’Brien KL; Campbell H; Black RE Global Burden of Childhood Pneumonia and Diarrhoea. Lancet 2013, 381, 1405–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vesikari T; Itzler R; Matson DO; Santosham M; Christie CD; Coia M; Cook JR; Koch G; Heaton P Efficacy of a Pentavalent Rotavirus Vaccine in Reducing Rotavirus-Associated Health Care Utilization across Three Regions (11 Countries). Int J Infect Dis 2007, 11 Suppl 2, S29–35. [DOI] [PubMed] [Google Scholar]
- 28.Yen C; Tate JE; Patel MM; Cortese MM; Lopman B; Fleming J; Lewis K; Jiang B; Gentsch J; Steele D; Parashar UD Rotavirus Vaccines: Update on Global Impact and Future Priorities. Hum. Vaccin. Immunother 2011, 7, 1282–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaman K; Dang DA; Victor JC; Shin S; Yunus M; Dallas MJ; Podder G; Vu DT; Le TPM; Luby SP; Le HT; Coia ML; Lewis K; Rivers SB; Sack DA; Schodel F; Steele AD; Neuzil KM; Ciarlet M Efficacy of Pentavalent Rotavirus Vaccine against Severe Rotavirus Gastroenteritis in Infants in Developing Countries in Asia: A Randomised, Double-Blind, Placebo-Controlled Trial. Lancet 2010, 376, 615–623. [DOI] [PubMed] [Google Scholar]
- 30.Madhi SA; Cunliffe NA; Steele D; Witte D; Kirsten M; Louw C; Ngwira B; Victor JC; Gillard PH; Cheuvart BB; Han HH; Neuzil KM Effect of Human Rotavirus Vaccine on Severe Diarrhea in African Infants. N Engl J Med 2010, 362, 289–298. [DOI] [PubMed] [Google Scholar]
- 31.Armah GE; Sow SO; Breiman RF; Dallas MJ; Tapia MD; Feikin DR; Binka FN; Steele AD; Laserson KF; Ansah NA; Levine MM; Lewis K; Coia ML; Attah-Poku M; Ojwando J; Rivers SB; Victor JC; Nyambane G; Hodgson A; Schodel F; Ciarlet M; Neuzil KM Efficacy of Pentavalent Rotavirus Vaccine against Severe Rotavirus Gastroenteritis in Infants in Developing Countries in Sub-Saharan Africa: A Randomised, Double-Blind, Placebo-Controlled Trial. Lancet 2010, 376, 606–614. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y; Ramelot TA; Huang P; Liu Y; Li Z; Feizi T; Zhong W; Wu FT; Tan M; Kennedy MA; Jiang X Glycan Specificity of P[19] Rotavirus and Comparison with Those of Related P Genotypes. J virol 2016, 90, 9983–9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang X; Liu Y; Tan M Histo-Blood Group Antigens as Receptors for Rotavirus, New Understanding on Rotavirus Epidemiology and Vaccine Strategy. Emerg Microbes Infect 2017, 6, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yih WK; Lieu TA; Kulldorff M; Martin D; McMahill-Walraven CN; Platt R; Selvam N; Selvan M; Lee GM; Nguyen M Intussusception Risk after Rotavirus Vaccination in U.S. Infants. N. Engl. J. Med 2014, 370, 503–512. [DOI] [PubMed] [Google Scholar]
- 35.Glass RI; Parashar UD Rotavirus Vaccines--Balancing Intussusception Risks and Health Benefits. N. Engl. J. Med 2014, 370, 568–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weintraub ES; Baggs J; Duffy J; Vellozzi C; Belongia EA; Irving S; Klein NP; Glanz JM; Jacobsen SJ; Naleway A; Jackson LA; DeStefano F Risk of Intussusception after Monovalent Rotavirus Vaccination. N. Engl. J. Med 2014, 370, 513–519. [DOI] [PubMed] [Google Scholar]
- 37.Settembre EC; Chen JZ; Dormitzer PR; Grigorieff N; Harrison SC Atomic Model of an Infectious Rotavirus Particle. EMBO J 2011, 30, 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu L; Crawford SE; Czako R; Cortes-Penfield NW; Smith DF; Le Pendu J; Estes MK; Prasad BV Cell Attachment Protein Vp8* of a Human Rotavirus Specifically Interacts with a-Type Histo-Blood Group Antigen. Nature 2012, 485, 256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu L; Ramani S; Czako R; Sankaran B; Yu Y; Smith DF; Cummings RD; Estes MK; Venkataram Prasad BV Structural Basis of Glycan Specificity in Neonate-Specific Bovine-Human Reassortant Rotavirus. Nat Commun 2015, 6, 8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang P; Xia M; Tan M; Zhong W; Wei C; Wang L; Morrow A; Jiang X Spike Protein Vp8* of Human Rotavirus Recognizes Histo-Blood Group Antigens in a Type-Specific Manner. J virol 2012, 86, 4833–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan M; Huang P; Xia M; Fang PA; Zhong W; McNeal M; Wei C; Jiang W; Jiang X Norovirus P Particle, a Novel Platform for Vaccine Development and Antibody Production. J virol 2011, 85, 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia M; Wei C; Wang L; Cao D; Meng XJ; Jiang X; Tan M Development and Evaluation of Two Subunit Vaccine Candidates Containing Antigens of Hepatitis E Virus, Rotavirus, and Astrovirus. Sci Rep 2016, 6, 25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groome MJ; Koen A; Fix A; Page N; Jose L; Madhi SA; McNeal M; Dally L; Cho I; Power M; Flores J; Cryz S Safety and Immunogenicity of a Parenteral P2-Vp8-P[8] Subunit Rotavirus Vaccine in Toddlers and Infants in South Africa: A Randomised, Double-Blind, Placebo-Controlled Trial. Lancet Infect Dis 2017, 17, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen XB; Cao DJ; Jones RW; Hoshino Y; Yuan LJ Tandem Truncated Rotavirus Vp8*Subunit Protein with T Cell Epitope as Non-Replicating Parenteral Vaccine Is Highly Immunogenic. Hum. Vaccin. Immunother 2015, 11, 2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burmeister WP; Buisson M; Estrozi LF; Schoehn G; Billet O; Hannas Z; Sigoillot C; Poulet H Structure Determination of Feline Calicivirus Virus-Like Particles in the Context of a Pseudo-Octahedral Arrangement. PLoS One 2015, 10, e0119289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plummer EM; Manchester M Viral Nanoparticles and Virus-Like Particles: Platforms for Contemporary Vaccine Design. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2011, 3, 174–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Q; Li S; Yu H; Xia N; Modis Y Virus-Like Particle-Based Human Vaccines: Quality Assessment Based on Structural and Functional Properties. Trends Biotechnol. 2013, 31, 654–663. [DOI] [PubMed] [Google Scholar]
- 48.White LJ; Hardy ME; Estes MK Biochemical Characterization of a Smaller Form of Recombinant Norwalk Virus Capsids Assembled in Insect Cells. J virol 1997, 71, 8066–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du J; Lan Z; Liu Y; Liu Y; Li Y; Li X; Guo T Detailed Analysis of Balb/C Mice Challenged with Wild Type Rotavirus Edim Provide an Alternative for Infection Model of Rotavirus. Virus Res. 2017, 228, 134–140. [DOI] [PubMed] [Google Scholar]
- 50.Doud MB; Koksal AC; Mi LZ; Song G; Lu C; Springer TA Unexpected Fold in the Circumsporozoite Protein Target of Malaria Vaccines. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 7817–7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L; Cao D; Wei C; Meng XJ; Jiang X; Tan M A Dual Vaccine Candidate against Norovirus and Hepatitis E Virus. Vaccine 2014, 32, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L; Xia M; Huang P; Fang H; Cao D; Meng XJ; McNeal M; Jiang X; Tan M Branched-Linear and Agglomerate Protein Polymers as Vaccine Platforms. Biomaterials 2014, 35, 8427–8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia M; Wei C; Wang L; Cao D; Meng XJ; Jiang X; Tan M A Trivalent Vaccine Candidate against Hepatitis E Virus, Norovirus, and Astrovirus. Vaccine 2016, 34, 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L; Huang P; Fang H; Xia M; Zhong W; McNeal MM; Jiang X; Tan M Polyvalent Complexes for Vaccine Development. Biomaterials 2013, 34, 4480–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan M; Jiang X The P Domain of Norovirus Capsid Protein Forms a Subviral Particle That Binds to Histo-Blood Group Antigen Receptors. J Virol 2005, 79, 14017–14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y; Huang P; Tan M; Liu Y; Biesiada J; Meller J; Castello AA; Jiang B; Jiang X Rotavirus Vp8*: Phylogeny, Host Range, and Interaction with Histo-Blood Group Antigens. J virol 2012, 86, 9899–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K Gctf: Real-Time Ctf Determination and Correction. J. Struct. Biol 2016, 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheres SH A Bayesian View on Cryo-Em Structure Determination. J Mol Biol 2012, 415, 406–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shoemaker GK; van Duijn E; Crawford SE; Uetrecht C; Baclayon M; Roos WH; Wuite GJ; Estes MK; Prasad BV; Heck AJ Norwalk Virus Assembly and Stability Monitored by Mass Spectrometry. Mol. Cell. Proteomics 2010, 9, 1742–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]