Summary
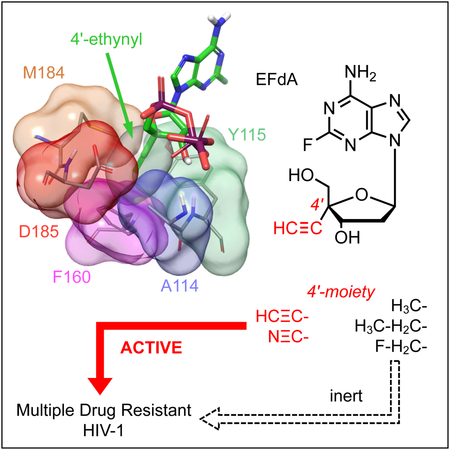
4′-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA/MK-8591), a nucleoside reverse transcriptase inhibitor (NRTI) under clinical trials, is a potent and promising long-acting anti-human immunodeficiency virus type 1 (HIV-1) agent. EFdA and its derivatives possess a modified 4′-moiety and potently inhibit the replication of a wide spectrum of HIV-1 strains resistant to existing NRTIs. Here, we report that EFdA and NRTIs with a 4′-ethynyl- or 4′-cyano-moiety exerted activity against HIV-1 with an M184V mutation and multiple NRTI-resistant HIV-1s, whereas NRTIs with other moieties (e.g., 4′-methyl) did not show this activity. Structural analysis indicated that EFdA and 4′-ethynyl-NRTIs (but not other 4′-modified NRTIs), formed strong van der Waals interactions with critical amino acid residues of reverse transcriptase. Such interactions were maintained even in the presence of a broad resistance-endowing M184V substitution, thus potently inhibiting drug-resistant HIV-1 strains. These findings also explain the mechanism for the potency of EFdA and provide insights for further design of anti-HIV-1 therapeutics.
Keywords: Drug resistance, 4′-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA/MK-8591), human immunodeficiency virus type 1 (HIV-1), reverse transcriptase, nucleoside reverse transcriptase inhibitor (NRTI)
Graphical Abstract
eTOC blurb
EFdA exerts potent antiviral activity against multiple-drug-resistant HIV-1 strains and is now undergoing clinical trials. Takamatsu et al. evaluated the antiviral activity and structural analysis of EFdA derivatives and report that 4′-ethynyl or 4′-cyano moiety are the key structural features for the potent antiviral activity against drug-resistant HIV-1 strains.
Introduction
Combined antiretroviral therapy (cART) is very effective in decreasing viral load and significantly improves the quality of life of patients with human immunodeficiency virus type 1 (HIV-1) and acquired immunodeficiency syndrome (AIDS). However, the emergence of HIV-1 variants that are resistant to multiple antiretroviral drugs is unavoidable after prolonged treatment of patients with AIDS with cART (Menéndez-Arias, 2013; Panel on Antiretroviral Guidelines for Adults and Adolescents with HIV, 2017). Therefore, it is essential to develop potent antiviral agents that are effective against HIV-1 variants resistant to all approved anti-HIV-1 therapeutics. 4’-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA or MK-8591) is one of the most promising anti-HIV-1 nucleoside reverse transcriptase inhibitors (NRTIs) that shows activity against various drug-resistant HIV-1 variants in vitro (Nakata et al., 2007; Kawamoto et al., 2008; Hattori et al., 2009; Murphey-Corb et al., 2012).
Recent clinical studies reported the pharmacokinetic (PK) profiles and antiviral efficacy of EFdA. Grobler et al. demonstrated that a 10-mg single dose administration in humans was able to achieve the exceeding projected efficacious concentrations for at least 7 days (Grobler et al., The Annual Conference on Retroviruses and Opportunistic Infections (CROI). Abstr. 98, 2016). In another study, treatment of cART-naïve HIV-1-infected patients with a single 10-mg dose of EFdA rapidly reduced the viral load; the mean viral load continued to decrease until day 10 after the treatment, with a mean reduction of 1.78 log10 and no evidence of recrudescence (Friedman et al., CROI. Abstr. 437LB, 2016). This study also reported that long-acting parenteral formulations of EFdA resulted in the continuous release of the drug for ≥6 months in rodents achieving high plasma levels. These findings suggest that unlike the current therapies that require at least one dose per day, EFdA can be administered orally at least once weekly (QW) or much longer with long-acting parenteral formulations (Grobler et al., CROI. Abstr. 98, 2016).
NRTIs are key components of cART that targets HIV-1 reverse transcriptase (RT) (Menéndez-Arias, 2013). Since the 1990s, various 4′-modified nucleoside analogs have been reported that show activity against HIV-1 by inhibiting HIV-1 RT (Maag et al., 1992; O-Yang et al., 1992; Kohgo et al., 1999a; Nomura et al., 1999; Kohgo et al., 1999b; Kohgo et al. 2004; Kawamoto et al., 2008). However, most of these analogs exerted cytotoxic effects and hence were not examined in clinical trials. We previously developed several 4′-ethynyl-2′-deoxynucleoside analogs (EdNs) that retained a 3′-hydroxyl (3′-OH) moiety in their sugar and showed activity against HIV-1 (Kodama et al., 2001). However, these EdNs also exerted toxic effects. We optimized these EdNs to successfully develop EFdA (Nakata et al., 2007). EFdA was generated by adding a fluorine atom at position 2 in the purine moiety of a potent but cytotoxic nucleoside analog, 4′-ethynyl-2′-deoxyadenosine (EdA). This 4′-modification successfully decreased the cytotoxicity of EdA, especially EdA’s human DNA polymerase γ inhibition, while maintaining EdA’s highly potent anti-HIV-1 activity (Nakata et al., 2007; Kawamoto et al., 2008). Moreover, we found that the intracellular half-life of EFdA-triphosphate (EFdA-TP), an active form of EFdA, was ≥17 h (Nakata et al., 2007). EFdA is currently under clinical trials in the United States and elsewhere (Grobler et al., CROI. Abstr. 98, 2016).
Kawamoto et al. reported that a moderate resistance of HIV-1 against EFdA (22-fold change compared with that against wild-type HIV-1 [HIV-1WT]) was associated with three amino acid substitutions (I142V, T165R, and M184V) in the RT-encoding gene (Kawamoto et al., 2008). Recently, we reported that the emergence of EFdA-resistant HIV-1 (HIV-1EFdAR) was significantly delayed when selection was performed in vitro. Moreover, we found that nine amino acid substitutions, namely, M41L, D67Δ, T69G, K70R, L74I, V75T, M184V, T215F, and K219Q (in HIV-1EFdAR), were associated with EFdA resistance (Maeda et al., 2014).
In the present study, we synthesized a panel of 4′-modified NRTIs (4′-NRTIs) structurally related to EFdA and examined their antiviral activity against various NRTI-resistant HIV-1 variants, including the highly multidrug resistant HIV-1EFdAR. We found that EFdA and some of the 4′-NRTIs maintained their potent activity against HIV-1 harboring drug-resistance-associated mutations. Moreover, we examined the mechanism based on the recently determined crystal structures of EFdA-TP complexed with RT (Salie et al., 2016).
Results
Activity of the nucleoside analogs against HIV-1WT and HIV-2EHO.
We designed and synthesized various 4′-NRTIs and determined the activity of different 4′-NRTIs, including EFdA (Figure 1), and reference NRTIs (zidovudine [AZT], abacavir [ABC], and tenofovir disoproxil fumarate [TDF]) against HIV-1WT. We first attempted to determine the antiviral activities of each compound against HIV-1WT and HIV-2EHO using methyl thiazol tetrazolium (MTT) assay, which evaluates the inhibitory effect of certain compounds based on its virally-induced cytopathicity. Results of MTT assays using MT-4 cells showed that all the examined 4′-NRTIs showed activity against HIV-1WT. Eight NRTIs showed potent activity, with 50% inhibitory concentration (IC50) values less than 10 nM. IC50 values of other compounds ranged from 24 to 276 nM (Table 1). Results of the MTT assay indicated that AZT, ABC, and TDF also showed activity against HIV-1WT, with IC50 values of 37, 262, and 203 nM, respectively (Table 1). EFdA and 4′-cyano-2-amino-2′-deoxyadenosine (CAdA) (Takamatsu et al., 2015) were found to be the most potent compounds, with IC50 values of 0.3 and 0.2 nM, respectively (Table 1). These 4′-NRTIs had moderate cell toxicity with a 50% cytotoxic concentration (CC50) value of <100 μM. Among them, EFdA had most favorable cytotoxicity profile with a selectivity index (SI; CC50/IC50) value of 64,000 (Table 1). We also examined the antiviral activity of 4′-NRTIs against HIV-2EHO and found that these 4′-NRTIs exerted potent activity against HIV-2 with similar IC50 values as against that of HIV-1WT (Table S1).
Figure 1.
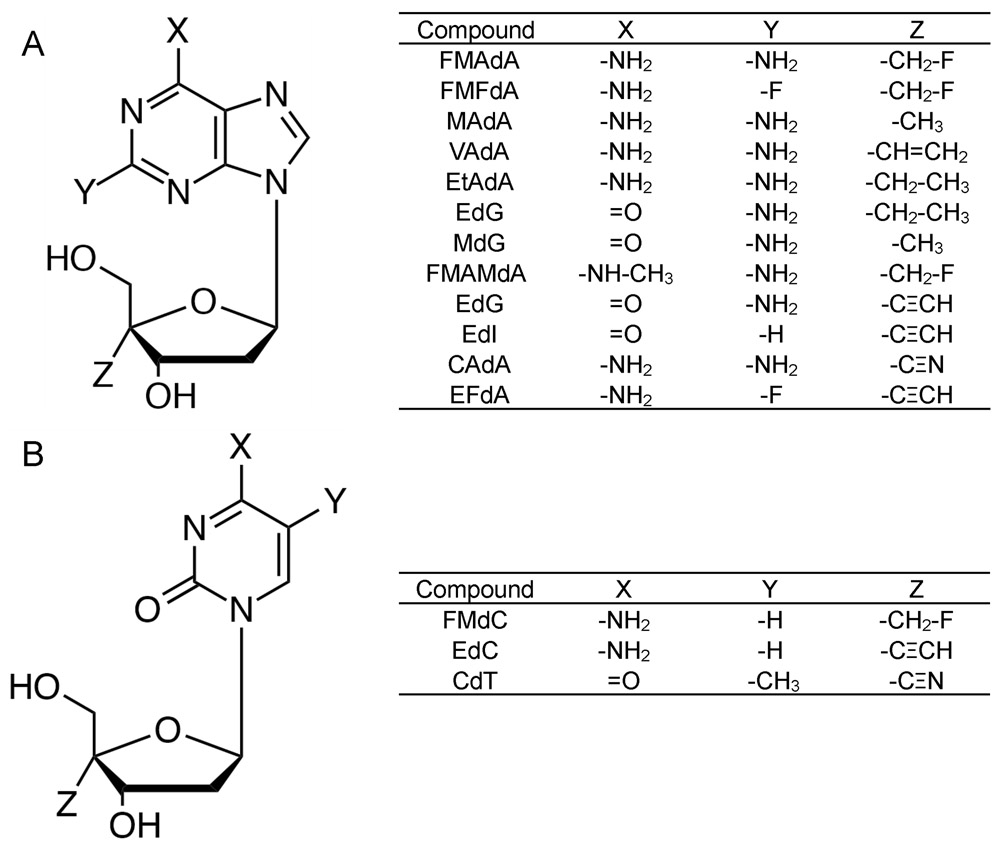
Structures of 4′-NRTIs. All purine (A) and pyrimidine (B) derivatives possess a 4′-modified moiety. FMAdA; 4′-fluoromethyl-2-amino-2′-deoxyadenosine, FMFdA; 4′-fluoromethyl-2-fluoro-2′-deoxyadenosine, MAdA; 4′-methyl-2-amino-2′-deoxyadenosine, VAdA; 4′-vinyl-2-amino-2′-deoxyadenosine, EtAdA; 4′-ethyl-2-amino-2′-deoxyadenosine, EtdG; 4′-ethyl-2′-deoxyguanosine, MdG; 4′-methyl-2′-deoxyguanosine, FMAMdA; 4′-fluoromethyl-2-amino-N6-methyl-2′-deoxyadenosine. EdG; 4′-ethynyl-2′-deoxyguanosine, EdI; 4′-ethynyl-2′-deoxyinosine, CAdA; 4′-cyano-2-amino-2′-deoxyadenosine, EFdA; 4′-ethynyl-2-fluoro-2′-deoxyadenosine, FMdC; 4′-fluoromethyl-2′-deoxycytidine, EdC; 4′-ethynyl-2′-deoxycytidine, CdT; 4′-cyano-2′-deoxythymidine.
Table 1.
Anti-HIV-1 activity and cytotoxicity of tested NRTIs
| Anti-HIV-1 IC50(nM) against | CC50 (μM) in MT-4 | S.I. with (CC50/IC50WT) |
|||||
|---|---|---|---|---|---|---|---|
| Compound | HIV-1WT | HIV-1K65R | HIV-167-75a | HIV-1M184V | HIV-1T215F | ||
| FMAdA | 3.0 ± 1.8 | 2.7 ±0.5 (0.9) | 6.9 ±3.6 (2.3) | > 1,000 (> 333.3) | 3.2 (1.1) | 22.2 ±4.1 | 7,400 |
| FMFdA | 24.8 ± 4.0 | 110.0 ±7.0 (4.4) | 299.5 ±126.9 (12) | > 1,000 (>40.3) | 274.6 ± 195.3 (11.1) | 6.6 ±0.5 | 260 |
| MAdA | 247.0 ±57.5 | 97.1 ±75.6 (0.4) | 209.0 ±40.0 (0.8) | > 1,000 (> 4) | 139.2 ±39.2 (0.6) | 4.0 ±0.3 | 15 |
| VAdA | 4.3 ±1.4 | 5.4 ±6.5 (1.3) | 7.8 ±1.3 (1.8) | > 1,000 (>232.6) | 7.9 (1.8) | 20.6 ±3.2 | 4,790 |
| EtAdA | 8.9 ±3.5 | 10.2 ± 13.7 (El) | 31.6 ±13.7 (3.6) | > 1,000 (> 112.4) | 25.9 ±6.1 (2.9) | 35.9 ± 3.1 | 4,030 |
| EtdG | 5.6 ±0.2 | 14 (2.5) | 25.6 ±7.6 (4.6) | > 1,000 (> 178.6) | 19.9 ±3.1 (3.6) | 47.0 ±3.2 | 8,390 |
| MdG | 170.8 ±35.6 | 189.2 ±75.2 (El) | 281.5 ±34.5 (1.6) | > 1,000 (> 5.9) | 184.8 ±72.0 (1.1) | 4.6 ±0.6 | 25 |
| FMAMdA | 164.8 ±8.6 | 132.9 ±7.2 (0.8) | 321.4 ±65.5 (2.0) | > 1,000 (> 6.1) | 189.0 ±68.5 (1.1) | > 100 | >600 |
| EdG | 1.6 ±0.2 | 0.1 ±0.06 (0.06) | 1.8 ± 0.1 (1.1) | 4.4 ±3.2 (2.8) | 2.0 ±0.6 (1.3) | 7 | 4,350 |
| CdT | 9.1 ±7.3 | 2.0 ±0.01 (0.2) | 8.6 ±5.9 (0.9) | 11.7±2.4 (1.3) | 3.0 (0.3) | > 10 | > 1,100 |
| FMdC | 32.9 ± 10.7 | 34.6 ± 3.6 (E0) | 28.4 ±6.2 (0.9) | > 1,000 (> 30.4) | 24.3 ±15.5 (0.7) | 4 | 120 |
| EdC | 25.2 ±4.3 | 10.2 ±4.0 (0.4) | 31.6 ± 9.9 (1.3) | 15.5 ± 1.7 (0.6) | 30.0 ± 13.9 (1.2) | N.D. | N.D. |
| EdI | 276.1 ±58.6 | 37.1 ±3.2 (0.1) | 294.3 ±48.3 (1.1) | 356.2 ±77.2 (1.3) | 297.7 ±96.3 (1.1) | N.D. | N.D. |
| CAdA | 0.2 ±0.1 | 0.6 ±0.3 (3) | 2.6 ±0.2 (13) | 4.4 ±3.2 (22) | 1.2 ±0.9 (6) | 3.5 ± 1.2 | 17,500 |
| EFdA | 0.3 ±0.2 | 0.8 ±0.3 (2.7) | 3.0 ±1.5 (10) | 3.7 ±1.7 (12.3) | 2.3 ±0.01 (1.7) | 19.2 ±6.0 | 64,000 |
| AZT | 37.4 ±7.5 | 13.5 ±1.0 (0.4) | 230.1 ±65.8 (6.2) | 16.8 ±12.8 (0.4) | 114.0 ±88.3 (3.0) | > 100 | > 2,670 |
| ABC | 262.9 ±7.6 | 1,512.2 ±704.6 (5.8) | 2,750.5 ±390.0 (10.5) | 1,525.6 ±431.1 (5.8) | 1,673.4 ±72.4 (6.4) | > 100 | >380 |
| TDF | 203.9 ±126.6 | 2,249.1 ± 1596.8 (1E0) | > 1,000 (> 4.9) | 422.8 ±112.6 (2.0) | N.D. | 39.4 ±7.3 | 190 |
Abbreviations: IC50 : 50% inhibitory concentration, CC50 : 50% cytotoxic concentration, S.I.: selectivity index. (CC50/ IC50), N.D.: not determined.
Anti-HIV-1 activity was determined using MT-4 cells by MTT assay. Each assay was conducted in duplicate or triplicate. The values represent mean values (± 1 S.D.) out of two or three independent experiments. See also Supp. Tables S1 and S2.
67Δ/T69G/K70R/L74I/V75T.
Determination of activity of 4′-NRTIs against NRTI-resistant HIV-1 variants.
Next, we examined the activity of the 4′-NRTIs against a drug-resistant HIV-1K65R variant because K65R is a TDF resistance-associated mutation that reportedly induces hypersensitivity to EFdA (Kawamoto et al., 2008; Brenner et al., 2009; Michailidis et al., 2013). As expected, seven of the 4′-NRTIs examined showed low IC50 values (fold increase from 0.06 to 0.9 compared with that against HIV-1WT) and none of them showed elevated level of resistance against HIV-1K65R (Table 1). EFdA and CAdA showed slightly elevated IC50 value compared with that against HIV-1WT with 3-fold changes, but they maintained potent antiviral activity with IC50 values of 1 nM or less.
We previously selected an HIV-1EFdAR variant harboring M41L, D67Δ, T69G, K70R, L74I, V75T, M184V, T215F, and K219Q mutations (Figure 2A) (Maeda et al., 2014). Therefore, we generated multiple HIV-1 clones containing some of the mutations present in the HIV-1EFdAR variant, such as HIV-1 clones harboring D67Δ, T69G, K70R, L74I, and V75T mutations (HIV-167-75); M184V mutation (HIV-1M184V); or T215F mutation (HIV-1T215F), by using site-directed-mutagenesis. EFdA showed activity against HIV-167-75, with an IC50 value of 3 nM, and all other EFdA-related analogs showed activity against HIV-167-75 with 0.8- to 13-fold higher IC50 values (Table 1). In contrast, reference NRTIs, ABC, and TDF, showed drastically reduced activity against HIV-167-75, with IC50 values of >1 μM in activity compared with that against HIV-1WT (Table 1). The EFdA-related derivatives also showed similar or slightly reduced activity (0.3~11.1 fold; Table 1) against HIV-1T215F. Among the reference NRTIs, AZT showed a 3-fold decrease in activity against HIVT215F. Previously, we observed that EFdA showed only a slightly decreased activity (~6-fold decrease) against HIV-1M184V (Kawamoto et al., 2008; Maeda et al., 2014). However, in the present study, nine examined 4′-NRTIs were virtually inert, with IC50 values of >1 μM (Table 1). In contrast, six 4′-NRTIs, including EFdA and CAdA, showed activity against HIV-1M184V, with IC50 values ranging from 4.4 to 356 nM (fold change between 0.6 and 22).
Figure 2.
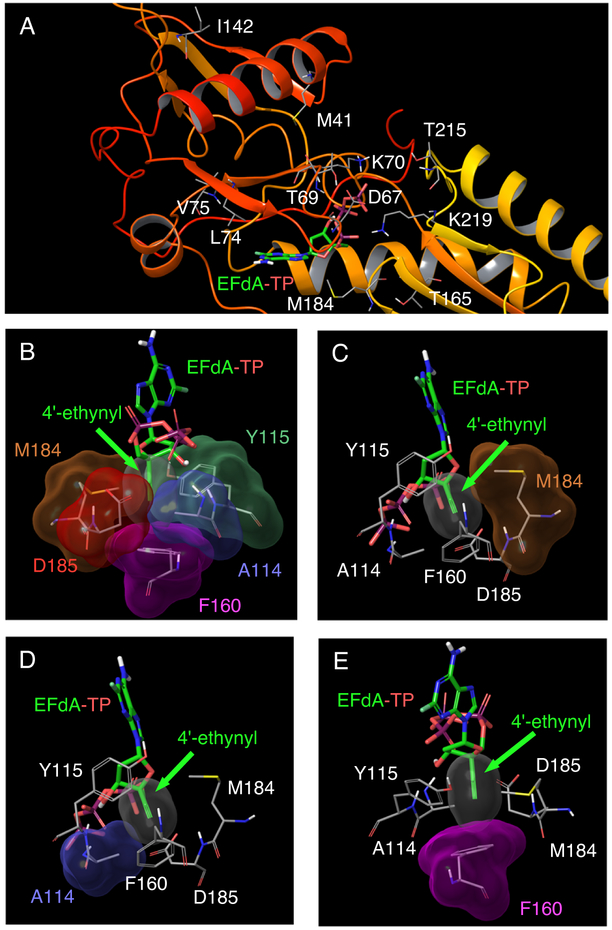
The structure of wild-type HIV-1 RT with EFdA-TP (PDB ID: 5J2M) (Salie et al., 2016). (A) Amino acids associated with NRTI resistance (M41, D67, T69, K70, L74, V75, I142, T165, M184, T215, and K219) are shown. Maeda et al. reported that the combination of M41L, D67Δ, T69G, K70R, L74I, V75T, M184V, T215F, and K219Q mutations (HIV-1EFdAR) induced EFdA resistance, while Kawamoto et al. reported that I142V, T165R, and M184V mutations were associated with EFdA resistance (Kawamoto et al., 2008; Maeda et al., 2014). (B) EFdA-TP in the active site cavity of HIV-1 RT. The hydrophobic pocket of wild-type HIV-1 RT active site and EFdA-TP in the N-site (pre-translocation mode) are shown. The 4′-ethynyl group of EFdA showed good vdW interactions with several residues, such as A114, Y115, F160, M184, and D185, in the active site cavity of RT. EFdA-TP in the N-site strongly interacted with these residues, thus preventing the shifting of EFdA to the P-site (post-translocation mode). See Fig. 3 for details of the mechanism underlying RT inhibition. (C–E) Detailed vdW interactions between the 4′-ethynyl group of EFdA and residues in the active site cavity of HIV-1 RT, namely, Met184 (C), Ala114 (D), and Phe160 (E). These amino acids showed good interaction with the 4′-ethynyl group of EFdA. Colors of vdW surfaces are as follows: 4′-ethynyl, red; A114, light blue; F160, magenta; and M184, brown.
The activity of 4′-NRTIs against HIV-1EFdAR was determined by performing p24 assay using MT-4 cells (Table S2). Some of the NRTI-resistant HIV-1 variants, such as HIV-1EFdAR, have less virally-induced cytopathicity against CD4+ cells, and therefore, we measured the antiviral activity based on the reduction of the supernatant HIV-1 p24 levels, as shown in Table 2. Nine 4′-NRTIs (at concentrations up to 1 μM) did not show any activity against HIV-1EFdAR (Table S2). The IC50 value of 4′-vinyl-2-amino-2′-deoxyadenosine (VAdA) was 804 nM, which was 190-fold higher than that against HIV-1WT, and therefore, HIV-1EFdAR was considered to be resistant against VAdA. 4′-Ethynyl-2′-deoxyguanosine (EdG), 4′-ethynyl-2′-deoxycytidine (EdC), 4′-cyano-2′-deoxythymidine (CdT), CAdA, and EFdA showed favorable activity against HIV-1EFdAR (Table S2). The IC50 value of EFdA for HIV-1EFdAR was 42 nM, while the fold decrease was 52-fold. Next, we examined the activity of these compounds against an HIV-1 clone namely HIV-167-75,184,215, harboring seven mutations, D67Δ T69G, K70R, L74I, V75T, M184V, and T215F, derived from HIV-1EFdAR and obtained almost similar results as those obtained for HIV-1EFdAR (Table S2, right column).
The five compounds that showed activity against HIV-1EFdAR (Table S2) also showed activity against HIV-167-75, HIV-1M184V, and HIV-1T215F (Table 1). Moreover, most compounds that did not show activity against HIV-1EFdAR also showed no activity against HIV-1M184V. These results suggest that M184V is a key mutation that induces resistance to the 4′-NRTIs examined in the present study. Based on the obtained results, we divided the 4′-NRTIs into two groups: (i) HIV-1M184V-active group, which includes 4′-NRTIs with the 4′-ethynyl or 4′-cyano moiety (e.g., EFdA, CAdA, and EdC), and (ii) HIV-1M184V-inactive group, which includes 4′-NRTIs with other 4′-moieties (e.g., 4′-fluoromethyl-2-amino-2′-deoxyadenosine [FMAdA], 4′-methyl-2-amino-2′-deoxyadenosine [MAdA], and VAdA) (Table 1 and Figure 1).
Structural details of wild-type HIV-1 RT complexed with EFdA-TP and its inhibitory mechanism.
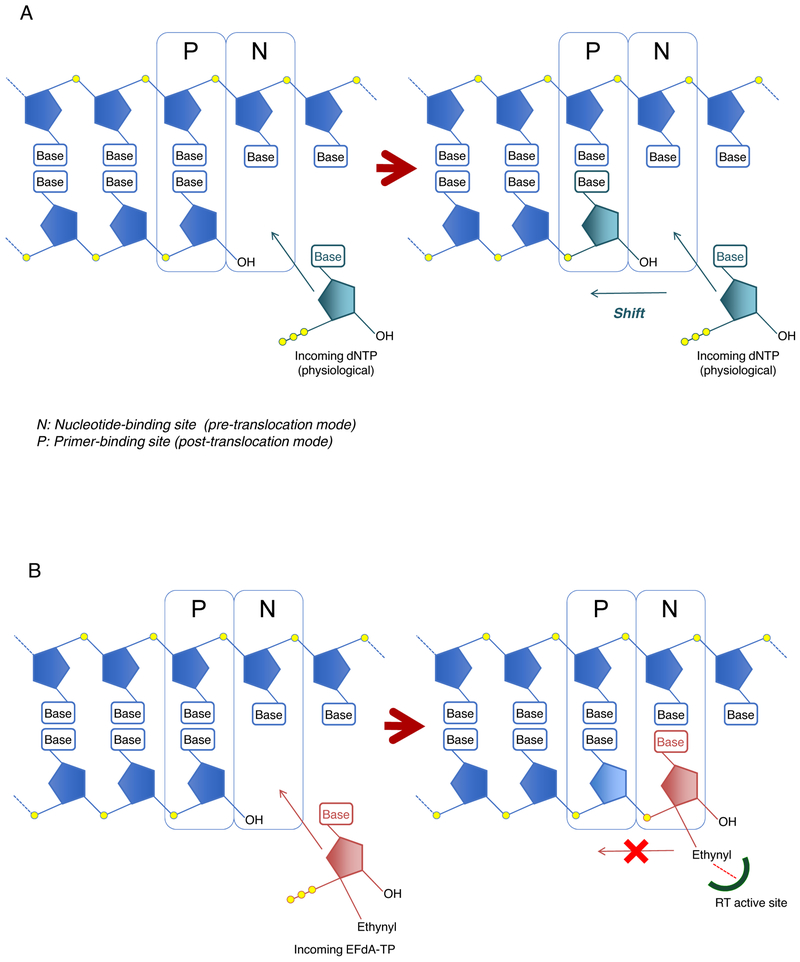
To elucidate the mechanism in which only 4′-NRTIs with a 4′-cyano-, or a 4′-ethynyl-moiety exhibit potent activity against HIV-1M184V and HIV-1EFdAR, we performed structural analysis by using a recently defined crystal structure of HIV-1 RT with a double-stranded DNA and EFdA-TP (PDB [RRID: SCR_012820] ID:5J2M) (Salie et al., 2016). The structure showed that the sugar moiety of EFdA-TP was located close to the hydrophobic pocket of HIV-1 RT and that the 4′-ethynyl of EFdA-TP was bound to a preformed hydrophobic pocket defined by conserved residues A114, Y115, F160, and M184 and an aliphatic part of D185 (Figures 2BߝE) (Salie et al., 2016). The 4′-ethynyl moiety of EFdA showed strong van der Waals (vdW) interactions with these residues (Figures 2B–E). Regarding the strong interaction of 4′-ethynyl of EFdA to the active site amino acids, Michailidis et al. discussed the mechanism underlying EFdA-induced RT inhibition. Briefly, they showed that after the incorporation of EFdA-monophosphate (EFdA-MP) at the 3′-terminus of HIV-1 RT, the RT mostly stayed in a pre-translocation binding mode (N-site) because of the strong interaction between EFdA-MP and the active site of RT (containing residues A114, Y115, F160, M184, and D185) and failed to shift to P-site (post-translocation mode), thus blocking the binding of an incoming dNTP and in turn terminating DNA polymerization. Hence, EFdA is defined as a translocation-deficient RT inhibitor (TDRTI) (Figure 3) (Michailidis et al., 2013; Salie et al., 2016).
Figure 3.
(A) Schematic representation of DNA polymerization and (B) a model of RT inhibition by EFdA. HIV-1 RT misincorporates EFdA-TP (b, red) efficiently like dNTP-TP (a, dark green) because EFdA-TP contains a normal 3′-OH moiety. After the incorporation of EFdA-MP into the N-site (pre-translocation mode), EFdA-MP interacts very strongly with amino acid residues A114, Y115, F160, M184, and D185 present in the active site cavity of RT and does not shift to the P-site (post-translocation mode). Hence, EFdA is defined as a translocation-deficient RT inhibitor (TDRTI) (Michailidis et al., 2009; Salie et al., 2016).
Direct and strong vdW interactions between 4′-moieties and amino acid residues, such as F160, in the active site cavity of RT is critical for the potency of the 4′-NRTIs.
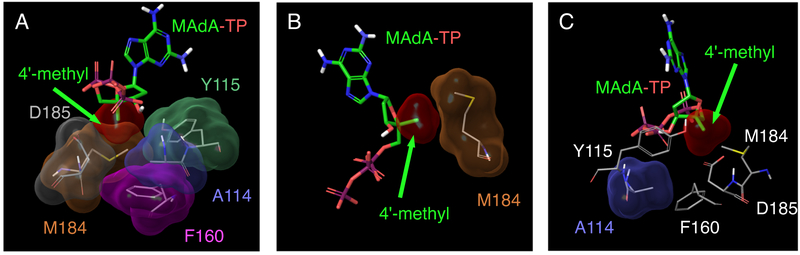
To compare the binding profile of EFdA with that of other 4′-NRTIs, we performed structural analyses starting from the crystal structure of wild-type RT complexed with EFdA-TP (PDB ID: 5J2M) (Salie et al., 2016) as a template. MAdA, which contains a 4′-methyl moiety, was examined and structurally compared with EFdA. As shown in Fig. 2, the 4′-ethynyl moiety of EFdA-TP showed strong vdW interactions with M184, A114, F160, and Y115 (Figures 2A–E). In contrast, the 4′-methyl moiety of MAdA-TP showed moderate interaction with M184 and much reduced interaction with A114, and no interaction with F160 (Figures 4A–C)
Figure 4.
(A) vdW interactions of MAdA-TP with amino acid residues A114, Y115, F160, M184, and D185 in the active site cavity of wild-type HIV-1 RT. (B) As with the 4′-ethynyl moiety of EFdA, the 4′-methyl moiety of MAdA-TP shows a good interaction with M184 in the active site cavity of RT. However, qualitative analysis suggests that 4′-ethynyl of EFdA shows improved interaction with M184 (Fig. 2C). (C) The 4′-methyl moiety of MAdA shows reduced interactions with A114 compared with the 4′-ethynyl moiety of EFdA (Fig. 2D).
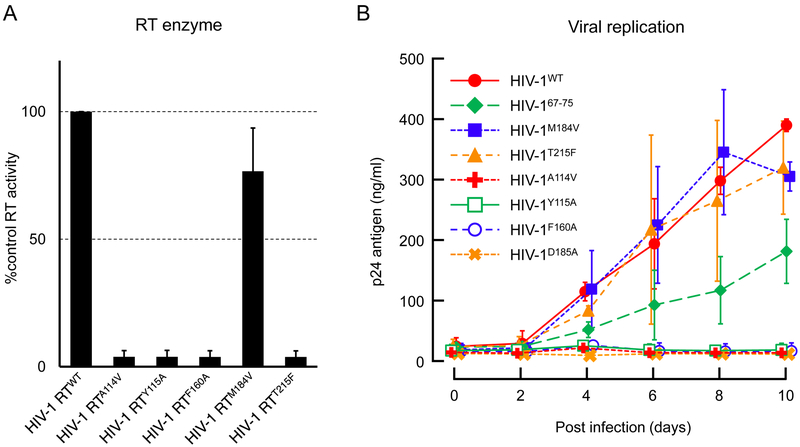
Amino acid residues present in the active site of RT are important for maintaining the activity of RT. We examined the importance of each of these amino acid residues in RT activity and HIV replication (Figure 5). The HIV-1 mutant RT enzymatic activity in the cytoplasm of HIV-1 infected cells was determined (Figure 5A). The replication ability of the viruses with RT mutation(s) was also determined by analyzing the changes in supernatant p24 Gag protein from HIV-1 infected MT-4 cells (Figure 5B). We found that one amino acid residue, F160, located at the bottom of the active site cavity of HIV-1 RT strongly interacted with EFdA. Substitution of F160 with Alanine (F160A) resulted in the complete loss of the enzymatic activity of HIV-1 RT (HIV-1 RTF160A) (Figure 5A). Similarly, HIV-1 expressing RTF160A showed negligible replication during the examination period of up to 10 days (<5% supernatant p24 value compared with that for wild-type RT) (Figure 5B). A114V (Van Cor-Hosmer et al. 2012), Y115A (Larder et al. 1989), and D185A mutations, which reduced the activity of HIV-1 RT by >90%, also critically decreased viral replication (Figures 5A, B). However, the RT activity of HIV-1 RTM184V was almost similar to that of wild-type RT (HIV-1 RTWT) and the virus with HIV-1 RTM184V maintained a substantial viral replication (Figures 5A, B). Next, we examined the effect of HIV-1 RT67-75 (containing D67Δ, T69G, K70R, L74I, and V75T mutations) and HIV-1 RTT215F on viral replication and found that these mutants only slightly affected viral replication kinetics (50% ~ 90% replication compared with that of HIV-1 expressing wild-type RT); moreover, replication was well maintained in HIV-1 harboring these mutants (Figure 5B).
Figure 5.
(A) The activity of HIV-1 RTs harboring different mutation was measured and compared with that of wild-type HIV-1 RT. HIV-1 RTM184V remained its RT enzyme activity while mutation in A114, Y115, F160, and D185 completely loss their enzyme activity. (B) Viral replication kinetics of HIV-1 variants harboring RT mutations were determined. HIV-1 variants with T215F and M184V mutations showed almost similar replication ability compared with HIV-1WT. The HIV-1 variants with A114V, Y115A, F160A, and D185A were unable to propagate apparently because of the drastically loss of RT enzyme activity. The plots represent mean values (± 1 S.D.) from two independent experiments.
Compared to MAdA, EFdA formed stronger vdW interactions with A114, Y115, F160, M184 and D185. EFdA also exhibited more potent activity than MAdA. One of the important differences between EFdA and MAdA is in the presence of a 4′-ethynyl in EFdA compared to a 4′-methyl in MAdA. The stronger vdW interactions of EFdA with the above residues is primarily because of the 4′-ethynyl substituent. These data suggested that stronger vdW interactions of a 4′-moiety (e.g., 4′-ethynyl) with A114, Y115, F160, M184, and D185 was critical for more potent inhibition of HIV-1 RT.
Structural mechanism underlying the potent activity of EFdA against HIV-1M184V.
We finally investigated the mechanism underlying the potent activity of NRTIs containing the 4′-ethynyl moiety (e.g., EFdA) against HIV-1M184V compared with that of NRTIs with other 4′-moieties (e.g., 4′-ethyl), which completely nullified their antiviral activity against HIV-1M184V. Figure 6A shows the superimposition of the interaction of 4′-ethynyl moiety of EFdA-TP with wild-type HIV-1 RT and HIV-1 RTM184V. The 4′-ethynyl moiety showed good vdW interaction with F160, M184, and other amino acids in the active site of HIV-1 RT (Figure 6A). We found that the 4′-ethynyl moiety showed slightly decreased vdW interactions with V184, but the moiety showed strong vdW interactions with other amino acids such as F160 and A114 even in the presence of a V184 substitution (Figures 6A and B). Notably, introduction of the V184 substitution in the active site cavity of RT did not change the conformation of other amino acid residues such as F160 and A114 (Figures 6B, D, and F). Next, we analyzed the interaction of FMAdA-TP (4′-fluoromethyl) and EtAdA-TP (4′-ethyl), which showed very potent activity against HIV-1WT but not against HIV-1 that contained M184V substitution (Table 1 and Table S2). Figure 6C and Figure 6E show superimposition of the interactions of FMAdA-TP and EtAdA-TP with wild-type HIV-1 RT and HIV-1 RTM184V. 4′-Fluoromethyl and 4′-ethyl moiety of FMAdA-TP and EtAdA-TP had good interaction with wild-type HIV-1 RT (M184), but these interactions were reduced with M184V substitution (Figures 6C and E). Moreover, these moieties showed less vdW interaction with F160 compared to that of 4′-ethynyl moiety of EFdA-TP (Figures 6A, C, and E). As described in Figure 5, amino acids A114, Y115, F160 and D185 are very important for RT activity, and NRTIs with the 4′-ethynyl moiety (e.g., EFdA) showed strong vdW interaction with these residues even in the presence of the V184 mutation (Figures 2 and 6A). Thus, NRTIs with the 4′-ethynyl moiety showed potent activity against HIV-1 RTM184V as well. In contrast, NRTIs with a 4′-moiety with the exception of 4′-ethynyl (and 4′-cyano) may not show strong interaction with residues such as F160, A114, and D185 (Figures 6C, E). Therefore, it is possible that these weaker NRTIs maintain their binding affinity by showing weak or moderate affinities toward multiple amino acids such as M184 in the active site cavity of RT. NRTIs containing the 4′-fluoromethyl moiety (e.g., FMAdA) and 4′-ethyl moiety (e.g., EtAdA) showed less interaction with V184 compared with wild-type M184 (Figures 6C, E), suggesting that decreased interaction with V184 considerably affects the antiviral activity of FMAdA and EtAdA.
Figure 6.
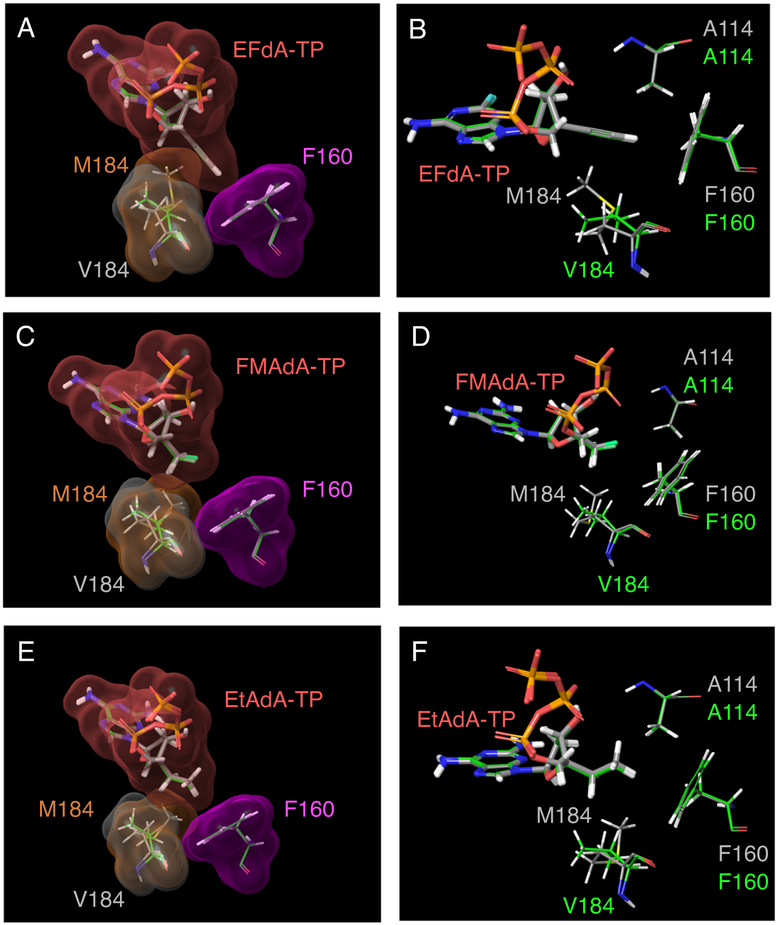
Superimposition of the interaction of the 4′-moiety of 4′-NRTI-TP with HIV-1 RTM184 (wild-type, shown in gray carbons) and HIV-1 RTV184 (shown in green carbons). The vdW surfaces are colored as follows: 4′-NRTI-TP, pink; F160, magenta; M184, brown; and V184, gray. (A) 4′-ethynyl moiety of EFdA-TP had predominantly interacts with F160 in both wild-type HIV-1 RT and HIV-1 RTM184V. It had also successfully maintained a strong interaction with both M184 and V184 in the active site cavity of RT. Fluorine is shown in cyan color. (C) 4′-fluoromethyl moiety of FMAdA-TP and (e) 4′-ethyl moiety of EtAdA-TP had less interaction with F160 compared to 4′-ethynyl moiety of EFdA-TP. These moieties had good interaction with M184 (wild-type), which is reduced in HIV-1 RTM184V. (B, D, F) The position of 4′-NRTI-TP with A114, F160, M184, and V184. The introduction of the V184 substitution in the active site cavity of RT did not change the conformation of other amino acid residues such as F160, A114, and D185.
Discussion
At present, EFdA/MK-8591 is under clinical trials in the US. A recent study reported promising PK data for EFdA and suggested QW oral administration or extended dosage regimen of EFdA with long-acting parenteral formulations (Grobler et al., CROI. Abstr. 98, 2016; Friedman et al., CROI. Abstr. 437LB, 2016). TDF is one of the most commonly used NRTIs for treating patients with AIDS; moreover, it is also used for treating HBV-infected patients. Furthermore, tenofovir alafenamide fumarate (TAF), a tenofovir pro-drug that is easily incorporated into HIV-1-infected cells and shows similar antiviral efficacy as TDF with less adverse effects on the kidneys at a low dose (Fernandez-Fernandez et al. 2011), is currently used for treating both HIV-1- and HBV-infected patients. However, it has been recently reported that TDF-resistant HIV strains have developed in addition to TDF-associated side effects, such as accelerated aging (shortening of telomere length) (Miller, 2004; Powderly et al., 2012; Leeansyah et al., 2013).
EFdA is potent against HIV-1 strains resistant to all the currently available NRTIs and is considered as a next-generation therapeutic. It was reported that EFdA showed hypersensitivity against K65R mutation, which is known as a TDF-resistant associated mutation (Nitanda et al., 2005; Kawamoto et al., 2008; Brenner et al., 2009). The K65R substitution causes minor changes in the efficacy of EFdA incorporation and of RT translocation, causes the hypersensitivity of EFdA against K65R mutation (Brenner et al., 2009). In the present study, we had assessed the anti-viral activity of a series of 4′-NRTIs against HIV-1K65R and found that these 4′-NRTIs also maintained their antiviral activity. Four compounds that contain 4′-ethynyl moiety (Figure 1), showed increased potency against HIV-1K65R compared to that against HIV-1WT (Table 1). Although EFdA showed slightly elevated IC50 value with 2.7-fold change, it still maintained its antiviral activity with an IC50 value of sub nano molar, indicating that 4′-ethynyl NRTIs including EFdA showed increased potency against HIV-1K65R. EFdA also retained its potency against the viruses containing not only M184V/I mutation, but also E138K mutation (Oliveira et al., 2017), which caused virological failure during the clinical trials that evaluated rilpivarine with emtricitabine as a first-line therapy (Van Eygen et al., 2016). Moreover, we previously showed that the emergence of EFdA resistance was considerably delayed compared to other NRTIs, including TDF (Maeda et al., 2014). In the present study, we investigated the structural mechanism underlying the activity of EFdA against HIV-1 variants, including drug-resistant HIV-1 variants, and found that EFdA and several NRTIs with the 4′-ethynyl moiety showed a strong vdW interaction with F160. We also found that F160 is one of the most critical amino acids for HIV-1 RT activity and thus the virus does not select F160 as a drug-resistant substitution. Moreover, this strong vdW interaction with active site residues A114, F160, and D185 was not reduced or lost even in the presence of drug resistant mutations, such as V184, also present in the active site cavity. Therefore, EFdA and other NRTIs that have a 4′-ethynyl moiety successfully maintain potent activity against HIV-1 variants, including drug-resistant HIV-1 variants.
We also examined the multidrug NRTI-resistant HIV-1 variant HIV-1EFdAR which harbors nine drug-resistant amino acid mutations in the gene encoding RT. We found that each of these nine mutations had a moderate effect on the activity of EFdA but had a substantial effect on the activities of conventional NRTIs, such as AZT and ABC (Table 1). Moreover, most of the selected amino acids, except M184, were not located in or around the active site cavity of RT (Figure 2A). This suggested that it was very difficult for the virus to obtain single substitutions that would have contributed to high resistance against EFdA. Rather, the virus accumulated minor conventional drug-resistant mutations which resulted in moderate resistance against EFdA (Table S2). Together, these findings suggest that EFdA is one of the most promising next-generation anti-HIV-1 agent with a favorable PK profile and improved activity against various drug-resistant HIV-1 strains.
In terms of the activity and drug-resistance profile of 4′-modified NRTIs against HIV-2, Wu et al. reported that EFdA (MK-8591) showed high antiviral activity against both wild-type HIV-2 and multi-NRTI-resistant HIV-2 variants (Wu et al., 2017). Smith et al. also reported that 4′-ethynyl-d4T (BMS-986001 or festinavir) showed increased potency against HIV-2, and 4′-ethynyl-d4T was also active against HIV-2M184V (Smith et al., 2015). The active site cavities of both HIV-1 and HIV-2 RTs are considered to be very similar (Boyer et al., 2006; Smith et al., 2015), but more detailed analyses are needed to explain the precise mechanism of drug-resistance of HIV-2 against EFdA and other 4′-modified derivatives.
We previously reported that two 4′-modified and EFdA-related NRTIs, namely, CAdA and CdG, potently inhibited both HBV and HIV-1 (Takamatsu et al., 2015). EFdA, a potent anti-HIV-1 inhibitor, only showed minor anti-HBV activity, whereas CAdA and CdG showed potent (sub nano molar IC50 values) activity against both HIV-1 and HBV. It has been reported that the reverse transcriptase (pol gene product) is the most stable in the changes during the evolution of retroviruses (McClure et al., 1988). Although the HBV RT and HIV-1 RT have less than 25% amino acid sequence identity (Das et al., 2001, Yasutake et al., 2018), key RT interactions with DNA and dNTP were well-conserved especially in the catalytic region of RTs, which include conserved domain A-G (Allen et al., 1998, Bartholomeusz et al., 2004). Moreover, A114 and Y115 form interactions with the incoming triphosphate moiety of dNTP (Huang et al., 1998, Boyer et al., 2000). A114 of HIV-1 RT is observed as A87 in HBV RT (Stuyver et al., 2001), while Y115 of HIV-1 RT corresponds to F88 in HBV RT (Stuyver et al., 2001). Interestingly, the Y115F mutation is reported to be associated with the HIV-1 drug (ABC and TDF) resistance (Margot et al., 2006) and is the only amino acid substitution that does not reduce the HIV-1 RT enzyme activity (Larder et al., 1989, Boyer et al., 2000). F160 of HIV-1 RT is a part of a helix and positionally equivalent to L180 of HBV RT (Das et al., 2001, Stuyver et al., 2001). CAdA and CdG possess a 4′-cyano moiety in their sugar groups. Modeling analysis of HIV-1 RT or HBV RT complexed with an NRTI-TP suggested that a shallow hydrophobic pocket in the polymerase active site of HBV RT accommodated the slightly shorter 4′-cyano moiety of CAdA-TP but could not accommodate the longer 4′-ethynyl moiety of EFdA-TP. In contrast, the deeper pocket of HIV-1 RT efficiently accommodates the 4′-moieties of both CAdA and CdG (Takamatsu et al., 2015). These data suggest that the type of 4′-moiety in NRTIs determines their affinity toward HIV-1 RT or HBV RT. In addition, as described, the present study showed that EFdA and related NRTIs with the 4′-ethynyl or 4′-cyano moiety showed very potent activity against HIV-1M184V and HIV-1EFdAR; however, related compounds with other moieties such as the 4′-methyl moiety did not show any activity. The result was also consistent to the previous studies for other 4′-modified NRTIs (Kohgo et al., 1999; Ohrui et al., 2011). Taken together, this finding suggests that the structure of the 4′-moiety of NRTIs determines the activity of the NRTIs not only against HIV-1 or HBV but also against drug-resistant HIV-1 variants.
Thus, EFdA and its derivatives with the 4′-ethynyl or 4′-cyano moiety warrant further clinical investigation as NRTIs potent against various multidrug-resistant HIV-1 variants. The results of the present study also suggest another strategy to design NRTIs that directly bind to residues that are critical for the function of HIV-1 RT (e.g., F160) or HBV RT even in the presence of drug-resistant mutations, such as (e.g., M184V). Such NRTIs will be effective against various drug-resistant viruses and will not induce resistance in HIV-1 and HBV because they directly and strongly bind to conserved amino acids in the active site of RT, that do not undergo mutation.
Star Methods
CONTACT FOR REAGENT AND RESOURCES
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kenji Maeda (kmaeda@ri.ncgm.go.jp).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cells.
MT-4 cells, originated from adult male human (RRID: CVCL_2632) authenticated using the STR profiling (ATCC, Manassas, VA), were grown in RPMI 1640-based culture medium with 10% fetal calf serum (FCS; Gemini Bio-Products, West Sacramento, CA) while HEK293T cells, originated from fetus female human (RRID: CVCL_0063), were cultured in DMEM supplemented with 10% FCS. The cells were grown and maintained in humidified atmosphere containing 5% Co2 at 37°C.
Viruses.
A wild-type laboratory HIV-1 isolate, HIV-1NL4-3 (Adachi et al., 1986), and HIV-2EHO (Rey et al., 1989) was used for performing the drug susceptibility assay.
METHOD DETAILS
Antiviral agents.
EFdA and CAdA (Figure 1) were synthesized as described previously (Kohgo et al., 2004). The synthesis of FMAdA and other 4′-modified EFdA-related derivatives are discussed elsewhere (O-Yang et al., 1992; Kitano et al., 1997; Kohgo et al., 1999a; Kohgo et al., 1999b, Ohrui et al., 2000; Kitano et al., 2000; Kohgo et al., 2004; Kohgo et al., 2018). AZT (ZDV) and ABC were purchased from Sigma-Aldrich (St. Louis, MO). TDF was purchased from BioVision (Milpitas, CA).
Recombinant Viruses.
Recombinant HIV-1 clones (HIV-1K65R, HIV-1A114V, HIV-1Y115A, HIV-1F160A, HIV-1M184V HIV-1D185A HIV-1T215F HIV-1D67del/T69G/K70R/L74I/V75T [HIV-167-75], and HIV-1D67del/T69G/K70R/L74I/V757T/M184V/T215F [HIV-167-75,184,215]) were propagated using HIV-1NL4-3-based infectious clones generated using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). Another recombinant HIV-1 clone HIV-1EFdAR was propagated using a recombinant HIV-1NL4-3-based infectious molecular clone, as reported previously (Maeda et al., 2014). Briefly, a DNA fragment containing 312 nucleic acids of the 3′-end of the gag-coding gene was inserted into the clone. Next, the entire protease-coding gene and 951 nucleic acids of the 5′-end of the RT-coding gene were amplified from a drug-selected virus population and were digested with ApaI (gag-coding region) and AgeI (RT-coding region) and were inserted into the infectious clone. Thus, the infectious clone contains the entire protease protein and the first 312 amino acids of RT derived from the selected virus population (Maeda et al., 2014). The nucleotide sequences of RT-coding gene were verified by the Sanger sequencing using BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and conducted at the CCR Genomics Core at the National Cancer Institute.
Anti-HIV and cytotoxicity assays.
Anti-HIV activity of EFdA and other EFdA-related NRTIs (Figure 1) was examined by performing the methyl thiazol tetrazolium (MTT) and p24 assays, as described previously (Kodama et al., 2001; Maeda et al., 2014). The potency of HIV inhibition by a compound was determined based on its inhibitory effect on virally induced cytopathicity (MTT assay) or on the production of p24 antigen (p24 assay) compared with that of drug-free controls. Briefly, for MTT assay, MT-4 cells were exposed to an HIV strain at fifty 50% tissue culture infectious dose. After viral exposure, the cell suspension was plated in each well of a 96-well flat microtiter culture plate (cell density, 5 χ 103 cells/well) containing various concentrations of the drug. After incubation for 5 days, the number of viable cells in each well (with or without drug) was measured using Cell Counting Kit-8 (Dojindo, Kumamoto, Japan), and 50% inhibitory concentration (IC50) value was determined. We also determined the anti-HIV-1 activity of EFdA against HIV-1EFdAR and HIV-167-75,184,215 by performing the p24 assay, as described previously (Kodama et al., 2001). Briefly, the amount of p24 antigen in supernatants of 5-day incubated HIV-1-infected cells was measured by performing a chemiluminescent enzyme immune assay with Lumipulse G1200 (Fujirebio, Tokyo, Japan), which measured the fluorescence (count per second [cps]). The fluorescence (cps) was converted to the amount of p24 antigen produced (ng/ml) by using a standard curve obtained using 2-fold serial dilutions of a standard concentration of the p24 antigen (ZeptoMetrix Corporation, Buffalo, NY). Cytotoxicity of a compound in MT-4 cells was also determined. Cells were plated in a 96-well plate at a density of 5 χ 103 cells in well and were continuously exposed to various concentrations of a compound throughout the entire period of the 7 days of culture. The number of viable cells in each well was determined using Cell Counting Kit-8, and 50% cytotoxicity concentration (CC50) values were determined. All the assays were conducted in triplicate of two or three independent experiments and the average values were determined.
RT activity and viral replication assays.
HIV-1 RT activity and viral replication assays were performed using HIV-1 mutants harboring RT mutations. HIV-1NL4-3 based plasmids (pHIV-1NL4-3) with RT mutations were transfected into HEK293T cells by using Attractene Transfection Reagent (Qiagen, Hilden, Germany), and the viruses were harvested at 72 h after the transfection. The viral p24 antigen was measured using Lumipulse G1200. RT activity of the HIV-1 mutants (for 100 ng p24 antigen) was measured by performing a colorimetric RT assay (Roche Applied Science, Penzberg, Germany), according to the manufacturer’s instructions. Percent control numbers was calculated by comparing to that of wild type RT, and the average value was given out of three independent assays. Replication of HIV-1 mutants was determined as described previously (Maeda et al., 2014), with minor modifications. Briefly, MT-4 cells (density, 1.5 χ 105 cells in 6 wells plate) were exposed to the HIV-1 mutants with the amount of 20 ng/ml of the p24 antigen and were cultured without the antiretroviral agents for 10 days. The amount of the p24 antigen was measured every 2 days. The average value of each data point was determined from two independent assays.
Molecular modeling.
Crystal structure of the ternary complexes of HIV-1 RTs with primer-templates (pt) and EFdA-triphosphates (EFdA-TP) reported by Salie etal. (PDB [RRID: SCR_012820] ID: 5J2M) was used as the starting structure (Salie et al., 2016). Correct bond orders of the residues, including zero-order bonds from the Mg2+ to the phosphate groups were assigned, and the termini were capped. A restrained minimization was performed (5J2Mprep). All subsequent minimizations were carried out from the 5J2Mprep structure. MAdA-TP, FMAdA-TP and EtAdA-TP-were built by appropriate modifications to EFdA in 5J2Mprep, and the structures were minimized to obtain respective wild-type complexes. M184 residue was mutated to valine and minimized to obtain respective HIV-1 RTM184V complexes. Maestro version 10.7.015 (Schrodinger, LLC. New York, NY) was used for molecular model building, visualization, analysis, and figure generation. The OPLS3 forcefield, as implemented in Maestro was used for all structure minimizations.
QUANTIFICATION AND STATISTICAL ANALYSIS
The IC50 values and CC50 values were calculated in each independent assay and the average values were determined. Statistical analysis (calculation of standard deviation) was performed by using Microsoft Excel.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| HIV-1NL4-3 |
Adachi et al., 1986 NIH AIDS Reagent Program |
Cat#114 |
| HIV-2EHO | Rey et al., 1989 | N/A |
| Biological Samples | ||
| One Shot MAX Efficiency DH5α-T1R Competent Cells | ThermoFisher Scientific | Cat#12297016 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4ʹ-Ethynyl-2-fluoro-2ʹ-deoxyadenosine (EFdA/MK-8591) | Nakata et al. 2007 | N/A |
| 4ʹ-cyano-2-amino-2ʹ-deoxyadenosine (CAdA) | Kohgo et al., 2004 | N/A |
| 4ʹ-fluoromethyl-2-amino-2ʹ-deoxyadenosine (FMAdA) | Kohgo et al., 2018 | N/A |
| 4ʹ-fluoromethyl-2-fluoro-2ʹ-deoxyadenosine (FMFdA) | This paper | N/A |
| 4ʹ-methyl-2-amino-2ʹ-deoxyadenosine (MAdA) | Kohgo et al., 2018 | N/A |
| 4ʹ-vinyl-2-amino-2ʹ-deoxyadenosine (VAdA) | Kohgo et al., 2018 | N/A |
| 4ʹ-ethyl-2-amino-2ʹ-deoxyadenosine (EtAdA) | Kohgo et al., 2018 | N/A |
| 4ʹ-ethyl-2ʹ-deoxyguanosine (EtdG) | Kohgo et al., 2018 | N/A |
| 4ʹ-methyl-2ʹ-deoxyguanosine (MdG) | Kohgo et al., 2018 | N/A |
| 4ʹ-fluoromethyl-2-amino-N6-methyl-2ʹ-deoxyadenosine (FMAMdA) | This paper | N/A |
| 4ʹ-ethynyl-2ʹ-deoxyguanosine (EdG) |
Ohrui et al. 2000. Kitano et al. 2000. |
N/A |
| 4ʹ-ethynyl-2ʹ-deoxyinosine (EdI) | Ohrui et al. 2000. | N/A |
| 4ʹ-fluoromethyl-2ʹ-deoxycytidine (FMdC) | Kitano et al., 1997 | N/A |
| 4ʹ-ethynyl-2ʹ-deoxycytidine (EdC) |
Kohgo et al., 1999a Kohgo et al., 1999b |
N/A |
| 4ʹ-cyano-2ʹ-deoxythymidine (CdT) | O-Yang et al., 1992 | N/A |
| Critical Commercial Assays | ||
| Cell Counting Kit-8 | Dojindo | Cat#CK04 |
| Lumipulse G1200 | Fujirebio Diagnostics, Inc. | https://www.fujirebio-us.com/Lumipulse/ |
| colorimetric RT assay | Roche Applied Science | Cat#11468120910 |
| Experimental Models: Cell Lines | ||
| MT-4 cells | NIH AIDS Reagent Program | Cat#120 RRID: CVCL_2632 |
| HEK293T cells | ATCC | ATCC CRL-3216 RRID: CVCL_0063 |
| Oligonucleotides | ||
| pNL4-3 | NIH AIDS Reagent Program | Cat#114 |
| Mutagenesis primers | See Table S3 for primer sequence | N/A |
| Recombinant DNA | ||
| HIV-1EFdAR | Maeda et al. 2014 | N/A |
| Software and Algorithms | ||
| Maestro version 10.7.015 | Schrödinger, LLC | https://www.schrodinger.com/maestro/ |
Significance.
Nucleoside reverse transcriptase inhibitors (NRTIs) are one of the backbone drugs of combined antiretroviral therapy (cART) regimens, which inhibit viral replication by competitive binding to the viral reverse transcriptase and terminate DNA extension. 4′-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA/MK-8591), which is currently under clinical trials, has a high genetic barrier against the emergence of drug-resistant variants and exerts potent antiviral activity against Human immunodeficiency virus type 1 (HIV-1) strains including highly multiple-drug-resistant variants. Here, we evaluated a series of 4′-modified EFdA derivatives and reported that EFdA and NRTIs with a 4′-ethynyl or 4′-cyano moiety exerted antiviral activity against HIV-1 with an M184V mutation, which is the most prevalent NRTI-associated mutation in treated patients, and multiple NRTI-resistant HIV-1s, whereas NRTIs with other moieties (e.g., 4′-methyl, 4′-ethyl) did not show this activity. This activity was due to the strong van der Waals interactions with critical amino acid residues A114, Y115, F160, and M184 present in the hydrophobic pocket of HIV-1 reverse transcriptase. Structural analysis revealed that these interactions were maintained even in the presence of a broad resistance-endowing M184V substitution. Moreover, we found that substitution of these residues resulted in the complete loss of HIV-1 reverse transcriptase enzymatic activity and HIV-1 harboring these mutations failed to replicate. Thus, the interaction of EFdA and 4′-ethynyl-NRTIs with conserved active site residues whose substitution is detrimental to the function of HIV-1 RT is key to their potent activity against drug-resistant HIV-1 strains. These findings shed important insight towards further design of clinically relevant antiviral therapeutics.
Highlights.
4′-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA/MK-8591) is a potent anti-HIV-1 agent.
EFdA and its derivatives possess a modified 4′-moiety.
4′-Ethynyl nucleoside analogs form strong interactions with reverse transcriptase.
These interactions are maintained even with drug resistance endowing mutations.
Acknowledgement
This work was supported in part by the Intramural Research Program of Center for Cancer Research, National Cancer Institute, NIH (grant number BC011486 and SC006738 to H.M.); by JSPS KAKENHI (grant number: JP16K08826 to K.M.), by AMED (grant number JP18fk0410015 to K.M. and JP18fk0310113 to H.M.); and by a grant from National Center for Global Health and Medicine Research Institute (grant number H29-1010 to K.M.). S.G.S was supported by grants R01AI112417 and R01AI076119 from the NIH. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH, Bethesda, MD (http://hpc.nih.gov). The Sanger sequencing was conducted at the CCR Genomics Core at the National Cancer Institute. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 NL4-3 Infectious Molecular Clone (pNL4-3) from Dr. Malcolm Martin. The authors thank Shin-ichiro Hattori and Kouki Matsuda for technical assistance. We thank Editage (http://www.editage.jp) for the English language editing.
Footnotes
Declaration of Interests
H.M. is listed as a coinventor in a patent, 4′-C-substituted-2-haloadenosine derivatives including 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) (US Patent 7,339,053 B2, March 4, 2008). All rights, title, and interest to the patent have been assigned to Yamasa Co, Chiba, Japan. All other authors declare that they do not have any competing interests related to this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, and Martin MA (1986). Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 59 284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MI, Deslauriers M, Andrews CW, Tipples GA, Walters KA, Tyrrell DL, Brown N, and Condreay LD (1998). Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 271670–1677. [DOI] [PubMed] [Google Scholar]
- Bartholomeusz A, Tehan BG, and Chalmers DK (2004). Comparisons of the HBV and HIV polymerase, and antiviral resistance mutations. Antivir Ther. 9 149–160. [PubMed] [Google Scholar]
- Boyer PL, Sarafianos SG, Arnold E, and Hughes SH (2000). Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc Natl Acad Sci U S A. 97 3056–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer PL, Sarafianos SG, Clark PK, Arnold E, and Hughes SH (2006). Why do HIV-1 and HIV-2 use different pathways to develop AZT resistance. Plos Pathogens. 2 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner BG, and Coutsinos D (2009). The K65R mutation in HIV-1 reverse transcriptase: genetic barriers, resistance profile and clinical implications. HIV Ther. 3 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Xiong X, Yang H, Westland CE, Gibbs CS, Sarafianos SG, and Arnold E (2001). Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J Virol. 75 4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fernandez B, Montoya-Ferrer A, Sanz AB, Sanchez-Nino MD, Izquierdo MC, Poveda J, Sainz-Prestel V, Ortiz-Martin N, Parra-Rodriguez A, Selgas R, et al. (2011). Tenofovir nephrotoxicity: 2011 Update. AIDS Res Treat. 2011 354908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori S, Ide K, Nakata H, Harada H, Suzu S, Ashida N, Kohgo S, Hayakawa H, Mitsuya H, and Okada S (2009). Potent activity of a nucleoside reverse transcriptase inhibitor, 4′-ethynyl-2-fluoro-2′-deoxyadenosine, against human immunodeficiency virus type 1 infection in a model using human peripheral blood mononuclear cell-transplanted NOD/SCID Janus kinase 3 knockout mice. Antimicrob Agents Chemother. 53 3887–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Chopra R, Verdine GL, and Harrison SC (1998). Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 282 1669–1675. [DOI] [PubMed] [Google Scholar]
- Kawamoto A, Kodama E, Sarafianos SG, Sakagami Y, Kohgo S, Kitano K, Ashida N, Iwai Y, Hayakawa H, Nakata H, et al. (2008). 2′-Deoxy-4′-C-ethynyl-2-halo-adenosines active against drug-resistant human immunodeficiency virus type 1 variants. Int J Biochem Cell Biol. 40, 2410–2420. [DOI] [PubMed] [Google Scholar]
- Kitano K, Miura S, Ohrui H, and Meguro H (1997). Synthesis of 4′-C-Fluoromethylnucleosides as Potential Antineoplastic Agents. Tetrahedron. 53 13315–13322. [Google Scholar]
- Kitano K, Sakata S, Kohgo S, Matsuoka M, Kodama E, Mitsuya H, and Ohrui H (2000). Synthesis of 4′-ethynyl-purine nucleosides possessing anti-HIV activity. Nucleic Acids Symp Ser. 44 105–106. [DOI] [PubMed] [Google Scholar]
- Kodama EI, Kohgo S, Kitano K, Machida H, Gatanaga H, Shigeta S, Matsuoka M, Ohrui H, and Mitsuya H (2001). 4′-Ethynyl nucleoside analogs: potent inhibitors of multidrug-resistant human immunodeficiency virus variants in vitro. Antimicrob Agents Chemother. 45 1539–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohgo S, Horie H, and Ohrui H (1999a). Synthesis of 4′-C-ethynyl-beta-D-arabino- and 4′-C-ethynyl-2′-deoxy-beta- D-ribo-pentofuranosyl pyrimidines, and their biological evaluation. Biosci Biotechnol Biochem. 1146–1149. [DOI] [PubMed] [Google Scholar]
- Kohgo S, Kodama E, Shigeta S, Saneyoshi M, Machida H, and Ohrui H (1999b). Synthesis of 4′-substituted nucleosides and their biological evaluation. Nucleic Acids Symp Ser. 42127–128. [DOI] [PubMed] [Google Scholar]
- Kohgo S, Yamada K, Kitano K, Iwai Y, Sakata S, Ashida N, Hayakawa H, Nameki D, Kodama E, Matsuoka M, et al. (2004). Design, efficient synthesis, and anti-HIV activity of 4′-C-cyano- and 4′-C-ethynyl-2′-deoxy purine nucleosides. Nucleosides Nucleotides Nucleic Acids. 23 671–690. [DOI] [PubMed] [Google Scholar]
- Kohgo S, Imoto S, Tokuda R, Takamatsu Y, Higashi-Kuwata N, Aoki M, Amano M, Kansui H, Onitsuka K, Maeda K, et al. (2018). Synthesis of 4′-Substituted Purine 2′-Deoxynucleosides and Their Activity against Human Immunodeficiency Virus Type 1 and Hepatitis B Virus. Chemistry Select. 3 3313–3317. [Google Scholar]
- Larder BA, Kemp SD, and Purifoy DJ (1989). Infectious potential of human immunodeficiency virus type 1 reverse transcriptase mutants with altered inhibitor sensitivity. Proc Natl Acad Sci U S A. 86 4803–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeansyah E, Cameron PU, Solomon A, Tennakoon S, Velayudham P, Gouillou M, Spelman T, Hearps A, Fairley C, Smit de V., et al. (2013). Inhibition of telomerase activity by human immunodeficiency virus (HIV) nucleos(t)ide reverse transcriptase inhibitors: a potential factor contributing to HIV-associated accelerated aging. J Infect Dis. 207 1157–1165. [DOI] [PubMed] [Google Scholar]
- Maag H, Rydzewski RM, McRoberts MJ, Crawford-Ruth D, Verheyden JP, and Prisbe EJ (1992). Synthesis and anti-HIV activity of 4′-azido- and 4′-methoxynucleosides. J Med Chem. 35 1440–1451. [DOI] [PubMed] [Google Scholar]
- Maeda K, Desai DV, Aoki M, Nakata H, Kodama EN, and Mitsuya H (2014). Delayed emergence of HIV-1 variants resistant to 4′-ethynyl-2-fluoro-2′-deoxyadenosine: comparative sequential passage study with lamivudine, tenofovir, emtricitabine and BMS-986001. Antivir Ther. 19 179–189. [DOI] [PubMed] [Google Scholar]
- Margot NA, Waters JM, and Miller MD (2006). In vitro human immunodeficiency virus type 1 resistance selections with combinations of tenofovir and emtricitabine or abacavir and lamivudine. Antimicrob Agents Chemother. 50 4087–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure MA, Johnson MS, Feng DF, and Doolittle RF (1988). Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci U S A. 85 2469–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez-Arias L (2013). Molecular basis of human immunodeficiency virus type 1 drug resistance: overview and recent developments. Antiviral Res. 98 93–120. [DOI] [PubMed] [Google Scholar]
- Michailidis E, Marchand B, Kodama EN, Singh K, Matsuoka M, Kirby KA, Ryan EM, Sawani AM, Nagy E, Ashida N, et al. (2009). Mechanism of inhibition of HIV-1 reverse transcriptase by 4′-ethynyl- 2-fuoro-2′-deoxyadenosine triphosphate, a translocation-defective reverse transcriptase inhibitor. J Biol Chem. 284 35681–35691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidis E, Ryan EM, Hachiya A, Kirby KA, Marchand B, Leslie MD, Huber AD, Ong YT, Jackson JC, Singh K, et al. (2013). Hypersusceptibility mechanism of Tenofovir-resistant HIV to EFdA. Retrovirology. 10 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD (2004). K65R, TAMs and tenofovir. AIDS Rev. 6 22–33. [PubMed] [Google Scholar]
- Murphey-Corb M, Rajakumar P, Michael H, Nyaundi J, Didier PJ, Reeve AB, Mitsuya H, Sarafianos SG, and Parniak MA (2012). Response of simian immunodeficiency virus to the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine in vitro and in vivo. Antimicrob Agents Chemother. 56 4707–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata H, Amano M, Koh Y, Kodama E, Yang G, Bailey CM, Kohgo S, Hayakawa H, Matsuoka M, Anderson KS, et al. (2007). Activity against human immunodeficiency virus type 1, intracellular metabolism, and effects on human DNA polymerases of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob Agents Chemother. 51 2701–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitanda T, Wang X, Kumamoto H, Haraguchi K, Tanaka H, Cheng YC, and Baba M (2005). Anti-human immunodeficiency virus type 1 activity and resistance profile of 2′,3′-didehydro-3′-deoxy-4′-ethynylthymidine in vitro. Antimicrob Agents Chemother. 49 3355–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Shuto S, Tanaka M, Sasaki T, Mori S, Shigeta S, and Matsuda A (1999). Nucleosides and nucleotides. 185. Synthesis and biological activities of 4′α-C-branched-chain sugar pyrimidine nucleosides. 42 2901–2908. [DOI] [PubMed] [Google Scholar]
- Ohrui H, Kohgo S, Kitano K, Sakata S, Kodama E, Yoshimura K, Matsuoka M, Shigeta S, and Mitsuya H (2000). Syntheses of 4′-C-ethynyl-β-D-arabino- and 4′-C-ethynyl-2′-deoxy-β-D-ribo-pentofuranosylpyrimidines and -purines and evaluation of their anti-HIV activity. J Med Chem. 43 4516–4525. [DOI] [PubMed] [Google Scholar]
- Ohrui H (2011). Development of modified nucleosides that have supremely high anti-HIV activity and low toxicity and prevent the emergence of resistant HIV mutants. Proc Jpn Acad Ser B Phys Biol Sci. 87:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M, Brenner BG, Xu H, Ibanescu RI, Mesplede T, and Wainberg MA (2017). M184I/V substitutions and E138K/M184I/V double substitutions in HIV reverse transcriptase do not significantly affect the antiviral activity of EFdA. J Antimicrob Chemother. 72 3008–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O-Yang C, Wu HY, Fraser-Smith EB and Walker AM (1992) Synthesis of 4′-cyanothymidine and analogs as potent inhibitors of HIV. Tetrahedron Lett. 33 37–40. [Google Scholar]
- Powderly WG (2012). Osteoporosis and bone health in HIV. Curr HIV/AIDS Rep. 9 218–222. [DOI] [PubMed] [Google Scholar]
- Rey MA, Krust B, Laurent AG, Guetard D, Montagnier L, and Hovanessian AG (1989). Characterization of an HIV-2-related virus with a smaller sized extracellular envelope glycoprotein. Virology. 173 258–267. [DOI] [PubMed] [Google Scholar]
- Salie ZL, Kirby KA, Michailidis E, Marchand B, Singh K, Rohan LC, Kodama EN, Mitsuya H, Parniak MA, and Sarafianos SG (2016). Structural basis of HIV inhibition by translocation defective RT inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA). Proc Natl Acad Sci U S A. 113 9274–9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Raugi DN, Wu VH, Leong SS, Parker KM, Oakes MK, Sow PS, Ba S, Seydi M, Gottlieb GS, and University of Washington-Dakar HIV-2 Study Group. (2015). The nucleoside analog BMS-986001 shows greater in vitro activity against HIV-2 than against HIV-1. Antimicrob Agents Chemother. 59 7437–7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuyver LJ, Locarnini SA, Lok A, Richman DD, Carman WF, Dienstag JL, and Schinazi RF (2001). Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology. 33 751–757. [DOI] [PubMed] [Google Scholar]
- Takamatsu Y, Tanaka Y, Kohgo S, Murakami S, Singh K, Das D, Venzon DJ, Amano M, Higashi-Kuwata N, Aoki M, et al. (2015). 4′-Modified nucleoside analogs: potent inhibitors active against entecavir-resistant hepatitis B virus. Hepatology. 62 1024–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cor-Hosmer SK, Daddacha W, Kelly Z, Tsurumi A, Kennedy EM, and Kim B (2012). The impact of molecular manipulation in residue 114 of human immunodeficiency virus type-1 reverse transcriptase on dNTP substrate binding and viral replication. Virology. 422 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eygen V, Thys K, Van Hove C, Rimsky LT, De Meyer S, Aerssens J, Picchio G, and Vingerhoets J (2016). Deep sequencing analysis of HIV-1 reverse transcriptase at baseline and time of failure in patients receiving rilpivirine in the phase III studies ECHO and THRIVE. J Med Virol. 88 798–806. [DOI] [PubMed] [Google Scholar]
- Wu VH, Smith RA, Masoum S, Raugi DN, Ba S, Seydi M, Grobler JA, Gottlieb GS, and University of Washington-Dakar HIV-2 Study Group. (2017). MK-8591 (4′-Ethynyl-2-Fluoro-2′-Deoxyadenosine) Exhibits Potent Activity against HIV-2 Isolates and Drug-Resistant HIV-2 Mutants in Culture. Antimicrob Agents Chemother. 61 e00744–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutake Y, Hattori SI, Hayashi H, Matsuda K, Tamura N, Kohgo S, Maeda K, and Mitsuya H (2018). HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir. Sci Rep. 8:1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.