Introduction
According to the statistics (2016 update) from the American Heart Association (AHA), 15.5 million people over 20 years old in the US have coronary heart disease, and every 42 seconds, an American suffers from myocardial infarction (MI) [1]. For patients admitted into hospitals with suspected acute coronary syndrome (ACS), electrocardiography (ECG) is an important risk-stratification and assessment tool to guide further treatment for MI, and ST-segment changes in ECG constitute the principle biomarker for such purpose. However, less than 25% of ACS patients present ST elevation (ST-elevation MI, or STEMI) and receive immediate medical attention. For the other 75% of myocardial infarctions, including non-ST elevation ACS (NSTE-ACS) or unstable angina (UA) [2], continuous ST-segment monitoring is crucial for early identification of transient myocardial ischemia (TMI, precursor of MI) and to prevent adverse clinical events.
Unfortunately, current ST-segment monitoring systems have yet to fulfil their designed purpose due to excessive false positive alarms. One study tracking a 16-bed intensive cardiac care (ICC) unit during a 31-day period discovered an average of 200 ST alarms per day, even with stricter trigger threshold at 200 μV being adopted in the facility instead of the recommended 100 μV, and over 90% of them are non-actionable alarms [3]. These nuisance alarms further contribute to the issue of alarm fatigue, which is ranked as one of the top 10 technology hazards by the Emergency Care Research Institute (ECRI) in 2014 [4]. Alarm fatigue is described as the sensory overload caused by the overwhelming visual and auditory alerts generated by bedside physiologic motors to caregivers, which may lead to missed critical clinical opportunities [3]. Due to alarm fatigue, a recent statement from AHA has decreased the class of recommendation (COR) for ST alarms from class I (should be performed) to class IIa (is reasonable to perform) [5]. Thus, there is an urgent unmet need for ST-segment monitoring algorithms with improved precision.
Recent advancement of deep learning has transformed many fields of study by taking advantage of big data and modern computing resources. The tremendous amount of digitized ECG data generated in clinical facilities meet well with the prerequisite for applying deep learning and have sparked various ECG research. One study adopted the convolutional neural network (CNN), one of the deep learning algorithms, to classify various types of ECG arrhythmia and has achieved cardiologist-level performance [6]. Another study used the CNN model to learn features in ECG that can screen patients with paroxysmal atrial fibrillation [7]. One more study took advantage of both convolutional and sequential models in deep learning to classify ECG signals from patients with coronary arterial disease from normal ECG [8], only to name a few. Inspired by these pioneering studies, the present work starts off to investigate the application of deep learning in detecting significant changes in ST segments, in an effort to improve the monitoring precision.
Expert cardiologists are able to identify ischemic changes in ST segments by visual inspection of ECG tracings in spite of the existence of moderate contamination of the waveform (body position change, motion artifact, numerous physiological confounders, etc.), where conventional ST-segment monitoring algorithms using numerical thresholding have usually failed. Addressing this circumstance, the present study adopts an image-based approach for sample representation to tackle the detection of ST changes as a computer visual task to leverage deep learning techniques, which have demonstrated close-to or even surpassed human performance [9]. In the present study, convolutional models are trained through a transfer-learning scheme from a publicly accessible long-term ECG database with expert annotation of ST events [10], and then are tested on an independent testing set in a simulated real-time fashion. Both qualitative and quantitative evaluations are performed to provide comprehensive examination of model performance. We further investigate various parameters during model building and their potential impact on the model performance, including finetuning parameters during training sample selection, and establishing a learning curve by varying number of ECG recordings in the training set.
Methods
Data source
The Long-Term ST Database from the Physionet is selected as the data source to generate training and testing samples [10, 11]. The database contains 86 whole-day Holter ECG recordings from 80 human subjects with 2- or 3-lead configuration. The signals are recorded at the sampling frequency of 250 Hz and at 12-bit resolution within the range between −10 to 10 millivolts. The database provides single-lead annotation information related to significant ST episodes (including ischemic and heart-rate related ST changes), significant ST shift (due to axis shift or conduction change), noisy and unreadable segments (as shown in Figure 1), based on three types of protocols to capture significant ST changes. The present study adopts annotation information from the protocol B [10], which defines significant ST changes to be exceeding 100 microvolts continuously for at least 30 seconds. For consistency, all recordings in the database that are with 2-lead configuration, are with significant ST episodes and are from subjects with a single recording are selected in the present study. ECG leads used in most of these recordings are the combination of one of precordial leads (V2, V3, V4 or V5) with modified limb lead III (MLIII). This results in 59 recordings from 59 distinct subjects for further analysis. We then randomly select 30 recordings as training data and the rest as testing data. A full list of recordings in training/testing sets can be found in Table 1.
Figure 1.

Conceptual timeline of annotated events in the LTST database. Events marked in green form up the ST condition. Events marked in red represent the non-ST condition. Events marked by black include noise, unreadable segments and sudden ST shifts, which are removed from analysis.
Table 1.
Full list of training/testing recordings used in the present study. Items shaded in grey are 30 recordings randomly selected as the training set. For testing set, the prevalence of each class is presented as (#ST samples: #non-ST samples).
| Training set | Testing set | ||||
|---|---|---|---|---|---|
| s20031 | s20301 | s20471 | s20021 (273:14785) |
s20211 (391:16197) |
s20411 (1211:15425) |
| s20051 | s20321 | s20481 | s20041 (3397:13761) |
s20241 (1161:15653) |
s20451 (960:16096) |
| s20061 | s20341 | s20491 | s20091 (460:15737) |
s20251 (141:16968) |
s20461 (265:16167) |
| s20071 | s20351 | s20561 | s20101 (359:14300) |
s20261 (1183:14899) |
s20511 (173:14010) |
| s20081 | s20361 | s20591 | s20121 (105:15218) |
s20281 (262:16796) |
s20521 (283:13202) |
| s20111 | s20371 | s20601 | s20131 (1493:13547) |
s20291 (722:15724) |
s20551 (1005:15985) |
| s20141 | s20401 | s20611 | s20151 (743:16240) |
s20311 (1803:15345) |
s20571 (139:10017) |
| s20161 | s20421 | s20631 | s20181 (601:16422) |
s20331 (346:17162) |
s20581 (50:15809) |
| s20171 | s20431 | s20641 | s20191 (72:17202) |
s20381 (347:16818) |
s20621 (4000:12322) |
| s20231 | s20441 | s20651 | s20201 (73:14979) |
s20391 (547:14315) |
|
Image sample generation
We take an image-based approach for sample generation, which has been successfully adopted for assessment of signal quality in ECG signals [12]. Specifically, we take the snapshots of 10-second ECG images from continuous single-lead ECG waveform as training/testing samples. In this way, monitoring ST changes is transformed into a computer vision task, which can be well approached using the convolutional neural network. Each 10-second ECG trace is firstly overlaid with a grid same as the standard ECG paper (40 ms per horizontal interval; 0.1 mV per vertical interval). Then the image is transformed into grayscale colormap to remove redundant color information that does not contribute to the classification task, and finally saved into an 8-bit jpeg file with image dimension of 600 px (W) by 450 px (H). These image samples are then further resized through bilinear interpolation to adhere to the input requirement of transfer learning using Google Inception V3.
Training/testing data preparation
For each ECG channel, we group ECG episodes associated with significant ST changes as the case condition (i.e. the ST condition), which includes ischemic ST and heart-rate related ST episodes (labeled in green in Figure 1). Then the remaining waveforms are grouped together as the control condition (i.e. the non-ST condition; labeled in red in Figure 1), by excluding segments annotated as unreadable, noise or ST shift. It’s worth noting that the database provides limited annotation information about events related to noise and ST shift, with single event time given rather than a duration. As an approximation, the 10-second data before and after the event time are excluded for these events.
Different sample selection schemes are designed for training and testing sets. For the training set, balanced numbers of image samples from ST and non-ST conditions are desirable for model training. To achieve this, 10,000 10-second image samples are randomly selected from non-ST ECG segments based on a uniform distribution for each recording as non-ST condition. For ST condition, the number of samples to be selected from each ST episode is determined by the total sample number (10,000) divided by the number of ST episodes in each recording. Within each ST episode, the corresponding number of image samples are selected by their temporal offsets with respect to the maxima of ST change, based on a Gaussian distribution with mean at the maxima of ST change and the standard deviation as a hyperparameter to tune. The maxima of ST change is determined by the maximal ST change within one episode, which is provided as a part of annotation information in LTST database [10]. Such design imposes higher weights for the selection of samples close to maxima of ST change, under the heuristic that more representative features related to ST change can be captured in this way. In total, there are 300,000 and 266,275 image samples for non-ST and ST conditions selected as training samples, respectively. For the testing set, consecutive 10-second image samples are selected for both conditions to approximate the real-time testing scenario. The prevalence of ST and non-ST conditions in the testing set can be found in Table 1.
Model training
We adopt a transfer learning scheme to attain CNN models from a comprehensively pretrained model rather than training the model from ground up. The underlying logic is recycling model parameters that capture common image features sharable across different computer vision tasks from a pretrained model in favor of an effective and efficient training process. The pretrained model used here is Google’s Inception V3, which has been trained from millions of images and 1000 classes from the ImageNet [13, 14]. We keep all the model parameters in the Inception model except the final layer, which is retrained by the training images in the present study. In this way, the pretrained model is adapted to identify image samples with significant ST changes. To retrain the final layer, the number of epochs is set at 2000. The training/validating/testing data separation in the training set follows an 80%−10%−10% split, which enables quick assessment of bias-variance tradeoff during the training process.
Model parameter investigation
To investigate the impact of training sample selection for the ST condition on the model performance, three models are trained from ST samples selected from different Gaussian distributions by tuning standard deviations (5, 10, and 30 seconds). Figure 2(a) shows the comparison of sample distributions from different standard deviations when selecting same number of samples. It shows the smaller the standard deviation, the closer selected samples are to the maxima of ST changes. The learning curve of the proposed model is also investigated by varying the number of recordings in the training set, from 5 to 30 with an increment of 5 recordings, during the model training. To achieve fair comparison, performance from all models is tested and compared based on the same testing set.
Figure 2.
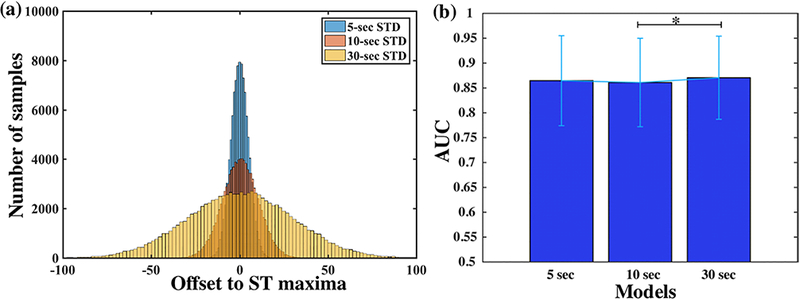
Impact of varying standard deviations (5, 10 and 30 seconds) for training sample selection on the model performance. Figure 2(a) Conceptual sample distributions with respect to maxima of ST change from different standard deviations; Figure 2(b) Comparison of classification performance (AUC) across different models. * indicates significant level of 0.01.
Performance evaluation
Model performance is evaluated both qualitatively and quantitively. Receiver operating characteristic (ROC) curves from all recordings in the testing set are firstly generated to provide qualitative evaluation of individual performance. Based on ROC curves, optimal probability cutoff points can be derived with maximal Youden’s index. Then various common performance metrics are calculated to provide quantitative evaluation at the group level, including sensitivity, specificity and area under the ROC curve (AUC). Since there are far more non-ST than ST samples in the testing set (see Table 1), support-weighted F1 score is also calculated to take inter-class prevalence into consideration, which is achieved by Scikit-learn library [15]. The Student t tests are performed to compare performance from different models, as well as their performance against the guess level.
Results
Figure 2(b) presents model performance from training samples selected from Gaussian distributions with different standard deviations. Each bar represents mean and standard deviation of AUCs across all 29 testing recordings. All three models yield comparable performance, with the 30-second model achieving the highest mean AUC at 87.05% (± 8.36%), followed by the 5- second model at 86.46% ± 9.05% and the 10-second model at 86.10% ± 8.89%. Statistical tests reveal significant difference in performance only between 10-second and 30-second models after Bonferroni correction for multiple comparisons (p < 0.01). Since 30-second model presents the best overall performance, subsequent model and results are based on the common standard of using 30 seconds as standard deviation for training sample selection.
Individual-level performance from each recording in the testing set is shown in Figure 3. Each curve in the figure depicts ROC curve from one recording, and the dashed line depicts the guess level. The figure provides a qualitative evaluation of model performance, with all recordings above the guess level. It also shows variation in performance across different recordings. Most recordings present ROC curves deflecting far away from the guess line, indicating high true positive (i.e. power) and low false positive rate are achievable with carefully selected threshold. However, there are also recordings presenting low power across all thresholds, with ROC curves close to the guess line.
Figure 3.
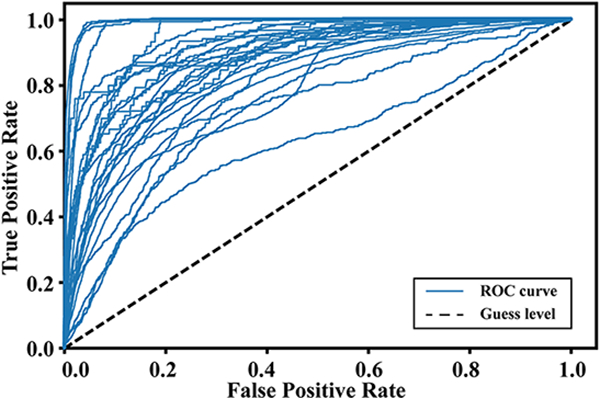
ROC curves from individual testing recordings achieved by the 30-second model. Dashed line denotes the guess level.
In addition to Figure 3, quantitative and group-level performance results are presented in Table-2 while maintaining comparable specificity at 80.34%. Besides, one-sample Student t tests show all metrics exceed the guess level with corrected significance level (p<<0.01). When examining the learning curve, all performance metrics show a general increasing trend along with more recordings in the training set, and best overall performance is achieved by the model using all 30 recordings in the training set. It also reveals that comparable performance can already be achieved with as few as 5 recordings in the training set, delivering a median AUC at 86.71% (range: 64.72–99.20%) and median F1 score at 84.27% (range: 52.85–97.82%).
Discussion
The image-based approach adopted in the study is inspired by the previous study on image- based ECG quality assessment [12]. The general idea is to convert one-dimensional ECG temporal dynamics into two-dimensional images so that image-based techniques and methods can be leveraged. In our study, taking 10-second snapshots of ECG waveforms transforms detection of ST changes into a computer vision task, where the convolutional neural network has demonstrated state-of-the-art performance. Then image features that differentiate ST from non- ST conditions can be extracted by convolutional layers in the CNN model for the classification of each 10-second image sample.
The sample representation of ECG signals as images also makes the implementation of transfer learning scheme with Google’s Inception V3 readily accessible, since current transfer-learning setup with Inception requires input as images to obtain the transfer-learned CNN model [13]. The transfer-learned model achieves median sensitivity at 82.64% to detect significant ST change, which is on par with previous studies using the same database (78.90%, 78.10% and 78.28%) [16–18], while maintaining a comparable specificity at 80.34%. Moreover, our approach using deep learning offers simple training process bypassing the complex rule-defining and feature engineering steps in conventional algorithms. By comparing to our previous feasibility study trained/tested with much fewer recordings [19], our approach presents stable performance in detecting significant ST change, which is further validated by the learning curve (as shown in Table 2).
Table 2.
Learning curve and quantitative evaluation of model performance at group level, including AUC, sensitivity, specificity and F1 score.
| # Training Recordings |
AUC (median: range) |
Sensitivity (median: range) |
Specificity (median: range) |
F1 Score (median: range) |
|---|---|---|---|---|
| 5 | 86.71: 64.72–99.20% |
82.08: 56.00–100% |
77.88: 36.81–96.41% |
84.27: 52.85–97.82% |
| 10 | 84.11: 57.63–98.47% |
80.92: 41.29–99.42% |
73.01: 47.74–97.58% |
83.32: 60.53–98.45% |
| 15 | 86.69: 63.12–99.26% |
80.20: 56.74–99.29% |
79.94: 61.45–96.39% |
84.92: 73.36–97.27% |
| 20 | 87.88: 65.14–99.30% |
80.56: 47.25–100% |
80.22: 57.41–95.96% |
85.82: 71.19–96.97% |
| 25 | 87.74: 63.73–99.27% |
81.92: 51.91–100% |
81.44: 61.74–95.97% |
87.31: 73.93–96.97% |
| 30 | 87.87: 63.07–99.28% |
82.64: 50.08–100% |
80.34: 59.49–96.08% |
87.38: 72.44–97.69% |
Our achieved performance also demonstrates the viability of transfer learning in biomedical research, especially when the original model has been trained from a large image database with most of its images irrelevant to the medical domain. The adopted transfer learning approach recycles model parameters from a pretrained model that can capture common image features regardless of classes and only trains the final layer or layers to equip the model with domain expertise. This could also have great implication to other image-based biomedical studies, such as computerized diagnostic classification using CT and MRI scans, to achieve an efficient and effective training process with transfer learning.
The duration of 10 seconds is selected in the present study with the following considerations. First, our image-based approach is inspired by the insight that clinicians usually read and identify pathological changes in ECG through visual pattern recognition. The selection of 10-second image samples aligns well with the current real-time clinical setup, where most of bedside physiological monitors offer 10-second ECG strips as the frontend presentation. The classification of ST change at 10-second resolution resembles the real-time clinical practice when clinicians visually evaluate those ECG strips from bedside monitors screen by screen.
Second, accurate classification of significant ST change at short-duration level can serve as groundwork to multiple succeeding goals. It’s been found that many false ST alarms in current in-hospital ECG monitors are induced by brief ST changes from turning, breathing, signal noise etc., and introducing a delay in monitoring algorithms can effectively reduce the number of alarms and mitigate alarm fatigue [3,20, 21]. Thus, the precise detection of short-duration ST change together with simple postprocessing steps, such as adding a delay, could provide great power in tackling the issue of excessive false positive alarms that plague the current ST monitoring software. On the other hand, the precise detection of short-duration ST change offers valuable information about temporal patterns of ST change that lead to downstream clinical endpoints, such as myocardial infarction. These temporal features could be further utilized by sequential models, e.g., recurrent neural network (RNN), to make prediction of the more clinically meaningful endpoints and to provide early warning.
When evaluating model performance at the individual level, we notice some testing recordings present considerably lower performance than most others. One plausible reason for this could be tied to one limitation of the LTST database that only single event time, instead of event duration, is provided for events of significant ST shift and noise. This greatly undermines the validity of true labels in the data especially for recordings with many episodes related to ST shift and ECG signal noise, such as those aforementioned ones. Future effort is needed to complete the annotation of these events in order to have them properly accounted for during model training and testing.
Another limitation of the present study is that the ST detection algorithm is built upon single-lead level, given that the database consists of ECG recordings with 2-or 3-lead configuration and annotation information is available at single-lead level. It has been found that true transient myocardial ischemia events typically have presence in contiguous leads (leads closed placed), and taking such information into account could improve detecting sensitivity [21]. Furthermore, some ischemic ST events are lead specific and can be only detectable through certain leads [22], so they might be missed by algorithms monitoring single or very few number of leads alone. Under our current framework using the image-based approach, one can easily add more information, such as ECG tracings from other leads, to fit into the image representation. Thus, one of our future directions is to establish an annotated ST database with in-hospital 12-lead ECG recordings, based on which a multi-lead prediction model can be built and evaluated under the same framework as proposed here. Lastly, one common hurdle of adopting deep learning in biomedical research is the lack of model transparency. Further investigation of model parameters to reveal underlying image features that contribute to the model decision is of great importance to the model understanding. Making the model findings transparent to clinicians may play an important role in clinical adoption and creation of clinical decision support tools.
Conclusions
The present study lays out a pipeline for using deep learning to improve the precision of ST- segment monitoring and to mitigate the issue of alarm fatigue. The combination of image-based approach and transfer-learning scheme adopted here provides efficiency and effectiveness in training CNN models for detection of ST changes, with both high sensitivity and specificity. Furthermore, robust performance has been demonstrated from models obtained with various number of recordings for training. The detection of ST changes at the short-duration level serves as a foundation for episode-level ST detection, and could also have great implication to the prediction of more clinical meaningful endpoints, such as MI, down the road.
Acknowledgments
Funding: This work in partially funded by R01NHLBI128679, R18H5022860, R01GM111378, UCSF Middle-Career scientist award, and UCSF Institute of Computational Health Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Sanchis-Gomar F, et al. , Epidemiology of coronary heart disease and acute coronary syndrome. Ann Transl Med, 2016. 4(13): p. 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amsterdam EA, et al. , 2014 AHA/ACC Guideline for the Management of Patients with Non-ST-Elevation Acute Coronary Syndromes: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol, 2014. 64(24): p. e139–e228. [DOI] [PubMed] [Google Scholar]
- 3.Drew BJ, et al. , Insights into the problem of alarm fatigue with physiologic monitor devices: a comprehensive observational study of consecutive intensive care unit patients. PLoS One, 2014. 9(10): p. e110274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ECRI, Top 10 health technology hazard for 2014. Health devices, 2013. 41: p. 1–13. [Google Scholar]
- 5.Sandau KE, et al. , Update to Practice Standards for Electrocardiographic Monitoring in Hospital Settings: A Scientific Statement From the American Heart Association. Circulation, 2017. 136(19): p. e273–e344. [DOI] [PubMed] [Google Scholar]
- 6.Rajpurkar P,HA, Haghpanahi M, Bourn C, Ng AY, Cardiologist-Level Arrhythmia Detection with Convolutional Neural Networks. arXiv:170701836, 2017. [Google Scholar]
- 7.Pourbabaee BRM, Khorasani K, Deep convolution neural networks and learning ECG features for screening paroxysmal atrial fibrillation patients. IEEE Transactions on Systems, Man, and Cybernetics: Systems, 2017. 99: p. 1–10. [Google Scholar]
- 8.Tan JH, et al. , Application of stacked convolutional and long short-term memory network for accurate identification of CAD ECG signals. Comput Biol Med, 2018. 94: p. 19–26. [DOI] [PubMed] [Google Scholar]
- 9.He KZX, Ren S, Sun J, Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. ICCV, 2015. [Google Scholar]
- 10.Jager F, et al. , Long-term ST database: a reference for the development and evaluation of automated ischaemia detectors and for the study of the dynamics of myocardial ischaemia. Med Biol Eng Comput, 2003. 41(2): p. 172–82. [DOI] [PubMed] [Google Scholar]
- 11.Goldberger AL, et al. , PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation, 2000. 101(23): p. E215–20. [DOI] [PubMed] [Google Scholar]
- 12.Shahriari Y, et al. , Electrocardiogram Signal Quality Assessment Based on Structural Image Similarity Metric. IEEE Transactions on Biomedical Engineering, 2018. 65(4): p. 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szegedy CVV; Loffe J; Shlens J; Wojna Z, Rethinking the Inception Architecture for Computer Vision. CVPR, 2016. [Google Scholar]
- 14.Russakovsky O, et al. , ImageNet Large Scale Visual Recognition Challenge. International Journal of Computer Vision (IJCV), 2015. 115(3): p. 211–252. [Google Scholar]
- 15.Pedregosa F, et al. , Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res, 2011. 12: p. 2825–2830. [Google Scholar]
- 16.Smrdel A and Jager F, Automated detection of transient ST-segment episodes in 24 h electrocardiograms. Med Biol Eng Comput, 2004. 42(3): p. 303–11. [DOI] [PubMed] [Google Scholar]
- 17.Minchole A, et al. Evaluation of a root mean squared based ischemia detector on the long-term ST database with body position change cancellation. in Computers in Cardiology, 2005. 2005. [Google Scholar]
- 18.Dranca L, Goni A, and Illarramendi A. Using DecisionTrees for Real-Time Ischemia Detection. in 19th IEEE Symposium on Computer-Based Medical Systems (CBMS’06) 2006. [Google Scholar]
- 19.Xiao R, et al. , A Deep Learning Approach to Examine Ischemic ST Changes in Ambulatory ECG Recordings. AMIA Informatics Summit, 2018. [PMC free article] [PubMed] [Google Scholar]
- 20.Tsimenidis C and Murray A, False alarms during patient monitoring in clinical intensive care units are highly related to poor quality of the monitored electrocardiogram signals. Physiol Meas, 2016. 37(8): p. 1383–91. [DOI] [PubMed] [Google Scholar]
- 21.Pelter MM, et al. , Evaluation of ECG algorithms designed to improve detect of transient myocardial ischemia to minimize false alarms in patients with suspected acute coronary syndrome. J Electrocardiol, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelter MM, Adams MG, and Drew BJ, Transient myocardial ischemia is an independent predictor of adverse in-hospital outcomes in patients with acute coronary syndromes treated in the telemetry unit. Heart & Lung: The Journal of Acute and Critical Care, 2003. 32(2): p. 71–78. [DOI] [PubMed] [Google Scholar]


