Abstract
The name “Actinomyces suis” was applied to each actinomycete isolate from swine actinomycosis by Grässer in 1962 and Franke in 1973. Nevertheless, this specific species was not included in the “Approved List of Bacterial Name” due to absence of the type cultures. Therefore, “Actinomyces suis” based on the description of Franke 1973 has been considered as “species incertae sedis”. We isolated a number of Actinomyces strains from swine. The representative strains of them was designated as Chiba 101 that was closely similar to the description in “Actinomyces suis” reported by Franke in 1973. Interestingly, it was found that the biological characteristics of these strains were also very similar to those of Actinomyces denticolens. Furthermore, the average nucleotide identity (ANI) value between strain Chiba 101 and the type-strain of Actinomyces denticolens (=DSM 20671T) was found to be 99.95%. Sequences of the housekeeping genes and 16S rRNA gene showed 100% homology. These results strongly suggested that “Actinomyces suis” Franke 1973 is the same species as Actinomyces denticolens. Since actinomycosis caused by Actinomyces denticolens have been demonstrated in horses recently, it is necessary to recognize that Actinomyces denticolens is the pathogenic actinomycetes in broader range of animals.
Keywords: Actinomyces denticolens, “Actinomyces suis” Franke 1973, actinomycosis, nomenclature
Although actinomycosis has been an almost forgotten classical disease, Actinomyces is one of the important pathogen that causes intrinsic infection in human and animals. In recent years, the cervicofacial abscesses caused by Actinomyces denticolens have been disclosed in horses [1, 5, 7, 8]. Actinomyces denticolens was first isolated as a new species from the dental plaque of cattle by Dent and Williams [6], which has been considered as non-pathogenic actinomycetes in general [25].
Apart from actinomycosis in horse, in the 1970’s, Azuma and Nakajima [3] had isolated eighteen strains of Actinomyces sp. that were assumed to be Actinomyces naeslundii and Actinomyces viscosus from lungs, submandibular lymph-nodes, tonsils, spleens and leg joints in fourteen pigs. There were no differences in biological properties except catalase production among those strains. Although four strains isolated from three tonsils and an arthritic leg joint in four pigs showed the same serotype, this serotype was clearly distinct from that of the other strains.
In the same period, eight Actinomyces strains were isolated from actinomycotic mammary glands of pigs by Franke [9]. Subsequently, Oomi et al. [23] isolated nine strains of Actinomyces sp. from several tissue samples including tonsils in pigs. The physiological, biochemical and serological characteristics of the isolates showed striking similarities to that of “Actinomyces suis” Franke 1973 as well as Actinomyces sp. Azuma and Nakajima 1973. Thus, strain Chiba 101 (=DSM 10177 =NBRC 111625) isolated from an arthritic leg joint by Azuma and Nakajima [3] was designated as the representative strain [23].
The name “Actinomyces suis” had already been reported by Grässer [10] but the descriptions were inadequate, and the cultures were not available. Since Franke [9] isolated Actinomyces strains similar to isolates of Grässer [10] from swine actinomycosis, he used the name “Actinomyces suis” for his isolates repeatedly, but the culture has not been available. Currently, “Actinomyces suis” is listed as “species incertae sedis” in Bergey’s Manual of Systematic Bacteriology 2nd edition [25]. On the other hand, Eubacterium suis [28] that was first isolated from sows with cystitis and pyelonephritis by Soltys and Spratling in 1957 was transferred to the genus Actinomyces as Actinomyces suis based on the phylogenetic evidence [15]. Therefore, Actinomyces suis (former Eubacterium suis) Ludwig et al. 1992 is approved by the International Code of Nomenclature of Bacteria (ICNB), but “Actinomyces suis” Franke 1973 is not. Currently, Actinomyces suis named by Ludwig et al. [15] has been reclassified into Actinobaculum suis [14]. This species causes urinary tract infection in sows without forming actinomycotic lesions in urinary tract [27].
Interestingly, we found that complete genome sequences of the strain Chiba 101 and the type-strain of Actinomyces denticolens (=DSM 20671T) had closely related each other [13]. We speculated that Actinomyces denticolens, strain Chiba 101 and “Actinomyces suis” Franke 1973 could possibly belong to the same species.
The purposes of this study were to clarify whether Actinomyces denticolens and strain Chiba 101 isolated by Azuma and Nakajima [3] are the same species, and to propose that Actinomyces denticolens is the same species as “Actinomyces suis” Franke 1973.
MATERIALS AND METHODS
Used strains
Four Actinomyces-like isolates of Higuchi 1197, Higuchi 1322, Higuchi 1328 and Higuchi 1354 isolated from swine tonsils (4 Higuchi strains), and 6 equine isolates of Otaki 887-1, Otaki 887-2, Otaki 888-1, Otaki 915 Otaki 946-3 and Otaki 946-6 identified as Actinomyces denticolens using 16S rRNA gene sequence (6 Otaki strains) [21] were used for comparison with Actinomyces denticolens DSM 20671T, strain Chiba 101 (=DSM 101773 =NBRC 111625) and the type-strain of Actinobaculum suis (=DSM 20639T) (formerly Actinomyces suis). Strain Chiba 101 was isolated using the gas jet method with the MVL medium [4] from an arthritic leg joint of a pig raised in Chiba Prefecture, Japan [3]. This isolation method was previously described [2]. The strain was cultured on 10% TMLV agar slants at 37°C with 5% CO2 and was stored in lyophilized ampules. The isolation methods of 4 Higuchi strains were described below.
Histopathology and immunohistochemistry
The tonsils of soft plate from eighteen fattening pigs were used. These tonsils were immediately stored at −80°C, and the frozen tonsils were cut into three parts from anterior to posterior. In order to isolate Actinomyces strains from the frozen tonsils, the symmetrical 1 mm thick sections were made from the frozen tonsils using a razor. After placing a piece of the tonsillar tissue on a glass slide, the cut sample was stored at −80°C. On the other hand, the remaining tonsils which mirrored the frozen tissues were cut into 2–3 mm thick, fixed with 20 percent formalin and methanol mixture (Sakura Finetek Japan, Tokyo, Japan), embedded in paraffin, and sliced at about 4 µm thick, followed by staining with hematoxylin and eosin (HE).
When actinomycotic abscesses were observed in the tonsillar crypts in HE specimens, tiny abscesses were further stained by Gram’s method and the clumps in the lesions were immunodetected with the method as described below. The immunohistochemistry was performed with a commercially available streptavidin-biotin-peroxidase conjugate (SAB) kit (Nichirei Biosciences Inc. Tokyo, Japan) in order to detect the positive-antigens in a serial section using rabbit hyperimmune serum against strain Chiba 101 [18] and rabbit hyperimmune serum against Actinomyces denticolens DSM 20671T [21] as primary antiserum.
In order to examine whether there was immunohistochemical cross-reactivities of the strain Chiba 101 antiserum between strain Chiba 101 and the bacterial antigens of Actinomyces species isolated from pigs, antigens were prepared from 7 Actinomyces species listed below: Actinomyces bovis ATCC 13683T, Actinomyces hyovaginalis BM 1192/5T, Actinomyces israelii ATCC 43320T, Actinomyces naeslundii ATCC 12104T, Actinomyces suimastitidis CCUG 39276T, Actinobacterium suis (former Actinomyces suis) DSM 20639T and Actinomyces viscosus ATCC 15987T. Furthermore, Actinomyces denticolens DSM 20671T was added. Strain Chiba 101 was used as the positive control. The Actinomyces denticolens DSM 20671T antiserum was used to verify whether there were immunohistochemical cross-reactivities between Actinomyces denticolens DSM 20671T and the 7 antigens of Actinomyces species as well as between Actinomyces denticolens DSM 20671T and strain Chiba 101. Actinomyces denticolens DSM 20671T was used as the positive control. The preparations of those tissue samples using these immunohistochemical examinations had already been described [17, 18, 21]. The immunohistochemical analysis was performed as described by Murakami et al. [21].
Isolation of Actinomyces from swine tonsils
When the actinomycotic abscesses were found in the HE specimens, the sulfa granule-like materials or the tissues were gouged out with a sterile needle from the same area of the frozen symmetrical tonsils. The immunoantigenic separation technique was performed to isolate Actinomyces from the homogenized samples [22]. The antiserum of strain Chiba 101 was used as the primary antibody for the isolation [18].
Physiological and biochemical analyses of swine and equine isolates
In this analysis, a commercially available ID-ASE color catalase kit (bioMérieux) for catalase activity, cytochrome oxidase test (bioMérieux) for oxidase activity and an API Rapid ID32A (bioMérieux) for diagnosis of biochemical properties were used to examine these strains. Thirteen strains used in this analysis were four Higuchi strains, six Otaki strains, Actinomyces denticolens DSM 20671T, strain Chiba 101 and the type-strain of Actinobaculum suis (=DSM 20639T) (formerly Actinomyces suis).
Analyses of average nucleotide identity (ANI) and house-keeping genes
Based on the homology of the data deciphered by whole-genome sequences by Kanesaki et al. [13], ANI between Actinomyces denticolens DSM 20671T (accession number BDIO01000001 to BDIO01000008) and the strain Chiba 101 (accession number AP017896) was calculated using the ANI Calculator (http://www.ezbiocloud.net/tools/ani). The selected house-keeping genes, atpA (ATP synthase F1, alpha subunit, ANA_0169), metG (methionyl-tRNA synthetase, ANA 1898), rpoB (DNA-directed RNA polymerase, beta subunit, ANA_1497), pgi (glucose-6-phosphate isomerase, ANA_0727), gltA (citrate synthase I, ANA_1674), gyrA (DNA gyrase, A subunit, ANA_2224), pheS (phenylalanyl-tRNA synthetase, alpha subunit, ANA_1034) and 2 genes for fimbrial proteins, Type-2 fimbriae fimA (ANA_0024 fimbrial structural subunit) and type-1 fimbriae fimP (ANA_2510 Type-1 fimbrial major subunit precursor) of strain Chiba 101 were compared to those of the Actinomyces denticolens DSM 20671T. The previously reported Actinomyces oris sequences [11] were used as the reference sequences [EU603149 (pgi), EU620779 (atpA), EU620895 (gltA), EU621011 (gyrA), EU621127 (metG) EU621243 (rpoB) and GQ354571 (pheS)].
Analysis of 16S rRNA gene sequences
DNA extraction of 4 Higuchi strains, Actinomyces denticolens DSM 20671T and strain Chiba 101 were carried out using GenTEL with high recovery (TaKaRa Bio, Kusatsu, Japan) according to the manufacturer’s protocol. The DNA pellet was washed in 70% ethanol and resuspended in 20 µl of TE buffer; the DNA extracts were stored at −20°C. For 16S rRNA sequence analysis, a partial 16S rRNA gene region (1,532 bp) was amplified with universal primers (Fwd primer: 5′-GAGTTTGATCCTGGCTCAG-3′ and Rev primer: 5′-AAGGAGGTGATCCAGCC-3′) using KAPATaq Extra DNA polymerase (Kapa Biosystems, Wilmington, NC, U.S.A.). The obtained PCR products were purified using a QIAquick PCR Purification Kit (Qiagen) with DNA cleanup protocol. The nucleotide sequences were determined by a DNA sequencing service (Fasmac, Atsugi, Japan), and the data were analyzed with Sequencher v5.1 software (Gene Codes Corp., Ann Arbor, MI, U.S.A.). The 16S rRNA gene sequences of Actinomyces denticolens DSM 20671T, strain Chiba 101 and 4 Higuchi strains were aligned by ClustalW program (MEGA 5) with those of other members of the family Actinomycetaceae. 16S rRNA gene sequence homologies were calculated manually. Phylogenetic trees based on 16S rRNA gene sequences were reconstructed by MEGA5 [26] using the maximum-likelihood method.
RESULTS
Histopathological and immunohistochemical findings
Seven swine tonsils out of eighteen pigs examined revealed a typical actinomycotic abscess in the tonsillar crypts. Those actinomycotic clumps accompanied by “club-shaped” structures were surrounded by a number of infiltrated neutrophils and macrophages in the tonsillar crypts (Fig. 1A). Two distinct types of microorganisms were observed in these foci by Gram staining. The clumps of six foci were consisted of slender and short-branching filaments (Fig. 2), whereas the remaining one was consisted of thick and rugged filaments. These lesions were similar to the morphology described in previous report [19].
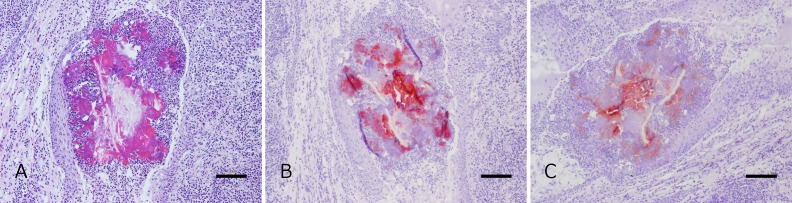
Fig. 1.
A: Actinomycotic abscess in a tonsillar crypt of a pig (No. 1197), from which the strain Higuchi 1197 was isolated. HE. Bar=100 µm. B: Antigens immunolabeled with strain Chiba 101 antiserum in a serial section of A. Bar=100 µm. SAB method. C: Antigens immunolabeled with Actinomyces denticolens DSM 20671T antiserum in a serial section of A. Bar=100 µm. SAB method.
Fig. 2.
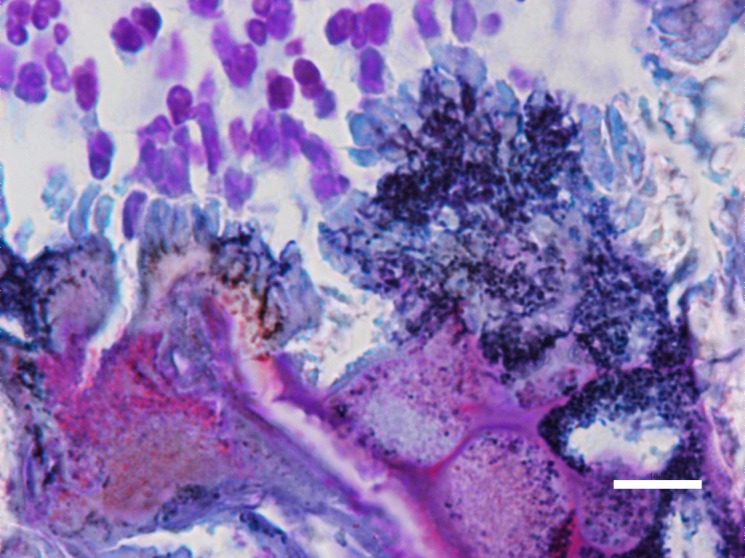
Slender and short-branching filaments in a serial section of A in Fig. 1. Bar=10 µm. Gram’s method.
Among them, the positive-antigens were clearly immunolabeled with 1:128,000 diluted both antisera on the clumps in the 6 foci (Fig. 1B and 1C) of 6 tonsils (one from each). The thick and rugged filaments of remaining one was not immunostained even with a 1:8,000 diluted both antisera.
Although the bacterial antigens of Actinomyces denticolens DSM 20671T and strain Chiba 101 were clearly immunolabeled with 1:128,000 diluted Actinomyces denticolens DSM 20671T antiserum and with 1:128,000 diluted strain Chiba 101 antiserum, those bacterial antigens of the other Actinomyces species were not immunostained even with a dilution of 1:64,000 in both antisera.
Bacteriological findings and biochemical traits of swine and equine isolates
Four Actinomyces-like strains (4 Higuchi strains) were isolated from each 4 of 6 foci immunodetected with both antisera. The isolation from the remaining one was not performed, because the characteristic microbial elements of Tonsilliphilus suis (former “Tonsillophilus suis”) such as flourishing stick like thalli were confirmed by Gram staining [20].
The Higuchi strains showed Gram-stain positive and diphtheroid rods. Except for α-galactosidase and raffinose acidification, biochemical traits of Actinomyces denticolens DSM 20671T were extremely similar to those of strain Chiba 101, four Higuchi strains and six Otaki strains, whereas biochemical properties of Actinobaculum suis DSM 20639T differed greatly from those of the other strains (Table 1).
Table 1. Biochemical properties of strain Chiba 101, Actinomyces denticolens DSM 20671T, ten isolates obtained from swine and equine tonsils and Actinobaculum suis (former Actinomyces suis) DSM 20639T.
| Property | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Catalase | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Oxidase | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Urease | - | - | - | - | - | - | - | - | - | - | - | - | + |
| arginine dihydrolase | - | - | - | - | - | - | - | - | - | - | - | - | - |
| α-galactosidase | + | - | - | - | - | - | - | - | - | - | - | - | - |
| β-galactosidase | + | + | + | + | + | + | + | + | + | - | + | w | - |
| β-galactosidase-6-phoshate | - | - | - | - | - | - | - | - | - | - | - | - | - |
| α-glucosidase | + | + | + | + | + | + | + | + | + | + | + | + | + |
| β-glucosidase | + | + | + | + | + | + | + | + | - | + | + | + | - |
| α-arabinosidase | - | - | - | - | - | - | - | - | - | - | - | - | - |
| β-glucuronidase | - | - | - | - | - | - | - | - | - | - | - | - | + |
| β-N-acetyl-glucosaminidase | - | - | - | - | - | - | - | - | - | - | - | - | - |
| mannose acidification | + | + | + | + | + | + | + | + | + | + | + | - | + |
| raffinose acidification | - | + | + | + | + | + | + | + | + | + | + | + | - |
| glutamic acid decarboxylase | - | - | - | - | - | - | - | - | - | - | - | - | - |
| α-fucosidase | - | - | - | - | - | - | - | - | - | - | - | - | - |
| nitrate reduction | + | + | + | + | + | + | + | + | + | + | + | + | - |
| indole production | - | - | w | - | - | - | - | w | - | - | - | - | - |
| alkaline phosphatase | - | - | - | - | - | - | - | - | - | - | - | - | + |
| arginine arylamidase | - | - | - | - | - | - | - | - | - | - | w | - | + |
| proline arylamidase | + | + | + | + | + | + | + | + | + | + | + | + | + |
| leucyl-glycine arylamidase | - | - | - | - | - | - | - | - | - | - | - | - | + |
| phenylalanine arylmidase | + | + | + | + | + | + | + | + | + | + | + | + | + |
| leucine arylamidase | + | + | + | + | + | + | + | + | + | + | + | + | + |
| pyroglutamic acid arylamidase | - | - | - | - | - | - | - | - | - | - | - | - | + |
| tyrosine arylamidase | + | + | + | + | + | + | + | + | + | + | + | + | + |
| alanine arylamidase | w | - | w | - | - | - | - | - | - | - | w | w | + |
| glycine arylamidase | - | - | - | - | - | - | - | - | - | - | - | - | + |
| histidine arylamidase | - | - | - | - | - | - | - | - | - | - | - | - | + |
| glutamyl glutamic acid arylamidase | - | - | - | - | - | - | - | - | - | - | - | - | - |
| serine arylamidase | - | - | - | - | - | - | - | - | - | - | - | - | + |
+: positive, -: negative, w: weakly positive. 1: Actinomyces denticolens DSM 20671T, 2: Strain Chiba 101, 3: Higuchi 1197, 4: Higuchi 1322, 5: Higuchi 1328, 6: Higuchi 1354, 7: Otaki 887-1, 8: Otaki 887-2, 9: Otaki 888-1, 10: Otaki 915, 11: Otaki 946-3, 12: Otaki 946-6, 13: Actinobaculum suis (former Actinomyces suis) DSM 20639 T.
Analyses of ANI, house-keeping genes and 16S rRNA gene sequences
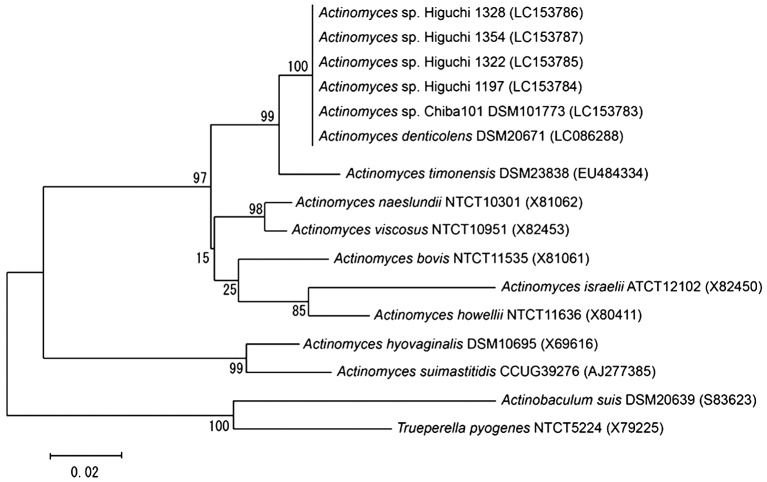
The ANI value found between Actinomyces denticolens DSM 20671T and strain Chiba 101 was 99.95%. This value was well above the recommended identification threshold for species delineation (96% ANI: [24]). In addition, the seven housekeeping genes and the 16S rRNA gene sequences were 100% identical between the two strains. Since both analyses of the ANI and the housekeeping gene sequences on strain Chiba 101 and Actinomyces denticolens DSM 20761T showed extremely high homologous, strain Chiba 101 should be identified as Actinomyces denticolens. In addition, 16S rRNA gene sequences of 4 Higuchi strains were 99.9% identical to that of Actinomyces denticolens DSM 20671T. The result of the phylogenetic analysis suggested that the 4 strains should also be identified as Actinomyces denticolens (Fig. 3).
Fig. 3.
Phylogenetic tree derived from 16S rRNA gene sequences, reconstructed using the maximum-likelihood method. The sequence of Trueperella pyogenes was used as an outgroup. Numbers at the branch points are percentages of 1,000 bootstrap replicates. Bar=0.02 substitutions per site.
DISCUSSION
As stated in the introduction, Actinobaculum suis was reclassified from Actinomyces suis, and Actinomyces suis had been transferred from Eubacterium suis before that. Wegienek and Reddy [28] reported the physiological and biochemical properties of Eubacterium suis (type strain Soltys 50052) and showed that it does not belong to the genus Actinomyces, whereas both physiological and biochemical properties of “Actinomyces suis” Franke 1973 and Actinomyces sp. Azuma and Nakajima 1973 showed striking similarities. The characteristics of Eubacterium suis were obviously different from that of both species (Table 2).
Table 2. Physiological and biochemical characteristics of “Actinomyces suis” Franke 1973, Actinomyces sp. Azuma and Nakajima 1973, Actinmyces denticolens Dent and Willians 1984 and Eubacterium suis Weglenek and Reddy 1982.
| Property | 1a) | 2b) | 3c) | 4d) | |
|---|---|---|---|---|---|
| Catalase production | 0/8* | 0/4 | 0/6 | 0/1 | |
| Nitrate reduction | 8/8 | 4/4 | 6/6 | 0/1 | |
| Hydrolysis of gelatin | 0/8 | 0/4 | nd | 0/1 | |
| Acid production | |||||
| Arabinose | 0/8 | 0/4 | 0/6 | 0/1 | |
| Dulcitol | 0/8 | 0/4 | nd | nd | |
| Fructose | 8/8 | 4/4 | nd | 0/1 | |
| Galactose | 8/8 | 4/4 | nd | nd | |
| Glucose | 8/8 | 4/4 | 6/6 | 0/1 | |
| Inulin | 8/8 | 4/4 | 6/6 | nd | |
| Lactose | 8/8 | 4/4 | 6/6 | 0/1 | |
| Maltose | 8/8 | 4/4 | 6/6 | 1/1 | |
| Mannitol | d | 0/4 | 3/6 | 0/1 | |
| Mannose | 8/8 | 4/4 | 5/6 | 0/1 | |
| Raffinose | 8/8 | 4/4 | 6/6 | 0/1 | |
| Salicin | 8/8 | 4/4 | 6/6 | 0/1 | |
| Sorbitol | 0/8 | 0/4 | 0/6 | 0/1 | |
| Xylose | 1/8 | 0/4 | 0/6 | 0/1 | |
*number of positive strains/number of strains examined, d: positive or negative, nd: no deta available. a) Data of eight Actinomyces strains compiled from actinomycotic mammary glands of pigs by Franke [9]. b) Data of four Actinomyces strains including strain Chiba 101 compiled from three tonsils and an arthritic leg joint of pigs by Azuma and Nakajima [3]. c) Data of six Actinomyces strains compiled from the dental pluqes of cattle by Dent and williams [6]. d) Data of strain Soltys 50052 compiled from cystitis or pyelonephritis of a pig by Weglenek and Reddy [28].
To ascertain the presence of the Actinomyces sp. isolated from pigs by Azuma and Nakajima [3], Oomi et al. [23] chemotaxonomically and serologically analyzed nine Actinomyces strains isolated from six tonsils, a dental plaque, a submandibular lymph-node and a mammary gland in pigs. As a result, catalase production and nitrate reduction of those strains were negative and positive, respectively. In addition, acid production traits of those strains were very similar to that of Actinomyces sp. Azuma and Nakajima 1973 and “Actinomyces suis” Franke 1973. In gel diffusion test using strain Chiba 101 antiserum absorbed with Actinomyces naeslundii ATCC 12104T and Actinomyces viscosus ATCC 15987T, although only one strain (the origin was not described) was not performed, eight strains of the nine strains mentioned above showed a fused precipitation line. Thus, it had been suggested that these strains were the same as Actinomyces sp. Azuma and Nakajima 1973 and “Actinomyces suis” Franke 1973. In addition, immunohistochemical examination revealed that strain Chiba 101 differed from other actinomycetes found in swine, and the antigens were immunodetected for the first time in the tonsillar crypt abscesses in fattening pigs and the actinomycotic mastitis in a sow [18].
In the past, the typical actinomycotic lesions had been observed in swine udders [12, 16], and subsequently, Grässer [10] and Franke [9] isolated so-called “Actinomyces suis” from the actinomycotic mastitis. However, it is extremely rare to find swine actinomycosis today. Nevertheless, typical actinomycotic lesions have been found commonly in swine tonsils [12, 18, 19]. Since strains of Actinomyces sp. related to strain Chiba 101 have been found to be naturally present in tonsils in pigs, it is reasonable to consider that the Actinomyces species are commonly colonized in porcine tonsillar crypts as part of the pig’s oral microbiota.
It had been speculated that the cause of the actinomycotic mastitis in sows was due to bites by suckling piglets [23, 29]. In order to confirm the pathogenicity of strain Chiba 101 and a tonsillar isolate obtained by Oomi et al. [23], Murakami et al. [20] carried out an experimental inoculation on mice, guinea-pigs and the mammary gland of a sow. As a result, the typical actinomycotic lesions were observed in these tissues of the mice and the sow. The Actinomyces sp. (the representative strain Chiba 101) is possibly a causative agent of actinomycosis in swine according to those findings.
On the other hand, the physiological and biochemical properties of six strains of Actinomyces denticolens isolated by Dent and Williams [6] were very similar to those of the strains of Azuma and Nakajima [3] as well as “Actinomyces suis” Franke 1973 (Table 2). In this study, strain Chiba 101 and Actinomyces denticolens DSM 20761T were immunohistochemically, genetically and phenotypically extremely similar to each other. Furthermore, the 16S rRNA gene sequences of four Higuchi strains isolated from the porcine tonsillar crypt abscesses showed very high homology to that of Actinomyces denticolens DSM 20761T. These results demonstrated that Actinomyces denticolens and strain Chiba 101 belong to the same species. Thus, it could be considered that both strains of Azuma and Nakajima [3] and Oomi et al. [23] are also the same species as Actinomyces denticolens. Based on these findings, we propose that Actinomyces denticolens is considered as a synonym of “Actinomyces suis” Franke 1973 in spite of absence of the type cultures.
In horses, actinomycosis has been thought as an extremely rare disease. However, in very recent years, the cervicofacial abscesses caused by Actinomyces denticolens have been found in horses repeatedly [1, 5, 7, 8]. Since the colonization of Actinomyces denticolens was found in equine tonsillar crypts, the tonsils might provide an intrinsic infection site for Actinomyces denticolens [21]. So far, Actinomyces denticolens has been thought to be a non-pathogenic Actinomyces species in general [25]. However, since Actinomyces denticolens develops visible clinical symptoms associated with actinomycosis in swine and horses, it is necessary to recognize Actinomyces denticolens as a causative agent of actinomycosis in animals.
Acknowledgments
The deceased Dr. Azuma had believed the existence of “Actinomyces suis” in swine and burned passion for its elucidation. We deeply appreciate Dr. Azuma and to achieve its elucidation. Additionally, we would like to thank for Miss K. Higuchi, M. Watanabe and T. Yokota, for their technical supports.
REFERENCES
- 1.Albini S., Korczak B. M., Abril C., Hüssy D., Limat S., Gerber V., Hermann M., Howald B., Miserez R.2008. Mandibular lymphadenopathy caused by Actinomyces denticolens mimicking strangles in three horses. Vet. Rec. 162: 158–159. doi: 10.1136/vr.162.5.158 [DOI] [PubMed] [Google Scholar]
- 2.Azuma R., Murakami S., Ogawa A., Okada Y., Miyazaki S., Makino T.2009. Arcanobacterium abortisuis sp. nov., isolated from a placenta of a sow following an abortion. Int. J. Syst. Evol. Microbiol. 59: 1469–1473. doi: 10.1099/ijs.0.004465-0 [DOI] [PubMed] [Google Scholar]
- 3.Azuma R., Nakajima Y.1973. Biological and serological properties of Actinomyces naeslundii and A. viscosus of animal origin. pp. 23–30. In: Third Symposium of Anaerobic Bacteria and Their Infectious Diseases, (Ishiyama, S. ed) Eisai Tokyo (in Japanese).
- 4.Azuma R., Suto T.1970. Validity of transfer of the taxonomical position of Corynebacterium pseudopyogenes from genus Corynebacterium to genus Actinomyces. pp. 493–505. In: Culture Collection of Microorganism. Proceedings of the 1st International Conference on Culture Collection. (Iizuka, H. and Hasegawa, T. eds.), University of Tokyo Press, Tokyo.
- 5.Beck A., Baird J. D., Slavić D.2011. Submandibular lymph node abscess caused by Actinomyces denticolens in a horse in Ontario. Can. Vet. J. 52: 513–514. [PMC free article] [PubMed] [Google Scholar]
- 6.Dent V. E., Williams R. A.1984. Actinomyces denticolens Dent & Williams sp. nov: a new species from the dental plaque of cattle. J. Appl. Bacteriol. 56: 183–192. doi: 10.1111/j.1365-2672.1984.tb01338.x [DOI] [PubMed] [Google Scholar]
- 7.Feary D. J., Abraham S., Woolford L., Trott D. J.2013. Identification of Actinomyces denticolens as a cause of a soft tissue abscess in a horse. Aust. Vet. J. 91: 416–417. doi: 10.1111/avj.12102 [DOI] [PubMed] [Google Scholar]
- 8.Fielding C. L., Magdesian K. G., Morgan R. A., Ruby R. E., Sprayberry K. A.2008. Actinomyces species as a cause of abscesses in nine horses. Vet. Rec. 162: 18–20. doi: 10.1136/vr.162.1.18 [DOI] [PubMed] [Google Scholar]
- 9.Franke F.1973. Untersuchungen zur atiologie der gesaugeaktiomykose des schweines. Zentrabl. Bakteriol. Prasitenkd. Infektionskr. Hyg I Abt Orig A. 223: 111–124. [PubMed] [Google Scholar]
- 10.Grässer R.1962. Mikroaerophile actinomyceten aus gesäugeaktinomykosen des schweines. Zentralbl Bakteriol Prasitenkd Infektionskr. Hyg. Abt. Orig. Bd 184: 478–492. [Google Scholar]
- 11.Henssge U., Do T., Gilbert S. C., Cox S., Clark D., Wickström C., Ligtenberg A. J., Radford D. R., Beighton D.2011. Application of MLST and pilus gene sequence comparisons to investigate the population structures of Actinomyces naeslundii and Actinomyces oris. PLoS One 6: e21430. doi: 10.1371/journal.pone.0021430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johne F.1882. Die Actinomykose oder Strahlenpilzerkrankung, eine neue Infectionskrankheit. Deutsche Zeitschrift F Thiermed U vergl Pathologie VII Bd. 141–192. [Google Scholar]
- 13.Kanesaki Y., Ishige T., Sekigawa Y., Kobayashi T., Torii Y., Yokoyama E., Ishiwata H., Hamada M., Tamura T., Azuma R., Murakami S.2017. Whole-genome sequences of two closely related bacteria, Actinomyces sp. strain Chiba 101 and Actinomyces denticolens DSM 20671T. Genome Announc. 5: e00126-17. doi: 10.1128/genomeA.00126-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawson P. A., Falsen E., Åkervall E., Vandamme P., Collins M. D.1997. Characterization of some Actinomyces-like isolates from human clinical specimens: reclassification of Actinomyces suis (Soltys and Spratling) as Actinobaculum suis comb. nov. and description of Actinobaculum schaalii sp. nov. Int. J. Syst. Bacteriol. 47: 899–903. doi: 10.1099/00207713-47-3-899 [DOI] [PubMed] [Google Scholar]
- 15.Ludwig W., Kirchhof G., Weizenegger M., Weiss N.1992. Phylogenetic evidence for the transfer of Eubacterium suis to the genus Actinomyces as Actinomyces suis comb. nov. Int. J. Syst. Bacteriol. 42: 161–165. doi: 10.1099/00207713-42-1-161 [DOI] [PubMed] [Google Scholar]
- 16.Magnusson H.1928. The commonest forms of actinomycosis in domestic animals and their etiology. Acta Pathol. Microbiol. Scand. 5: 170–245. doi: 10.1111/j.1600-0463.1928.tb05317.x [DOI] [Google Scholar]
- 17.Murakami S., Ogawa A., Azuma R., Ohba T., Murata R.2011. Aborted lesions of a pig associated with Arcanobacterium abortisuis and the immunohistochemical features. J. Vet. Med. Sci. 73: 797–799. doi: 10.1292/jvms.10-0469 [DOI] [PubMed] [Google Scholar]
- 18.Murakami S., Azuma R., Koeda T., Oomi H., Watanabe T., Fujiwara H.1998. Immunohistochemical detection for Actinomyces sp. in swine tonsillar abscess and granulomatous mastitis. Mycopathologia 141: 15–19. doi: 10.1023/A:1006820611103 [DOI] [PubMed] [Google Scholar]
- 19.Murakami S., Azuma R., Oomi H., Koeda T., Fujiwara H.1997. Incidence of tonsillar lesions caused by Tonsillophilus suis and Actinomyces sp infection in swine. Zentralbl. Veterinarmed. A 44: 611–618. doi: 10.1111/j.1439-0442.1997.tb01147.x [DOI] [PubMed] [Google Scholar]
- 20.Murakami S., Azuma R., Oomi H., Watanabe T., Suzuki S., Koeda T., Fujiwara H.1999. Experimental actinomycosis caused by Actinomyces-like bacteria in mice and a sow. Zentralbl. Veterinarmed. A 46: 533–543. doi: 10.1046/j.1439-0442.1999.00242.x [DOI] [PubMed] [Google Scholar]
- 21.Murakami S., Otaki M., Hayashi Y., Higuchi K., Kobayashi T., Torii Y., Yokoyama E., Azuma R.2016. Actinomyces denticolens colonisation identified in equine tonsillar crypts. Vet. Rec. Open 3: e000161. doi: 10.1136/vetreco-2015-000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsvik O., Popovic T., Skjerve E., Cudjoe K. S., Hornes E., Ugelstad J., Uhlén M.1994. Magnetic separation techniques in diagnostic microbiology. Clin. Microbiol. Rev. 7: 43–54. doi: 10.1128/CMR.7.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oomi H., Azuma Y., Ishiwata H., Asahara T., Watanabe T., Murakami S.1994. Bacteriological study on another pathogenic agent “Actinomyces suis” Grässer 1957 in swine tonsils. p. 233. Proc. Int. Pig Vet. Sci. 13th Congress, Bangkok. [Google Scholar]
- 24.Richter M., Rosselló-Móra R.2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106: 19126–19131. doi: 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaal K. P., Yassin A. F.2012. Family I Actinomycetaceae pp. 36–126. In: Bergey’s Manual of Systemic Bacteriology, Vol. 5, 2nd ed. (Goodfelow, M., Kampfer, P., Busse, H.-J., Trujillo, M. E., Suzuki, K., Ludwing, W. and Whitmon, W. B. eds.), Springer, New York. [Google Scholar]
- 26.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor D. J.2012. Actinobaculum (Eubacterium) suis pp. 866–867. In: Diseases of Swine, 10th ed. (Zimmerman, J. J., Karriker, L. A., Ramirez, A., Schwartz, K. J. and Stevenson, G. eds.), Blakwell Publishing, Ames. [Google Scholar]
- 28.Wegienek J., Reddy C. A.1982. Nutritional and metabolic features of Eubacterium suis. J. Clin. Microbiol. 15: 895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Von Englett H. K.1961. Gesäugeaktinomykose beim schwein. Dtsch. Tierarztl. Wochenschr. 68: 110–113. [Google Scholar]