Abstract
Innate immune cells are key player in immune response to influenza virus infection. Influenza infected monocytes exacerbate the disease pathology. However, monocytes differ in susceptibilities to influenza virus infection. Herein, susceptibilities of U937 and THP-1 monocytic cells to PR8 virus infection, the associated cellular factor- sialic acid (SA) receptor distribution and viral factor were determined. Moreover, amino acid sequences in hemagglutinin (HA) receptor binding domain (RBD) of PR8 virus that determine SA preferences were analysed. PR8 infected U937 cells express significantly higher numbers of nucleoprotein positive cells suggesting U937 cells being more susceptible to influenza virus than THP-1 cells. Lectin staining suggested similar pattern of SA receptor distribution in both cells. Interestingly, sequence analysis of RBD suggested their preferences for alpha 2,3 SA receptors suggesting RBD sequences are not always determining for SA preferences. Furthermore, the resistance barrier on THP-1 cells was overcome by H5N1 NS gene. In conclusion, the study demonstrated that decreased susceptibility of THP-1 cells to PR8 virus could not be related to the SA receptor distribution, and H5N1 NS gene was sufficient to determine tropism for THP-1 cells. Hence, mechanistic basis of NS gene on cell tropism and contribution of other internal genes remained to be determined.
Keywords: THP-1 and U937 monocytic cells, Infectivity, Sialic acid receptors, Influenza A/Puerto Rico/8/34 H1N1 virus, H5N1 virus, Nonstructural (NS) gene
Innate immune cells like macrophages, monocytes, dendritic cells, NK cells and neutrophils play a critical role in immunopathogenesis of influenza virus infection [1]. As such, infiltration of mononuclear cells like monocytes, and macrophages in the lungs during influenza virus infection was correlated with disease severity [1–3]. These mononuclear cells produce inflammatory cytokines; TNF-α, IFNs, IL-6, IP-10 and RANTES [2, 3]. In severe influenza, enhanced amount of inflammatory cytokine produced is responsible for tissue damage [1]. Also, influenza infected individuals demonstrated a dysregulated monocyte function, in particular decreased IFN signal, a major effector arm against influenza infection [3]. Moreover, it could not be denied that the infiltrating monocytes get infected and further contribute to the production of soluble factors and cytokines that exacerbate the disease pathology. Also, these wandering monocytes might act as a vehicle to transport influenza virus which might contribute for extrapulmonary manifestations as observed with H5N1 virus infection.
For efficient influenza virus infection, a number of barriers must be overcome; an important being recognition of the sialic acid (SA) receptors by influenza virus hemagglutinin (HA) protein. Human influenza A virus preferentially binds with α-2,6 linked SAs while avian influenza A viruses preferentially binds with α-2,3 linked SAs [4, 5]. Moreover, key amino acids in the receptor binding domain (RBD) of influenza virus HA protein were shown to determine receptor preferences [6, 7].
Previously, it has been demonstrated that monocytes could be infected with influenza virus [8, 9]. Moreover, among the two monocytic cell lines, U937 cells were more susceptible to PR8 influenza A virus infection compared to THP-1 cells [8]. In addition, THP-1 cells were shown to be unsusceptible to PR8 virus infection [9]. It still remained elusive whether the difference in the susceptibility is due to difference in sialic acid (SA) receptor distribution in those cells.
Role of influenza virus NS gene on cell tropism is limited; however most of these studies employed susceptible cells and demonstrated an increase in infectivity [10–12]. To better understand the role of influenza NS gene on tropism, cells relatively resistant to influenza virus infection like macrophages [3] and monocytes [8, 9] needed to be employed. Previously, it has been shown that U937 monocytes, but not THP-1 monocytes, were susceptible to PR8 virus infection [8]. Nevertheless, it remained to be determined whether this difference is related to the distribution of SA receptors on cells, or the amino acid sequences on RBD of PR8 HA protein. Also, the possibility of H5N1 virus NS gene to restore infectivity of PR8 virus for THP-1 cells needed to be explored.
This study constructed recombinant PR8 virus using reverse genetics technique as described previously [8]. Briefly, eight recombinant pHW2000 plasmids carrying each genomic segment of PR8 virus (1 μg each) in 100 μl of 1X Opti-MEM (Gibco, Life Technologies, NY) is mixed with 18 μl of TransIT®-LTI reagent (MirusBio, Madison, WI) in 100 μl of 1X Opti-MEM. The reaction mixture was used to transfect human embryonic kidney (HEK)-293T cells co-cultured with Madin-Darby canine kidney (MDCK) cells in the ratio of 1:1. The inoculated culture was incubated at 37 °C in a CO2 incubator. The transfection mixture was replaced with 1X Opti-MEM at 7 h and subsequently with 1X Opti-MEM containing 2 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) treated trypsin (TPCK-trypsin) at 24 h. The infected culture was daily observed for virus induced cytopathic effects. The recombinant PR8 virus obtained was propagated and titrated in MDCK cells by a plaque assay as described previously [8].
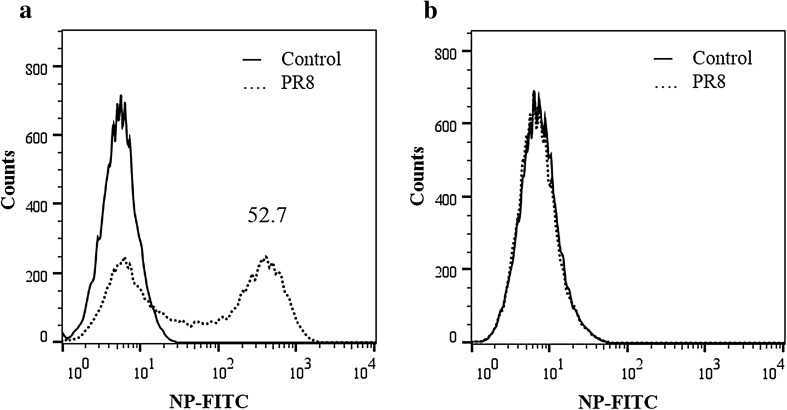
Herein, THP-1 and U937 human monocytic cells were infected with the PR8 virus at the m.o.i. of 3. The infected cell culture was incubated in 1X RPMI containing 2% Fetal Bovine Serum and 1 μg/ml TPCK-trypsin for 24 h at 37 °C. At 24 h post-infection, the cells were assessed for the expression of influenza nucleoprotein (NP) by flow cytometry as described previously [8]. It was found that U937 cells were significantly more susceptible to PR8 virus infection when compared with THP-1 cells (P = 0.001; two-tailed t test). At m.o.i. of 3, 52% cells were positive for influenza nucleoprotein (NP) antigen as determined by flow cytometry (Fig. 1). This is in agreement with the previous studies [8, 9] wherein THP-1 cells were shown to be resistant to PR8 virus infection. Previously, the difference in susceptibility has been inferred to the maturation stage of the cells (U937 cells more mature than THP-1 cells) and constitutive expression of virus restriction factor i.e. interferon inducible transmembrane protein 3 in THP-1 cells [8]. However, other cellular factors like influenza virus SA receptor distribution in those cells; and viral factors like SA binding preferences of the HA protein and contribution of NS gene was not explored.
Fig. 1.
Flow cytometry for determining susceptibility of monocytic cells to PR8 virus infection. U937 or THP-1 cells infected with PR8 virus at m.o.i. of 3. At 24 h post infection, cells were analysed for influenza NP by flow cytometry. Histogram shows U937 cells (a) or THP-1 cells (b) that are NP + . Compared to THP-1 cells, U937 cells are significantly more susceptible to PR8 virus infection (two-tailed t test: P = 0.001)
A key step in influenza virus infection is their ability to bind with SA receptors on cell surface. This study determined the distribution of SA receptors on the surface of U937 and THP-1 cells by flow cytometry. Briefly, 106 monocytic cells washed with 1 ml of phosphate buffer saline (PBS). The cell surface was blocked with 1% bovine serum albumin for 30 min on ice followed by a wash step. Fluorescence isothiocyanate (FITC) labelled Maackia amurensis lectin 1 (MAA1-FITC) (Vector Laboratories) or FITC labelled Sambucus nigra lectin (SNA-FITC) (Vector Laboratories) that recognises α-2,3 SA or α-2,6 SA, respectively were diluted 1:100 in PBS. The cell pellets were stained with either MAA1-FITC or SNA-FITC. The reaction tubes were kept on ice for 30 min in the dark. The stained cells were washed twice, resuspended in 1% paraformaldehyde and kept in the dark at 4 °C until analysed using a BD FACSCalibur cytometer (BD Biosciences, San Jose, CA). Trypsinized MDCK cells were similarly stained and used as the positive control for SA receptors (Fig. 2). The result showed that both U937 and THP-1 monocytic cells exhibited similar staining pattern with MAA1 and SNA (Fig. 2a, b and d). Similar results with MDCK cells (Fig. 2c), that were known to harbor both α-2,3 SA or α-2,6 SA receptors, confirmed specificity of the reaction. This suggest that these monocytic cells harbors similar no of α-2,3 SA or α-2,6 SA receptors on the cell surface.
Fig. 2.
Cellular and viral factors determining the influenza virus sialic acid receptor preferences. Flow cytometry for determining cell surface sialic acid receptors distribution on monocytic or MDCK cells (a–d). Monocytic cells and MDCK cells are stained for α-2,3 sialic acid using MAA1-FITC or α-2,6 sialic acid using SNA-FITC and analysed by flow cytometry. Histogram shows U937 (a), THP-1 (b) or MDCK (c) cells that are positive for MAA1 or SNA. Bar diagram (d) shows average percentage (Mean ± SD) of monocytic cells positive for α-2,3 sialic acid or α-2,6 sialic acid from three independent experiments. Number of U937 and THP-1 cells positive for α-2,3 sialic acid or α-2,6 sialic acid receptors are comparable (ANOVA: ns, P > 0.050). Sequence alignment of hemagglutinin (HA) protein of H3N2 and PR8 (H1N1) virus (e). Only amino acids in the receptor binding domain of influenza HA protein are shown
Key amino acids in the RBD of the influenza virus HA protein was shown to determine the α-2,6 SA or α-2,3 SA receptor preferences [6, 7]. Previously, influenza viruses that have lysine (L) and serine (S) at position 226 and 228, respectively binds with α-2,6 SA receptors. On the other hand, the viruses that recognises α-2,3 SA receptors have glutamine (Q) and glycine (G) at position 226 and 228, respectively [6, 7]. Moreover, glutamic acid (E) at position 190 increases HA affinity for α-2,3 SA receptors while aspartic acid (D) at 190 confer binding to α-2,6 SA receptors [6]. The present study aligned HA protein sequence of influenza A/Pureto Rico/8/1937 (H1N1) and A/Aichi/2/1968 (H3N2) viruses using BioEdit Sequence Alignment Editor 7.2.6. Key mutations in the RBD of the HA protein specific to H3N2 virus was shown (Fig. 2e). It was found that PR8 virus have 226Q and 228G suggesting their preferences for α-2,3 SA receptors. However, the staining pattern contradicted the sequence based SA preferences suggesting RBD sequences might not always determine for the receptor preferences.
Previously, NS gene of influenza viruses has been shown to determine host range restriction [10–12]. Also, reassortant viruses harbouring H5N1 NS gene showed higher infectivity than those from other subtypes [13]. Cell types that are abortive for influenza virus infection are the best model to demonstrate cell tropism. Herein, THP-1 cells were infected with reassortant PR8 virus harbouring H5N1 NS gene (rgH5N1-NS virus). Construction of rgH5N1-NS virus was done as described for reassortant PR8 virus, except the NS gene of PR8 virus was replaced with that from influenza A/Thailand/1 (KAN-1A)/2004 (H5N1) virus. The infected cells were similarly processed and measured for infectivity by flow cytometry. As shown in Fig. 3, rgH5N1-NS virus significantly enhanced infectivity in THP-1 cells when compared with rgPR8 virus (P = 0.010; two-tailed t test) suggesting that NS gene modulate virus infectivity and determined cell tropism. This could be explained by a previous observation wherein NS gene post-transcriptionally regulate HA on viral envelope and thus increase levels of HA protein on viral envelope [14].
Fig. 3.
Flow cytometry for determining infectivity in THP-1 monocytic cells infected with NS reassortant PR8 virus. THP-1 cells infected with PR8 virus or rgH5N1-NS virus at m.o.i. of 3. At 24 h post infection, cells were analysed for influenza NP by flow cytometry. Numbers indicate percentages of cells in each gate. Uninfected cells are used as control staining. Histogram shows the average percentage (mean ± SD) of NP+ THP-1 cells from 3 independent experiments (two-tailed t test: P = 0.010)
In conclusion, this and the other studies [8, 15] showed monocytic cells differ in their susceptibilities to influenza virus infection. Moreover, the present study demonstrates that THP-1 cells could be used as a model to identify viral factors that determine cell tropism. The differences in susceptibilities of U937 and THP-1 cells to PR8 virus could not be explained by SA receptor distribution. However, NS gene of H5N1 virus could overcome the cellular resistance of THP-1 cells for PR8 virus. Further studies on viral factors and possibly, cellular factors that contribute for the difference in susceptibilities to influenza virus infections need to be explored.
Acknowledgements
PPL was supported by Siriraj Graduate Studentship for international students. Eight recombinant pHW2000 plasmids carrying each genomic segment of PR8 virus for construction of reassortant viruses was kindly provided by Prof. Robert G. Webster, St. Jude Children’s Research Hospital, Memphis, TN, USA.
References
- 1.Cole SL, Ho LP. Contribution of innate immune cells to pathogenesis of severe influenza virus infection. Clin Sci (Lond) 2017;131(4):269–283. doi: 10.1042/CS20160484. [DOI] [PubMed] [Google Scholar]
- 2.Herold S, Steinmueller M, von Wulffen W, Cakarova L, Pinto R, Pleschka S, et al. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med. 2008;205(13):3065–3077. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duan M, Hibbs ML, Chen W. The contributions of lung macrophage and monocyte heterogeneity to influenza pathogenesis. Immunol Cell Biol. 2017;95(3):225–235. doi: 10.1038/icb.2016.97. [DOI] [PubMed] [Google Scholar]
- 4.Nicholls JM, Chan RW, Russell RJ, Air GM, Peiris JS. Evolving complexities of influenza virus and its receptors. Trends Microbiol. 2008;16(4):149–157. doi: 10.1016/j.tim.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 6.Nobusawa E, Ishihara H, Morishita T, Sato K, Nakajima K. Change in receptor-binding specificity of recent human influenza A viruses (H3N2): a single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology. 2000;278(2):587–596. doi: 10.1006/viro.2000.0679. [DOI] [PubMed] [Google Scholar]
- 7.Vines A, Wells K, Matrosovich M, Castrucci MR, Ito T, Kawaoka Y. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol. 1998;72(9):7626–7631. doi: 10.1128/jvi.72.9.7626-7631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamichhane PP, Boonnak K, Changsom D, Noisumdaeng P, Sangsiriwut K, Pattanakitsakul SN, et al. H5N1 NS genomic segment distinctly governs the influenza virus infectivity and cytokine induction in monocytic cells. Asian Pac J Allergy Immunol. 2018;36(1):58–68. doi: 10.12932/AP0870. [DOI] [PubMed] [Google Scholar]
- 9.Ana-Sosa-Batiz F, Vanderven H, Jegaskanda S, Johnston A, Rockman S, Laurie K, et al. Influenza-specific antibody-dependent phagocytosis. PLoS ONE. 2016;11(4):e0154461. doi: 10.1371/journal.pone.0154461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen H, Wang Z, Lenz E, Pleschka S, Rautenschlein S. Reassortment of NS segments modifies highly pathogenic avian influenza virus interaction with avian hosts and host cells. J Virol. 2013;87(10):5362–5371. doi: 10.1128/JVI.02969-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Twu KY, Kuo RL, Marklund J, Krug RM. The H5N1 influenza virus NS genes selected after 1998 enhance virus replication in mammalian cells. J Virol. 2007;81(15):8112–8121. doi: 10.1128/JVI.00006-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma W, Brenner D, Wang Z, Dauber B, Ehrhardt C, Hogner K, et al. The NS segment of an H5N1 highly pathogenic avian influenza virus (HPAIV) is sufficient to alter replication efficiency, cell tropism, and host range of an H7N1 HPAIV. J Virol. 2010;84(4):2122–2133. doi: 10.1128/JVI.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipatov AS, Andreansky S, Webby RJ, Hulse DJ, Rehg JE, Krauss S, et al. Pathogenesis of Hong Kong H5N1 influenza virus NS gene reassortants in mice: the role of cytokines and B- and T-cell responses. J Gen Virol. 2005;86(Pt 4):1121–1130. doi: 10.1099/vir.0.80663-0. [DOI] [PubMed] [Google Scholar]
- 14.Yan X, Wang Q, Zhang Z, Xie Y, Zhang H, Razi M, et al. Involvement of non-structural proteins (NS) in influenza A infection and viral tropism. Biochem Biophys Res Commun. 2012;428(1):62–67. doi: 10.1016/j.bbrc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoeve MA, Nash AA, Jackson D, Randall RE, Dransfield I. Influenza virus A infection of human monocyte and macrophage subpopulations reveals increased susceptibility associated with cell differentiation. PLoS ONE. 2012;7(1):e29443. doi: 10.1371/journal.pone.0029443. [DOI] [PMC free article] [PubMed] [Google Scholar]