Abstract
Long non-coding RNAs (lncRNAs) are a class of cellular transcripts, which are involved in various biological processes. There is conflicting data regarding to the origin of these non-coding molecules and lncRNAs are thought to be the origin of viral genome. Here we sought to find the homology between human lncRNAs and viruses. For this purpose, the lncRNAdb database was searched for human lncRNAs. The lncRNAs’ sequences were aligned with virus taxa using NCBI’s BLAST tool. The phylogenic study was performed with maximum-likelihood based algorithm. The database contains 152 human lncRNAs. As a result, 63 (41.44%) of the lncRNAs have homologies with viruses. Of which, 50 (79.36%) have homology with Stealth virus. Other viruses with homology to lncRNAs were nuclear integrating DNA/RNA viruses. Moreover, 35 of 64 (23.03%) of cancer-associated lncRNAs have sequence homology with the same viruses. In phylogenetic analyses, lncRNAs with no homology to viruses were found to be the ancestor of those with homology to viruses and cancer-irrelevant lncRNAs were found to be the ancestor of cancer-related transcripts. In conclusion, lncRNAs could be the origin of nuclear integrating viruses and the nuclear integrating viruses may evolved from the non-coding regions. The results imply the role of lncRNAs with homology to viruses in human cancers.
Electronic supplementary material
The online version of this article (10.1007/s13337-018-0486-9) contains supplementary material, which is available to authorized users.
Keywords: lncRNAs, Non-coding transcripts, Viruses, Homology, Stealth virus
Introduction
During the last decade, high through put sequencing approaches and genome profiling uncovered several kinds of non-coding RNAs (ncRNAs). The Encyclopedia of DNA Elements (ENCODE) project has indicated that most region of human transcriptome are non-coding and role as housekeeping and/or regularly elements [17]. Based on their diversity, the ncRNAs have been classified into two main groups. First group with short length (≤ 200 nucleotide) are highly conserved and regulate transcriptional and post-transcriptional pathways. Small ncRNAs are classified into different sub-groups, including microRNAs (miRNAs), small nuclear RNAs (snRNAs), piwiRNAs (piRNAs), and small interfering RNAs (siRNAs) [9]. Second group with long length (≥ 200 nucleotide) such as long-ncRNAs (lncRNAs) are poorly conserved and have regulatory roles but they are not completely understood [2, 24]. Based on genomic location, lncRNAs are classified into large intergenic and intronic lncRNAs (lincRNAs) [9]. Based on orientation of transcription from coding regions, lncRNAs divide into sense/antisense lncRNAs. A well known example of antisense lncRNAs is HOTAIR, which is an long non-codding RNA with oncogenic properties in different cancer [5]. Long non-coding transcripts are further classify according to their targeting mechanisms and their mechanism of functioning [9]. Studies are ongoing to uncover accurate function and structure of lncRNAs due to their essential roles in wide range of human disorders [24].
LncRNAs are expressed in a wide variety of organisms, and have the same biogenesis process and post-transcriptional modification as protein-coding transcripts. It is well known that lncRNAs play a critical role in the regulation of gene expression and are important in biological process such as cell cycle, proliferation, and apoptosis. According to ncRNA sequences and their conservancy, it could be possible to predict lncRNAs between and across species [8]. It is thought that lncRNAs originated from different processes, including transposable elements (TEs) disruption, fusion of broken sequences, mutations and genetic loss or excision. TEs take place within most of mature lncRNAs, while they rarely found in protein-coding transcripts [8, 7]. Four important classes of TEs are short interspersed elements (SINEs), long interspersed elements (LINEs), long terminal repeat/endogenous retroviral (LTR/ERV) elements, and DNA transposons with their own function and evolutionary history [24, 7].
Studies have been demonstrated that lncRNAs expression could be affected by viral infection and revealed their significant correlation with virus replication and diseases outcome [7]. During last decades, virus evolution is well argued and believed that they may originated from non-coding cellular transcripts. In the present study, the sequence of known human lncRNAs were investigated for any homology to viruses. As a result, a conserved region was found within lncRNAs, which has homology with nuclear viruses. Furthermore, phylogenic study revealed an evolutionary association between lncRNAs and viruses. It was also found that the homology between viruses and cancer-associated lncRNAs was significant that in other group of lncRNAs. Our results for first time indicate that there is a homology between lncRNAs and viruses at nucleotide level, and viruses may derived from ncRNAs.
Materials and methods
Search for human lncRNAs’ homology to viruses
The lncRNAdb database containing annotated mammalian lncRNAs was searched for human lncRNAs [20]. Basic Local Alignment Search Tool (BLAST) was employed for sequence similarity search against GenBank [1]. For this purpose, nucleotide BLAST (BLASTn) parameters were changed. Briefly, viruses (taxid: 10239) and Homo sapiens (taxid: 9606) were included and excluded, respectively. The later was excluded to remove any human data related to viral integration sites. Program selection was optimized for BLASTn algorithm (Somewhat similar sequences). The expected threshold changed to 40 to reduce chance of nucleotide mismatch. Finally, Only 50 first results with E-value ≤ 1e−4 cut off were included to further analysis. Moreover, Lnc2Cancer database was searched to find any association of the human to [18].
Phylogenic study
Full-length sequences of the LncRNAs with homology to viral sequences or those related to cancer have chosen for ClustalW Multiple Sequence Alignment (MSA) with CLC Sequence Viewer 6 (CLC bio, a QIAGEN Company). The MSA PHYLIP output results were chosen for phylogenic tree reconstruction using MEGA 5 with Maximum Likelihood algorithm after gap cleansing with Gap Strip/Squeeze v2.1.0 server (Operated by Los Alamos National Security, LLC, for the U.S. Department of Energy’s National Nuclear Security Administration) [23]. The chance of random clustering (sampling) error was reduced by using Maximum Likelihood algorithm [19]. The constructed rooted trees were validated with bootstrap replicate value of 1000.
Probing non-coding RNAs in human RNAseq database and RNA structure alignment
For detection of human lncRNAs, the Human reference RNA sequence (refseq_rna) database was scavenged using Stealth virus conserved sequence as query. The BLAST search setting was changed as the same. Furthermore, RNA structures of lncRNAs were aligned with the conserved sequence of Stealth virus by using LocARNA server [13–22]. For that, default server setting was adopted. Briefly, the lncRNAs with homology to viruses were aligned and a consensus sequence was retrieved. The consensus sequence was aligned structurally to find a matched secondary structure between conserved regions of Stealth virus and lncRNAs with virus homology.
Statistical analysis
Statistical analyses were performed using SPSS16.0. The homology of lncRNAs and cancer-associated lncRNAs with viruses were analyzed by Chi Square test. Strength of association was also analyzed by Phi and Cramer’s V test. P value less than 0.05 was considered as significant.
Results and discussion
LncRNAs have homology with viruses
LncRNAs regulate diverse biological processes in eukaryotic cells and impact on viral infection. On the other hand, viral infections may lead to the differential expression of cellular lncRNAs and this change seems to exist as a common pathological phenomenon. There is growing studies regarding to specify lncRNAs and viral interaction, but it is not well clarified. Additionally, there is no known data exploring the homology of lncRNAs and viruses and their evolutionary pathways. Since viruses are obligate intracellular pathogens and evolutionary adopted with host cells and lncRNAs are important intracellular molecules, here we hypothesized that viruses may originated from lncRNAs or vice versa. For that, the lncRNAdb database has searched for human lncRNAs and aligned with virus taxa using NCBI’s BLAST tool. Phylogenic analyses were performed by maximum-likelihood based algorithm.
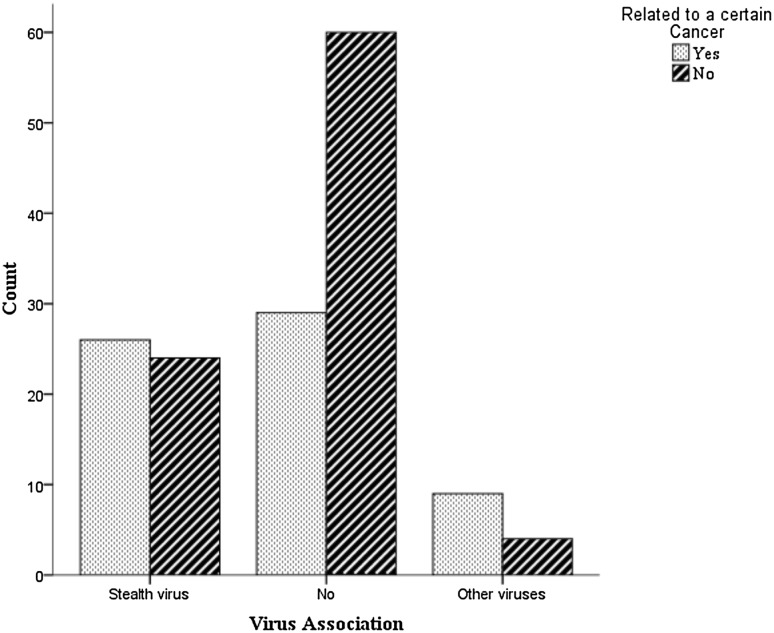
The lncRNAdb database was contained 152 human lncRNAs. According to lnc2cancer server, 42.11% (64/152) of the lncRNAs were associated to cancers. Alignment of lncRNAs to GenBank viral sequences using BLAST showed 41.44% (63/152) homology with viruses, in which 79.36% (50/63) was related to Stealth virus. Other viruses with lncRNAs homology were nuclear integrating DNA/RNA viruses such as human endogenous retroviruses (HERV), human immunodeficiency virus-1 (HIV-1), and human papillomaviruses (HPV) (Fig. 1). The homology was observed in one region within all studied lncRNAs. Moreover, 54.69% (35/64) of cancer-associated lncRNAs had sequence homology with viruses (9.242(2), p = 0.01). Strength of test was also significant (0.247, p = 0.01). Of those, 74.29% (26/35) had homology with Stealth virus and 25.71% (9/35) with other viruses. Furthermore, there were no significant differences between cancer related and non-cancer related lncRNAs (Full data are provided in Supplemented file).
Fig. 1.
Distribution of virus associated lncRNAs
Cytopathic Stealth virus has been identified as a DNA virus, which is discriminated from other nuclear viruses by its unusual cytopathic effect and different type of diagnostics methods [10, 16]. It has been investigated in several immunological and psychiatric disorders including chronic fatigue syndrome (CFS) [13], Alzheimer disease, dementia, bipolar disorder [12], acute encephalopathy [12], depression [10], autism [11], and fibromyalgia syndrome (FMS) [10].
Driving Stealth virus entity from herpes viruses with respect to long term mutualistic relationship of herpes viruses with their host would support cellular origination of the virus [21]. Interestingly, Stealth virus can achieve plenty of host genetic materials from replication process to other compensatory mechanisms and transfer to bystander cells as biological magnet [15]. Additionally, high degree of similarity between Stealth virus and cellular counterparts in comparison with other herpesvirus family members [14] supports cellular-based evolutionary pathway. Despite of viral immune invasion, Stealth virus does not recognize by host cell immune system [12], proving evolutionary host cell-derived origination. In addition, anti-herpesvirus antibodies do not recognize Stealth virus [12], which may confirm difference genetic contents of Stealth from other herpesviruses. These findings support the idea that Stealth virus is derived from host cells and our results imply that this virus may originate from lncRNAs [15].
An evidence of viral origination from lncRNAs
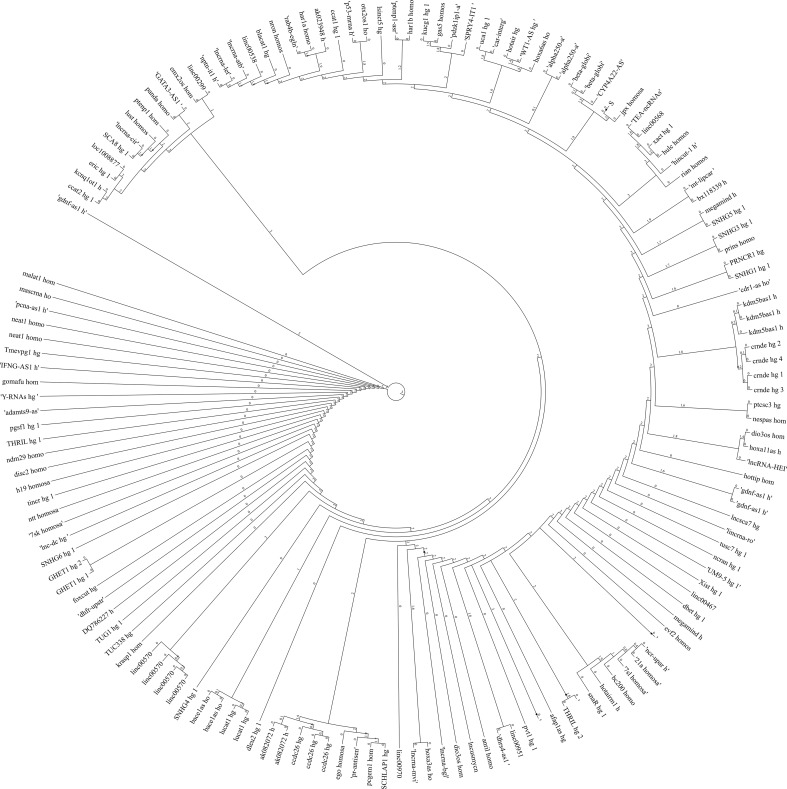
Phylogenic and evolutionary analyses of lncRNAs with homolog to viruses are shown in Fig. 2. The result indicated that lncRNAs with no homology to viruses were found to be the ancestor of virus-homolog ones. Furthermore, cancer-irrelevant lncRNAs were found to be the ancestor of cancer-related transcripts (Fig. 3). It was found that there was a conserved region with ~ 280 bp among lncRNAs with homology to Stealth virus (supplemented file). Alignment of the conserved region against GenBank with the same setting have shown more than 30% query cover with viral integration sites (E-value ≤ 2e−80) if Homo Sapience taxid was included, and more than 32% query covers with Stealth virus, HIV-1, HPV, Simian Virus 40 (SV-40), and human T lymphotropic virus (HTLV) (E ≤ 1e−60) if Homo Sapience taxid was excluded.
Fig. 2.
Cladogram analysis of Stealth virus related and non-related as well as other viruses-related (O) lncRNAs. The age of the phylogenic distance is illustrated on the branches. Furthermore, the results of lncRNAs are provided in a supplemented file
Fig. 3.
Cladogram analyses of cancer-related and non-related lncRNAs. The age of the phylogenic distance is illustrated on the branches. Furthermore, the results of lncRNAs are provided in a supplemented file
The results of phylogenic study indicate that the lncRNAs with no homology to viruses to be the ancestor of virus-homolog ones and cancer-irrelevant lncRNAs to be the ancestor of cancer-related transcripts. This results along with parasitic and cell dependence properties of viruses, as well as common ancestrally relationships within them [6], support common cellular ancestor, non-coding RNA transcripts (like lncRNAs) for both nuclear integrating DNA/RNA viruses such as HTLV, HIV, HPV, and SV40. Moreover, these findings suggest same nucleus origin for nuclear viruses and produce results corroborating the hypothesis of DNA viruses’ evolution from RNA, while it is also consistent with RNA virus emergence in nucleus, where lncRNAs transcribe [4]. Although, the results have no conflict with RNA world idiom [3], but it can put a light into same world in which non-coding RNA transcripts are ancestors of nuclear viruses. Importantly, if lncRNAs be considered as separated genetic fragments of host, the results of our study support escape theory of viral origination [4], indicating that capsidless elements like non-coding transcripts would be transfer from host reservoirs into novel viruses or vice versa [15]. Capsid structure is much of interest since most of eukaryotic and prokaryotic viruses have similar evolutionary structural features [14]. However, with respect to parasitic life of viruses, it simply implies common ancestry outcome based on capsid architecture and does not support scape theory of viral origination.
Stealth virus sequence pointed out the human non-codding transcripts
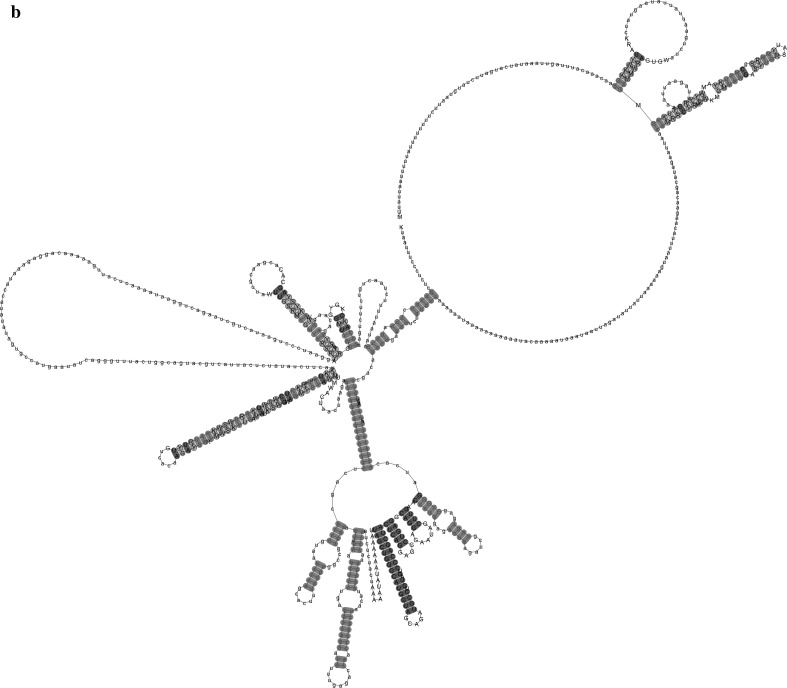
By searching human transcriptome, 180 lncRNA were found. The identity of matched transcripts was ranged from 84 to 90% and e-value of 3e−73 to 2e−88. The list of lncRNAs are provided in the supplemented file. The results are shown in the Fig. 4. As shown, both sequences are compatible in several places the stems of the RNA secondary structure. The result of the structure alignment indicates evolutionary structure of lncRNAs resulted from genetic combination of different foreign nucleotide elements.
Fig. 4.
a Shows RNA structure alignment of Stealth virus and lncRNAs’ conserved region. b illustrate compatible base paring in a secondary structure. Compatible base pairs are colored, where the hue shows the number of different types C-G, G-C, A-U, U-A, G-U or U-G of compatible base pairs. In this way, the hue shows sequence conservation of the base pair. The saturation decreases with the number of incompatible base pairs. Thus, it indicates the structural conservation of the base pair. Red colors indicate one compatible base pair. Green colors demonstrate three compatible base pairs (color figure online)
In conclusion, our results clearly indicate that: (1) lncRNAs have a significant homology with viruses in one part, (2) lncRNAs with viral homology significantly associated with cancer, (3) lncRNAs with no homology to viruses proceeding to ones with homology to viruses, and (4) cancer-irrelevant lncRNAs are the ancestor of cancer-related transcripts. These findings implicate an association between lncRNAs and viruses, further comprehensive investigations are warranted in this regard. The results just obtained from one lncRNAs database and the similarity searched was limited to viral taxa, thus investigation in wide databases with similarity search through other taxa would be interesting.
Electronic supplementary material
Below is the link to the electronic supplementary material.
References
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 3.Forterre P. The two ages of the RNA world, and the transition to the DNA world: a story of viruses and cells. Biochimie. 2005;87:793–803. doi: 10.1016/j.biochi.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006;117:5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014;30:439–452. doi: 10.1016/j.tig.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay LA, Bourque G, et al. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013;9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:924–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin WJ. Stealth viruses as neuropathogens. CAP Today. 1994;8:67–70. [PubMed] [Google Scholar]
- 11.Martin WJ. Stealth virus isolated from an autistic child. J Autism Dev Disord. 1995;25:223–224. doi: 10.1007/BF02178507. [DOI] [PubMed] [Google Scholar]
- 12.Martin WJ. Simian cytomegalovirus-related stealth virus isolated from the cerebrospinal fluid of a patient with bipolar psychosis and acute encephalopathy. Pathobiology. 1996;64:64–66. doi: 10.1159/000164010. [DOI] [PubMed] [Google Scholar]
- 13.Martin WJ. Detection of rna sequences in cultures of a stealth virus isolated from the cerebrospinal fluid of a health care worker with chronic fatigue syndrome. Pathobiology. 1997;65:57–60. doi: 10.1159/000164104. [DOI] [PubMed] [Google Scholar]
- 14.Martin WJ. Cellular sequences in stealth viruses. Pathobiology. 1998;66:53–58. doi: 10.1159/000027996. [DOI] [PubMed] [Google Scholar]
- 15.Martin WJ. Stealth virus culture pigments: a potential source of cellular energy. Exp Mol Pathol. 2003;74:210–223. doi: 10.1016/S0014-4800(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 16.Martin WJ, Glass RT. Acute encephalopathy induced in cats with a stealth virus isolated from a patient with chronic fatigue syndrome. Pathobiology. 1995;63:115–118. doi: 10.1159/000163942. [DOI] [PubMed] [Google Scholar]
- 17.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 18.Ning S, Zhang J, Wang P, Zhi H, Wang J, Liu Y, et al. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016;44:D980–D985. doi: 10.1093/nar/gkv1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quek XC, Thomson DW, Maag JLV, Bartonicek N, Signal B, Clark MB, et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson S. Cytomegalovirus: the stealth virus. Pract Midwife Engl. 2016;19:28–29. [PubMed] [Google Scholar]
- 22.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA tools: a web server integrating IntaRNA, ExpaRNA and LocARNA. Nucleic Acids Res. 2010;38:W373–W377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Will S, Joshi T, Hofacker IL, Stadler PF, Backofen R. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA. 2012;18:900–914. doi: 10.1261/rna.029041.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput Biol. 2007;3:680–691. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.