Abstract
Hepatitis B virus (HBV) and Hepatitis C virus (HCV) infection often lead to hepatocellular carcinoma (HCC), which is mostly detected in advanced stage. Hence, its early detection is of paramount importance using a biomarker having sensitivity and specificity both. The present study highlights differentially expressed host proteins in response to HBV/HCV infection at different stages. Comparative proteomic study was done by two-dimensional gel electrophoresis followed by mass spectrometry. Sera from each of chronically infected, liver cirrhosis and HCC in HBV or HCV infection along with controls were selected. Analysis of functional association between differentially expressed proteins with viral hepatitis was extensively carried out. Forty-three differentially expressed spots (≥ 1.5 fold; P < 0.05) on two-dimensional gel electrophoresis were corresponded to 28 proteins by mass spectrometry in variable liver diseases. Haptoglobin protein levels were decreased upon disease progression to HCC due to HBV infection. The other proteins expressed differentially are ceruloplasmin, serum paraoxonase 1, retinol binding protein and leucine rich alpha 2 proteins in plasma maybe associated to HBV HCC. Whereas, upregulation of C4a/C4b showed it as a reliable marker in patients with end stage liver disease related to HCV infection. ApolipoproteinA1 levels in liver diseases in both HBV and HCV infection corresponding to healthy controls may be a common marker for early diagnosis and disease monitoring. Protein interaction studies by extensive pathway analysis using bioinformatics tools such as EnrichNet application and STRING revealed significant associations with specific infections.
Electronic supplementary material
The online version of this article (10.1007/s13337-018-0484-y) contains supplementary material, which is available to authorized users.
Keywords: Hepatitis B virus, Hepatitis C virus, Hepatocellular carcinoma, Protein biomarkers
Introduction
Hepatocellular carcinoma (HCC) is a primary malignant neoplasm derived from hepatocytes, accounting for about 90% of all liver cancers [11]. HCC is the sixth most common cancer worldwide, being the fifth in men and the eighth in women. It accounts for approximately 5.7% of all new cancer cases [44]. Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) infections are the major causes of acute and chronic hepatitis worldwide and, lead to liver cirrhosis and HCC [12]. In India, 40–45 million HBV carriers outweigh 10 million HCV carriers [1]. Multiple factors such as genetic, epigenetic and proteomic changes are reported in HCC patients.
Alpha feto protein (AFP) is the established diagnostic and prognostic biomarker of HCC which is neither sensitive nor specific [10]. Many times, patients with advanced stage of liver disease at the time of diagnosis show normal level of AFP. It is difficult to detect HCC at an early stage due to inadequacy of etiology specific biomarkers [37]. Proteomic profiling approach for HCC looks promising including advanced mass spectrometry (MS) technology along with bioinformatics tools [27, 35] Two-dimensional gel electrophoresis (2DE) followed by MS has been extensively employed to unveil differentially expressed proteins at different stages of the disease. A functional proteomic approach has been used previously to study differential protein expressions, but most studies are either HBV [29, 32] or HCV [13, 31] specific and, have reported alterations in proteins at the end stage of the disease [6, 41]. Very few studies have emphasized on both etiology and liver disease stages [27, 28]. The present study is therefore designed to explore differentially expressed host proteins in response to HBV/HCV infection at different stages: Chronic hepatitis, Liver Cirrhosis and HCC in comparison to healthy controls by using two dimensional gel electrophoresis (2-DE) and mass spectrometry. The study also involves identification and extensive analysis of the host protein interaction networks.
Materials and methods
Subject/Case recruitment
This Study was approved by Institutional Ethics Committee (IEC-I), Seth GS Medical College and King Edward Memorial Hospital and, informed consent was obtained from each individual. Patients (N = 24) were recruited from outpatient department (OPD) and inpatient department (IPD) of Gastroenterology in King Edward Memorial Hospital, Mumbai during 2014–2017. Healthy controls (N = 4) aged between 18 and 65 years were included in this study with no history of HBV, HCV and HIV. Total 12 HBsAg positive and 12 Anti HCV positive patients aged between 18 and 65 years were included in the study. They were categorized into groups viz chronic hepatitis, liver cirrhosis and hepatocellular carcinoma based on guidelines of the Asian Pacific Association for the Study of the Liver. Their disease status was assessed by serological, biochemical, clinical, radiological and pathological (whenever biopsy possible) findings. Clinical and pathological reports were obtained from the medical records. Patients with history of HIV, acute liver disease, renal failure, autoimmune diseases, Wilson’s disease, Cholangio carcinoma and Lamellar cell carcinoma were excluded from the study.
Sample collection
10 ml of venous blood was drawn from each participant and allowed to clot for 2 h. Blood samples were centrifuged at 3000 rpm for 15 min. Serum was separated and aliquots were stored at − 80 °C until analysis.
Laboratory tests
HBV, HCV, HIV serological tests- HBeAg, Anti-HBe, HBcIgG, Anti HCV, Anti-HIV ELISAs were performed according to manufacturer’s instructions. Liver function tests and liver enzyme were measured on Cobas C111.
Sample lysis and processing
Serum sample was mixed with binding buffer (20 mM sodium phosphate, 0.15 M sodium chloride, pH 7.4) and subjected to sonication at an amplitude of 20% for 5 s pulse on and 30 s pulse off to allow disruption of cells and release of total proteins. High abundant proteins like albumin and immunoglobulin G (IgG) were removed from 50 μl of serum using albumin IgG depletion spintrap columns (GE Healthcare) to increase protein resolution by unmasking low molecular weight proteins. Depleted serum sample was precipitated using acetone and pellets were dissolved in 100 μl 2D lysis buffer (7 M Urea, 2 M Thiourea, 4% CHAPS, 40 mM DTT). Protein concentration of depleted sera was measured by Bradford’s protein assay using bovine serum albumin as a standard.
Two-dimensional gel electrophoresis and spot analysis
200 μg of albumin IgG depleted proteins along with rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS, 0.5% IPG buffer, 1% Bromophenol Blue) were loaded onto 4–7 NL 13 cm IPG strips (GE Healthcare) for isoelectric focusing (IEF). IEF was carried out at 20 °C in the Ettan™ IPGphor™ 3 unit (GE Healthcare) with current limited to 75 IA/strip achieving up to 52,000 Vh for 19:00 h. The focused strips were equilibrated and subjected to a second-dimension separation on 12.5% polyacrylamide gels. The gels were stained using a silver staining procedure compatible for mass spectrometry and scanned with the ImageScanner III scanner. The images were analysed using the high-throughput 2-DE software ImageMaster™ 2D Platinum 7.0. It calculated the spot intensity by integrating the absorbance over the spot area. Differential analysis was carried out using statistical tools of the software. Expression of proteins were considered significant if 1.5–12 fold change and analysis of variance (ANOVA) < 0.05 were seen in case and control group.
In-gel digestion
In-gel digestion on silver stained gels was carried out using modified trypsin (Promega). Briefly, the protein spots were washed with water and de-stained with potassium ferric cyanide and sodium thiosulphate. The gel particles were then washed with 50 mM NH4HCO3/acetonitrile 1:1 (v/v). Further, reduction and alkylation of gel particles were carried out by 10 mM dithiotretiol and 55 mM iodoacetamide in 50 mM NH4HCO3. The gel particles were allowed to shrink in acetonitrile and again dried using vaccum centrifuge. Freshly prepared enzyme solution (25 mM NH4HCO3 with 5 ng/lL of trypsin) was added on to gel particles and incubated at 37 °C for 30 min. Gel particles were covered with 25 mM NH4HCO3 and incubated overnight at 37 °C. The peptides were extracted from gel pieces by adding series of solvents: 0.1% TFA in 50% ACN, 0.1% TFA in 60% ACN and 0.1% TFA in 80% ACN (100 μl each). All the three fractions were pooled and vacuum dried. The samples were stored at − 20 °C. Samples were reconstituted in 5–10 μl of 0.1% TFA in 50% acetonitrile before mass spectrometric analysis.
Protein identification by MALDI TOF/TOF
The mass spectrometric analysis was carried out on matrix associated laser desorption ionization—time of flight (MALDI-TOF) instrument (Bruker Daltonik GmbH) at ACTREC, Kharghar, Navi Mumbai. Briefly 2 μl of the extracted protein was mixed with 2 μl (equal volumes) of a-cyano-4 hydroxy-cinnamic acid (CHCA) matrix and, loaded on ground steel MALDI target plate and allowed to dry at room temperature. The MALDI-TOF MS target was subsequently introduced into Bruker Daltonik GmbH instrument for automated measurement. Initial manual/visual study of the mass spectra was done using Flex Analysis 2.4 software (Bruker Daltonik GmbH, Germany). For automated data analysis, raw spectra were processed using the Biotools Version 3.0 software (Bruker Daltonik GmbH, Germany). The instrument was calibrated with standard peptide mix before sample analysis with default settings. The instrument was operated in reflectron mode using Nd: YAG 335 nm laser, with a mass range of 800–4000 Da. MS peak list was subjected to MASCOT version 2.1 (http://www.martixscience.com) search engine for protein identification. The parameters used for Mascot search were as follows: trypsin digestion, single missed cleavage, oxidation of methionine and carbamidomethylation of cysteine as variable and fixed modifications respectively. Hundred ppm mass tolerance was selected for MS and Swiss-Prot database for Homosapiens taxonomy.
Association between differentially expressed proteins and their interaction pathways
Functional associations of differentially expressed proteins identified from sera of cases with different liver disease conditions were analysed with online EnrichNet application using Reactome and GO by incorporating the protein accession number. The significance of overlap between protein sets was found with one-side Fisher’s exact test (q < 0.05) and along with network similarity scores (XD-scores). The threshold values were assessed by EnrichNet with a regression fit equivalent Fisher q value of 0.05 and upper boundary of 95% confidence for linear fitting. The trend of these identified pathways among development of liver diseased stages was determined by Reactome expression analysis. Host protein interaction networks for the differentially expressed proteins were analysed by STRING tool version 10.5.
Results
Subject/cases recruitment and laboratory tests
The demographic and laboratory parameters of enrolled participants are summarized in Supplementary Table 1. The mean age was 45.85 ± 11.36 years and majority of them were between 35 and 50 years (57.1%). Most of them were males (64.3%). Liver enzymes and bilirubin levels were increased in HCC patients compared to the other groups.
Two dimensional gel electrophoresis and spot analysis
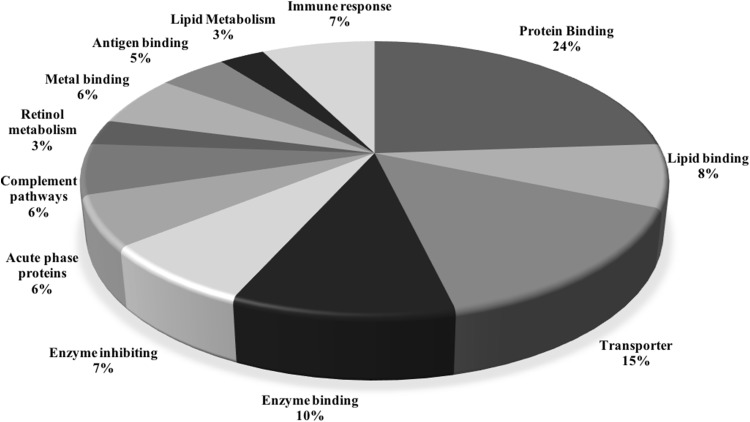
The differential protein spots identified after 2DE and MALDI-TOF analysis are shown in Fig. 1 and Supplementary Table 2 along with their molecular weights, accession number, fold change, location on the 2DE gel and their biological roles. They were classified into 12 broad categories depending upon their multiple functions, namely; protein binding, lipid binding, transporter, enzyme binding, enzyme inhibition, acute phase proteins, complement activation and, pathways: retinol metabolism, metal binding proteins, antigen binding proteins, lipid metabolism and immune responses. Percentage distribution of these proteins in different categories is depicted in Fig. 2.
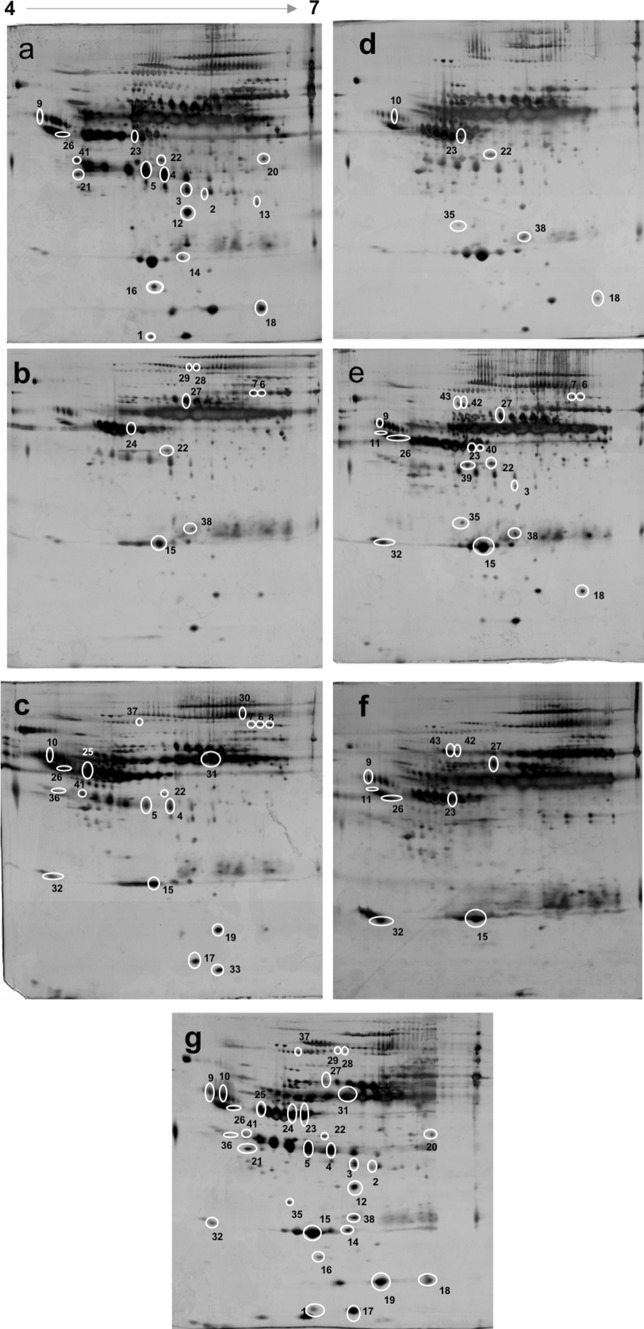
Fig. 1.
Representative serum 2-DE images from different liver pathologies of viral hepatitis. Serum protein expression in chronic hepatitis, liver cirrhosis and hepatocellular carcinoma due to HBV (a–c); HCV (d–f) respectively and, healthy individuals (g). 200 μg of proteins were resolved using 2-DE and visualized by silver staining. White circle mark around the protein spots indicate spots with differential expression (1.5–12 fold change and analysis of variance (ANOVA) < 0.05) between case and control group. Spot numbers are in accordance with Supplementary Table 2 for relevant information about the proteins
Fig. 2.
Percentage distribution of functionally classified differentially expressed proteins in chronic hepatitis, liver cirrhosis and HCC cases due to HBV or HCV. Major changes were observed in protein binding, transporter and enzyme binding proteins
Comparison of differential protein spots in HBV liver diseases
Several differential spots were identified after comparison between healthy and liver disease conditions as well as intergroup comparison. Figure 1 illustrates 2DE images of protein profiles in HBV/HCV liver diseases along with healthy controls. All the cases (chronic hepatitis, LC, HCC) showed decreased expression of apolipoprotein A1 and transthyretin but increased expression of beta actin when compared with healthy individuals. Chronic hepatitis B showed increased levels of haptoglobin, ficolin-3, antichymotrypsin, Ig heavy chain mu intermediate segment, zinc-alpha-2-glycoprotein, serum paraoxonase/arylesterase 1 and alpha-2-HS-glycoprotein while alpha1 antitrypsin was decreased while comparing with healthy controls. The cirrhotic cases showed increased expression of alpha1 antitrypsin, haemopexin, complement factor B (C3/C5 convertase), alpha 2 macroglobulin as compared to healthy subjects. Serum amyloid A, alpha microglobulin, leucine rich alpha 2, serum paraoxonase/arylesterase 1, ceruloplasmin, antichymotrypsin, Ig J chain, alpha1 antitrypsin, alpha-2-HS-glycoprotein, complement factor B (C3/C5 convertase) and albumin expression were upregulated in HBV positive HCC cases. Haptoglobin MW 43,156 and 17,069 Da were increased by 3.17 and 3.38 fold (p = 0.00002 and p = 0.013) in chronic hepatitis B as compared to cirrhosis and HCC respectively. Transthyretin, retinol binding protein and complement factor B (C3/C5 convertase) were downregulated in HCC when compared to chronic hepatitis B.
Differentially expressed proteins in HCV related liver diseases
Beta actin was increased in chronic hepatitis C, cirrhosis and HCC by 12, 8.78, 4.46 fold respectively (P = 0.00008, 0.0012, 0.02) when matched with controls. Chronic hepatitis C patients exhibited increased expression of haptoglobin, alpha 1 microglobulin and complement C4A/C4B when compared with healthy controls. Alpha-2-HS-glycoprotein, vitamin D binding protein and complement factor B (C3/C5 convertase) were increased in cirrhotic patients whereas, apolipoprotein A1 was decreased in this group. HCV positive HCC patients showed upregulation of Ig J chain, apolipoprotein A1, alpha-2-HS-glycoprotein and complement C4A/C4B but, downregulation of transthyretin against normal. Serum amyloid P was increased by threefold (P = 0.012) in chronic hepatitis C and alpha1 antitrypsin was increased by 7.37 fold (p = 0.026) in cirrhotic patients when compared with HCV HCC.
Differentially expressed proteins between HBV and HCV related liver diseases
Alpha-2-HS-glycoprotein was significantly increased in chronic hepatitis B (fold 3.97, P = 0.04) when compared with chronic hepatitis C infection. We observed significant increase in haemopexin precursor (fold 3.11, P = 0.006), apolipoprotein A4 (fold 4.72, p = 0.005) but decrease in antichymotrypsin (fold 2.09, p = 0.04) in cirrhotic HBV when matched with cirrhotic HCV. Alpha 1 antitrypsin (fold 3.17, p = 0.04) and antichymotrypsin (fold 3.82, p = 0.006) were increased in HBV related HCC than that with HCV.
Association between differentially expressed proteins and their interaction pathways
Pathway analysis of all differentially expressed proteins in the HBV and HCV related liver diseases using EnrichNet application using Reactome and Biological process GO database identified various affected pathways. Based on Fisher test q value < 0.05 and XD scores (more than 0.91 and 1.07 threshold values as estimated by the application for Reactome and Biological process respectively), pathways identified are as follows: HDL mediated lipid transport, lipid metabolism, chylomicron mediated lipid transport, phospholipid efflux, positive regulation of cholesterol efflux and lipid homeostasis as given in Supplementary Tables-3 and 4.
The STRING tool was used to analyse protein–protein interactions for all identified proteins in both HBV or HCV related liver diseases including chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. Enrichment analysis for KEGG pathways and first five biological pathways (GO) identified various pathways as shown in Supplementary Tables 5 and 6 based on the protein–protein interaction for P < 0.05, minimum required interaction score > highest confidence (0.900).
Discussion
Viral hepatitis due to HBV or HCV has variable clinical presentations: acute, chronic, fulminant flare, fibrosis, cirrhosis and HCC. Our present study was designed to examine the alterations in serum protein levels of HBV or HCV infected patients with various liver complications by proteomics. By combining isoelectric focusing with SDS PAGE (two dimensional gel electrophoresis), the sera proteins were separated to an extent which allowed us to discern subtle differences.
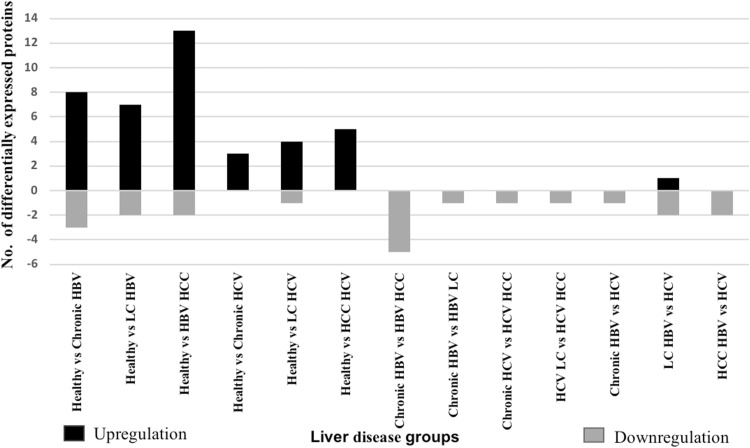
On resolving the proteins by 2-DE, 43 differentially expressed spots were corresponded to 28 identified proteins as given in Supplementary Table 2. Figure 3 illustrates number of differentially expressed upregulated and downregulated proteins. They are classified into 12 broad categories based on their function as given in Fig. 2. Major alterations were observed in protein binding (16%), transporter (10%) and enzyme binding (7%) proteins. As viruses are known to manipulate metabolic processes of the host for viral genome replication, number of host metabolic proteins are expected to be altered.
Fig. 3.
Number of differentially expressed proteins along with their up/down regulation in HBV or HCV related liver diseases
As given in Fig. 1, we found Alpha-2-HS-glycoprotein (AHSG) the most upregulated protein in all the liver diseased conditions due to HBV and HCV. Alpha-2-HS-glycoprotein is produced predominantly by the liver. AHSG is a transforming growth factor-β (TGF-β) type II receptor mimic and cytokine antagonist [21]. Increased Alpha-2-HS-glycoprotein is not showing any trend with HBV or HCV chronicity which is in accordance with previous reports [19, 36].
Apolipoprotein A1 (ApoA1), mainly secreted by hepatocytes, represents the major protein component of high-density lipoprotein. ApoA1 secretion may be disrupted by HBV infection. Wang et al. [40] suggested that HBV can inhibit apoA1 mRNA and decreased protein levels in hepatoma cells. Our experiments have shown that the level of apoA1 was downregulated more than two folds in HBV related liver diseases when compared to healthy individuals as reported earlier [18]. Ma et al. suggested that ApoA-1 can inhibit tumor cell proliferation through cell cycle arrest and promote apoptosis by inhibiting mitogen-activated protein kinase (MAPK) pathway. ApoA-1 might decrease angiogenesis by impairing with extracellular matrix degradation properties of tumor cells. But, the level of apoA1 was increased in HCV related HCC which is in concordance with findings from previous studies [7, 9]. Taken together, our findings indicate that decreased serum ApoA-1 can be useful for HBV HCC detection [22].
Transthyretin (also called as prealbumin), a protein with short half-life, is synthesized and secreted by liver cells and choroid plexus of the brain. Transportation of thyroxine (T4) and retinol (vitamin A) are the two significant physiological functions of TTR. Considering the fact that the liver is a source of transthyretin, its synthesis varies in liver diseases such as hepatitis and cancer [28]. Transthyretin has been found to be at significantly reduced levels in various liver diseases. In our study, a more acidic variant of the two transthyretin isoforms was decreased in chronic hepatitis, liver cirrhosis and HCC due to HBV as reported previously [31].
Leucin rich alpha 2 glycoprotein is an acute-phase protein and oncogene associated protein. It is capable of accelerating angiogenesis by activating Smad1/5/8 signalling pathway through binding to the TGF-β accessory receptor endoglin [38]. Wang et al. [39] revealed its up-regulation and increased mobility of HCC cells, suggesting its potential role in the progression. In our study it is 4.4 fold expressed in patients with HBV HCC which is in accordance with earlier findings and may be used as candidate marker for HCC diagnosis [5].
We observed 5.0 and 5.4 fold increase in levels of serum paraoxonase (arylesterase) 1 in chronic hepatitis and HCC due to HBV, respectively. PON1 is a calcium-dependent hydrolase protein synthesized mainly in the liver and secreted into the circulatory system [26]. Huang et al. [17] and Zhang et al. [42] have reported its active role in the regulation of oxidative stress, fibrosis and hepatic cell apoptosis in chronic liver diseases and, can be used as diagnostic biomarker for distinguishing early HCC from LC patients.
Ceruloplasmin (Cp) is a glycoprotein and metal (copper) binding circulating protein which exhibits oxidase activity and oxidizes Fe+2 to Fe+3 that can then be stored as ferritin when transported into cells by transferrin. Through this ferroxidase activity, ceruloplasmin plays an important role in iron metabolism. Changes in Cp leads to iron accumulation in the liver and finally to liver damage. Pousset et al. [25] have shown prolonged presence of Cp is due to higher amount N-glycan sialylation in murine model and thus increase in its concentration. Moderately increased levels of Cp have been reported to occur in human HCC and to be of prognostic value with elevated plasma Cp concentrations corresponding to more rapidly progressing tumours. Our findings in HBV-HCC cases, levels are increased sevenfold (Supplementary Table 2) when compared to controls as in concordance with these findings which can be exclusively used for HBV HCC diagnosis [15].
Alpha 1 Antitrypsin and alpha-1-antichymotrypsin, acute-phase proteins, acting as protease inhibitors protecting tissues from the enzymatic action of inflammatory cells. Mondal et al. [23] have reported significantly high AAT levels was in HCV LC and HCV HCC than control but less in HCV Chronic. Also, as reported by Kimhofer et al. [20], we have found increased levels of Alpha 1 Antitrypsin and alpha-1-antichymotrypsin in liver cirrhosis and HCC patients in comparison with chronic infection. Also, these proteins were differentially expressed in HBV HCC and HCV HCC. In similar study by Sarvari et al. has shown 42 differentially expressed spots which corresponded to 16 proteins in variable liver diseased conditions in HBV and HCV infection in Iranian population. In discordance to their study, we have found Alpha-2-HS-glycoprotein differentially expressed in chronic hepatitis B and C infection. In addition, several differentially expressed proteins are identified in each group in present study which may reflect on ethnicity of the heterogeneous population and also different strains of circulating viruses. They have concluded that upregulation of leucine rich alpha 2 in HBV HCC compared with HCV HCC may have potential to distinguish viral HCC which is in accordance with our findings [27].
Enrichment analysis using Reactome and Biological process (GO) databases highlighted significance of the identified proteins in several cellular pathways as summarized in Supplementary Tables 3 and 4 which were mainly associated with lipid metabolism. The prediction of possible involvement of identified proteins in pathways associated with HCC development influenced by HBV or HCV constituted 20% of the altered proteins in this study. The liver is highly metabolically active organ and, one of its key functions is to control the balance of lipid throughout the body. The present study confirms functional association of APOA1, APOA4, APOE, PON1, A2M and ALB with lipid transport, lipid metabolism, regulation and lipid homeostasis. Apart from lipid transport apolipoproteins play a wider role in cancers and are known to interact with diverse receptors to elicit cellular events as demonstrated for APOE to cause sustained proliferation of cells with progressive pathogenesis of HBV-related liver disease [4, 30]. Similarly, several studies have shown that lipid metabolic events play a key role in HCV life cycle in promoting liver disease pathogenesis [33, 34].
In addition to pathway analysis using EnrichNet, we further studied various protein–protein interactions using STRING version 10.5 based on evidence and molecular action at medium confidence level and, confidence network edges for strength of data support at highest confidence level as shown in Supplementary Fig. 1.
Vitamin absorption and digestion pathways were found to be significant by EnrichNet analysis using KEGG. We also found complement and coagulation cascades as the most significantly enriched signalling pathways. This pathway involves 3 proteins (A2M, CFB, SERPINA1) which were up-regulated in the present study in HBV liver cirrhosis and HBV HCC when compared with controls. This cascade involving 3 proteins (C4B, CFB, SERPINA1) was also increased in HCV related chronic hepatitis, LC and HCC patients along with disease progression. It has been reported that complement and coagulation pathways are the most perturbed pathways in various cancers [3].
As a part of innate immunity, complement system plays a central role in clearance of immune complexes and apoptotic cells, anticancer defense and, the pathogenesis of variety of liver disorders including viral hepatitis, liver injury and repair. Zhang et al. demonstrated increasing evidence of complement component and complement activated product to promote tumour cell growth, tumour angiogenesis, and immunosuppression in HCC using bioinformatics analysis. The up-regulation of proteins in complement cascade in our study also supported the latest reports [14, 43].
Highest confident string analysis (confidence score > 0.90) showed a functional link between SERPINA1 and AHSG (combined confidence score = 0.952) with their increased levels in HBV/HCV related HCC as given in Supplementary Table 2. Another interesting functional link was observed between APOA1 and SAA1 (combined confidence score = 0.938). APOA1 aids HDL to exert anti-cancer effects by mediating cellular cholesterol and phospholipid efflux by limiting the availability of essential lipids for rapidly proliferating cells [2]. SAA1, acute-phase apolipoprotein and minor constituent of HDL is elevated dramatically in acute inflammatory viral infection, which causes major change in apolipoprotein composition of HDL [24]. APOA1 is substantially reduced and replaced by SAA1, thus altering the role of HDL helping cancer cell proliferation as depicted in supplementary Fig. 2 [8, 24]. Another link was between APOA1 and APCS (combined confidence score = 0.945) in HCV related liver diseases. APCS may participate in promoting cholesterol efflux and lipid metabolism [16]. We have found highest functional link between complement components in HCV liver diseases. Increased levels of C4A/B and CFB in liver cirrhosis and HCC due to HCV suggest immune activation due to HCV infection, and/or tumor antigens [3].
In conclusion, our study made an attempt to identify differentially expressed serum proteins in HBV and HCV related diseases using proteomic approach. Haptoglobin protein levels were decreased upon disease progression to HCC in HBV infection in correspondence with controls. The other proteins expressed differentially are ceruloplasmin, serum paraoxonase 1, retinol binding protein and leucine rich alpha 2 proteins in plasma maybe associated to HBV HCC. Whereas, upregulation of C4a/C4b showed it as a reliable marker in patients with end stage liver disease related to HCV infection. Transthyretin level was decreased among HCC than cirrhotic in HBV. Exclusively in HCV LC, increase in Vitamin D binding protein and complement C3/C5 may be used for early diagnosis of HCV HCC. ApolipoproteinA1 levels in liver diseases in both HBV and HCV infection corresponding healthy controls may be candidate marker for early diagnosis and disease monitoring. Pathway analysis, protein–protein interactions and functional link between 28 differentially expressed proteins look promising to understand changes occurring due to viral hepatitis and to develop diagnostic assay. However, further study on larger basis may be undertaken to validate our findings.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 2 (DOCX 13812 kb)
Acknowledgements
The authors thank Lady Tata Memorial Trust, Mumbai for providing Research Scholarship to Ms. Kruti Dalal and Haffkine institute for utilization of proteomic setup. The study was supported by intramural funding of Indian Council of Medical Research (ICMR).
Compliance with ethical standards
Conflict of interest
Authors have no conflict of interest.
Contributor Information
Avinash Kale, Phone: +91-22-2653 2134, Email: avinash.kale@cbs.ac.in.
Aruna Shankarkumar, Phone: +91-22-2411161, Email: arp21@rediffmail.com.
References
- 1.Acharya SK. Epidemiology of hepatocellular carcinoma in India. J Clin Exp Hepatol. 2014;4:S27–S33. doi: 10.1016/j.jceh.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen C, Murray K, Ragonesi N, Doerr A, Cintron-Rivera L, Dupree L. HDL-associated proteins and LDL differentially modulate chronic myelogenous leukemia cell viability. J Clin Lipidol. 2017;11:793–794. doi: 10.1016/j.jacl.2017.04.036. [DOI] [PubMed] [Google Scholar]
- 3.Awan FM, Naz A, Obaid A, Ali A, Ahmad J, Anjum S, et al. Identification of circulating biomarker candidates for hepatocellular carcinoma (HCC): an integrated prioritization approach. PLoS ONE (Internet). 2015;10. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4586137/. [DOI] [PMC free article] [PubMed]
- 4.Borlak J, Singh P, Gazzana G. Proteome mapping of epidermal growth factor induced hepatocellular carcinomas identifies novel cell metabolism targets and mitogen activated protein kinase signalling events. BMC Genom (Internet). 2015;16. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4357185/. [DOI] [PMC free article] [PubMed]
- 5.Chaerkady R, Thuluvath PJ, Kim M-S, Nalli A, Vivekanandan P, Simmers J, et al. 18O labeling for a quantitative proteomic analysis of glycoproteins in hepatocellular carcinoma. Clin Proteomics. 2008;4:137–155. doi: 10.1007/s12014-008-9013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corona. Differential proteomic analysis of hepatocellular carcinoma. Int J Oncol (Internet). 2009 [cited 2017 Dec 28];36. http://www.spandidos-publications.com/ijo/36/1/93. [PubMed]
- 7.Derbyshire E, Hungin A, Nickerson C. UEG Week 2015 poster presentations. United Eur Gastroenterol J. 2015;2015(3):146–687. [Google Scholar]
- 8.Digre A, Nan J, Frank M, Li J-P. Heparin interactions with apoA1 and SAA in inflammation-associated HDL. Biochem Biophys Res Commun. 2016;474:309–314. doi: 10.1016/j.bbrc.2016.04.092. [DOI] [PubMed] [Google Scholar]
- 9.Dillon ST, Bhasin MK, Feng X, Koh DW, Daoud SS. Quantitative proteomic analysis in HCV-induced HCC reveals sets of proteins with potential significance for racial disparity. J Transl Med. 2013;11:239. doi: 10.1186/1479-5876-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehsani Ardakani MJ, Safaei A, Arefi Oskouie A, Haghparast H, Haghazali M, Mohaghegh Shalmani H, et al. Evaluation of liver cirrhosis and hepatocellular carcinoma using protein–protein interaction networks. Gastroenterol Hepatol Bed Bench. 2016;9:S14–S22. [PMC free article] [PubMed] [Google Scholar]
- 11.European association for the Study of the Liver. European Organisation for Research and Treatment of Cancer EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Fang M, Zhao Y-P, Zhou F-G, Lu L-G, Qi P, Wang H, et al. N-glycan based models improve diagnostic efficacies in hepatitis B virus-related hepatocellular carcinoma. Int J Cancer. 2010;127:148–159. doi: 10.1002/ijc.25030. [DOI] [PubMed] [Google Scholar]
- 13.Ferrín G, Ranchal I, Llamoza C, Rodríguez-Perálvarez ML, Romero-Ruiz A, Aguilar-Melero P, et al. Identification of candidate biomarkers for hepatocellular carcinoma in plasma of HCV-infected cirrhotic patients by 2-D DIGE. Liver Int. 2014;34:438–446. doi: 10.1111/liv.12277. [DOI] [PubMed] [Google Scholar]
- 14.Ferrín G, Rodríguez-Perálvarez M, Aguilar-Melero P, Ranchal I, Llamoza C, Linares CI, et al. Plasma protein biomarkers of hepatocellular carcinoma in HCV-infected alcoholic patients with cirrhosis. PLoS ONE. 2015;10:e0118527. doi: 10.1371/journal.pone.0118527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonçalves LDR, Campanhon IB, Domingues RR, Paes Leme AF, Soares da Silva MR. Comparative salivary proteome of hepatitis B- and C-infected patients. PLoS ONE (Internet). 2014;9. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4244100/. [DOI] [PMC free article] [PubMed]
- 16.Xi D, Zhao J, Liu J, Xiong H, He W, Hu J, et al. The impact of serum amyloid P-component on gene expression in RAW264.7 mouse macrophages. BioMed Res Int (Internet). 2016;2016. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4864538/. [DOI] [PMC free article] [PubMed]
- 17.Huang C, Wang Y, Liu S, Ding G, Liu W, Zhou J, et al. Quantitative proteomic analysis identified paraoxonase 1 as a novel serum biomarker for microvascular invasion in hepatocellular carcinoma. J Proteome Res. 2013;12:1838–1846. doi: 10.1021/pr3011815. [DOI] [PubMed] [Google Scholar]
- 18.Katrinli S, Ozdil K, Sahin A, Ozturk O, Kir G, Baykal AT, et al. Proteomic profiling of HBV infected liver biopsies with different fibrotic stages. Proteome Sci (Internet). 2016 [cited 2017 Oct 5];15. http://proteomesci.biomedcentral.com/articles/10.1186/s12953-017-0114-4. [DOI] [PMC free article] [PubMed]
- 19.Kawakami T, Hoshida Y, Kanai F, Tanaka Y, Tateishi K, Ikenoue T, et al. Proteomic analysis of sera from hepatocellular carcinoma patients after radiofrequency ablation treatment. Proteomics. 2005;5:4287–4295. doi: 10.1002/pmic.200401287. [DOI] [PubMed] [Google Scholar]
- 20.Kimhofer T, Fye H, Taylor-Robinson S, Thursz M, Holmes E. Proteomic and metabonomic biomarkers for hepatocellular carcinoma: a comprehensive review. Br J Cancer. 2015;112:1141–1156. doi: 10.1038/bjc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Gu X, Fang M, Ji J, Yi C, Gao C. The diagnostic value of serum fucosylated fetuin A in hepatitis B virus-related liver diseases. Clin Chem Lab Med CCLM [Internet]. 2016 [cited 2017 Dec 28];54. https://www.degruyter.com/view/j/cclm.2016.54.issue-4/cclm-2015-0307/cclm-2015-0307.xml. [DOI] [PubMed]
- 22.Ma X-L, Gao X-H, Gong Z-J, Wu J, Tian L, Zhang C-Y, et al. Apolipoprotein A1: a novel serum biomarker for predicting the prognosis of hepatocellular carcinoma after curative resection. Oncotarget. 2016;7:70654. doi: 10.18632/oncotarget.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mondal G, Saroha A, Bose PP, Chatterjee BP. Altered glycosylation, expression of serum haptoglobin and alpha-1-antitrypsin in chronic hepatitis C, hepatitis C induced liver cirrhosis and hepatocellular carcinoma patients. Glycoconj J. 2016;33:209–218. doi: 10.1007/s10719-016-9658-2. [DOI] [PubMed] [Google Scholar]
- 24.Ni X-C, Yi Y, Fu Y-P, He H-W, Cai X-Y, Wang J-X, et al. Serum amyloid A is a novel prognostic biomarker in hepatocellular carcinoma. Asian Pac J Cancer Prev APJCP. 2014;15:10713–10718. doi: 10.7314/APJCP.2014.15.24.10713. [DOI] [PubMed] [Google Scholar]
- 25.Pousset D, Piller V, Bureaud N, Piller F. High levels of ceruloplasmin in the serum of transgenic mice developing hepatocellular carcinoma. Eur J Biochem. 2001;268:1491–1499. doi: 10.1046/j.1432-1327.2001.02015.x. [DOI] [PubMed] [Google Scholar]
- 26.Précourt L-P, Amre D, Denis M-C, Lavoie J-C, Delvin E, Seidman E, et al. The three-gene paraoxonase family: physiologic roles, actions and regulation. Atherosclerosis. 2011;214:20–36. doi: 10.1016/j.atherosclerosis.2010.08.076. [DOI] [PubMed] [Google Scholar]
- 27.Sarvari J, Mojtahedi Z, Taghavi SAR, Kuramitsu Y, Shamsi Shahrabadi M, Ghaderi A, et al. Differentially expressed proteins in chronic active hepatitis, cirrhosis, and HCC related to HCV infection in comparison with HBV infection: a proteomics study. Hepat Mon (Internet). 2013;13. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3776151/. [DOI] [PMC free article] [PubMed]
- 28.Sarvari J, Mojtahedi Z, Kuramitsu Y, Fattahi MR, Ghaderi A, Nakamura K, et al. Comparative proteomics of sera from HCC patients with different origins. Hepat Mon (Internet). 2014;14. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3909643/. [DOI] [PMC free article] [PubMed]
- 29.Shamsi Shahrabadi M. Differential expression of haptoglobin isoforms in chronic active hepatitis, cirrhosis and HCC related to HBV infection. Oncol Lett (Internet). 2011 [cited 2017 Dec 28]; http://www.spandidos-publications.com/10.3892/ol.2011.321. [DOI] [PMC free article] [PubMed]
- 30.Shen Y, Li M, Ye X, Bi Q. Association of apolipoprotein E with the progression of hepatitis B virus-related liver disease. Int J Clin Exp Pathol. 2015;8:14749–14756. [PMC free article] [PubMed] [Google Scholar]
- 31.Sugimoto K, Shiraki K, Takei Y, Ito M, Nobori T, Suzuki H, et al. Serum protein isoform profiles indicate the progression of hepatitis C virus-induced liver diseases. Int J Mol Med. 2013;31:943–950. doi: 10.3892/ijmm.2013.1267. [DOI] [PubMed] [Google Scholar]
- 32.Sun W, Xing B, Sun Y, Du X, Lu M, Hao C, et al. Proteome analysis of hepatocellular carcinoma by two-dimensional difference gel electrophoresis novel protein markers in hepatocellular carcinoma tissues. Mol Cell Proteomics. 2007;6:1798–1808. doi: 10.1074/mcp.M600449-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Syed GH, Amako Y, Siddiqui A. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab TEM. 2010;21:33–40. doi: 10.1016/j.tem.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Targett-Adams P, Boulant S, Douglas MW, McLauchlan J. Lipid metabolism and HCV infection. Viruses. 2010;2:1195–1217. doi: 10.3390/v2051195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai T-H, Song E, Zhu R, Di Poto C, Wang M, Luo Y, et al. LC-MS/MS-based serum proteomics for identification of candidate biomarkers for hepatocellular carcinoma. Proteomics. 2015;15:2369–2381. doi: 10.1002/pmic.201400364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voelkl J, Pakladok T, Lin Y, Viereck R, Lebedeva A, Kukuk D, et al. Up-regulation of hepatic alpha-2-HS-glycoprotein transcription by testosterone via androgen receptor activation. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. 2014;33:1911–1920. doi: 10.1159/000362968. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Zhang A, Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatol Baltim Md. 2013;57:2072–2077. doi: 10.1002/hep.26130. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Abraham S, McKenzie JAG, Jeffs N, Swire M, Tripathi VB, et al. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature. 2013;499:306–311. doi: 10.1038/nature12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C-H, Li M, Liu L-L, Zhou R-Y, Fu J, Zhang CZ, et al. LRG1 expression indicates unfavorable clinical outcome in hepatocellular carcinoma. Oncotarget. 2015;6:42118. doi: 10.18632/oncotarget.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Hao J, Liu X, Wang H, Zeng X, Yang J, et al. The mechanism of apoliprotein A1 down-regulated by Hepatitis B virus. Lipids Health Dis. 2016;15:64. doi: 10.1186/s12944-016-0232-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu W, Li J, Liu Y, Zhang C, Meng X, Zhou Z. Comparative proteomic studies of serum from patients with hepatocellular carcinoma. J Invest Surg. 2012;25:37–42. doi: 10.3109/08941939.2011.603816. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S, Jiang K, Zhang Q, Guo K, Liu Y. Serum fucosylated paraoxonase 1 as a potential glycobiomarker for clinical diagnosis of early hepatocellular carcinoma using ELISA Index. Glycoconj J. 2015;32:119–125. doi: 10.1007/s10719-015-9576-8. [DOI] [PubMed] [Google Scholar]
- 43.Zhang C, Peng L, Zhang Y, Liu Z, Li W, Chen S, et al. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med Oncol Northwood Lond Engl (Internet). 2017;34. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5400790/. [DOI] [PMC free article] [PubMed]
- 44.Zhu RX, Seto W-K, Lai C-L, Yuen M-F. Epidemiology of hepatocellular carcinoma in the Asia–Pacific region. Gut Liver. 2016;10:332–339. doi: 10.5009/gnl15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 2 (DOCX 13812 kb)