Abstract
Several studies reported a complex interplay between viral infections and neural cells leading to multiple sclerosis. A role for some viral infections has been proposed in MS. In this study, DNA sequences of human herpesvirus 8 (HHV-8) were searched in the peripheral blood of 54 patients with multiple sclerosis and 130 healthy subjects using nested-PCR assay to amplify ORF26 locus. Furthermore, HHV-8 positive samples were subjected to a nested-PCR to amplify K1 gene of HHV-8 followed by direct nucleotide sequencing. HHV-8 genome was detected in 18.5% (10/54) and 3% (4/130) of MS patients and controls, respectively, and the difference reached statistically significant level (P = 0.0017). Genotyping analysis revealed that genotype C was common (88.9%) in all study subjects, followed by genotype A. Our results showed higher detection of HHV-8 DNA in MS patients than control group.
Electronic supplementary material
The online version of this article (10.1007/s13337-018-0481-1) contains supplementary material, which is available to authorized users.
Keywords: Multiple sclerosis, Human herpesvirus 8, Genotypes
Multiple sclerosis (MS) is a demyelinating and neurodegenerative disease of the brain [16]. Although the precise mechanism which triggers the disease is still inconclusive, both environmental and genetic factors are involved in the etiology of MS [10]. A role for some viral infections, such as herpesviruses and human endogenous retroviruses, has been proposed in MS [10, 11].
Human herpesvirus 8 (HHV-8) is considered as an etiological agent of Kaposi’s sarcoma and two kind of lymphoma, including primary effusion lymphoma and Multicentric Castleman disease [1, 5]. Detection of HHV-8 in brain tissue also indicates the neurotropic nature of this virus. Indeed, it has been recently documented that HHV-8 can infect neurons and oligodendrocytes in parenchymal brain tissues [18]. Growing evidence also indicate that HHV-8 is able to invade the CNS, consequently leading to the neural diseases. In this regard, HHV-8 has been detected in encephalitis among HIV-positive and -negative subjects, CNS lymphomas, and amyotrophic lateral sclerosis [15]. Moreover, few studies suggest the possible role of HHV-8 in MS pathogenesis [12–14].
Globally, phylogenetic studies based on the HHV-8 ORF K1 sequences identified five major genotypes, including: A and C, which are mainly found in USA and Eurasia; B and A5, spread in Africa; D, is mostly common in the Pacific islands and Australia; and E that is dominant among the Amerindian populations [3, 6, 19]. It is hypothesized that distinct HHV-8 genotypes may exert different pathogenic properties. To support this notion, it is shown that genotype A of HHV-8 progress more rapidly in classic KS patients [7]. Transmission of HHV-8 A5 genotype seems to occur more easily than the B genotype from mother-to-child in African population [8]. Furthermore, it is shown that diverse HHV-8 genotypes exert different tropism toward cell lines [8]. However, no studies investigated whether distinct HHV-8 genotypes may show different tendency towards neurons.
To our knowledge, this is the first report investigating the association between HHV-8 and MS in Iranian population. Also, this study is the first report evaluating HHV-8 genotypes distribution in MS patients in the world. The aim of this study was to both estimate the genoprevalence of HHV-8 in peripheral blood mononuclear cells (PBMCs) and characterize the HHV-8 genotypes of MS patients and healthy individuals.
Fifty-four patients with relapsing–remitting multiple sclerosis and 130 healthy individuals were included in this study. All patients were attending and had been diagnosed based on McDonald’s criteria and assessed for clinical disability (Expanded Disability Status Scale: EDSS) by clinicians at the MS Research Centre, Neuroscience Institute, Tehran, Iran [11]. All study participants signed the informed consent form to donate 5 mL blood samples. This study was approved by the Local Research Ethics Committee of Tehran University of Medical Sciences (TUMS) with Approval No. 95-02-27-32301.
DNA extraction was performed by the High Pure Viral Nucleic Acid Kit (Roche, Mannheim, Germany) according to manufacturer’s instructions. The extracted DNA was eluted in 50 μL of elution buffer and stored at − 70 °C until the use. To detect HHV-8 DNA, a nested PCR was performed to amplify a 172 bp fragment of ORF26. Then, HHV-8 positive samples were subjected to a semi-nested PCR to amplify a 344 bp of HHV-8 ORF K1. The sequence of primers, the PCR reaction conditions, and the PCR thermal regime was summarized in Supplementary Table 1.
The PCR amplification products of ORF K1 were sequenced as described previously [19]. By MEGA6.06 software, phylogenetic tree was made using the Maximum Likelihood method based on the Kimura 2-parameter model with 1000 boot strap value.
Statistical analysis was performed by EPI Info 7 Statistical Analysis System Software (Atlanta, GA, USA) using the χ2 test or the Fisher exact test. When the P values were less than 0.05, they were considered statistically significant.
The prevalence of HHV-8 in fifty-four patients with relapsing–remitting multiple sclerosis (15 male and 39 female) and 130 healthy individuals (80 male and 50 female) were investigated in this study. The mean age was 35.4 ± 7.6 and 41 ± 7.2 in MS patients and healthy individuals, respectively.
HHV-8 genome was detected in 18.5% (10/54) and 3% (4/130) of MS patients and healthy controls, respectively, and the difference reached statistically significant level (P = 0.0017). As shown in Supplementary Table 2, however, no significant differences were found between HHV-8 positivity and almost all variables, except for having family history of autoimmune diseases in MS patients (P = 0.042). The rate of HHV-8 infection was 45.4% among MS patients that have family history of autoimmune disorders, whereas this rate was 11.6% in those patients with no family history of autoimmune diseases. Although statistically no significant differences were found for most variables, the HHV-8 prevalence was higher in males, age group < 40 years, and those have family history of MS disease (Supplementary Table 2). In healthy subjects, no significant differences were found between HHV-8 positivity and demographic variables (P > 0.05).
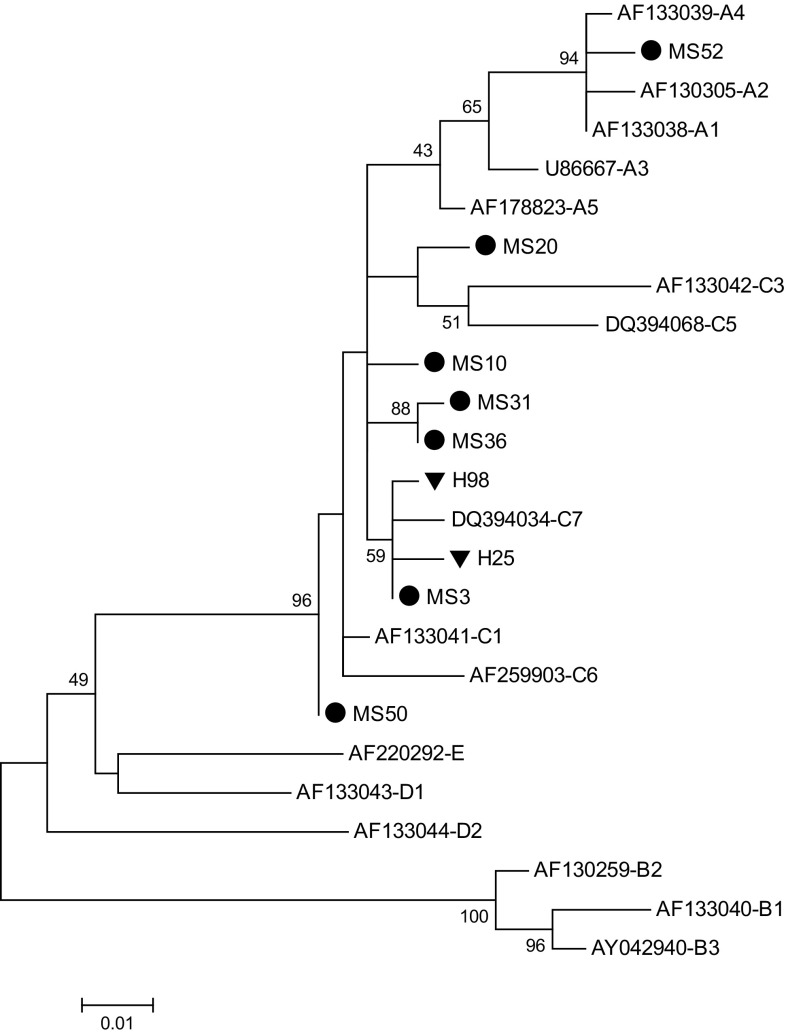
Seven out of 10 HHV-8 positive samples from MS patients and 2 out of 4 HHV-8 positive samples in healthy individuals were successfully amplified for the K1 gene and PCR products subjected for sequencing analysis (Fig. 1). In overall, genotype C was common (8 out of 9; 88.9%) in all study subjects and the remaining (1 out of 9; 10.1%) was genotype A. According to the classification pioneered by Cook et al. [2], 7 out of 8 samples infected with C genotype were classified in C″ subgroup and only one samples fall into C′ subgroup. Sample with A genotype was belonged to A′ subgroup.
Fig. 1.
Phylogenetic analysis of HHV-8 K1 gene on samples of multiple sclerosis (MS) patients and healthy subjects (designated with MS and H, respectively) were conducted in MEGA6.06, using the Maximum Likelihood method based on the Kimura 2-parameter model. All reference sequences were extracted from GenBank. Numbers above the branches indicate the bootstrap values
The analysis of the HHV-8 DNA in PBMCs of MS patients and healthy subjects revealed that 18.5% and 3% of them were positive, respectively (P = 0.0017). This finding is in agreement with few studies in this area that have detected HHV-8 in MS patients [9, 12–14]. Indeed, Merelli et al. [9] have shown that HHV-8 DNA is present in 80% and 37.5% of brain tissue autopsies among MS patients and controls, respectively. Looking at the HHV-8 mRNA in brain tissue, Opsahl and Kennedy [13] have shown the HHV-8 mRNA detection in 50% of MS patients and 33.3% of controls.
Our results indicate that genotypes C and A were present in our study groups though genotype C was more common (Fig. 1). This result is accordance to earlier studies in Iranian Kaposi’s sarcoma patients that have shown that genotype C is prominent genotype in Iran [6, 19]. Evolutionarily, B genotype is appeared around 100,000 years ago in Africa although D and E genotypes arose about 60,000 years ago on the Pacific Islands and Amerindian populations, respectively. A and C genotypes appear to emerge about 35,000 years ago in Eurasia [4, 19, 20]. Several studies have revealed that genotype C is more prevalent that genotype A in Asia and Middle East [6, 20]. However, our result has not supported this concept that distinct HHV-8 genotypes may exert different tendency towards neurons. It has been previously indicated that genotype A replicates faster and shows more tropism to epithelial cells rather than PBMCs when compared to genotype C [8].
In this study, most of the genotype C (88.9%) fall into the C″ subgroup and only 10.1% belong to C′ subgroup that is consistent with prevoius studies from Middle Eastern/North African countries [6, 19, 20]. It is thought that C″ subgroup is more ancient than C′ subgroup which seems to arise in the Middle Eastern/North African regions although the C′ subgroup might be representative of European and North Asian migrations [6, 17].
In conclusion, our results showed higher detection of HHV-8 genome in MS patients than control group. However, future studies with larger sample sizes and evaluation of immune status of HHV-8 are warranted.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study has been funded and supported by Tehran University of Medical Sciences (TUMS), Grant Nos. 95-02-27-32301 and 93-03-27-26793.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Cook PM, Whitby D, Calabro ML, Luppi M, Kakoola DN, Hjalgrim H, Ariyoshi K, Ensoli B, Davison AJ, Schulz TF. Variability and evolution of Kaposi’s sarcoma-associated herpesvirus in Europe and Africa. International Collaborative Group, Aids. 1999;13:1165–1176. doi: 10.1097/00002030-199907090-00004. [DOI] [PubMed] [Google Scholar]
- 3.da Ramos SS, Ferraz da Silva AP, Bacchi MM, Bacchi CE, de Elgui OD. KSHV genotypes A and C are more frequent in Kaposi sarcoma lesions from Brazilian patients with and without HIV infection, respectively. Cancer Lett. 2011;301:85–94. doi: 10.1016/j.canlet.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Hayward GS, Zong JC. Modern evolutionary history of the human KSHV genome. Curr Top Microbiol Immunol. 2007;312:1–42. doi: 10.1007/978-3-540-34344-8_1. [DOI] [PubMed] [Google Scholar]
- 5.Jalilvand S, Shoja Z, Mokhtari-Azad T, Nategh R, Gharehbaghian A. Seroprevalence of Human herpesvirus 8 (HHV-8) and incidence of Kaposi’s sarcoma in Iran. Infect. Agent. Cancer. 2011;6:5. doi: 10.1186/1750-9378-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jalilvand S, Tornesello ML, Buonaguro FM, Buonaguro L, Naraghi ZS, Shoja Z, Ziaee AA, Hamkar R, Shahmahmoodi S, Nategh R, Mokhtari-Azad T. Molecular epidemiology of human herpesvirus 8 variants in Kaposi’s sarcoma from Iranian patients. Virus Res. 2012;163:644–649. doi: 10.1016/j.virusres.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 7.Mancuso R, Biffi R, Valli M, Bellinvia M, Tourlaki A, Ferrucci S, Brambilla L, Delbue S, Ferrante P, Tinelli C, Clerici M. HHV8 a subtype is associated with rapidly evolving classic Kaposi’s sarcoma. J Med Virol. 2008;80:2153–2160. doi: 10.1002/jmv.21322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matteoli B, Broccolo F, Scaccino A, Cottoni F, Angeloni A, Faggioni A, Ceccherini-Nelli L. In vivo and in vitro evidence for an association between the route-specific transmission of HHV-8 and the virus genotype. J Med Virol. 2012;84:786–791. doi: 10.1002/jmv.23246. [DOI] [PubMed] [Google Scholar]
- 9.Merelli E, Bedin R, Sola P, Barozzi P, Mancardi GL, Ficarra G, Franchini G. Human herpes virus 6 and human herpes virus 8 DNA sequences in brains of multiple sclerosis patients, normal adults and children. J Neurol. 1997;244:450–454. doi: 10.1007/s004150050121. [DOI] [PubMed] [Google Scholar]
- 10.Mostafa A, Jalilvand S, Shoja Z, Nejati A, Shahmahmoodi S, Sahraian MA, Marashi SM. Multiple sclerosis-associated retrovirus, Epstein-Barr virus, and vitamin D status in patients with relapsing remitting multiple sclerosis. J Med Virol. 2017;89:1309–1313. doi: 10.1002/jmv.24774. [DOI] [PubMed] [Google Scholar]
- 11.Nejati A, Shoja Z, Shahmahmoodi S, Tafakhori A, Mollaei-Kandelous Y, Rezaei F, Hamid KM, Mirshafiey A, Doosti R, Sahraian MA, Mahmoudi M, Shokri F, Emery V, Marashi SM. EBV and vitamin D status in relapsing-remitting multiple sclerosis patients with a unique cytokine signature. Med Microbiol Immunol. 2016;205:143–154. doi: 10.1007/s00430-015-0437-7. [DOI] [PubMed] [Google Scholar]
- 12.Olut AI, Ozunlu H, Tan E, Kocagoz T. Human herpesvirus-8 DNA in patients with certain demyelinating disorders. Mikrobiyoloji bulteni. 2005;39:169–174. [PubMed] [Google Scholar]
- 13.Opsahl ML, Kennedy PG. Investigating the presence of human herpesvirus 7 and 8 in multiple sclerosis and normal control brain tissue. J Neurol Sci. 2006;240:37–44. doi: 10.1016/j.jns.2005.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietropaolo V, Fioriti D, Mischitelli M, Anzivino E, Santini M, Millefiorini E, Di Rezze S, Degener AM. Detection of human herpesviruses and polyomaviruses DNA in a group of patients with relapsing-remitting multiple sclerosis. The new microbiologica. 2005;28:199–203. [PubMed] [Google Scholar]
- 15.Sola P, Bedin R, Casoni F, Barozzi P, Mandrioli J, Merelli E. New insights into the viral theory of amyotrophic lateral sclerosis: study on the possible role of Kaposi’s sarcoma-associated virus/human herpesvirus 8. Eur Neurol. 2002;47:108–112. doi: 10.1159/000047961. [DOI] [PubMed] [Google Scholar]
- 16.Stangel M, Kuhlmann T, Matthews PM, Kilpatrick TJ. Achievements and obstacles of remyelinating therapies in multiple sclerosis. Nature Reviews Neurology. 2017;13:742. doi: 10.1038/nrneurol.2017.139. [DOI] [PubMed] [Google Scholar]
- 17.Tornesello ML, Biryahwaho B, Downing R, Hatzakis A, Alessi E, Cusini M, Ruocco V, Katongole-Mbidde E, Loquercio G, Buonaguro L, Buonaguro FM. Human herpesvirus type 8 variants circulating in Europe, Africa and North America in classic, endemic and epidemic Kaposi’s sarcoma lesions during pre-AIDS and AIDS era. Virology. 2010;398:280–289. doi: 10.1016/j.virol.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Tso FY, Sawyer A, Kwon EH, Mudenda V, Langford D, Zhou Y, West J, Wood C. Kaposi’s sarcoma-associated herpesvirus infection of neurons in HIV-positive patients. J Infect Dis. 2017;215:1898–1907. doi: 10.1093/infdis/jiw545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varmazyar S, Shoja Z, Kakavand-Ghalehnoei R, Shahmahmoodi S, Marashi SM, Jalilvand S. Molecular typing of human herpesvirus 8 among HIV positive in comparison to HIV-negative individuals in Iran. J Med Virol. 2017;89:703–709. doi: 10.1002/jmv.24644. [DOI] [PubMed] [Google Scholar]
- 20.Zong JC, Ciufo DM, Alcendor DJ, Wan X, Nicholas J, Browning PJ, Rady PL, Tyring SK, Orenstein JM, Rabkin CS, Su IJ, Powell KF, Croxson M, Foreman KE, Nickoloff BJ, Alkan S, Hayward GS. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi’s sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73:4156–4170. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.