Abstract
Vaccination with canine parvovirus-2 (CPV-2) modified live attenuated vaccine remains an effective control strategy for preventing parvovirus induced enteritis in dogs. Virus shedding is a common phenomenon few days after vaccination, possessing a diagnostic dilemma for accurate differentiation of CPV-2 vaccine and wild type field strains. Though several molecular approaches are available for differentiation, the present study focuses on a simple, rapid, cost-effective differentiating infected from vaccinated animals strategy employing ARMS-PCR for differentiation of CPV-2 vaccine and wild type field strains. The ARMS-PCR was initially validated using positive controls of recombinant plasmids, further used for screening six commercial CPV-2 vaccines and 24 archived CPV-2 positive field samples as well as to check fecal shedding of vaccine virus for 10 days post-vaccination in three vaccinated dogs. Sequencing of randomly selected CPV-2 commercial vaccine strains and archived field samples confirmed authenticity of the developed ARMS-PCR assay.
Keywords: CPV-2 vaccine, CPV-2 field strains, ARMS-PCR, Genetic DIVA
Canine parvoviral enteritis is the most common viral gastroenteric disease encountered in dogs worldwide [1, 9]. The disease is often fatal in young puppies with clinical manifestation of fever, anorexia, lethargy, vomiting and hemorrhagic gastroenteritis. Canine parvovirus-2 (CPV-2) has undergone mutations within VP2 gene giving rise to different antigenic variants; viz, CPV-2a, CPV-2b and CPV-2c [1, 2]. The original CPV-2 strain, which no longer circulates in the field, is still being used in most of the commercial vaccine formulations [10]. Vaccination with modified live attenuated vaccine is the most preferred way for controlling parvoviral infection in dogs. Fecal shedding of vaccine virus few days to couple of weeks post-vaccination is a common phenomenon [4, 6, 8]. There are instances of canine parvovirus outbreaks in vaccinated dogs probably due to interference of maternal antibodies in pups or lack of protective antibody titers against heterologous CPV antigenic types [4, 10]. Thus, exact determination of CPV-2 vaccine and field wild type strains is urgent requirement in CPV-2 diagnostics and for development of effective control strategies [8].
Several approaches have been employed over the years like PCR–RFLP (7) [7], MGB probe based real-time PCR [6] and HRM based real-time PCR [1]. Though, all the approaches have been successful in differentiating vaccine and field strains, they have suffered from some limitations. Requirement of sophisticated instruments, inherent cost of reagents and multi-step approach are some of the limiting factors of these assays. Thus, a simplified genetic differentiating infected from vaccinated animals (DIVA) approach based on Amplification Refractory Mutation System (ARMS-PCR) which overcomes these short comings was developed in this study for simultaneous detection and differentiation of CPV-2 vaccine and field strains.
Samples in the present study included 6 commercial vaccines consisting of original CPV-2 strain (VANGUARD®, Megavac-6, CANIGEN DHPPi/L, CANVAC® 8 DHPPiL, CaniShot® DHPPL, Nobivac®DHPPi), 24 archived clinical samples which included all the CPV-2 antigenic variants previously sequenced (CPV-2a, CPV-2b and CPV-2c) and post-vaccinated rectal swabs from three vaccinated dogs. Recombinant plasmid DNA containing full length VP2 gene from a commercial vaccine (CaniShot® DHPPL) and a field strain (Isolate KN-1/2015; Accession number: KX425922) were used as controls.
DNA from 24 archived clinical samples (obtained fromVirus Repository, CADRAD) was extracted using QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) as per manufacturer’s recommendations.
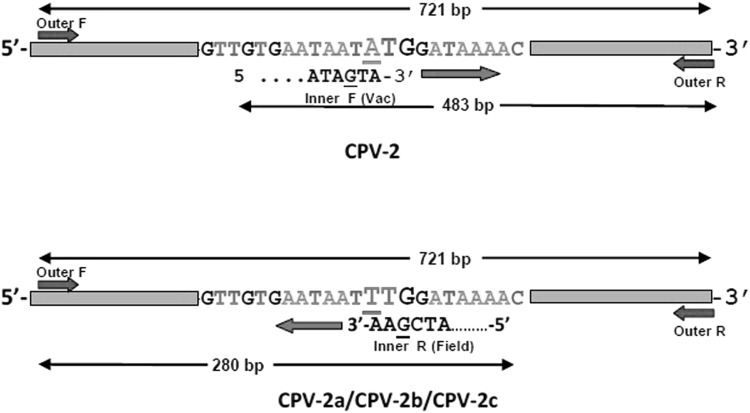
Mutation at A259T nucleotide position within VP-2 gene of parvoviral genome was targeted to differentiate CPV-2 (ATG) and field strains (TTG). The outer primers were designed within the conserved region of parvoviral genome, while inner forward and inner reverse primer were designed for vaccine (CPV-2) and field strains respectively. ARMS-PCR strategy has been illustrated in Fig. 1 with primer details in Table 1. The inner primers were specifically designed based on alignment report of sequenced CPV-2 vaccine strains and field isolates which included all the CPV-2 antigenic variants for avoiding non-specific binding and giving false negative results (Supplementary text 1).
Fig. 1.
Schematic representation of CPV ARMS-PCR strategy for differentiating vaccine (CPV-2) and field strains (CPV-2a/CPV-2b/CPV-2c) showing primer binding region and amplified product formed. The bold case (bigger font size) represents the target codon (position 259–261) and primer binding site. Deliberate mismatch of the nucleotide at − 2 from 3′ end position within primers is underlined
Table 1.
Primers used for ARMS-PCR differentiating vaccine and field strains showing primer sequence, position within the VP2 gene, amplicon size for vaccine and field strains (deliberate mismatch within the inner specific CPV-2 primer of vaccine and field strains has been underlined and in bold)
| Primer code | Sequence (5′–3′) | Primer position within VP2 gene | Amplicon size (in bp) |
|---|---|---|---|
| CPV-OF | ATGAGTGATGGAGCAGTTCAACC | 1–23 | 721 |
| CPV-OR | CATCATCTGGATCTGTACCATG | 721–699 | |
| CPV-IF (vac) | AGAAGAGTGGTTGTGAATAGTA | 238–260 | 721 + 483 |
| CPV-IR (field) | CGTTAACTGCAGTTTTATCGAA | 282–260 | 721 + 280 |
ARMS-PCR was optimized with 2X Dream Taq Master Mix (7.5 µl), outer primers (CPV-2 OF and CPV-2 OR) at 10 µM each while the inner primers [CPV-2 IF (vac) and CPV-2 IR (field)] at 5 µM each, parvoviral DNA at 50-100 ng and the volume was made up to 15 µl with nuclease free water (NFW). Cycling conditions consisted of initial denaturation at 95 °C for 5 min followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 54 °C for 30 s and extension at 72 °C for 40 s, with a final extension step at 72 °C for 10 min. DNA from healthy unvaccinated dogs as well as non-template control (NFW as template) were run in all PCR experiments. PCR-amplified products were resolved on 1.5% agarose gel in Tris–acetate–EDTA (TAE) buffer (1X), stained with EZ-vision® (Amresco, USA) and visualized under UV light in a gel documentation system (Molecular Imager® Gel Doc™ XR + System, Bio-Rad, USA).
The specificity of the primers was checked with positive control DNA/cDNA of other putative canine pathogens (canine adenovirus-1, canine adenovirus-2, Leptospira spp., cDNA from canine distemper virus) obtained from repository maintained at the institute.
Analytical sensitivity of ARMS-PCR assay was determined with known quantity of serially diluted parvoviral DNA both from commercial vaccines and CPV-2 field strain.
The developed ARMS-PCR was subsequently validated to screen six commercial vaccines, used routinely in our country and 24 archived CPV-2 clinical samples obtained from the field. The assay was also extended to screen rectal swabs from 3 vaccinated dogs that were vaccinated with CPV-2 vaccines for a period of 10 days post-vaccination.
ARMS-PCR amplification with recombinant plasmid (positive controls) resulted in amplicon of size 721 bp and 483 bp in CPV-2 vaccine strain, while the field strain virus resulted in amplified products of 721 bp and 280 bp in CPV-2 field strains (either CPV-2a/CPV-2b/CPV-2c). ARMS-PCR did not show any amplification from DNA of healthy unvaccinated dogs and non-template control (Fig. 2). The primers for vaccine and field CPV strains were specific and showed no cross-reactivity. The analytical sensitivity was found out to be 5 × 10−6ng/µl of parvoviral DNA in both CPV-2 vaccine and field strains.
Fig. 2.
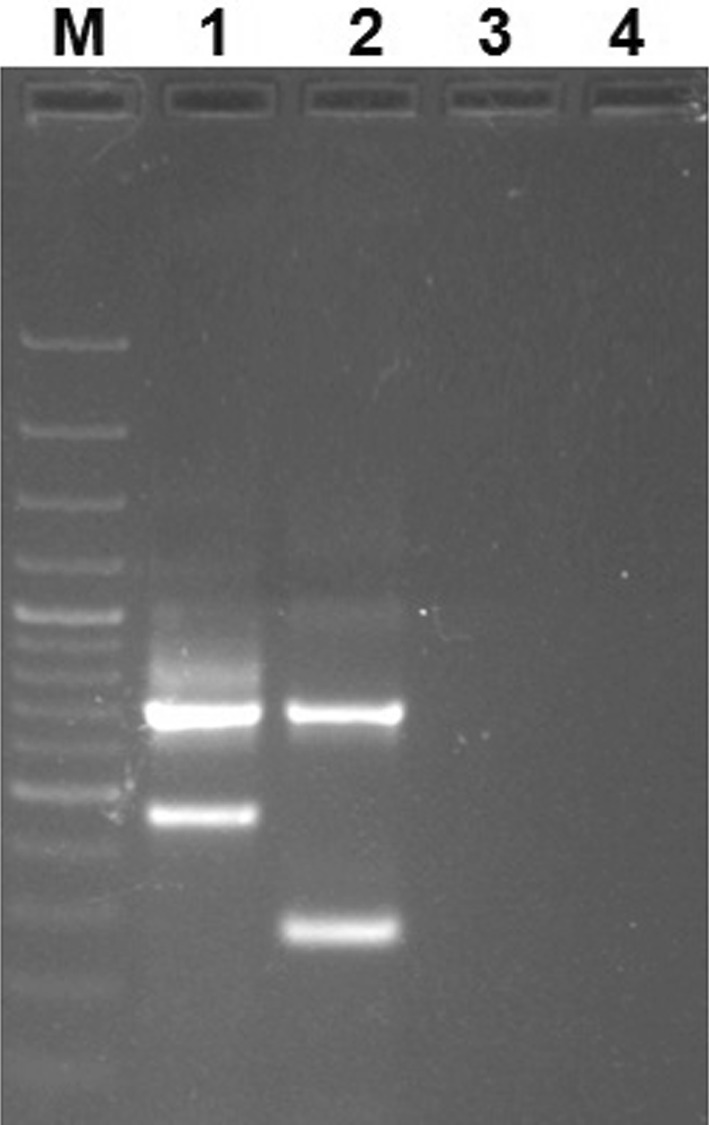
Agarose gel (1.5%) showing PCR amplified product from positive controls (recombinant plasmids containing full length VP2 gene of CPV-2 vaccine and field strains). Lane M: 100 bp plus DNA ladder (Thermo Scientific, USA), Lane 1: PCR amplified product with CPV-2 vaccine (CaniShot® DHPPL), Lane 2: PCR amplified product with CPV-2 field strain (Isolate KN-1/2015; NCBI GenBank Accession number: KX425922), Lane 3: PCR amplification from healthy unvaccinated dog, Lane4: Non-template control
ARMS-PCR when validated in six commercial vaccines produced amplified products of 721 bp and 483 bp, while all 24 CPV-2 field strains resulted in amplified products of 721 bp and 280 bp size (Fig. 3). Sequencing results of randomly selected vaccine and wild type field strains gave expected nucleotide change at 3045 position corresponding to either CPV-2 vaccine or field strains. The CPV-2 field strains have been deposited in NCBI GenBank with following Accession numbers: KX425922, KX469434, KX219735, KX878988, KX425921 and KX425920.
Fig. 3.
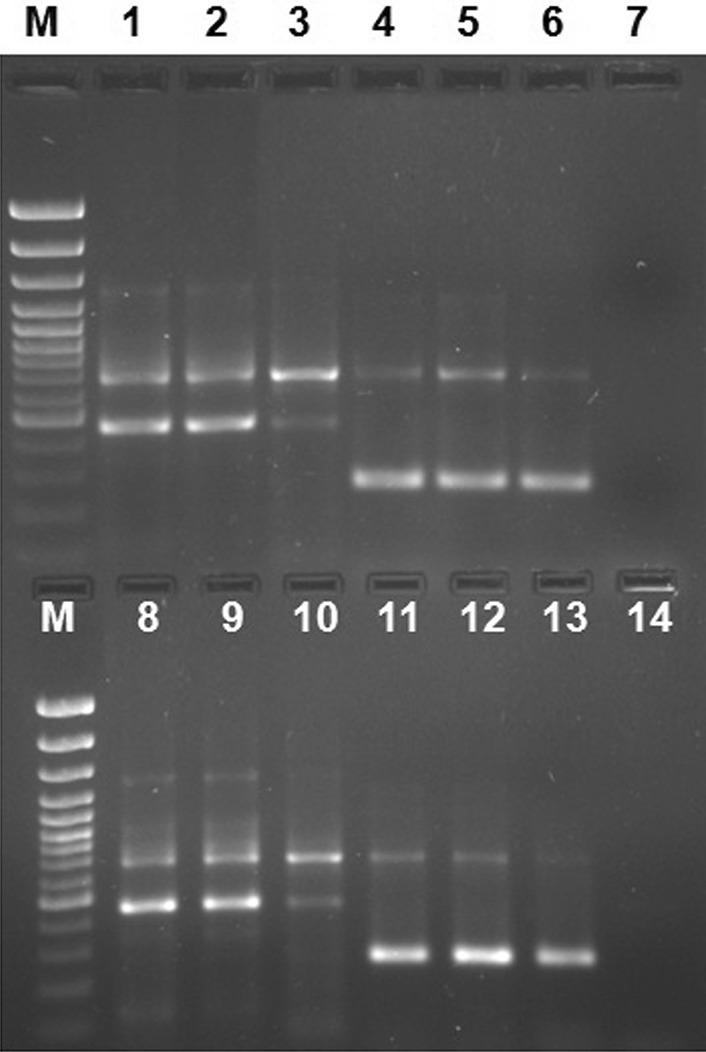
Agarose gel (1.5%) showing PCR amplified product from CPV-2 vaccine and field strains. Lane M: 100 bp plus DNA ladder (Thermo Scientific, USA), Lane 1–3, 8–10: CPV-2 vaccine strains, Lane 4–6, 11–13: CPV-2 field strains, Lane 7: PCR amplification from healthy unvaccinated dog, Lane 14: Non-template control
Fecal shedding of vaccine strain was observed 3 days post vaccination in two dogs while in feces of the third dog the presence of vaccine was observed from the fourth day. The shedding of the vaccine strain continued for 7 day post vaccination in one dog while the other dogs, shedding was observed up to the 8th day post-vaccination.
Vaccination with modified live vaccine (MLV) remains pivotal in preventing parvoviral enteritis in domestic dog population. Most of the commercial vaccines that are available either use CPV-2 or CPV-2b types in their vaccine formulations [10]. In India, most commercial vaccines that are used have CPV-2 strain in their vaccine formulation, while CPV-2b based vaccines are rarely being used. Molecular epidemiological studies from different parts of this country reveal that new CPV-2a is the predominant antigenic type, with sporadic occurrence of other antigenic variants (CPV-2b/CPV-2c) [10]. These may be the reason why use of CPV-2b vaccine is not popular in India.
Vaccination with CPV-2 MLV causes transient viraemia and shedding of vaccine virus occurs a few days to a couple of weeks post-vaccination. The problem is complicated in situations where clinical signs appear (may be infection with virulent CPV-2 field strains before attaining protective antibody titer) after vaccination which posses diagnostic dilemma to differentiate the vaccine and the field strains [3–6, 8]. Real-time PCR has also been used to estimate titer of viral load in fecal samples and correlate with vaccination or infection; as it is hypothesized that vaccine induced fecal shedding of virus is excreted in a lower titer [6]. A differentiation based on this real-time PCR result is highly specific but it needs sophisticated instruments which are beyond the reach of resource limited labs [5]. Virus shedding is also observed in sub-clinical infections in apparently healthy dogs as a result of lesser mucosal immunity, even though antibody titer may be present in the animals.
In this study, ARMS-PCR has been successful in differentiating only CPV-2 vaccine strains with other CPV-2 field strains (CPV-2a/CPV-2b/CPV-2c) based on the nucleotide change at 259 position. Studies have reported that some carnivore protoparvovirus and very limited CPV-2a strains (FJ222824, GU212792, KM236572, U22186 and U22192) have A nucleotide at 259 position which sometimes may lead misdiagnosis but during the present study we could not encounter this problem. In future, if new CPV-2 antigenic variants (CPV-2b/CPV-2c) are used in vaccine formulations this ARMS-PCR technique can be developed after identifying conserved SNPs in vaccine and field strains.
In conclusion, the present study using ARMS-PCR has been successful in differentiation of CPV-2 vaccine and wild type field strains, providing a simplified genetic DIVA strategy. This technique can be extended in other diagnostic laboratories worldwide for differentiation of CPV-2 and other circulating CPV-2 field strains (CPV-2a/CPV-2b/CPV-2c) when only CPV-2 strain is used in vaccine formulations.
Acknowledgements
All the authors are thankful to Direct, ICAR-Indian Veterinary Research Institute, Izatanagar, India, for providing research facilities. The first author is also thankful to Indian Council of Medical Research (ICMR) for awarding fellowship during the Ph.D. program.
Ethical approval
The present study does not involve use of human or animal subjects for experimentation and use of any invasive method for sample collection; hence animal ethics approval of the institute was not sought. The consent of animal owner was sought while collecting diarrhoeic samples/rectal swabs from dogs.
Conflict of interest
All the authors declare no conflict of interest.
Contributor Information
Vikas Gupta, Email: vet.gvikas@gmail.com.
Sukdeb Nandi, Email: sukdebnandi@yahoo.in.
References
- 1.Bingga G, Liu Z, Zhang J, Zhu Y, Lin L, Ding S, Guo P. High resolution melting curve analysis as a new tool for rapid identification of canine parvovirus type 2 strains. Mol Cell Probes. 2014;28:271–278. doi: 10.1016/j.mcp.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Chander V, Chakravarti S, Gupta V, Nandi S, Singh M, Badasara SK, Sharma C, Mittal M, Dandapat S, Gupta VK. Multiplex amplification refractory mutation system PCR (ARMS-PCR) provides sequencing independent typing of canine parvovirus. Infect Genet Evol. 2016;46:59–64. doi: 10.1016/j.meegid.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 3.Decaro N, Elia G, Desario C, Roperto S, Martella V, Campolo M, Lorusso A, Cavalli A, Buonavoglia C. A minor groove binder probe real-time PCR assay for discrimination between type 2-based vaccines and field strains of canine parvovirus. J Virol Methods. 2006;136:65–70. doi: 10.1016/j.jviromet.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decaro N, Crescenzo G, Desario C, Cavalli A, Losurdo M, Colaianni ML, Ventrella G, Rizzi S, Aulicino S, Lucente MS, Buonavoglia C. Long-term viremia and fecal shedding in pups after modified-live canine parvovirus vaccination. Vaccine. 2014;32:3850–3853. doi: 10.1016/j.vaccine.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decaro N, Buonavoglia C. Canine parvovirus postvaccination shedding: interference with diagnostic assays and correlation with host immune status. Vet J. 2017 doi: 10.1016/j.tvjl.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freisl M, Speck S, Truyen U, Reese S, Proksch AL, Hartmann K. Faecal shedding of canine parvovirus after modified-live vaccination in healthy adult dogs. Vet J. 2017;219:15–21. doi: 10.1016/j.tvjl.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Hirasawa T, Yono K, Mikazuki K. Detection and genomic analysis of canine parvovirus by the polymerase chain reaction. J Vet Med B. 1995;43:545–554. doi: 10.1111/j.1439-0450.1996.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 8.Meggiolaro MN, Ly A, Rysnik-Steck B, Silva C, Zhang J, Higgins DP, Muscatello G, Norris JM, Krockenberger M, Slapeta J. MT-PCR panel detection of canine parvovirus (CPV-2): vaccine and wild-type CPV-2 can be difficult to differentiate in canine diagnostic fecal samples. Mol Cell Probes. 2017;33:20–23. doi: 10.1016/j.mcp.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miranda C, Thompson G. Canine parvovirus: the worldwide occurrence of antigenic variants. J GenVirol. 2016;97:2043–2057. doi: 10.1099/jgv.0.000540. [DOI] [PubMed] [Google Scholar]
- 10.Mittal M, Chakravarti S, Mohapatra JK, Chug PK, Dubey R, Upmanuyu V, Narwal PS, Kumar A, Churamani CP, Kanwar NS. Molecular typing of canine parvovirus strains circulating from 2008 to 2012 in an organized kennel in India reveals the possibility of vaccination failure. Infect Genet Evol. 2014;23:1–6. doi: 10.1016/j.meegid.2014.01.015. [DOI] [PubMed] [Google Scholar]