Abstract
In contrast to the genetic component in mammary carcinogenesis, epigenetic alterations are particularly important for the development of sporadic breast cancer (BC) comprising over 90% of all BC cases worldwide. Most of the DNA methylation processes are physiological and essential for human cellular and tissue homeostasis, playing an important role in a number of key mechanisms. However, if dysregulated, DNA methylation contributes to pathological processes such as cancer development and progression. A global hypomethylation of oncogenes and hypermethylation of tumor-suppressor genes are characteristic of most cancer types. Moreover, histone chemical modifications and non-coding RNA-associated multi-gene controls are considered as the key epigenetic mechanisms governing the cellular homeostasis and differentiation states. A number of studies demonstrate dietary plant products as actively affecting the development and progression of cancer. “Nutri-epigenetics” focuses on the influence of dietary agents on epigenetic mechanisms. This approach has gained considerable attention; since in contrast to genetic alterations, epigenetic modifications are reversible affect early carcinogenesis. Currently, there is an evident lack of papers dedicated to the phytochemicals/plant extracts as complex epigenetic modulators, specifically in BC. Our paper highlights the role of plant natural compounds in targeting epigenetic alterations associated with BC development, progression, as well as its potential chemoprevention in the context of preventive medicine. Comprehensive measures are stated with a great potential to advance the overall BC management in favor of predictive, preventive, and personalized medical services and can be considered as “proof-of principle” model, for their potential application to other multifactorial diseases.
Keywords: Breast cancer, Breast cancer prevention, Epigenetic modulations, Phytochemicals, Natural modulators, DNA methylations, Histones chemical modifications, RNA mechanisms, Predictive preventive personalized medicine, Individual outcomes, Patient stratification, Individualized patient profile, Innovation
Introduction
Approximately two million new cases and a half of million deaths registered annually; breast cancer (BC) has reached an epidemic scale in the early twenty-first century [1]. Poor survival rates in young asymptomatic patient cohorts such as the premenopausal triple-negative breast cancer (TNBC) require new strategies in the overall BC management, focused on innovative screening programs in young subpopulations, predictive diagnosis, effective targeted prevention, and customized treatments [2–5]. Due to the high incidence of BC worldwide, there is an urgency in oncology research to better understand the causes of this disease with the aim to improve clinical strategies and allow the progress in the predictive, preventive, and personalized medicine.
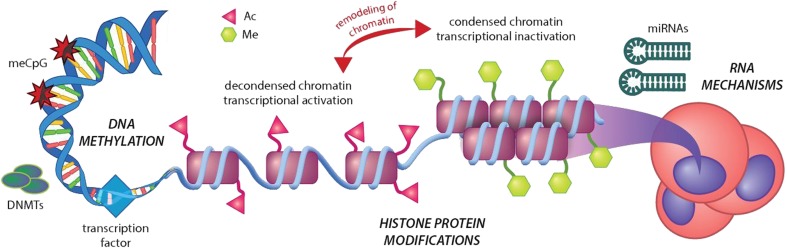
Mammary carcinogenesis and metastatic disease comprise both genetic and epigenetic components. In contrast to the genetic (inborn, non-modifiable) component, epigenetic alterations occur permanently being linked with environmental and lifestyle (modifiable) risk factors which are particularly important for the development of sporadic BC comprising over 90% of all BC cases worldwide [1]. DNA methylation, histone chemical modifications, and non-coding RNA (ncRNA)-associated multi-gene silencing are considered as key epigenetic mechanisms governing the cellular homeostasis and differentiation states (Fig. 1). Most of DNA methylation processes are physiological and essential for human cellular and tissue homeostasis, playing an important role in many key mechanisms. However, being dysregulated, DNA methylation contributes to pathologic process such as cancer development and progression. A global hypomethylation of oncogenes is characteristic for most of cancer types contributing to the genomic instability by activating retrotransposons and otherwise silent genomic regions [6]. On the other hand, hypermethylation of tumor-suppressor gene promoters plays a specific role in carcinogenesis [7, 8]. The overall impact of the epigenetic regulation on the human body is multifactorial being piloted by several endogenous and exogenous factors such as age, gender, environmental stress factors, lifestyle, diet, and stage of cancer.
Fig. 1.
The relevance of epigenetics in the carcinogenesis. Epigenetic mechanisms of carcinogenesis consist of three distinct processes: DNA methylations, histone protein methylations, and acetylations and RNA mechanisms including aberrant activity of siRNA, miRNA, and lncRNA. Ac, acetylation; DNMTs, methyltransferases; Me, methylation
A number of studies demonstrated that dietary natural plant products (both whole products and their compounds) affect the development and progression of cancer disease including breast cancer [9–13]. Phytochemicals, as widely available bioactive substances, have demonstrated apparent anticancer potential specifically by targeting aberrant epigenetic changes [14, 15]. Cancer risk prediction is rapidly continuing to evolve in medicinal practice. In this regard, clinical research can define specific epigenetic modulations related to the identification of high-risk individuals [6–8]. Consequently, epigenetic profiling of individuals can serve in the development of modern clinical strategies with crucial interventions in the primary, secondary, and tertiary prevention of BC. Furthermore, the integration of relevant epigenetic data associated with complex medical informatics and personalized medicine is highly recommended in the BC treatment and management. Medicinal approaches fall into the progressive concept of advanced health care tailored to the person [1]. “Nutri-epigenetics” focuses on the influence of dietary agents on epigenetic mechanisms [16]. This approach has gained considerable attention; since in contrast to genetic alterations, epigenetic modifications are reversible and affect early carcinogenesis. Currently, there is an evident lack of papers dedicated to the phytochemicals/plant extracts as complex epigenetic modulators specifically in BC. Our paper highlights the role of plant natural compounds in targeting epigenetic alterations associated with BC development, progression, as well as its potential chemoprevention.
Source of data
Data from the available biomedical literature were collected and analyzed. Relevant studies published in the English-language literature were retrieved by the use of epigenetics or breast cancer or predictive preventive and personalised medicine or DNA methylation or histone chemical modifications or siRNA or miRNA or lncRNA or phytochemicals or plant natural substances or plant food or diet as either a keyword or MeSH (medical subject heading) term in searches of the PubMed bibliographic database. We focused primarily on the most recent scientific papers from the years 2013–2018.
1. Aberrant DNA methylation is a hallmark of cancer
The comparison of DNA methylation levels in cancer tissue with the DNA methylation in an analogous normal tissue or DNA methylation in a variety of normal tissues revealed that cancer cells are very often associated with a global DNA hypomethylation [17, 18]. It is well described that aberrant DNA methylation affects a plethora of signaling pathways within transcription of relevant genes, and it leads to abnormalities in the differentiation of cells [18–20]. The prevailing epigenetic modification in the eukaryotic nucleus of mammals is the methylation of DNA at the 5-position of cytosine. This process is catalyzed by DNA methyltransferases (DNMTs) using S-adenosyl methionine as a universal donor of methyl group. Several types of DNMTs were described in eukaryotic cells. DNMT1 is responsible for maintaining DNA methylation status in the cell. DNMT2 plays a crucial role in the process, typically the recognition of DNA damage, DNA repair, or DNA recombination. DNMT3B, DNMT3A, and DNMT3L are usually upregulated in in malignant cells; these are responsible for the hypermethylation of tumor-suppressor genes’ promoters [21]. Sequences of DNA rich on CG repeats, known as CpG islands, are heavily methylated and act to maintain chromosomal stability. CpG islands within the promoter regions of a gene are generally unmethylated. Global DNA hypomethylation in CpG islands can lead to the genomic instability commonly observed in malignant cells; on the other hand, increased methylation of CpG islands within the gene promoters can lead to transcriptional silencing of tumor suppressors [15, 22–25].
Phytochemicals with demethylation activity in preclinical cancer research
Decrease in the total promoter methylation status of tumor-suppressor genes has a great potential as molecular target for cancer therapy. Aberrant DNA methylation patterns that are frequently observed in sporadic BC represent reversible changes, which are responsive to environmental factors, including dietary habits [26]. Plant compounds such as phenolics that can act as hypomethylating agents [25] can reverse epigenetic silencing of tumor-suppressor genes. For instance, the epigenetic silencing of glutathione S-transferase P1 is linked to the pathogenesis of BC and other cancer types. In this regard, Kumar et al. [25] assessed the epigenetic potential of curcumin on the methylation pattern of this tumor-suppressor gene in MCF-7 breast cancer cells. They demonstrated that curcumin completely reversed the glutathione S-transferase P1 promoter hypermethylation, and moreover, this modification led to re-expression of glutathione S-transferase P1. In another in vitro study, curcumin decreased the promoter methylation of Ras-association domain family protein 1A (RASSF1A) (TSG) and increased its mRNA and protein levels in breast cancer MCF-7 cells [27]. Moreover, curcumin decreased DNA methylation activity of nuclear extract through the downregulation of the mRNA and protein levels of DNMT1, which may be linked with curcumin-induced disruption of NF-κB/Sp1 complex bound to the promoter region of DNMT1. Taken together, curcumin seems to be an excellent nontoxic hypomethylating agent with a possible potential against BC. Mirza et al. [28] hypothesized that epigallocatechin gallate (EGCG), genistein, withaferin A, curcumin, resveratrol, and guggulsterone are potential demethylators. Therefore, the effect of these plant compounds on DNMTs expressions and methylation status of the panel of genes in MCF-7 and MDA-MB 231 cells was evaluated. The treatment with various polyphenols showed an apparent decrease in the transcript levels of DNMT1, DNMT3A, and DNMT3B in both cell lines. Moreover, these plant compounds lowered the protein levels of epigenetic regulators histone deacetylase 1 and MeCP2. Several in vitro studies showed the epigenetic modulator potential of sulforaphane in BC. Lewinska et al. [29] evaluated sulforaphane as an epigenetic modulator in BC cells (MCF-7, MDA-MB-231, and SK-BR-3). Sulforaphane caused cell cycle arrest and senescence of cancer cells. Oncostatic characteristics of sulforaphane were accompanied by global DNA hypomethylation, decreased levels of DNA methyltransferases (DNMT1, DNMT3B), and diminished pools of N6-methyladenosine (m6A) RNA methylation. In another study, combination of sulforaphane with (−)-epigallocatechin gallate as a demethylating agent was identified as an effective approach for re-expression of estrogen receptor in hormone negative BC [30]. Moreover, in the study of diet by Lubecka-Pietruszewska et al. [31], sulforaphane caused hypomethylation of PTEN and RARbeta2 promoters with subsequent gene upregulation in MCF-7 and MDA-MB-231 cells. TNBC is characterized by poor prognosis and a DNA hypomethylation profile. An in vitro study by Kala et al. [32] demonstrated that resveratrol and pterostilbene administered at a dose (close to physiologically relevance) shows synergistic growth inhibition on HCC1806 and MDA-MB-157 cells, a TNBC cell lines [32]. Authors concluded that this result provided a novel nutrient control strategy that may contribute to future clinical strategies against relapsed/refractory metastatic TNBC. Szarc Vel Szic et al. [33] reported that DNA hypermethylation of corresponding CpG sites in PLAU, ADAM8, TNSF12, GSTM1, and ME3 genes is linked with receptor tyrosine-protein kinase erbB-2 amplification (HER2)/estrogen receptor (ER)/progesterone receptor (PR) status in TNBC. Withaferin A (WA), a plant-derived steroidal lactone, induced DNA hypermethylation of multiple CpG sites which may contribute to epigenetic reprogramming of genes involved in aggressiveness of TNBC towards less aggressive luminal-like BC hallmarks. Verification of gene expression alterations caused by WA was performed in MDA-MB-231 and MCF-7 cells. There are several other recent papers describing plant extracts or isolated phytochemicals, such as green tea polyphenols, wild yam root extract, kazinol Q, genistein, and wild thyme, as demethylating agents in vitro [34–38].
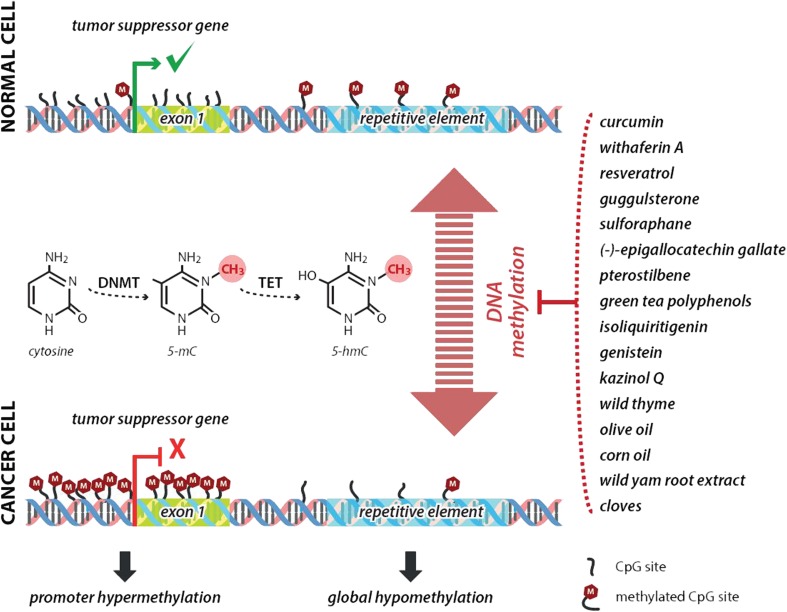
Several papers reported the analyses of DNA methylation changes in BC animal models. Rodriguez-Miquel et al. [39] evaluated the role of nutritional factors, especially dietary lipids, in the etiology of rat mammary carcinogenesis. Authors analyzed the global and gene specific (RASSF1A, TIMP3) DNA methylation levels in mammary glands and mammary tumors after the administration of olive oil– or corn oil–enriched diet, respectively. Olive oil, rich in a plethora of beneficial bioactive compounds and a monounsaturated fats, elevated the levels of global DNA methylation in rat mammary gland and cancer tissue. Corn oil, rich in sterols, polyunsaturated and monounsaturated fatty acids, increased DNA methyltransferase activity in both tissues; moreover, it increased the promoter methylation of the tumor-suppressor genes RASSF1A and TIMP3. These results demonstrated differential effects of the high-fat diets on epigenetic changes in carcinogen-induced rat mammary carcinogenesis. In this regard, our recent chemoprevention study [40] analyzed the effect of clove buds on the methylation status in the promoter regions of RASSF1A (three CpG islands) and TIMP3 (six CpG islands) genes in rat breast carcinoma cells in vivo. Strong chemopreventive effects of cloves in rats were accompanied with significant effects on the methylation levels of RASSF1A and TIMP3 promoters in different CpG islands. However, epigenetic mechanisms of cloves in our study could represent only one of the plethora mechanisms by which this spice prevented NMU-induced rat mammary gland carcinogenesis in our study. Moreover, this result requires more complex experimental validation (e.g., the use of wider spectrum of promoters assessed). In another study, Wang et al. [41] described new epigenetic function of isoliquiritigenin, a chalcone-type molecule derived from licorice root. This natural compound lowered the methylation levels of WIF1 promoter as the consequence of decreased DNMT1 activity and thus prevented mouse mammary cancerogenesis. Epigenetically silenced functions of WIF1 tumor-suppressor gene have been also described in other cancer types [41]. The role of phytochemicals or plant foods in the influencing of methylation status of genes’ promotors is summarized in Fig. 2.
Fig. 2.
Mechanism of enhanced tumor-suppressor gene expression through increased promoter demethylation using isolated plant-derived compounds or plant foods. DNMTs, DNA methyltransferases; TET, ten-eleven translocation protein; 5mC, 5-methylcytosine; 5hmC, 5-hydroxymethylcytosine
Clinical research: dietary intervention and DNA methylation patterns in breast cancer
Despite preclinical research, only limited clinical data evaluating the effects of phytochemicals on DNA methylation patterns in BC disease are available. It is well described that global DNA hypomethylation in the tissue and blood has been associated with increased cancer risk; on the other hand, healthier lifestyle patterns have been linked with increased levels of global DNA methylation. Recently, Greenlee et al. [42] assessed the long-term effects of a short-term culturally based dietary intervention on increasing fruits/vegetables, decreasing fat, and changing biomarkers associated with BC recurrence risk among Hispanic BC survivors. Investigators enrolled 70 women with a history of stage 0–III BC. After 12 months, the intervention group demonstrated increase in plasma lutein (+ 20.4% vs. − 11.5%; P < 0.01) and borderline significant increase in global DNA methylation (+ 0.8% vs. − 0.5%; P = 0.06). Authors concluded that dietary intervention positively altered the biomarkers linked with BC recurrence risk. Another clinical study examined the linkage between changes in lifestyle modifications (diet, weight loss, including eating ≥ 2 servings of fruit and ≥ 3 servings of vegetables), metabolic markers, and global epigenetic biomarkers using white blood cells [43]. Study participants were Hispanic, African American, and Afro-Caribbean overweight and sedentary female BC survivors (n = 24). Data were measured at baseline and after 6 and 12 months of intervention. Investigators evaluated DNA methylation of long interspersed nucleotide element 1 (LINE-1) and satellite 2 by pyrosequencing and MethyLight, respectively, and global DNA methylation by the luminometric methylation assay (LUMA). DNA methylation of LINE-1 was increased after 6 months (75.5% vs. 78.5% [P < 0.0001]) and 12 months (75.5% vs. 77.7% [P < 0.0001]) in comparison with baseline. Importantly, 12-month changes in dietary measures such as vegetable intake were positively associated with changes in LUMA DNA methylation, as was intake of fruit positively associated with changes in LINE-1 DNA methylation [43]. Hypermethylation of RARB (retinoic acid receptor-beta), BRCA1, and RASSF1A promoters was linked with the downregulation of transcript levels in the relevant gene in primary BC. In this regard, Pirouzpanah et al. [44] assessed the hypermethylation status in 146 dissected BC tissue samples in Iranian women using methylation-specific PCR. A validated 136-item food frequency questionnaire was used as a method for the estimation of dietary nutrients. The intake of folate and cobalamin was inversely associated with methylated RARB and BRCA1. On the other hand, higher dietary intake of riboflavin and pyridoxine was linked with increased methylation in the RARB promoter. Moreover, authors concluded with evidence for age-dependent effects of nutrients on promoter methylation status.
Table 1 summarizes the role of phytochemicals targeting epigenetic changes linked with DNA methylation patterns in breast carcinogenesis.
Table 1.
Natural substances targeting DNA methylations associated with breast cancer
| Phytochemicals/plant foods | Mechanism of intervention | Type of BC research | References |
|---|---|---|---|
| Curcumin | Decrease in promoter methylation status (RASSF1A, glutathione S-transferase P1) or DNA methylation activity | MCF-7 cells | [25, 27] |
| EGCG, genistein, withaferin A, curcumin, resveratrol, guggulsterone | Decrease in DNMT1, DNMT3a, and DNMT3b expressions | MCF-7 and MDA-MB 231 cells | [28] |
| Sulforaphane | Global DNA hypomethylation, decrease in DNMT1 and DNMT3 activity, and m6A RNA methylation | MCF-7 MDA-MB 231, and SK-BR-3 cells | [29] |
| Sulforaphane + EGCG | ERα promoter hypomethylation and hyperacetylation | MDA-MB 231 cells | [30] |
| Sulforaphane | PTEN and RARbeta2 promoter hypomethylation | MCF-7 and MDA-MB 231 cells | [31] |
| Resveratrol + pterostilbene | Decrease in DNMTs activity | HCC1806 and MDA-MB-157 breast cancer cells | [32] |
| Withaferin A | Epigenetic reprogramming through DNA hypermethylation | MDA-MB-231, MCF-7 cells | [33] |
| Green tea polyphenols +5-aza-2′-deoxycytidine | Decrease in HDAC1, DNMT1, DNMT3b, and MBD4 activity | MDA-MB-231, MCF-7 cells | [34] |
| Kazinol Q | Inhibition of DNMT1 activity | MDA-MB-231 cells | [36] |
| Genistein | Decrease in global DNA methylation, DNMT activity, and DNA methylation of several promoters (ATM, APC, PTEN, SERPINB5) | MCF-7 and MDA-MB-231 cells | [37] |
| Wild yam root extract | Activation of GATA3 gene in ER- cells | MCF-7 and MDA-MB-231 cells | [35] |
| Wild thyme | Inhibition of DNMT and HDAC activities | MDA-MB-231 cells | [38] |
| Isoliquiritigenin | Demethylation of WIF1 promoter, decrease in DNMT1 activity | Mouse BC model | [41] |
| Olive oil | Increase in global DNA methylation | Rat BC model | [39] |
| Corn oil | Increase in DNMT methylation and promoter methylation of RASSF1A and TIMP3 | Rat BC model | [39] |
| Clove buds (mixture of phenolic acids, flavonol glucosides, tannins and phenolic volatile oils – eugenol, acetyl eugenol) | Increase in total RASSF1A promoter methylation | Rat BC model | [40] |
| Fruits/vegetables | Increase in global DNA methylation | Hispanic BC survivors | [42] |
| Vegetables | Increase in DNA methylation of LINE-1 after 6 and 12 months | Hispanic, African American, and Afro-Caribbean overweight and sedentary BC survivors | [43] |
| Folate + cobalamin | Hypomethylation of RARB and BRCA1 promoters | BC tissue samples of Iranian women | [44] |
| Riboflavin + pyridoxine | Hypermethylation of RARB promoter | BC tissue samples of Iranian women | [44] |
EGCG, epigallocatechin gallate; DNMT, DNA methyltransferase; ER, estrogen receptor; HDAC, histone deacetylase; MBD, methyl-CpG-binding domain
2. Chemoprevention by histone chemical modifications
Histones are proteins that order and package DNA into nucleosomes. Tightly packed DNA regions called heterochromatins are less accessible for transcription factors and are thus inactive in the term of gene expression, whereas lightly packed euchromatin regions are active. Modifications of histones are covalent post-translational changes of histone amino acid residues located in their N- and C-terminal tails [45, 46]. They include acetylation, methylation, phosphorylation, ubiquitylation, sumoylation, and ADP ribosylation and are achieved by diverse histone-modifying enzymes (HATs, HDACs, HMTs, HDMs, and others) grouped in several classes. Elevated levels of global histone acetylation and methylation have been found in majority of luminal-like breast tumors and have been associated with a favorable prognosis. Moderate and low levels of lysine acetylation (H3K9ac, H3K18ac, H4K12ac) and lysine and arginine di-/three-methylations (H3K4me2, H4K20me3, H4R3me2) were found in BC subtypes with poorer prognosis, including basal carcinomas and HER2-positive tumors [47, 48]. Study of Müller et al. [49] revealed significantly different expression of class I histone deacetylases (HDAC) in human BC. HDAC1 expression was elevated in hormone receptor–positive tumors, while HDAC2 and HDAC3 were expressed significantly higher in low differentiated tumors and correlated with negative hormone receptor status. Moreover, high HDAC2 expression was associated with an overexpression of HER2 and nodal metastasis.
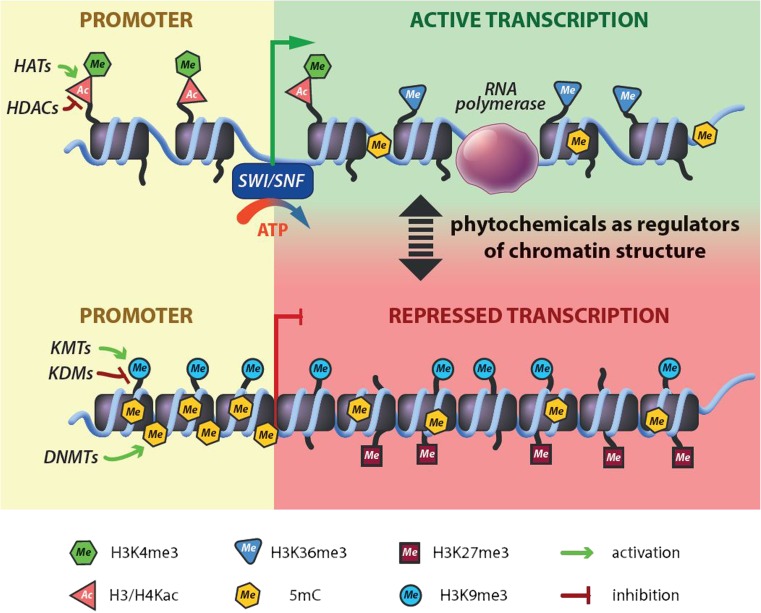
Aberrant histone modifications and changed expression of histone-modifying enzymes are caused by genetic mutations in chromatin regulators enzymes and by epigenetic changes. Dietary phytochemicals, like epigallocatechin-3-gallate (EGCG), curcumin, garcinol, antioxidant vitamins, and flavones, are able to modulate epigenetic modifications and thus are actively involved in chemoprevention [50–54]. The mechanisms of the chemical modifications of histones caused by phytochemicals are shown in Fig. 3.
Fig. 3.
The role of plant compounds in the chemical modification of histones. Phytochemicals affect the functions of KMTs, KDMs, HDACs, HATs, and DNMTs and thus regulate genes’ transcription activity. SWI/SNF (SWItch/Sucrose Non-Fermentable), is ATP-dependent nucleosome remodeling protein complex. It is able to restructure the nucleosome to make its DNA accessible during transcription, replication, and DNA repair. DNMTs, DNA methyltransferases; HATs, histone acetyltransferases; HDACs, histone deacetylases; KDMs, lysine demethylases; KMTs, lysine methyltransferases
Histone acetylation
Histone chemical modifications including acetylations are commonly analyzed in BC. Clinical analyses of Elsheikh et al. [48] discovered low or absent H4K16ac in the majority of BC cases in humans (78.9%), proposing that this change may denote an early sign of breast carcinoma. Histone acetylation is enzymatic addition of an acetyl group (COCH3) from acetyl coenzyme A. This process is involved in the regulation of many cellular processes such gene silencing, cell cycle progression, apoptosis, differentiation, and DNA repair [55–57]. The modifying enzymes are called histone acetyltransferases (HATs) and histone deacetylaces (HDACs).
Epigallocatechin-3-gallate (EGCG) is a major component of green tea polyphenols. It has important anticancer activity via epigenetic mechanisms and play important role in induction of apoptosis, anti-oxidation and inhibition of proliferation, angiogenesis, and metastasis [58–60]. Li et al. [61] studied the role of EGCG treatment in ERα-negative MDA-MB-231 breast cancer cells. ERα-negative BC is clinically aggressive and has a poor prognosis because it normally does not respond to conventional anti-hormone therapies. They proved that EGCG can induce re-expression of endogenous estrogen receptor α (ERα). The reactivation is regulated via chromatin remodeling of the ERα promoter by altering histone acetylation and methylation status. EGCG was found to decrease binding of the transcription repressor complex in the promoter region of ERα Rb/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 contributing to ERα transcriptional activation. The effect was even enhanced when EGCG was combined with the histone deacetylase (HDAC) inhibitor, trichostatin A (TSA). Meeran et al. [62] analyzed epigenetic impact of EGCG on the hTERT expression in human breast cancer cells MCF-7 (ER+) and MDA-MB-231 (ER−). EGCG treatment of these cell lines significantly inhibited HATs activities, but not HDACs activities. They found an EGCG-induced time dependent decrease of transcriptional active chromatin markers such as H3ac, H3K9ac, and H4ac. Deacetylation induced by EGCG further recruits hTERT transcriptional repressors such as E2F-1 and MAD1, contributing to inhibition of hTERT expression; what resulted in cellular apoptosis induction is similar in both ER (+) and ER (−) human breast cancer cells but not in normal cells [62]. Besides the described impact of EGCG on the histone acetylation status, this metabolite exerted its anticancer effect also by correcting other epigenetic alterations in cancer cells such as polycomb-group (PcG) protein–dependent histone methylation, DNA methylation, class-I HDACs-dependent deacetylation of histones, and some non-histone proteins and ubiquitination [63].
A similar histone acetylation level reduction was reported in the case of curcumin and garcinol in human breast proliferating MCF7 cells. H3 and H4 acetylation was reduced after exposure to curcumin, garcinol, or garcinol derivate LTK 14, although pan-acetylation of H4 indicated differential and dose-dependent effects of these compounds on histone modifications in MCF7 cells [64]. Increased H3 acetylation was found in the study of Weng et al. [50] who analyzed newly identified flavone, 3,4′-dimethoxy-3′,5,7-trihydroxyflavone from the plant Myoporum bontioides in human MCF-7 breast cancer cells. This phytochemical downregulates the expression of histone deacetylase 2 (HDAC2) and HDAC4, leading to increased histone H3 acetylation and p21 upregulation. It is involved in the cell cycle regulation and growth inhibition of MCF-7 cells and 3,4′-dimethoxy-3′,5,7-trihydroxyflavone thus possess the similar antiproliferative activity as the EGCG and genistein [50]. Secoiridoids, a family of complex polyphenols found in extra virgin olive oil also appear to influence the histone acetylation. Study of the unique BC cell line JIMT-1, specific for inherently exhibition of cross-resistance to multiple HER1/2-targeted drugs, showed histone hyperacetylation. As these phenols should efficiently circumvent de novo resistance to HER1/2 inhibitors, they could represent novel anti-breast cancer molecules [65].
Several other studies were conducted in the term of histone acetylation modulation by phytochemical compounds. The analyses revealed their antitumor activity as they act as HDAC inhibitors. Among them, the attention has been paid to diallyl disulphide (DADS), the major organosulfur compound found in garlic oil [66]; isothiocyanates, found in cruciferous vegetables [67]; luteolin (3′,4′,5,7-tetrahydroxyflavone), a member of the flavonoid family found in various fruits and vegetables [68] and to triterpenoid 5β,19-epoxy-19-methoxycucurbita-6,23-dien-3β,25-diol [69].
Histone methylation
Histone methylation is defined as the transfer of one, two, or three methyl groups (me1, me2, or me3) to histone proteins, which can be methylated on lysine (K) and arginine (R) residues [70]. The most commonly observed methylation is on lysine residues of histone tails H3 and H4. The transfer of S-adenosyl-l-methionine is catalyzed by histone methyltransferases (HMTs) which are specific for lysine and arginine residues [71]. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated and how many methyl groups are attached. It seems that histone methylation marks are coupled with the pathology of BC, as Rivenbark et al. [70] showed differential levels of histone methylation in several of the cancer cell lines when compared to a normal mammary epithelial cell line. BT549 and SUM102 cells exhibited low levels of H3K4me3 and increased levels of H3K36me3. In addition, alterations in methylation of histone lysines such as H3K4m3, H3K9m3, and H4K20m3 have been frequently associated with breast carcinoma [71]. Moreover, the loss of histone H4K20 trimethylation has been documented as a marker of poor prognosis in breast carcinoma patients and is linked with the invasiveness of breast carcinoma cells in a HER2-independent manner [72].
The role of plant natural substances on the histone methylation patterns has been documented in several in vitro and in vivo studies [10, 73, 74]. For example, garcinol also affects the processes of histone methylation and the trimethylation of H4K20 (H4K20me3) in MCF7 breast cancer cells. Authors found that this is due to the induced expression of SUV420H2, an enzyme that specifically trimethylate H4K20 [64]. Dagdemir et al. [75] showed that soy phytoestrogens (daidzein, genistein, equol, 17β-estradiol, and suberoylanilide hydroxamic acid) are able to modulate gene transcription via the acetylation and demethylation of histones in six genes (BRCA1, EZH2, ERα, ERβ, P300, and SRC3) in MCF-7 and MDA-MB 231 cancer cells. Genistein represents natural compound with a multi-targeted biological/molecular activities, including epigenetic histone chemical modifications in carcinoma cells. In another experimental study, genistein reactivated expression of ERα through the histone modification in the ERα promoter and the inhibition of histone deacetylase inhibitors in MDA-MB-231 and MDA-MB-157 cells [74]. Results of Li et al. [76] demonstrated that genistein suppressed cell proliferation in human breast precancerous and cancer cells; on the other hand, it showed only slight effect on normal breast epithelial cells. Moreover, genistein administration upregulated the expression of p21(WAF1) (p21) and p16(INK4a) (p16) tumor-suppressor genes and downregulated the expression of BMI1 and c-MYC tumor promoting genes. Genistein induced histone chemical modifications in p21 and p16 promoters and affected the binding of the c-MYC-BMI1 complex to the p16 promoter resulting in the stimulation of such tumor-suppressor genes. As well, the authors administered genistein (250 mg/kg of diet) to mice, and these doses corresponded to daily consumption of soybean products in humans. The results confirmed significant impact of genistein on the tumor growth from the very early stages.
There are only few experimental BC studies describing the role of whole plant-based foods in the chemical modulations of histones. We have found only two papers assessing the effect of dietary factors on histone chemical modifications in rat mammary carcinogenesis. The results of our group showed that dietary administered clove buds significantly and dose-dependently increased the levels of H4K20m3 (and also H4K16ac) in chemically induced cancers [40]. We did not find the changes in chemical modifications of three methylations of H3K4 and H3K9. Our results could be considered as a one of plethora mechanisms by which phytochemicals from cloves may prevent nitrosomethylurea-induced mammary gland carcinogenesis in female rats, however, our results await further experimental validation (e.g., with wider spectrum of histone chemical modifications or gene promoters analyzed). Results from similar rat model also pointed to significant effect of extra virgin olive oil on H4K20m3 levels in mammary tumors or glands [39].
As indicated above (summarization in Table 2), histone-modifying methods and enzymes interact with each other as well as with other chromatin related proteins and other epigenetic mechanisms, which makes the issue of histone modifications complex and extensive. Therefore, more research is needed to find out exact pathways of phytochemical impact and to find out proper benefits in utilizing in the cancer risk reduction.
Table 2.
Natural substances inducing histone chemical modifications associated with breast cancer
| Phytochemicals/plant foods | Mechanism of intervention | Type of BC research | References |
|---|---|---|---|
| EGCG | Alteration of histone acetylation and methylation status, ERα reactivation | MDA-MB-231 cells | [61] |
| EGCG | Inhibition of HATs activities, hTERT promoter hypomethylation and histone deacetylations (acetyl-H3, acetyl-H3K9, and acetyl-H4) | MCF-7, MDA-MB-231 cells | [62] |
| Curcumin and garcinol, | Reduction in H3K18ac (garcinol), elevated global levels of H4K16ac and H4K20m3 (garcinol), increased global levels of H3K18ac and H4K16ac (curcumin) | MCF-7 cells | [64] |
| 3,4′-dimethoxy-3′,5,7-trihydroxyflavone from Myoporum bontioides L. | Downregulation of HDAC2 and HDAC4 expression, increase of H3ac | MCF-7 cells | [50] |
| Secoiridoids | Histone hyperacetylation | JIMT-1 cells | [65] |
| Diallyl disulphide/garlic oil | HDAC inhibition | MCF-7 cells | [66] |
| Luteolin (flavonoid) | HDAC inhibition | MCF-7/6 and MDA-MB231–1833 cells | [68] |
| Triterpenoid 5β,19-epoxy-19-methoxycucurbita-6,23-dien-3β,25-diol from Momordica charantia L. | Downregulation of HDAC1, increase in H3ac | MCF-7 | [69] |
| Soy phytoestrogens (isoflavones) | Histones demethylation and acetylation (H3K27me3, H3K9me3, H3K4me3, H4K8ac, and H3K4ac) | MCF-7 and MDA-MB 231 cells | [75] |
| Genistein | Histones modification in theERα promoter, reactivation of ERα expression | Mouse BC model using MDA-MB-231 cells | [74] |
| Genistein | Histone modifications in the promoters of p21 and p16 genes | HMECs, SH, SHR, MDA-MB-231 | [76] |
| Clove buds (mixture of phenolic acids, flavonol glucosides, tannins and phenolic volatile oils – eugenol, acetyl eugenol) | Increase in lysine trimethylations and acetylations (H4K20me3, H4K16ac) | Rat BC model | [40] |
| Extra virgin olive oil | Decrease in H4K20m3 levels in mammary tumors or glands | Rat BC model | [39] |
EGCG, epigallocatechin gallate; ER, estrogen receptor; HDAC, histone deacetylase; HAT, histone acetyltransferase
3. Plant-derived compounds in breast cancer prevention and therapy: RNA level of intervention
Recent experimental and clinical BC studies focused on influencing the epigenome have provided convincing evidence that plant-derived compounds participate in regulation of genes through affecting non-coding RNAs (ncRNAs), especially microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) [77–84]. miRNAs together with short (or small)-interfering RNAs (siRNAs) are two main types of small ncRNAs, which regulate gene expression through the RNA interference (RNAi) mechanism [85]. Currently, it is well known that RNAi plays important regulatory role in the process of normal cell development and differentiation, as well as in the process of carcinogenesis. RNAi may also be potential therapeutic device to shut down upregulated oncogenes, mutated tumor-suppressor (TS) genes, and other genes involved in tumor progression within targeted gene therapy in cancer patients [86, 87]. Recent studies suggest that synthetic siRNAs and endogenous miRNAs may influence transcriptional gene silencing by promoting histone deacetylation, DNA, and histone methylation [88, 89]. However, CpG methylation of selected miRNAs and inhibition of histone deacetylases may lead to changes in miRNA expression in cancer cells [90]. As well, phytochemicals may act as inhibitors of DNA methyltransferases and histone deacetylases, and in this way, they can reactivate for example methylation-silenced TS genes or miRNAs controlling TS genes [89, 91, 92].
RNA interference is a natural mechanism of RNA-dependent gene silencing in which small RNA molecules inhibit expression and translation of protein-coding genes bearing either fully or partially complementary sequence [93, 94]. The classical RNAi pathway is triggered by long double-stranded RNA (dsRNAs) precursors such as long dsRNA or short hairpin RNA (shRNA). These precursors are processed into siRNAs by the RNase III enzyme termed Dicer [86, 95, 96]. The siRNAs then bind to an enzyme-containing molecule known as RNA-induced silencing complex (RISC). The siRNA-RISC or miRNA-RISC complex leads to the recognition and ultimately the cleavage or translational repression of complementary single-stranded RNAs (mRNAs or viral genomic/antigenomic RNAs) [86, 95, 96]. Taken together, siRNAs as well as miRNAs are able degrade mRNA and inhibit its expression by very similar mechanisms. Plethora preclinical studies showed that plant-derived compounds modulating the RNA-based epigenetic mechanisms may have considerable importance in BC prevention and therapy.
Short-interfering RNAs
Despite the fact that RNAi seems to be promising in in vitro cancer research, it presents some limitations in vivo. Short RNAs have low stability due to RNase degradation; therefore, their nontoxic delivery into deep tissues is still problematic. Other limitations are also low potential to overpass the membranes, stimulation of innate immune response, or the error rate of “off-target” interactions [97, 98]. The first study, which overcame these limitations and brought good evidence for clinical utility of RNAi-based therapeutics in patients with advanced cancer disease, used lipid nanoparticle approach [99]. To date, siRNA nanoparticles were used in some preclinical studies of various types of BC [100–104] and have paved the way for clinical trials [105].
Regarding the plant-based compounds, siRNA mechanisms and BC, there are only limited data available in databases. Physapubenolide (PB) is a cytotoxic withanolide present in Physalis angulata that is used as a traditional Chinese medicine. Ma et al. [106] evaluated the role of TIGAR (TP53-inducible glycolysis and apoptosis regulator) and ROS in PB-induced apoptosis and autophagosome formation in MDA-MB-231 and MCF-7 cancer cells. Investigators described that the downregulation of TIGAR by siRNA was augmented with low concentrations of PB. These effects were associated with induced apoptosis and autophagosome formation, which led to the anti-tumor effect of PB in vitro [106]. Another study showed that phosphatase and tensin homolog (PTEN) is a key target of bergapten (psoralen derivative present in many fruits and vegetables) action against BC cells for the induction of autophagy. The mechanism of action involved the p38MAPK/NF-Y pathway, site-direct mutagenesis of NF-Y element, and NF-Y siRNA regulation [107]. Anticancer effects were also exhibited by honokiol, a phytochemical from Magnolia spp., which caused a reduction in cell migration of murine BC cells through the COX-2 siRNA mechanism [108]. There are also other in vitro studies using plant-derived compounds pointing to siRNA mechanism influencing mammary carcinogenesis [109–111].
MicroRNAs
Several recent preclinical and clinical BC studies demonstrated that isolated phytochemicals or their mixture influence the expression of oncogenic or tumor-suppressive miRNAs, which, in turn, modulate the important signaling pathways in cancer cells and in this way affect the pathogenic properties of cancer cells and their sensitivity towards anticancer drugs. Stilbenoid resveratrol belongs among intensively evaluated molecules in BC research [112]. In the study of Hagiwara et al. [78], resveratrol upregulated several tumor-suppressive miRNAs (miR-16, miR-141, miR-143, and others) in MDA-MB-231 breast cancer cell line resulting in the induction of an anti-tumor effect against the cancer stem-like cells phenotype in cancer cells. In other experimental study, resveratrol inhibited DNA-methyltransferase DNMT3b in rat mammary tumors by upregulation of miR-129, miR-204, and miR-489 [113]. Other in vitro studies showed that tumor-suppressive miR-34a is an essential component of the antiproliferative activities of phytochemicals such as I3C and artemisinin [82]. Curcumin regulated the expression of several miRNAs in MCF-7 (miR-19a, miR-19b, miR-181b), MDA-MB-231, and MDA-MB-435 breast cancer cells (miR-34a) [114–116]. Currently, drug resistance is considered a main factor influencing BC clinical outcome. Dysregulated miRNAs contribute significantly to autophagy and chemoresistance. Therefore, targeting autophagy-related miRNAs is a novel strategy to reverse drug resistance. In the study of Wang et al. [117], natural compound isoliquiritigenin induced the chemosensitization drug-resistant BC cells through inhibition of the novel autophagy-related miR-25 in both in vitro and in vivo assays. Isolated phytochemicals were able to affect the levels of miRNAs in other experimental in vitro studies as well [79, 80, 83].
In addition, the experimental approach using mixtures of phytochemicals seems to be very promising in the positive modulation of miRNAs expression regarding the BC cells. Rhodes et al. [77] evaluated the anticancer activity of glyceollin mixture using the MDA-MB-231 xenograft model. Glyceollins significantly increased the expression of selected tumor-suppressive miRNAs (miR-22, miR-29b, miR-29c, miR-30d, miR-34a, miR-181c, miR-181d, and miR-195) and significantly decreased the expression of several oncogenic miRNAs (miR-21, miR-185, and miR-224). Banerjee et al. [118] showed in their experimental study in BC that the anticancer effects of pomegranate polyphenol extract were partially the result of downregulation of two miRNAs, miR-155 and miR-27a, using MDA-MB-231 and BT-474 cell lines, and BT474 xenografts in nude mice. In the recent study, blueberry delayed the tumor latency and reduced tumor volume and multiplicity in 17β-estradiol-mediated mammary carcinogenesis in female ACI rats partially through modulation of miR-18a and miR-34c levels [119]. In the study of Su et al. [119], the Chinese traditional medicine Antrodia cinnamomea showed significant anti-CSCs effects in several cancer cell lines, including MDA-MB-231 cells, through significant downregulation of selected oncogenic miRNAs in BC stem cells [120]. The clinical study of Guo et al. [84] assessed the linkage of soy intake with the expression of microRNAs (miRNAs) and genes in the tumor tissue of 272 women with TNBC. Thirteen of the 14 miRNAs, including tumor-suppressive miR-29a-3p, manifested overexpression in women with high soy intake. Authors concluded that long-term soy food intake is associated with the specific expression of selected miRNAs and has protective effect against BC risk in TNBC patients.
Long non-coding RNAs
Comprehensive research has shown that lncRNAs demonstrate crucial role in the modulation of gene expression at the level of chromatin, transcriptional, and posttranscriptional modifications [121, 122]. In the case of epigenetic modifications, previous experimental and clinical evaluations have showed that DNA methylation and modifications of histone result in deregulation of lncRNA expression in mammary tumors [123–127]. Regarding transcriptional modulation, lncRNAs interfere with the expression of high number of gene via interacting with chromatin at numerous different locations across multiple chromosomes [128]. Posttranscriptional regulation through lncRNAs includes modulation of mRNA stability, cell cycle distribution, and cell differentiation [127, 129]. To date, several recent preclinical and clinical oncological studies have shown that specific lncRNAs have a significant association with mammary carcinogenesis [124, 126, 130–143]. Some lncRNAs have great potential to be reliable biomarkers in the diagnosis, prognosis, and prediction of BC, or could be effectively used within anticancer therapy of this disease.
So far, few BC studies have dealt with the influence of plant natural substances on expression and/or function of lncRNAs. Chen et al. [81] demonstrated that genistein and calycosin were able to inhibit the expression of the oncogenic HOTAIR lncRNA in ER+ breast cancer cell line MCF-7. In other study using the xenograft BC model (MDA-MB-231-Luc-GFP breast cancer cells in athymic mice), the levels of HOTAIR lncRNAs were downregulated by using of anthocyanin delphinidin-3-glucoside [144]. Further studies in this field of research are necessary. Results of these studies may provide important information regarding the influence of phytochemicals on lncRNAs, which are significantly associated with BC and thus may help to improve prevention and therapeutic strategies in this disease. Qu et al. [145] described that compound Kushen injection (CKI), a complex mixture of plant secondary metabolites present in traditional Chinese medicine, shows significant anticancer effects in MCF-7 lines. Authors found that many lncRNAs were expressed as a response to CKI treatment.
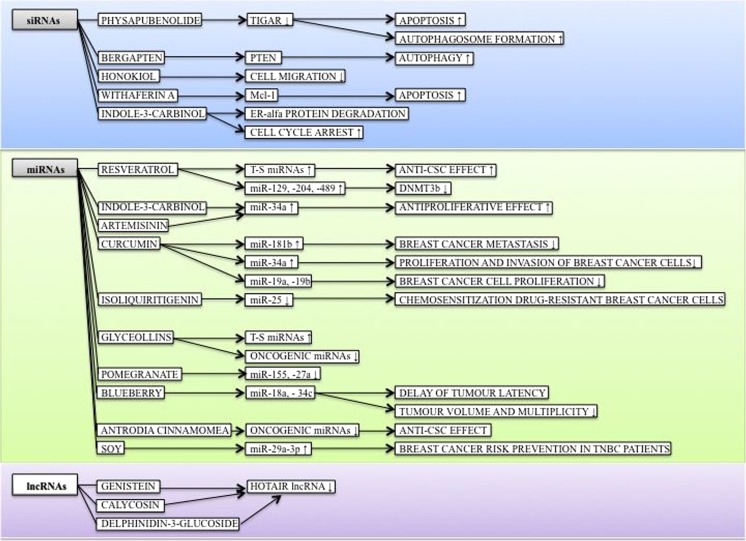
The mechanism of action of plant-derived compounds or plant foods involving siRNA, miRNA, and lncRNA regulation in breast carcinogenesis is summarized in Fig. 4.
Fig. 4.
Mechanism of action of phytochemicals involving non-coding RNA regulations within breast carcinogenesis. See text for indicated changes as each displayed axis represents the findings of presented studies. Arrows demonstrate specific effect of phytochemicals in female breast cancer model; ↑induction/upregulation, ↓inhibition/reduction. DNMT3b, DNA(cytosine-5-)-methyltransferase 3 beta; Mcl-1, induced myeloid leukemia cell differentiation protein; PTEN, phosphatase and tensin homolog; TIGAR, TP53-inducible glycolysis and apoptosis regulator; T-S, tumor-suppressive
4. Concluding remarks and outlook
Dietary habits and phytochemicals are of particular interest in terms of effective cancer prevention and targeted therapy [146, 147]. Many of plant-derived natural compounds demonstrated significant anticancer properties through multiple cell signaling pathways and mechanisms. Regarding BC, its chemoprevention and targeted therapies, an application of the dietary phytochemicals, is demonstrated as cost-effective and readily applicable clinical approach. The pivotal role of epigenetic mechanisms (DNA methylations, histone chemical modifications, and non-coding RNAs) has been demonstrated: epigenetic modifications by phytochemicals may play an important role in the targeted cancer chemoprevention. The demonstrated low toxicity of phytochemicals is crucial for their broad application. Noteworthy, most of the bioactive plant compounds demonstrate more than one epigenetic target. Consequently, a dual beneficial effect can be expected by upregulating tumor-suppressor genes and downregulating oncogenes. Further, a multidrug administration (combination of several plant compounds) may be more effective compared to the application of individual phytochemicals. This approach might lead to substantial progress made in the development of effective chemopreventive and therapeutic modalities. In this regard, Tables 1 and 2, and Fig. 4 summarize currently available information within BC research.
Despite the extensive research focused on human epigenome, there is still insufficient experimental data published on the epigenetic alterations induced by plant compounds. Moreover, most of the data in this topic are of preclinical nature, which are linked with several limitations within clinical practice. The future research focused on phytochemicals influencing the human epigenome should be directed within the several issues: (a) Novel data demand more detailed insight on the molecular mechanisms responsible for these effects. (b) The next essential step is to determine an effective (individual) dose of dietary phytochemicals. The doses used in preclinical research in vitro and in vivo is to be taken with caution, because such high concentrations of phytochemicals might not be achievable in humans, and consequently might lack clinical benefits. In this respect, the synthesis of chemical analogues of active substances could be very helpful. (c) It might be useful to extend the spectrum of relevant phytochemicals by currently untested isolated molecules, extracts, and/or whole plant products. (d) Assessing the combined effects of several plant compounds targeting several relevant epigenetic pathways could be a promising strategy for future epigenetic experiments. (e) Modifying sub-optimal epigenomes by phytochemicals in order to regulate the expression of key inflammatory genes may be an effective approach by immunotherapy. (f) Investigating epigenetic mechanisms induced by plant compounds linked with the cancer stem cells survival could provide important tool for oncologist in the management of cancer disease, regarding the relapse or multidrug resistance. Moreover, “epigenetic therapy” might have a great potential to overcome drug resistance and/or to re-sensitize cancer cells towards chemotherapy. (g) An improved bioavailability of phytochemicals (for example, by utilizing nanomaterial complexes) should be furtherly considered. (h) An administration of plant compounds either for chemoprevention or treatment of the clinically manifested BC represents an attractive approach enhancing the efficacy of conventional therapies. (i) The plant compounds demonstrate cell-, organ-, and organism-specific effects; therefore, better understanding of the target mechanisms and individual characteristics is a big challenge for scientists to develop personalized supplements. Currently preferred diagnostic approaches are in many cases unable to identify early stages of cancer initiation/promotion that impairs clinical outcomes [148]. Innovative screening programs, multiomic diagnostics including evaluation of epigenetic changes (such as global DNA and gene promoters methylation status, histone chemical modifications, and miRNAs expressions), and individualized patient profiling and stratification are crucial clinical approaches to target and start personalized preventive programs in high-risk individuals [149, 150]. Each epigenetic mechanism is predicted to have many molecular targets [151] and therefore may be regulated by plethora of specific phytochemicals or plant foods. The applicability of major phytochemicals/foods as an “epi-drugs” against cancer disease is already experimentally established and has great potential to open a new area of individualized healthcare in the clinical medicine [152]. The above stated measures have a great potential to advance the overall BC management in favor of predictive, preventive, and personalized medical services and can be considered as the “proof-of principle” model for their potential application to other multifactorial diseases.
Authors’ contribution
PK contributed to conception of the idea, literature search, manuscript drafting and editing and SU, ZD, AK, PS, MS, KK, MK, MP, and BZ contributed to literature search and manuscript drafting. VV sketched and drew the figures. OG and TKK contributed to conception of the idea and edited the manuscript. AZ, PZ, DB, and JD revised the manuscript with critical reviews and comments and all the authors approved the final version.
Funding information
This work was supported by the Scientific Grant Agency of the Ministry of Education of the Slovak Republic under the contracts no. VEGA 1/0108/16, 1/0018/16 and the Slovak Research and Development Agency under the contract no. APVV-16-0021.
Conflict of interest
The authors declare that they have no conflict of interest.
Statement of informed consent
Patients have not been involved in the study.
Statement of human and animal rights
No experiments have been performed including patients and/or animals.
Footnotes
Sona Uramova and Peter Kubatka are co-first/equal authorship.
References
- 1.Golubnitschaja O, Debald M, Yeghiazaryan K, Kuhn W, Pešta M, Costigliola V, Grech G. Breast cancer epidemic in the early 21st century: evaluation of risk factors, cumulative questionnaires and recommendations for preventive measures. Tumor Biol. 2016;37:12941–12957. doi: 10.1007/s13277-016-5168-x. [DOI] [PubMed] [Google Scholar]
- 2.Polivka J, Jr, Kralickova M, Polivka J, Jr, Kaiser C, Kuhn W, Golubnitschaja O. Mystery of the brain metastatic disease in breast cancer patients: improved patient stratification, disease prediction and targeted prevention on the horizon? EPMA J. 2017;8:119–127. doi: 10.1007/s13167-017-0087-5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golubnitschaja O. Feeling cold and other underestimated symptoms in breast cancer: anecdotes or individual profiles for advanced patient stratification? EPMA J. 2017;8:17–22. doi: 10.1007/s13167-017-0086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zubor P, Gondova A, Polivka J, Jr, Kasajova P, Konieczka K, Danko J, et al. Breast cancer and Flammer syndrome: any symptoms in common for prediction, prevention and personalised medical approach? EPMA J. 2017;8:129–140. doi: 10.1007/s13167-017-0089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bubnov R, Polivka J, Jr, Zubor P, Koniczka K, Golubnitschaja O. “Pre-metastatic niches” in breast cancer: are they created by or prior to the tumour onset? “Flammer syndrome” relevance to address the question. EPMA J. 2017;8:141–157. doi: 10.1007/s13167-017-0092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, Camargo AA, Stevenson BJ, Ecker JR, Bafna V, Strausberg RL, Simpson AJ, Ren B. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng JM, Yu J. Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int J Mol Sci. 2015;16:2472–2496. doi: 10.3390/ijms16022472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhavan-Niaki H, Samadani AA. DNA methylation and cancer development: molecular mechanism. Cell Biochem Biophys. 2013;67(2):501–513. doi: 10.1007/s12013-013-9555-2. [DOI] [PubMed] [Google Scholar]
- 9.Li Ya, Li Sha, Meng Xiao, Gan Ren-You, Zhang Jiao-Jiao, Li Hua-Bin. Dietary Natural Products for Prevention and Treatment of Breast Cancer. Nutrients. 2017;9(7):728. doi: 10.3390/nu9070728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla S, Meeran SM, Katiyar SK. Epigenetic regulation by selected dietary phytochemicals in cancer chemoprevention. Cancer Lett. 2014;355:9–17. doi: 10.1016/j.canlet.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapira N. The potential contribution of dietary factors to breast cancer prevention. Eur J Cancer Prev. 2017;26:385–395. doi: 10.1097/CEJ.0000000000000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giacosa A, Barale R, Bavaresco L, Gatenby P, Gerbi V, Janssens J, Johnston B, Kas K, la Vecchia C, Mainguet P, Morazzoni P, Negri E, Pelucchi C, Pezzotti M, Rondanelli M. Cancer prevention in Europe: the Mediterranean diet as a protective choice. Eur J Cancer Prev. 2013;22:90–95. doi: 10.1097/CEJ.0b013e328354d2d7. [DOI] [PubMed] [Google Scholar]
- 13.Simopoulos AP. The traditional diet of Greece and cancer. Eur J Cancer Prev. 2004;13:219–230. doi: 10.1097/01.cej.0000130011.99148.07. [DOI] [PubMed] [Google Scholar]
- 14.Dalasanur Nagaprashantha Lokesh, Adhikari Ramesh, Singhal Jyotsana, Chikara Shireen, Awasthi Sanjay, Horne David, Singhal Sharad S. Translational opportunities for broad-spectrum natural phytochemicals and targeted agent combinations in breast cancer. International Journal of Cancer. 2017;142(4):658–670. doi: 10.1002/ijc.31085. [DOI] [PubMed] [Google Scholar]
- 15.Khan S, Shukla S, Sinha S, Meeran SM. Epigenetic targets in cancer and aging: dietary and therapeutic interventions. Expert Opin Ther Targets. 2016;20:689–703. doi: 10.1517/14728222.2016.1132702. [DOI] [PubMed] [Google Scholar]
- 16.Shankar E, Kanwal R, Candamo M, Gupta S. Dietary phytochemicals as epigenetic modifiers in cancer: promise and challenges. Semin Cancer Biol. 2016;40-41:82–99. doi: 10.1016/j.semcancer.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, Absher DM, Wold BJ, Myers RM. Dynamic DNA methylation across diverse human cell lines and tissues. Gen Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veland N, Hardikar S, Zhong Y, Gayatri S, Dan J, Strahl BD, et al. The arginine methyltransferase PRMT6 regulates DNA methylation and contributes to global DNA hypomethylation in cancer. Cell Rep. 2017;21:3390–3397. doi: 10.1016/j.celrep.2017.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heichman KA, Warren JD. DNA methylation biomarkers and their utility for solid cancer diagnostics. Clin Chem Lab Med. 2012;50:1707–1721. doi: 10.1515/cclm-2011-0935. [DOI] [PubMed] [Google Scholar]
- 20.Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5:553–568. doi: 10.2217/epi.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang HJ, Petrelli NJ, Zhang X, O'Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 23.Ehrlich M, Jiang G, Fiala E, Dome JS, Yu MC, Long TI, Youn B, Sohn OS, Widschwendter M, Tomlinson GE, Chintagumpala M, Champagne M, Parham D, Liang G, Malik K, Laird PW. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene. 2002;21:6694–6702. doi: 10.1038/sj.onc.1205890. [DOI] [PubMed] [Google Scholar]
- 24.Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CPE, van Dijk CM, Tollenaar RAEM, van den Berg D, Laird PW. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar U, Sharma U, Rathi G. Reversal of hypermethylation and reactivation of glutathione S-transferase pi 1 gene by curcumin in breast cancer cell line. Tumour Biol. 2017;39:101042831769225. doi: 10.1177/1010428317692258. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Long X. Association of BRCA1 promoter methylation with sporadic breast cancers: evidence from 40 studies. Sci Rep. 2015;5:17869. doi: 10.1038/srep17869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du L, Xie Z, Wu LC, Chiu M, Lin J, Chan KK, et al. Reactivation of RASSF1A in breast cancer cells by curcumin. Nutr Cancer. 2012;64:1228–1235. doi: 10.1080/01635581.2012.717682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirza S, Sharma G, Parshad R, Gupta SD, Pandya P, Ralhan R. Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins. J Breast Cancer. 2013;16:23–31. doi: 10.4048/jbc.2013.16.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewinska A, Adamczyk-Grochala J, Deregowska A, Wnuk M. Sulforaphane-induced cell cycle arrest and senescence are accompanied by DNA hypomethylation and changes in microRNA profile in breast cancer cells. Theranostics. 2017;7:3461–3477. doi: 10.7150/thno.20657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meeran SM, Patel SN, Li Y, Shukla S, Tollefsbol TO. Bioactive dietary supplements reactivate ER expression in ER-negative breast cancer cells by active chromatin modifications. PLoS One. 2012;7:e37748. doi: 10.1371/journal.pone.0037748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lubecka-Pietruszewska K, Kaufman-Szymczyk A, Stefanska B, Cebula-Obrzut B, Smolewski P, Fabianowska-Majewska K. Sulforaphane alone and in combination with clofarabine epigenetically regulates the expression of DNA methylation-silenced tumour suppressor genes in human breast cancer cells. J Nutrigenet Nutrigenomics. 2015;8:91–101. doi: 10.1159/000439111. [DOI] [PubMed] [Google Scholar]
- 32.Kala R, Shah HN, Martin SL, Tollefsbol TO. Epigenetic-based combinatorial resveratrol and pterostilbene alters DNA damage response by affecting SIRT1 and DNMT enzyme expression, including SIRT1-dependent γ-H2AX and telomerase regulation in triple-negative breast cancer. BMC Cancer. 2015;15:672. doi: 10.1186/s12885-015-1693-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szarc Vel Szic K, Declerck K, Crans RAJ, Diddens J, Scherf DB, Gerhäuser C, vanden Berghe W. Epigenetic silencing of triple negative breast cancer hallmarks by withaferin A. Oncotarget. 2017;8:40434–40453. doi: 10.18632/oncotarget.17107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyagi T, Treas JN, Mahalingaiah PK, Singh KP. Potentiation of growth inhibition and epigenetic modulation by combination of green tea polyphenol and 5-aza-2′-deoxycytidine in human breast cancer cells. Breast Cancer Res Treat. 2015;149:655–668. doi: 10.1007/s10549-015-3295-5. [DOI] [PubMed] [Google Scholar]
- 35.Aumsuwan P, Khan SI, Khan IA, Avula B, Walker LA, Helferich WG, Katzenellenbogen BS, Dasmahapatra AK. Evaluation of wild yam (Dioscorea villosa) root extract as a potential epigenetic agent in breast cancer cells. In Vitro Cell Dev Biol Anim. 2015;51:59–71. doi: 10.1007/s11626-014-9807-5. [DOI] [PubMed] [Google Scholar]
- 36.Weng JR, Lai IL, Yang HC, Lin CN, Bai LY. Identification of kazinol Q, a natural product from Formosan plants, as an inhibitor of DNA methyltransferase. Phytother Res. 2014;28:49–54. doi: 10.1002/ptr.4955. [DOI] [PubMed] [Google Scholar]
- 37.Xie Q, Bai Q, Zou LY, Zhang QY, Zhou Y, Chang H, Yi L, Zhu JD, Mi MT. Genistein inhibits DNA methylation and increases expression of tumor suppressor genes in human breast cancer cells. Genes Chromosom Cancer. 2014;53:422–431. doi: 10.1002/gcc.22154. [DOI] [PubMed] [Google Scholar]
- 38.Bozkurt E, Atmaca H, Kisim A, Uzunoglu S, Uslu R, Karaca B. Effects of Thymus serpyllum extract on cell proliferation, apoptosis and epigenetic events in human breast cancer cells. Nutr Cancer. 2012;64(8):1245–1250. doi: 10.1080/01635581.2012.719658. [DOI] [PubMed] [Google Scholar]
- 39.Rodríguez-Miguel C, Moral R, Escrich R, Vela E, Solanas M, Escrich E. The role of dietary extra virgin olive oil and corn oil on the alteration of epigenetic patterns in the rat DMBA-induced breast cancer model. PLoS One. 2015;10:e0138980. doi: 10.1371/journal.pone.0138980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kubatka P, Uramova S, Kello M, Kajo K, Kruzliak P, Mojzis J, Vybohova D, Adamkov M, Jasek K, Lasabova Z, Zubor P, Fialova S, Dokupilova S, Solar P, Pec M, Adamicova K, Danko J, Adamek M, Busselberg D. Antineoplastic effects of clove buds (Syzygium aromaticum L.) in the model of breast carcinoma. J Cell Mol Med. 2017;21:2837–2851. doi: 10.1111/jcmm.13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang N, Wang Z, Wang Y, Xie X, Shen J, Peng C, You J, Peng F, Tang H, Guan X, Chen J. Dietary compound isoliquiritigenin prevents mammary carcinogenesis by inhibiting breast cancer stem cells through WIF1 demethylation. Oncotarget. 2015;6:9854–9876. doi: 10.18632/oncotarget.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenlee H, Ogden Gaffney A, Aycinena AC, Koch P, Contento I, Karmally W, Richardson JM, Shi Z, Lim E, Tsai WY, Santella RM, Blaner WS, Clugston RD, Cremers S, Pollak S, Sirosh I, Crew KD, Maurer M, Kalinsky K, Hershman DL. Long-term diet and biomarker changes after a short-term intervention among Hispanic breast cancer survivors: the ¡Cocinar Para Su Salud! randomized controlled trial. Cancer Epidemiol Biomark Prev. 2016;25:1491–1502. doi: 10.1158/1055-9965.EPI-15-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delgado-Cruzata L, Zhang W, McDonald JA, Tsai WY, Valdovinos C, Falci L, et al. Dietary modifications, weight loss, and changes in metabolic markers affect global DNA methylation in Hispanic, African American, and Afro-Caribbean breast cancer survivors. J Nutr. 2015;4:783–790. doi: 10.3945/jn.114.202853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirouzpanah S, Taleban FA, Mehdipour P, Atri M. Association of folate and other one-carbon related nutrients with hypermethylation status and expression of RARB, BRCA1, and RASSF1A genes in breast cancer patients. J Mol Med (Berl) 2015;93:917–934. doi: 10.1007/s00109-015-1268-0. [DOI] [PubMed] [Google Scholar]
- 45.Füllgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- 46.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 47.Chervona Y, Costa M. Histone modifications and cancer: biomarkers of prognosis? Am J Cancer Res. 2012;2:589–597. [PMC free article] [PubMed] [Google Scholar]
- 48.Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA, Grainge MJ, Ball GR, Abdelghany MK, Martinez-Pomares L, Heery DM, Ellis IO. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 49.Müller BM, Jana L, Kasajima A, Lehmann A, Prinzler J, Budczies J, Winzer KJ, Dietel M, Weichert W, Denkert C. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer--overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer. 2013;13:215. doi: 10.1186/1471-2407-13-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weng JR, Bai LY, Lin WY, Chiu CF, Chen YC, Chao SW, et al. A flavone constituent from Myoporum bontioides induces M-phase cell cycle arrest of MCF-7 breast cancer cells. Molecules. 2017;22:472. doi: 10.3390/molecules22030472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mirabella AC, Foster BM, Berke T. Chromatin deregulation in disease. Chromosoma. 2016;125:75–93. doi: 10.1007/s00412-015-0530-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bathaie SZ, Tmanoi F. Mechanism of the anticancer effect of phytochemicals (the enzymes), 1st edn. Elsevier Inc.; 2015.
- 53.Busch CH, Burkard M, Leischner CH, Lauer UM, Frank J, Venturelli S. Epigenetic activities of flavonoids in the prevention and treatment of cancer. Clin Epigenetics. 2015;7:64. doi: 10.1186/s13148-015-0095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics. 2011;3:503–518. doi: 10.2217/epi.11.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Messier TL, Gordon JAR, Boyd JR, Tye CE, Browne G, Stein JL, Lian JB, Stein GS. Histone H3 lysine 4 acetylation and methylation dynamics define breast cancer subtypes. Oncotarget. 2016;7:5094–5109. doi: 10.18632/oncotarget.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen I, Poreba E, Kamieniarz K, Schneider R. Histone modifiers in cancer. Friends or foes? Genes Cancer. 2011;2:631–647. doi: 10.1177/1947601911417176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 58.Yiannakopoulou ECH. Effect of green tea catechins on breast carcinogenesis: a systematic review of in-vitro and in-vivo experimental studies. Eur J Cancer Prev. 2014;23:84–89. doi: 10.1097/CEJ.0b013e328364f23e. [DOI] [PubMed] [Google Scholar]
- 59.Thangapazham R, Singh A, Sharma A, Warren J, Gaddipati J, Maheshwari R. Green tea polyphenols and its constituent epigallocatechin gallate inhibits proliferation of human breast cancer cells in vitro and in vivo. Cancer Lett. 2007;245:232–241. doi: 10.1016/j.canlet.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 60.Mittal A, Pate MS, Wylie RC, Tollefsbol TO, Katiyar SK. EGCG down-regulates telomerase in human breast carcinoma MCF-7 cells, leading to suppression of cell viability and induction of apoptosis. Int J Oncol. 2004;24:703–710. [PubMed] [Google Scholar]
- 61.Li Y, Yuan YY, Meeran SM, Tollefsbol TO. Synergistic epigenetic reactivation of estrogen receptor-α (ERα) by combined green tea polyphenol and histone deacetylase inhibitor in ERα-negative breast cancer cells. Mol Cancer. 2010;9:274. doi: 10.1186/1476-4598-9-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meeran SM, Patel SH, Chan TH, Tollefsbol TO. A novel prodrug of epigallocatechin-3-gallate: differential epigenetic hTERT repression in human breast cancer cells. Cancer Prev Res (Phila) 2011;4:1243–1254. doi: 10.1158/1940-6207.CAPR-11-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thakur VS, Deb G, Babcook MA, Gupta S. Plant phytochemicals as epigenetic modulators: role in cancer chemoprevention. AAPS J. 2014;16:151–163. doi: 10.1208/s12248-013-9548-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Collins HM, Abdelghany MK, Messmer M, Yue B, Deeves SE, Kindle KB, Mantelingu K, Aslam A, Winkler GS, Kundu TK, Heery DM. Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells. BMC Cancer. 2013;13:37. doi: 10.1186/1471-2407-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oliveras-Ferraros C, Fernández-Arroyo S, Vazquez-Martin AV. Crude phenolic extracts from extra virgin olive oil circumvent de novo breast cancer resistance to HER1/HER2-targeting drugs by inducing GADD45-sensed cellular stress, G2/M arrest and hyperacetylation of histone H3. Int J Oncol. 2011;38:1533–1547. doi: 10.3892/ijo.2011.993. [DOI] [PubMed] [Google Scholar]
- 66.Altonsy MO, Habib TN, Andrews SC. Diallyl disulfide-induced apoptosis in a breast-cancer cell line (MCF-7) may be caused by inhibition of histone deacetylation. Nutr Cancer. 2012;64:1251–1260. doi: 10.1080/01635581.2012.721156. [DOI] [PubMed] [Google Scholar]
- 67.Nian H, Delage B, Ho E, Dashwood RH. Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds. Environ Mol Mutagen. 2009;50:213–221. doi: 10.1002/em.20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Attoub S, Hassan AH, Vanhoecke B, Iratni R, Takahashi T, Gaben AM, Bracke M, Awad S, John A, Kamalboor HA, al Sultan MA, Arafat K, Gespach C, Petroianu G. Inhibition of cell survival, invasion, tumor growth and histone deacetylase activity by the dietary flavonoid luteolin in human epithelioid cancer cells. Eur J Pharmacol. 2011;651:18–25. doi: 10.1016/j.ejphar.2010.10.063. [DOI] [PubMed] [Google Scholar]
- 69.Weng JR, Bai LY, Lin WY. Identification of a triterpenoid as a novel PPARγ activator derived from Formosan plants. Phytother Res. 2017;31:1722–1730. doi: 10.1002/ptr.5900. [DOI] [PubMed] [Google Scholar]
- 70.Rivenbark AG, Coleman WB, Stahl BD. Histone methylation patterns in human breast cancer. FASEB J. 2009;23:S38.1. [Google Scholar]
- 71.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yokoyama Y, Matsumoto A, Hieda M, Shinchi Y, Ogihara E, Hamada M, Nishioka Y, Kimura H, Yoshidome K, Tsujimoto M, Matsuura N. Loss of histone H4K20 trimethylation predicts poor prognosis in breast cancer and is associated with invasive activity. Breast Cancer Res. 2014;16:R66. doi: 10.1186/bcr3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pudenz M, Roth K, Gerhauser C. Impact of soy isoflavones on the epigenome in cancer prevention. Nutrients. 2014;6:4218–4272. doi: 10.3390/nu6104218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Meeran SM, Patel SN, Chen H, Hardy TM, Tollefsbol TO. Epigenetic reactivation of estrogen receptor-α (ERα) by genistein enhances hormonal therapy sensitivity in ERα-negative breast cancer. Mol Cancer. 2013;12:9. doi: 10.1186/1476-4598-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dagdemir A, Durif J, Ngollo M, Bignon YJ, Bernard-Gallon D. Histone lysine trimethylation or acetylation can be modulated by phytoestrogen, estrogen or anti-HDAC in breast cancer cell lines. Epigenomics. 2013;5:51–63. doi: 10.2217/epi.12.74. [DOI] [PubMed] [Google Scholar]
- 76.Li Y, Chen H, Hardy TM, Tollefsbol TO. Epigenetic regulation of multiple tumor-related genes leads to suppression of breast tumorigenesis by dietary genistein. PLoS One. 2013;8:e54369. doi: 10.1371/journal.pone.0054369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rhodes LV, Tilghman SL, Boue SM. Glyceollins as novel targeted therapeutic for the treatment of triple-negative breast cancer. Oncol Lett. 2012;3:163–171. doi: 10.3892/ol.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hagiwara K, Kosaka N, Yoshioka Y, Takahashi RU, Takeshita F, Ochiya T. Stilbene derivatives promote Ago2- dependent tumour- suppressive microRNA activity. Sci Rep. 2012;2:314. doi: 10.1038/srep00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahmad A, Ali S, Ahmed A, Ali AS, Raz A, Sakr WA, Rahman KMW. 3, 3′- Diindolylmethane enhances the effectiveness of Herceptin against HER-2/neu- expressing breast cancer cells. PLoS One. 2013;8:e54657. doi: 10.1371/journal.pone.0054657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li X, Xie W, Xie C, Huang C, Zhu J, Liang Z, Deng F, Zhu M, Zhu W, Wu R, Wu J, Geng S, Zhong C. Curcumin modulates miR-19/PTEN/AKT/p53 axis to suppress bisphenol A-induced MCF-7 breast cancer cell proliferation. Phytother Res. 2014;28:1553–1560. doi: 10.1002/ptr.5167. [DOI] [PubMed] [Google Scholar]
- 81.Chen J, Lin C, Yong W, Ye Y, Huang Z. Calycosin and genistein induce apoptosis by inactivation of HOTAIR/p-Akt signaling pathway in human breast cancer MCF-7 cells. Cell Physiol Biochem. 2015;35:722–728. doi: 10.1159/000369732. [DOI] [PubMed] [Google Scholar]
- 82.Hargraves KG, He L, Firestone GL. Phytochemical regulation of the tumor suppressive microRNA, miR-34a, by p53-dependent and independent responses in human breast cancer cells. Mol Carcinog. 2016;55:486–498. doi: 10.1002/mc.22296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de la Parra C, Castillo-Pichardo L, Cruz-Collazo A, Cubano L, Redis R, Calin GA, Dharmawardhane S. Soy isoflavone genistein-mediated downregulation of miR-155 contributes to the anticancer effects of genistein. Nutr Cancer. 2016;68:154–164. doi: 10.1080/01635581.2016.1115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guo X, Cai Q, Bao P, Wu J, Wen W, Ye F, Zheng W, Zheng Y, Shu XO. Long-term soy consumption and tumor tissue MicroRNA and gene expression in triple-negative breast cancer. Cancer. 2016;122:2544–2551. doi: 10.1002/cncr.29981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yeung ML, Jeang KT. MicroRNAs and cancer therapeutics. Pharm Res. 2011;28:3043–3049. doi: 10.1007/s11095-011-0526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abdelrahim M, Safe S, Baker C, Abudayyeh A. RNAi and cancer: implications and applications. J RNAi Gene Silenc. 2006;2:136–145. [PMC free article] [PubMed] [Google Scholar]
- 87.Mansoori B, Sandoghchian Shotorbani S, Baradaran B. RNA interference and its role in cancer therapy. Adv Pharm Bull. 2014;4:313–321. doi: 10.5681/apb.2014.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145–2156. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kong AN, Zhang C, Su ZY. Targeting epigenetics for cancer prevention by dietary cancer preventive compounds--the case of miRNA. Cancer Prev Res. 2013;6:622–624. doi: 10.1158/1940-6207.CAPR-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 91.Papoutsis AJ, Lamore SD, Wondrak GT, Selmin OI, Romagnolo DF. Resveratrol prevents epigenetic silencing of BRCA-1 by the aromatic hydrocarbon receptor in human breast cancer cells. J Nutr. 2010;140:1607–1614. doi: 10.3945/jn.110.123422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vaid M, Prasad R, Singh T, Jones V, Katiyar SK. Grape seed proanthocyanidins reactivate silenced tumor suppressor genes in human skin cancer cells by targeting epigenetic regulators. Toxicol Appl Pharmacol. 2012;263:122–130. doi: 10.1016/j.taap.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Petrocca F, Lieberman J. Promise and challenge of RNA interference-based therapy for cancer. J Clin Oncol. 2011;29:747–754. doi: 10.1200/JCO.2009.27.6287. [DOI] [PubMed] [Google Scholar]
- 94.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 96.Parvani JG, Jackson MW. Silencing the roadblocks to effective triple-negative breast cancer treatments by siRNA nanoparticles. Endocr Relat Cancer. 2017;24:R81–R97. doi: 10.1530/ERC-16-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75–86 100. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Young SW, Stenzel M, Yang JL. Nanoparticle-siRNA: a potential cancer therapy? Crit Rev Oncol Hematol. 2016;98:159–169. doi: 10.1016/j.critrevonc.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 99.Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- 100.Deng ZJ, Morton SW, Ben-Akiva E, Dreaden EC, Shopsowitz KE, Hammond PT. Layer-by-layer nanoparticles for systemic codelivery of an anticancer drug and siRNA for potential triple-negative breast cancer treatment. ACS Nano. 2013;7:9571–9784. doi: 10.1021/nn4047925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feng Q, Yu MZ, Wang JC, Hou WJ, Gao LY, Ma XF, Pei XW, Niu YJ, Liu XY, Qiu C, Pang WH, du LL, Zhang Q. Synergistic inhibition of breast cancer by co-delivery of VEGF siRNA and paclitaxel via vapreotide-modified core-shell nanoparticles. Biomaterials. 2014;35:5028–5038. doi: 10.1016/j.biomaterials.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 102.Liu Y, Zhu YH, Mao CQ, Dou S, Shen S, Tan ZB, Wang J. Triple negative breast cancer therapy with CDK1 siRNA delivered by cationic lipid assisted PEG-PLA nanoparticles. J Control Release. 2014;192:114–121. doi: 10.1016/j.jconrel.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 103.Parvani JG, Gujrati MD, Mack MA, Schiemann WP, Lu ZR. Silencing β3 integrin by targeted ECO/siRNA nanoparticles inhibits EMT and metastasis of triple-negative breast cancer. Cancer Res. 2015;75:2316–2325. doi: 10.1158/0008-5472.CAN-14-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Su S, Tian Y, Li Y, Ding Y, Ji T, Wu M, Wu Y, Nie G. “Triple-punch” strategy for triple negative breast cancer therapy with minimized drug dosage and improved antitumor efficacy. ACS Nano. 2015;9:1367–1378. doi: 10.1021/nn505729m. [DOI] [PubMed] [Google Scholar]
- 105.Schultheis B, Strumberg D, Santel A, Vank C, Gebhardt F, Keil O, Lange C, Giese K, Kaufmann J, Khan M, Drevs J. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J Clin Oncol. 2014;32:4141–4148. doi: 10.1200/JCO.2013.55.0376. [DOI] [PubMed] [Google Scholar]
- 106.Ma T, Zhang Y, Zhang C, Luo JG, Kong LY. Downregulation of TIGAR sensitizes the antitumor effect of physapubenolide through increasing intracellular ROS levels to trigger apoptosis and autophagosome formation in human breast carcinoma cells. Biochem Pharmacol. 2017;143:90–106. doi: 10.1016/j.bcp.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 107.De Amicis F, Aquila S, Morelli C, Guido C, Santoro M, Perrotta I, et al. Bergapten drives autophagy through the up-regulation of PTEN expression in breast cancer cells. Mol Cancer. 2015;14:130. doi: 10.1186/s12943-015-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Singh T, Katiyar SK. Honokiol, a phytochemical from Magnolia spp., inhibits breast cancer cell migration by targeting nitric oxide and cyclooxygenase-2. Int J Oncol. 2011;38:769–776. doi: 10.3892/ijo.2011.899. [DOI] [PubMed] [Google Scholar]
- 109.Hahm ER, Lee J, Singh SV. Role of mitogen-activated protein kinases and Mcl-1 in apoptosis induction by withaferin a in human breast cancer cells. Mol Carcinog. 2014;53:907–916. doi: 10.1002/mc.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Marconett CN, Sundar SN, Poindexter KM, Stueve TR, Bjeldanes LF, Firestone GL. Indole-3-carbinol triggers aryl hydrocarbon receptor-dependent estrogen receptor (ER)alpha protein degradation in breast cancer cells disrupting an ERalpha-GATA3 transcriptional cross-regulatory loop. Mol Biol Cell. 2010;21:1166–1177. doi: 10.1091/mbc.E09-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nguyen HH, Aronchik I, Brar GA, Nguyen DH, Bjeldanes LF, Firestone GL. The dietary phytochemical indole-3-carbinol is a natural elastase enzymatic inhibitor that disrupts cyclin E protein processing. Proc Natl Acad Sci U S A. 2008;105:19750–19755. doi: 10.1073/pnas.0806581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kisková T, Jendželovský R, Rentsen E, Maier-Salamon A, Kokošová N, Papčová Z, Mikeš J, Orendáš P, Bojková B, Kubatka P, Svoboda M, Kajo K, Fedoročko P, Jäger W, Ekmekcioglu C, Kassayová M, Thalhammer T. Resveratrol enhances the chemopreventive effect of celecoxib in chemically induced breast cancer in rats. Eur J Cancer Prev. 2014;23:506–513. doi: 10.1097/CEJ.0000000000000083. [DOI] [PubMed] [Google Scholar]
- 113.Qin W, Zhang K, Clarke K, Weiland T, Sauter ER. Methylation and miRNA effects of resveratrol on mammary tumors vs. normal tissue. Nutr Cancer. 2014;66:270–277. doi: 10.1080/01635581.2014.868910. [DOI] [PubMed] [Google Scholar]
- 114.Li Q, Eades G, Yao Y, Zhang Y, Zhou Q. Characterization of a stem-like subpopulation in basal-like ductal carcinoma in situ (DCIS) lesions. J Biol Chem. 2014;289:1303–1312. doi: 10.1074/jbc.M113.502278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kronski E, Fiori ME, Barbieri O, Astigiano S, Mirisola V, Killian PH, Bruno A, Pagani A, Rovera F, Pfeffer U, Sommerhoff CP, Noonan DM, Nerlich AG, Fontana L, Bachmeier BE. miR181b is induced by the chemopreventive polyphenol curcumin and inhibits breast cancer metastasis via down-regulation of the inflammatory cytokines CXCL1 and −2. Mol Oncol. 2014;8:581–595. doi: 10.1016/j.molonc.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guo J, Li W, Shi H, Xie X, Li L, Tang H, et al. Synergistic effects of curcumin with emodin against the proliferation and invasion of breast cancer cells through upregulation of miR-34a. Mol Cell Biochem. 2013;82:103–111. doi: 10.1007/s11010-013-1723-6. [DOI] [PubMed] [Google Scholar]
- 117.Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie T, Zhang J, Peng C, Lin Y, Chen J. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget. 2014;5:7013–7026. doi: 10.18632/oncotarget.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Banerjee N, Talcott S, Safe S, Mertens-Talcott SU. Cytotoxicity of pomegranate polyphenolics in breast cancer cells in vitro and vivo: potential role of miRNA-27a and miRNA-155 in cell survival and inflammation. Breast Cancer Res Treat. 2012;136:21–34. doi: 10.1007/s10549-012-2224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jeyabalan J, Aqil F, Munagala R, Annamalai L, Vadhanam MV, Gupta RC. Chemopreventive and therapeutic activity of dietary blueberry against estrogen-mediated breast cancer. J Agric Food Chem. 2014;62:3963–3971. doi: 10.1021/jf403734j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Su YK, Shih PH, Lee WH, Bamodu OA, Wu ATH, Huang CC, Tzeng YM, Hsiao M, Yeh CT, Lin CM. Antrodia cinnamomea sensitizes radio−/chemo-therapy of cancer stem-like cells by modulating microRNA expression. J Ethnopharmacol. 2017;207:47–56. doi: 10.1016/j.jep.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 121.Dong J, Xu J, Wang X, Jin B. Influence of the interaction between long noncoding RNAs and hypoxia on tumorigenesis. Tumour Biol. 2016;37:1379–1385. doi: 10.1007/s13277-015-4457-0. [DOI] [PubMed] [Google Scholar]
- 122.Zhang M, Gu H, Xu W, Zhou X. Down–regulation of lncRNA MALAT1 reduces cardiomyocyte apoptosis and improves left ventricular function in diabetic rats. Int J Cardiol. 2016;203:214–216. doi: 10.1016/j.ijcard.2015.10.136. [DOI] [PubMed] [Google Scholar]
- 123.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lo PK, Zhang Y, Wolfson B, Gernapudi R, Yao Y, Duru N, Zhou Q. Dysregulation of the BRCA1/long non- coding RNA NEAT1 signaling axis contributes to breast tumorigenesis. Oncotarget. 2016;7:65067–65089. doi: 10.18632/oncotarget.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lu L, Zhu G, Zhang C, Deng Q, Katsaros D, Mayne ST, Risch HA, Mu L, Canuto EM, Gregori G, Benedetto C, Yu H. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat. 2012;136:875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 126.Shen X, Xie B, Ma Z, Yu W, Wang W, Xu D, Yan X, Chen B, Yu L, Li J, Chen X, Ding K, Cao F. Identification of novel long non-coding RNAs in triple-negative breast cancer. Oncotarget. 2015;6:21730–21739. doi: 10.18632/oncotarget.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vennin C, Spruyt N, Robin YM, Chassat T, Le Bourhis X, Adriaenssens E. The long non-coding RNA 91H increases aggressive phenotype of breast cancer cells and up-regulates H19/IGF2 expression through epigenetic modifications. Cancer Lett. 2017;385:198–206. doi: 10.1016/j.canlet.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 128.Wang L, Chen Z, An L, Wang Y, Zhang Z, Guo Y, Liu C. Analysis of long non-coding RNA expression profiles in non-small cell lung cancer. Cell Physiol Biochem. 2016;38:2389–2400. doi: 10.1159/000445591. [DOI] [PubMed] [Google Scholar]
- 129.Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015;17:40. doi: 10.1186/s13058-015-0542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lei B, Xu SP, Liang XS, Li YW, Zhang JF, Zhang GQ, Pang D. Long non-coding RNA MVIH is associated with poor prognosis and malignant biological behavior in breast cancer. Tumour Biol. 2016;37:5257–5264. doi: 10.1007/s13277-015-4360-8. [DOI] [PubMed] [Google Scholar]
- 131.Wang G, Liu C, Deng S, Zhao Q, Li T, Qiao S, Shen L, Zhang Y, Lü J, Meng L, Liang C, Yu Z. Long noncoding RNAs in regulation of human breast cancer. Brief Funct Genomics. 2016;15:222–226. doi: 10.1093/bfgp/elv049. [DOI] [PubMed] [Google Scholar]
- 132.Xu SP, Zhang JF, Sui SY, Bai NX, Gao S, Zhang GW, Shi QY, You ZL, Zhan C, Pang D. Downregulation of the long non- coding RNA EGOT correlates with malignant status and poor prognosis in breast cancer. Tumour Biol. 2015;36:9807–9812. doi: 10.1007/s13277-015-3746-y. [DOI] [PubMed] [Google Scholar]
- 133.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, Brogi E, Egeblad M, Spector DL. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang YS, Chang CC, Lee SS, Jou YS, Shih HM. Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget. 2016;7:43256–43266. doi: 10.18632/oncotarget.9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, Wei Y, Ma G, Park PK, Zhou J, Zhou Y, Hu Z, Zhou Y, Marks JR, Liang H, Hung MC, Lin C, Yang L. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks Ikappa B phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 137.Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y, et al. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2015;37:1437–1444. doi: 10.1007/s13277-015-4521-9. [DOI] [PubMed] [Google Scholar]
- 138.Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, Buiga R, Pop V, Irimie A, Fodde R, Bedrosian I, Martens JW, Foekens JA, Berindan-Neagoe I, Calin GA. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4:1748–1762. doi: 10.18632/oncotarget.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sorensen KP, Thomassen M, Tan Q, Bak M, Cold S, Burton M, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat. 2013;142:529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 141.Wang L, Li J, Zhao H, Hu J, Ping Y, Li F, Lan Y, Xu C, Xiao Y, Li X. Identifying the crosstalk of dysfunctional pathways mediated by lncRNAs in breast cancer subtypes. Mol BioSyst. 2016;12:711–720. doi: 10.1039/c5mb00700c. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple- negative breast cancer. Nat Struct Mol Biol. 2016;26:522–530. doi: 10.1038/nsmb.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang Z, Weaver DL, Olsen D, deKay J, Peng Z, Ashikaga T, Evans MF. Long non-coding RNA chromogenic in situ hybridisation signal pattern correlation with breast tumour pathology. J Clin Pathol. 2016;69:76–81. doi: 10.1136/jclinpath-2015-203275. [DOI] [PubMed] [Google Scholar]
- 144.Yang X, Luo E, Liu X, Han B, Yu X, Peng X. Delphinidin-3-glucoside suppresses breast carcinogenesis by inactivating the Akt/HOTAIR signaling pathway. BMC Cancer. 2016;16:423. doi: 10.1186/s12885-016-2465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qu Z, Cui J, Harata-Lee Y, Aung TN, Feng Q, Raison JM, Kortschak RD, Adelson DL. Identification of candidate anti-cancer molecular mechanisms of compound Kushen injection using functional genomics. Oncotarget. 2016;7:66003–66019. doi: 10.18632/oncotarget.11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bubnov RV, Spivak MY, Lazarenko LM, Bomba A, Boyko NV. Probiotics and immunity: provisional role for personalized diets and disease prevention. EPMA J. 2015;6(1):14. doi: 10.1186/s13167-015-0036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cauchi JP, Camilleri L, Scerri C. Environmental and lifestyle risk factors of breast cancer in Malta-a retrospective case-control study. EPMA J. 2016;7:20. doi: 10.1186/s13167-016-0069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Golubnitschaja O, Yeghiazaryan K, Costigliola V, Trog D, Braun M, Debald M, Kuhn W, Schild HH. Risk assessment, disease prevention and personalised treatments in breast cancer: is clinically qualified integrative approach in the horizon? EPMA J. 2013;4(1):6. doi: 10.1186/1878-5085-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fröhlich H, Patjoshi S, Yeghiazaryan K, Kehrer C, Kuhn W, Golubnitschaja O. Premenopausal breast cancer: potential clinical utility of a multi-omics based machine learning approach for patient stratification. EPMA J. 2018;9(2):175–186. doi: 10.1007/s13167-018-0131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smokovski I, Risteski M, Polivka J, Jr, Zubor P, Konieczka K, Costigliola V, Golubnitschaja O. Postmenopausal breast cancer: European challenge and innovative concepts. EPMA J. 2017;8(2):159–169. doi: 10.1007/s13167-017-0094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sacco K, Grech G. Actionable pharmacogenetic markers for prediction and prognosis in breast cancer. EPMA J. 2015;6(1):15. doi: 10.1186/s13167-015-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Golubnitschaja O, Baban B, Boniolo G, Wang W, Bubnov R, Kapalla M, Krapfenbauer K, Mozaffari MS, Costigliola V. Medicine in the early twenty-first century: paradigm and anticipation - EPMA position paper 2016. EPMA J. 2016;7:23. doi: 10.1186/s13167-016-0072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]