Abstract
Neurons of the CNS elaborate highly branched dendritic arbors that host numerous dendritic spines, which serve as the postsynaptic platform for most excitatory synapses. The actin cytoskeleton plays an important role in dendrite development and spine formation, but the underlying mechanisms remain incompletely understood. Tropomodulins (Tmods) are a family of actin-binding proteins that cap the slow-growing (pointed) end of actin filaments, thereby regulating the stability, length, and architecture of complex actin networks in diverse cell types. Three members of the Tmod family, Tmod1, Tmod2, and Tmod3 are expressed in the vertebrate CNS, but their function in neuronal development is largely unknown. In this study, we present evidence that Tmod1 and Tmod2 exhibit distinct roles in regulating spine development and dendritic arborization, respectively. Using rat hippocampal tissues from both sexes, we find that Tmod1 and Tmod2 are expressed with distinct developmental profiles: Tmod2 is expressed early during hippocampal development, whereas Tmod1 expression coincides with synaptogenesis. We then show that knockdown of Tmod2, but not Tmod1, severely impairs dendritic branching. Both Tmod1 and Tmod2 are localized to a distinct subspine region where they regulate local F-actin stability. However, the knockdown of Tmod1, but not Tmod2, disrupts spine morphogenesis and impairs synapse formation. Collectively, these findings demonstrate that regulation of the actin cytoskeleton by different members of the Tmod family plays an important role in distinct aspects of dendrite and spine development.
SIGNIFICANCE STATEMENT The Tropomodulin family of molecules is best known for controlling the length and stability of actin myofilaments in skeletal muscles. While several Tropomodulin members are expressed in the brain, fundamental knowledge about their role in neuronal function is limited. In this study, we show the unique expression profile and subcellular distribution of Tmod1 and Tmod2 in hippocampal neurons. While both Tmod1 and Tmod2 regulate F-actin stability, we find that they exhibit isoform-specific roles in dendrite development and synapse formation: Tmod2 regulates dendritic arborization, whereas Tmod1 is required for spine development and synapse formation. These findings provide novel insight into the actin regulatory mechanisms underlying neuronal development, thereby shedding light on potential pathways disrupted in a number of neurological disorders.
Keywords: actin, cytoskeleton, dendritic arborization, dendritic spines, pointed end
Introduction
Actin is the major cytoskeletal component in dendritic spines (Fifková and Delay, 1982), where it provides structural support and spatially organizes postsynaptic components (Hotulainen and Hoogenraad, 2010). During synapse development, highly motile actin-based filopodia are replaced by relatively stable, mushroom-shaped spines that contain the synaptic components required for receiving presynaptic input (Yuste and Bonhoeffer, 2004). This morphological transition is characterized by the conversion of longitudinally arranged F-actin filaments to a highly branched F-actin network that predominates in the spine head (Hotulainen and Hoogenraad, 2010; Korobova and Svitkina, 2010). The actin cytoskeleton also plays a crucial role in dendrite development. For example, the formation of elaborate dendritic arbors is achieved, in part, through the dendritic growth cone, a motile, actin-based structure present at the tips of developing dendrites. Dendritic growth cones are crucial for dendrite extension as well as the formation of collateral dendritic branches. Despite intense studies, the molecules and mechanisms that regulate actin organization and remodeling during dendrite development and spine morphogenesis are not fully understood.
Actin filaments are polarized with a fast-growing (barbed) end and a slow-growing (pointed) end—the former favors assembly, whereas the latter favors disassembly. Tropomodulins (Tmod) belong to a conserved family of actin-binding proteins that are best known for capping the pointed end of actin filaments (Yamashiro et al., 2012; Fowler and Dominguez, 2017). By blocking monomer exchange at filament ends and inhibiting filament elongation or depolymerization, Tmod regulates the stability, length, and architecture of complex actin networks in diverse cell types (Yamashiro et al., 2012). Of the four Tmod isoforms, Tmod1, Tmod2, and Tmod3 are expressed in the CNS (Sussman et al., 1994; Watakabe et al., 1996; Conley et al., 2001; Cox et al., 2003). Tmod1 is expressed in many types of terminally differentiated cells (including neurons), Tmod2 is expressed solely in neurons, and Tmod3 is ubiquitously expressed (Yamashiro et al., 2012).
Altered expression of Tmod1 and Tmod2 is observed in several neurological diseases, suggesting that the regulation of actin dynamics by Tmod is critical for brain function (Sussman et al., 1994; Iwazaki et al., 2006; Yang et al., 2006; Chen et al., 2007; Sun et al., 2011). In support of this, Tmod2 knock-out mice exhibit synaptic and behavioral deficits, including altered learning and memory (Cox et al., 2003). However, genetic knockdown of Tmod2 causes protein levels of Tmod1 to be upregulated eightfold (Cox et al., 2003), making isoform-specific interpretation of Tmod function challenging. In addition, the cellular mechanisms that account for these phenotypes are unclear. Tmod1 and Tmod2 are present in growth cones of elongating neurites in cultured rat hippocampal neurons, and are suggested to play roles in neurite formation in N2a cells (Fath et al., 2011) and PC12 cells (Moroz et al., 2013; Guillaud et al., 2014). A recent study (Gray et al., 2016) using GFP-Tmod1 and GFP-Tmod2 overexpression indicates that Tmod1 and Tmod2 regulate dendritic branching and spine morphology in cultured rat hippocampal neurons, but the underlying actin mechanisms remain to be elucidated. At present, Tmod remains poorly characterized in neurons, with fundamental information such as the subcellular distribution and isoform-specific pattern of protein expression lacking. Therefore, if and how Tmod regulates F-actin during postsynaptic development and/or synapse formation remain unknown.
In this study, we investigated the expression profile, subcellular distribution, and function of Tmod1 and Tmod2 in hippocampal neurons. We find that both Tmod1 and Tmod2 are expressed in developing rat hippocampal neurons, but with distinct temporal profiles. Using a combination of high-resolution imaging and loss-of-function analyses, we find that Tmod1 and Tmod2 regulate spine formation and dendritic development, respectively. Our study provides evidence that Tmod plays an important and isoform-specific role in postsynaptic development underlying the formation of neuronal circuitry.
Materials and Methods
DNA constructs
DNA constructs of EGFP-Tmod1 and EGFP-Tmod2 were made by subcloning the full-length rat sequences (Tmod1, NCBI reference sequence NP_037176.2; Tmod2, NCBI reference sequence NP_113801.1) in frame downstream of the full-length EGFP sequence in an EGFP-C1 vector (Clontech). For knock-down experiments, short hairpins against rat Tmod1 (shTmod1, 5′-CACAGAAGTTCAGTCTGATAA-3′, directed against-3′ UTR; shTmod1-B, CCAGAACTTGAAGAGGTTAAT, directed against coding sequence) and Tmod2 (shTmod2, 5′-CCAGTTGTTCTGGAACTTT-3′, directed against the coding sequence; shTmod2-B, 5′-GTCAACCTCAACAACATTAAG-3′, directed against the coding sequence) were subcloned, using Bg1II and XhoI sites, into a pSUPER backbone also encoding EGFP to allow visualization of transfected cells. The hairpin sequence directed against shLuciferase (shLucif) is 5′-CGTACGCGGAATACTTCGA-3′. For fluorescence recovery after photobleaching (FRAP) experiments, EGFP-actin (pCS2+ EGFP–γ-actin) was used.
Immunostaining
For immunostaining of primary hippocampal cultures, neurons were fixed with 4% (w/v) paraformaldehyde (PFA) in PBS for 15 min at room temperature. Fixed neurons were washed and permeabilized with 0.2% Triton X-100 (w/v) in PBS for 15 min. Neurons were blocked with PBS containing 4% BSA, 1% goat serum and 0.1% Triton X-100 for 1 h and incubated with the following primary antibodies overnight at 4°C. The antibodies used are as follows: anti-Tmod1 (affinity-purified custom rabbit polyclonal R1749bl3c (Fowler et al., 1993); anti-Tmod2 (catalog #ab67407, Abcam; custom-made Tmod2 antiserum (Fath et al., 2011); anti-postsynaptic density-95 (PSD-95; catalog #MA1-046, Thermo Fisher Scientific); and anti-SV2 (Developmental Studies Hybridoma Bank). Cells were then washed and labeled with anti-rabbit or anti-mouse Alexa Fluor 488/546 antibody (A-11030/A-11035, Thermo Fisher Scientific) for 45 min at room temperature. Actin filaments were stained with phalloidin conjugated to Alexa Fluor 488 (A12379) or 568 (A12380), purchased from Thermo Fisher Scientific.
Neuronal culture, transfection, and imaging
Sprague Dawley timed-pregnant adult rats (8–10 weeks of age) were purchased from Charles River Laboratories. Primary hippocampal neurons were prepared from embryonic day 18 (E18) rat embryos of both sexes and plated on 25 mm coverslips pretreated with 0.1 mg/ml poly-d-lysine (EMD Millipore) at a density of ∼400,000 cells/dish. Neurons were plated and maintained in Neurobasal medium supplemented with B-27 and GlutaMAX (Invitrogen) and maintained at 37°C and 5% CO2. Cells were transfected using the calcium phosphate transfection kit (Clontech) at 10 d in vitro (DIV10) or DIV13 and imaged between DIV21 and DIV23. Each experiment was replicated at least three times from independent batches of cultures.
All animals were housed and treated in accordance with the Emory University Institutional Animal Care and Use Committee guidelines.
Microscopy and imaging
Laser-scanning confocal imaging was performed using a Nikon C1 Confocal System based on a Nikon Eclipse TE300 inverted microscope (Nikon Instruments). A 60×/1.4 numerical aperture (NA) Plan Apo oil-immersion objective was used to acquire the three-dimensional (3-D) stack of images of a dendritic region, which was then projected into a 2-D image (maximum intensity) for visualization and analysis.
Stimulated emission depletion super-resolution microscopy
The majority of images were acquired using a commercially available multicolor stimulated emission depletion (STED) microscope (model TCS SP8 STED 3×, Leica Microsystems) equipped with a white light laser source operated in pulsed mode (repetition rate, 78 MHz) for fluorophore excitation and two STED lasers for fluorescence inhibition, as follows: one STED laser with a wavelength of 775 nm operated in pulsed mode with pulse trains synchronized to the white light laser pulses; and two continuous-wave STED laser with wavelengths of 592 and 660 nm. Excitation wavelengths between 470 and 670 nm were selected from the white light laser emission spectrum via an acousto-optical beam splitter. All laser beams were focused in the cell sample with a wavelength-corrected, 1.40 NA oil objective (HC PL APO 100×/1.40 OIL STED WHITE). The samples were scanned using the field-of-view beam scanner at uniform scanning rates and within specific scanning areas, which are further specified below. The fluorescence from a given sample were detected by GaAsP (gallium arsenide phosphide) hybrid detectors. The recorded fluorescence signal in the STED imaging mode was time gated using the white light laser pulses as internal trigger signals. A small number of images were obtained using a Leica SP8 STED 3× system (Leica Microsystems), equipped with a white light laser and a 592 and 775 nm STED depletion lasers. A 100× 1.4 NA oil-immersion objective lens (HC PL APO STED WHITE, Leica Microsystems) was used for imaging (Yu et al., 2015). Deconvolution on these images was performed using Huygens Professional software (Scientific Volume Imaging).
All acquired or reconstructed images were processed and visualized using ImageJ (imagej.nih.gov/ij/). Brightness and contrast were linearly adjusted for the entire images.
Structured illumination microscopy
Three-dimensional structured illumination microscopy (SIM) was performed on an inverted Nikon N-SIM Eclipse Ti-E Microscope System equipped with the Perfect Focus System (PFS), 100×/1.49 NA oil-immersion objective, and an electron multiplying charge-coupled device camera (DU-897, Andor Technology). Images were reconstructed in Nikon Elements Software. Images were analyzed using ImageJ software and NIS-Elements AR Analysis software (Nikon).
Data analysis
SIM analysis.
To quantify subspine localization, dendritic spines were separated into the following three compartments: the spine neck, the distal most part of the spine head (area 1), and the proximal-most part of the spine head (area 2, closest to spine neck). To equally divide the spine head into two equal compartments (area 1 and area 2), the height of the spine head was measured in ImageJ and a horizontal line corresponding to half the value of the measured height was placed through the spine head. Using the “freehand selection” function in ImageJ, a region of interest (ROI) was drawn around the circumference of the given region of the spine head, as demarcated by phalloidin staining. For each region (area 1, area 2, or spine neck) the raw integrated density value was measured for both the phalloidin channel and the Tmod channel. A ratio derived from the raw integrated density value of Tmod/phalloidin was generated for each area, and the mean ratio values were used to generate graphs.
Spine analysis.
The 3-D images of spines were rendered from confocal z-stacks using ImageJ. For spine analysis, filopodia were defined as thin protrusions without a distinguishable head, and spines were defined as protrusions with a length of <4 μm and an expanded, distinguishable head. Spine and filopodia numbers were counted manually to calculate the density (number per 100 μm length of the parent dendrite). Spine head width was measured as the spine diameter (the longest possible axis), and neck length was measured from the proximal edge of the spine head to the edge of the dendrite. For spines with no discernible necks, a minimum value of 0.2 μm was used. To quantify synaptic density, the cluster number of SV2 and PSD-95 signals per unit neurite length were counted using ImageJ.
Western blotting
For developmental immunoblots, hippocampal cultures at DIV4, DIV7, DIV14, and DIV21 were homogenized in lysis buffer containing 20 mm Tris HCl, pH8, 137 mm NaCl, 10% glycerol, 1% TX-100, and 2 mm EDTA, supplemented with 1 mm phenylmethylsulfonyl fluoride and a protein inhibitor cocktail (catalog #P2850, Sigma-Aldrich). Hippocampi were collected from the brains of three independent littermates at E18, postnatal day 8 (P8), P14, and P21, and adult Sprague Dawley rats of both sexes. Hippocampi were snap frozen in liquid nitrogen before homogenization in lysis buffer. Lysates were denatured in 1× Laemmli sample buffer and boiled for 3 min. Fifteen micrograms of protein, as determined by Bradford assay, was loaded and fractioned by SDS-PAGE in a 12% Tris-glycine acrylamide gels (Invitrogen), and subsequently was transferred to nitrocellulose membrane. Membranes were treated with 5% milk in PBS containing 0.05% Triton X-100 and then incubated with primary antibody overnight at 4°C, as follows: anti-Tmod1 (affinity-purified custom rabbit polyclonal; Fowler et al., 1993); anti-Tmod2 (catalog #ab67407, Abcam; and custom-made Tmod2 antiserum (Fath et al., 2011). Bound antibodies were detected by HRP-conjugated secondary antibody (Jackson ImmunoResearch) and visualized by chemiluminescence using Pierce ECL (Thermo Fisher Scientific). Gels were quantified using the gel analysis function of ImageJ software (National Institutes of Health).
Live cell extraction
Hippocampal neurons were extracted for 1 min at room temperature in cytoskeleton buffer (10 mm MES, pH 6.1, 90 mm KCl, 2 mm EGTA, 3 mm MgCl2, 0.16 m sucrose containing 0.025% saponin, 0.1 mm ATP, and 1 μm unlabeled phalloidin). Neurons were immediately fixed in 4% PFA in PBS and stained with phalloidin and Tmod1 or Tmod2 according to the protocol detailed above. Identical fluorescent labeling conditions and acquisition parameters were used in extracted and nonextracted neurons. To quantify the relative level of Tmod in dendrites and spines of extracted and nonextracted neurons, images of Tmod were merged with phalloidin-labeled F-actin to identify dendritic spines and corresponding dendrites. The fluorescence intensity of Tmod in spines within a dendritic region of ∼100 μm was averaged and compared with the mean fluorescence intensity of the Tmod along the entire length of the corresponding dendritic region. Thirty cells were examined from at least three independent batches of culture.
Fluorescence recovery after photobleaching
The FRAP assay was performed on a Nikon A1R laser-scanning confocal microscope platform. The platform was equipped with the PFS, multiple laser sources with acousto-optical tunable filter control, motorized x–y stage, a modular incubation chamber with temperature and CO2 control, and a full line of photomultiplier tube detectors. Neurons grown on coverslips were mounted in a custom live-cell chamber. A 60× PlanApo N TIRF oil-immersion objective (1.49 NA) was used for all image acquisitions. Time-lapse images were acquired through four stages. For stage 1, six consecutive control images were acquired with a 2 s interval between frames. For stage 2, a single spine head within a preselected ROI was photobleached with 100% power of the 488 nm laser line from a 40 mW argon laser for 500 ms with the pixel dwell set at 3.9 μs. For stage 3, immediately after photobleaching, a 5 s imaging sequence was acquired with no delay between frames, yielding 19 frames in total. For stage 4, a 5 min imaging sequence was taken with a 2 s interval between frames, generating 151 frames in total. The following imaging settings were used for stages 1, 3, and 4: 2% 488 nm laser power, 1.2 μs pixel dwell, and 0.07 μm/pixel resolution.
Experimental design and statistical analysis
All data from this study were collected from at least three replicates of independently prepared samples. Parametric data were statistically analyzed using a one- or two-tailed unpaired Student's t test or a two-way repeated-measures ANOVA with a Sidak's multiple-comparison post hoc test. A Kruskal–Wallis one-way ANOVA with a Dunn's multiple-comparison test was used to analyze nonparametric data. GraphPad Prism (version 7, GraphPad Software) and Excel (Microsoft) were used for statistical analysis. Detailed statistical results, including p values, are provided in the corresponding figure legends. Unless otherwise specified, data are presented as the mean ± SEM, with in-text values stated. Asterisks indicate a p value ≤0.05, and nonsignificant is denoted by “n.s.”
Results
Expression of Tmod1 and Tmod2 in hippocampal neurons
Previous studies suggest that the expression of Tmod1 (Sussman et al., 1994) and Tmod2 (Watakabe et al., 1996) in rat brains increases during development. To better assess the expression profile of Tmods in the hippocampus, we performed immunoblotting of rat hippocampal lysates from E18, P8, P12, P23, and adult rat brains, which approximately correspond to the stages before, during, and after synapse formation (Markus and Petit, 1987; Harris et al., 1992; Fiala et al., 1998). We found that both Tmod1 and Tmod2 are expressed in rat hippocampus, but with different profiles (Fig. 1A). Tmod1 is undetectable at E18, but its expression starts to become apparent at approximately P8. Substantial Tmod1 expression is detected at approximately P12 and continues to increase steadily until adulthood. By contrast, Tmod2 is highly expressed at E18 and its level further increases at P8 and then remains relatively steady until adulthood. Since the formation of spine synapses commences near the end of the second postnatal week (P14; Markus and Petit, 1987; Harris et al., 1992; Fiala et al., 1998), the expression profile of Tmod1 suggests that it may play a role in spine development and synapse formation. The early onset of Tmod2 expression, on the other hand, suggests that it may be involved in the early stages of neuronal development, such as dendritic development.
Figure 1.
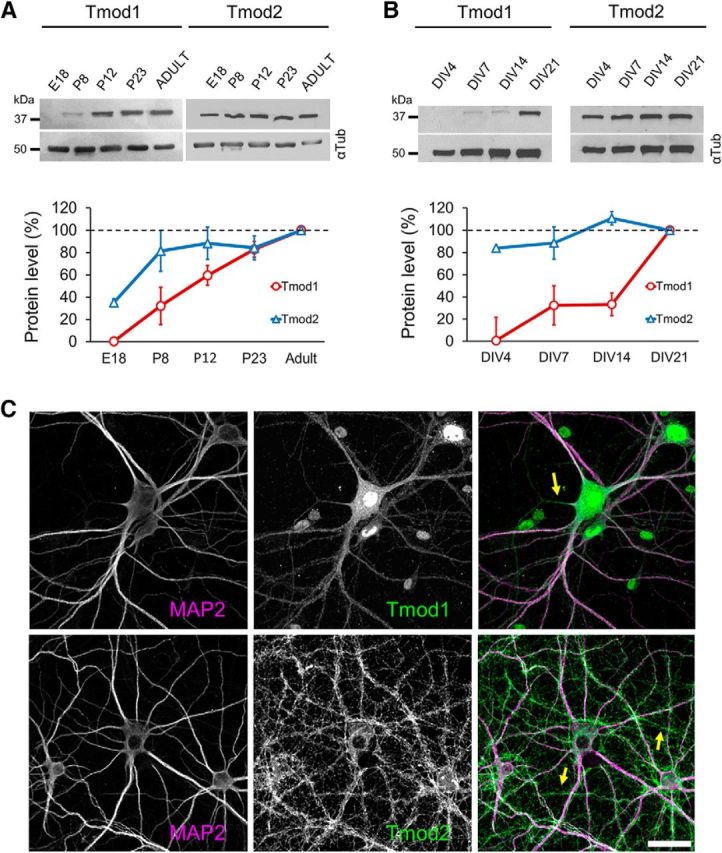
Expression of Tmod1 and Tmod2. A, B, Representative Western blots of Tmod1 and Tmod2 in rat hippocampal tissue (A) and primary hippocampal neurons (B) depict the changes in Tmod expression over time. All bands in gel are cropped for clarity. Quantification from three separate replicates is shown in the corresponding line graphs (N = 3 for each time point). The mean value for each time point was normalized to the corresponding tubulin loading control (αTub), and then to the “Adult” (A) or “DIV21” (B) value. C, Immunostaining of endogenous Tmod1 and Tmod2 (green), together MAP2 (magenta) in DIV21 cultured hippocampal neurons. Arrows indicate axons. Scale bar, 30 μm.
We next examined the expression of both Tmod isoforms in primary rat hippocampal neurons at specific DIV (Fig. 1B). Primary hippocampal neurons exhibit defined stages of postsynaptic development, thereby allowing us to align Tmod protein expression with dendrite development and synapse formation. Hippocampal neurons in culture develop axon–dendrite polarity at approximately DIV5, with substantial growth and arborization of dendrites occurring until DIV14 (Dotti et al., 1988). From DIV14 to DIV18, relatively stable dendritic arbors undergo small but continuous growth until maturation. In parallel with dendrite development, synapse formation in cultured hippocampal neurons initiates on dendritic arbors at approximately DIV7 to DIV9 and peaks at approximately DIV11 to DIV14. The formation of mature dendritic spines and synapses is observed mostly from approximately DIV18 onward (Ziv and Smith, 1996; Friedman et al., 2000; Grabrucker et al., 2009). Consistent with immunoblots from hippocampal tissue, Tmod2 is expressed very early in cultured hippocampal neurons (DIV4), and its protein level remains relatively steady before, during, and after the processes of synapse formation and dendrite development (Fig. 1B). In contrast, Tmod1 expression remains low before DIV14, but is substantially elevated at DIV21. Together, these immunoblot data demonstrate that Tmod1 and Tmod2 exhibit different temporal profiles during hippocampal development, suggesting that Tmod1 and Tmod2 may regulate distinct processes of postsynaptic development.
We next performed immunofluorescence staining to examine the expression and subcellular distribution of Tmod proteins in cultured primary hippocampal neurons. Since we detected little Tmod3 signaling in hippocampal neurons using a previously verified Tmod3-specific antibody (Fischer et al., 2003), we focused on Tmod1 and Tmod2. We found that both Tmod1 and Tmod2 are abundantly expressed in the somatodendritic compartment of DIV21 neurons, as evidenced by their colocalization with dendrite-enriched microtubule-associated protein 2 (MAP2; Fig. 1C). Tmod signals are also detected in neuronal processes lacking MAP2, suggesting that Tmod is also present in in axons (Fig. 1C, arrows). Tmod1 signals were also observed in the nucleus, as reported previously (Kong and Kedes, 2004). Together with the developmental expression profile, these immunofluorescence data support the notion that Tmod1 and Tmod2 are present in the dendritic compartment and may function in postsynaptic development.
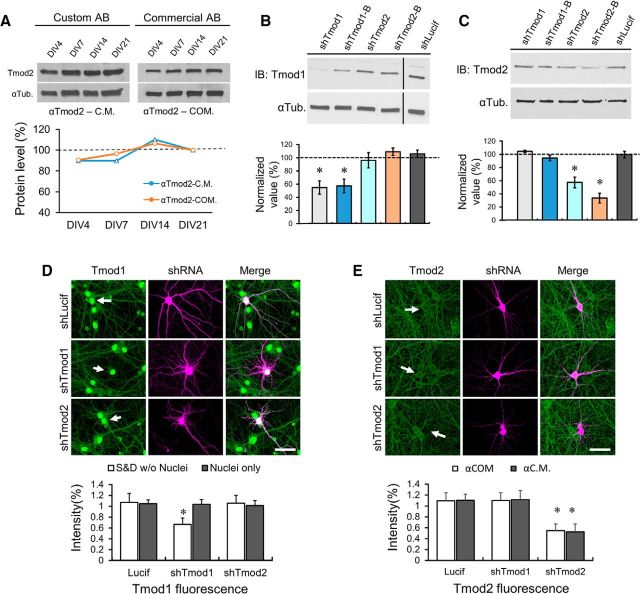
It is important to note that the specificity of the custom Tmod1 antibody used in this study (Fowler et al., 1993) has been extensively verified using immunofluorescence staining and immunoblotting of tissues from a Tmod1 knock-out mouse (Fischer et al., 2003; Nowak et al., 2009; Gokhin et al., 2010; Moyer et al., 2010). For Tmod2, we mostly used a commercial antibody (Commercial AB/αTmod2-COM), which generates a near-identical immunoblot expression profile as a custom-made Tmod2 antibody from the laboratory of V.M.F. (Custom AB/αTmod2-C.M.; Fig. 2A). This custom Tmod2-specific antibody was previously verified using an immunoblot of tissues from a Tmod2 knock-out mouse (Cox et al., 2003), as well as purified Tmod1-4 proteins (Fath et al., 2011). Furthermore, we show that knocking down endogenous Tmod1 or Tmod2 did not affect the endogenous level of Tmod2 or Tmod1, respectively, in Cath.A-differentiated (CAD) neuroblastoma cells by immunoblotting (Fig. 2B,C) or in hippocampal neurons by immunofluorescence (Fig. 2D,E). The knockdown efficiency of these short hairpin RNAs (shRNAs) on endogenous Tmod1 and Tmod2 is ∼50%, as assessed by immunoblotting or immunofluorescence (Fig. 2B–E). It is interesting to note that the nuclear Tmod1 signal was not changed after knockdown (Fig. 2D), suggesting that nuclear Tmod1 may be much more stable than the cytosolic fraction or may represent additional components in the nucleus (Nowak et al., 2009). Unlike previous studies (Cox et al., 2003; Fath et al., 2011), we did not observe compensatory changes in the endogenous level of Tmod1 or Tmod2 when either Tmod isoform was targeted by shRNA in culture (Fig. 2B–E). Therefore, the shTmod1 and shTmod2 hairpins we developed are specific and robust in knocking down endogenous Tmod1 or Tmod2 in primary hippocampal neurons. These results also demonstrate that the Tmod1 and Tmod2 antibodies used in this study are specific for the intended isoforms. We therefore believe that the expression profiles detected by immunoblotting and immunofluorescence represent the true, endogenous expression patterns of Tmod1 and Tmod2 in neuronal tissues.
Figure 2.
Antibody specificity and shRNA knockdown of endogenous Tmods. A, Representative immunoblot depicting levels of endogenous Tmod2 expression in primary hippocampal neurons, as detected using αTmod2-C.M., which has been previously verified for specificity (Fowler et al., 1993), and αTmod2-COM, which was used throughout this study. The corresponding line graph from the immunoblots is shown below. The value for each time point was normalized to the corresponding tubulin loading control (αTub), and then to the DIV21 value. B, C, Representative immunoblot images reveal robust and specific reduction of Tmod1 and Tmod2 protein levels in CAD neuroblastoma cells expressing knock-down constructs (48 h). Black vertical bar in Tmod1 blot (left) delineates the boundary between two nonadjacent lanes from the same gel (processed identically). All bands in gel are cropped for clarity. Bar graphs depict mean Tmod1 (B) and Tmod2 (C) knockdown levels in CAD cells from three independent replicates. Data are normalized to Tmod protein levels in shLucif-expressing cells. Statistical analysis was performed using an unpaired Student's t test. Error bars represent the SEM. *Statistical difference from the control. For immunoblotting (IB): Tmod1: t(df) = 2.44(6), pshTmod1 = 2.5E-3, pshTmod1-B = 1.6E-4; Tmod2: t(df) = 2.78(4), pshTmod2 = 6.8E-3, pshTmod2-B = 0.023. D, E, Immunostaining of endogenous Tmod1 (D) and Tmod2 (E) by custom and commercial antibodies in DIV21 cultured hippocampal neurons expressing shLucif, shTmod1, or shTmod2. Neurons were transfected at DIV13 and imaged at DIV21. Statistical analysis was performed using an unpaired Student's t test comparing the mean Tmod1 or Tmod2 fluorescence intensity to that of neighboring nontransfected cells (control) in the same field of view. Error bars represent SEM. Tmod1 fluorescence: t(df) = 2.00(59), pshTmod1 = 3.36E-15; Tmod2 fluorescence: t(df) = 2.02(38), pshTmod2αC.M. = 2.03E-16, t(df) = 1.97(66), pshTmod2αCOM = 1.64E-24. Arrows depict transfected neurons. S&D, somatodendritic; αCOM, αTmod2-COM; αC.M., αTmod2-C.M. Scale bar, 35 μm.
Tmod1 and Tmod2 in dendrite development
Both Tmod1 and Tmod2 are present in the dendritic shaft of DIV21 hippocampal neurons in culture, but Tmod2 appears to be more concentrated along the plasma membrane of some dendritic segments (Fig. 3A), suggesting that it may be associated with the cortical actin cytoskeleton. When examined by super resolution STED microscopy, the localization of Tmod2 to the subcortical region is apparent (Fig. 3B). To investigate the roles of Tmod1 and Tmod2 in dendrite development, we performed loss-of-function analysis in hippocampal cultures during dendrite development using the specific shRNAs described above. In culture, hippocampal neurons establish axon–dendrite polarity at approximately DIV5, with substantial growth and arborization of dendrites until approximately DIV14 (Dotti et al., 1988). We therefore transfected hippocampal neurons with shTmod1 or shTmod2 on DIV10 and examined them on DIV21. Control neurons expressing shLucif displayed complex dendritic arbors that possessed many secondary and tertiary branches (Fig. 3C). By contrast, neurons expressing shTmod2 exhibited a large reduction in both the length (54 ± 23.7%) and branch number (55 ± 24.9%) of the dendritic arbor (Fig. 3C,D). Sholl analysis (Sholl, 1953) further depicts the marked reduction in dendritic arborization in neurons expressing shTmod2 (Fig. 3E). Intriguingly, hippocampal neurons expressing shTmod1 appeared to be normal, exhibiting no significant change in their dendritic branches, compared with the control cells. These results indicate that Tmod2, but not Tmod1, plays an important role in dendrite growth and arborization during development.
Figure 3.
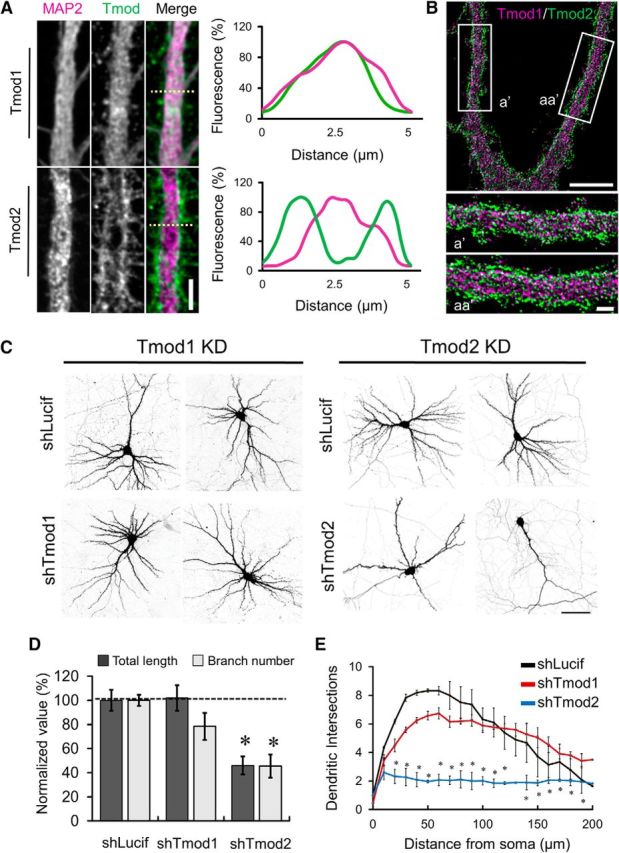
Regulation of dendrite development by Tmod. A, Representative confocal immunofluorescence images of dendritic regions in neurons stained for Tmod1 or Tmod2 and MAP2. Scale bar, 4.5 μm. Right, Representative line profiles from regions in corresponding images (dashed line). B, Representative STED images of dendrites from DIV21 hippocampal neurons costained with Tmod1 and Tmod2. Scale bar: top, 5 μm; bottom, 1 μm. C, Representative confocal images of DIV21 neurons expressing shLucif, shTmod1, or shTmod2. Images were inverted in grayscale for presentation. D, Bar graph shows changes in total dendritic length and branch number, as labeled. For each condition, values are normalized to shLucif. Error bars represent SEM. *Statistical significance using an unpaired, Student's t test. ShTmod2: t(df) = 2.22(10), ptotal length = 9.43E-4, t(df) = 2.26(9), pbranch number = 5.0E-4. E, Line graph depicts the results of Sholl analysis from control, shTmod1-expressing, and shTmod2-expressing neurons. Statistical analysis was performed using a two-way ANOVA with a Dunn's multiple-comparison test to determine statistical differences. *Statistical significance compared with shLucif. Tmod2: F(DFn, DFd) = F(2,105) = 137.2, p20–130 μm = 0.0002, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0026, and 0.407. Error bars represent the SEM.
Tmod1 and Tmod2 in dendritic spines
Both Tmod1 and Tmod2 are present in the dendritic compartments of DIV21 hippocampal neurons, including postsynaptic dendritic spines (Fig. 4A). Here, dendritic spines are highlighted with a low concentration of fluorescent phalloidin (Gu et al., 2008), a phallotoxin that binds F-actin with high affinity, as well as PSD-95, a key postsynaptic component of excitatory synapses. When comparing the fluorescence intensity of Tmod1 and Tmod2 in dendritic spines and the corresponding shaft region (spine/shaft ratio), we find that Tmods are slightly enriched in dendritic spines, with mean ± SD spine/shaft ratios of 132.24 ± 16.68% and 119.59 ± 5.96%, respectively, for Tmod1 and Tmod2. These results suggest that Tmod1 and Tmod2 are slightly enriched in postsynaptic spines.
Figure 4.
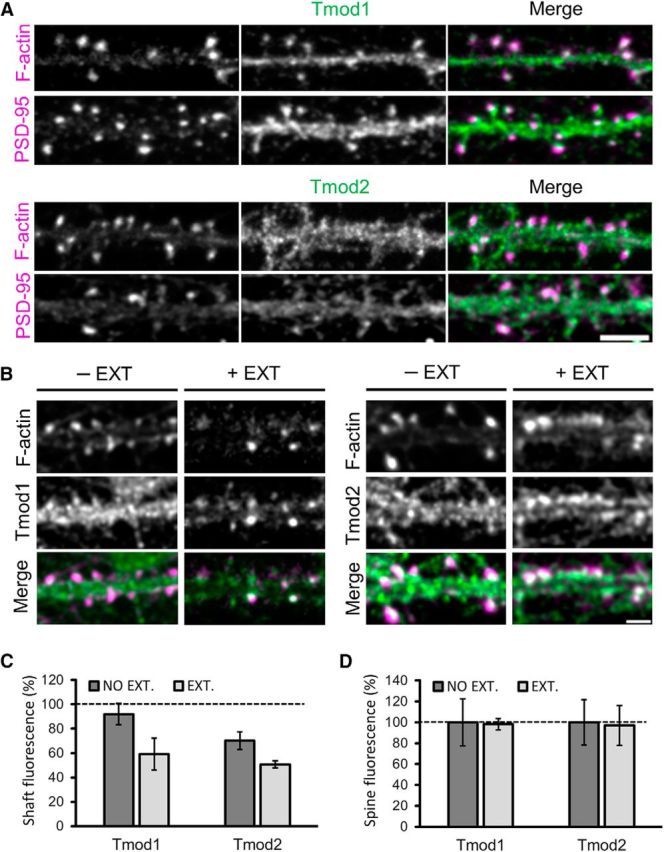
Tmods in dendritic spines of hippocampal neurons in culture. A, Immunostaining of endogenous Tmod1 and Tmod2 (green), together with phalloidin-labeled dendritic spines and PSD-95 (magenta). Scale bar, 5 μm. B, Representative immunofluorescence images of Tmod1 and Tmod2 in select dendritic regions of extracted and nonextracted hippocampal neurons. Scale bar, 2.5 μm. C, Bar graph depicts the mean fluorescence intensity of Tmod1 and Tmod2 in the dendritic shaft of nonextracted (control) and extracted neurons. The fluorescence intensity values of Tmod1 and Tmod2 in the shaft are normalized to the mean corresponding value in the spine. An unpaired Student's t test was used to compare the nonextracted vs extracted mean intensity values for Tmod1 and Tmod2. *Statistical difference from the control. For shaft fluorescence: t(df) = 2.44(6), pTmod1-ext. = 0.08, t(df) = 2.575(5), pTmod2-ext. = 0.06. Error bars represent the SEM. D, Bar graphs depict the mean fluorescence intensity of Tmod1 and Tmod2 in dendritic spines of nonextracted (control) and extracted neurons. Extracted values are normalized to the nonextracted values of Tmod1 and Tmod2, respectively. Nspines = 196 and 233, for Tmod1 and Tmod2, respectively. Nshaft = 180 and 173, for Tmod1 and Tmod2, respectively.
To test whether Tmod1 and Tmod2 in spines are diffusible and/or associated with F-actin, we performed live-cell extraction using the mild detergent saponin (Lee et al., 2013; Lei et al., 2017). Brief exposure of live neurons to saponin is known to remove freely diffusible cytosolic molecules without affecting relatively stable structures such as the cytoskeleton and cytoskeleton-associated proteins (Lee et al., 2013; Lei et al., 2017). To estimate the cytoskeleton-associated fraction of Tmod in distinct postsynaptic regions, we used identical fluorescent labeling conditions and acquisition parameters for extracted versus nonextracted neurons and compared the fluorescence intensities of Tmod in specific regions. Live cell extraction largely reduced the signal of Tmod1 and Tmod2 in the dendritic shaft, but not in dendritic spines (Fig. 4B). Quantification of Tmod immunoreactivity in the dendritic shaft demonstrates that the mean ± SD fluorescence intensity of Tmod in extracted cells is 64.40 ± 26.12% (n = 180) and 67.20 ± 6.00% (n = 173) of that of nonextracted cells for Tmod1 and Tmod2, respectively (Fig. 4C). On the other hand, the levels of endogenous Tmod1 and Tmod2 detected in dendritic spines of extracted neurons are 98.24 ± 24.27% (n = 196), and 96.97 ± 29.86% (n = 233) of that of nonextracted cells, respectively (Fig. 4D). These data indicate that the majority of Tmod molecules in spines are associated with stable structures such as F-actin and/or the PSD.
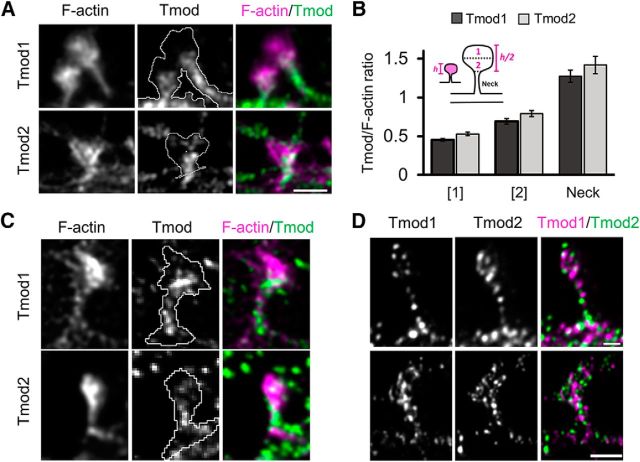
It is of great interest to note that the Tmod signals in dendritic spines do not appear to completely overlap with phalloidin-labeled F-actin (Fig. 4A). Rather, Tmod1 and Tmod2 appear to be restricted to the spine neck and center of the spine head. To obtain a more detailed analysis of the spatial organization of Tmod in dendritic spines, we turned to SIM, which uses overlapping moiré patters to give a resolution of ∼115 nm (Gustafsson, 2000). SIM images revealed that Tmod1 and Tmod2 are largely absent from the distal-most and lateral sides of the spine, instead localizing to the center-most region of the spine head and the spine neck (Fig. 5A). To quantify the apparent subspine localization of Tmod1 and Tmod2, dendritic spines were separated into the following three compartments: the spine neck, the distal-most part of the spine head (area 1), and proximal-most part of the spine head (area 2; closest to spine neck). For each region (area 1, area 2, or spine neck) the raw integrated density value was measured for both the F-actin channel and the Tmod channel and a subsequent Tmod/F-actin ratio was generated (Fig. 5B). Quantitative analysis using SIM images reveals that Tmod1 and Tmod2 are indeed enriched in the spine neck and lower part of the spine head (area 2). STED microscopy, which provides an optimal lateral resolution of ∼30–50 nm (Meyer et al., 2008), was used to further confirm the subspine localization of Tmod1 and Tmod2 (Fig. 5C). Interestingly, while both Tmod1 and Tmod2 are present in the same spine, they do not significantly overlap, as revealed by STED super-resolution microscopy (Fig. 5D), suggesting that these two Tmod isoforms may have distinct functions in dendritic spines.
Figure 5.
Subspine localization of Tmod1 and Tmod2. A, 3D-SIM images of Tmod1/Tmod2 and F-actin in dendritic spines demonstrate the subspine localization of Tmods to the neck and base of the spine head. B, Bar graphs represent mean raw integrated density Tmod/F-actin ratios in the proximal and distal region of the spine head as well as the spine neck. C, STED image showing the distribution of Tmod1 or Tmod2 (green) and F-actin (magenta) in spines. D, STED image showing the distributions of Tmod1 (green) and Tmod2 (magenta) in the same dendritic protrusion. Scale bars: top, 0.5 μm; bottom, 1 μm.
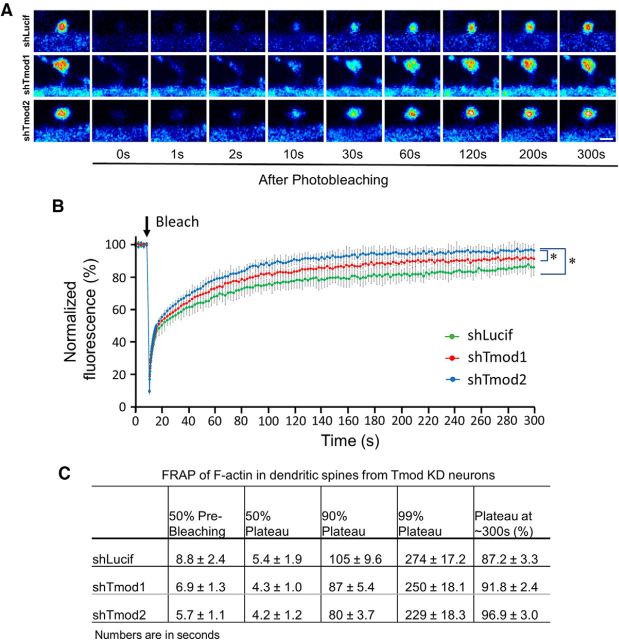
Previous studies have shown that dendritic spines contain different pools of F-actin, namely a dynamic and a stable pool (Star et al., 2002; Honkura et al., 2008; Frost et al., 2010). Importantly, the dynamic pool of F-actin appears to be at the distal tip and lateral edges of the dendritic spine head, and the stable pool of F-actin is located at the base of the spine head (Honkura et al., 2008). We therefore hypothesized that the localization of Tmod in the neck and base of the spine head, a region enriched in stable F-actin, may be a mechanism used to regulate the stability of F-actin in dendritic spines. To test this hypothesis, we used FRAP to study the turnover of the actin cytoskeleton in dendritic spines from neurons depleted of Tmod1 and Tmod2 (Fig. 6). In control cells expressing shLucif, EGFP-actin fluorescence after photobleaching reached 50% of prebleaching levels in ∼9 s, consistent with our previous findings (Lei et al., 2017). In contrast, EGFP–actin signals in spines of shTmod1- and shTmod2-expressing neurons recovered significantly quicker, achieving 50% recovery in ∼7 and ∼ 6 s, for Tmod1 and Tmod2, respectively (Fig. 6B,C). Furthermore, the plateau of EGFP-actin recovery at 300 s was ∼87% in control cells, indicating that the proportion of the stable F-actin that had not yet recovered was ∼13%. By contrast, the plateau values for shTmod1- and shTmod2-expressing cells are ∼92% and ∼97% by 300 s, respectively, suggesting that the proportion of the stable F-actin pool was significantly decreased after knockdown of either Tmod isoform. These results thus support the notion that Tmod molecules play a role in stabilizing the F-actin structures in dendritic spines.
Figure 6.
FRAP analysis of spine actin turnover in Tmod1 or Tmod2 knock-down (KD) neurons. A, Representative image sequences showing the fluorescence recovery of EGFP-actin after single-spine photobleaching in neurons expressing shLucif, shTmod1, or shTmod2. Scale bar, 1 μm. B, FRAP curves for control or shTmod-expressing neurons. The fluorescent signals in spines are normalized to the prebleaching mean and corrected for acquisition-based bleaching using the signals from the adjacent shaft regions. N = 15, 24, and 20 for shLucif, shTmod1, or shTmod2 groups, respectively. Error bars represent the SD. *Statistical difference from the control (shLucif). Unpaired Student's t test was performed selectively, and only the statistical results on data at 100, 200, and 300 s are shown here. For shTmod1: t(df) = 2.03(37), p = 3.85E-09, 9.73E-11, and 6.79E-07; shTmod2: t(df) = 2.03(33), p = 5.34E-14, 1.66E-13, and 3.90E-10. C, Quantification table showing the EGFP-actin recovery time for 50% prebleaching levels, 50% plateau levels, 90% and 99% plateau levels, and the final recovery percentages in dendritic spines. Results are shown as the mean ± SEM.
Tmod1 is required for spine development and synapse formation
In culture, hippocampal neurons undergo extensive dendritic growth and branching from approximately DIV8 to DIV14 (Dotti et al., 1988). From DIV14 to DIV18, relatively stable dendritic arbors undergo small but continuous growth until maturation. Robust formation of spine synapses initiates on dendritic arbors at approximately DIV7 to DIV9, peaks at approximately DIV11 to DIV14, and is largely complete by the end of 3 weeks (Ziv and Smith, 1996; Friedman et al., 2000; Grabrucker et al., 2009). To examine the role of Tmod in spine development and synapse formation without secondary effects due to dendritic retraction, we introduced shTmod1 and shTmod2 into hippocampal neurons at DIV13 and examined the spines and synapses on DIV21. Since ∼48 h is needed for the expression of short hairpins and effective knockdown of endogenous Tmod proteins, the experimental setup allows us focus on the effects of Tmod loss on spine and synapse development with minimal effects on dendritic development. In control DIV21 neurons expressing shLucif, most dendritic protrusions are characterized as mushroom-shaped spines with a distinct head and neck (Fig. 7A). By contrast, the knockdown of Tmod1 caused a substantial reduction in the density of mushroom-shaped spines and a large increase in filopodia-like protrusions, while Tmod2 knockdown slightly increased the spine density (Fig. 7A,B). Furthermore, we find that the average width of the spine head in shTmod1-expressing neurons was significantly reduced when compared with the shLucif- and shTmod2-expressing neurons (Fig. 7C). No change in spine neck length was observed in shTmod1- or shTmod2-expressing neurons (Fig. 7D).
Figure 7.
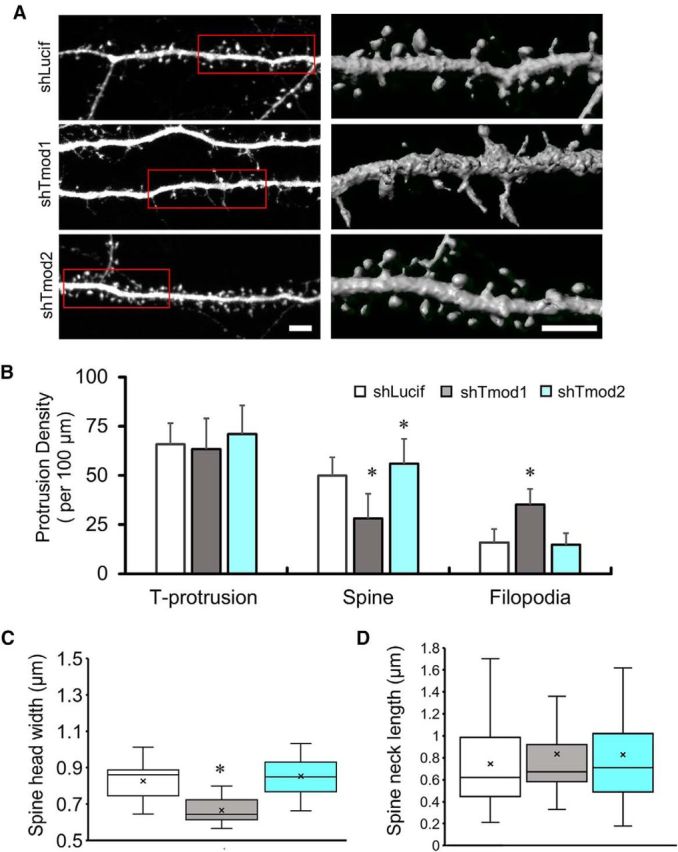
Knockdown of Tmod1 and Tmod2 on spine formation. A, Representative images of selected dendritic regions from control (shLucif) and shTmod-expressing cells showing reduced spine density in shTmod1-expressing neurons. Left panels are 2-D images from confocal stacks using maximum intensity projection. The regions enclosed by the red rectangles are 3-D rendered and shown on the right. Scale bars, 5 μm. B, Quantification of the densities of total protrusions (T-protrusion), spines, and filopodia. Error bars represent the SD. An unpaired Student's t test was used to compare different Tmod knock-down groups to the control group (shLucif). *Statistical difference from the control. For shTmod1: t(df) = 1.986(93, pSpine = 1.72E-15, pFilopodia = 6.26E-22; shTmod2: t(df) = 1.984(98), pT-protrusion = 0.051, pSpine = 0.007. C, D, Box-and-whisker plots show changes in spine head width (C) and spine neck length (D) in Tmod knock-down and control neurons. The box represents the 25th and 75th percentiles, with the median and mean shown as a line and a single x, respectively. The error bars represent the maximum and the minimum values in the distribution. Statistical analysis was done using an unpaired Student's t test. Spine head width: t(df) = 2.07(24), pTmod1 = 3.09E-11.
The observed reduction in the spine density by Tmod1 knockdown suggests that synapse formation might likewise be impaired. We tested this by examining the density of the presynaptic marker SV2 and postsynaptic marker PSD-95. We found that a majority of dendritic spines in control neurons were apposed by SV2 puncta and contained PSD-95 signal. In contrast, neurons expressing shTmod1 showed a significant reduction in the number of SV2-paired or PSD95-containing spines (Fig. 8A,B). We found that the knockdown of Tmod2 slightly increased the SV2 puncta density, which is consistent with the spine density data (Fig. 7). We next examined the miniature EPSCs (mEPSCs) by whole-cell patch-clamp recordings. Tmod1 knockdown consistently resulted in a reduction in the frequency and amplitude of mEPSCs (Fig. 8C), supporting the conclusion that synapse formation is impaired by Tmod1 knockdown. Interestingly, Tmod2 knockdown did not affect the amplitude, but increased the frequency of mEPSCs (Fig. 8C), which is consistent with the observed increase in spine/synapse number. Together, these results indicate that Tmod1, not Tmod2, is required for spine development and synapse formation.
Figure 8.
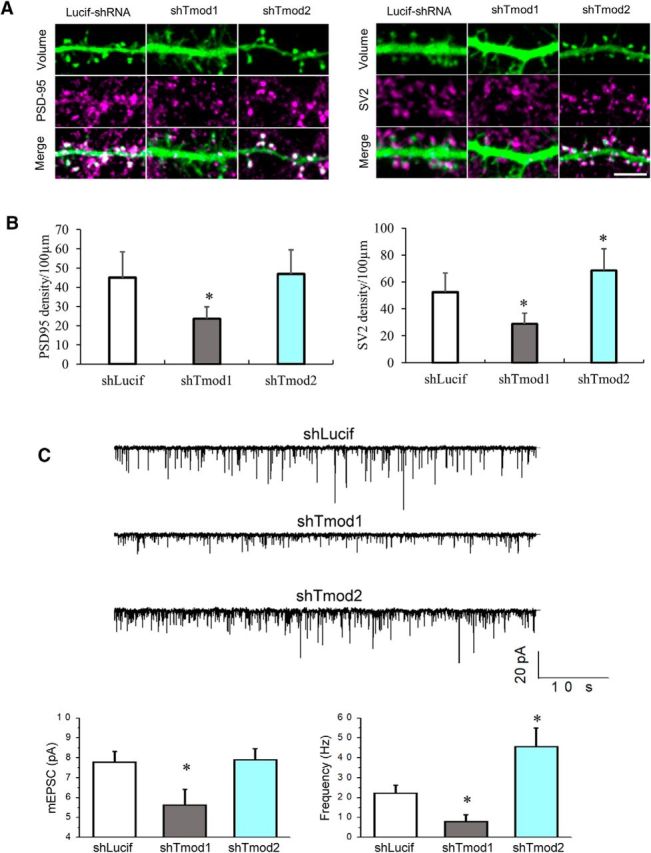
Tmod1 loss impairs synapse formation. A, shLucif and shTmod1 or shTmod2 neurons were stained for PSD-95 (left) and SV2 (right). Scale bar, 5 μm. B, Bar graphs depict changes in the density of PSD-95 and SV2 in knock-down and control groups (shLucif). Statistical analysis was performed using an unpaired Student's t test. *Statistical difference from the control. For PSD95: t(df) = 2.01(47), pshTmod1 = 4.31E-09; SV2: t(df) = 2.01(45), pshTmod1 = 2.53E-08, 2.02(44), pshTmod2 = 0.001. C, mEPSCs in DIV21 hippocampal neurons transfected with shLucif, shTmod1, and shTmod2. Each group contains at least five cells from two to three batches of cell culture. Sample traces are shown in the top portion and the bar graphs depict the average mEPSC amplitude and frequency. *Statistical difference from the shLucif group using unpaired Student's t test. For mEPSC amplitude: t(df) = 2.18(12), pshTmod1 = 0.04, pshTmod2 = 0.89; mEPSC frequency: t(df) = 2.18(12), pshTmod1 = 0.04, pshTmod2 = 0.02.
Discussion
Tropomodulin molecules are best known for controlling the length and stability of actin filaments in the sarcomere of striated muscle and the membrane skeleton of diverse cell types (Yamashiro et al., 2012). To date, fundamental knowledge about the endogenous distribution and role of Tmods during dendrite development and synapse formation remains largely unknown. In this study, we performed a series of experiments to examine the expression profile, subcellular distribution, and function of endogenous Tmod1 and Tmod2 in hippocampal neurons. Our results show that Tmod1 and Tmod2 have isoform-specific roles in postsynaptic development. Tmod1 is critical for dendritic spine development and synapse formation, whereas Tmod2 regulates dendrite growth and arborization. The distinct roles for Tmod1 and Tmod2 in spine and dendrite development are consistent with their distinct expression profiles. Collectively, our data propose a model whereby isoform-specific regulation of F-actin stability by Tropomodulin molecules regulates dendritic arbor development, spine head expansion, and synapse formation and transmission.
Our results show that Tmod2, but not Tmod1, plays a role in dendrite branching during development, which is consistent with its early onset of expression. Mechanistically, Tmod2 may regulate F-actin in motile actin-based dendritic growth cones, which enables dendrite extension as well as the formation of collateral dendritic branches (Ulfhake and Cullheim, 1988; Fiala et al., 1998; Niell et al., 2004). Tmod1 and Tmod2 are expressed in axonal growth cones of young primary hippocampal neurons (Fath et al., 2011), and they regulate neurite formation in PC12 and N2A cells (Gray et al., 2017). Alternatively, Tmod2 may function to support and/or maintain the cytoskeletal structures underlying dendritic arbors. The cortical distribution of Tmod2 suggests that Tmod2 may be associated with the membrane-associated periodic skeleton (MPS) in dendrites (Zhong et al., 2014; D'Este et al., 2015, 2016; Han et al., 2017). The dendritic MPS bears striking molecular similarity to the spectrin-actin “membrane skeleton” found in diverse metazoan cells, of which Tmod plays a crucial role. It is therefore plausible that Tmod2 may function in capping the pointed ends of short actin filaments in periodic F-actin rings that wrap around the circumference of the dendrite (Fig. 9). Clearly, future studies are needed to test this hypothesis. In addition to the MPS, Tmod2 may associate with the longitudinal F-actin cytoskeleton underneath the dendritic shaft membrane (lateral cytoskeleton). Though the contribution of the MPS and the lateral cytoskeleton to dendrite development is not known, Tmod2 may regulate dendrite development by stabilizing F-actin filaments in cortical F-actin structures during neuronal development. Although a fraction of Tmod1 is detected in the dendritic shaft, its late onset of expression may preclude its involvement in dendritic arborization. Further experiments are required to better elucidate the functions of Tmod2, and potentially Tmod1, in dendrite development and maintenance.
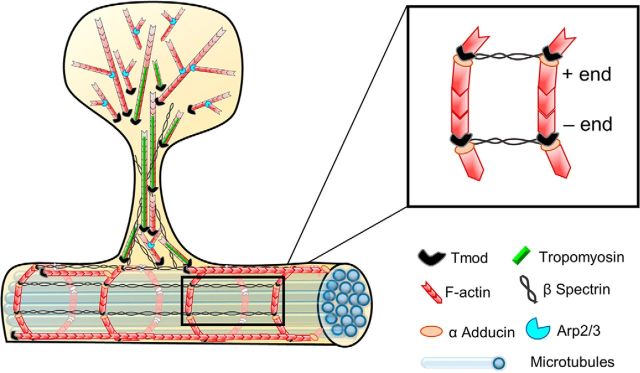
Figure 9.
Schematic representation of proposed Tmod1 and Tmod2 function in the postsynaptic compartment. Based on both our experimental data and ultrastructural and kinetic studies from other investigators, we propose that a fraction of actin filaments in the spine head is oriented with the barbed ends of the filaments toward the membrane and the pointed ends toward the spine center, or core. Tmod molecules are enriched in the spine neck and “core” of the spine head, where they cap the pointed ends of actin filaments. By contrast, Tmod molecules are rarely detected in the peripheral region of the spine head, which is characterized by newly nucleated filaments whose pointed ends are associated with the Arp2/3 complex, leading to a high number of branched filaments. βII/βIII spectrin tetramers, together with short actin filaments, form an MPS in dendrites, the spine neck, and the base of the spine head. Like βII/βIII-spectrin, Tmod immunofluorescence is detected in the spine head, but does not overlap with the PSD. The presence of Tmod in this subspine region is highly indicative of TM expression, though which TM isoform coats actin filaments in the stable core is unknown. In dendrites, short actin filaments capped by adducin at the barbed end and by Tmod2 at the pointed end, may be organized into evenly spaced rings near the plasma membrane that are connected by longitudinally oriented spectrin tetramers. Although our immunofluorescence data show that Tmod1 and Tmod2 have a largely nonoverlapping distribution in dendritic spines and along dendritic shafts, both Tmod1 and Tmod2 are presented here as “Tmod” for the sake of clarity and simplicity.
A significant observation of this study is the subspine localization of Tmod1 and Tmod2 (Fig. 5). Tropomodulin molecules are best known for capping the pointed ends of filament and inhibiting depolymerization, thereby stabilizing actin filaments. Previous studies have shown that the peripheral region of spine heads contain branched F-actin structures that undergo dynamic turnover (Fifková and Delay, 1982; Landis and Reese, 1983; Korobova and Svitkina, 2010). By contrast, the core of the spine head and spine neck contain relatively stable F-actin. The existence of two distinct kinetic pools of F-actin may allow the spine to maintain a stable core for structural integrity, while simultaneously undergoing polymerization-driven structural rearrangements that underlie synapse development and plasticity. The localization of Tmod1 and Tmod2 to a subspine region characterized by stable filaments, as well as our FRAP data indicating that Tmod depletion increases F-actin dynamics, suggests that Tmod regulates the stable pool of the spine F-actin, likely by capping of the pointed end (Fig. 9). In addition, Tmod may promote F-actin stability in spines by capping pointed ends in the actin-βII/III spectrin membrane lattice, which is present in the base and neck dendritic spines (Sidenstein et al., 2016). Intriguingly, this detergent-resistant (Efimova et al., 2017) membrane lattice is discontinued at synaptic sites, entering the spine head but not reaching the PSD (Sidenstein et al., 2016). Consistently, we find that Tmod distribution in spines is (1) enriched in the neck and base of the spine head (2) exhibits a nonoverlapping distribution with the PSD, and (3) is overwhelmingly resistant to detergent extraction. Therefore, it is plausible that Tmod proteins could be a component of the subcortical lattice in dendritic spines that is required for maintaining dendritic spine shape as well as the formation of a constricted spine neck (Efimova et al., 2017).
The formation of dendritic spines and mature excitatory synapses is accompanied by a significant increase in the size of the stable pool of F-actin in dendritic spines (Koskinen et al., 2014), which is consistent with the conversion of highly motile filopodia into stable mushroom-shaped structures (Dailey and Smith, 1996; Ziv and Smith, 1996). Dynamic rearrangements of the underlying F-actin cytoskeleton from linear bundles into a highly branched network accounts for the structural changes underlying spine morphogenesis (Marrs et al., 2001; Hotulainen et al., 2009; Korobova and Svitkina, 2010). We find that depletion of Tmod1 during synaptogenesis reduces spine density in mature neurons, consistent with previous studies showing that the overexpression of GFP-Tmod1 promotes dendritic protrusions (Gray et al., 2016). Our expression profile and loss-of-function analyses suggest that Tmod1 is required for the morphological remodeling underlying the filopodia-to-spine transition. We propose that the capping of filament pointed ends by Tmod1, together with the concerted efforts of other actin regulatory proteins, leads to filament stabilization and/or net polymerization, creating a stable F-actin core in the spine neck and core. It is likely that this stable cytoskeletal structure serves as a platform for Arp2/3-mediated expansion (Hotulainen et al., 2009), consistent with the observed reduction in spine head diameter in shTmod1-expressing neurons.
It is puzzling to see that Tmod2, which is present in spines and expressed during synaptogenesis, does not appear to be required for spine development and synapse formation. Instead, Tmod2 knockdown slightly increased spine/synapse number and mEPSCs. These results are consistent with previous studies showing that (1) Tmod2 knockout resulted in enhanced LTP (Cox et al., 2003) and (2) Tmod2 expression is upregulated in LTD to allow spine shrinkage (Hu et al., 2014). While the overexpression of Tmod2 was found to increase spine density (Hu et al., 2014), it remains to be seen whether the effect reflects a physiological role of Tmod2. It should be noted that unlike the Tmod2 knock-out study (Cox et al., 2003), Tmod2 knockdown in this study did not cause noticeable changes in Tmod1 expression (Fig. 2B,D). This is likely due to the short knock-down period (8–11 d) in cultured hippocampal neurons, relative to the in vivo period. Other factors, including the nucleotide composition of the shTmod sequence, the level of knockdown, and the specific cell type used, could also contribute to the lack of compensatory upregulation of Tmod1 observed in previous studies (Fath et al., 2011). Nevertheless, future experiments are needed to understand the precise functions of Tmod1 and Tmod2 in spine and synapse formation, spine maintenance, and synaptic plasticity.
The observed isoform-specific role of Tmod1 and Tmod2 in dendrite development and synapse function is likely tightly regulated by tropomyosin (TM), an α-helical coiled-coil protein that binds along the sides of F-actin filaments. Tight binding of Tmod to F-actin pointed ends requires tropomyosin (Weber et al., 1999), and divergence in Tmod1 and Tmod2 function is likely mediated through differential binding to TPM3 and TPM4, two TM isoforms expressed in the postsynaptic compartment (Had et al., 1994; Guven et al., 2011). Tmod1 and Tmod2 have differential affinities for TM isoforms (Kostyukova, 2008; Uversky et al., 2011; Colpan et al., 2016), which are known to alter the spectrum of actin-regulatory proteins that can bind to F-actin (Gunning et al., 2008, 2015). The spatiotemporal association of Tmod1 or Tmod2 with distinct TM-coated filaments may enable Tmod1 and Tmod2 to adopt distinct roles during various stages of dendrite development and synapse development. In support of this, we show that Tmod1 and Tmod2 have largely nonoverlapping distributions in spines and differentially regulate F-actin dynamics. The inherent biochemical and structural differences between Tmod isoforms (Yamashiro et al., 2012), coupled with differential affinity for TM-coated actin filaments, are likely mechanisms used to fine-tune the isoform-specific regulation of F-actin by Tmod1 and Tmod2.
Our finding that Tmod1 and Tmod2 are essential for dendrite development and synapse formation adds to our limited body of knowledge about Tmod function in neurons. These studies shed light on the mechanisms by which F-actin is regulated in neurons and provide a platform for future studies to uncover the precise mechanisms by which Tmod modifies the cytoskeleton in neurons and contributes to neuronal function.
Footnotes
The authors declare no competing financial interests.
This research project was supported in part by research grants from the National Institutes of Health to J.Q.Z. (GM-083889, MH-104632, and MH-108025), O.F.O. (5F31-NS-092437-03), V.M.F. (EY-017724), and H.C.H. (EY-014852 and AR-067786); as well as by Emory University Integrated Cellular Imaging Microscopy Core of the Emory Neuroscience National Institute of Neurological Disorders and Stroke Core Facilities Grant 5P30-NS-055077. We thank Dr. Kenneth Myers for his technical expertise and help throughout the project. We also thank Drs. Sharada Tilve, Daniela Malide, and Herbert Geller at National Heart, Lung, and Blood Institute for their help with super-resolution STED imaging.
References
- Chen A, Liao WP, Lu Q, Wong WS, Wong PT (2007) Upregulation of dihydropyrimidinase-related protein 2, spectrin alpha II chain, heat shock cognate protein 70 pseudogene 1 and tropomodulin 2 after focal cerebral ischemia in rats—a proteomics approach. Neurochem Int 50:1078–1086. 10.1016/j.neuint.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Colpan M, Moroz NA, Gray KT, Cooper DA, Diaz CA, Kostyukova AS (2016) Tropomyosin-binding properties modulate competition between tropomodulin isoforms. Arch Biochem Biophys 600:23–32. 10.1016/j.abb.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM (2001) Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics 73:127–139. 10.1006/geno.2000.6501 [DOI] [PubMed] [Google Scholar]
- Cox PR, Fowler V, Xu B, Sweatt JD, Paylor R, Zoghbi HY (2003) Mice lacking tropomodulin-2 show enhanced long-term potentiation, hyperactivity, and deficits in learning and memory. Mol Cell Neurosci 23:1–12. 10.1016/S1044-7431(03)00025-3 [DOI] [PubMed] [Google Scholar]
- Dailey ME, Smith SJ (1996) The dynamics of dendritic structure in developing hippocampal slices. J Neurosci 16:2983–2994. 10.1523/JNEUROSCI.16-09-02983.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Este E, Kamin D, Göttfert F, El-Hady A, Hell SW (2015) STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Rep 10:1246–1251. 10.1016/j.celrep.2015.02.007 [DOI] [PubMed] [Google Scholar]
- D'Este E, Kamin D, Velte C, Göttfert F, Simons M, Hell SW (2016) Subcortical cytoskeleton periodicity throughout the nervous system. Sci Rep 6:22741. 10.1038/srep22741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA (1988) The establishment of polarity by hippocampal neurons in culture. J Neurosci 8:1454–1468. 10.1523/JNEUROSCI.08-04-01454.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimova N, Korobova F, Stankewich MC, Moberly AH, Stolz DB, Wang J, Kashina A, Ma M, Svitkina T (2017) βIII spectrin is necessary for formation of the constricted neck of dendritic spines and regulation of synaptic activity in neurons. J Neurosci 37:6442–6459. 10.1523/JNEUROSCI.3520-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath T, Fischer RS, Dehmelt L, Halpain S, Fowler VM (2011) Tropomodulins are negative regulators of neurite outgrowth. Eur J Cell Biol 90:291–300. 10.1016/j.ejcb.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Popov V, Harris KM (1998) Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci 18:8900–8911. 10.1523/JNEUROSCI.18-21-08900.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fifková E, Delay RJ (1982) Cytoplasmic actin in neuronal processes as a possible mediator of synaptic plasticity. J Cell Biol 95:345–350. 10.1083/jcb.95.1.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RS, Fritz-Six KL, Fowler VM (2003) Pointed-end capping by tropomodulin3 negatively regulates endothelial cell motility. J Cell Biol 161:371–380. 10.1083/jcb.200209057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VM, Dominguez R (2017) Tropomodulins and leiomodins: actin pointed end caps and nucleators in muscles. Biophys J 112:1742–1760. 10.1016/j.bpj.2017.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VM, Sussmann MA, Miller PG, Flucher BE, Daniels MP (1993) Tropomodulin is associated with the free (pointed) ends of the thin filaments in rat skeletal muscle. J Cell Biol 120:411–420. 10.1083/jcb.120.2.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman HV, Bresler T, Garner CC, Ziv NE (2000) Assembly of new individual excitatory synapses: time course and temporal order of synaptic molecule recruitment. Neuron 27:57–69. 10.1016/S0896-6273(00)00009-X [DOI] [PubMed] [Google Scholar]
- Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA (2010) Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron 67:86–99. 10.1016/j.neuron.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhin DS, Lewis RA, McKeown CR, Nowak RB, Kim NE, Littlefield RS, Lieber RL, Fowler VM (2010) Tropomodulin isoforms regulate thin filament pointed-end capping and skeletal muscle physiology. J Cell Biol 189:95–109. 10.1083/jcb.201001125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabrucker A, Vaida B, Bockmann J, Boeckers TM (2009) Synaptogenesis of hippocampal neurons in primary cell culture. Cell Tissue Res 338:333–341. 10.1007/s00441-009-0881-z [DOI] [PubMed] [Google Scholar]
- Gray KT, Suchowerska AK, Bland T, Colpan M, Wayman G, Fath T, Kostyukova AS (2016) Tropomodulin isoforms utilize specific binding functions to modulate dendrite development. Cytoskeleton (Hoboken) 73:316–328. 10.1002/cm.21304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KT, Kostyukova AS, Fath T (2017) Actin regulation by tropomodulin and tropomyosin in neuronal morphogenesis and function. Mol Cell Neurosci 84:48–57. 10.1016/j.mcn.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Firestein BL, Zheng JQ (2008) Microtubules in dendritic spine development. J Neurosci 28:12120–12124. 10.1523/JNEUROSCI.2509-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaud L, Gray KT, Moroz N, Pantazis C, Pate E, Kostyukova AS (2014) Role of tropomodulin's leucine rich repeat domain in the formation of neurite-like processes. Biochemistry 53:2689–2700. 10.1021/bi401431k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P, O'Neill G, Hardeman E (2008) Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev 88:1–35. 10.1152/physrev.00001.2007 [DOI] [PubMed] [Google Scholar]
- Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP (2015) Tropomyosin-master regulator of actin filament function in the cytoskeleton. J Cell Sci 128:2965–2974. 10.1242/jcs.172502 [DOI] [PubMed] [Google Scholar]
- Gustafsson MG. (2000) Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc 198:82–87. 10.1046/j.1365-2818.2000.00710.x [DOI] [PubMed] [Google Scholar]
- Guven K, Gunning P, Fath T (2011) TPM3 and TPM4 gene products segregate to the postsynaptic region of central nervous system synapses. Bioarchitecture 1:284–289. 10.4161/bioa.1.6.19336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Had L, Faivre-Sarrailh C, Legrand C, Méry J, Brugidou J, Rabié A (1994) Tropomyosin isoforms in rat neurons: the different developmental profiles and distributions of TM-4 and TMBr-3 are consistent with different functions. J Cell Sci 107:2961–2973. [DOI] [PubMed] [Google Scholar]
- Han B, Zhou R, Xia C, Zhuang X (2017) Structural organization of the actin-spectrin-based membrane skeleton in dendrites and soma of neurons. Proc Natl Acad Sci U S A 114:E6678–E6685. 10.1073/pnas.1705043114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B (1992) Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci 12:2685–2705. 10.1523/JNEUROSCI.12-07-02685.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H (2008) The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron 57:719–729. 10.1016/j.neuron.2008.01.013 [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Hoogenraad CC (2010) Actin in dendritic spines: connecting dynamics to function. J Cell Biol 189:619–629. 10.1083/jcb.201003008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Llano O, Smirnov S, Tanhuanpää K, Faix J, Rivera C, Lappalainen P (2009) Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol 185:323–339. 10.1083/jcb.200809046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yu D, Gu QH, Yang Y, Tu K, Zhu J, Li Z (2014) miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat Commun 5:3263. 10.1038/ncomms4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwazaki T, McGregor IS, Matsumoto I (2006) Protein expression profile in the striatum of acute methamphetamine-treated rats. Brain Res 1097:19–25. 10.1016/j.brainres.2006.04.052 [DOI] [PubMed] [Google Scholar]
- Kong KY, Kedes L (2004) Cytoplasmic nuclear transfer of the actin-capping protein tropomodulin. J Biol Chem 279:30856–30864. 10.1074/jbc.M302845200 [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T (2010) Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell 21:165–176. 10.1091/mbc.e09-07-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen M, Bertling E, Hotulainen R, Tanhuanpää K, Hotulainen P (2014) Myosin IIb controls actin dynamics underlying the dendritic spine maturation. Mol Cell Neurosci 61:56–64. 10.1016/j.mcn.2014.05.008 [DOI] [PubMed] [Google Scholar]
- Kostyukova AS. (2008) Tropomodulin/tropomyosin interactions regulate actin pointed end dynamics. Adv Exp Med Biol 644:283–292. 10.1007/978-0-387-85766-4_21 [DOI] [PubMed] [Google Scholar]
- Landis DM, Reese TS (1983) Cytoplasmic organization in cerebellar dendritic spines. J Cell Biol 97:1169–1178. 10.1083/jcb.97.4.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Vitriol EA, Shim S, Wise AL, Velayutham RP, Zheng JQ (2013) Dynamic localization of G-actin during membrane protrusion in neuronal motility. Curr Biol 23:1046–1056. 10.1016/j.cub.2013.04.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei W, Myers KR, Rui Y, Hladyshau S, Tsygankov D, Zheng JQ (2017) Phosphoinositide-dependent enrichment of actin monomers in dendritic spines regulates synapse development and plasticity. J Cell Biol 216:2551–2564. 10.1083/jcb.201612042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus EJ, Petit TL (1987) Neocortical synaptogenesis, aging, and behavior: lifespan development in the motor-sensory system of the rat. Exp Neurol 96:262–278. 10.1016/0014-4886(87)90045-8 [DOI] [PubMed] [Google Scholar]
- Marrs GS, Green SH, Dailey ME (2001) Rapid formation and remodeling of postsynaptic densities in developing dendrites. Nat Neurosci 4:1006–1013. 10.1038/nn717 [DOI] [PubMed] [Google Scholar]
- Meyer L, Wildanger D, Medda R, Punge A, Rizzoli SO, Donnert G, Hell SW (2008) Dual-color STED microscopy at 30-nm focal-plane resolution. Small 4:1095–1100. 10.1002/smll.200800055 [DOI] [PubMed] [Google Scholar]
- Moroz N, Guillaud L, Desai B, Kostyukova AS (2013) Mutations changing tropomodulin affinity for tropomyosin alter neurite formation and extension. PeerJ 1:e7. 10.7717/peerj.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer JD, Nowak RB, Kim NE, Larkin SK, Peters LL, Hartwig J, Kuypers FA, Fowler VM (2010) Tropomodulin 1-null mice have a mild spherocytic elliptocytosis with appearance of tropomodulin 3 in red blood cells and disruption of the membrane skeleton. Blood 116:2590–2599. 10.1182/blood-2010-02-268458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niell CM, Meyer MP, Smith SJ (2004) In vivo imaging of synapse formation on a growing dendritic arbor. Nat Neurosci 7:254–260. 10.1038/nn1191 [DOI] [PubMed] [Google Scholar]
- Nowak RB, Fischer RS, Zoltoski RK, Kuszak JR, Fowler VM (2009) Tropomodulin1 is required for membrane skeleton organization and hexagonal geometry of fiber cells in the mouse lens. J Cell Biol 186:915–928. 10.1083/jcb.200905065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA. (1953) Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat 87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Sidenstein SC, D'Este E, Böhm MJ, Danzl JG, Belov VN, Hell SW (2016) Multicolour multilevel STED nanoscopy of Actin/Spectrin organization at synapses. Sci Rep 6:26725. 10.1038/srep26725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star EN, Kwiatkowski DJ, Murthy VN (2002) Rapid turnover of actin in dendritic spines and its regulation by activity. Nat Neurosci 5:239–246. 10.1038/nn811 [DOI] [PubMed] [Google Scholar]
- Sun Y, Dierssen M, Toran N, Pollak DD, Chen WQ, Lubec G (2011) A gel-based proteomic method reveals several protein pathway abnormalities in fetal down syndrome brain. J Proteomics 74:547–557. 10.1016/j.jprot.2011.01.009 [DOI] [PubMed] [Google Scholar]
- Sussman MA, Sakhi S, Tocco G, Najm I, Baudry M, Kedes L, Schreiber SS (1994) Neural tropomodulin: developmental expression and effect of seizure activity. Brain Res Dev Brain Res 80:45–53. 10.1016/0165-3806(94)90088-4 [DOI] [PubMed] [Google Scholar]
- Ulfhake B, Cullheim S (1988) Postnatal development of cat hind limb motoneurons. II: in vivo morphology of dendritic growth cones and the maturation of dendrite morphology. J Comp Neurol 278:88–102. 10.1002/cne.902780106 [DOI] [PubMed] [Google Scholar]
- Uversky VN, Shah SP, Gritsyna Y, Hitchcock-DeGregori SE, Kostyukova AS (2011) Systematic analysis of tropomodulin/tropomyosin interactions uncovers fine-tuned binding specificity of intrinsically disordered proteins. J Mol Recognit 24:647–655. 10.1002/jmr.1093 [DOI] [PubMed] [Google Scholar]
- Watakabe A, Kobayashi R, Helfman DM (1996) N-tropomodulin: a novel isoform of tropomodulin identified as the major binding protein to brain tropomyosin. J Cell Sci 109:2299–2310. [DOI] [PubMed] [Google Scholar]
- Weber A, Pennise CR, Fowler VM (1999) Tropomodulin increases the critical concentration of barbed end-capped actin filaments by converting ADP.P(i)-actin to ADP-actin at all pointed filament ends. J Biol Chem 274:34637–34645. 10.1074/jbc.274.49.34637 [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Gokhin DS, Kimura S, Nowak RB, Fowler VM (2012) Tropomodulins: pointed-end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton (Hoboken) 69:337–370. 10.1002/cm.21031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JW, Czech T, Felizardo M, Baumgartner C, Lubec G (2006) Aberrant expression of cytoskeleton proteins in hippocampus from patients with mesial temporal lobe epilepsy. Amino Acids 30:477–493. 10.1007/s00726-005-0281-y [DOI] [PubMed] [Google Scholar]
- Yu P, Agbaegbu C, Malide DA, Wu X, Katagiri Y, Hammer JA, Geller HM (2015) Cooperative interactions of LPPR family members in membrane localization and alteration of cellular morphology. J Cell Sci 128:3210–3222. 10.1242/jcs.169789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T (2004) Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci 5:24–34. 10.1038/nrn1300 [DOI] [PubMed] [Google Scholar]
- Zhong G, He J, Zhou R, Lorenzo D, Babcock HP, Bennett V, Zhuang X (2014) Developmental mechanism of the periodic membrane skeleton in axons. Elife 3:e04581. 10.7554/eLife.04581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ (1996) Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 17:91–102. 10.1016/S0896-6273(00)80283-4 [DOI] [PubMed] [Google Scholar]