Sir,
We appreciate the interest taken by Dr Fratta and his team (Sivakumar et al., 2018) in our recent work documenting the impact of TDP-43 on the splicing of hnRNP A1 and its potential relevance in the pathogenesis of amyotrophic lateral sclerosis (ALS) as outlined in our paper ‘TDP-43 regulates the alternative splicing of hnRNP A1 to yield an aggregation-prone variant in amyotrophic lateral sclerosis’ (Deshaies et al., 2018). Indeed, their letter highlights the complexity of RNA binding protein (RBP) functional networks. Using analyses of RNA-Seq data not validated by additional biochemical methods, the authors claim that TDP-43 does not bind to exon 7B or its flanking introns. Moreover, the primary claim is that TDP-43 mutations increase inclusion of hnRNP A1 exon 7B (i.e. gain-of-function), while TDP-43 loss-of function decreases exon 7B inclusion. The accompanying RNA-Seq and iCLIP analyses supporting these assertions are interesting but raise many questions.
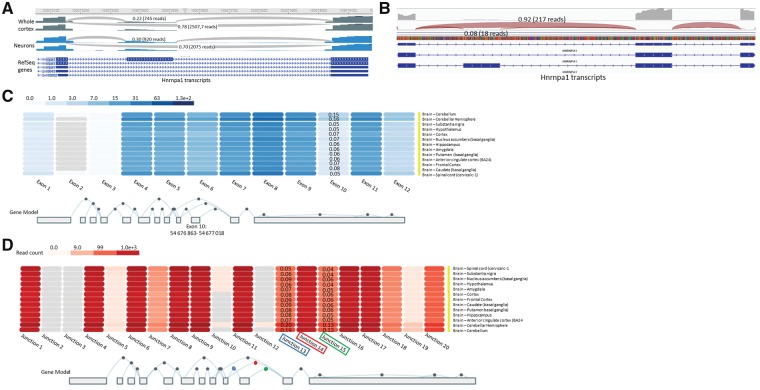
First and foremost, we acknowledged in our paper that TDP-43 binding sites in HNRNPA1 were not detected in available iCLIP data performed in mice. In addition, there are a number of limitations to RNA-Seq data that should be considered. Most relevant, the small changes in splicing expressed by the delta PSI (per cent spliced in) values presented by Sivakumar et al. (2018) (their Fig. 1A and B) would require subsequent validation with RT-PCR and in vitro work, as we have done. This is especially important if one considers the variability in exon 7B PSI values obtained in wild-type mice across the six mouse datasets analysed (ranging 16–25%). Additionally, TDP-43 antisense oligonucleotide knockdown (ASO KD) samples appear to have the same PSI values as wild-type mice in the LCDmut embryonic Day (e)14.5, LCDmut 6-month-old (mo), and Q331K 5-month-old (mo) datasets. Thus, the analyses of these mouse datasets would appear inconclusive. We acknowledge that we have used a human transformed cell line for our in vitro studies that is admittedly not a cell type of relevance for the disease. However, the RNA-Seq datasets analysed by Sivakumar and colleagues derive from mouse brains that are a complex mixture of cell types. Thus, events preferentially relevant to neurons may be diluted out in such an experiment. This consideration is supported by our published immunohistochemistry of human spinal cords, which demonstrated robust detection of hnRNP A1 and A1B in the motor neurons of those sections, but relatively little labelling of other cell types in the parenchyma. Further support derives from data available through the Brain Cell RNASeq Browser, which indicates that exon 7B inclusion is an event more often observed in neurons than in other neural cell types in wild-type mouse brain (30% of transcripts compared to 20% in whole cortex; Fig. 1A) (Zhang et al., 2014).
Figure 1.
Exon and junction expression of HNRNPA1 exon 7B. (A) Representative RNA-seq read traces (Brain CellRNAseq Browser; Jiaqian Wu Lab) with Sashimi plots for Hnrnpa1 in wild-type mice whole cortex and neurons. (B) Representative RNA-seq read traces from wild-type human amygdala. (C) HNRNPA1 exon expression in brain tissues using GTEx RNA-seq data (www.gtexportal.org). HNRNPA1 transcripts were collapsed into a single transcript. In this representation, exon 10 corresponds to exon 7B. Values correspond to exon 10 usage compared to exons 9 and 11. (D) HNRNPA1 junction expression in brain tissues using GTEx RNA-seq data (www.gtexportal.org). HNRNPA1 transcripts were collapsed into a single transcript. In this representation, junction 13 corresponds to junction between exon 9/10 (ie. Exon 7/7B); junction 15 between exon 10/11 (ie. Exon 7B/8); and junction 14 between exon 9/11 (ie. Exon 7/8). Values correspond to junctions 13 or 15 usage compared to junction 14.
For the analysis of induced pluripotent stem cell (iPSC)-derived human motor neurons, the PSI data provided in Sivakumar et al’s Fig. 1C and D do not indicate whether there is sufficient coverage of this gene to support the claim asserted. This is made all the more relevant by the apparent disagreement in the frequency of exon 7B inclusion in adult brain versus these neurons. Data in human brain available through GTEx (Carithers et al., 2015) indicate that exon 7B inclusion, and thus usage of junctions 7/7B and 7B/8, is observed at a much lower frequency (7.3% in cortex, 4.5% in spinal cord) than observed by our colleagues in iPSC-derived motor neurons (45%; Fig. 1B–D). Perhaps it would be prudent here to also highlight that the field has yet to reach a consensus as whether in vitro patient-derived motor neurons are representative of the aged stage and/or anatomical location of the adult human motor neurons affected in ALS patients (Sances et al., 2016; Guo et al., 2017). Thus, while studies in induced motor neurons are undoubtedly of value in our efforts to better understand ALS pathogenesis, perhaps some caution is required before generalizing these data.
Sivakumar and colleagues’ analyses also indicate that there is potential TDP-43 binding in exon 8 of human HNRNPA1, and additional conserved sites in the 3′ UTR of mouse and human HNRNPA1. Using a mini-gene sequence including human exon 7 and 7B and a universal 3′ splice site acceptor from adenovirus, we observed a direct influence of TDP-43 on exon 7B inclusion. Thus, our system did not permit exploration of this potential exon 8 binding site. Nevertheless, in vivo, it remains possible that TDP-43 binding in exon 8 may affect exon 7B inclusion since the 5′ splice site of exon 7 is looking to choose between the 3′ splice site of exon 7B and the 3′ splice site of exon 8 (we have shown previously that the 5′ splice site of exon 7B is weakened by secondary structures, so it is likely not involved in the original selection and thus possibly used by default) (Blanchette and Chabot, 1997). Thus, if the 3′ splice site of exon 8 is normally strengthened by TDP-43 binding in exon 8, loss of TDP-43 may still promote increased exon 7B inclusion. Lastly, while cell type/model differences may certainly be relevant, using a luciferase-based reporter assay, we did not observe an impact of TDP-43 on the 3′ UTR of HNRNPA1. Thus, the significance of these potential TDP-43 binding sites remains to be established.
With regards to the impact of ALS-associated TDP-43 mutations, we did not explore this for the very reason stated by Sivakumar and colleagues: that TDP-43 is exquisitely dosage-sensitive. Indeed, it is an issue relevant to the majority of TDP-43 functional studies in the literature that renders their interpretation difficult. Thus, given our lack of access to samples bearing TDP-43 mutations at physiologically relevant levels (i.e. single copy), we did not explore this and instead focused on endogenous TDP-43 function. However, the provided analysis of TDP-43Q331K knock-in mice is worth discussing. RNA-Seq of laser microdissected motor neurons from these mice indicated there was no evidence of misregulation of known TDP-43 targets since TARDBP, SORT1 and MAPT were not differentially expressed or spliced in these cells. In contrast, however, RNA-Seq of the homogenates of the remaining lumbar spinal cord (from which the motor neurons had been laser captured), demonstrated alterations in TARDBP and SORT1 splicing that was consistent with TDP-43 gain-of-function. Indeed, White et al. (2018) concluded that TDP-43Q331K expression actually caused splicing differences within spinal interneurons, not the motor neurons, as supported by a decrease in parvalbumin-positive interneurons. We would again reiterate here that in our admittedly limited analysis of post-mortem tissues, hnRNP A1/A1B labelling was predominantly observed within motor neurons.
In the RNA-Seq analysis of TDP-43Q331K mice provided in the letter by Sivakumar et al., it is noteworthy that only minor differences (~3%) in exon 7B inclusion were documented, and this was restricted to 20-month-old homozygous animals, i.e. old animals with non-physiological expression of the TDP-43 mutation. There were no significant differences in exon 7B inclusion between wild-type and heterozygous TDP-43Q331K mice, which would be the genotype most reflective of the human condition. Lastly, these mice do not exhibit evidence of TDP-43 aggregation or mislocalization. While these animals do exhibit cognitive impairment, there is no impact on motor neuron number or morphology and they have normal neuromuscular junctions (White et al., 2018). Thus, they do not replicate the situation observed in the majority of ALS patients where TDP-43 is nuclear depleted/cytoplasmic accumulated, nor do they parallel our loss of function studies.
Similarly, Sivakumar and colleagues report minor changes in exon 7B PSI in heterozygous LCDmut mice (~3–4%), which also do not feature the pathological disease hallmark of TDP-43 cytoplasmic inclusions accompanied with TDP-43 nuclear depletion. This feature is also absent in RRM2mut mice bearing the mutation F210I, which are reportedly a loss-of-function model. However, while RRM2 has been previously demonstrated to be less relevant to RNA binding (Lukavsky et al., 2013; Kuo et al., 2014; Furukawa et al., 2016), the F210I mutation is reported to reduce RNA binding capacity in vitro (Fratta et al., 2018). Remarkably, despite an apparently reduced ability to bind RNA, heterozygous RRM2mut mice (on a hybrid background) aged up to 2 years do not display any motor phenotype or pathology. This contrasts with reports that TDP-43 loss-of-function (Iguchi et al., 2013; Yang et al., 2014) or disruption of the nuclear localization signal (Winton et al., 2008; Igaz et al., 2011; Alfieri et al., 2014) can result in a progressive neurodegenerative phenotype impacting motor neurons.
Ultimately, our work and that of Sivakumar and colleagues highlights that there is a previously unappreciated connection between TDP-43, hnRNP A1B and ALS. Our work emphasizes that widespread dysregulation of RBPs is possibly an important consideration in the pathogenesis of ALS and reinforces that the field should not have a TDP-43-centric view of the disease, but rather consider RBPs imbalance more broadly as a core disease-initiating and/or driving mechanism, as recently suggested (Conlon et al., 2018). Indeed, the ensuing mechanism that splicing changes affecting one or a few RBPs may act as drivers of RBP aggregation (Conlon et al., 2018) is consistent with our data. In addition, detectable splicing changes and highly insoluble protein levels may only become evident in the later stages of disease. Given that our analysis of patient samples is obligatorily at end-stage, this remains a possibility. Nonetheless, it is clear that we are in collective agreement that unravelling the complex interplay between RBPs in ALS should be a priority to improve our understanding of the disease process occurring in patients and aid the development of disease-modifying therapies.
Data availability
Data analysed is available at: Brain cell RNAseq browser (mouse): http://jiaqianwulab.org/braincell/RNASeq.html; GTEX (human): https://gtexportal.org/home/gene/HNRNPA1; exon expression for HNRNPA1 (ENSG00000135486.13) Data Source: GTEx Analysis Release V7 (dbGaP Accession phs000424.v7.p2).
Funding
This work was supported by an NSERC Discovery grant and the ALS Society of Canada (CVV). B.C. was supported by CIHR grant MOP-136948 and is the Pierre C. Fournier Chair in Functional Genomics. H.S. is supported by a FRQS Doctoral award. CVV is a FRQS Senior Scholar.
Competing interests
The authors report no competing interests.
References
- Alfieri JA, Pino NS, Igaz LM. Reversible behavioral phenotypes in a conditional mouse model of TDP-43 proteinopathies. J Neurosci 2014; 34: 15244–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Chabot B. A highly stable duplex structure sequesters the 5' splice site region of hnRNP A1 alternative exon 7B. RNA 1997; 3: 405–19. [PMC free article] [PubMed] [Google Scholar]
- Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, et al. A novel approach to high-quality postmortem tissue procurement: the gtex project. Biopreserv Biobank 2015; 13: 311–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon EG, Fagegaltier D, Agius P, Davis-Porada J, Gregory J, Hubbard I, et al. Unexpected similarities between C9ORF72 and sporadic forms of ALS/FTD suggest a common disease mechanism. Elife 2018; 7, pii: e37754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies JE, Shkreta L, Moszczynski AJ, Sidibé H, Semmler S, Fouillen A, et al. TDP-43 regulates the alternative splicing of hnRNP A1 to yield an aggregation-prone variant in amyotrophic lateral sclerosis. Brain 2018; 141: 1320–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Sivakumar P, Humphrey J, Lo K, Ricketts T, Oliveira H, et al. Mice with endogenous TDP-43 mutations exhibit gain of splicing function and characteristics of amyotrophic lateral sclerosis. EMBO J 2018; 37, pii: e98684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, Suzuki Y, Fukuoka M, Nagasawa K, Nakagome K, Shimizu H, et al. A molecular mechanism realizing sequence-specific recognition of nucleic acids by TDP-43. Sci Rep 2016; 6: 20576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Fumagalli L, Prior R, Van Den Bosch L. Current advances and limitations in modeling ALS/FTD in a dish using induced pluripotent stem cells. Front Neurosci 2017; 11: 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Lee EB, Chen-Plotkin A, Swanson E, Unger T, et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest 2011; 121: 726–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi Y, Katsuno M, Niwa J, Takagi S, Ishigaki S, Ikenaka K, et al. Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain 2013; 136 (Pt 5): 1371–82. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Chiang CH, Wang YT, Doudeva LG, Yuan HS. The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids. Nucleic Acids Res 2014; 42: 4712–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, et al. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol 2013; 20: 1443–9. [DOI] [PubMed] [Google Scholar]
- Sances S, Bruijn LI, Chandran S, Eggan K, Ho R, Klim JR, et al. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat Neurosci 2016; 19: 542–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar P, De Giorgio F, Ule AM, Neeves J, Nair RR, Bentham M, et al. TDP-43 mutations increase HNRNP A1-7B through gain of splicing function. Brain 2018; doi: 10.1093/brain/awy260. [DOI] [PubMed] [Google Scholar]
- White MA, Kim E, Duffy A, Adalbert R, Phillips BU, Peters OM, et al. TDP-43 gains function due to perturbed autoregulation in a Tardbp knock-in mouse model of ALS-FTD. Nat Neurosci 2018; 21: 552–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem 2008; 283: 13302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang H, Qiao T, Yang B, Aliaga L, Qiu L, et al. Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA 2014; 111: E1121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014; 34: 11929–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data analysed is available at: Brain cell RNAseq browser (mouse): http://jiaqianwulab.org/braincell/RNASeq.html; GTEX (human): https://gtexportal.org/home/gene/HNRNPA1; exon expression for HNRNPA1 (ENSG00000135486.13) Data Source: GTEx Analysis Release V7 (dbGaP Accession phs000424.v7.p2).