Abstract
AIM
To investigate whether promoter methylation is responsible for the silencing of formin 2 (FMN2) in colorectal cancer (CRC) and to analyze the association between FMN2 methylation and CRC.
METHODS
We first identified the expression levels and methylation levels of FMN2 in large-scale human CRC expression datasets, including GEO and TCGA, and analyzed the relationship between the expression and methylation levels. Then, the methylation levels in four CpG regions adjacent to the FMN2 promoter were assessed by MethylTarget™ assays in CRC cells and in paired colorectal tumor samples and adjacent nontumor tissue samples. Furthermore, we inhibited DNA methylation in CRC cells with 5-Aza-2’-deoxycytidine and assessed the expression of FMN2 by qRT-PCR. Last, the association between FMN2 methylation patterns and clinical indicators was analyzed.
RESULTS
A statistically significant downregulation of FMN2 expression in large-scale human CRC expression datasets was found. Subsequent analysis showed that a high frequency of hypermethylation occurred in the FMN2 gene promoter in CRC tissues; operating characteristic curve analysis revealed that FMN2 gene methylation had a good capability for discriminating between CRC and nontumor tissue samples (AUC = 0.8432, P < 0.0001). MethylTarget™ assays showed that CRC cells and tissues displayed higher methylation of these CpG regions than nontumor tissue samples. Correlation analysis showed a strong inverse correlation between methylation and FMN2 expression, and the inhibition of DNA methylation with 5-Aza significantly increased endogenous FMN2 expression. Analysis of the association between FMN2 methylation patterns and clinical indicators showed that FMN2 methylation was significantly associated with age, N stage, lymphovascular invasion, and pathologic tumor stage. Notably, the highest methylation of FMN2 occurred in tissues from cases of early-stage CRC, including cases with no regional lymph node metastasis (N0), cases in stages I and II, and cases with no lymphovascular invasion, but the methylation level began to decrease with tumor progression. Additionally, FMN2 promoter hypermethylation was more common in patients > 60 years old and in colon cancer tissue.
CONCLUSION
FMN2 promoter hypermethylation may be an important early event in CRC, most likely playing a critical role in cancer initiation, and can serve as an ideal diagnostic biomarker in elderly patients with early-stage colon cancer.
Keywords: Formin 2, Colorectal cancer, Methylation, Methylation-associated silencing, Early-stage cancer
Core tip: Colorectal cancer (CRC) is the leading cause of cancer death in the world. We identified a statistically significant downregulation of formin 2 (FMN2) expression in large-scale human CRC expression datasets and our clinical samples. Then, we first showed that a high frequency of hypermethylation occurred in the FMN2 gene promoter, which is responsible for the downregulation of FMN2 expression. Additionally, the highest methylation of FMN2 occurred in tissues from cases of early-stage CRC and patients > 60 years old. FMN2 hypermethylation may be an important early event in CRC and can serve as an ideal diagnostic biomarker in elderly patients with early-stage CRC.
INTRODUCTION
Colorectal cancer (CRC) is a common malignancy and a major cause of incidence and mortality in many countries, especially more developed countries[1]. Over the years, researchers have identified CRC as a gradual process that lasts for several years, since it begins as a single mutation in a cell until it becomes a detectable malignancy[2]. Therefore, the main secondary preventive strategy for CRC is the early detection of preneoplastic or neoplastic lesions in the large bowel[3].
Formin 2 (FMN2) is a member of the formin homology protein family; the encoded protein is thought to have essential roles in the organization of the actin cytoskeleton and in cell polarity[4]. More recently, FMN2 was reported to be involved in cancer, for example, serving as a potential oncogene in leukemia[5]. FMN2 can enhance the expression of the cell cycle inhibitor p21 by preventing its degradation. In addition, FMN2 is induced by the activation of other oncogenes, hypoxia, and DNA damage[6]. To date, only two studies have explored the relationship between FMN2 and CRC. The first study was in 2012; to identify the genetic determinants of colon tumorigenesis, Liu et al[7] and colleagues carried out a genome-wide association study of azoxymethane-induced colon tumorigenesis and subsequently confirmed through fine mapping that FMN2 is associated with colon tumor susceptibility. Another study[8] assessed the expression of FMN2 in tumor and adjacent nontumor tissue by immunohistochemistry, with results showing that FMN2 was predominantly localized in the cytoplasm of tumor cells; that the rate of positive FMN2 protein expression in the CRC and paracarcinoma tissues was 53.73% (180/335) and 80.90% (271/335), respectively, with a significant difference (P < 0.05); and that the level of the FMN2 protein in CRC patients was associated with tumor differentiation, TNM stage, and lymph node metastasis. However, the mechanism leading to the downregulation of FMN2 expression in CRC has not been studied. In this article, we tried to show that the high frequency of hypermethylation occurring in the promoter of the FMN2 gene in CRC tissues is responsible for the silencing of FMN2, and we further explored the association between FMN2 methylation and clinical indicators. We found that the highest level of FMN2 methylation occurred in early-stage CRC tissues; thus, FMN2 may provide a new biomarker for the secondary prevention of CRC.
MATERIALS AND METHODS
Cell lines and tissue samples
Human CRC cell lines (SW620 and SW480) were cultured in L15 medium (KeyGEN BioTECH, Nanjing, China) supplemented with 10% fetal bovine serum (FBS) (Biological Industries, Israel), and HCT116 and HT-29 cells were cultured in McCoy‘s 5A medium (KeyGEN BioTECH, Nanjing, China) supplemented with 10% FBS. Cells were grown in a 5% CO2 cell culture incubator at 37 °C. Clinical samples were obtained from patients treated at the Third Xiangya Hospital of Central South University (Hunan, China) under informed consent and approval by the Ethics Committee of Central South University (more details about the clinical samples can be found in Table 1).
Table 1.
Clinical materials used in this article
| No. | Sex | Age | Lymph nodes | TNM | Pathology | Differentiation | MSI | Organ |
| 1 | M | 67 | 5 (14) | T4aN2aM0 | Adenocarcinoma | Moderate | No | Rectum |
| 2 | M | 53 | 0 (22) | T2N0M0 | Adenocarcinoma | Moderate | - | Rectum |
| 3 | M | 72 | 4 (20) | T4aN2aM0 | Adenocarcinoma | Poor | - | Right colon |
| 4 | M | 52 | 0 (16) | T3N0M0 | Adenocarcinoma | Moderate | - | Rectum |
| 5 | M | 52 | 3 (25) | T3N1bM0 | Adenocarcinoma | Moderate | - | Sigmoid colon |
| 6 | F | 57 | 0 (15) | T4aN0M0 | Adenocarcinoma | Moderate | - | Left colon |
| 7 | F | 51 | 0 (14) | T4aN0M0 | Adenocarcinoma | Moderate | No | Rectum |
| 8 | F | 66 | 19 (27) | T3N2bM0 | Adenocarcinoma | Poor | No | Rectum |
| 9 | F | 71 | 0 (16) | T4bN0M1b | Adenocarcinoma | Moderate | No | Rectum |
RNA purification and reverse transcription-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, United States). The reverse transcription reaction was performed using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, Japan), and this kit includes reagents for reverse transcription and for the removal of genomic DNA (DNase I treatment). cDNA was amplified with KOD SYBR® qPCR mix (TOYOBO, Japan) on a LightCycler® 480II system (Roche) according to the manufacturer’s instructions (FMN2 primers: forward, GCGAACGCTGTTGGAGAAG and reverse, CTGATTACACGGTTCCCTGAAG).
Treatment with 5-aza-2’-deoxycytidine
Cells were grown in appropriate culture conditions. For demethylation treatment, colorectal cells were treated with 5-aza-2’-deoxycytidine (5-Aza) (Sigma, United States) for 96 h (5 μmol/L), with daily replacement of the drug and medium. Untreated cells were used as a control group.
DNA extraction and bisulfate treatment
DNA was isolated with an Easypure Genomic DNA kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. The DNA concentration was assessed by spectrophotometry and confirmed by gel electrophoresis; DNA was stored at -20 °C. An EZ DNA Methylation Gold™ Kit (Zymo Research Corporation, CA, United States) was used to convert all unmethylated cytosine to uracils.
DNA methylation analysis
MethylTarget™ assays (targeted bisulfite sequencing) developed by Genesky BioTech (Shanghai, China) were carried out as previously described[9,10]. Briefly, CpG islands adjacent to the promoter region of the FMN2 gene were analyzed, and based on these CpG islands, four CpG regions from CpG islands in FMN2 were sequenced (the details, including the relative distance from the transcriptional start site, amplification primers, and product size, of these CpG regions can be found in Tables 2 and 3). Genomic DNA was converted with bisulfite, and PCR was performed to amplify the targeted DNA sequences. The products were sequenced on an Illumina MiSeq benchtop sequencer (Illumina, CA, United States).
Table 2.
Details of the four CpG regions in the CpG islands of formin 2
| Target | TSS | Start | End | Length | Target strand | Distance2TSS |
| FMN2_1 | 240255184 | 240255353 | 240255531 | 179 | + | 169 |
| FMN2_2 | 240255184 | 240255809 | 240256007 | 199 | + | 625 |
| FMN2_3 | 240255184 | 240256241 | 240256483 | 243 | + | 1057 |
| FMN2_4 | 240255184 | 240256757 | 240257017 | 261 | + | 1573 |
Start: The starting position of the product on the reference genome; TSS: The mRNA transcription initiation site; End: The end position of the product on the reference genome; Length: the product length; Target strand: The product orientation; Distance2TSS: The distance from the product to the TSS.
Table 3.
Primers used for the MethylTarget™ assays
| Primer name | Primer |
| FMN2_1_F | GAGGGTYGGGATGGTTTGAG |
| FMN2_1_R | CCCCCRCTCCCCTTCTTT |
| FMN2_2F | GAGTGTGYGGATTTTTTTGAGGT |
| FMN2_2R | AAATATCTAAAAACAAATCCTCTTACTCC |
| FMN2_3F | GATTTGTTYGAGAGTTTGGTTGT |
| FMN2_3R | AAACRCATCCTCAAAAACATCCT |
| FMN2_4F | TTTTTGAGTYGAGGGTTTAGAATTG |
| FMN2_4R | CACRTTCTAAAAACCATCCRCAAC |
Bioinformatics
Cosmic[11], a database cataloguing somatic mutations in human cancer, was used to assess methylation mutations in human cancer tissues. Datasets for CRC, including GEO GSE20842, GSE8671, and GSE4183, as well as TCGA[12], were used to assess the expression level of FMN2. Data about DNA methylation, the patients, and the samples were downloaded from the TCGA database (https://cancergenome.nih.gov/).
Statistical analysis
GraphPad Prism 7 software was used to analyze the data. The methylation and expression levels in normal, adenoma, and cancerous tissues, as well as the methylation level in different clinical materials, were assessed by two-tailed unpaired Student’s t-tests; box-whisker plots depict the means, 1st and 3rd quartiles, and minimum/maximum. The error bars in the figures represent SDs. Spearman’s correlation coefficient (r) was used to determine the correlation. Overall survival (OS) and disease-free survival (DFS) were estimated using the Kaplan-Meier method. Receiver operating characteristic curves were constructed based on the level of FMN2 methylation. The data were considered nonsignificant for P > 0.05.
RESULTS
Analysis of FMN2 expression in large-scale human CRC expression datasets
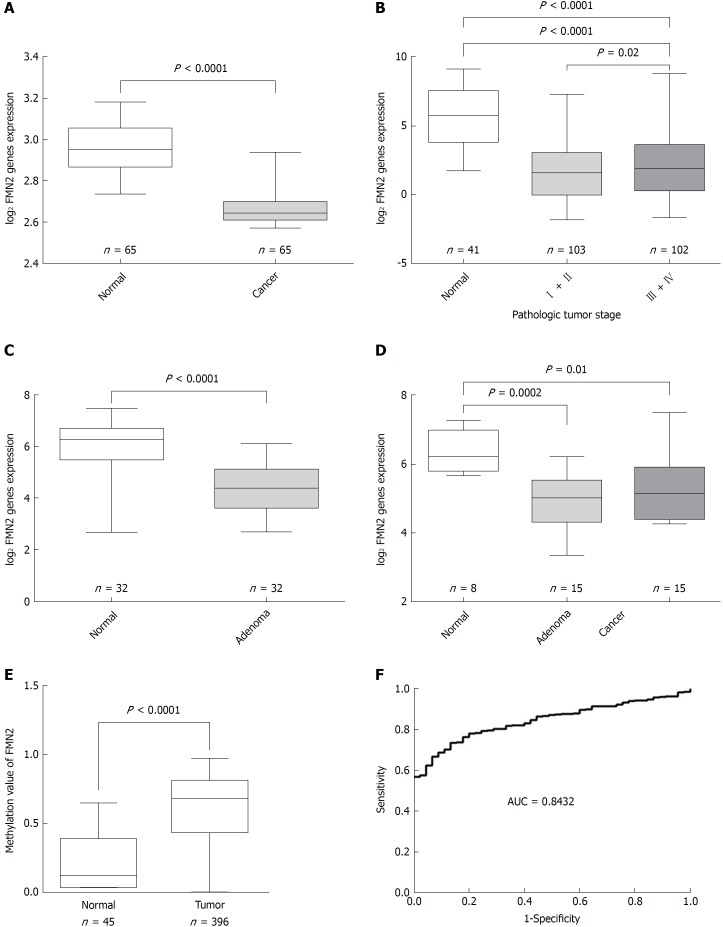
Previous studies demonstrated that FMN2 is underexpressed in CRC. To further confirm the expression characteristics of FMN2 in CRC, we expanded our analysis to published, large-scale human CRC expression datasets. GSE20842, which contains 65 paired samples of tumor and mucosal tissue samples from 65 CRC patients, was used to assess FMN2 expression; we found a statistically significant downregulation of FMN2 expression from normal colorectal tissue to carcinoma tissue (P < 0.0001) (Figure 1A). An identical pattern was observed in human CRC samples from TCGA (P < 0.0001); however, it is noteworthy that we found that the expression level of FMN2 was significantly lower in early-stage CRC tissues (stages I + II vs stages III + IV, P = 0.02) (Figure 1B), which indicates that FMN2 downregulation may be an important early event in CRC. To confirm this finding, GSE8671, which contains 32 colorectal adenoma and corresponding normal colonic mucosa samples, was downloaded to analyze FMN2 expression; we found a statistically significant downregulation of FMN2 expression from normal colorectal tissue to adenoma tissue (P < 0.0001) (Figure 1C). In addition, we downloaded the GSE4183 dataset, which contains colonic biopsies of 15 patients with CRC, 15 patients with adenoma, and 8 healthy normal controls, to simultaneously assess the reduction of FMN2 in adenomas and cancer tissues. As shown in Figure 1D, adenoma tissues displayed a greater reduction in expression (P = 0.0002) than tumor tissues (P = 0.01).
Figure 1.
Colorectal tissues show decreased formin 2 expression and a high frequency of hypermethylation in the formin 2 promoter. A: Normalized expression of formin 2 (FMN2) mRNA in carcinoma and adjacent normal tissues, presented as box-whisker plots (unpaired t-test, GEO: GSE20842); B: Normalized expression of FMN2 mRNA in tissue from different stages of carcinoma (according to the AJCC Cancer Staging Manual) and in normal tissue, presented as box-whisker plots (unpaired t-test, colorectal cancer samples from TCGA); C: Normalized expression of FMN2 mRNA in adenoma and corresponding normal colonic mucosal tissues, presented as box-whisker plots (unpaired t-test, GEO: GSE8671); D: Normalized expression of FMN2 mRNA in normal, adenoma, and carcinoma tissues, presented as box-whisker plots (unpaired t-test, GEO: GSE4183); E: The FMN2 gene shows an increased methylation level in colorectal cancer tissues compared with normal tissues (unpaired t-test, colorectal cancer samples from TCGA); F: Receiver operating characteristic curve analysis was used to assess the clinical diagnostic utility of FMN2 DNA methylation for the prediction of colorectal cancer.
High frequency of hypermethylation occurs in the FMN2 gene promoter in CRC tissues
Somatic mutation in cancer is an important reason for the aberrant expression of genes, so the Catalogue of Somatic Mutations in Cancer (COSMIC), the world’s largest and most comprehensive resource for exploring the impact of somatic mutations in human cancer, was used to analyze the somatic mutations of FMN2 in CRC tissues. The result indicated that methylation was the main somatic mutation and that FMN2 gene promoter hypermethylation occurred in 37.37% of CRC tissues. In addition, COSMIC indicated that FMN2 methylation mainly occurred in CRC tissues and was the most common somatic mutation among all human tumor tissues and that FMN2 was one of the top 20 genes with an extremely high frequency of hypermethylation in CRC. Then, FMN2 gene promoter DNA methylation profiles were downloaded from TCGA; the results revealed a statistically significant hypermethylation of FMN2 in tumor tissues compared with adjacent nontumor tissue samples (P < 0.0001) (Figure 1E). We next assessed the accuracy of the FMN2 methylation signature for the detection of CRC via receiver operating characteristic curve analysis, and the analysis revealed a good capability for discriminating between CRC and nonneoplastic tissue specimens (AUC = 0.8432, 95%CI: 0.8022-0.8841; P < 0.0001) (Figure 1F).
Analysis of the features of methylation-associated FMN2 silencing in CRC tissue and cells
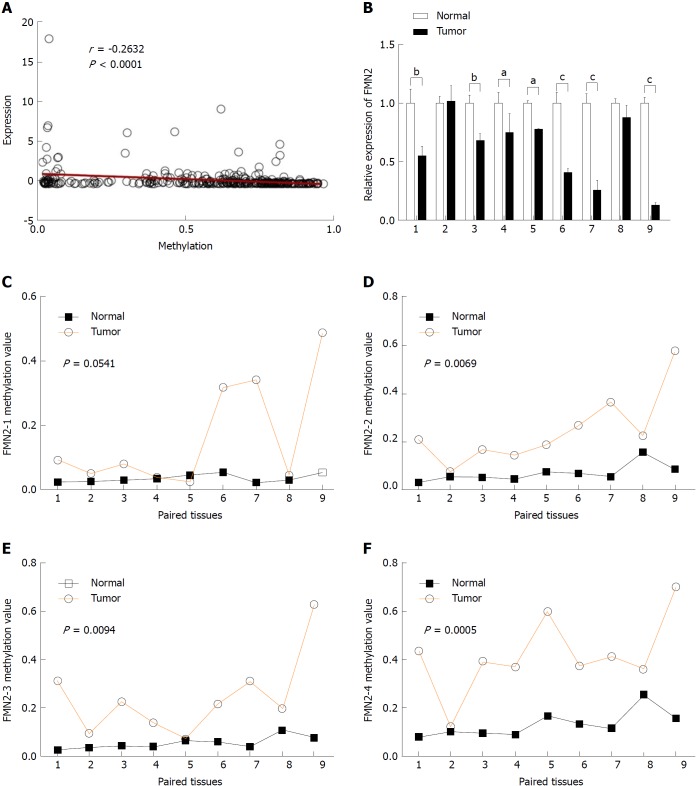
The majority of research indicates that promoter hypermethylation is a key mediator underlying the downregulation of gene expression. The high frequency of hypermethylation in the FMN2 gene promoter and the significant inverse correlation between methylation levels and the expression of FMN2 in 372 CRC tissues (Figure 2A) remind us that methylation is the main cause of FMN2 gene silencing. To investigate this topic, nine paired colorectal tumor samples and adjacent nontumor tissue samples (for details, see Table 1) were selected for RT-qPCR and MethylTarget™ assays. We first examined FMN2 expression using RT-qPCR in these paired samples. A significant reduction in FMN2 expression was observed in seven of the nine CRC tissues (Figure 2B). Then, based on the CpG islands adjacent to the FMN2 promoter region, four CpG regions (FMN2-1, FMN2-2, FMN2-3, and FMN2-4, the details of which can be found in Table 2) from CpG islands were amplified and sequenced. The result demonstrated that CRC tissues revealed a stronger methylation pattern than noncancerous tissues and identified three CpG regions, namely, FMN2-2, FMN2-3, and FMN2-4, with a statistically significant difference (P = 0.0069, P = 0.0094, and P = 0.0005, respectively) (Figure 2C-F; Table 2). Notably, tumor tissues (cases 1, 3-7, and 9) exhibiting lower FMN2 expression than the corresponding noncancerous tissues displayed an increase in methylation at these islands compared with the noncancerous tissues, whereas CRC cases 2 and 8, with moderate FMN2 expression, revealed a methylation pattern similar to that of the corresponding noncancerous tissues. Correlation analysis revealed a strong inverse correlation between the methylation of island 2 (r = -0.86), island 4 (r = -0.71), and island 3 (r = -0.78) and FMN2 expression (data not shown). In addition, we assessed the methylation level of each CG site in CpG island regions and found that 8 of 14 CG sites in FMN2-1 and all CG sites in FMN2-2 (13/13), FMN2-3 (23/23), and FMN2-4 (27/27) revealed a significantly stronger methylation pattern in the tumor tissues than in the corresponding noncancerous tissues (Table 4).
Figure 2.
Hypermethylation of formin 2 correlates with decreased expression of formin 2 during colorectal cancer. A: The expression of the formin 2 (FMN2) gene is significantly inversely correlated with the methylation level (Pearson correlation analysis); B: Quantitative real-time RT-PCR for FMN2 was carried out on nine colorectal cancer surgical specimens (filled bars) and paired noncancerous tissues (unfilled bars). The expression levels in the tumor samples were independently calculated relative to those in the nontumor samples, which are normalized to 1. Vertical bar, SD; aP < 0.05, bP < 0.03, cP < 0.01 (unpaired t-test); C-F: Results of MethylTarget™ assays on nine paired noncancerous tissues and colorectal cancer tissues. Four CpG regions (FMN2-1, FMN2-2, FMN2-3, and FMN2-4) from CpG islands adjacent to the FMN2 promoter were sequenced (unpaired t-test. More details about the four CpG regions can be found in Table 2).
Table 4.
Methylation level of each CG site in the CpG island regions
| Target | Position | Type | P value (t-test) | OR (L95-U95) (logistic) | C mean | N mean |
| FMN2_1 | 25 | CG | 0.007774578 | 2.1731 (0.9323-5.0653) | 0.17 | 0.04 |
| FMN2_1 | 27 | CG | 0.01061292 | 1.5266 (0.8980-2.5953) | 0.16 | 0.03 |
| FMN2_1 | 30 | CG | 0.1359111 | 1.3535 (0.7970-2.2986) | 0.15 | 0.03 |
| FMN2_1 | 32 | CG | 0.01875771 | 1.7416 (0.8662-3.5016) | 0.16 | 0.04 |
| FMN2_1 | 35 | CG | 0.03998355 | 1.2722 (0.8959-1.8068) | 0.18 | 0.05 |
| FMN2_1 | 38 | CG | 0.01419169 | 1.6993 (0.8462-3.4125) | 0.16 | 0.03 |
| FMN2_1 | 46 | CG | 0.003990128 | 2.2454 (0.9140-5.5161) | 0.15 | 0.02 |
| FMN2_1 | 90 | CG | 0.1134924 | 1.3103 (0.6401-2.6825) | 0.16 | 0.03 |
| FMN2_1 | 108 | CG | 0.06252571 | 1.2532 (0.7873-1.9948) | 0.17 | 0.04 |
| FMN2_1 | 114 | CG | 0.1134924 | 1.1858 (0.8641-1.6273) | 0.18 | 0.05 |
| FMN2_1 | 117 | CG | 0.05030852 | 1.3499 (0.8867-2.0550) | 0.19 | 0.04 |
| FMN2_1 | 123 | CG | 0.01061292 | 2.2247 (0.9162-5.4017) | 0.16 | 0.03 |
| FMN2_1 | 126 | CG | 0.09391197 | 1.2852 (0.7471-2.2110) | 0.16 | 0.03 |
| FMN2_1 | 134 | CG | 0.01875771 | 2.4645 (0.6195-9.8050) | 0.16 | 0.02 |
| FMN2_2 | 27 | CG | 0.000493624 | 1.3442 (1.0433-1.7321) | 0.28 | 0.07 |
| FMN2_2 | 51 | CG | 0.01419169 | 1.2762 (0.9670-1.6843) | 0.17 | 0.04 |
| FMN2_2 | 63 | CG | 0.007774578 | 1.4062 (0.9872-2.0030) | 0.37 | 0.25 |
| FMN2_2 | 70 | CG | 0.000781571 | 1.3753 (1.0378-1.8226) | 0.24 | 0.05 |
| FMN2_2 | 74 | CG | 0.000493624 | 1.3079 (1.0283-1.6636) | 0.31 | 0.07 |
| FMN2_2 | 78 | CG | 0.000287947 | 1.3676 (1.0603-1.7639) | 0.25 | 0.05 |
| FMN2_2 | 81 | CG | 0.00185109 | 1.3630 (0.9664-1.9224) | 0.21 | 0.05 |
| FMN2_2 | 107 | CG | 0.000493624 | 1.3025 (0.9942-1.7063) | 0.24 | 0.06 |
| FMN2_2 | 131 | CG | 0.007774578 | 1.2706 (0.9662-1.6708) | 0.15 | 0.03 |
| FMN2_2 | 137 | CG | 0.002756067 | 1.3002 (0.9986-1.6928) | 0.21 | 0.05 |
| FMN2_2 | 143 | CG | 0.00123406 | 1.3743 (1.0297-1.8341) | 0.20 | 0.04 |
| FMN2_2 | 164 | CG | 0.000287947 | 1.3527 (1.0108-1.8101) | 0.29 | 0.07 |
| FMN2_2 | 170 | CG | 0.00123406 | 1.2431 (1.0019-1.5424) | 0.28 | 0.08 |
| FMN2_3 | 24 | CG | 0.005635541 | 1.3854 (0.9994-1.9207) | 0.20 | 0.05 |
| FMN2_3 | 31 | CG | 0.005635541 | 1.4241 (0.9731-2.0841) | 0.23 | 0.04 |
| FMN2_3 | 47 | CG | 0.002756067 | 1.3776 (0.9695-1.9574) | 0.24 | 0.06 |
| FMN2_3 | 50 | CG | 0.003990128 | 1.3787 (1.0219-1.8599) | 0.22 | 0.05 |
| FMN2_3 | 57 | CG | 0.00123406 | 1.3592 (1.0212-1.8091) | 0.25 | 0.06 |
| FMN2_3 | 60 | CG | 0.003990128 | 1.5630 (0.9433-2.5897) | 0.19 | 0.04 |
| FMN2_3 | 68 | CG | 0.00185109 | 1.3474 (1.0273-1.7673) | 0.23 | 0.06 |
| FMN2_3 | 75 | CG | 0.00123406 | 1.3787 (0.9962-1.9082) | 0.26 | 0.06 |
| FMN2_3 | 80 | CG | 0.00123406 | 1.3582 (0.9518-1.9380) | 0.24 | 0.05 |
| FMN2_3 | 92 | CG | 0.00185109 | 1.3665 (0.9746-1.9159) | 0.23 | 0.05 |
| FMN2_3 | 98 | CG | 0.00123406 | 1.3200 (0.9414-1.8509) | 0.27 | 0.06 |
| FMN2_3 | 105 | CG | 0.000781571 | 1.4801 (0.9296-2.3566) | 0.22 | 0.04 |
| FMN2_3 | 107 | CG | 0.007774578 | 1.2402 (1.0141-1.5168) | 0.21 | 0.05 |
| FMN2_3 | 119 | CG | 0.00123406 | 1.4326 (0.9810-2.0921) | 0.19 | 0.04 |
| FMN2_3 | 131 | CG | 0.000493624 | 1.7790 (0.9589-3.3006) | 0.22 | 0.04 |
| FMN2_3 | 147 | CG | 0.007774578 | 1.3343 (0.9443-1.8856) | 0.20 | 0.04 |
| FMN2_3 | 153 | CG | 0.000493624 | 1.5405 (0.9677-2.4523) | 0.22 | 0.05 |
| FMN2_3 | 158 | CG | 8.23E-05 | 1.9417 (0.8981-4.1979) | 0.31 | 0.08 |
| FMN2_3 | 170 | CG | 4.11E-05 | 1714240.0000 (0.0000-Inf) | 0.32 | 0.08 |
| FMN2_3 | 174 | CG | 4.11E-05 | 11577516733.0000 (0.0000-Inf) | 0.34 | 0.08 |
| FMN2_3 | 183 | CG | 0.000287947 | 1.6836 (0.9224-3.0731) | 0.28 | 0.06 |
| FMN2_3 | 187 | CG | 0.000287947 | 1.4526 (0.9861-2.1399) | 0.27 | 0.06 |
| FMN2_3 | 203 | CG | 0.000781571 | 1.4980 (0.9815-2.2861) | 0.25 | 0.05 |
| FMN2_4 | 36 | CG | 0.003990128 | 1.1409 (1.0248-1.2702) | 0.41 | 0.14 |
| FMN2_4 | 42 | CG | 0.002756067 | 1.2068 (1.0353-1.4068) | 0.38 | 0.12 |
| FMN2_4 | 47 | CG | 0.000781571 | 1.2264 (1.0309-1.4589) | 0.37 | 0.10 |
| FMN2_4 | 61 | CG | 0.005386639 | 1.1552 (1.0206-1.3075) | 0.38 | 0.10 |
| FMN2_4 | 66 | CG | 0.0003108 | 243.9222 (0.0000-Inf) | 0.40 | 0.09 |
| FMN2_4 | 69 | CG | 8.23E-05 | 1.7294 (0.6520-4.5874) | 0.42 | 0.09 |
| FMN2_4 | 72 | CG | 0.003990128 | 1.2209 (1.0296-1.4477) | 0.4 | 0.13 |
| FMN2_4 | 80 | CG | 4.11E-05 | 2515571.0000 (0.0000-Inf) | 0.48 | 0.18 |
| FMN2_4 | 84 | CG | 0.00185109 | 1.2366 (1.0241-1.4931) | 0.39 | 0.12 |
| FMN2_4 | 94 | CG | 0.002756067 | 1.2121 (1.0363-1.4178) | 0.38 | 0.12 |
| FMN2_4 | 98 | CG | 0.003990128 | 1.1618 (1.0236-1.3186) | 0.36 | 0.11 |
| FMN2_4 | 110 | CG | 0.00185109 | 1.1712 (1.0253-1.3378) | 0.44 | 0.19 |
| FMN2_4 | 123 | CG | 0.000781571 | 1.2218 (1.0321-1.4463) | 0.40 | 0.10 |
| FMN2_4 | 127 | CG | 0.00185109 | 1.2220 (1.0324-1.4464) | 0.39 | 0.11 |
| FMN2_4 | 131 | CG | 8.23E-05 | 1.3179 (1.0259-1.6931) | 0.38 | 0.08 |
| FMN2_4 | 136 | CG | 0.003990128 | 1.1979 (1.0209-1.4054) | 0.34 | 0.09 |
| FMN2_4 | 150 | CG | 4.11E-05 | 1217702.0000 (0.0000-Inf) | 0.54 | 0.22 |
| FMN2_4 | 152 | CG | 0.003990128 | 1.1644 (1.0196-1.3297) | 0.39 | 0.12 |
| FMN2_4 | 166 | CG | 0.000164541 | 1.3446 (0.9896-1.8269) | 0.45 | 0.16 |
| FMN2_4 | 168 | CG | 0.001710676 | 1.2557 (1.0089-1.5628) | 0.44 | 0.16 |
| FMN2_4 | 176 | CG | 4.11E-05 | 26059.6300 (0.0000-Inf) | 0.52 | 0.21 |
| FMN2_4 | 186 | CG | 0.000781571 | 1.1735 (1.0224-1.3469) | 0.54 | 0.2 |
| FMN2_4 | 189 | CG | 0.003990128 | 1.1348 (1.0221-1.2600) | 0.49 | 0.18 |
| FMN2_4 | 213 | CG | 0.00185109 | 1.1572 (1.0241-1.3076) | 0.47 | 0.14 |
| FMN2_4 | 221 | CG | 0.002756067 | 1.1534 (1.0276-1.2947) | 0.45 | 0.13 |
| FMN2_4 | 234 | CG | 0.00185109 | 1.1981 (1.0181-1.4099) | 0.32 | 0.08 |
| FMN2_4 | 236 | CG | 0.000493624 | 1.2631 (1.0149-1.5720) | 0.36 | 0.08 |
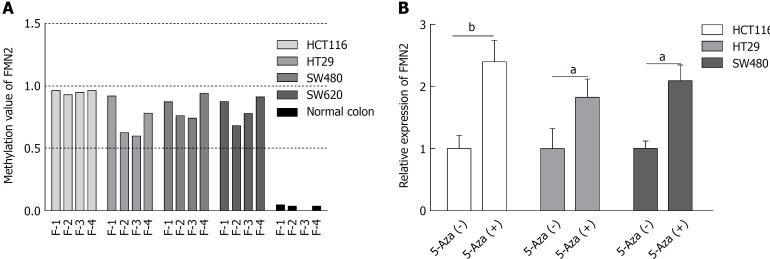
We next wondered whether promoter hypermethylation at these loci and FMN2 expression are causally related. To answer this question, four CRC cell lines (HCT116, HT29, SW480, and SW620) were selected for MethylTarget™ assays. The result revealed a high level of DNA methylation in all CRC cell lines, whereas only limited methylation was observed in normal colon tissue from a healthy individual (Figure 3A). Then, we treated HCT116, HT29, and SW480 cells with 5-Aza; the inhibition of DNA methylation by 5-Aza significantly increased the endogenous FMN2 expression (Figure 3B).
Figure 3.
Promoter hypermethylation and formin 2 expression are causally related. A: Results of MethylTarget™ assays on four colorectal cancer cell lines and normal colon cells from a healthy individual; B: The inhibition of DNA methylation with 5-Aza significantly increased the endogenous FMN2 expression (unpaired t-test).
FMN2 promoter hypermethylation is an early event in CRC
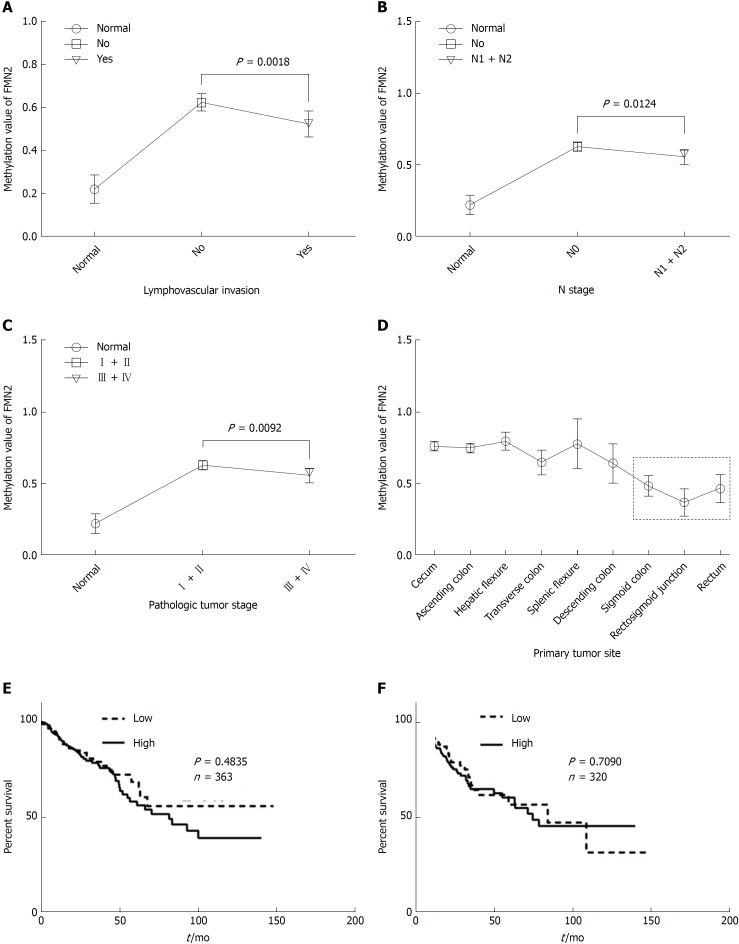
To analyze the clinicopathological significance of FMN2 promoter methylation, we divided the patients into a high-methylation group and a low-methylation group according to the median value of methylation. The association between clinicopathological factors and FMN2 methylation was analyzed (Table 5). High FMN2 methylation was associated with age (P = 0.0006), N stage (P = 0.0293), pathologic tumor stage (P = 0.0161), and lymphovascular invasion (P = 0.0180). Notably, the association analysis showed that the highest methylation level of FMN2 occurred in tissues from cases of early-stage CRC, including cases with no regional lymph node metastasis (N0), cases in stages I and II, and cases with no lymphovascular invasion; the methylation level began to decrease with tumor progression. To understand our conclusions more intuitively, we then performed unpaired t-tests based on the pathological data. An identical pattern is shown in Figure 4A-C; DNA methylation of the FMN2 gene increased from normal tissues to cancer tissues, reaching the highest level in early-stage cancer (N0, stages I and II, and no lymphovascular invasion) and decreasing stepwise in advanced-stage carcinomas. However, it is worth noting that the methylation level of FMN2 in advanced-stage carcinomas is still significantly higher than that in normal tissue (P < 0.0001). Additionally, we analyzed the methylation level of FMN2 according to the primary tumor site, and the result showed that there were statistically significant differences among the means (P < 0.0001) and that tumors located in the sigmoid colon and rectum had a relatively low FMN2 methylation level (Figure 4D). Furthermore, we found that FMN2 promoter hypermethylation was not associated with OS or DFS (Figure 4E and F). All these results indicated that FMN2 promoter hypermethylation mainly occurred in elderly and early-stage patients with colon cancer.
Table 5.
Relationship between formin 2 methylation levels and clinicopathological data
| Factor | No. |
FMN2 |
P value | |
| Low, n (%) | High, n (%) | |||
| Age (yr) | ||||
| < 60 | 136 | 66 (45.83) | 70 (28.69) | 0.00061 |
| ≥ 60 | 252 | 78 (54.17) | 174 (71.31) | |
| Gender | ||||
| Male | 210 | 72 (50.00) | 138 (56.56) | 0.2105 |
| Female | 178 | 72 (50.00) | 106 (43.44) | |
| Height (cm) | ||||
| < 170 | 138 | 52 (48.60) | 86 (47.25) | 0.8250 |
| ≥ 170 | 151 | 55 (51.40) | 96 (52.75) | |
| Weight (kg) | ||||
| < 80 | 156 | 56 (49.12) | 100 (51.55) | 0.6812 |
| ≥ 80 | 152 | 58 (50.88) | 94 (48.45) | |
| T | ||||
| T1 | 11 | 5 (3.47) | 6 (2.47) | 0.4830 |
| T2 | 54 | 16 (11.11) | 38 (15.71) | |
| T3 | 270 | 106 (73.61) | 164 (67.77) | |
| T4 | 51 | 17 (11.81) | 34 (14.05) | |
| N | ||||
| N0 | 212 | 69 (47.92) | 143 (59.34) | 0.02931 |
| N1 + N2 | 173 | 75 (52.08) | 98 (40.66) | |
| M | ||||
| M0 | 264 | 98 (80.33) | 166 (85.13) | 0.2651 |
| M1 | 53 | 24 (19.67) | 29 (14.87) | |
| Stage | ||||
| I + II | 197 | 62 (45.26) | 135 (58.19) | 0.01611 |
| III + IV | 172 | 75 (54.74) | 97 (41.81) | |
| Lymphovascular invasion | ||||
| Yes | 231 | 77 (60.63) | 154 (72.99) | 0.01801 |
| No | 107 | 50 (39.37) | 57 (27.01) | |
| Vascular invasion | ||||
| Yes | 78 | 31 (24.80) | 47 (22.71) | 0.6627 |
| No | 254 | 94 (75.20) | 160 (77.29) | |
| Perineural invasion | ||||
| Yes | 59 | 20 (24.39) | 39 (26.53) | 0.7225 |
| No | 170 | 62 (75.61) | 108 (73.47) | |
| Tumor status | ||||
| Tumor-free | 246 | 86 (71.07) | 160 (77.29) | 0.2094 |
| Tumor-present | 82 | 35 (28.93) | 47 (22.71) | |
| KRAS mutation | ||||
| Yes | 28 | 8 (42.11) | 20 (54.05) | 0.3972 |
| No | 28 | 11 (57.89) | 17 (45.95) | |
Indicates a statistically significant result. TNM stage was defined according to the AJCC Cancer Staging Manual. Tumor status: The state or condition of an individual's neoplasm at a particular point in time.
Figure 4.
Formin 2 promoter hypermethylation mainly occurs in early-stage colon cancer patients. A: Formin 2 (FMN2) promoter methylation level in colorectal cancer (CRC) tissues with (yes) and without (no) lymphovascular invasion, presented as column means with 95%CI, mean connected. (unpaired t-test; normal: n = 45, CRC tissues with and without lymphovascular invasion: n = 231 and n = 107, respectively); B: FMN2 promoter methylation level in CRC tissues with (N1 + N2) and without (N0) regional lymph node metastasis, presented as column means with 95%CI, mean connected (unpaired t-test, Normal: n = 45, CRC tissues with and without regional lymph node metastasis: n = 212 and n = 173, according to the AJCC Cancer Staging Manual); C: FMN2 promoter methylation level in CRC tissues from different stages, presented as column means with 95%CI, mean connected (unpaired t-test; Normal: n = 45, Stages I + II: n = 197, Stages III + IV: n = 172, according to the AJCC Cancer Staging Manual); D: FMN2 promoter methylation level in different organ sites, presented as column means with 95%CI, mean connected; E and F: FMN2 hypermethylation has no effect on the overall survival or disease-free survival of CRC patients.
DISCUSSION
In this article we first reported that a high frequency of hypermethylation occurred in the promoter of the FMN2 gene in CRC tissues and that hypermethylation was responsible for the silencing of FMN2, which suggests that the epigenetic silencing of FMN2 may be an important event in CRC. To the best of our knowledge, although FMN2 was one of the top 20 genes with an extremely high frequency of hypermethylation in CRC (COSMIC, https://cancer.sanger.ac.uk/cosmic), this study is the first and only to investigate the regulation of the FMN2 gene by methylation.
Previously, studies and investigations of publicly available datasets in FireBrowse (http://firebrowse.org/) have demonstrated differential FMN2 RNA expression in human tumors, depending on the tumor type. For example, FMN2 is overexpressed in approximately 95% of pre-B acute lymphoblastic leukemias[5] but is underexpressed in kidney renal clear cell carcinoma (FireBrowse); the mechanism underlying these phenomena has not been studied. In our research, we identified that FMN2 is underexpressed in CRC tissues and were the first to explore the underlying mechanism. Epigenetic modifications of DNA, such as DNA promoter hypermethylation, have critical roles in mediating gene expression in mammalian development and human disease[13]. Methylation-mediated silencing of some genes in CRC has been reported in previous studies; for example, Lgr5 methylation, by effecting Lgr5 expression and CRC stem cell differentiation, may serve as a novel prognostic marker in CRC patients[14]. SMYD3 promoter hypomethylation suppressed SMYD3 expression and was associated with the risk of CRC[15]. Based on the fact that a high frequency of hypermethylation occurred in the FMN2 gene promoter in CRC tissues, we found that both the downregulation of FMN2 expression and the high frequency of hypermethylation occurred at the earliest stages of carcinogenesis; furthermore, these parameters had a significant inverse correlation in both our study and in published, large-scale human CRC electronic datasets. We also carried out MethylTarget™ assays in paired CRC tissues and inhibited DNA methylation with 5-Aza in CRC cells, which identified that hypermethylation and FMN2 expression are causally related.
It is particularly important to determine which CpG islands adjacent to the promoter affect the expression of a gene, because this knowledge can provide directions for future research and potential targets for treatment. For example, Tavazoie SF[16] found that the Mest/miR-335 promoter contained three CpG islands upstream of the transcriptional start site; among these CpG islands, island 3 demonstrated a strong inverse correlation between methylation and miR-335 expression (r2 = -0.81). The methylation status of island 3 plays a key role in inhibiting miR-335 expression. An identical pattern was observed in our study; we found that although the methylation status of FMN2-2, -3 and -4 was significantly increased in the eight CRC tissues, the methylation of FMN2-1 was not significantly higher than that in the paired nontumor tissues. All these findings suggest that during CRC progression, FMN2 expression may be mainly mediated by the methylation of a specific CpG island region in the FMN2 promoter.
In addition, we found that the hypermethylation of FMN2 was significantly associated with age, N stage, pathologic tumor stage, and lymphovascular invasion. Notably, the association analysis showed that the highest methylation level of FMN2 occurred in tissues from cases of early-stage CRC, including cases with no regional lymph node metastasis (N0), cases in stages I and II, and cases with no lymphovascular invasion and that the methylation level began to decrease with tumor progression. Identical results have been reported in previous research. In endometrial tumorigenesis, Schneider-Stock[17] found that DNA methylation of the APC gene increased from atypical hyperplasia (23.5%) to endometrial carcinoma, reaching its highest level of 77.4% in early-stage cancer (FIGO stages I and II) and decreasing stepwise to 24.2% in advanced-stage carcinomas (FIGO stages III and IV). Epigenetic inactivation of the candidate tumor suppressor USP44 is a frequent and early event in colorectal neoplasia[18], and P16 methylation is an early event in cervical carcinogenesis[19]. All these studies, along with ours, have identified that DNA hypermethylation can be an early event in tumorigenesis. Hypermethylation most likely plays a critical role in cancer initiation, and creates an environment conducive to the overwhelming accumulation of simultaneous genetic and epigenetic mutations[20]. Additionally, we identified that patients younger than 60 years and patients with tumors located in the sigmoid colon and rectum exhibit relatively low FMN2 methylation levels, which indicates that the methylation-associated silencing of FMN2 may be involved in specific patients. In addition, Grady[21] identified that the patterns of DNA methylation in the colon vary by anatomical location, patient gender, and patient age.
In summary, we identified that the RNA expression of FMN2 is reduced in CRC tissues and were the first to reveal that the hypermethylation of specific CpG islands adjacent to the FMN2 promoter is the mechanism underlying FMN2 silencing. Although the FMN2 methylation is significantly stronger in tumor tissues than in paired nontumor tissues, the methylation level begins to decrease with tumor progression, which suggests that DNA hypermethylation is an early event in CRC tumorigenesis and can serve as a biomarker for the detection of CRC.
ARTICLE HIGHLIGHTS
Research background
Colorectal cancer (CRC) is a critical contributor to cancer mortality and morbidity, and some critical genes and pathways were important in the initiation and progression of CRC. Recent studies have shown that formin 2 (FMN2) may be down-regulated in CRC. Whether FMN2 is abnormally expressed in CRC and what causes its abnormal expression are unclear. Revealing the role of FMN2 in CRC may provide potential therapeutic targets.
Research motivation
Epigenetic modifications of DNA, especially promoter hypermethylation, have critical roles in mediating some gene expression in the initiation and progression of CRC. Whether abnormal DNA methylation changes can occur in the promoter of the FMN2 gene in CRC and whether such changes can be responsible for the silencing of FMN2 are poorly known. So the main topic in this article is to try to solve this question that epigenetic silencing of FMN2 may be an important event in CRC. All can provide the basis and direction for future research.
Research objectives
This study focuses on whether FMN2 is underexpressed in CRC, and whether methylation changes occur in CpG islands located in the promoter region of FMN2. If changes occur, what are the characteristics of fragments and CpG sites in these CpG islands? What is the core fragment of the change? Does methylation change silence the expression of FMN2, and is there a correlation between methylation changes in FMN2 and clinical indicators of CRC? The answers to these questions can better explain the role of FMN2 in CRC.
Research methods
Large-scale human CRC expression datasets, including GEO and TCGA, were used to assess the expression levels and methylation levels of FMN2 in CRC. Then, the mRNA levels of FMN2 in our own clinical samples was analyzed by real-time quantitative polymerase chain reaction, and the methylation levels in four CpG regions adjacent to the FMN2 promoter were assessed by MethylTarget™ assays in CRC cells and in paired colorectal tumor samples and adjacent nontumor tissue samples. Furthermore, we performed demethylation treatment in CRC cells with 5-Aza-2’-deoxycytidine and assessed the expression of FMN2 by qRT-PCR. Last, the association between FMN2 methylation patterns and clinical indicators was analyzed.
Research results
FMN2 is underexpressed in CRC, and the most obvious low expression occurs in early colon cancer tissues. Subsequent analysis coming from large-scale human CRC datasets showed that FMN2 was one of the top 20 genes with an extremely high frequency of hypermethylation in CRC, methylation was the main somatic mutation, and FMN2 gene promoter hypermethylation occurred in 37.37% of CRC tissues. Our own experiments confirmed that CRC cells and tissues displayed higher methylation levels in CpG regions of FMN2 than nontumor tissue samples. Correlation analysis showed a strong inverse correlation between methylation and FMN2 expression. Treatment of CRC cells with demethylation reagent, 5-Aza-2’-deoxycytidine, can significantly increase endogenous FMN2 expression. The highest methylation of FMN2 occurred in tissues from cases of early-stage CRC, including cases with no regional lymph node metastasis (N0), cases in stages I and II, and cases with no lymphovascular invasion. Additionally, FMN2 promoter hypermethylation was more common in patients > 60 years and in colon cancer tissue.
Research conclusions
In this article we confirmed the low expression of FMN2 in CRC and first reported that a high frequency of hypermethylation occurred in the promoter of the FMN2 gene in CRC tissues and that hypermethylation was responsible for the silencing of FMN2. It is worth noting that we found that the low expression and hypermethylation of FMN2 occurred more prominently in early colon cancer tissues, which suggests that DNA hypermethylation leading to the low expression of the FMN2 gene may be an important early event in CRC, most likely playing a critical role in cancer initiation, and can serve as an ideal diagnostic biomarker in elderly patients with early-stage colon cancer.
Research perspectives
In the era of big data, the rational use of tumor databases such as TCGA, GEO, and COSMIC can provide us with valuable information. The emergence of some new technologies can provide more accurate data for our experiments. For example, MethylTarget™ assays developed by Genesky BioTech used in this article can be used very well for methylation analysis. Involvement of the methylation-associated silencing of FMN2 in colorectal carcinogenesis may be a valuable research direction. Future research can select more clinical specimens to explore whether it can be used as a good clinical diagnostic index, and can also develop demethylation treatment. The article on the relationship between FMN2 and CRC is rare, so further study investigating whether FMN2 participates in the process of colorectal carcinogenesis and its underlying mechanisms are very important.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: This study was reviewed and approved by the Ethics Committee of Central South University. Informed consent was obtained from all individual participants included in the study.
Conflict-of-interest statement: To the best of our knowledge, no conflict of interest exists.
Data sharing statement: No additional data are available.
Peer-review started: August 20, 2018
First decision: October 11, 2018
Article in press: November 7, 2018
P- Reviewer: Bordonaro M, Leon J, Noda H S- Editor: Wang XJ L- Editor: Wang TQ E- Editor: Yin SY
Contributor Information
Dao-Jiang Li, Department of Gastrointestinal Surgery, the Third Xiangya Hospital, Central South University, Changsha 410013, Hunan Province, China.
Zhi-Cai Feng, Department of Burns and Plastic Surgery, the Third Xiangya Hospital, Central South University, Changsha 410013, Hunan Province, China.
Xiao-Rong Li, Department of Gastrointestinal Surgery, the Third Xiangya Hospital, Central South University, Changsha 410013, Hunan Province, China.
Gui Hu, Department of Gastrointestinal Surgery, the Third Xiangya Hospital, Central South University, Changsha 410013, Hunan Province, China. xy3zzz@csu.edu.cn..
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Zeuner A, Todaro M, Stassi G, De Maria R. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692–705. doi: 10.1016/j.stem.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Roncucci L, Mariani F. Prevention of colorectal cancer: How many tools do we have in our basket? Eur J Intern Med. 2015;26:752–756. doi: 10.1016/j.ejim.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Faix J, Grosse R. Staying in shape with formins. Dev Cell. 2006;10:693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Charfi C, Voisin V, Levros LC Jr, Edouard E, Rassart E. Gene profiling of Graffi murine leukemia virus-induced lymphoid leukemias: identification of leukemia markers and Fmn2 as a potential oncogene. Blood. 2011;117:1899–1910. doi: 10.1182/blood-2010-10-311001. [DOI] [PubMed] [Google Scholar]
- 6.Yamada K, Ono M, Perkins ND, Rocha S, Lamond AI. Identification and functional characterization of FMN2, a regulator of the cyclin-dependent kinase inhibitor p21. Mol Cell. 2013;49:922–933. doi: 10.1016/j.molcel.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu P, Lu Y, Liu H, Wen W, Jia D, Wang Y, You M. Genome-wide association and fine mapping of genetic loci predisposing to colon carcinogenesis in mice. Mol Cancer Res. 2012;10:66–74. doi: 10.1158/1541-7786.MCR-10-0540. [DOI] [PubMed] [Google Scholar]
- 8.Chen Wenping, Shi Hai, Zhou Yi, Li Xiaohua, Chu Dake, Liu Zhaoxu. Expression and significance of Formin2 protein in colorectal cancer. Chinese J of Dig Surge. 2015:335–338. [Google Scholar]
- 9.Zhou S, Zhang Y, Wang L, Zhang Z, Cai B, Liu K, Zhang H, Dai M, Sun L, Xu X, et al. CDKN2B methylation is associated with carotid artery calcification in ischemic stroke patients. J Transl Med. 2016;14:333. doi: 10.1186/s12967-016-1093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pu W, Wang C, Chen S, Zhao D, Zhou Y, Ma Y, Wang Y, Li C, Huang Z, Jin L, et al. Targeted bisulfite sequencing identified a panel of DNA methylation-based biomarkers for esophageal squamous cell carcinoma (ESCC) Clin Epigenetics. 2017;9:129. doi: 10.1186/s13148-017-0430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 14.Su S, Hong F, Liang Y, Zhou J, Liang Y, Chen K, Wang X, Wang Z, Wang Z, Chang C, et al. Lgr5 Methylation in Cancer Stem Cell Differentiation and Prognosis-Prediction in Colorectal Cancer. PLoS One. 2015;10:e0143513. doi: 10.1371/journal.pone.0143513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B, Pan R, Zhou C, Dai J, Mao Y, Chen M, Huang T, Ying X, Hu H, Zhao J, et al. SMYD3 promoter hypomethylation is associated with the risk of colorectal cancer. Future Oncol. 2018;14:1825–1834. doi: 10.2217/fon-2017-0682. [DOI] [PubMed] [Google Scholar]
- 16.Png KJ, Yoshida M, Zhang XH, Shu W, Lee H, Rimner A, Chan TA, Comen E, Andrade VP, Kim SW, et al. MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev. 2011;25:226–231. doi: 10.1101/gad.1974211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ignatov A, Bischoff J, Ignatov T, Schwarzenau C, Krebs T, Kuester D, Costa SD, Roessner A, Semczuk A, Schneider-Stock R. APC promoter hypermethylation is an early event in endometrial tumorigenesis. Cancer Sci. 2010;101:321–327. doi: 10.1111/j.1349-7006.2009.01397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sloane MA, Wong JW, Perera D, Nunez AC, Pimanda JE, Hawkins NJ, Sieber OM, Bourke MJ, Hesson LB, Ward RL. Epigenetic inactivation of the candidate tumor suppressor USP44 is a frequent and early event in colorectal neoplasia. Epigenetics. 2014;9:1092–1100. doi: 10.4161/epi.29222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang LW, Pan HS, Lin YH, Seow KM, Chen HJ, Hwang JL. P16 methylation is an early event in cervical carcinogenesis. Int J Gynecol Cancer. 2011;21:452–456. doi: 10.1097/IGC.0b013e31821091ea. [DOI] [PubMed] [Google Scholar]
- 20.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 21.Kaz AM, Wong CJ, Dzieciatkowski S, Luo Y, Schoen RE, Grady WM. Patterns of DNA methylation in the normal colon vary by anatomical location, gender, and age. Epigenetics. 2014;9:492–502. doi: 10.4161/epi.27650. [DOI] [PMC free article] [PubMed] [Google Scholar]