Abstract
Background: Breast cancer (BrC) is the most frequent neoplasm in women. New biomarkers, including aberrant DNA methylation, may improve BrC management. Herein, we evaluated the detection and prognostic performance of seven genes’ promoter methylation (APC, BRCA1, CCND2, FOXA1, PSAT1, RASSF1A and SCGB3A1). Methods: Methylation levels were assessed in primary BrC tissues by quantitative methylation-specific polymerase chain reaction (QMSP) and in circulating cell-free DNA (ccfDNA) by multiplex QMSP from two independent cohorts of patients (Cohort #1, n = 137; and Cohort #2, n = 44). Receiver operating characteristic (ROC) curves were constructed, and log-rank test and Cox regression were performed to assess the prognostic value of genes’ methylation levels. Results: The gene-panel APC, FOXA1, RASSF1A, SCGB3A1 discriminated normal from cancerous tissue with high accuracy (95.55%). In multivariable analysis, high PSAT1-methylation levels [>percentile 75 (P75)] associated with longer disease-free survival, whereas higher FOXA1-methylation levels (>P75) associated with shorter disease-specific survival. The best performing panel in ccfDNA (APC, FOXA1 and RASSF1A) disclosed a sensitivity, specificity and accuracy over 70%. Conclusions: This approach enables BrC accurate diagnosis and prognostic stratification in tissue samples, and allows for early detection in liquid biopsies, thus suggesting a putative value for patient management.
Keywords: breast cancer, DNA methylation, epigenetic biomarker, Cell-free DNA, liquid biopsy, diagnosis, prognosis
1. Introduction
Breast cancer (BrC) is the most common and lethal cancer in women worldwide, corresponding to 25% of all cancers in females [1]. Implementation of mammography-based BrC screening increase the proportion of cancers detected at an early-stage, contributing to a decrease in BrC-related mortality [2]. Nevertheless, this screening strategy is hampered by frequent false positive results, leading to overdiagnosis. Furthermore, its usefulness in women with dense breast tissue remains controversial [3,4]. Although grade, stage, histological, and molecular subtype are currently used to risk-stratify BrC patients, divergent outcomes and therapeutic responses are common [5]. Furthermore, currently used prognostic and predictive biomarkers, such as hormone receptor or Erb-b2 receptor tyrosine kinase 2 (ERBB2) status have a limited power to predict recurrence and therapeutic response [6]. Hence, despite all improvements in early detection, patients’ stratification and treatment, BrC remains the foremost cause of cancer-related mortality among women, mostly due to disease recurrence and/or metastasis development [1]. In recent years, several biomarkers for early diagnosis have been proposed. Yet, despite their less invasive nature [7,8,9], improved tumor characterization [10,11,12,13] or better patient stratification [9,14] have been proposed, but with limited success.
Because aberrant DNA methylation is considered a cancer-associated event, characterization of tumor-specific methylome has become the focus of multiple studies [15]. Interestingly, aberrant promoter methylation of several tumor suppressor genes was found in BrC precursor lesions, indicating that DNA methylation is an early event in breast carcinogenesis [16,17,18,19]. Moreover, DNA methylation has been proposed as a valuable cancer detection and prognosis biomarker owing to its link with tissue-specific gene silencing [9,20,21,22,23]. Tumor-specific DNA methylation may also be detected in circulating cell-free DNA (ccfDNA) from liquid biopsies [24], and its potential for early cancer detection was already reported [23,25,26,27], representing a minimal-invasive test [28]. Herein, we aimed to define a DNA methylation-based test to improve or complement early detection strategies and to enable better BrC patients’ prognostic stratification. Thus, methylation levels of seven gene promoters [Adenomatosis polyposis coli (APC), BRCA1, DNA repair associated (BRCA1), Cyclin D2 (CCND2), Fork-head box A1 (FOXA1), Phosphoserine Aminotransferase 1 (PSAT1), Ras association domain family 1 isoform A (RASSF1A) and Secretoglobin family 3A member 1 (SCGB3A1)] previously reported as dysregulated in BrC and conveying diagnostic and/or prognostic information [7,8,9,14,29] were firstly assessed in tissue for confirmation of cancer-specificity and prognostic significance. Then, the best performing gene panel was tested in plasma ccfDNA to determine its BrC detection performance.
2. Experimental Section
2.1. Patients and Samples Collection
Two independent cohorts of BrC patients were included in this study. Cohort #1 comprised 137 patients, primarily submitted to surgery, from 1996 to 2001, at the Portuguese Oncology Institute of Porto (IPO Porto), with frozen tissue available. For control purposes, normal breast tissue (NBr) was collected from reduction mammoplasty of contralateral breast of BrC in patients without BrC hereditary syndrome. After surgical resection and examination, samples were immediately frozen at −80 °C. Relevant clinical and pathological data was retrieved from the patients’ clinical charts. Five μm frozen sections were cut and stained by hematoxilin-eosin for histological evaluation by an experienced pathologist.
Cohort #2 was composed of 44 BrC patients, primarily diagnosed between 2015 and 2017 at IPO Porto, which voluntarily provided blood samples prior any treatment. For control purposes, blood samples were also obtained from 39 asymptomatic controls (AC). The blood samples were collected in two EDTA tubes and centrifuged at 2000 rpm for 10 min at 4 °C for plasma separation. Plasma was immediately frozen at −80 °C until further use. Relevant clinical data was collected from clinical records.
This study was approved by the institutional review board (Comissão de Ética para a Saúde—CES 120/2015) of IPO Porto, Portugal. All patients and controls enrolled had signed an informed consent.
2.2. Immunohistochemistry
Immunohistochemistry (IHC) allowed for identification of BrC molecular subtype of each case in Cohort #1, using corresponding formalin-fixed paraffin-embedded tissue. Commercially available antibodies for Estrogen Receptor (ER) (Clone 6F11, mouse, Leica, Newcastle, UK), Progesterone Receptor (PR) (Clone 16, mouse, Leica, Newcastle, UK), ERBB2 (Clone 4B5, rabbit, Roche, Tucson, AZ, USA) and Ki67 (Clone MIB-1, mouse, Dako, Glostrup, Denmark) were used. IHC was carried out in BenchMark ULTRA (Ventana, Roche, Tucson, AZ, USA) using ultraView Universal DAB Detection Kit (Ventana, Roche, Tucson, AZ, USA) according to manufacturer’s instructions.
IHC staining was evaluated by an experienced pathologist according to College of American Pathologists’ recommendations. Each case was categorized according to European Society for Medical Oncology (ESMO) guidelines [6]. Cutoff values were set for Ki67 (high proliferative rate if ≥15% positive cells) and PR (high expression if ≥25% positive cells).
2.3. DNA Extraction
Genomic DNA was extracted from tumor and normal tissues by the phenol–chlorophorm method at pH 8, as previously described [30]. Samples were first submitted to overnight digestion in a bath at 55°C, using buffer solution SE (75 mM NaCl; 25 mM EDTA), SDS 10% and proteinase K, 20 mg/mL (Sigma-Aldrich®, Schnelldorf, Germany). After digestion, extraction was performed with phenol/chloroform (Sigma-Aldrich®, Schnelldorf, Germany, Merck, Darmstadt, Germany) followed by precipitation with 100% ethanol.
CcfDNA was extracted from 2 mL of plasma using QIAamp MinElute ccfDNA (Qiagen, Hilden, Germany), according to manufacturers’ recommendations. The ccfDNA was eluted in 20 µL of ultra-clean water (Qiagen, Hilden, Germany).
2.4. Bisulfite Treatment and Whole Genome Amplification (WGA)
Bisulfite conversion was performed using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA), according to manufacturer’s instructions. One µg of DNA obtained from fresh frozen sections was used. Modified DNA was eluted with 60 μL of sterile distilled water. In plasma samples, 20 μL of ccfDNA was used for bisulfite modification. Modified ccfDNA was eluted in 10 μL of sterile distilled water. For control purposes, 1 μg of CpGenome™ Universal Methylated DNA (Millipore, Temecula, CA, USA) was also modified, according to the method described above and eluted in 20 μL of M-elution buffer. All samples were stored at −80 °C until further use. Whole-genome amplification of sodium bisulfite modified ccfDNA was carried out using the EpiTect Whole Bisulfitome Kit (Qiagen, Hilden, Germany) according to manufacturer’s recommendations. The amplified ccfDNA final volume was 65 μL.
2.5. Quantitative Methylation-Specific Polymerase Chain Reaction (QMSP)
Modified DNA was used as template for QMSP. Overall, seven gene promoters (APC, BRCA1, CCND2, FOXA1, PSAT1, RASSF1A and SCGB3A1) were assessed in BrC tissue samples. The primers used specifically amplify methylated bisulfite converted complementary sequences and are listed in Supplementary File 1 (Table S1). β-actin (ACTβ) was used as reference gene to normalize for DNA input in each sample [9]. Reactions were performed in 96-well plates using Applied Biosystems 7500 RealTime System (Applied Biosystems, Perkin Elmer, CA, USA) using 2 μL of modified DNA and 5 μL of 2X KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems, Wilmington, MA, USA). All the samples were run in triplicates and the melting curves were obtained for each case/gene. Owing to the limited amount of ccfDNA plasma samples, three gene promoters were selected (APC, FOXA1, RASSF1A) in addition to the reference gene (ACTβ) for assessment of methylation using multiplex QMSP with TaqMan probes having different fluorescent reporters and Xpert Fast Probe (GRISP, Porto, Portugal), whereas SCGB3A1 methylation levels were assessed in a separated QMSP reaction (Supplementary File 1—Table S2).
Modified CpGenome™ Universal Methylated DNA was used in each plate to generate a standard curve, allowing for quantification, as well as to ascertain polymerase chain reaction (PCR) efficiency. All plates disclosed efficiency values between 90–100%. For each gene, relative methylation levels in each sample were determined by the ratio of the mean quantity obtained by QMSP analysis for each gene and the respective value of the internal reference gene (ACTβ), multiplied by 1000 for easier tabulation (methylation level = target gene/reference gene × 1000).
2.6. Statistical Analysis
The frequency, median and interquartile range of promoter methylation levels of normal tissue/control samples and plasma samples were determined. Non-parametric tests were performed to determine statistical significance in all the comparisons made. Specifically, Kruskall-Wallis test was used for comparisons between three or more groups, whereas Mann-Whitney U test was used for comparisons between two groups.
For each gene, receiving operating characteristic (ROC) curves were built to assess respective performance as tumor biomarker. Moreover, specificity, sensitivity, positive predictive value (PPV), negative predictive value (NPV), accuracy were determined. For this purpose, the cut-off established was the highest value obtained by the ROC curve analysis [sensitivity + (1 − specificity)]. To categorize samples as methylated or unmethylated, a cut-off value was chosen based on Youden’s J index obtained by the ROC curve analysis for each gene [31,32]. For combination of markers, the cases were considered positive if at least one of the individual markers was positive. Logistic regression models were built in order to evaluate the potential of confounding factors as age in our BrC detection model.
Spearman nonparametric correlation test was used to assess the association of methylation levels and age. Disease-specific survival curves and disease-free survival curves (Kaplan–Meier with log rank test) were computed for standard clinicopathological variables and for categorized methylation status. A Cox-regression model comprising all significant variables (multivariable model) was computed to assess the relative contribution of each variable to the follow-up status.
Two-tailed p-values were derived from statistical tests, using a computer assisted program (SPSS Version 20.0, Chicago, IL, USA), and results were considered statistically significant at p < 0.05, with Bonferroni’s correction for multiple tests, when applicable. Graphics were assembled using GraphPad 6 Prism (GraphPad Software, La Jolla, CA, USA).
3. Results
3.1. Clinical and Pathological Data of Tissue Cohort
Relevant clinical and pathological data are presented in Supplementary File 1 (Tables S3 and S4). Although patients’ age did not differ between the two cohorts, a significant difference was observed between BrC patients and controls age (Cohort #1: p = 0.007, Cohort #2: p = 0.001).
3.2. Assessment of BrC and NBr Tissue Samples Methylation Levels
To assess cancer-specificity, promoters’ methylation levels of APC, BRCA1, CCND2, FOXA1, PSAT1, RASSF1A and SCGB3A1 were evaluated in Cohort #1 (BrC and NBr tissue samples). Overall, BrC samples displayed higher APC, CCND2, FOXA1, PSAT1, RASSF1A, and SCGB3A1 methylation levels than NrB samples (p < 0.001 for all genes, Supplementary File 1—Table S5), whereas no differences were found for BRCA1, which was not further tested (Supplementary File 1—Table S5).
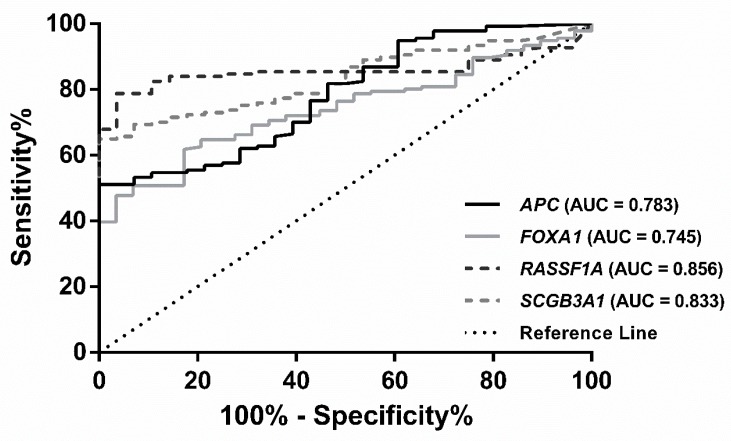
Subsequently, ROC curve analysis was performed, and an empirical cutoff value was determined for each gene (APC: 16.99, CCND2: 0.4171 for, FOXA1: 21.57, PSAT1: 48.05, RASSF1A: 114.5 and SCGB3A1: 67.18). All genes displayed an Area under the Curve (AUC) superior to 0.70 (Supplementary File 1—Table S6). APC and SCGB3A1 disclosed 100% specificity for cancer detection, whereas PSAT1 showed the highest sensitivity (91.97%). RASSF1A demonstrated the best individual performance, with 78.83% sensitivity and 96.43% specificity (Table 1). Several gene combinations were tested, and the best detection performance was achieved for the panel comprising APC, FOXA1, RASSF1A and SCGB3A1, displaying 97.81% sensitivity, 78.57% specificity and 94.50% accuracy (Table 1, Figure 1). Due to age’s difference between patients and controls, a multivariable model was constructed using logistic regression with the most informative genes and age. In this model, age did not show a significant impact in BrC detection (p = 0.2227). Moreover, biomarker performance was carried out restricted to BrC patient’s with similar age to controls (p = 0.136, Mann-Whitney for age). Similar results were obtained in biomarker performance (Supplementary File 1—Table S7).
Table 1.
Performance of promoter gene methylation as biomarkers for detection of breast cancer (Brc) in tissue samples.
| Genes | Sensitivity % | Specificity % | PPV a % | NPV b % | Accuracy % |
|---|---|---|---|---|---|
| APC | 51.09 | 100.0 | 100.0 | 29.47 | 59.39 |
| CCND2 | 72.26 | 92.86 | 98.02 | 40.63 | 75.76 |
| FOXA1 | 62.04 | 82.14 | 94.44 | 30.67 | 65.45 |
| PSAT1 | 91.24 | 50.00 | 89.93 | 53.85 | 84.24 |
| RASSF1A | 78.83 | 96.43 | 99.08 | 48.21 | 81.82 |
| SCGB3A1 | 64.96 | 100.0 | 100.0 | 36.84 | 70.91 |
|
APC/FOXA1
RASSF1A/SCGB3A1 |
97.81 | 78.57 | 95.71 | 88.00 | 94.55 |
a PPV—Positive Predictive Value; b NPV—Negative Predictive Value.
Figure 1.
Receiver operating characteristic (ROC) curve of the four-gene panel (APC, FOXA1, RASSF1A and SCGB3A1) in Breast Cancer tissues from Cohort #1.
3.3. Association between Promoter Methylation Levels, Molecular Subtypes and Standard Clinicopathological Parameters in Cohort #1
No significant differences in promoter methylation levels were apparent according to molecular subtype (Supplementary File 1—Figure S1), tumor grade, pathological stage or ERBB2 status in tissue samples. Nevertheless, in BrC patients, but not in controls, a significant correlation was found between CCND2 and RASSF1A methylation levels and age (R = 0.194, p = 0.023 and R = 0.223, p = 0.009, respectively). Additionally, a significant association was found between histological subtype and APC and SCGB3A1 methylation levels: special subtype carcinomas disclosed the lowest SCGB3A1 methylation levels in comparison to all the other histological subtypes (p = 0.016) and lower APC methylation levels comparing with invasive lobular carcinomas (p = 0.0293).
Additionally, FOXA1 and RASSF1A methylation levels associated with hormone receptor status. ER+ and PR+ BrC displayed significantly lower FOXA1 methylation levels than ER− and PR− BrC (p = 0.0084) or ER+ BrC (p = 0.0319). Contrarily, ER+ and PR+ BrC showed higher RASSF1A methylation levels than ER+ BrC (p = 0.0017). No statistical differences were observed for the remainder genes and for Cohort #2.
3.4. Survival Analysis
Survival analysis was only carried out for Cohort #1, due to the short-time of follow-up of Cohort #2. In the former Cohort (#1), 10 years of follow-up was considered for analysis. During this period, 37 patients (27.0%) had deceased, 24 of which were due to BrC (17.5% of all cases). At the time of the last follow-up, eight patients (5.8%) were alive with cancer and 92 patients (67.2%) were alive with no evidence of cancer.
Clinicopathological features were grouped according to: Grade (G1 & G2 vs. G3), pathological tumor size and extension (pT) stage (pT1, pT2 and pT3 & pT4), pathological regional lymph node status (pN) stage (N0 & N1 vs. N2 & N3) and stage (I, II and III & IV).
Higher tumor grade and pN stage and low PSAT1 methylation levels categorized by P75significantly associated with worse disease-free survival (DFS) in Cox regression univariable analysis (Table 2). Nonetheless, in multivariable analysis, however, only PSAT1 methylation levels and pN stage remained independent predictors of DFS (Table 2).
Table 2.
Cox regression models assessing the potential of clinical and epigenetic variables in the prediction of disease-free survival for 122 patients with BrC and disease-specific survival for 127 patients with BrC.
| Disease-Free Survival | Variable | HR a | CI b (95%) | p Value |
|---|---|---|---|---|
| Univariable | Grade | |||
| G1 | 1 | |||
| G2 c & G3 | 2.054 | 1.029–4.098 | 0.041 | |
| pN Stage | ||||
| N0 d & N1 | 1 | |||
| N2 & N3 | 3.894 | 1.940–7.812 | <0.001 | |
| PSAT1 | ||||
| >P75 e | 1 | |||
| ≤P75 | 3.707 | 1.133–12.127 | 0.030 | |
| Multivariable | Grade | |||
| G1 | 1 | |||
| G2 & G3 | 1.490 | 0.717–3.096 | 0.286 | |
| pN Stage | ||||
| N0 & N1 | 1 | |||
| N2 & N3 | 4.345 | 2.114–8.930 | <0.001 | |
| PSAT1 | ||||
| >P75 e | 1 | |||
| ≤P75 | 3.613 | 1.077–12.123 | 0.038 | |
| Disease-Specific Survival | Variable | HR a | CI b (95%) | p Value |
| Univariable | Grade | |||
| G1 | 1 | |||
| G2 & G3 | 2.725 | 1.155–6.428 | 0.022 | |
| pN Stage | ||||
| N0 & N1 | 1 | |||
| N2 & N3 | 4.061 | 1.814–9.089 | 0.001 | |
| FOXA1 | ||||
| ≤P75 f | 1 | |||
| >P75 | 2.678 | 1.200–5.978 | 0.016 | |
| Multivariable | Grade | |||
| G1 | 1 | |||
| G2 & G3 | 2.005 | 0.082–4.866 | 0.124 | |
| pN Stage | ||||
| N0 & N1 | 1 | |||
| N2 & N3 | 4.855 | 1.981–10.611 | <0.001 | |
| FOXA1 | ||||
| ≤P75 f | 1 | |||
| >P75 | 2.710 | 1.161–6.324 | 0.021 |
a HR—Hazard Ratio; b CI—Confidence Interval; c G—Grade; d N—Regional lymph node status; e P75—Percentile 75 of methylation levels of PSAT1; f P75—Percentile 75 of methylation levels of FOXA1.
Concerning disease-specific survival (DSS), in univariable model, pN stage and grade significantly associated with prognosis. Moreover, BrC patients with high FOXA1 promoter methylation (above P75) levels disclosed shorter DSS (Table 2). In the Cox regression multivariable model, only FOXA1 methylation levels and pN stage retained significance for DSS prediction (Table 2).
3.5. Biomarker Detection Performance in ccfDNA Liquid Biopsies (Cohort #2)
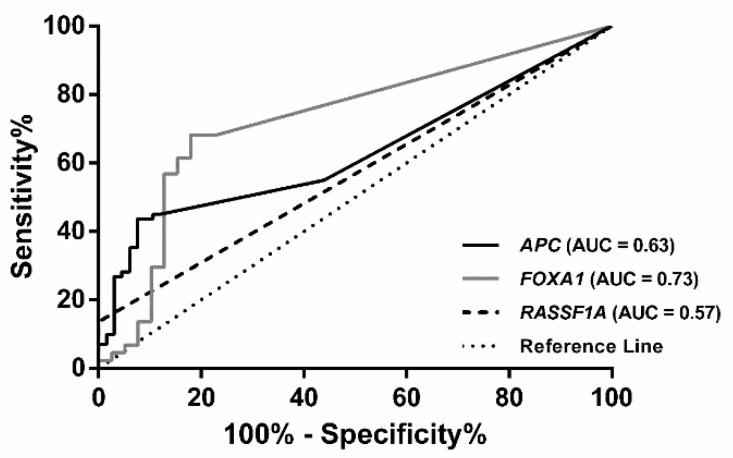
The 4-gene panel (APC, FOXA1, RASSF1A, and SCGB3A1) identified in Cohort #1 was tested in ccfDNA extracted from plasma samples of Cohort #2. APC, FOXA1 and RASSF1A promoter methylation levels significantly differed between BrC patients and AC (p = 0.008, p < 0.001 and p = 0.017, respectively), whereas no significant differences were found for SCGB3A1 (p = 0.127) (Supplementary File 1—Table S8). Thus, SCGB3A1 was not further analyzed. An empirical cutoff value was determined for each gene using ROC curve analysis (APC: 3.446, FOXA1: 64.38 and RASSF1A: 30.00). FOXA1 disclosed the best individual performance, with 68.18% sensitivity and 82.05% specificity (Table 3). Nevertheless, the three-gene panel achieved 81.82% sensitivity and 76.92% specificity (Table 3, Figure 2). Similar to Cohort #1, a biomarker performance analysis restricted by the maximum age of the controls was performed (p = 0.777, Mann-Whitney for age). The biomarker performance was similar (Supplementary File 1—Table S9).
Table 3.
Performance of promoter gene methylation as biomarkers for detection of BrC in plasmas samples.
| Genes | Sensitivity % | Specificity % | PPV a % | NPV b % | Accuracy % |
|---|---|---|---|---|---|
| APC | 27.27 | 94.87 | 85.71 | 53.62 | 59.04 |
| FOXA1 | 68.18 | 82.05 | 81.08 | 69.57 | 74.70 |
| RASSF1A | 13.64 | 100.0 | 100.0 | 50.65 | 54.22 |
| APC/FOXA1/RASSF1A | 81.82 | 76.92 | 80.00 | 78.95 | 79.52 |
a PPV—Positive Predictive Value; b NPV—Negative Predictive Value.
Figure 2.
Receiver operating characteristic (ROC) curve of the three-gene panel (APC, FOXA1 and RASSF1A) in plasma samples from breast cancer patients from Cohort #2.
4. Discussion
BrC remains the major cause of cancer-related death among women worldwide. Mammographic-based screening has contributed to a 28–45% reduction in BrC mortality [4,33,34,35], disclosing 70% sensitivity, and 92% specificity for BrC detection [3]. Owing to its limitations, the need for novel detection methods, with improved accuracy and allowing for stratification of BrC aggressiveness has been emphasized [35]. In recent years, the methylome has emerged as the basis for diagnostic and prognostic biomarkers, which might be used in DNA extracted from liquid biopsies [36,37]. Considering published studies on gene promoter methylation in BrC, we aimed to define the best gene panel for detection and prognosis in tissue samples, as well as BrC detection in ccfDNA.
From the seven most promising candidates, six (APC, CCND2, FOXA1, PSAT1, RASSF1A and SCGB3A1) confirmed its cancer-specificity, discriminating normal from cancerous tissues, although with variable performance, paralleling previous observations from our team and others [8,9,38]. Interestingly, a panel combining APC, FOXA1, RASSF1A and SCGB3A1 disclosed the highest accuracy for BrC detection (94%). APC, RASSF1A and SCGB3A1 promoter methylation have been previously tested in a diagnostic setting of fine-needle aspiration biopsy samples [8,9], whereas FOXA1 expression has been associated with BrC subtype and prognosis [39,40], but not diagnosis. This result compares well with other gene promoter methylation panels that have been reported, disclosing 60–80% sensitivity and 78–100% specificity, and differences in performance are most likely related to biological sample type (tissue vs. bodily fluids) and methylation assessment methods [41].
Since a major goal of this study was to define a panel for BrC detection, ideally its performance should be homogenous regardless of molecular subtype. Thus, we used IHC for tumor subtyping, although acknowledging its limitations in triple negative breast cancer (TNBC)/basal-type classification and luminal A vs. luminal B discrimination [42,43,44]. Interestingly, no association was found between gene promoter methylation and BrC molecular subtype, suggesting that the gene panel might be effective across molecular subtypes. Some studies have associated DNA methylation and specific molecular subtypes, but these have used a similar proportion of all subtypes or have only analyzed a specific subtype [13,45,46], or even used different methods [10,11,47,48]. Our results, however, are based on a consecutive series of cases, which were not selected according to subtype, and, thus, ERBB2-like and TNBC tumor subtypes are, naturally, in a smaller proportion, which might limit statistical analysis. Nevertheless, APC and SCGB3A1 promoter methylation levels associated with specific histological subtypes, confirming previous observations [49]. Interestingly, FOXA1 and RASSF1A promoter methylation levels associated with hormone receptor status. Although the reason for these associations is unclear, similar results for RASSF1A have been reported [50,51]. On the other hand, the higher FOXA1 promoter methylation observed in hormone receptor negative BrC is in accordance with reported FOXA1 hypermethylation in basal BrC cell lines [29,52].
Because liquid biopsies represent a promising method for minimally-invasive early cancer detection [24,28], we tested the selected gene panel in ccfDNA. Interestingly, three genes retained diagnostic significance (APC, FOXA1 and RASSF1A), whereas SCGB3A1 did not discriminate BrC patients from controls. These results are in accordance with another study [53], and might be due to differences in sample number and methylation assessment methods [54]. Moreover, the frequency of gene methylation in Cohort #2 was similar to that previous reported in ccfDNA (Table 4) [25,54,55,56,57,58,59,60,61,62,63].
Table 4.
Frequency of positive cases [n(%)] for methylation levels of cancer-related genes in ccfDNA.
| Genes/Panel | Controls (Healthy Donors) | Patients | References | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| HIC-1/RARβ2/RASSF1A a | 0/10 | 0% | 18/20 | 90% | [60] |
| APC | 0/38 | 0% | 8/47 | 17% | [55] |
| GSTP1 | 0/38 | 0% | 12/47 | 26% | |
| RARβ2 | 3/38 | 8% | 12/47 | 26% | |
| RASSF1A | 2/38 | 5% | 15/47 | 32% | |
| APC/GSTP1/RARβ2/RASSF1A | 5/38 | 13% | 29/47 | 62% | |
| ATM | 0/14 | 0% | 13/50 | 26% | [58] |
| RASSF1A | 0/14 | 0% | 7/50 | 14% | |
| ATM/RASSF1A | 0/14 | 0% | 18/50 | 36% | |
| RARβ2 | 8/125 | 6% | 103/119 | 87% | [54] |
| RASSF1A | 6/125 | 5% | 39/119 | 33% | |
| SCGB3A1 | 0/125 | 0% | 36/119 | 30% | |
| Twist | 10/125 | 8% | 65/119 | 55% | |
| RARβ2/RASSF1A/SCGB3A1/Twist | 23/125 | 18% | 117/119 | 98% | |
| ITIH5 | 7/135 | 6% | 19/138 | 14% | [25] |
| DKK3 | 2/135 | 2% | 41/138 | 30% | |
| RASSF1A | 25/135 | 26% | 64/138 | 47% | |
| ITIH5/DKK3/RASSF1A | 42/135 | 31% | 92/138 | 67% | |
| CDH1 | 0/25 | 0% | 24/50 | 48% | [57] |
| RASSF1A | 0/25 | 0% | 32/50 | 64% | |
| CDH1/RASSF1A | 0/25 | 0% | 38/50 | 76% | |
| SFN | 143/245 | 58% | 197/268 | 74% | [59] |
| P16 | 41/245 | 17% | 60/268 | 33% | |
| hMLH1 | 35/245 | 14% | 75/268 | 28% | |
| HOXD13 | 6/245 | 2% | 37/268 | 14% | |
| PCDHGB7 | 116/245 | 48% | 149/268 | 56% | |
| RASSF1A | 25/245 | 10% | 46/248 | 17% | |
| SFN/P16/hMLH1/HOXD13/PCDHGB7/RASSF1A b | 68/245 | 28% | 213/268 | 80% | |
| ESR1 | 35/74 | 47% | 80/106 | 75% | [56] |
| 14-3-3-σ | 28/74 | 38% | 69/106 | 65% | |
| ESR1/14-3-3-σ b | 33/74 | 45% | 86/106 | 81% | |
| GSTP1 | 2/87 | 2% | 4/101 | 4% | [62] |
| RARβ2 | 0/87 | 0% | 7/101 | 7% | |
| RASSF1A | 4/87 | 5% | 12/101 | 12% | |
| GSTP1/RARβ2/RASSF1A | 6/87 | 7% | 22/101 | 22% | |
| DAPK1 | 0/12 | 0% | 23/26 | 88% | [63] |
| RASSF1A | 1/12 | 8% | 18/26 | 69% | |
| DAPK1/RASSF1A | 1/12 | 8% | 25/26 | 96% | |
| APC | 1/19 | 5% | 23/79 | 30% | [61] |
| ESR1 | 2/19 | 11% | 16/79 | 20% | |
| RASSF1A | 0/19 | 0% | 28/79 | 35% | |
| APC/ESR1/RASSF1A | 3/19 | 16% | 42/79 | 53% | |
| APC | 2/39 | 5% | 12/44 | 27% | --- |
| FOXA1 | 7/39 | 18% | 30/44 | 68% | |
| RASSF1A | 0/39 | 0% | 6/44 | 14% | |
| APC/FOXA1/RASSF1A | 9/39 | 23% | 36/44 | 82% | |
a No information about single gene methylation; b The cut-off used in the panel was different the one used in the single gene analysis.
Nonetheless, the three gene-panel identified BrC with sensitivity, specificity and accuracy higher than 75%. Thus, this panel disclosed a better combination of sensitivity and specificity than most published studies using plasma or serum samples (Table 5), excepting those of Skvortosova et al. (three-gene panel in plasma) and Kim et al. (four-gene panel in serum) [25,54,55,56,57,58,59,60,61,62,63]. Nevertheless, the same authors tested a very limited set of samples (20 BrC, 15 fibroadenomas and 10 healthy donors). Importantly, we used a 4-color multiplex assay that, when compared with the most widely reported two-color multiplex assays represents a faster method and requires less amounts of DNA, thus facilitating its use in clinical routine [36,37,64,65,66]. Hence, this gene-panel may constitute an appealing alternative to conventional diagnostic methods, due to its less-invasive characteristics and to also detect also women with a dense breast.
Table 5.
Comparison of sensitivity and specificity of previously published panels with values obtained.
| Panels | Sensitivity (%) | Specificity (%) | Specimen Type | Methods | References |
|---|---|---|---|---|---|
| HIC-1/RARβ2/RASSF1A | 90 | 100 | Plasma | MSP a | [60] |
| APC/GSTP1/RARβ2/RASSF1A | 62 | 87 | Plasma | QMSP b | [55] |
| ATM/RASSF1A | 36 | 100 | Plasma | QMSP b | [58] |
|
RARβ2/RASSF1A/SCGB3A1/
Twist |
98.3 | 81.6 | Serum | Two-steps QMSP b | [54] |
| ITIH5/DKK3/RASSF1A | 67 | 72 | Serum | QMSP b | [25] |
| CDH1/RASSF1A | 76 | 90 | Serum | MSP a | [57] |
| SFN/P16/hMLH1/HOXD13/PCDHGB7/RASSF1A | 79.6 | 72.4 | Serum | QMSP b | [59] |
| ESR1/14-3-3-σ | 81 | 55 | Serum | QMSP b | [56] |
| GSTP1/RARβ2/RASSF1A | 22 | 93 | Serum | One-step MSP a | [62] |
| DAPK1/RASSF1A | 96 | 71 | Serum | MSP a | [63] |
| APC/ESR1/RASSF1A | 53 | 84 | Serum | QMSP b | [61] |
| APC/FOXA1/RASSF1A | 81.82 | 76.92 | Plasma | Multiplex QMSP b | --- |
a MSP—Methylation-Specific Polimerase Chain Reaction; b QMSP—Quantitative Methylation-Specific Polimerase Chain Reaction.
Although BrC displays high mortality and recurrence rate, clinical course is heterogeneous and perfecting disease prognostication might improve patient management. Interestingly, lower PSAT1 promoter methylation independently predicted for worse DFS. The potential of PSAT1 methylation to predict BrC recurrence has been previously reported in early diagnosed luminal-type BrC. Futhermore, a correlation between high PSAT1 methylation levels, on the one hand, and low PSAT1 mRNAs levels and better outcome, on the other, were disclosed [14]. Interestingly, high PSAT1 expression were associated with poor outcome in nasopharyngeal carcinoma [67]. These data are in accordance with our findings. Furthermore, high FOXA1 methylation levels independently predicted shorter DSS, a finding that, to best of our knowledge, has not been reported, thus far. Remarkably, FOXA1 expression was previously shown to associate with good prognosis and response to endocrine therapy in BrC patients [39,40], and, thus, promoter methylation is the most likely mechanism underlying FOXA1 downregulation in BrC. In Cohort #1, RASSF1A methylation levels did not show prognostic value, which is in accordance with some previous publications [68,69,70]. Nonetheless, other studies have found RASSF1A hypermethylation as a poor prognosis marker in BrC, associating with shorter DFS and DSS [9,22,71]. This discrepancy might be due to differences in sample type and methodologies. Because a meta-analysis suggested that RASSF1A methylation is, indeed, associated with worse DFS and DSS [72], additional studies are needed to definitively establish the prognostic value of RASSF1A promoter methylation in BrC.
The retrospective nature of Cohort #1, the limited sample size of Cohort #2 and the age differences between BrC patients and controls in both series constitute the main limitations of our study. Nonetheless, it should be emphasized that the use of a multiplex assay for a three-gene panel that is able to accurately detect BrC in ccfDNA, regardless of tumor subtype, constitutes a step forward in this field and allow for a swifter translation into routine clinical practice. Indeed, owing to its characteristics, this panel might not only be useful for BrC detection, but also for disease monitoring which deserves further exploration.
Acknowledgments
We acknowledge all the patients and healthy donors who accepted to participate in this study. We thank all the Nursing Staff from Breast Cancer Clinic and Laboratory Medicine from IPO Porto that actively participated in patients and controls’ enrollment.
Supplementary Materials
The following are available online at http://www.mdpi.com/2077-0383/7/11/420/s1, Supplementary File 1 which includes: Table S1: Primers sequences and QMSP conditions for each gene studied in tissues samples; Table S2: Primers and probe sequences for each gene studied in plasma samples; Table S3: Clinical—pathological data of normal breast tissue, and (NBr) breast cancer (BrC) patient’s (Cohort #1); Table S4: Clinical and pathological data of asymptomatic controls (AC) and Breast Cancer (BrC) patients (Cohort #2); Table S5: Frequency of positive cases [n(%)] and distribution of methylation levels of cancer-related genes in tissues from Cohort #1 [gene/ACTB × 1000 median (Interquartile range (IQR))]; Table S6: Area Under the Curve (AUC) of Receiver Operating Characteristic (ROC) Curve for each gene in tissues from Cohort#1; Table S7: Performance of promoter gene methylation as biomarkers for detection of BrC in tissue samples from patients with age below 70 years (controls n = 27 and tumors n = 108); Table S8: Frequency of positive cases [n(%)] for methylation levels of cancer-related genes in ccfDNA of Cohort #2.; Table S9: Performance of promoter gene methylation as biomarkers for detection of BrC in plasmas samples from patients with age below 66 years (controls n = 39 and tumors n = 25); Figure S1: Boxplots of APC (a), BRCA1 (b), CCND2 (c), FOXA1 (d), PSAT1 (e), RASSF1A (f) and SCGB3A1 (g) methylation levels in the breast cancer molecular subtypes and normal breast tissues.
Author Contributions
S.S. and S.P.N. performed fresh-frozen sections, DNA extraction and QMSP, analyzed data and drafted the manuscript. M.C. and R.H. assisted in the histopathological evaluation of tissue samples and IHC evaluation. P.L. performed the FFPE sections for IHC for molecular subtype determination. M.F. assisted in molecular analyses. L.A. assisted in the statistical analyses. S.P.d.S. and M.F.-S. collected clinical follow-up data. F.C. and P.A. assisted in patients’ enrolment for plasma cohort. R.H. and C.J. designed and supervised the study and revised the manuscript. All the authors read and approved the final manuscript.
Funding
This work was supported by a grant from Research Center of Portuguese Oncology Institute of Porto (PI 74-CI-IPOP-19-2016). SoS is supported by a PhD fellowship IPO/ESTIMA-1 NORTE-01-0145-FEDER-000027.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Tabar L., Vitak B., Chen H.H., Yen M.F., Duffy S.W., Smith R.A. Beyond randomized controlled trials: organized mammographic screening substantially reduces breast carcinoma mortality. Cancer. 2001;91:1724–1731. doi: 10.1002/1097-0142(20010501)91:9<1724::AID-CNCR1190>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 3.Pisano E.D., Gatsonis C., Hendrick E., Yaffe M., Baum J.K., Acharyya S., Conant E.F., Fajardo L.L., Bassett L., D’Orsi C., et al. Diagnostic performance of digital versus film mammography for breast-cancer screening. N. Engl. J. Med. 2005;353:1773–1783. doi: 10.1056/NEJMoa052911. [DOI] [PubMed] [Google Scholar]
- 4.Warner E. Clinical practice. Breast-cancer screening. N. Engl. J. Med. 2011;365:1025–1032. doi: 10.1056/NEJMcp1101540. [DOI] [PubMed] [Google Scholar]
- 5.Sorlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Nat. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senkus E., Kyriakides S., Ohno S., Penault-Llorca F., Poortmans P., Rutgers E., Zackrisson S., Cardoso F., Committee E.G. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015;26(Suppl. 5):v8–v30. doi: 10.1093/annonc/mdv298. [DOI] [PubMed] [Google Scholar]
- 7.Jeronimo C., Costa I., Martins M.C., Monteiro P., Lisboa S., Palmeira C., Henrique R., Teixeira M.R., Lopes C. Detection of gene promoter hypermethylation in fine needle washings from breast lesions. Clin. Cancer Res. 2003;9:3413–3417. [PubMed] [Google Scholar]
- 8.Jeronimo C., Monteiro P., Henrique R., Dinis-Ribeiro M., Costa I., Costa V.L., Filipe L., Carvalho A.L., Hoque M.O., Pais I., et al. Quantitative hypermethylation of a small panel of genes augments the diagnostic accuracy in fine-needle aspirate washings of breast lesions. Breast Cancer Res. Treat. 2008;109:27–34. doi: 10.1007/s10549-007-9620-x. [DOI] [PubMed] [Google Scholar]
- 9.Martins A.T., Monteiro P., Ramalho-Carvalho J., Costa V.L., Dinis-Ribeiro M., Leal C., Henrique R., Jeronimo C. High RASSF1A promoter methylation levels are predictive of poor prognosis in fine-needle aspirate washings of breast cancer lesions. Breast Cancer Res. Treat. 2011;129:1–9. doi: 10.1007/s10549-010-1160-0. [DOI] [PubMed] [Google Scholar]
- 10.Bediaga N.G., Acha-Sagredo A., Guerra I., Viguri A., Albaina C., Ruiz Diaz I., Rezola R., Alberdi M.J., Dopazo J., Montaner D., et al. DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res. 2010;12:R77. doi: 10.1186/bcr2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Győrffy B., Bottai G., Fleischer T., Munkácsy G., Budczies J., Paladini L., Børresen-Dale A.L., Kristensen V.N., Santarpia L. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int. J. Cancer. 2016;138:87–97. doi: 10.1002/ijc.29684. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.S., Fackler M.J., Lee J.H., Choi C., Park M.H., Yoon J.H., Zhang Z., Sukumar S. Basal-like breast cancer displays distinct patterns of promoter methylation. Cancer Biol. Ther. 2010;9:1017–1024. doi: 10.4161/cbt.9.12.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens J.W., Nimmrich I., Koenig T., Look M.P., Harbeck N., Model F., Kluth A., Bolt-de Vries J., Sieuwerts A.M., Portengen H., et al. Association of DNA methylation of phosphoserine aminotransferase with response to endocrine therapy in patients with recurrent breast cancer. Cancer Res. 2005;65:4101–4117. doi: 10.1158/0008-5472.CAN-05-0064. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 16.Van Hoesel A.Q., Sato Y., Elashoff D.A., Turner R.R., Giuliano A.E., Shamonki J.M., Kuppen P.J., van de Velde C.J., Hoon D.S. Assessment of DNA methylation status in early stages of breast cancer development. Br. J. Cancer. 2013;108:2033–2038. doi: 10.1038/bjc.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muggerud A.A., Ronneberg J.A., Warnberg F., Botling J., Busato F., Jovanovic J., Solvang H., Bukholm I., Borresen-Dale A.L., Kristensen V.N., et al. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res. 2010;12:R3. doi: 10.1186/bcr2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fackler M.J., McVeigh M., Evron E., Garrett E., Mehrotra J., Polyak K., Sukumar S., Argani P. DNA methylation of RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and invasive lobular breast carcinoma. Int. J. Cancer. 2003;107:970–975. doi: 10.1002/ijc.11508. [DOI] [PubMed] [Google Scholar]
- 19.Widschwendter M., Jones P.A. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- 20.Heyn H., Esteller M. DNA methylation profiling in the clinic: Applications and challenges. Nat. Rev.Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 21.Jeronimo C., Henrique R. Epigenetic biomarkers in urological tumors: A systematic review. Cancer Lett. 2014;342:264–274. doi: 10.1016/j.canlet.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 22.Muller H.M., Widschwendter A., Fiegl H., Ivarsson L., Goebel G., Perkmann E., Marth C., Widschwendter M. DNA methylation in serum of breast cancer patients: An independent prognostic marker. Cancer Res. 2003;63:7641–7645. [PubMed] [Google Scholar]
- 23.Gobel G., Auer D., Gaugg I., Schneitter A., Lesche R., Muller-Holzner E., Marth C., Daxenbichler G. Prognostic significance of methylated RASSF1A and PITX2 genes in blood- and bone marrow plasma of breast cancer patients. Breast Cancer Res. Treat. 2011;130:109–117. doi: 10.1007/s10549-010-1335-8. [DOI] [PubMed] [Google Scholar]
- 24.Stewart C.M., Tsui D.W.Y. Circulating cell-free DNA for non-invasive cancer management. Cancer Genet. 2018 doi: 10.1016/j.cancergen.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kloten V., Becker B., Winner K., Schrauder M.G., Fasching P.A., Anzeneder T., Veeck J., Hartmann A., Knuchel R., Dahl E. Promoter hypermethylation of the tumor-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res. 2013;15:R4. doi: 10.1186/bcr3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radpour R., Barekati Z., Kohler C., Lv Q., Burki N., Diesch C., Bitzer J., Zheng H., Schmid S., Zhong X.Y. Hypermethylation of tumor suppressor genes involved in critical regulatory pathways for developing a blood-based test in breast cancer. PloS ONE. 2011;6:e16080. doi: 10.1371/journal.pone.0016080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avraham A., Uhlmann R., Shperber A., Birnbaum M., Sandbank J., Sella A., Sukumar S., Evron E. Serum DNA methylation for monitoring response to neoadjuvant chemotherapy in breast cancer patients. Int. J. Cancer. 2012;131:E1166–E1172. doi: 10.1002/ijc.27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasculli B., Barbano R., Parrella P. Epigenetics of breast cancer: Biology and clinical implication in the era of precision medicine. Semin. Cancer biol. 2018;51:22–35. doi: 10.1016/j.semcancer.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Locke W.J., Zotenko E., Stirzaker C., Robinson M.D., Hinshelwood R.A., Stone A., Reddel R.R., Huschtscha L.I., Clark S.J. Coordinated epigenetic remodelling of transcriptional networks occurs during early breast carcinogenesis. Clin. Epigenet. 2015;7:52. doi: 10.1186/s13148-015-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson H., Stirling D. DNA extraction from tissue. Methods Mol. Biol. 2003;226:33–34. doi: 10.1385/0-89603-627-8:33. [DOI] [PubMed] [Google Scholar]
- 31.Schisterman E.F., Perkins N.J., Liu A., Bondell H. Optimal cut-point and its corresponding Youden Index to discriminate individuals using pooled blood samples. Epidemiology. 2005;16:73–81. doi: 10.1097/01.ede.0000147512.81966.ba. [DOI] [PubMed] [Google Scholar]
- 32.Youden W.J. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Schuur E.R., DeAndrade J.P. Breast Cancer: Molecular Mechanisms, Diagnosis, and Treatment. In: de Mello A.R., Tavares Á., Mountzios G., editors. International Manual of Oncology Practice. Springer International Publishing; Cham, Switzerland: 2015. pp. 155–200. [Google Scholar]
- 34.Schnitt S.J., Lakhani S.R. Breast Cancer. In: Stewart B., Wild C.P., editors. World Cancer Report 2014. International Agency for Research on Cancer; Lyon, France: 2014. pp. 362–373. [Google Scholar]
- 35.Independent UK Panel on Breast Cancer Screening The benefits and harms of breast cancer screening: An independent review. Lancet. 2012;380:1778–1786. doi: 10.1016/S0140-6736(12)61611-0. [DOI] [PubMed] [Google Scholar]
- 36.Olkhov-Mitsel E., Zdravic D., Kron K., van der Kwast T., Fleshner N., Bapat B. Novel multiplex MethyLight protocol for detection of DNA methylation in patient tissues and bodily fluids. Sci. Rep. 2014;4:4432. doi: 10.1038/srep04432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Q., Chen H.Y., Bai E.Q., Luo Y.X., Fu R.J., He Y.S., Jiang J., Wang H.Q. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet. Cytogenet. 2010;202:1–10. doi: 10.1016/j.cancergencyto.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 38.Bu D., Lewis C.M., Sarode V., Chen M., Ma X., Lazorwitz A.M., Rao R., Leitch M., Moldrem A., Andrews V., et al. Identification of breast cancer DNA methylation markers optimized for fine-needle aspiration samples. Cancer Epidemiol. Biomark. Prev. 2013;22:2212–2221. doi: 10.1158/1055-9965.EPI-13-0208. [DOI] [PubMed] [Google Scholar]
- 39.Mehta R.J., Jain R.K., Leung S., Choo J., Nielsen T., Huntsman D., Nakshatri H., Badve S. FOXA1 is an independent prognostic marker for ER-positive breast cancer. Breast Cancer Res. Treat. 2012;131:881–890. doi: 10.1007/s10549-011-1482-6. [DOI] [PubMed] [Google Scholar]
- 40.Albergaria A., Paredes J., Sousa B., Milanezi F., Carneiro V., Bastos J., Costa S., Vieira D., Lopes N., Lam E.W., et al. Expression of FOXA1 and GATA-3 in breast cancer: The prognostic significance in hormone receptor-negative tumours. Breast Cancer Res. 2009;11:R40. doi: 10.1186/bcr2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parrella P. Epigenetic Signatures in Breast Cancer: Clinical Perspective. Breast Care. 2010;5:66–73. doi: 10.1159/000309138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maisonneuve P., Disalvatore D., Rotmensz N., Curigliano G., Colleoni M., Dellapasqua S., Pruneri G., Mastropasqua M.G., Luini A., Bassi F., et al. Proposed new clinicopathological surrogate definitions of luminal A and luminal B (HER2-negative) intrinsic breast cancer subtypes. Breast Cancer Res. 2014;16:R65. doi: 10.1186/bcr3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prat A., Cheang M.C., Martin M., Parker J.S., Carrasco E., Caballero R., Tyldesley S., Gelmon K., Bernard P.S., Nielsen T.O., et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal a breast cancer. J. Clin. Oncol. 2013;31:203–209. doi: 10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coates A.S., Winer E.P., Goldhirsch A., Gelber R.D., Gnant M., Piccart-Gebhart M., Thürlimann B., Senn H.-J., André F., Baselga J. Tailoring therapies—Improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann. Oncol. 2015;26:1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stefansson O.A., Jonasson J.G., Olafsdottir K., Hilmarsdottir H., Olafsdottir G., Esteller M., Johannsson O.T., Eyfjord J.E. CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics. 2011;6:638–649. doi: 10.4161/epi.6.5.15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sunami E., Shinozaki M., Sim M.S., Nguyen S.L., Vu A.T., Giuliano A.E., Hoon D.S. Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors. Breast Cancer Res. 2008;10:R46. doi: 10.1186/bcr2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stirzaker C., Zotenko E., Clark S.J. Genome-wide DNA methylation profiling in triple-negative breast cancer reveals epigenetic signatures with important clinical value. Mol. Cell Oncol. 2016;3:e1038424. doi: 10.1080/23723556.2015.1038424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stefansson O.A., Moran S., Gomez A., Sayols S., Arribas-Jorba C., Sandoval J., Hilmarsdottir H., Olafsdottir E., Tryggvadottir L., Jonasson J.G., et al. A DNA methylation-based definition of biologically distinct breast cancer subtypes. Mol. Oncol. 2015;9:555–568. doi: 10.1016/j.molonc.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tisserand P., Fouquet C., Barrois M., Gallou C., Dendale R., Stoppa-Lyonnet D., Sastre-Garau X., Fourquet A., Soussi T. Lack of HIN-1 methylation defines specific breast tumor subtypes including medullary carcinoma of the breast and BRCA1-linked tumors. Cancer Biol. Ther. 2003;2:559–563. doi: 10.4161/cbt.2.5.511. [DOI] [PubMed] [Google Scholar]
- 50.Benevolenskaya E.V., Islam A.B., Ahsan H., Kibriya M.G., Jasmine F., Wolff B., Al-Alem U., Wiley E., Kajdacsy-Balla A., Macias V., et al. DNA methylation and hormone receptor status in breast cancer. Clin. Epigenet. 2016;8:17. doi: 10.1186/s13148-016-0184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Widschwendter M., Siegmund K.D., Müller H.M., Fiegl H., Marth C., Müller-Holzner E., Jones P.A., Laird P.W. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 52.Gong C., Fujino K., Monteiro L., Gomes A., Drost R., Davidson-Smith H., Takeda S., Khoo U., Jonkers J., Sproul D., et al. FOXA1 repression is associated with loss of BRCA1 and increased promoter methylation and chromatin silencing in breast cancer. Oncogene. 2014;34:5012–5024. doi: 10.1038/onc.2014.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sturgeon S.R., Balasubramanian R., Schairer C., Muss H.B., Ziegler R.G., Arcaro K.F. Detection of promoter methylation of tumor suppressor genes in serum DNA of breast cancer cases and benign breast disease controls. Epigenetics. 2012;7:1258–1267. doi: 10.4161/epi.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J.-H., Shin M.-H., Kweon S.-S., Park M.H., Yoon J.H., Lee J.S., Choi C., Fackler M.J., Sukumar S. Evaluation of promoter hypermethylation detection in serum as a diagnostic tool for breast carcinoma in Korean women. Gynecol. Oncol. 2010;118:176–181. doi: 10.1016/j.ygyno.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 55.Hoque M.O., Feng Q., Toure P., Dem A., Critchlow C.W., Hawes S.E., Wood T., Jeronimo C., Rosenbaum E., Stern J., et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J. Clin. Oncol. 2006;24:4262–4269. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- 56.Martínez-Galán J., Torres B., del Moral R., Muñoz-Gámez J.A., Martín-Oliva D., Villalobos M., Núñez M.I., Luna J.D.D., Oliver F.J., Almodóvar J.M.R.D. Quantitative detection of methylated ESR1 and 14-3-3-σ gene promoters in serum as candidate biomarkers for diagnosis of breast cancer and evaluation of treatment efficacy. Cancer Biol. Ther. 2008;7:958–965. doi: 10.4161/cbt.7.6.5966. [DOI] [PubMed] [Google Scholar]
- 57.Nandi K., Yadav P., Mir R., Khurana N., Agarwal P., Saxena A. The Clinical Significance of Rassf1a and Cdh1 Hypermethylation in Breast Cancer Patients. Int. J. Sci. Res. 2008 in press. [Google Scholar]
- 58.Papadopoulou E., Davilas E., Sotiriou V., Georgakopoulos E., Georgakopoulou S., Koliopanos A., Aggelakis F., Dardoufas K., Agnanti N.J., Karydas I. Cell-free DNA and RNA in Plasma as a New Molecular Marker for Prostate and Breast Cancer. Ann. N. Y. Acad. Sci. 2006;1075:235–243. doi: 10.1196/annals.1368.032. [DOI] [PubMed] [Google Scholar]
- 59.Shan M., Yin H., Li J., Li X., Wang D., Su Y., Niu M., Zhong Z., Wang J., Zhang X., et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget. 2016;7:18485–18494. doi: 10.18632/oncotarget.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skvortsova T.E., Rykova E.Y., Tamkovich S.N., Bryzgunova O.E., Starikov A.V., Kuznetsova N.P., Vlassov V.V., Laktionov P.P. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. Br. J. Cancer. 2006;94:1492–1495. doi: 10.1038/sj.bjc.6603117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van der Auwera I., Elst H., Van Laere S., Maes H., Huget P., Van Dam P., Van Marck E., Vermeulen P., Dirix L. The presence of circulating total DNA and methylated genes is associated with circulating tumour cells in blood from breast cancer patients. Br. J. Cancer. 2009;100:1277. doi: 10.1038/sj.bjc.6605013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto N., Nakayama T., Kajita M., Miyake T., Iwamoto T., Kim S.J., Sakai A., Ishihara H., Tamaki Y., Noguchi S. Detection of aberrant promoter methylation of GSTP1, RASSF1A, and RARβ2 in serum DNA of patients with breast cancer by a newly established one-step methylation-specific PCR assay. Breast Cancer Res. Treat. 2012;132:165–173. doi: 10.1007/s10549-011-1575-2. [DOI] [PubMed] [Google Scholar]
- 63.Ahmed I.A., Pusch C.M., Hamed T., Rashad H., Idris A., El-Fadle A.A., Blin N. Epigenetic alterations by methylation of RASSF1A and DAPK1 promoter sequences in mammary carcinoma detected in extracellular tumor DNA. Cancer Genet. Cytogenet. 2010;199:96–100. doi: 10.1016/j.cancergencyto.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 64.Roperch J.P., Incitti R., Forbin S., Bard F., Mansour H., Mesli F., Baumgaertner I., Brunetti F., Sobhani I. Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC Cancer. 2013;13:566. doi: 10.1186/1471-2407-13-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fackler M.J., McVeigh M., Mehrotra J., Blum M.A., Lange J., Lapides A., Garrett E., Argani P., Sukumar S. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res. 2004;64:4442–4452. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 66.Melnikov A.A., Scholtens D.M., Wiley E.L., Khan S.A., Levenson V.V. Array-based multiplex analysis of DNA methylation in breast cancer tissues. J. Mol. Diagn. 2008;10:93–101. doi: 10.2353/jmoldx.2008.070077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liao K.-M., Chao T.-B., Tian Y.-F., Lin C.-Y., Lee S.-W., Chuang H.-Y., Chan T.-C., Chen T.-J., Hsing C.-H., Sheu M.-J. Overexpression of the PSAT1 Gene in Nasopharyngeal Carcinoma Is an Indicator of Poor Prognosis. J. Cancer. 2016;7:1088–1094. doi: 10.7150/jca.15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klajic J., Fleischer T., Dejeux E., Edvardsen H., Warnberg F., Bukholm I., Lonning P.E., Solvang H., Borresen-Dale A.L., Tost J., et al. Quantitative DNA methylation analyses reveal stage dependent DNA methylation and association to clinico-pathological factors in breast tumors. BMC Cancer. 2013;13:456. doi: 10.1186/1471-2407-13-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharma G., Mirza S., Yang Y.H., Parshad R., Hazrah P., Datta Gupta S., Ralhan R. Prognostic relevance of promoter hypermethylation of multiple genes in breast cancer patients. Cell Oncol. 2009;31:487–500. doi: 10.3233/CLO-2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cho Y.H., Shen J., Gammon M.D., Zhang Y.J., Wang Q., Gonzalez K., Xu X., Bradshaw P.T., Teitelbaum S.L., Garbowski G., et al. Prognostic significance of gene-specific promoter hypermethylation in breast cancer patients. Breast Cancer Res. Treat. 2012;131:197–205. doi: 10.1007/s10549-011-1712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buhmeida A., Merdad A., El-Maghrabi J., Al-Thobaiti F., Ata M., Bugis A., Syrjänen K., Abuzenadah A., Chaudhary A., Gari M. RASSF1A methylation is predictive of poor prognosis in female breast cancer in a background of overall low methylation frequency. Anticancer Res. 2011;31:2975–2981. [PubMed] [Google Scholar]
- 72.Jiang Y., Cui L., Chen W.D., Shen S.H., Ding L.D. The prognostic role of RASSF1A promoter methylation in breast cancer: A meta-analysis of published data. PloS ONE. 2012;7:e36780. doi: 10.1371/journal.pone.0036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.