Abstract
STUDY QUESTION
In PGS, does chromosomal constitution differ among trophectoderm (TE) biopsy sites and between them and the inner cell mass (ICM)?
SUMMARY ANSWER
The ploidy concordance between ICM and TE was independent of whether the biopsy site in the TE was near to or far from the ICM.
WHAT IS KNOWN ALREADY
TE biopsies are considered less harmful to developing embryos than blastomere biopsies. Removal of multi-cellular samples permits high-resolution next-generation sequencing (Veriseq NGS) to detect aneuploidy present in a minority of cells (mosaicism of diploid and aneuploid cells). However, the prevalence of ploidy discrepancies between different TE biopsy sites and the ICM, as well as confined mosaicism (aneuploidy only in a particular area), has not been established.
STUDY DESIGN, SIZE, DURATION
Biopsies were taken from a site opposite to the ICM (TE1), near the ICM (TE2) and within the ICM of the same embryo in 33 donated blastocysts obtained from 12 volunteer patients. The samples were analyzed by the Veriseq NGS to assess ploidy concordance.
PARTICIPANTS/MATERIALS, SETTING, METHODS
The mean age of the patients was 34.4 years, and samples from all three biopsy sites were achieved in 29 frozen thawed blastocysts. The aneuploid percentage in each sample was interpreted by Veriseq NGS at the finest resolution involving the number of reads after filtering, sample overall noise score, and average quality/alignment scores according to the Veriseq quality control assessment. Ploidy concordance was then assessed between different TE fractions, and between the TE and ICM.
MAIN RESULTS AND THE ROLE OF CHANCE
The euploid rates were similar in the TEs and ICM, and no preferential allocation of euploid lineage within a blastocyst was demonstrated. Whether the biopsy site in the TE was near to or far from the ICM, the chromosomal consistency rate was similar [TE1-to-ICM, 86.2% (25/29) versus TE2-to-ICM, 89.7% (26/29); P = 1.0], suggesting that the cells with different chromosomal components may spread randomly throughout the TE. The following two types of inconsistent PGS conclusions between TE and ICM due to confined mosaicism were observed: (i) euploid TE with mosaic ICM (3%) (1/29); and (ii) mosaic TE with euploid ICM (3%) (1/29) or with aneuploid ICM (7%) (2/29). Thus, the overall rate of confined mosaicism was 14% (4/29).
LARGE SCALE DATA
N/A.
LIMITATION, REASONS FOR CAUTION
The approach used in the present study was affected by biopsy manipulation limitations involving possible cell contamination and the technical challenge of comprehensive chromosomal screening (CCS) procedures.
WIDER IMPLICATIONS OF THE FINDINGS
The rate of confined mosaicism in the blastocysts was estimated in this preliminary study, thus, specifying the incidence of biological sampling biases. The results also verified the random distribution of different cell lineages, and the representative value of a single biopsied sample from the TE.
STUDY FUNDING AND CONFLICT OF INTEREST(S)
No external funding was obtained; all the authors declare no conflicts of interest regarding this study.
Keywords: preimplantation genetic screening, confined mosaicism, next-generation sequencing, chromosome concordance, aneuploid percentage
Introduction
Chromosomal mosaicism of an embryo, which is defined as the presence of two or more distinct cell lineages within the embryo, has recently led to a discussion involving the competence of normal pregnancies (Munné et al., 2016). Aneuploidy (imbalance of chromosome dosage) has been long-shown to be one of the main factors affecting the success rate of IVF (Greco et al., 2014). Aneuploidy is also known to be a common cause of miscarriage during early gestation (Maxwell et al., 2016). Pure aneuploidies are mostly driven by errors occurring during maternal meiosis of oocytes, the prevalence of which rises with increasing maternal age (Demko et al., 2016). Unlike the meiotic aneuploidy uniformly presenting in whole embryos and affecting further development adversely, low-rate aneuploidy (mosaicism) has been reported with diverse clinical penetrance and could still result in a live birth (Greco et al., 2015).
The primary origin of embryonic mosaicism has been attributed to mitotic errors occurring during the post-zygotic stage (Delhanty et al., 1997; Munné et al., 2002). Improper segregation of chromosomes caused by mitotic non-disjunction is the most frequent mechanism underlying embryonic mosaicism (Gueye et al., 2014; Taylor et al., 2014; Capalbo et al., 2017), followed by other rare causes, such as anaphase lagging (Coonen et al., 2004; Capalbo et al., 2013) and endo-replication or endo-deletion (Fox and Duronio, 2013). Because the time of occurrence of mitotic errors and the fate of aneuploid lineages determine the mosaicism in a preimplantation embryo, the clinical outcomes of mosaics may change with many variables, including aneuploidy distribution, affected chromosome length, involved chromosomes and aneuploid percentage (Capalbo et al., 2017). However, the actual causes of these mitotic errors in the early-stage embryos were not clearly defined and some researchers believe that they could be a consequence of environmental effects during IVF (Munné et al., 2016; Fragouli et al., 2017; Wells et al., 2017).
Currently, PGS involving Day 5 trophectoderm (TE) biopsies and comprehensive chromosome screening (CCS) has become more frequent in reproductive medicine. Compared to Day 3 blastomere biopsies in cleavage-stage embryos, TE biopsies are expected to provide more accurate information and less damage (Schoolcraft et al., 2010; Gutierez-Mateo et al., 2011; Yang et al., 2012; Huang et al., 2013). Through the PGS examination of chromosome materials in blastocysts, the aneuploidies generated from both the meiotic and mitotic origins can be detected. Mosaicism occurs, but to a lesser extent in blastocysts than cleavage-stage embryos (Johnson et al., 2010). Mosaics with a diploid and aneuploid mixture (low-rate aneuploidy) account for ~10% of total blastocysts (Fragouli et al., 2011).
Mosaicism can be categorized into the general or confined mosaicism based on the distribution area of chromosomal discrepancy. General mosaicism is defined as the presence of two or more cell lines throughout the entire embryo, and could be the result of a very early mitotic event. In contrast, confined mosaicism is restricted to a particular area of an embryo, and could be due to events during later development (Taylor et al., 2014). A clinical example is confined placental mosaicism (CPM), which shows different chromosomal constitutions between the fetus and the placenta, with an estimated incidence of 1–2% (Kalousek et al., 1992). Applying a similar philosophy to a blastocyst scale, the chromosomal discrepancy between the inner cell mass (ICM) and TE can also be defined as confined mosaicism, which would affect the accuracy of PGS diagnosis.
Accordingly, the current detection of mosaicism is challenged by two facets. The biological facet involves the distribution of different cell lineages in a blastocyst, the incidence of confined mosaicism, and the possibility of reciprocal aneuploidies in sampled cells. Whether or not the cells with different chromosomal components are distributed randomly throughout the TE or unevenly allocated in an area of the embryo is unclear. Information pertaining to confined mosaicism in human blastocysts is important for the representative value of a biopsied sample and is currently limited; thus, further studies are warranted. With respect to the technical facet, PCR artifacts derived from the CCS process and the masking which occurs as a result of statistical smoothing algorithms during data analysis are taken into consideration (Scott and Galliano, 2016; Chen et al., 2017; Treff and Franasiak, 2017).
Regarding the issue of confined mosaicism, comparisons of the ploidies between the TE and ICM have been reported. In a small study involving 10 embryos with array comparative genomic hybridization (aCGH) screening, the concordance between TE and ICM was reported to be as high as 100% (Fragouli et al., 2008). Using the same approach, Johnson et al. (2010) demonstrated that the concordance of TE and ICM was 96.1% in 51 blastocysts, which were mostly euploid (80%) and recruited from patients with an average age of 31 years. Of note, the methodological limitations of aCGH and the mostly euploid samples may affect the results of such comparisons of TE and ICM. Because aCGH is unable to detect low-rate aneuploidy or trivial segmental aneuploidy, some confined mosaicisms could be missed (Lai et al., 2017). Furthermore, chromosomal mosaicism tends to occur in aneuploid embryos rather than euploids, and includes aneuploid TE with euploid ICM, inconsistent anomalies between the TE and ICM, or euploid TE with aneuploid ICM (Liu et al., 2012). Thus, a more precise CCS system and analysis of embryos with different ploidies are needed.
In 2014, next-generation sequencing (NGS) technology was gradually applied to PGS (Wells et al., 2014; Fiorentino et al., 2014a and 2014b), and it allows the embryo aneuploidy to be detected in single cells using a rapid low-pass whole genome sequencing methodology (Wells et al., 2014). With proper adaption of NGS to the diagnosis of TE ploidy, blastocysts can be classified as aneuploid, euploid or low-rate aneuploid (diploid/aneuploid mosaic), and an embryo is further scored by the percentage of aneuploidy to determine its transfer priority (Fiorentino et al., 2016). With use of the PGS/NGS, the biological factors affecting detection of mosaicism can be re-evaluated in greater detail. Thus, we conducted an investigation of chromosomal discrepancies among different biopsied sites (TE opposite to the ICM, TE near to the ICM and the ICM itself). Using a high-resolution PGS/NGS platform (Veriseq NGS), the aneuploidy of each biopsied fraction was displayed as a percentage, and the distribution of karyotypically distinct cells was assessed according to this ratio. Based on the present study, the distribution of cells with different chromosomal components in distinct biopsy sites of the TE and ICM was evaluated. The proportion of inconsistent PGS conclusions between the TE and ICM due to confined mosaicism was analyzed, and the type of confined mosaicism was determined.
Materials and Methods
Study design
The study was approved by the Ethics Reviewing Committee of National Taiwan University Hospital. Thirty-three donated blastocysts were obtained from 12 volunteer patients who underwent PGS. Written informed consent was obtained from all participants. The donated blastocysts were biopsied at the following three sites: TE opposite to the ICM, TE near the ICM and the ICM. Then the biopsies were analyzed using a Veriseq NGS platform, and chromosomal discrepancies among the three fractions were analyzed.
Study subjects
All patients were from the outpatient department of a private fertility center (Hsinchu, Taiwan). The patients underwent individualized stimulation protocols for IVF/ICSI (Wang et al., 2016; Hwang et al., 2018). The blastocysts used in the study were surplus embryos from patients who achieved successful live births. Full consent was required prior to obtaining donated blastocysts for research. The embryos were cryopreserved on culture Day 5 and had not undergone previous aneuploidy diagnosis. They were morphologically graded according to the Gardner and Schoolcraft system (Gardner and Schoolcraft, 1999). Only the embryos with an ICM grading ≥ B and with a distinctly cellular TE grading ≥ C were thawed for the study.
Embryo thawing and disaggregation
The embryos were vitrified using the Cryotech Vitrification kit (Cryotech, Tokyo, Japan). The studied embryos were thawed using a Cryotech Warming kit, then cultured in one-step human embryo culture media (Global, LifeGlobal, Guilford, CT, USA) until full expansion was reached. During the biopsy procedure, 5–10 cells for TE and ~10 cells for ICM were aspirated using a biopsy pipette (Origio, Måløv, Denmark), then mechanically separated by shearing force between the biopsy and holding pipettes. The blastocysts were biopsied at sites opposite to the ICM (TE1), near the ICM (TE2) and within the ICM (Supplementary Fig. S1). The biopsied fractions were then washed twice in sterile 1X phosphate-buffered saline (PBS) solution (Cell Signaling Technologies, Danvers, MA, USA) containing 1% (w/v) polyvinylpyrrolidone (PVP; Sigma, St. Louis, MO, USA). The washed fractions were gently transferred into a 0.2-ml PCR tube with 2.5 μl of PBS/PVP solution and stored at −20°C for whole genome amplification (WGA).
Cell lysis, WGA and product quantification
The biopsies were thawed for WGA, and underwent the lysis, random fragmentation, and subsequently amplification by using the Sureplex WGA method (Sureplex; Illumina, San Diego, CA, USA) following the manufacturer’s protocol. Succinctly, the biopsies were lysed with Sureplex cell extraction master mix; and the released DNAs were fragmented and initially amplified using Sureplex pre-amplification cocktail; and finally the Sureplex amplification cocktail was added for the amplification. A dsDNA high-sensitivity (HS) Assay Kit (Qubit®; Life Technologies, Waltham, MA, USA) was used to quantify the concentration of 10× diluted amplified products.
NGS analysis
The amplified product from each sample proceeded to the library preparation according to the manufacturer’s guidelines of VeriSeq PGS (Illumina). In brief, the diluted DNA was fragmented, tagged, and then underwent a dual-indexed sequencing. Then the products were purified by using a size selection and normalized to equalize the quantity of each sample. The final products were pooled, denatured and sequenced using the Miseq Reagent Kit (v.3; Illumina) on a Miseq System (Illumina). The generated bioinformatics data were analyzed using the BlueFuse Multi Software (Illumina). The samples were distinguished if a median chromosomal copy number deviated from the default copy number, and a possible gain (trisomy) or loss (monosomy) of autosomal chromosomes would be displayed as a copy number >2 or <2, respectively. Additional details regarding the Veriseq NGS procedures and determinations of copy number analyzed on BlueFuse Multi Software have been published previously (Fiorentino et al., 2014b). Of the segmental aneuploidy, it needed manual identification when a chromosome fragment deviated from the default copy number of euploidy (automated calls generated by BlueFuse Multi). Based on the previous scientific reports and the individual resolution of each sample, chromosomal imbalances above 10 Mb were defined as segmental aneuploidy in the study (Sermon et al., 2016; Vera-Rodríguez et al., 2016).
Internal validation of mosaicism and ploidy determination
The PGS conclusion of each sample was based on the aneuploid percentage, which was adjusted by a validation curve derived from a mixing experiment with aneuploid/diploid amniotic stem cell lines (Supplementary Table S1). Before establishing the validation curve, we conducted an experiment to define the false positive rate for predicting mosaicism when no mosaicism exists by using the amniotic stem cell line with normal karyotype (AF02, Bioresource Collection and Research Center, Hsinchu, Taiwan) (Tsai et al., 2007). Each tested sample contained five cells (to model a TE biopsy level of cells) and underwent the same Sureplex WGA and following Veriseq NGS analysis as applied to the studied samples. The data showed that no false-positive condition was observed in the 10 replicates, and thus the false positive rate in each chromosome was 0% (0/10). Of the validation curve, two cell lines were mixed: AF01 and AF02 (Supplementary Table S1). The trisomy 21 cell line (AF01), was serially diluted with the diploid cell line (AF02) in the following proportion: 100, 80, 60, 50, 40, 20, 10 and 0% of aneuploidy, in which the total number of cells was 10. The mosaic model was amplified and tested on the Veriseq NGS platform in triplicate. According to the obtained data of simulating chromosomal mosaicism, the lowest aneuploid percentage detectable was ~20% (mean 2.19 copies, standard deviation, SD = 0.035), while the highest mosaicism identifiable was 80% (mean 2.83 copies, SD = 0.095). Thus, the euploid range was defined as below 20% aneuploidy; aneuploid range was above 80% aneuploidy; and the range between 20 and 80% aneuploidy was diploid/aneuploid mosaic.
Determinations of confined mosaicism
Confined mosaicism is defined as mosaicism in either the TE or the ICM. Blastocysts having euploid TE with mosaic ICM, as well as mosaic TE with euploid ICM or with aneuploid ICM, might be transferred and could result in a PGS discrepancy in clinical practice. Additionally, the blastocysts of aneuploid TE with mosaic ICM may represent a biological source of false positives, with the potential to affect the clinical outcome of IVF treatment and lower the cumulative live birth rate. By contrast, blastocysts with inconsistent anomalies between the TE and ICM would not be transferred. Therefore, aneuploid-to-aneuploid mosaicism will not be discussed in this study.
Statistical analysis
The count data are presented as percentages, and the continuous data as averages with SDs. Comparisons of percentage distributions between the groups were analyzed by Chi-square or Fisher’s exact test. Significant differences were defined as a P-value <0.05. All of the analyses were generated using scientific GraphPad software (Prism; GraphPad Software, La Jolla, CA, USA).
Results
Patient profile
The patient profile was shown in Supplementary Table SII. Overall, 12 women were 26–43 years of age (mean age, 34.4 years), and included 2 patients with severe male factor, 4 at advanced maternal age (≥36 years), 2 with repeated implantation failure, 3 donated-oocyte recipients for single embryo transfer and 1 with an inherited chromosomal abnormality. A total of 33 surplus embryos were obtained, and 99 fractions were biopsied for the study. The unsuccessful extraction rate is 4.04% (4/99).
Similar euploid rates among three distinct biopsies
Thirty-three blastocysts were biopsied at three distinct positions: TE1 (opposite to the ICM), TE2 (near the ICM) and within the ICM (Supplementary Fig. S1). An overview result of ploidy is shown in Table I. For 29 embryos, we successfully extracted the DNAs from all three fractions; amplification failed in the TEs or ICM of four embryos. Of the Veriseq NGS result per fraction, neither TEs nor ICM displayed significantly higher euploid rates: TE1, 32% (10/31); TE2, 30% (10/33); and ICM, 29% (9/31) (P = 0.72). In the studied embryos, the majority were the group with more than one aneuploidy, and the second was euploid group.
Table I.
Overview of results per biopsied fraction.
| TE1 | TE2 | ICM | |
|---|---|---|---|
| Number of embryos | 33 | ||
| Number of analyzed biopsies | 31 | 33 | 31 |
| Failed amplifications | 2 | 0 | 2 |
| Euploid | 10 (32%) | 10 (30%) | 9 (29%) |
| Mosaica | 5 (16%) | 2 (6%) | 3 (10%) |
| Aneuploidb | 16 (52%) | 21 (64%) | 19 (61%) |
| Single | 5 (31%) | 6 (29%) | 7 (37%) |
| >1 aneuploidy | 10 (63%) | 13 (62%) | 10 (53%) |
| Segmentalc | 1 (6%) | 2 (10%) | 2 (11%) |
TE, trophectoderm; ICM, inner cell mass.
aMosaic was defined as between 20 and 80% aneuploidy.
bAneuploid was defined as above 80% aneuploidy.
cSegmental aneuploidy was defined as an affected length above 10 Mb.
Similar consistency of ploidy among different TE fractions and ICM
The concordance assessment of ploidy between two TE fractions, and between each TE fraction and ICM, are shown in Table II. No remarkable difference was observed between the TE biopsy sites in their overall ploidy consistency with the ICM: TE1-to-ICM, 86% (25/29) versus TE2-to-ICM, 90% (26/29), P= 1.0. Too few mosaic embryos were available to draw meaningful conclusions. The consistency of the aneuploid pattern, was similar among the different biopsy sites: TE1-to-TE2, 38% (11/29) versus TE1-to-ICM, 35% (10/29) versus TE2-to-ICM, 35% (10/29), P = 0.95.
Table II.
Concordance assessment of ploidy between fractions.
| TE1–TE2 | TE1–ICM | TE2–ICM | |
|---|---|---|---|
| Consistency of ploidy | 90% (26/29) | 86% (25/29) | 90% (26/29) |
| Euploid | 31% (9/29) | 28% (8/29) | 28% (8/29) |
| Aneuploid | 52% (15/29) | 52% (15/29) | 59% (17/29) |
| Mosaic | 7% (2/29) | 7% (2/29) | 3% (1/29) |
| Consistency of aneuploid patterna | 38% (11/29) | 35% (10/29) | 35% (10/29) |
TE, trophectoderm; ICM, inner cell mass.
aThe analyzed data include aneuploid and mosaic fractions.
No preferential allocation of euploid lineage
Table III shows the detailed PGS results in each biopsy site from 29 blastocysts. Nine patients had more than one embryo biopsied (75%) (9/12): four of them had one embryo with confined mosaicism between TE and ICM (33%) (4/12); two had only pure aneuploid embryos (17%) (2/12); and three had both the pure euploid/aneuploid embryos (25%) (3/12). The other three patients had only one embryo for the biopsy, and their embryos were all pure aneuploid (25%) (3/12). The ploidy of embryos shows different within the individuals. According to the aneuploid percentage in each fraction of the three diploid/aneuploid mosaic blastocysts (189-4 C, 717-6 C and 500-4 C), the euploid lineage displayed no preferential allocation. Moreover, of the embryos containing aneuploid lineages, a predominant aneuploidy was often observed among all the fractions within the same blastocyst. Thus, the chromosomal aneuploidies were mostly consistent rather than reciprocal in a particular embryo, with the exception of embryo 854-1 C.
Table III.
Per fraction next generation sequencing (NGS) results of all embryos.
| Patient number | Embryo number | Morphologya | Distantb trophectoderm TE1 | Closeb trophectoderm TE2 | Inner cell mass ICM | Confined mosaicismc |
|---|---|---|---|---|---|---|
| 189 | 189-1 C | 5BC | +2, +16p(70%) | +2 | +2 | No |
| 189-3 C | 5AB | −10(30%), −18(60%) | +18p, −18q | +18 | Yes | |
| 189-4 C | 5BC | +1pq(20%) | +1pq(60%) | +1pq(50%) | No | |
| 934 | 934-4 C | 5AB | +15, −21 | +15, −21 | +15, −21 | No |
| 934-6 C | 5BB | Eu | Eu | Eu | No | |
| 717 | 717-1 C | 5BC | Eu | Eu | Eu | No |
| 717-5 C | 5BB | Eu | Eu | Eu | No | |
| 717-6 C | 5BC | −5q(30%) | Eu | −5q(40%) | No | |
| 717-8 C | 5BB | Eu | Eu | Eu | No | |
| 717-11 C | 5BC | Eu | Eu | Eu | No | |
| 803 | 803-3 C | 4BC | +17 | +17 | +17 | No |
| 783 | 783-2 C | 5BB | −4p, +14, +19, −20 | −4p, +14, +19, −20 | −4p, +14, +19, −20 | No |
| 191 | 191-1 C | 5BB | −8 | −8 | −8 | No |
| 191-2 C | 5BB | Eu | Eu | Eu | No | |
| 851 | 851-2 C | 5BC | +20p(70%), −X | −X | −X | No |
| 581 | 581-1 C | 3BB | −9p, +9q(70%), +22 | −18, +22 | −9(30%), −18(60%), +22 | No |
| 581-2 C | 3BC | +7, +12, −13, −22 | +7, +12, −13, −22 | +7, +12, −13, −22 | No | |
| 581-3 C | 5BB | +13, +22 | +13, +22 | −9q(50%), +13, +22 | No | |
| 581-4 C | 5BB | −3p(20%), −14(50%), −15, +20 | −3p, −15, +20 | +9(60%), −15, +20 | No | |
| 581-5 C | 5BB | −7(30%), −18(20%), −22 | +10q(60%), −22 | −8(50%), −22 | No | |
| 854 | 854-1 C | 5BC | +18(30%) | −18(30%) | Eu | Yes |
| 854-4 C | 5BB | Eu | Eu | Eu | No | |
| 412 | 412-1 C | 5BB | +16 | +16 | +16 | No |
| 412-2 C | 5BB | −8 | −8 | −8, +22 | No | |
| 500 | 500-2 C | 5AB | +15p | +15p | +15p | No |
| 500-4 C | 5BB | Eu | Eu | −1(40%) | Yes | |
| 643 | 643-1 C | 4BB | +11q(70%), −Xq(40%) | −2q(30%), +11q | +11q | Yes |
| 643-2 C | 4BB | −X | −X, −10(25%) | −X | No | |
| 643-4 C | 5BC | Eu | Eu | Eu | No |
Eu, euploid. Where aneuploidy is present the chromosomes affected at the biopsy site are shown by number, if aneuploidy is present in all cells (defined as >80% aneuploidy) no percentage is shown but where the aneuploidy is mosaic (defined as 20–80% aneuploidy) the % aneuploidy is shown in parentheses.
aThe morphology of blastocysts is graded after Gardner and Schoolcraft (1999).
bWith respect to the inner cell mass (see Supplementary Fig. S1).
cConfined mosaicism is defined as inconsistent PGS conclusions between the TE and ICM within the same embryo. Embryos with inconsistent chromosomal ploidies but consistent PGS conclusions of the different biopsies, are not included.
PGS discrepancy caused by confined mosaicism
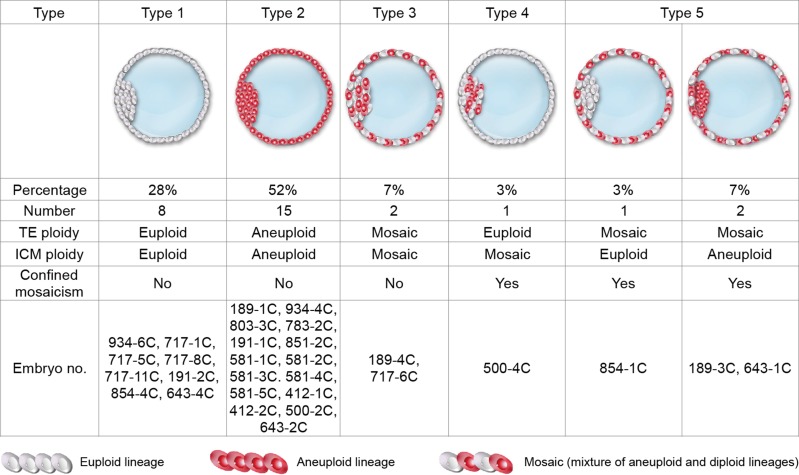
An overview diagrams of embryos with different types of mosaicism is summarized in Fig. 1. Eight euploid embryos and 15 aneuploid embryos resulted in consistent PGS conclusions for the TE and ICM fractions (Types 1 and 2). Of note, the actual chromosomal constitutions of each fraction within an embryo may not have been completely uniform in the above-mentioned 15 aneuploids because the aneuploid type or percentage of aneuploidy could be different (Table III), but no discrepancy of PGS conclusions in the same embryo occurred. Two embryos contained both the mosaic TE and mosaic ICM, and thus were categorized as general mosaicism: 189-4 C and 717-6 C, 7% (2/29) (Type 3). Approximately 86% of the population (25/29) had consistent PGS conclusions between the TE and ICM. Inconsistent PGS conclusions between the TE and ICM caused by confined mosaicism included (i) euploid TE with mosaic ICM: 500-4 C, 3% (1/29) (Type 4); and (ii) mosaic TE with euploid ICM: 854-1 C, 3% (1/29); or with aneuploid ICM: 189-3 C and 643-1 C, 7% (2/29) (Type 5). Based on Fig. 1, two types of inconsistent PGS conclusions between the TE and ICM due to confined mosaicism were observed (Types 4 and 5), and thus the overall rate of confined mosaicism was 14% (4/29). The example copy number plot for each type of mosaicism depicted in Fig. 1 was displayed in Supplementary Fig. S2.
Figure 1.
The types of concordance and non-concordance between trophectoderm (TE) and the inner cell mass (ICM) observed in the study.
Discussion
A concordance assessment between the three different biopsy sites in the same blastocyst was performed using the high-resolution Veriseq NGS. Based on the concordance analysis of TEs-to-ICM, no preferential allocation of the euploid lineage within a blastocyst was found, and thus the euploid rate was similar between TEs and ICM. Also, biopsy sites in the TE near to or far from the ICM, showed a similar chromosomal consistency rate, suggesting that the cells with different chromosomal components may spread randomly throughout the TE. Therefore, single biopsy samples from the TE are equally representative whether taken near to or far from the ICM. In addition, two types of PGS discrepancy between TE and ICM due to confined mosaicism were observed in the study, and their combined prevalence was 14%.
According to the present results, two forms of confined mosaicism could lead to the different reproductive issues: (i) confined mosaicism involving mosaic TE with euploid ICM (3%), which may result in normal embryos not being considered useful for transfer and being discarded; and (ii) confined mosaicism involving euploid TE with mosaic ICM (3%) or mosaic TE with aneuploid ICM (7%), which may lead to abnormal embryos being selected for transfer and to adverse clinical outcomes. The above-mentioned embryos were clearly of the aneuploid lineage, but detection could have been missed due to sampling bias. Aneuploid-to-aneuploid mosaicism was not discussed in this study, because the embryo would never be selected for transfer. General mosaicism involving both euploid and aneuploid lineages throughout the entire blastocyst was also observed (7%), and transfer of these embryos in clinical practice should be a concern due to the unknown effect during pregnancy. Furthermore, there were two embryos with higher percentages of mosaic aneuploidy in the TE (60–70% aneuploidy), and both embryos contained an aneuploid ICM (>80% aneuploidy), suggesting that the embryo with a higher percentage of mosaic aneuploidy in the TE biopsy should be ranked with lower priority in the clinical transfer because the chromosomal constitution of these embryos more likely had a severely abnormal pattern. Since the mosaic embryos are currently considered useful for transfer and are transferred resulting in healthy births (Greco et al., 2015), the correlation between the aneuploid percentage and transfer outcomes in mosaic embryos needs a further investigation.
Some authors considered the preferential allocation of euploid lineage in TE (Mantikou et al., 2012; Bazrgar et al., 2013). In the present study, no signs of preferential allocation of a euploid lineage were noted in the blastocysts, which implies a possible random distribution of cell lineages within an embryo. Therefore, the site of biopsy in the TE layer is not crucial for concordance with the ICM, while the incidence of confined mosaicism between the TE near to and far from the ICM were similar (14 versus 10%, P = 1.0).
Biologically, an early-stage aneuploid event occurring during mitosis could adversely affect embryo development and lead to arrest; however, the capacity of a cleavage-stage embryo containing an aneuploid lineage to develop normally could depend on the type of aneuploidy, the proportion of aneuploid blastomeres or occurrence of a correction. Mechanisms during mitosis by which chromosomal mosaicism can occur are: (i) non-disjunction: failure of sister chromatid separation; (ii) anaphase lagging: failure of a single chromatid incorporating into the nucleus; (iii) endo-replication: duplication of a chromosome without cell division; and (iv) trisomy rescue: trisomy rescue of meiotic errors occurring during the mitotic stage through anaphase lagging. In situation (i), a pair of cells with monosomy and triosomy would be created; in situation (ii), a pair of cells with monosomy and disomy would be created; in situation (iii), a pair of cells with trisomy and disomy would be created; and in situation (iv), a pair of cells with trisomy and uniparental disomy would be created (Taylor et al., 2014). Nevertheless, some embryos with such aneuploid cells could still grow into the blastocysts (Bielanska et al., 2002). A predominant aneuploidy is often observed within all the biopsied fractions in the same blastocyst according to the results shown in our study. This predominant aneuploidy could be a consequence of an early aneuploid event occurring in the meiotic origin or in the very beginning of embryo cleavage, and then be tolerated by several cell-cycle checkpoints. During the following mitotic process, more malsegregations could be introduced, and thus divergent aneuploid patterns involving a predominant aneuploidy are formed in the blastocyst stage (Maurer et al., 2015). The aneuploid–aneuploid mosaicism could be observed in this condition. In contrast, if an aneuploid event occurs during the later stages in rare cells of an embryo, and the cell death or reduced proliferation of this aneuploid lineage restricted distribution to a further extent in later development, the blastocyst stage could involve a simply low-rate aneuploid pattern, which diploid–aneuploid mosaicism could be observed.
The fate of mosaicism is an uncertain issue with respect to preimplantation and prenatal testing. Although the incidence of mosaicism in preimplantation embryos is 15–90% depending on the embryo stages or detection tools, the incidence of mosaicism in post-implantation embryos was much lower (Taylor et al., 2014). Accordingly, the mosaicism rate is 1–2% in chronic villous sampling, which is mostly confined to the placenta (Battaglia et al., 2014; Malvestiti et al., 2015). In amniocentesis, the mosaicism rate has been reported to be merely 0.2%, which could not be confirmed at birth (Winsor et al., 1999). Additionally, a mouse model of chromosomal mosaicism displayed a possible mechanism of aneuploidy rescue by lineage-specific depletion of aneuploid cells: ICM, eliminated by apoptosis; TE, showing severe proliferative defects (Bolton et al., 2016). These data implied a possible selection against mosaic aneuploidy during the development.
Using the NGS technology in PGS, the identification of diploid/aneuploid mosaicism in a multi-cellular biopsied sample was significantly improved; however, the accuracy of mosaicism detection in PGS has been challenged by several biological biases, including the possibility of sampling and reciprocal errors (Scott and Galliano, 2016). The sampling error concerned the representative value of a simple TE biopsy in a blastocyst, and the reciprocal error was related to the possible compromising results of a trisomy/monosomy mixture. In our study, the incidence of inconsistent PGS conclusions between TE and ICM due to sampling error in embryos with confined mosaicism was 13.8%, and the possibility of reciprocal error was 3.4%. Up to 86.2% of the studied embryos exhibited consistent PGS conclusions between the TE and ICM. According to the live births report of mosaic embryo transfer (Greco et al., 2015), the outcomes were highly diverse in individuals. The prevalence of a confined pattern may exist in these mosaic embryos, and those diagnosed as mosaic in TE could contain a euploid, a mosaic or an aneuploid ICM. The results of present study could correlate with this consideration.
It is noteworthy that the possibility of cell contamination between TE and ICM could not be completely avoided due to the limitations of biopsy manipulation procedures. Of the biological limitations, it is quite possible that when ICM aneuploidy or mosaicism was noted and discrepant from the TE biopsy that those two sampled sites simply did not contain the abnormality, since only two TE biopsy sites were evaluated in this study. The term confined mosaicism implies that the entirety of the embryo was evaluated, but It was not performed here, since we would like to simulate the clinical biopsy procedures. Of the technical limitations, it is critical to temper the interpretation of results as it is well established that intermediate copy numbers can originate from phenomena other than actual mosaicism (i.e. technical artifacts). Using the same platform, Goodrich et al. (2016, 2017) detected aneuploidy in mosaic samples, demonstrating the potential of Veriseq NGS-based detection methods to detect aneuploidy in mosaicism within a biopsy. They emphasized the importance of preclinical evaluation of testing criteria prior to clinical implementation, and a balance between sensitivity and specificity to improve detection of mosaicism within preimplantation embryos. As to the internal validation of mosaicism, caution should be used in extrapolating results from standard dilutions of immortalized cell lines to primary cells. The primary cells would be more appropriate for the preclinical validation of Veriseq NGS system to detect mosaicism than the amniotic stem cell lines used in the present study. Moreover, it would be interesting to use SNPs to investigate the detected aneuploidy segment whether the aneuploidy seen is preferentially from one or the other allele or random. Besides, the preliminary result of confined mosaicism rate in the study is affected by the small sample size, and it is mandatory to increase the number of studied sample regarding to statistical power before drawing solid conclusions. To date, it is not clear what reproductive potential is conferred in the circumstance of confined mosaicism, and thus it is worthy of an investigation for the clinical effect.
In conclusion, the current study demonstrated that no preferential allocation of the euploid lineage within a particular blastocyst was observed. Regardless of the biopsy site in the TE near to or far from the ICM, leverage to chromosomal consistency was not shown. Additionally, the incidence of confined mosaicism between the TE and ICM was 14%, and it may cause discrepant conclusions in PGS.
Supplementary Material
Authors’ roles
T.C. designed the study concept and wrote the article; J.H. conducted the CCS and analyzed the data; M.L., C.H., H.W. and H.L. recruited the patients; Y.C. kindly provided the cell lines and the method of mosaicism simulating experiment; S.C. reviewed the article, revised the final version; and all the authors approved the submitted version.
Funding
No external funding was obtained for this study.
Conflict of interest
All the authors declare no conflicts of interest regarding this study.
References
- Battaglia P, Baroncini A, Mattarozzi A, Baccolini I, Capucci A, Spada F, Pompilii E, Pittalis MC. Cytogenetic follow-up of chromosomal mosaicism detected in first-trimester prenatal diagnosis. Prenat Diagn 2014;34:739–747. [DOI] [PubMed] [Google Scholar]
- Bazrgar M, Gourabi H, Valojerdi MR, Yazdi PE, Baharvand H. Self-correction of chromosomal abnormalities in human preimplantation embryos and embryonic stem cells. Stem Cells Dev 2013;22:2449–2456. [DOI] [PubMed] [Google Scholar]
- Bielanska M, Tan SL, Ao A. Chromosomal mosaicism throughout human preimplantation development in vitro: incidence, type, and relevance to embryo outcome. Hum Reprod 2002;17:413–419. [DOI] [PubMed] [Google Scholar]
- Bolton H, Graham SJ, Van der Aa N, Kumar P, Theunis K, Fernandez Gallardo E, Voet T, Zernicka-Goetz M. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun 2016;7:11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capalbo A, Bono S, Spizichino L, Biricik A, Baldi M, Colamaria S, Ubaldi FM, Rienzi L, Fiorentino F. Sequential comprehensive chromosome analysis of polar bodies, blastomeres and trophoblast: insights into female meiotic errors and chromosomal segregation in the preimplantation window of embryo development. Hum Reprod 2013;28:509–518. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Ubaldi FM, Rienzi L, Scott R, Treff N. Detecting mosaicism in trophectoderm biopsies: current challenges and future possibilities. Hum Reprod 2017;32:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HF, Chen SU, Ma GC, Hsieh ST, Tsai HD, Yang YS, Chen M. Preimplantation genetic diagnosis and screening: current status and future challenges. J Formos Med Assoc 2017;117:94–100. [DOI] [PubMed] [Google Scholar]
- Coonen E, Derhaag JG, Dumoulin JCM, van Wissen LCP, Bras M, Janssen M, Evers LHJ, Geraedts E. Anaphase lagging mainly explains chromosomal mosaicism in human preimplantation embryos. Hum Reprod 2004;19:316–324. [DOI] [PubMed] [Google Scholar]
- Delhanty JD, Harper JC, Ao A, Handyside AH, Winston RM. Multicolour FISH detects frequent chromosomal mosaicism and chaotic division in normal preimplantation embryos from fertile patients. Hum Genet 1997;99:755–760. [DOI] [PubMed] [Google Scholar]
- Demko ZP, Simon AL, McCoy RC, Petrov DA, Rabinowitz M. Effects of maternal age on euploidy rates in a large cohort of embryos analyzed with 24-chromosome single-nucleotide polymorphism-based preimplantation genetic screening. Fertil Steril 2016;105:1307–1313. [DOI] [PubMed] [Google Scholar]
- Fiorentino F, Biricick A, Bono S, Greco E, Minasi MG, Ruberti A, Spinella F Clinical outcome derived after transfer of embryos with chromosomal mosaicism. European Society of Human Reproduction and Embryology 2016, ESHRE abstract of oral presentation (O-028).
- Fiorentino F, Biricik A, Bono S, Spizzichino L, Cotroneo E, Cottone G, Kokocinski F, Michel CE. Development and validation of a next-generation sequencing (NGS)-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil Steril 2014. a;101:1375–1382. [DOI] [PubMed] [Google Scholar]
- Fiorentino F, Bono S, Biricik A, Nuccitelli A, Cotroneo E, Cottone G, Kokocinski F, Michel CE, Minasi MG, Greco E. Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum Reprod 2014. b;29:2802–2813. [DOI] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development 2013;140:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T, Gordon A, Wells D. Cytogenetic analysis of blastocysts with use of FISH, CHG, and aCGH: scientific data and technical evaluation. Hum Reprod 2011;26:480–490. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Simpkins M, Cutts G, Spath K, Babariya D, Kubikova N, Rubistello L, Munne S, Wells D. Factors affecting embryonic mosaicism. Hum Reprod 2017;32:i49. [Google Scholar]
- Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod 2008;23:2596–2608. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Schoolcraft WB. Culture and transfer of human blastocysts. Curr Opin Obstet Gynecol 1999;11:307–311. [DOI] [PubMed] [Google Scholar]
- Goodrich D, Tao X, Bohrer C, Lonczak A, Xing T, Zimmerman R, Zhan Y, Scott RT Jr, Treff NR. A randomized and blinded comparison of qPCR and NGS-based detection of aneuploidy in a cell line mixture model of blastocyst biopsy mosaicism. J Assist Reprod Genet 2016;33:1473–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich D, Xing T, Tao X, Lonczak A, Zhan Y, Landis J, Zimmerman R, Scott RT Jr, Treff NR. Evaluation of comprehensive chromosome screening platforms for the detection of mosaic segmental aneuploidy. J Assist Reprod Genet 2017;34:975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco E, Bono S, Ruberti A, Lobascio AM, Greco P, Biricik A, Spizzichino L, Greco A, Tesarik J, Minasi MG et al. Comparative genomic hybridization selection of blastocysts for repeated implantation failure treatment: a pilot study. Biomed Res Int 2014;2014:457913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco E, Minasi MG, Fiorentino F. Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. N Engl J Med 2015;373:2089–2090. [DOI] [PubMed] [Google Scholar]
- Gueye NA, Devkota B, Taylor D, Pfundt R, Scott RT Jr, Treff NR. Uniparental disomy in the human blastocyst is exceedingly rare. Fertil Steril 2014;101:232–236. [DOI] [PubMed] [Google Scholar]
- Gutierez-Mateo C, Colls P, Sanchez-Garcia J, Escudero T, Prates R, Ketterson K, Wells D, Munné S. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril 2011;95:953–958. [DOI] [PubMed] [Google Scholar]
- Huang CC, Chang LJ, Tsai YY, Hung CC, Fang MY, Su YN, Chen HF, Chen SU. A feasible strategy of preimplantation genetic diagnosis for carriers with chromosomal translocation: using blastocyst biopsy and array comparative genomic hybridization. J Formos Med Assoc 2013;112:537–544. [DOI] [PubMed] [Google Scholar]
- Hwang JL, Chen SU, Chen HJ, Chen HF, Yang YS, Chang CH, Seow KM, Tzeng CR, Lin YH. Feasibility of corifollitropin alfa/GnRH antagonist protocol combined with GnRH agonist triggering and freeze-all strategy in polycystic ovary syndrome patients. J Formos Med Assoc 2018;117:535–540. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Cinnioglu C, Ross R, Filby A, Gemelos G, Hill M, Ryan A, Smortrich D, Rabinowitz M, Murray MJ. Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Mol Hum Reprod 2010;16:944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousek DK, Barrett IJ, Gartner AB. Spontaneous abortion and confined placental mosaicism. Hum Genet 1992;88:642–646. [DOI] [PubMed] [Google Scholar]
- Lai HH, Chuang TH, Wong LK, Lee MJ, Hsieh CL, Wang HL, Chen SU. Identification of mosaic and segmental aneuploidies by next-generation sequencing in preimplantation genetic screening can improve clinical outcomes compared to array-comparative genomic hybridization. Mol Cytogenet 2017;10:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang W, Sun X, Liu L, Jin H, Li M, Witz C, Williams D, Griffith J, Skorupski J et al. DNA microarray reveals that high proportions of human blastocysts from women of advanced maternal age are aneuploid and mosaic. Biol Reprod 2012;87:148. [DOI] [PubMed] [Google Scholar]
- Malvestiti F, Agrati C, Grimi B, Pompilii E, Izzi C, Martinoni L, Gaetani E, Liuti MR, Trotta A, Maggi F et al. Interpreting mosaicism in chorionic villi: results of a monocentric series of 1001 mosaics in chorionic villi with follow-up amniocentesis. Prenat Diagn 2015;35:1117–1127. [DOI] [PubMed] [Google Scholar]
- Mantikou E, Wong KM, Repping S, Mastenbroek S. Molecular origin of mitotic aneuploidies in preimplantation embryos. Biochim Biophys Acta 2012;1822:1921–1930. [DOI] [PubMed] [Google Scholar]
- Maurer M, Ebner T, Puchner M, Mayer RB, Shebl O, Oppelt P, Duba HC. Chromosomal aneuploidies and early embryonic developmental arrest. Int J Fertil Steril 2015;9:346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell SM, Colls P, Hodes-Wertz B, McCulloh DH, McCaffrey C, Wells D, Munné S, Grifo JA. Why do euploid embryos miscarry? A case-control study comparing the rate of aneuploidy within presumed euploid embryos that resulted in miscarriage or live birth using next-generation sequencing. Fertil Steril 2016;106:1414–1419. [DOI] [PubMed] [Google Scholar]
- Munné S, Grifo J, Wells D. Mosaicism: ‘survival of the fittest’ versus ‘no embryo left behind. Fertil Steril 2016;105:1146–1149. [DOI] [PubMed] [Google Scholar]
- Munné S, Sandalinas M, Escudero T, Màrquez C, Cohen J. Chromosome mosaicism in cleavage-stage human embryos: evidence of a maternal age effect. Reprod Biomed Online 2002;4:223–232. [DOI] [PubMed] [Google Scholar]
- Schoolcraft W, Treff N, Ferry K, Stevens J, Katz-Jaffe M, Scott R. First clinical application of SNP microarray based 24 chromosome aneuploidy screening of human blastocysts. Fertil Steril 2010;94:2017–2021.20188357 [Google Scholar]
- Scott RT, Galliano D. The challenge of embryonic mosaicism in preimplantation genetic screening. Fertil Steril 2016;105:1150–1152. [DOI] [PubMed] [Google Scholar]
- Sermon K, Capalbo A, Cohen J, Coonen E, De Rycke M, De Vos A, Delhanty J, Fiorentino F, Gleicher N, Griesinger G et al. The why, the how and the when of PGS 2.0: current practices and expert opinions of fertility specialists, molecular biologists, and embryologists. Mol Hum Reprod 2016;22:845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TH, Gitlin SA, Patrick JL, Crain JL, Wilson JM, Griffin DK. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum Reprod Update 2014;20:571–581. [DOI] [PubMed] [Google Scholar]
- Treff NR, Franasiak JM. Detection of segmental aneuploidy and mosaicism in the human preimplantation embryo: technical considerations and limitations. Fertil Steril 2017;107:27–31. [DOI] [PubMed] [Google Scholar]
- Tsai MS, Hwang SM, Chen KD, Lee YS, Hsu LW, Chang YJ, Wang CN, Peng HH, Chang YL, Chao AS et al. Functional network analysis of the transcriptomes of mesenchymal stem cells derived from amniotic fluid, amniotic membrane, cord blood, and bone marrow. Stem Cells 2007;25:2511–2523. [DOI] [PubMed] [Google Scholar]
- Vera-Rodríguez M, Michel CE, Mercader A, Bladon AJ, Rodrigo L, Kokocinski F, Mateu E, Al-Asmar N, Blesa D, Simón C et al. Distribution patterns of segmental aneuploidies in human blastocysts identified by next-generation sequencing. Fertil Steril 2016;105:1047–1055. [DOI] [PubMed] [Google Scholar]
- Wang HL, Lai HH, Chuang TH, Shih YW, Huang SC, Lee MJ, Chen SU. A patient friendly corifollitropin alfa protocol without routine pituitary suppression in normal responders. PLoS One 2016;11:e0154123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D, Babariya D, Alfarawati S, Spath K, Kubikova N, Munne S, Fragouli E Frequency and clinical relevance of mosaic segmental aneuploidy in blastocyst stage human embryos. 2017. ESHRE abstract of oral presentation (O-112).
- Wells D, Kaur K, Grifo J, Glassner M, Taylor JC, Fragouli E, Munne S. Clinical utilization of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J Med Genet 2014;51:553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor EJ, Tomkins DJ, Kalousek D, Farrell S, Wyatt P, Fan YS, Carter R, Wang H, Dallaire L, Eydoux P et al. Cytogenetic aspects of the Canadian early and mid-trimester amniotic fluid trial (CEMAT). Prenat Diagn 1999;19:620–627. [DOI] [PubMed] [Google Scholar]
- Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, Peck AC, Sills ES, Salem RD. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet 2012;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.