ABSTRACT
Although GCN5L1 (also known as BLOC1S1) facilitates mitochondrial protein acetylation and controls endosomal-lysosomal trafficking, the mechanisms underpinning these disparate effects are unclear. As microtubule acetylation modulates endosome-lysosome trafficking, we reasoned that exploring the role of GCN5L1 in this biology may enhance our understanding of GCN5L1-mediated protein acetylation. We show that α-tubulin acetylation is reduced in GCN5L1-knockout hepatocytes and restored by GCN5L1 reconstitution. Furthermore, GCN5L1 binds to the α-tubulin acetyltransferase αTAT1, and GCN5L1-mediated α-tubulin acetylation is dependent on αTAT1. Given that cytosolic GCN5L1 has been identified as a component of numerous multiprotein complexes, we explored whether novel interacting partners contribute to this regulation. We identify RanBP2 as a novel interacting partner of GCN5L1 and αTAT1. Genetic silencing of RanBP2 phenocopies GCN5L1 depletion by reducing α-tubulin acetylation, and we find that RanBP2 possesses a tubulin-binding domain, which recruits GCN5L1 to α-tubulin. Finally, we find that genetic depletion of GCN5L1 promotes perinuclear lysosome accumulation and histone deacetylase inhibition partially restores lysosomal positioning. We conclude that the interactions of GCN5L1, RanBP2 and αTAT1 function in concert to control α-tubulin acetylation and may contribute towards the regulation of cellular lysosome positioning.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: GCN5L1, α-TAT1, RanBP2, Nup358, Microtubule acetylation, Lysosome positioning
Summary: GCN5L1 interacts with α-tubulin acetyltransferase. In parallel, the nucleoporin RanBP2 facilitates binding of GCN5L1 to α-tubulin to orchestrate microtubule acetylation.
INTRODUCTION
GCN5L1 (also known as BLOC1S1) was identified as a protein with sequence homology to the nuclear acetyl transferase general control of amino acid synthesis 5 (Gcn5) (Driessen et al., 1997; Inoue et al., 1996). Subsequent studies showed that the genetic depletion of GCN5L1 reduced mitochondrial protein acetylation (Fukushima et al., 2016; Scott et al., 2012; Webster et al., 2013, 2014), and that GCN5L1 was enriched in mitochondria and interacted with canonical substrates of the established mitochondrial deacetylase (SIRT3) (Scott et al., 2012). Interestingly, short-term GCN5L1 depletion resulted in the induction of restricted mitochondrial autophagy (mitophagy) in parallel with enhanced mitochondrial protein turnover and a reduction in overall mitochondrial content and cellular oxidative metabolism (Webster et al., 2013). In contrast, primary GCN5L1-knockout MEF cells retained their mitochondrial content with a concurrent induction of mitophagy and mitochondrial biogenesis regulatory programs (Webster et al., 2014). Additional studies found that GCN5L1 modulated mitochondrial control of fat oxidation (Fukushima et al., 2016; Thapa et al., 2017) and mitochondrial-linked regulation of gluconeogenesis (Wang et al., 2017). Recently, the first validated lysine target of GCN5L1 was identified, namely, acetylation of the mitochondrial-associated kinesis Kif1Bα-binding protein (KBP), which required cooperation between GCN5L1 and the mitochondrial acetyl-CoA-generating enzyme L-threonine dehydrogenase (TDH) (Donato et al., 2017).
In parallel studies, GCN5L1 was found to interact with the multiprotein biogenesis of lysosome-related organelles complex-1 (BLOC-1) and was alternatively named as a BLOC-1 subunit (i.e. BLOS1 or BLOC1S1) (Starcevic and Dell'Angelica, 2004). Mutations in numerous BLOC-1 complex subunits, but not in GCN5L1 itself, result in the development of the Hermansky–Pudlak syndrome (Chiang et al., 2003; Li et al., 2003), a syndrome linked with abnormal vesicular trafficking to lysosome and related organelles such as melanosomes. Interestingly, Drosophila GCN5L1 deficiency produced eye pigment defects that implicated this protein in the control of melanosome trafficking (Cheli et al., 2010). Although the germline knockout of GCN5L1 in mice is embryonic lethal (Webster et al., 2014; Zhang et al., 2014), E14.5 embryos showed defects in eye pigmentation (Zhang et al., 2014) and the exploration of epidermal growth factor receptor (EGFR) endolysosomal trafficking demonstrated that GCN5L1 interacted with sorting nexin 2 (SNX2) and an endosomal sorting complex component TSG101, to modulate EGFR trafficking (Zhang et al., 2014). More recently, GCN5L1 was found to associate with BORC, a distinct multiprotein complex that regulates lysosome positioning (Pu et al., 2015). Although BORC has some overlapping proteins with the BLOC-1 complex, most subunits are unique. Nevertheless, the genetic knockdown of GCN5L1 phenocopied the effect of depletion of other BORC subunits and resulted in reduced dissemination of lysosomes to the periphery of cells (Pu et al., 2015).
Together these studies point to differential roles of GCN5L1 in distinct subcellular compartments that span from acetylation dependent effects on mitochondrial homeostasis and metabolism to impaired endosomal lysosome trafficking and function. Given that the acetylation of microtubules plays an important role in endosome-lysosome trafficking (Perdiz et al., 2011; Xu et al., 2017), we reasoned that a common link between the diverse effects/phenotypes of GCN5L1 may, in part, be due to the role of GCN5L1-mediated acetylation in modulating organelle positioning.
In this study, we confirm that the absence of GCN5L1 results in impaired centrifugal movement of lysosomes. Furthermore, we show that GCN5L1 modulates lysosomal positioning via the regulation of microtubule acetylation in concert with the canonical α-tubulin acetyltransferase αTAT1. Finally, we show that this regulation is dependent on the RAN-binding protein 2 (RanBP2), a component of nuclear pore complexes also known as Nup358, which facilitates the interaction of GCN5L1 with α-tubulin. This study identifies a novel mechanism underpinning the role of GCN5L1 in protein acetylation due to its interaction with αTAT1, and expands our understanding of its role in the regulatory control of lysosome positioning. Furthermore, this study uncovers a potential mechanism for the previously identified role of RanBP2 in the modulation of organelle positioning.
RESULTS
GCN5L1 regulates the acetylation of α-tubulin
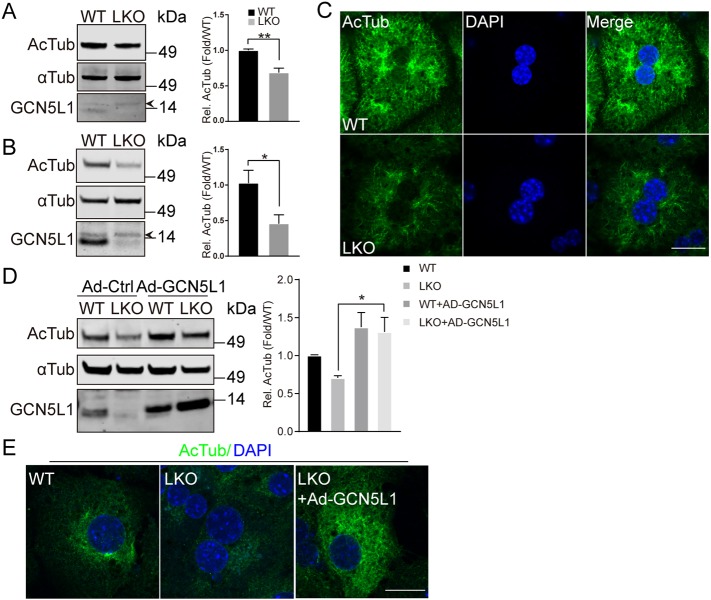
To explore whether GCN5L1 levels effect microtubule acetylation, we used liver homogenates from wild-type (WT) and liver-specific GCN5L1 (LKO) knockout mice (Wang et al., 2017). LKO mice show a significant reduction in α-tubulin acetylation on its cognate functional K40 residue (LeDizet and Piperno, 1991) (Fig. 1A). This reduction in K40 acetylation of α-tubulin was even more readily apparent in LKO primary hepatocytes (Fig. 1B). Immunofluorescence images also showed reduced α-tubulin acetylation without evidence of changes in microtubule distribution in primary LKO hepatocytes (Fig. 1C). Furthermore, adenovirus-mediated overexpression of GCN5L1 rescued LKO hepatocyte α-tubulin acetylation as evident by immunoblotting and immunofluorescence experiments (Fig. 1D,E). To reduce the potential for a non-specific effect of GCN5L1 overexpression, the levels of α-tubulin acetylation was assayed in response to an incremental 4-fold reduction in the multiplicity of infection (MOI) of adenoviral infection. All doses of GCN5L1 induced α-tubulin acetylation (Fig. S1A), supporting the idea that this is a functional effect. Taken together, these findings support that GCN5L1 plays a regulatory role in the modulation of microtubule acetylation.
Fig. 1.
GCN5L1 deficiency impairs the acetylation of α-tubulin. (A,B) Reduced acetylated α-tubulin (Ac-Tub) levels are detected by immunoblot analysis in liver homogenates (A) and primary hepatocytes lysates (B) from GCN5L1-knockout (LKO) mice compared to WT mice. The antibody directed against α-tubulin (αTub) was used as loading control and the antibody directed against GCN5L1 to show the mouse genotype. Of note, a non-specific band (depicted with an arrowhead) was evident at a higher molecular mass than the specific GCN5L1 band. The accompanying histogram represents the relative (Rel) quantified ratio of ac-Tub to α-Tub normalized to the WT samples from three pairs of mouse livers. (C) GCN5L1-knockout causes reduced Ac-Tub levels in primary hepatocytes. Primary hepatocytes from WT and LKO mice were immunostained for endogenous Ac-Tub (green). DNA was visualized by DAPI staining (blue). Scale bar: 20 µm. (D,E) Overexpression of exogenous GCN5L1 restored Ac-Tub levels in LKO hepatocytes. WT and LKO hepatocytes were infected with control adenovirus (Ad-Ctrl) or with adenovirus coding for WT GCN5L1 (Ad-GCN5L1). (D) Ac-Tub levels were analyzed by immunoblotting. A quantification of the relative levels from three independent experiments is also shown. (E) Immunostaining as in C. Scale bar: 20 µm. Representative images are shown from three independent experiments. Data were expressed as mean±s.e.m. *P<0.05; **P<0.01 (unpaired Student's t-test).
GCN5L1 modulation of α-tubulin acetylation is independent of acetyl-CoA levels
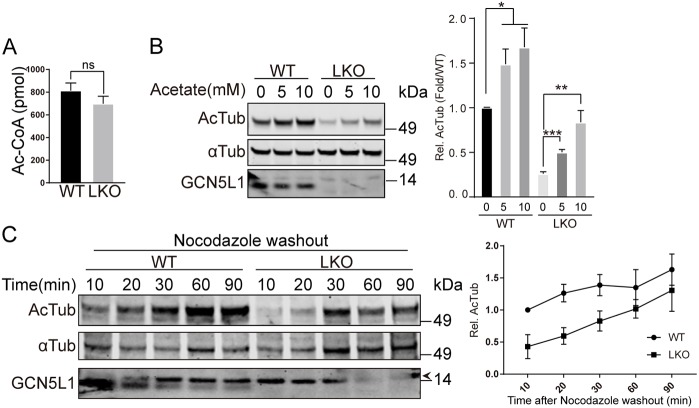
The concept that non-enzymatic acetylation of proteins is operational in mitochondria (Wagner and Hirschey, 2014) and in the cytosol (Moussaieff et al., 2015) is being actively investigated. Given this, and the known requirement of mitochondrial GNC5L1 to partner with an acetyl-CoA-generating enzyme to facilitate acetylation on the mitochondrial membrane (Donato et al., 2017), we then determined whether cellular levels of acetyl-CoA in hepatocytes could underpin the effect of GCN5L1 on α-tubulin acetylation. However, the levels of acetyl-CoA were not different when comparing WT and LKO primary hepatocytes as measured by using high-pressure liquid chromatography (HPLC) and liquid chromatography mass spectrometry (LC-MS) acetyl-CoA assays (Fig. 2A; Fig. S2A). Interestingly, this finding did not preclude acetyl-CoA level-dependent acetylation of α-tubulin, given that acetate and ethanol, which both increase hepatic intracellular acetyl-CoA levels (Cai et al., 2011; Cederbaum, 2012), increased the acetylation of α-tubulin irrespective of the presence or absence of GCN5L1 (Fig. 2B; Fig. S2B). Here, despite the low basal level of α-tubulin acetylation in the LKO hepatocytes, their parallel dose-dependent increase in acetylation supports the idea that the GCN5L1 effect is not-dependent on cellular acetyl-CoA levels within the physiological range. To validate that GCN5L1 did not modulate the kinetics of microtubule acetylation, but rather the absolute extent of α-tubulin acetylation, we employed nocodazole to depolymerize and deacetylate microtubules (Chabin-Brion et al., 2001; Perdiz et al., 2011). The kinetics of the restoration of α-tubulin acetylation was then observed following the washout of nocodazole. Again, we show that the kinetics of α-tubulin acetylation was similar in the presence or absence of GCN5L1, but that the absolute level of acetylation was lower in the GCN5L1-knockout hepatocytes (Fig. 2C). Taken together, these data suggest that the effect of GCN5L1 on microtubule acetylation within the cytosol requires a ‘catalytic’ rather than a non-enzymatic mass action effect.
Fig. 2.
GCN5L1 modulation of α-tubulin acetylation is independent of cellular acetyl-CoA levels. (A) The whole-cell levels of acetyl-CoA (Ac-CoA) were similar in GCN5L1 LKO and WT hepatocytes. Ac-CoA content was quantified by HPLC as described in the Materials and Methods. (B,C) The supplementation of media with acetate (B) increased acetylated α-tubulin (Ac-Tub) levels in WT and LKO hepatocytes and levels of acetylation of α-tubulin (αTub) were assessed using immunoblot analysis and quantification. α-Tubulin was used as a loading control. (C) Ac-Tub recovery kinetics in response to nocodazole was similar in WT and LKO hepatocytes. αTub acetylation was assessed after an initial exposure to nocodazole for 5 h and then, following washout, repolymerization is associated with reacetylation of α-tubulin. The extent of acetylation was assessed over a 90 min period by following the level of the acetylation of α-tubulin by immunoblot analysis. The accompanying line chart represents the relative quantified ratio of Ac-Tub to αTub normalized to the initial WT samples. Results are mean±s.e.m. of three independent experiments. α-Tubulin was used as loading control. Representative images are shown from three independent experiments. *P<0.05; **P<0.01; ***P<0.001; ns, not significant (unpaired Student's t-test).
GCN5L1 interacts with the canonical α-tubulin acetyltransferase αTAT1
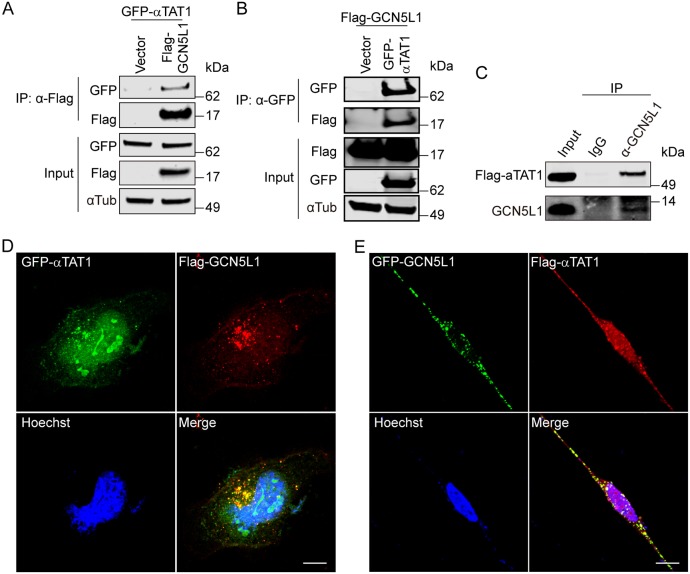
Since GCN5L1 does not contain a canonical acetyltransferase catalytic domain, and nor does it modulate α-tubulin acetylation in an acetyl-CoA dose-dependent manner or facilitate the α-tubulin acetylation in vitro (Fig. S3A), we explored potential GCN5L1 partners that would facilitate α-tubulin acetylation. Since αTAT1 is a major α-tubulin acetyltransferase, we tested whether it interacts with GCN5L1 (Akella et al., 2010; Kalebic et al., 2013). Bidirectional co-immunoprecipitation studies were performed following overexpression of GFP-tagged αTAT1 and Flag-tagged GCN5L1 in HEK293 cells. Immunoprecipitation with an anti-Flag antibody (Fig. 3A), or with an antibody directed against GFP (Fig. 3B) reveal a clear interaction between overexpressed GCN5L1 and αTAT1. To further validate this interaction, an antibody directed against endogenous GCN5L1 was used to immunoprecipitate Flag-tagged αTAT1 (Fig. 3C). To confirm this interaction, confocal microscopy was performed in HeLa cells following the overexpression of GFP–αTAT1 and Flag–GCN5L1 (Fig. 3D), or Flag–αTAT1 and GFP–GCN5L1 (Fig. 3E). Following cell fixation and immunostaining, the overlay of reciprocal GFP and Flag images show significant overlap between GCN5L1 and αTAT1 in the cytoplasm.
Fig. 3.
GCN5L1 interacts with the canonical α-tubulin acetyltransferase αTAT1. (A,B) Co-immunoprecipitation studies revealed an association between GCN5L1 and αTAT1. 293T cells were co-transfected with GFP–αTAT1 plus control vector (Vector) or Flag–GCN5L1 (A), or co-transfected with Flag–GCN5L1 plus control vector (Vector) or GFP–αTAT1 (B), then anti-Flag (A), or anti-GFP (B) immunoprecipitates (IP) were subjected to immunoblot analysis with anti-Flag, anti-GFP and anti-α-tubulin antibody. α-Tubulin was used as loading control. (C) The interaction between GCN5L1 and αTAT1 was confirmed by the immunoprecipitation of endogenous GCN5L1 with Flag–αTAT1 versus an IgG control. (D,E) Confocal microscopy showed colocalization of GCN5L1 with αTAT1. HeLa cells were co-transfected with GFP–αTAT1 (green) and Flag–GCN5L1 (red) (D), or co-transfected with GFP–GCN5L1 (green) and Flag–αTAT1 (red) (E), then analyzed for the localization using anti-Flag antibodies. GFP fluorescence was visualized directly. DNA was visualized by DAPI staining (blue). Scale bars: 10 µm. The confocal images are a representative image of three independent experiments.
GCN5L1 modulation of α-tubulin acetylation is dependent on α-tubulin acetyltransferase
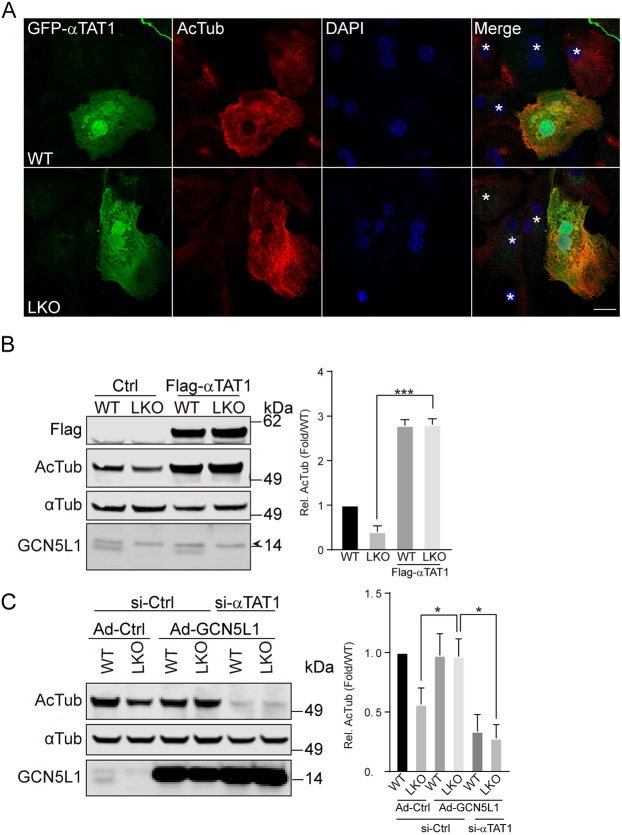
To evaluate whether this interaction between GCN5L1 and αTAT1 is functional, a Flag-tagged αTAT1 expression construct was transfected into WT and GCN5L1 KO hepatocytes. Confocal microscopy shows that the αTAT1-transfected cells have higher levels of acetylated α-tubulin in the WT and LKO cells (Fig. 4A). In contrast, in non-transfected cells (shown with an asterisk in the merged panels), the levels of acetylated α-tubulin are higher in the WT compared to the KO cells (Fig. 4A). To quantify these differences, the levels of acetylated α-tubulin was assessed by immunoblot analysis. As shown previously, non-transfected LKO cells had lower levels of acetylated α-tubulin compared to non-transfected WT cells, and the overexpression of αTAT1 increased acetylated tubulin levels in both WT and KO cells (Fig. 4B). We then explored whether the genetic knockdown of αTAT1 could abrogate the rescue of α-tubulin acetylation following reconstitution of GCN5L1 in LKO hepatocytes. This concept was validated by the observation that the α-tubulin acetylation in LKO cells was not restored upon GCN5L1 adenovirus infection when siRNA targeting αTAT1 was concurrently transfected (Fig. 4C). Similarly, WT cells transduced with GCN5L1 adenovirus had diminished levels of acetylated tubulin in the presence of αTAT1 siRNA (Fig. 4C).
Fig. 4.
GCN5L1 modulation of α-tubulin acetylation is dependent on αTAT1. (A,B) Overexpression of αTAT1 rescued the reduced levels of Ac-Tub in LKO hepatocytes as detected by immunostaining (A) and immunoblot analysis (B). WT and LKO hepatocytes were transfected with GFP–αTAT1 (green) (A) or Flag–αTAT1 (B) and analyzed for endogenous Ac-Tub (red) by immunostaining (A) or immunoblot (B) using specific antibodies. DNA was visualized by DAPI staining (blue), and non-transfected cells are shown with an asterisk (*) in the merge panels (A). Scale bar: 10 µm. The antibody directed against Flag was used to confirm the transfection of Flag–αTAT1 and α-tubulin was used as loading control (B). (C) Knockdown αTAT1 abolished the rescue effect on acetylated α-tubulin levels seen upon overexpression of GCN5L1 in LKO hepatocytes. Control adenovirus (Ad-Ctrl) or adenovirus coding for WT GCN5L1 (Ad-GCN5L1) infected hepatocytes were mock-transfected (siCtrl) or transfected with αTAT1 siRNA (si-αTAT1), then the Ac-Tub levels were determined by immunoblotting with specific antibodies. α-Tubulin was used as loading control. The relative quantitation of representative immunoblot images are shown for three independent experiments. Results are mean±s.e.m. *P<0.05, ***P<0.001 (unpaired Student's t-test).
Identification of potential novel interacting proteins with GCN5L1
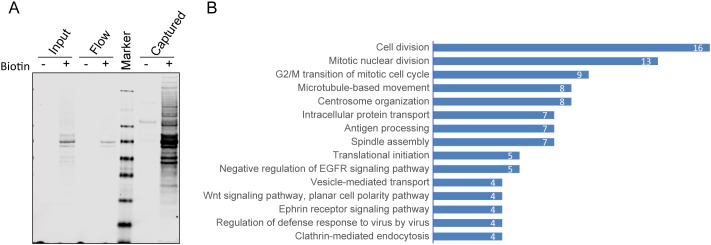
Given that cytosolic GCN5L1 has been found to be operational in numerous multiprotein complexes (Pu et al., 2015; Starcevic and Dell'Angelica, 2004; Zhang et al., 2014) we employed the proximity-dependent biotin identification assay BioID (Roux et al., 2012), to identify potential novel interacting partners that could play additional regulatory roles in GCN5LI-dependent α-tubulin acetylation. To identify proteins that interacted with GCN5L1, a biotin-ligase (BirA)-tagged GCN5L1 (GCN5L1–BirA) was transiently expressed in 293T cells. In the presence of biotin supplementation, proteins biotinylated by GCN5L1–BirA were captured with streptavidin beads and verified on immunoblot analysis (Fig. 5A). MS experiments were then performed, to identify potential interacting proteins. In total, 324 proteins were identified with high confidence, including most subunits of BLOC1 (Falcón-Pérez et al., 2002), including BLOC1S2, BLOC1S4, BLOC1S6 and Snapin (Table S1). These proteins have previously been identified as GCN5L1-interacting proteins (Starcevic and Dell'Angelica, 2004). Interestingly, this proximity-dependent biotin identification assay identified all of the shared subunits with BLOC1, and LOH12CR1 of the BORC complex, as previously described (Pu et al., 2015). To approximate the relative abundance of proteins recovered in the MS analyses, and to correct for overall recovery between experiments, the peptide spectrum match (PSM) values for each protein were calculated (Pisitkun et al., 2012) and the top 123 most-enriched proteins (PSM>1) were analyzed by functional annotation clustering using DAVID (Fig. 5B; Table S2). Intriguingly, numerous proteins identified were enriched in pathways that were linked to microtubule-related functions, such as mitosis, cytoskeleton organization and microtubule-linked trafficking. These data further support that GCN5L1 plays a role in microtubule-related functions.
Fig. 5.
Identification of potential novel interacting proteins with GCN5L1. (A) Enrichment of BioID proximity labeled proteins in GCN5L1–BirA-transfected cells. 293T cells were transfected with biotin ligase-fused GCN5L1 expression plasmids (GCN5L1–BirA), and the biotinylated proteins were purified from cells expressing GCN5L1–BriA, with or without added biotin. The crude cell lysates (input), unbound protein (flow) and streptavidin resin captured protein (captured) were analyzed by immunoblotting with dye-labeled streptavidin. (B) Functional annotation clustering of streptavidin-purified proteins identified by mass spectrometry from cells expressing GCN5L1–BirA. The top 123 most-enriched proteins (PSM>1) were analyzed by functional annotation clustering using DAVID and show high correlation with microtubules related functions. The results represent three independent experiments.
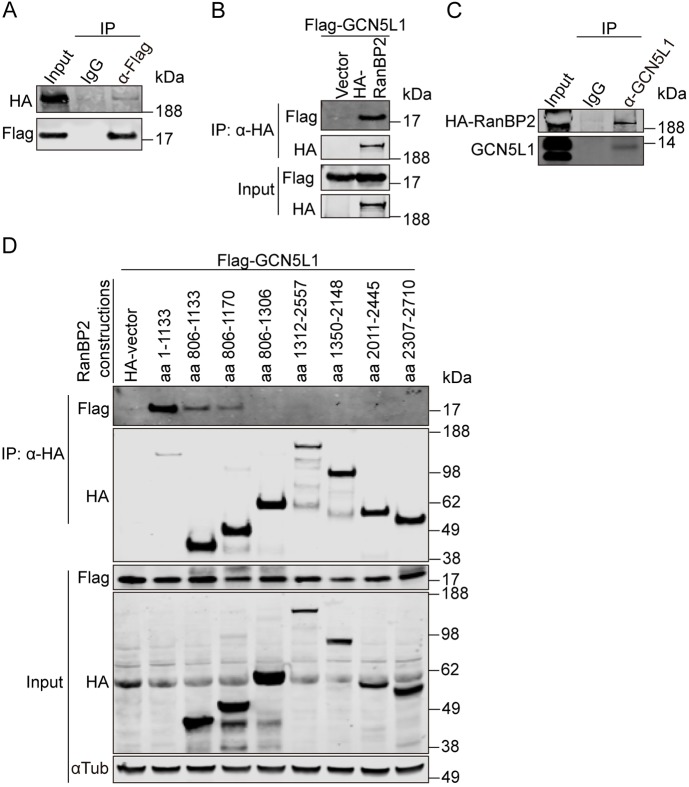
RanBP2 directly interacts with GCN5L1
An interesting candidate interacting protein was the nuclear pore complex protein RanBP2. Although this protein is most highly enriched on the cytosolic surface of the nuclear pore and regulates nuclear protein transport (Dickmanns et al., 2015), it also functions as an allosteric activator of kinesin-1 (Cho et al., 2009) and modulates microtubule-linked processes, including mitosis, α-tubulin acetylation and mitochondrial localization and function (Cho et al., 2007; Ganley et al., 2011; Joseph and Dasso, 2008). Given these putative overlapping functional pathways, we validated the interaction between GCN5L1 with RanBP2 by immunoprecipitation analysis. Anti-Flag immunoprecipitation of cell lysis from 293T cells overexpressing Flag–GCN5L1 and HA–RanBP2 expression vectors with subsequent immunoblot analysis showed a modest affinity between GCN5L1 and the full-length RANBP2 (Fig. 6A). The inverse interaction using anti-HA immunoprecipitation similarly showed the interaction of RanBP2–HA with GCN5L1–Flag (Fig. 6B). As an additional control to exclude a non-specific interaction between HA-RanBP2 and endogenous GCN5L1, immunoprecipitation was assessed in response to using either an anti-IgG antibody or an antibody directed at GCN5L1. Here too, an interaction between overexpressed RanBP2 and endogenous GCN5L1 is found (Fig. 6C). RanBP2 is a large protein (3224 amino acids) with multiple functional domains (Kassube et al., 2012; Lin et al., 2013; Werner et al., 2012). To pinpoint the interacting region, multiple length C-terminal deletion mutants of RanBP2 (Wälde et al., 2012) were employed in immunoprecipitation studies. All the C-terminal deletion mutants of RanBP2 bound the Flag-tagged GCN5L1 (Fig. S4A), suggesting that GCN5L1 interacted with the N-terminal region (amino acids 1–1133) of RanBP2. The interaction of GCN5L1 with RanBP2 was most robust when the 1133 amino acid N-terminal region of RanBP2 was co-expressed. This affinity was reduced when the initial 805 amino acids was excluded from the 1133 N-terminal fragment of RanBP2 and co-expressed with GCN5L1 (Fig. 6D). Interestingly the N-terminal region of RanBP2 (amino acids 1–900) had previously been shown to be required for the association of RanBP2 with microtubules (Joseph and Dasso, 2008). In this study, we find that RanBP2 directly interacted with α-tubulin. This was performed in 293T cells following the overexpression of HA-tagged RanBP2 (amino acids 1–1133) and α-tubulin. Subsequent immunoprecipitation with an antibody directed at the HA tag showed the interaction between RanBP2 and α-tubulin (Fig. S4B).
Fig. 6.
RanBP2 directly interacts with GCN5L1. (A,B) GCN5L1 co-immunoprecipitated with RanBP2. (A) Flag–GCN5L1 and HA–RanBP2-transfected 293T cells lysates were subjected to immunoprecipitation (IP) using anti-Flag antibodies, and the immunoprecipitates were probed for the presence of HA–RanBP2 by immunoblotting with anti-HA antibody. Normal mouse IgG (IgG) was used as control. (B) 293T cells were transfected with the indicated expression plasmids, then the anti-HA immunoprecipitates were subjected to anti-Flag and anti-HA immunoblot analysis. (C) This interaction was confirmed by the immunoprecipitation of endogenous GCN5L1 with HA–RanBP2 versus an IgG control. (D) The N-terminal region of RanBP2 is required for the association with GCN5L1. Different HA-tagged fragments of RanBP2 were transiently expressed in 293T cells with Flag–GCN5L1 and the cells lysates were subjected to immunoprecipitation using anti-HA antibody. The immunoprecipitates were probed with anti-Flag antibodies to identify the region of RanBP2 that is required for interaction with GCN5L1. Representative immunoblots of three independent experiments are shown.
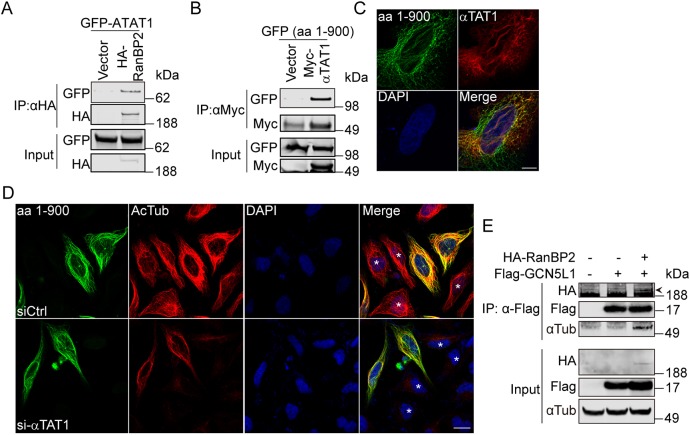
RanBP2 recruits GCN5L1–αTAT1 complex to facilitate α-tubulin acetylation
Given the interaction of RanBP2 with GCN5L1, we then explored whether RanBP2 also binds to αTAT1. Here, bidirectional co-immunoprecipitation studies show that αTAT1 binds to the N-terminal of RanBP2 (Fig. 7A,B), and confocal microscopy showed that this domain of RanBP2 is distributed along the microtubular network in concert with αTAT1 (Fig. 7C). To confirm the role of RanBP2 in this complex, we show that the overexpression of RanBP2 (amino acids 1–900) mediated an increase in α-tubulin acetylation and that the concurrent knockdown of αTAT1 abolished this acetylation effect (Fig. 7D; Fig. S5A). The non-transfected cells in the same image panels, as seen with DAPI staining, serve as a negative control. Collectively, these results indicate that RanBP2 recruits the GCN5L1–αTAT1 complex to promote α-tubulin acetylation. We then confirmed that the N-terminal region of RanBP2 (amino acids 1–900) was sufficient to bind to microtubules (Fig. 7D; Fig. S5B), and to increase the acetylation of α-tubulin (Fig. 7D; Fig. S5C). The requirement of this domain was confirmed by the finding that a truncated fragment RanBP2 (amino acids 1–600) disrupted microtubule binding and α-tubulin acetylation (Fig. S5B,C).
Fig. 7.
RanBP2 recruits the GCN5L1–αTAT1 complex to facilitate α-tubulin acetylation. (A) RanBP2 co-immunoprecipitated with αTAT1. 293T cells were transfected with the indicated expression plasmids, then anti-HA immunoprecipitates were subjected to anti-HA and anti-GFP immunoblot analysis. (B) The N-terminal region of RanBP2 was required for the association with αTAT1. The GFP-tagged N-terminal fragment of RanBP2 [amino acids (aa) 1–900] was expressed in 293T cells with control vector or Myc–αTAT1 and the cells lysates were subjected to immunoprecipitate using anti-Myc antibody. The immunoprecipitates were probed with anti-GFP and anti-Myc antibody. (C) Confocal microscopy showed colocalization of RanBP2 (aa 1–900) with αTAT1. HeLa cells were co-transfected with GFP-tagged N-terminal RanBp2 (aa 1–900) (green) and Flag–αTAT1 (red); then the localization was analyzed with anti-Flag antibodies. GFP fluorescence was visualized directly. DNA was visualized by DAPI staining (blue). Scale bar: 10 µm. (D) Knockdown αTAT1 abolished the RanBP2-induced α-tubulin acetylation (Ac-Tub). GFP-tagged N-terminal RanBp2 (aa 1–900)-transfected 293T cells were mock-transfected (siCtrl) or transfected with αTAT1 siRNA (si-αTAT1) and the Ac-Tub levels (red) were visualized by immunostaining. Non-transfected cells are marked with an asterisk (*) in the merge panels. Scale bar: 20 µm. (E) RanBP2 recruited GCN5L1 to microtubules. 293T cells were transfected with indicated expression vectors, then the anti-Flag immunoprecipitates were subjected to anti-GFP, anti-Flag and anti-α-tubulin immunoblot analysis. The specific band for HA–RanBP2 (depicted with an arrowhead) was evident at a higher molecular mass than a non-specific band. Representative immunoblots of three independent experiments.
Given the finding that GCN5L1 played a role in modeling α-tubulin acetylation, we then assessed whether GCN5L1 itself interacts with α-tubulin. However, neither conventional nor pre-emptive crosslinking co-immunoprecipitation studies could verify a direct interaction (Fig. 7E). We then explored whether an indirect association exists between GCN5L1 and α-tubulin by employing RanBP2 as a structural intermediary. Here, following co-expression of Flag–GCN5L1 and the HA–RanBP2 fragment (amino acids 1–1810), subsequent immunoprecipitation with an antibody recognizing Flag (GCN5L1) pulled down both RanBP2 and α-tubulin (Fig. 7E). Only the co-expression of GCN5L1 and RanBP2 had the ability to immunoprecipitate α-tubulin (Fig. 7E). Collectively, the results suggest that GCN5L1 is recruited to microtubules by RanBP2, which then coordinately facilitate interaction with, and acetylation of, α-tubulin by αTAT1.
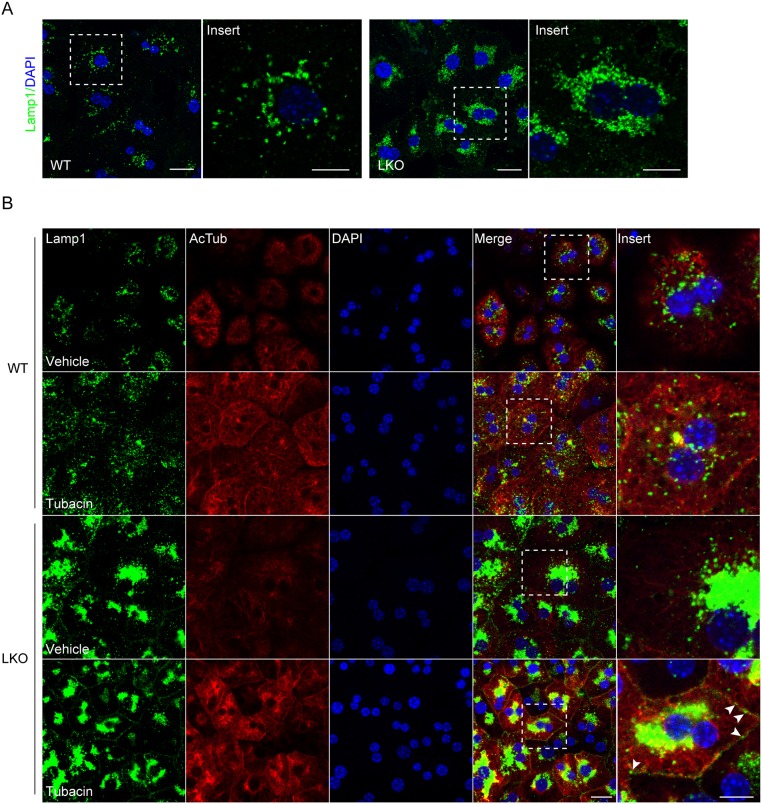
GCN5L1-dependent α-tubulin acetylation contributes to lysosomal centrifugal movement
As a subunit of BORC, knockdown of GCN5L1 had previously been shown to impair the centrifugal movement of lysosomes in HeLa cells by decreasing the recruitment of kinesin-1 molecules to lysosomes (Pu et al., 2015). To validate this in primary hepatocytes, we examined the intracellular distribution of lysosomes in WT and LKO primary hepatocytes using an antibody directed against lysosome-associated membrane protein-1 (LAMP-1). GCN5L1-KO hepatocytes showed increased levels of LAMP-1 compared to WT (Fig. S6A). Concurrently, we found the lysosomes were distributed throughout the cytoplasm in WT cells, but were tightly clustered in the perinuclear region in LKO hepatocytes (Fig. 8A). This abnormal distribution of lysosomes in LKO cells was restored by adenovirus-driven overexpression of GCN5L1 (Fig. S6B). As the acetylation of α-tubulin promotes kinesin-1 binding to microtubules with increased cargo delivery to the peripheral region of cells (Reed et al., 2006), we employed tubacin, a selective inhibitor of the α-tubulin deacetylase HDAC6 (Haggarty et al., 2003), to assess its effect on lysosomal distribution. Here, we find that tubacin increases microtubule acetylation in both WT and LKO hepatocytes (Fig. 8B). In parallel, tubacin promoted the peripheral distribution of lysosomes in both genotypes, although the response in the KO cells was only partial (Fig. 8B). This partial response may reflect the excess of accumulated perinuclear lysosomes in the LKO cells. Together, these experiments support the idea that GCN5L1-dependent α-tubulin acetylation contributes to the centrifugal movement of lysosomes.
Fig. 8.
GCN5L1-dependent α-tubulin acetylation contributes to lysosomal centrifugal distribution. (A) Lysosomes show abnormal perinuclear clusters in LKO hepatocytes compared to WT hepatocytes. Lysosome distribution was visualized by immunostaining for Lamp1 (green), a lysosome membrane protein, in WT and LKO hepatocytes. The marked regions are magnified in the adjacent panel. (B) The HDAC6 inhibitor (tubacin) increased the acetylation of tubulin in WT and GCN5L1 LKO hepatocytes. This intervention partially restored the peripheral distribution of lysosomes in LKO hepatocytes. Lamp1 (green) and acetylated α-tubulin (Ac-Tub; red) were immunostained in WT and LKO hepatocytes treated with or without tubacin. Magnified views of the marked regions reveal that some lysosomes are delivered to the cell surface region in LKO hepatocytes (arrowheads). DNA was visualized by DAPI staining (blue). Scale bars: 10 µm.
DISCUSSION
In this present study, we show that a novel multiprotein complex comprising GCN5L1, RanBP2 and αTAT1 functions in concert to acetylate α-tubulin, and that this acetylation modification plays an important role in lysosome positioning in the cell. Moreover, we identify that GCN5L1 and RanBP2-mediated microtubule acetylation is dependent on αTAT1 and that RanBP2 appears to function as the intermediary linking GCN5L1 to α-tubulin.
Prior to this study, the only functionally characterized target of GCN5L1 acetylation was the kinesin motor-binding protein KBP (Donato et al., 2017). In that study, performed in murine embryonic stem cells (mESCs), the genetic depletion of GCN5L1 promoted mitochondrial biogenesis and mESC differentiation (Donato et al., 2017). Interestingly, GCN5L1-mediated acetylation of KBP required the presence of the mitochondrial acetyl-CoA generating enzyme L-threonine dehydrogenase (TDH) (Donato et al., 2017). This finding was intriguing, given that a GCN5L1 includes a prokaryote-conserved acetyltransferase substrate- and acetyl-CoA-binding regions (Scott et al., 2018), but not a canonical catalytic domain (Scott et al., 2012). In our study, we show that GCN5L1 facilitated acetylation of α-tubulin is dependent on the αTAT1 acetyltransferase, and that in the absence of GCN5L1 α-tubulin acetylation is diminished. Interestingly, although the nuclear acetyltransferase GCN5 does contain an acetyltransferase catalytic domain, it requires additional partners to function as a canonical enzyme (Grant et al., 1999). Putting this together, GCN5L1 may function as a molecular component of an acetyltransferase machinery, and, at least in the cytosol, can partner with αTAT1 and RanBP2 to confer this activity. At the same time, and in contrast to the requirement of TDH to generate the acetyl-CoA for GCN5L1 mediated acetylation of KBP (Donato et al., 2017), the high abundance of acetyl-CoA in the liver may preclude the requirement for its generation for GCN5L1 functioning in this tissue.
The role of the acetylation of α-tubulin in enhancing microtubular strength (Xu et al., 2017) and its function in intracellular organelle trafficking is well established (Geeraert et al., 2010; Reed et al., 2006). Additionally, the major enzymes regulating α-tubulin acetylation and deacetylation, respectively, have been identified as αTAT1 and HDAC6 (Akella et al., 2010; Hubbert et al., 2002; Shida et al., 2010). The genetic depletion of GCN5L1 has been found to disrupt endolysosomal trafficking (Zhang et al., 2014), lysosome positioning (Pu et al., 2015) and lysosome function (Gaidt et al., 2017), and here we find that GCN5L1 directly interacts with αTAT1 and that the absence of GCN5L1 inhibits α-tubulin acetylation. Taken together, these data support the role of GCN5L1 as a novel regulatory protein for the function of αTAT1 in modulating α-tubulin acetylation and subsequent lysosome positioning/distribution.
Traditionally RanBP2, which is located on the cytoplasmic filaments of the nuclear pore complex, contributes to the transport of proteins into or out of the nucleus (Dickmanns et al., 2015). However, additional studies strongly suggested that RanBP2 may play a role in intracellular trafficking as evident by a requirement of RanBP2 in the regulation of interphase microtubules (Pichler et al., 2002; Joseph and Dasso, 2008), in microtubule–kinetochore interactions (Joseph et al., 2004) and in the appropriate intracellular distribution of mitochondria (Cho et al., 2007). In this study, we find that GCN5L1 and RanBP2 interact with each other and that RanBP2 facilitates recruitment of GCN5L1 to α-tubulin for subsequent acetylation.
Advancing proteomics studies have identified a greater diversity in protein subcellular location than had been previously appreciated (Thul et al., 2017), and this emerging recognition is pertinent to both our findings with respect to GCN5L1 and RanBP2. Proteomic analysis of GCN5L1-binding partners and subsequent DAVID analysis revealed cellular processes operating in distinct subcellular locations. However, whether the mechanism of action of GCN5L1 and its binding partners are the same or specialized in different subcellular locations needs to be explored. GCN5L1 has been found to modulate the acetylation of proteins in the mitochondria, on the outer mitochondrial membrane and, as shown here, in the cytosol (Donato et al., 2017; Scott et al., 2012). Additionally, its functional link with αTAT1 in the cytoplasm and the requirement for an acetyl-CoA-generating enzyme for its function on the outer mitochondrial membrane further support its role in protein acetylation. In mitochondria, the abundance of acetyl-CoA, and the presence of a GCN5L1 acetyl-CoA-binding region suggests that GCN5L1 could function as an adaptor protein to spatially align acetyl-CoA with interacting lysine residues. This prospect will be supportive of the observation that non-enzymatic acetylation occurs within mitochondria. However, these concepts will need to be characterized further.
Historically the function of lysosomes was thought to be restricted to the degradation and recycling of cellular waste. However, in the past few decades, we have begun to appreciate that these organelles play an integral role in a diffuse array of cellular processes, including roles in autophagosome and endosome biology, cellular secretion, cell membrane repair, nutrient sensing, signal transduction and energy metabolism (Korolchuk et al., 2011; Settembre et al., 2013). To orchestrate these functions, the regulation of lysosomal positioning within the cell is fundamental, as is the role of microtubules in the modulation of bound molecular motors, which support the spatial-dependent role for distinct lysosomal functions (Matteoni and Kreis, 1987; Mrakovic et al., 2012). Concordant with these observations, a case for a potential multi-organellar role for GCN5L1 can be made, as we found profound lysosomal distribution defects in knockout cells compared to WT cells. In parallel, microtubules are subject to numerous post-translational modifications, and a change in tubulin acetylation is generally proposed to modify tubulin stability (Sadoul and Khochbin, 2016; Wloga et al., 2017). Additionally, the acetylation of tubulin may have distinct functions in different tissues, and proteomics analysis now suggests that tubulin acetylation is not restricted to the K40 residue (Sadoul and Khochbin, 2016). Furthermore, how changes in acetylation modify the function of bound molecular motor proteins additionally requires further validation. Despite these uncertainties, the identification of the more intricate machinery regulating αTAT1 through GCN5L1 and RanBP2 sheds additional insight into the complexity of this regulation. Furthermore, given that GCN5L1 has been shown to modulate the stability of a kinesin motor-binding protein (Donato et al., 2017), the role of GCN5L1 in microtubular function and its effects on organelle biology will be an intriguing arena for ongoing studies.
MATERIALS AND METHODS
Animal studies
All animal protocols were in accordance with Institutional Guidelines and approved by the Institutional Animal Care and Use Committee of the National Heart, Lung and Blood Institute of the National Institutes of Health, USA. The mice were maintained on a 12-h-light–12-h-dark cycle and housed at 3–5 mice per cage with free access to water and normal chow diet. All mice were generated in the C56BL/6 background and backcrossed for >10 generations (Wang et al., 2017). GCN5L1 liver knockout mice (LKO) were generated by crossing GCN5L1 flox/flox mice with albumin-Cre flox mice and the littermate GCN5L1 flox/flox mice were used as the controls (WT). For immunoblot analysis, 8-week-old male mice were anesthetized, then livers were removed and homogenized in RIPA buffer, 40 μg total protein was used for each sample.
Cell culture
All cell lines were obtained from the American Type Culture Collection (Manassas, Virginia) and were tested routinely tested for contamination. Primary hepatocytes were isolated from normal chow fed mice at the age of 8–12 weeks as previously described (Liu et al., 2011). Briefly, mice were anesthetized with isoflurane, and the liver was perfused with Krebs–Ringer buffer with glucose and EGTA (100 mM) for 3 min, followed by continuous perfusion with the same buffer containing CaCl2 (1.4 mM) and collagenase (7000 IU/mouse, type I, Worthington) for 8 min. The livers were extracted, and dissociated cells were filtrated through a 100-μm cell strainer and purified by Percoll (Sigma-Aldrich). Hepatocytes were plated (6×105 cells per well in a six-well plate) onto collagen (Sigma-Aldrich)-coated plates and cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin (P/S). HeLa cells and 293T cells were cultured in DMEM containing 10% fetal bovine serum, 1% P/S and 2 mM glutamine at 37°C and 5% CO2.
For overexpression studies, primary hepatocytes and HeLa cells were transfected with Lipofectamine 3000 (Life Technologies) following the manufacturer's instructions. 293T cells were transfected with PolyJet In Vitro DNA Transfection Reagent (SignaGen Laboratories). Cells were analyzed 48 h after transfection. For knockdown experiments, cells were transfected with indicated 25 nM siRNAs (ON-TARGETplus SMART-pool or ON-TARGETplus Non-Targeting SMARTpool; GE Dharmacon) with Lipofectamine RNAiMAX Reagent and analyzed 48 h later.
Adenovirus production and transduction
GCN5L1 overexpression adenoviruses were produced using the Adeasy Adenoviral System (Agilent) as described previously (Wang et al., 2017). For primary hepatocyte infection, adenovirus overexpressing either empty vector (control) or GCN5L1 were added to cells at a dose of 20 plaque-forming units/cell and analyzed 48 h later.
Determination of acetyl-coenzyme A in primary hepatocytes
Acetyl-CoA levels were measured by HPLC or LC-MS as previously described with slight modification (Shurubor et al., 2017; Yang et al., 2017). Briefly, 109 primary hepatocytes were lysed in buffer A (100 mM sodium phosphate monobasic, 75 mM sodium acetate, pH 4.6). After adding 10% 5-sulfosalicylic acid to reach the final concentration of 5%, the lysates were centrifuged (17,000 g) for 10 min at 4°C, then the supernatants were analyzed by HPLC or LC-MS.
Proximity labeling experiments using BioID
GCN5L1–BirA-transfected 293T cells were grown in 15 cm tissue culture plates for 24 h, then incubated with or without 50 µM biotin (Sigma-Aldrich) for 15–17 h. The cells were then harvested, and total protein levels equalized by protein estimation using a BCA protein assay kit (Pierce). The cell pellets were lysed in 1 ml RIPA lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM EGTA, 0.1% SDS), supplemented with protease cocktail inhibitor (Roche). The lysates were centrifuged for 30 min at 20,000 g at 4°C. Next, the biotinylated proteins were purified from the supernatant by incubating with Sera-Mag streptavidin-coated magnetic particles (GE healthcare life sciences) for 4 h at 4°C. The beads were separated using a magnet and washed twice with RIPA lysis buffer followed by two washes with TAP lysis buffer (50 mM HEPES-KOH pH 8.0, 100 mM KCl, 10% glycerol, 2 mM EDTA, 0.1% NP-40) and three washes with 50 mM ammonium bicarbonate pH 8.0. Proteins on beads were reduced with 10 mM Tris(2-carboxyethyl)phosphine hydrochloride, and alkylated with 20 mM chloroacetamide, then digested using 5 μg/ml trypsin in 2 M urea at 37°C. The supernatant was harvested, and beads were washed twice with 50 mM ammonium bicarbonate pH 8.0, and washes were pooled together with the supernatant. The pooled tryptic digest was lyophilized and resuspended in 8 M urea, 50 mM ammonium bicarbonate and acidified with 1% (v/v) trifluoroacetic acid (TFA) prior to desalting on SDB-XC (Empore) StageTips as previously described.
Mass spectrometry and data analysis
The resulting peptides from BioID were separated on a nanoLC system and simultaneously detected in data-dependent analysis mode on an LTQ Orbitrap Fusion machine (Thermo Fisher Scientific, San Jose, CA). By searching the resulting LC-MS/MS raw data against the SwissProt Mouse protein database, the peptide and protein IDs were assigned using Mascot V2.5 (Matrix Science Inc., Boston, MA) on the Proteome Discoverer 2.1 platform (Thermo Fisher Scientific). All samples were analyzed in triplicates from three independent experiments. Protein inclusion criterion required a protein be present in at least two of the three experiments. After proteins were compiled, keratins, histones and endogenously biotinylated carboxylases were discarded before calculating the total peptide spectrum match (PSM) for each individual experiment and the normalized PSM for each protein. The average normalized PSM/observable peptide number (OPN) (av n-PSM/OPN) was then calculated as previously described (Pisitkun et al., 2012).
Fluorescence microscopy
Primary hepatocytes or HeLa cells were plated on 12 mm diameter, #1 thickness coverslips coated with rat tail type I collagen (neuvitro) and grown to 80% confluency before transfection. At 24 h after transfection, coverslips were washed in PBS and fixed with 4% paraformaldehyde (PFA) in PBS for 10 min. Coverslips were washed twice for 5 min in PBS and permeabilized for 15 min in 0.2% Triton X-100. Cells were blocked for 60 min in 1% BSA, 10% goat normal serum and 0.3 M glycine in PBS and stained for 60 min with primary antibodies (Table S3) in 10% goat normal serum in PBS at room temperature. The secondary antibody was an Alexa-Fluor-488- or 594-conjugated antibody used at a 1:1000 dilution in PBS for 1 h. Coverslips were again washed twice with PBS and mounted on slides using Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific). Images were obtained on an inverted confocal laser-scanning microscope (LSM880; CarlZeiss) fitted with a 63×, 1.4 numerical aperture (NA) objective.
Immunoprecipitation
For immunoprecipitation, transfected 293T cells were lysed in 50 mM Tris-HCl pH 7.4, 137 mM NaCl, 10 mM MgCl2, 10% glycerol and 1% Triton X-100 containing protease inhibitors and centrifuged at 17,000 g for 10 min at 4°C. Supernatants (1–2 mg of total protein in 1 ml) were incubated with 2 µg primary antibody (Table S3) overnight at 4°C. 20 µl Sera-Mag Streptavidin-Coated Magnetic Particles (GE healthcare life sciences) were added to each sample and incubated at room temperature for 1 h. The beads were separated using a magnet and washed four times in TBS containing 0.1% Triton X-100, then the bound proteins were eluted in SDS sample buffer and analyzed by SDS-PAGE and immunoblotting.
In vitro tubulin acetylation assay
In vitro tubulin acetylation assays were performed as previously described (Conacci-Sorrell et al., 2010). Briefly, cytosolic extract from HeLa cells (30 µg) were incubated with in vitro translated GCN5L1 (IVT-GCN5L1) or IVT vector for 1 h in the presence of acetylation buffer (50 mM Tris-HCl pH 8.0, 50 mM EDTA, 10% glycerol, 20 mM acetyl CoA, 10 mM Na butyrate, 10 mMTSA, 1 mMGTP) at 37°C, then the reactants were assayed by immunoblotting.
Other methods
The recombinant DNA constructs and antibodies used in this work are described in Table S3.
Statistical analysis
Results are displayed as the mean±s.e.m. A comparison of groups was performed using two-tailed unpaired Student's t-test. A value of P<0.05 was considered to indicate statistical significance.
Supplementary Material
Acknowledgements
We thank the NHLBI Light Microscopy, Biophysics and Biochemistry Cores for the use of equipment and technical expertise, and Mary C. Dasso (National Institute of Child Health and Human Development, NIH, Bethesda, MD) for the GFP-tagged RanBP2 plasmids.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.W., M.N.S.; Methodology: K.W., L.W., Y.C., M.P., K.S., S.W., R.H.K., I.S., M.G.; Formal analysis: K.W., Y.C., M.P., M.G.; Investigation: K.W., L.W., K.S.; Resources: S.W., R.H.K., I.S.; Writing - original draft: K.W., M.N.S.; Writing - review & editing: R.H.K.; Supervision: M.P., M.N.S.; Funding acquisition: M.N.S.
Funding
M.N.S. is supported by the Division of Intramural Research of the National Heart, Lung and Blood Institute of the National Institutes of Health (ZIA-HL006047-07). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.221036.supplemental
References
- Akella J. S., Wloga D., Kim J., Starostina N. G., Lyons-Abbott S., Morrissette N. S., Dougan S. T., Kipreos E. T. and Gaertig J. (2010). MEC-17 is an alpha-tubulin acetyltransferase. Nature 467, 218-222. 10.1038/nature09324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Sutter B. M., Li B. and Tu B. P. (2011). Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426-437. 10.1016/j.molcel.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederbaum A. I. (2012). Alcohol metabolism. Clin. Liver Dis. 16, 667-685. 10.1016/j.cld.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabin-Brion K., Marceiller J., Perez F., Settegrana C., Drechou A., Durand G. and Poüs C. (2001). The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell 12, 2047-2060. 10.1091/mbc.12.7.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli V. T., Daniels R. W., Godoy R., Hoyle D. J., Kandachar V., Starcevic M., Martinez-Agosto J. A., Poole S., DiAntonio A., Lloyd V. K. et al. (2010). Genetic modifiers of abnormal organelle biogenesis in a Drosophila model of BLOC-1 deficiency. Hum. Mol. Genet. 19, 861-878. 10.1093/hmg/ddp555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang P.-W., Oiso N., Gautam R., Suzuki T., Swank R. T. and Spritz R. A. (2003). The Hermansky-Pudlak syndrome 1 (HPS1) and HPS4 proteins are components of two complexes, BLOC-3 and BLOC-4, involved in the biogenesis of lysosome-related organelles. J. Biol. Chem. 278, 20332-20337. 10.1074/jbc.M300090200 [DOI] [PubMed] [Google Scholar]
- Cho K.-I., Cai Y., Yi H., Yeh A., Aslanukov A. and Ferreira P. A. (2007). Association of the kinesin-binding domain of RanBP2 to KIF5B and KIF5C determines mitochondria localization and function. Traffic 8, 1722-1735. 10.1111/j.1600-0854.2007.00647.x [DOI] [PubMed] [Google Scholar]
- Cho K.-I., Yi H., Desai R., Hand A. R., Haas A. L. and Ferreira P. A. (2009). RANBP2 is an allosteric activator of the conventional kinesin-1 motor protein, KIF5B, in a minimal cell-free system. EMBO Rep. 10, 480-486. 10.1038/embor.2009.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conacci-Sorrell M., Ngouenet C. and Eisenman R. N. (2010). Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell 142, 480-493. 10.1016/j.cell.2010.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickmanns A., Kehlenbach R. H. and Fahrenkrog B. (2015). Nuclear pore complexes and nucleocytoplasmic transport: from structure to function to disease. Int. Rev. Cell Mol. Biol. 320, 171-233. 10.1016/bs.ircmb.2015.07.010 [DOI] [PubMed] [Google Scholar]
- Donato V., Bonora M., Simoneschi D., Sartini D., Kudo Y., Saraf A., Florens L., Washburn M. P., Stadtfeld M., Pinton P. et al. (2017). The TDH-GCN5L1-Fbxo15-KBP axis limits mitochondrial biogenesis in mouse embryonic stem cells. Nat. Cell Biol. 19, 341-351. 10.1038/ncb3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen C. A. G. G., Winkens H. J., Kuhlmann L. D., Janssen B. P. M., van Vugt A. H. M., Deutman A. F. and Janssen J. J. M. (1997). Cloning and structural analysis of the murine GCN5L1 gene. Gene 203, 27-31. 10.1016/S0378-1119(97)00486-1 [DOI] [PubMed] [Google Scholar]
- Falcón-Pérez J. M., Starcevic M., Gautam R. and Dell'Angelica E. C. (2002). BLOC-1, a novel complex containing the Pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J. Biol. Chem. 277, 28191-28199. 10.1074/jbc.M204011200 [DOI] [PubMed] [Google Scholar]
- Fukushima A., Alrob O. A., Zhang L., Wagg C. S., Altamimi T., Rawat S., Rebeyka I. M., Kantor P. F. and Lopaschuk G. D. (2016). Acetylation and succinylation contribute to maturational alterations in energy metabolism in the newborn heart. Am. J. Physiol. Heart Circ. Physiol. 311, H347-H363. 10.1152/ajpheart.00900.2015 [DOI] [PubMed] [Google Scholar]
- Gaidt M. M., Ebert T. S., Chauhan D., Ramshorn K., Pinci F., Zuber S., O'Duill F., Schmid-Burgk J. L., Hoss F., Buhmann R. et al. (2017). The DNA inflammasome in human myeloid cells is initiated by a STING-cell death program upstream of NLRP3. Cell 171, 1110-1124.e18. 10.1016/j.cell.2017.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley I. G., Wong P.-M., Gammoh N. and Jiang X. (2011). Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol. Cell 42, 731-743. 10.1016/j.molcel.2011.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeraert C., Ratier A., Pfisterer S. G., Perdiz D., Cantaloube I., Rouault A., Pattingre S., Proikas-Cezanne T., Codogno P. and Poüs C. (2010). Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J. Biol. Chem. 285, 24184-24194. 10.1074/jbc.M109.091553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. A., Eberharter A., John S., Cook R. G., Turner B. M. and Workman J. L. (1999). Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274, 5895-5900. 10.1074/jbc.274.9.5895 [DOI] [PubMed] [Google Scholar]
- Haggarty S. J., Koeller K. M., Wong J. C., Grozinger C. M. and Schreiber S. L. (2003). Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl Acad. Sci. USA 100, 4389-4394. 10.1073/pnas.0430973100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.-F. and Yao T.-P. (2002). HDAC6 is a microtubule-associated deacetylase. Nature 417, 455-458. 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- Inoue M., Isomura M., Ikegawa S., Fujiwara T., Shin S., Moriya H. and Nakamura Y. (1996). Isolation and characterization of a human cDNA clone (GCN5L1) homologous to GCN5, a yeast transcription activator. Cytogenet. Cell Genet. 73, 134-136. 10.1159/000134324 [DOI] [PubMed] [Google Scholar]
- Joseph J. and Dasso M. (2008). The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett. 582, 190-196. 10.1016/j.febslet.2007.11.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J., Liu S. T., Jablonski A. S., Yen T. J. and Dasso M. (2004). The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14, 611-617. 10.1016/j.cub.2004.03.031 [DOI] [PubMed] [Google Scholar]
- Kalebic N., Sorrentino S., Perlas E., Bolasco G., Martinez C. and Heppenstall P. A. (2013). alphaTAT1 is the major alpha-tubulin acetyltransferase in mice. Nat. Commun. 4, 1962 10.1038/ncomms2962 [DOI] [PubMed] [Google Scholar]
- Kassube S. A., Stuwe T., Lin D. H., Antonuk C. D., Napetschnig J., Blobel G. and Hoelz A. (2012). Crystal structure of the N-terminal domain of Nup358/RanBP2. J. Mol. Biol. 423, 752-765. 10.1016/j.jmb.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V. I., Saiki S., Lichtenberg M., Siddiqi F. H., Roberts E. A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F. M. et al. (2011). Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453-460. 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M. and Piperno G. (1991). Detection of acetylated alpha-tubulin by specific antibodies. Methods Enzymol. 196, 264-274. 10.1016/0076-6879(91)96025-M [DOI] [PubMed] [Google Scholar]
- Li W., Zhang Q., Oiso N., Novak E. K., Gautam R., O'Brien E. P., Tinsley C. L., Blake D. J., Spritz R. A., Copeland N. G. et al. (2003). Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat. Genet. 35, 84-89. 10.1038/ng1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. H., Zimmermann S., Stuwe T., Stuwe E. and Hoelz A. (2013). Structural and functional analysis of the C-terminal domain of Nup358/RanBP2. J. Mol. Biol. 425, 1318-1329. 10.1016/j.jmb.2013.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Fergusson M. M., Wu J. J., Rovira I. I., Liu J., Gavrilova O., Lu T., Bao J., Han D., Sack M. N. et al. (2011). Metabolism Wnt signaling regulates hepatic metabolism. Sci. Signal. 4, ra6 10.1126/scisignal.2001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteoni R. and Kreis T. E. (1987). Translocation and clustering of endosomes and lysosomes depends on microtubules. J. Cell Biol. 105, 1253-1265. 10.1083/jcb.105.3.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A., Rouleau M., Kitsberg D., Cohen M., Levy G., Barasch D., Nemirovski A., Shen-Orr S., Laevsky I., Amit M. et al. (2015). Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 21, 392-402. 10.1016/j.cmet.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Mrakovic A., Kay J. G., Furuya W., Brumell J. H. and Botelho R. J. (2012). Rab7 and Arl8 GTPases are necessary for lysosome tubulation in macrophages. Traffic 13, 1667-1679. 10.1111/tra.12003 [DOI] [PubMed] [Google Scholar]
- Perdiz D., Mackeh R., Poüs C. and Baillet A. (2011). The ins and outs of tubulin acetylation: more than just a post-translational modification? Cell. Signal. 23, 763-771. 10.1016/j.cellsig.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Pichler A., Gast A., Seeler J. S., Dejean A. and Melchior F. (2002). The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109-120. 10.1016/S0092-8674(01)00633-X [DOI] [PubMed] [Google Scholar]
- Pisitkun T., Gandolfo M. T., Das S., Knepper M. A. and Bagnasco S. M. (2012). Application of systems biology principles to protein biomarker discovery: urinary exosomal proteome in renal transplantation. Proteomics Clin. Appl. 6, 268-278. 10.1002/prca.201100108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Schindler C., Jia R., Jarnik M., Backlund P. and Bonifacino J. S. (2015). BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell 33, 176-188. 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed N. A., Cai D., Blasius T. L., Jih G. T., Meyhofer E., Gaertig J. and Verhey K. J. (2006). Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16, 2166-2172. 10.1016/j.cub.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Roux K. J., Kim D. I., Raida M. and Burke B. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801-810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoul K. and Khochbin S. (2016). The growing landscape of tubulin acetylation: lysine 40 and many more. Biochem. J. 473, 1859-1868. 10.1042/BCJ20160172 [DOI] [PubMed] [Google Scholar]
- Scott I., Webster B. R., Li J. H. and Sack M. N. (2012). Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem. J. 443, 655-661. 10.1042/BJ20120118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I., Wang L., Wu K., Thapa D. and Sack M. N. (2018). GCN5L1/BLOS1 links acetylation, organelle remodeling, and metabolism. Trends Cell Biol. 28, 346-355. 10.1016/j.tcb.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Fraldi A., Medina D. L. and Ballabio A. (2013). Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14, 283-296. 10.1038/nrm3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T., Cueva J. G., Xu Z., Goodman M. B. and Nachury M. V. (2010). The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. USA 107, 21517-21522. 10.1073/pnas.1013728107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurubor Y. I., D'Aurelio M., Clark-Matott J., Isakova E. P., Deryabina Y. I., Beal M. F., Cooper A. J. L. and Krasnikov B. F. (2017). Determination of coenzyme A and acetyl-coenzyme A in biological samples using HPLC with UV detection. Molecules 22, 1388 10.3390/molecules22091388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M. and Dell'Angelica E. C. (2004). Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 279, 28393-28401. 10.1074/jbc.M402513200 [DOI] [PubMed] [Google Scholar]
- Thapa D., Zhang M., Manning J. R., Guimaraes D., Stoner M., O'Doherty R. M., Shiva S. and Scott I. (2017). Acetylation of mitochondrial proteins by GCN5L1 promotes enhanced fatty acid oxidation in the heart. Am. J. Physiol. Heart Circ. Physiol. 313, H265-H274. 10.1152/ajpheart.00752.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul P. J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L. M. et al. (2017). A subcellular map of the human proteome. Science 356, eaal3321 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- Wagner G. R. and Hirschey M. D. (2014). Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell 54, 5-16. 10.1016/j.molcel.2014.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wälde S., Thakar K., Hutten S., Spillner C., Nath A., Rothbauer U., Wiemann S. and Kehlenbach R. H. (2012). The Nucleoporin Nup358/RanBP2 promotes nuclear import in a cargo- and transport receptor-specific manner. Traffic 13, 218-233. 10.1111/j.1600-0854.2011.01302.x [DOI] [PubMed] [Google Scholar]
- Wang L., Scott I., Zhu L., Wu K., Han K., Chen Y., Gucek M. and Sack M. N. (2017). GCN5L1 modulates cross-talk between mitochondria and cell signaling to regulate FoxO1 stability and gluconeogenesis. Nat. Commun. 8, 523 10.1038/s41467-017-00521-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster B. R., Scott I., Han K., Li J. H., Lu Z., Stevens M. V., Malide D., Chen Y., Samsel L., Connelly P. S. et al. (2013). Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J. Cell Sci. 126, 4843-4849. 10.1242/jcs.131300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster B. R., Scott I., Traba J., Han K. and Sack M. N. (2014). Regulation of autophagy and mitophagy by nutrient availability and acetylation. Biochim. Biophys. Acta 1841, 525-534. 10.1016/j.bbalip.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A., Flotho A. and Melchior F. (2012). The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol. Cell 46, 287-298. 10.1016/j.molcel.2012.02.017 [DOI] [PubMed] [Google Scholar]
- Wloga D., Joachimiak E., Louka P. and Gaertig J. (2017). Posttranslational modifications of Tubulin and Cilia. Cold Spring Harb. Perspect. Biol. 9, a028159 10.1101/cshperspect.a028159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Schaedel L., Portran D., Aguilar A., Gaillard J., Marinkovich M. P., Théry M. and Nachury M. V. (2017). Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356, 328-332. 10.1126/science.aai8764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Ma Y., Li N., Cai H. and Bartlett M. G. (2017). Development of a novel method for the determination of Acyl-CoA compounds by liquid chromatography mass spectrometry to probe the metabolism of fatty acids. Anal. Chem. 89, 813-821. 10.1021/acs.analchem.6b03623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., He X., Zhang L., Yang L., Woodman P. and Li W. (2014). Biogenesis of lysosome-related organelles complex-1 subunit 1 (BLOS1) interacts with sorting nexin 2 and the endosomal sorting complex required for transport-I (ESCRT-I) component TSG101 to mediate the sorting of epidermal growth factor receptor into endosomal compartments. J. Biol. Chem. 289, 29180-29194. 10.1074/jbc.M114.576561 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.