ABSTRACT
Integrins are α/β heterodimers that interconvert between inactive and active states. In the active state the α/β cytoplasmic domains recruit integrin-activating proteins and separate the transmembrane and cytoplasmic (TMcyto) domains (unclasped TMcyto). Conversely, in the inactive state the α/β TMcyto domains bind integrin-inactivating proteins, resulting in the association of the TMcyto domains (clasped TMcyto). Here, we report the isolation of integrin cytoplasmic tail interactors using either lipid bicelle-incorporated integrin TMcyto domains (α5, αM, αIIb, β1, β2 and β3 integrin TMcyto) or a clasped, lipid bicelle-incorporated αMβ2 TMcyto. Among the proteins found to preferentially bind clasped rather than the isolated αM and β2 subunits was L-plastin (LCP1, also known as plastin-2), which binds to and maintains the inactive state of αMβ2 integrin in vivo and thereby regulates leukocyte adhesion to integrin ligands under flow. Our findings offer a global view on cytoplasmic proteins interacting with different integrins and provide evidence for the existence of conformation-specific integrin interactors.
KEY WORDS: LCP1, Integrin inactivator, Leukocyte adhesion, Integrin interactome, Proteomics
Summary: LCP1 forms a ternary complex with αMβ2 integrin and negatively regulates integrin-mediated cell adhesion by maintaining αMβ2 integrin in an inactive conformation.
INTRODUCTION
Integrins are α/β heterodimeric cell surface receptors that establish cell–cell and cell–extracellular matrix (ECM) interactions required for migration, proliferation, differentiation and survival. The α and β integrin subunits are composed of a large extracellular domain, a single transmembrane segment of ∼25 amino acids and a short cytoplasmic tail of 10–70 amino acids. Integrin binding to ligand leads to integrin clustering, the assembly of a protein network termed adhesome, and eventually the transduction of biochemical and biophysical signals (also called outside-in signaling).
A hallmark of integrins is their ability to adopt different affinities toward their ligand. The affinity switch from an unbound, low-affinity conformation to the bound, high-affinity conformation (called integrin activation or inside-out signaling) is believed to be mediated by the two adaptor protein families, talins and kindlins, which bind to specific sites in the β integrin cytoplasmic domain and to lipids of the nearby plasma membrane. The consequence of talin and kindlin binding is the dissociation of the transmembrane and cytoplasmic (TMcyto) domains of the α and β subunits, leading to the separation (unclasping) of the proximal legs of the α/β integrin ectodomain, followed by a conformational change in the extracellular domain that allows high-affinity ligand binding (Campbell and Humphries, 2011; Kim et al., 2011; Shattil et al., 2010). Although it is evident that the high-affinity conformation can be reversed, it is not entirely clear how this is achieved at the molecular level. Several proteins such as sharpin, filamin family members and ICAP, as well as phosphorylation of the β integrin tail, have been shown to displace talin and/or kindlin family proteins, leading to the association of integrin α/β TMcyto subunits through the interaction between GxxxG dimerization motifs in the outer half of the plasma membrane (called the outer plasma membrane clasp) and a salt bridge in the juxtamembrane region at the inner half of the plasma membrane (called the inner plasma membrane clasp) (Lau et al., 2009; Yang et al., 2009). Both the outer and the inner plasma membrane clasps constitute weak interactions, suggesting that proteins may exist that lock and/or maintain integrins in an inactive state. Such integrin inactivators could represent a regulatory mechanism that efficiently prevents unwanted integrin activation, which is particularly important for cells of hematopoietic origin (Schmidt et al., 2013). Furthermore, proteins interacting with inactive integrins would allow cells to distinguish between active and inactive integrins in other processes including integrin biosynthesis and intracellular integrin trafficking, which affect the levels of integrins at the cell surface and modulate signaling pathways (Bouvard et al., 2013; De Franceschi et al., 2015).
In the present paper, we developed a screening strategy to identify proteins that preferentially bind clasped α/β integrin tails. Among the proteins that favored binding of clasped αMβ2 integrin tails was L-plastin. Plastins are a family of three actin-bundling proteins with distinct expression patterns: T-plastin (also known as plastin-3) is broadly expressed, including in all cells with replicative potential (Lin et al., 1993), I-plastin (also known as plastin-1) is restricted to intestine and kidney cells, while L-plastin (LCP1, also known as plastin-2) expression occurs in leukocytes and in many malignant human cell lines of non-hematopoietic origin (Lin et al., 1988). Plastins consist of two amino-terminal EF-hands, implicated in Ca2+ binding, and two tandem actin-binding domains each composed of two calponin homology domains. Although plastins are primarily involved in regulation of the actin cytoskeleton, they possess additional properties and are involved in several cellular functions such as cell migration, neutrophil function, DNA repair and endocytosis (Delanote et al., 2005). LCP1 has been linked to integrins, yet the precise mechanism of how it regulates integrin function is unclear. LCP1 has been shown to interact with β integrin subunits (Le Goff et al., 2010), and to activate αMβ2 integrin in polymorphonuclear neutrophils (PMNs) (Jones et al., 1998) and αVβ3 integrin in K562 cells (Wang et al., 2001). In contrast, however, LCP1−/− neutrophils exhibit no integrin activation defect but fail to mount an efficient integrin adhesion-dependent respiratory burst (Chen et al., 2003). Here, we show that LCP1 forms a ternary complex with αMβ2 integrin and negatively regulates integrin-mediated cell adhesion by maintaining αMβ2 integrin in an inactive conformation.
RESULTS
α and β integrin tail interactors isolated with bicelle-incorporated TMcyto domains
Due to the short cytoplasmic domains of integrin α and β subunits, direct integrin interactors such as talins and kindlins are in close proximity to the plasma membrane with which they interact to reinforce integrin tail binding and induce signaling (Anthis et al., 2009; Goult et al., 2010, 2009; Liu et al., 2011; Moore et al., 2012; Perera et al., 2011; Saltel et al., 2009). Previous studies employed yeast two-hybrid or pull-down experiments with either single α or β integrin cytoplasmic domains to identify specific binding partners (Legate and Fassler, 2009; Morse et al., 2014; Raab et al., 2010). However, these approaches do not detect binding partners whose interaction with the plasma membrane is crucial for integrin tail binding. To solve this issue we incorporated the recombinant integrin β1 TMcyto domain into bicelles as a membrane-mimetic environment. Bicelles form discoidal nanostructures that have been used to study the structure of different integrin TMcyto domains through the use of nuclear magnetic resonance (NMR) spectroscopy (Lau et al., 2008a,b, 2009; Lu et al., 2012; Surya et al., 2013). The bicelles used in this work were composed of the long-chain phospholipid 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 6-cyclohexyl-1-hexylphosphocholine (Cyclofos-6) as the short-chain phospholipid that stabilizes the edge of the bilayer discs (Fig. S1A). As biological membranes contain negatively charged lipids (Leventis and Grinstein, 2010) we replaced 10% of DMPC with phosphatidylserine (POPS) and confirmed the phospholipid ratio in the bicelles through the use of liquid chromatography-mass spectrometry (LC-MS) (Fig. S1A–C). In line with previous studies (Lu et al., 2012; van Dam et al., 2004), changing the ratio of DMPC to Cyclofos-6, referred to as the q-value, gave rise to bicelles with hydrodynamic radii ranging from 3.16±0.08 nm (mean±s.e.m.; q=0.25) to 19.95±0.64 nm (q=4), as measured using dynamic light scattering (DLS) (Fig. S1D).
Next, we expressed His-tagged α5 and β1 integrin TMcyto domains in E. coli, purified them and determined their incorporation into bicelles by measuring their hydrodynamic radius via DLS. Although α5 and β1 integrin TMcyto domains were successfully incorporated into bicelles with a q-value of 0.25, their direct incorporation into bigger bicelles (q=2) failed (Fig. S1E). As in silico modeling suggests that the bicelle radius must be >8 nm to support membrane interactions of kindlin and talin proteins bound to β3 integrin, we tested the possibility of increasing the radius by mixing small and large bicelles. Indeed, incubation of q=0.25 bicelles with q=4 bicelles to produce a q=2* (asterisk indicates the generation of q=2 bicelles by incubating q=0.25 bicelles with q=4 bicelles) mixture resulted in a homogenous solution of bicelles with a hydrodynamic radius of 8.88±0.51 nm (Fig. S1D). Using this approach we incorporated α5 or β1 integrin TMcyto domains into q=0.25 bicelles and added an equal volume of q=4 bicelles, resulting in q=2* bicelles containing our protein of interest with a hydrodynamic radius of ∼10 nm (Fig. S2; Table S1).
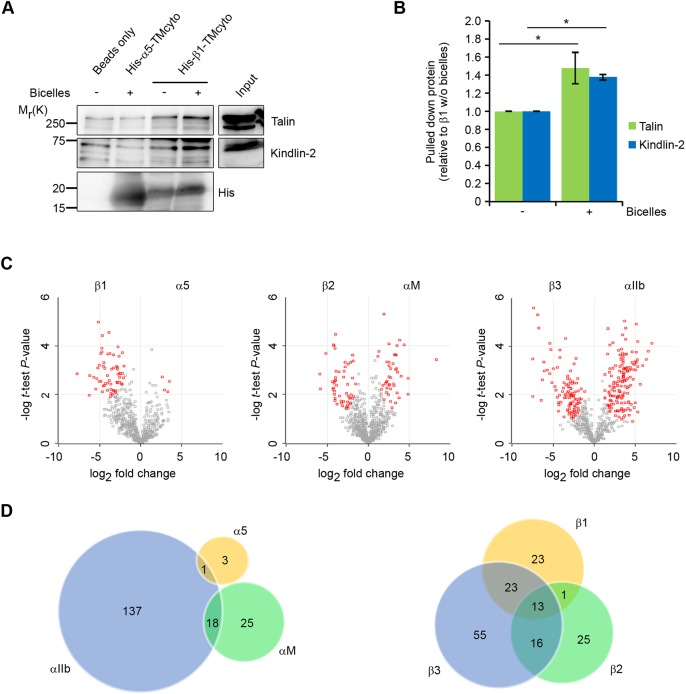
To test whether negatively charged lipids in β1 integrin TMcyto-containing bicelles increase the association of proteins to β1 tails in pull-down experiments, we incubated His-tagged TMcyto domains of β1 and α5 integrins either incorporated into bicelles or left without bicelles with mouse fibroblast cell lysates and pulled down talin and kindlin-2, known to establish plasma membrane interactions for optimal integrin binding (Anthis et al., 2009; Goult et al., 2010, 2009; Liu et al., 2011; Moore et al., 2012; Perera et al., 2011). Due to the mild washing conditions, we observed a very weak binding of both proteins to α5 integrin TMcyto and empty beads. Importantly, however, β1 integrin TMcyto domain incorporated into bicelles bound significantly more talin (48%) and kindlin-2 (38%) compared to the β1 integrin TMcyto domain alone (Fig. 1A,B). These data show that the incorporation of integrin TMcyto domains into negatively charged bicelles can increase the affinity of proteins for the integrin cytoplasmic domains and thereby could promote the identification of novel integrin tail interactors in pull-down assays. To identify interacting partners of different integrin TMcyto domains, we recombinantly expressed α5, αM and αIIb as well as β1, β2 and β3 integrin TMcyto domains, incorporated them into q=0.25 bicelles, mixed those with q=4 bicelles and performed a pull-down with hypotonic cell lysates derived from mouse bone marrow-derived macrophages (BMDM). After pull-down, the interacting proteins were resolved using SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and then identified through the use of liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Fig. S3A).
Fig. 1.
Interactome analysis of individual, lipid-incorporated α and β integrin TMcyto domains. (A) Pull-down of talin and kindlin-2 using recombinant His-tagged α5–TMcyto or β1–TMcyto proteins with or without bicelle incorporation, from cell lysates. (B) Quantification of talin and kindlin-2 binding to His-tagged β1–TMcyto proteins with or without bicelle incorporation from western blots, using ImageJ (mean±s.e.m., n=3, *P<0.05). (C) Volcano plots of fold-change of protein intensities versus t-test P-value for binding to distinct α and β integrin subunits; statistically significant (P<0.05) interactors are labeled in red. (D) Overlap of proteins identified as interactors with the different α and β TMcyto domains illustrated as a Venn diagram.
Our analysis identified around 1500 proteins (Table S2). Label-free quantification revealed good reproducibility and consistency between replicates (Fig. S3B). Four, 43 and 156 proteins were significantly enriched in the α5, αM and αIIb cytoplasmic tail interactome, respectively, (Fig. 1C,D; Table S3), while 60, 55 and 107 proteins were enriched in β1, β2 and β3 pull-downs, respectively (Fig. 1C,D; Table S3). Interestingly, no common interactors were identified for all three integrin α cytoplasmic tails, while the β1, β2 and β3 subunits had 13 common interactors, including talin-1 and filamin-1 (also known as filamin-A) (Fig. 1D; Table S3). These results are in line with the presence of conserved motifs shared among the β1, β2 and β3 integrin cytoplasmic domains and highly divergent motifs in α integrin tails. These data demonstrate the feasibility of screening approaches based on bicelle-incorporated proteins and show qualitative differences between the interactomes of different integrin subunits.
Identification of conformation-specific αMβ2 integrin interactors
Various proteins bind to either α or β cytoplasmic tails to regulate the integrin activity state (Legate and Fassler, 2009). However, filamin-1 was very recently shown to simultaneously bind inactive, clasped αIIbβ3 integrins in order to maintain the inactive state (Liu et al., 2015). A shortcoming of earlier screening approaches is the use of single α or β integrin cytoplasmic domains, which do not recruit interactors that require both integrin tails for binding. To detect such integrin interactors we developed a strategy to mimic the heterodimeric integrin cytoplasmic tail conformation in its inactive, clasped state. Our method is based on the Jun–Fos dimerization domains (fused to the ectoplasmic N-termini of the TM domains) as ‘velcro’ to mediate heterodimer formation. Since a previous fluorescence resonance energy transfer (FRET) study revealed a spatial separation of the αM and β2 subunit cytoplasmic tails in the plasma membrane upon chemokine-induced integrin activation (Lefort et al., 2009), we decided to construct a Jun–Fos-dimerized αMβ2 TMcyto domain to clasp the αM and β2 TMcyto domains and used it as bait to search for interactors of integrins in the inactive state. To this end we expressed recombinant proteins consisting of an amino-terminal tag sequence (His or FLAG for purification and pull-down) followed by a cysteine–glycine (CG) linker, the Jun–Fos dimerization domains, a 4× glycine linker (GGGG) and the αM or β2 integrin TMcyto domains (Fig. 2A,B). The CG linker was inserted to ensure stability, a parallel orientation and a correct stagger of the coiled coil sequences within the dimer, while the GGGG linker was inserted to provide flexibility of the integrin tails. The proteins were expressed and purified from bacteria (Fig. 2B, lanes 2–4) and when incubated together, His–Fos–αM–TMcyto and FLAG–Jun–β2–TMcyto formed heterodimers in a 1:1 ratio (Fig. 2B, lane 1). DLS measurements revealed that the incorporation of αM or β2 TMcyto into bicelles increased their size from 8.88±0.51 nm (empty bicelles) to 10.05±0.26 nm or 10.25±0.31 nm, and after incorporation of the Jun–Fos-dimerized αMβ2 TMcyto to 11.17±0.49 nm (Fig. S2).
Fig. 2.
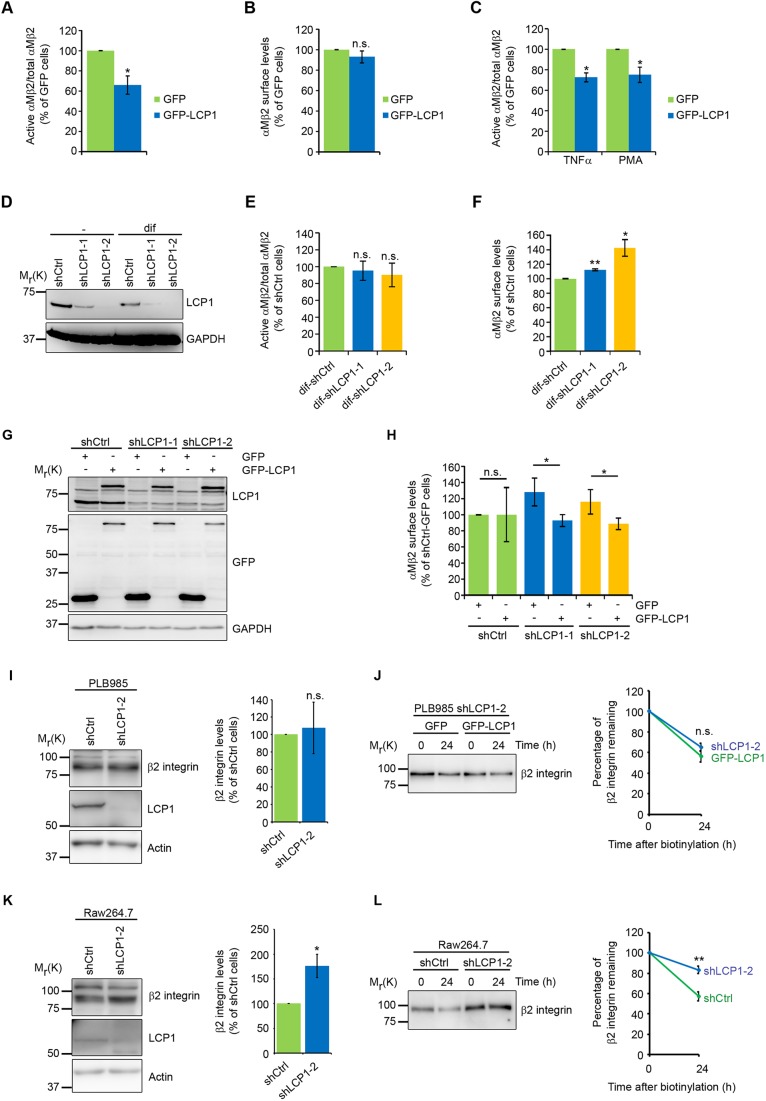
Interactor screen with clasped α/β integrin TMcyto baits. (A) Schematic representation of the integrin TMcyto constructs. CG, cysteine-glycine motif to stabilize the dimer; Fos (Dim) and Jun (Dim), dimerization domain of Fos and Jun proteins; GGGG, glycine stretch to increase the flexibility of the integrin cytoplasmic tails. (B) Recombinant His–Fos–αM–TMcyto, His–Jun–β2–TMcyto, and 3×FLAG–Jun–β2–TMcyto were expressed and purified with Ni-NTA or FLAG–M2 beads as monomers (lane 2, 3, 4). Co-incubation of His–Fos–αM–TMcyto and 3×FLAG–Jun–β2–TMcyto followed by Ni-NTA reveals dimer formation (lane 1). (C) Volcano plot showing fold-change of protein intensities versus t-test P-value for cytosolic proteins interacting with αM–TMcyto versus αMβ2-TMcyo domains. Proteins with significantly increased binding to either αM or αMβ2 (P<0.05) are labeled in red. Filamin-2 (FLNC) and LCP1 are marked. (D) Western blot analysis of talin and LCP1 pulled down with individual αM–TMcyto and β2–TMcyto or clasped αMβ2–TMcyto domains embedded in bicelles. (E) Quantification of LCP1 and talin western blot band intensities shown in D (mean±s.e.m., n=3, *P<0.05). (F) Quantification of GST or GST–LCP1 binding to immobilized His–Fos–αM–TMcyto, His–Jun–β2–TMcyto and the clasped His–Fos–αM–TMcyto/3×FLAG–Jun–β2–TMcyto dimer using solid-phase ligand-binding assays. Bound proteins were detected with anti-LCP1 antibodies. GST was used as control for nonspecific binding (mean±s.e.m., n=3, **P<0.01). (G) Western blot showing the pull-down of MBP–β2–cyto with His–αM–cyto in the presence of GST–LCP1. (H) Western blot and densitometric analysis of β2 integrin co-immunoprecipitations for GFP and β2 integrin from lysates of PMA- or DMSO-treated PLB985 cells expressing GFP–LCP1. β2 integrin served as a loading control to quantify GFP–LCP1 binding (mean±s.e.m., n=3, *P<0.05). (I) Binding of GST–LCP1 to His–Fos–αM–TMcyto or His–Fos–αM–TMcyto/3×FLAG–Jun–β2–TMcyto dimer was assessed in the absence or presence of recombinant talin-1 head. A representative SDS-PAGE of three independent experiments is shown.
The Jun–Fos-dimerized αMβ2 TMcyto domains incorporated into bicelles were used to pull down proteins from BMDM lysates. The interactome was subjected to LC-MS/MS and the identified proteins were quantified using the label-free quantification algorithm of the MaxQuant software and compared with the interactome from individual αM and β2 TMcyto domains. We identified 1561 proteins, and application of the statistical t-test between β2 and αMβ2 interactors revealed 222 proteins that increased binding to associated αMβ2 integrins (Fig. 2C, Table S4), among them filamin-2 (also known as filamin-C), and moesin, which has been shown to displace talin-1 and inactivate integrins (Vitorino et al., 2015). Taken together, these results highlight the existence of cytoplasmic proteins that interact predominantly or exclusively with different αMβ2 integrin structural states (corresponding to clasped inactive state or non-clasped activated state heterodimers).
L-plastin (LCP1) maintains αMβ2 integrins in an inactive state
The hematopoietic-specific α-actinin family member L-plastin (LCP1) (Lebart et al., 2004) was among the proteins with the strongest increase in binding to the clasped αMβ2 TMcyto domain (Fig. 2C). LCP1 is an actin-bundling protein that has been implicated in the formation and/or maintenance of integrin-associated adhesion structures (Jones et al., 1998; Morley, 2012; Wang et al., 2001). As LCP1 has been shown to interact with the cytoplasmic domain of β1 and β2 integrins (Le Goff et al., 2010), we first validated the interaction with bicelle-embedded, Jun–Fos-dimerized αMβ2 and individual β2 TMcyto domains in pull-down experiments. In line with our proteomics data, the Jun–Fos-dimerized αMβ2 TMcyto bait pulled down significantly more LCP1 than the β2 TMcyto bait. Conversely, talin was predominantly pulled down by the β2 TMcyto bait (Fig. 2D,E). Furthermore, purified recombinant glutathione S-transferase (GST)-tagged LCP1 also showed higher binding to clasped Jun–Fos-dimerized αMβ2 TMcyto compared to the individual αM and β2 TMcyto domains in ELISA assays (Fig. 2F). To further support the formation of a ternary complex between αM, β2 and LCP1 we performed pull-down experiments with recombinant GST, GST–LCP1, His–αM-cytoplasmic tail (His–αM–cyto), and maltose-binding protein–β2 cytoplasmic tail (MBP–β2–cyto) (Fig. S4A,B). These experiments revealed that His–αM–cyto was only able to pull down MBP–β2–cyto in the presence of GST–LCP1 but not in the presence of GST (Fig. 2G). GFP–LCP1 co-immunoprecipitated with β2 integrins from neutrophil-like PLB985 cell lysates and, importantly, this interaction was significantly reduced after agonist-induced integrin activation by phorbol 12-myristate 13-acetate (PMA) (Fig. 2H), indicating that LCP1 also preferentially interacts with inactive β2 integrins in cells. Finally, we tested whether LCP1 competes with talin for integrin binding similar to the integrin inactivator filamin-1 (Calderwood et al., 2001). To this end, we examined the binding of GST–LCP1 to dimeric αMβ2 integrin TMcyto in the absence or presence of excess amounts of purified talin-1 head domain and found that the talin-1 head reduced GST–LCP1 binding to Jun–Fos-dimerized αMβ2 TMcyto domains (Fig. 2I). Taken together, these findings indicate that LCP1 forms a ternary complex with cytosolic domains of αMβ2 integrin and competes with talin-1 for integrin tail binding.
LCP1 regulates β2 integrin-mediated functions and activity
The biochemical data suggests that LCP1 stabilizes the inactive conformation of αMβ2 integrins. In order to test this hypothesis we overexpressed GFP-tagged LCP1 in differentiated neutrophil-like PLB985 cells (Fig. S5A) and analyzed the levels of active αMβ2 integrin by fluorescence-activated cell sorting (FACS) using antibodies that recognize activation-induced epitopes. GFP–LCP1 but not GFP alone exerted a significant inhibitory effect on αMβ2 integrin activation (Fig. 3A) without altering the total cell-surface αMβ2 integrin levels (Fig. 3B). Interestingly, PMA- or tumor necrosis factor α (TNFα)-induced integrin activation failed to fully rescue the integrin activation defect in GFP–LCP1-overexpressing PLB985 cells (Fig. 3C).
Fig. 3.
LCP1 regulates αMβ2 integrin-mediated functions and activity. (A) Quantification of αMβ2 integrin activation in PLB985 cells overexpressing GFP–LCP1, determined by means of FACS using an antibody against an activation-induced epitope. Active αMβ2 integrin antibody binding levels were normalized to total cell-surface levels of αMβ2 integrin; values are normalized to GFP-expressing cells. (mean±s.e.m., n=5, *P<0.05). (B) αMβ2 integrin cell-surface expression in GFP–LCP1 overexpressing PLB985 cells relative to GFP-expressing PLB985 cells (values are normalized to GFP-expressing cells; mean±s.e.m.; n=5; n.s., not significant). (C) Quantification of active αMβ2 integrin cell-surface levels in GFP–LCP1-overexpressing PLB985 cells using FACS after TNFα- and PMA-induced integrin activation (active αMβ2 integrin levels were normalized to total cell-surface levels of αMβ2 integrin; values are normalized to GFP-expressing cells; mean±s.e.m., n=5, *P<0.05). (D) Western blot analysis of shRNA-mediated depletion of LCP1 in differentiated (dif) and undifferentiated PLB985 cells. Cells were infected with shCtrl or two different LCP1 shRNAs (shLCP1-1 and shLCP1-2). (E) Quantification of αMβ2 integrin activation in LCP1-depleted differentiated PLB985 cells using FACS (active αMβ2 integrin levels were normalized to total cell-surface levels of αMβ2 integrin; values are normalized to differentiated cells expressing shCtrl; mean±s.e.m.; n=3; n.s., not significant). (F) FACS analysis of αMβ2 cell-surface expression on LCP1-depleted differentiated PLB985 cells after LCP1 knockdown (values are normalized to shCtrl cells; mean±s.e.m., n=3, *P<0.05, **P<0.01). (G) Western blot of LCP1 and GFP in differentiated LCP1-depleted PLB985 cells after GFP–LCP1 re-expression. (H) Re-expression of shRNA-resistant GFP–LCP1 in LCP1-depleted PLB985 cells (shLCP1-1, shLCP1-2) restores αMβ2 integrin cell-surface levels. Expression of cell-surface αMβ2 integrin was determined using FACS (values are normalized to shCtrl–GFP; mean±s.e.m.; n=3; *P<0.05; **P<0.01, n.s., not significant). (I,K) Western blot and densitometric analysis of cell lysates from shCtrl and LCP1-depleted PLB985 (I) and Raw264.7 (K) cells for total β2 integrin protein levels. Actin served as a loading control to quantify β2 integrin levels (mean±s.e.m.; n=3; *P<0.05; n.s.; not significant). (J,L) Degradation of cell-surface β2 integrin was determined in PLB985 (J) and Raw264.7 (L) cells through biotinylation of cell-surface proteins and incubation for 0 h or 24 h, followed by streptavidin pull-down, western blot analysis and quantification (mean±s.e.m.; J, n=3; L, n=4; **P<0.01; n.s., not significant).
Since LCP1 overexpression locks αMβ2 integrin in an inactive state, we expected that LCP1 depletion would render αMβ2 integrins more active. To deplete LCP1 we retrovirally expressed two different short hairpin RNAs (shRNA) in differentiated PLB985 and macrophage-like Raw264.7 cells (Fig. 3D; Fig. S5B), which reduced LCP1 levels by ∼85% and ∼70%, respectively. Surprisingly, LCP1 depletion did not change the levels of active αMβ2 in differentiated PLB985 cells (Fig. 3E) but instead increased the cell-surface levels of αMβ2 integrin in differentiated PLB985 (Fig. 3F) and Raw264.7 (Fig. S5C) cells without significantly altering αM and β2 integrin mRNA levels (Fig. S5D,E). Importantly, the increased αMβ2 integrin levels could be normalized after re-expression of shRNA-resistant wild-type LCP1 (Fig. 3G,H). LCP1 could potentially change cell-surface levels of β2 integrin without altering mRNA levels by affecting the total amount of β2 integrin protein in the cell or through altered surface presentation. We therefore analyzed β2 integrin protein levels and the stability of cell-surface β2 integrins in control and LCP1-depleted PLB985 and Raw264.7 cells. LCP1 levels had no effect on β2 integrin stability and protein levels in PLB985 cells (Fig. 3I,J) suggesting that changes in β2 integrin surface levels are due to altered protein trafficking. In contrast, LCP1 depletion in Raw264.7 cells reduced β2 integrin degradation and as consequence increased the total β2 integrin protein levels (Fig. 3K,L).
In line with a role of LCP1 as negative regulator of integrin activation, LCP1-depleted Raw264.7 cells showed increased adhesion to the integrin αMβ2 ligands fibrinogen and intercellular cell adhesion molecule-1 (ICAM-1) without prior agonist-induced integrin activation (Fig. 4A–D), even after sorting for equal αMβ2 integrin levels to control Raw264.7 cells (Fig. 4E,F). Interestingly, the increased adhesion of LCP1-depleted Raw264.7 cells was lost upon stimulation with PMA, which is a strong integrin activator (Fig. 4A,B), but not upon stimulation with TNFα (Fig. 4C,D). Similar results were obtained with LCP1-depleted differentiated PLB985 cells, which adhered more strongly to fibrinogen, and this effect was reversed on re-expression of LCP1 (Fig. 4G–I). Since phosphorylation of LCP1 on serine-5 and serine-7 regulates its actin bundling activity and localization (Morley, 2012), we next tested the effect of LCP1 phosphorylation on cell adhesion in PLB985 cells and its ability to interact with clasped αMβ2 integrin. To this end, we substituted serine-5 and serine-7 of LCP1, the major phosphorylation sites located in its regulatory headpiece domain (Lin et al., 1998; Shinomiya et al., 1995), with alanine (LCP1 SS/AA) or glutamic acid (LCP1 SS/EE). The serine-to-glutamic acid exchange strongly reduced the binding of GFP-tagged LCP1 (GFP–LCP1 EE) to Jun–Fos-dimerized αMβ2 TMcyto bait proteins compared to wild-type LCP1 and LCP1 SS/AA (Fig. 4H). As a result, expression of GFP–LCP1 EE in LCP1-depleted PLB985 cells failed to reduce cell adhesion to fibrinogen (Fig. 4I).
Fig. 4.
LCP1 regulates αMβ2 integrin-mediated cell adhesion. (A–D) Quantification of cell adhesion of LCP1-depleted Raw264.7 cells on fibrinogen (10 µg/ml) (A,C) and ICAM-1 (4 µg/ml) (B,D) for 30 min with or without PMA (A,B) or TNFα (C,D) treatment; values are normalized to control cells (shCtrl) (mean±s.e.m.; n=3; *P<0.05; **P<0.01; n.s., not significant). (E) FACS analysis of β2 integrin cell-surface expression on LCP1-depleted Raw264.7 cells sorted for equal β2 surface levels (values are normalized to shCtrl cells; mean±s.d., n=3, n.s. not significant). (F) Quantification of cell adhesion of control and LCP1-depleted Raw264.7 cells with equal αMβ2 integrin surface levels for 30 min on fibrinogen (10 µg/ml); values are normalized to control cells (shCtrl) (mean±s.e.m., n=4, *P<0.05). (G) Western blot of LCP1 and GFP in differentiated LCP1-depleted PLB985 cells expressing GFP, or GFP-tagged wild-type or mutant LCP1 variants. (H) Western blot analysis to determine GFP–LCP1 binding to αMβ2–TMcyto domains with antibodies against GFP. Cell lysates of differentiated LCP1-depleted PLB985 cells expressing GFP, or GFP-tagged wild-type or mutant LCP1 variants, incubated with His–αM–TMcyto or clasped His–αM–TMcyto/3×FLAG–β2–TMcyto domains bound to Talon beads. Binding of recombinant integrin tails to the resin was verified using Coomassie Blue staining. A representative experiment of three independent experiments is shown. (I) Quantification of cell adhesion of differentiated control and LCP1-depleted PLB985 cells expressing GFP, or GFP-tagged wild-type or mutant LCP1 variants for 30 min on fibrinogen (100 µg/ml); values are normalized to control cells (shCtrl) (mean±s.d., n=5, *P<0.05).
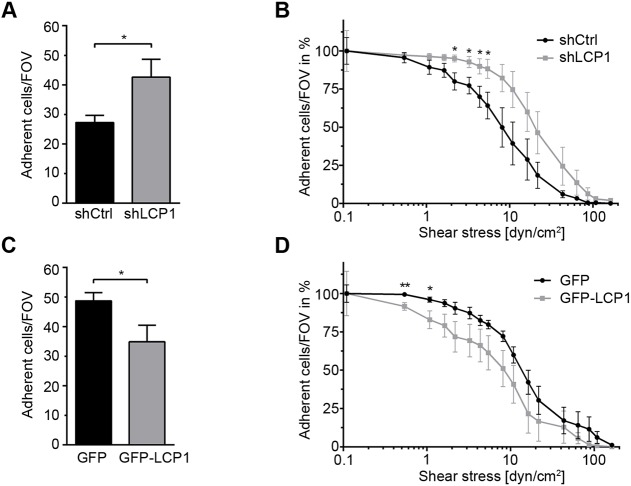
To test the influence of LCP1 on adhesion of PLB985 cells under flow conditions, we analyzed adhesion of LCP1-depleted and GFP–LCP1 overexpressing PLB985 cells using a microflow chamber. The experiments revealed that the number of adherent LCP1 knockdown cells per field of view (FOV) in fibrinogen-coated microflow chambers is significantly increased compared to shCtrl cells at baseline condition (Fig. 5A). By increasing shear stress levels in the flow chamber, control adherent cells detached faster compared to adherent LCP1-depleted PLB985 cells, indicating that the reduction in LCP1 levels induced resistance against shear forces (Fig. 5B). Conversely, GFP–LCP1-overexpressing cells adhered less compared to control cells at low shear (Fig. 5C) and detached faster after increasing the shear stress (Fig. 5D). Taken together, these results suggest that LCP1 negatively regulates adhesion strength and resistance to shear forces by maintaining αMβ2 integrin in an inactive conformation.
Fig. 5.
LCP1 regulates αMβ2 integrin-mediated adhesion under shear flow. LCP1-overexpressing (GFP–LCP1) and LCP1-depleted (shLCP1) differentiated PLB985 cells and their controls (GFP and shCtrl, respectively) were allowed to attach to fibrinogen-coated flow chambers for 30 min, and then flow was initiated and flow rates increased every 30 s until all adherent cells detached. (A,C) Quantification of adherent (A) shLCP1 versus shCtrl cells/FOV and (C) GFP–LCP1 versus GFP cells/FOV at baseline conditions. (B,D) Percentage of adherent (B) shLCP1 versus shCtrl cells/FOV and (D) GFP–LCP1 versus GFP cells/FOV exposed to different shear stress levels (mean±s.e.m., n=7–8, **P<0.01, *P<0.05).
DISCUSSION
Integrins rely on the recruitment of proteins to their cytoplasmic tails to regulate their activity state, establish a connection to the actin cytoskeleton, initiate intracellular signaling and regulate their intracellular trafficking (Legate and Fassler, 2009; Margadant et al., 2011). Previous interaction studies identified cytoplasmic interactors of isolated α and β integrin subunits but most approaches are unable to detect interactors that require hetero-association of both integrin tails for binding. Therefore, we generated bait proteins for pull-down assays that consisted of clasped α/β integrin TMcyto domains. To ensure that integrin-interacting proteins requiring TMcyto–protein as well as plasma membrane–lipid interactions did not escape our detection, we embedded the clasped α/β TMcyto domains into bicelles. We used quantitative MS to compare the interactome of individual α and β integrin subunits (representing the unclasped, active conformation of an integrin transmembrane and cytoplasmic domain) with the clasped α/β integrin TMcyto domain (representing the inactive TMcyto-associated heterodimeric state) and found several integrin interactors that preferably bound to either single or clasped integrin TMcyto domains.
One important modification of our proteomic approach is the incorporation of integrin TMcyto domains into bicelles as a membrane-mimicking system to stabilize and support protein interactions with the integrin cytoplasmic tails. Integrins contain only short cytoplasmic domains and recruit their intracellular interactors in close proximity or even in direct contact with the plasma membrane. A number of intracellular integrin interactors, including talins and kindlins, contain membrane-binding sites that increase the association constant upon binding integrin tails and charged lipids (Anthis et al., 2009; Goult et al., 2010, 2009; Liu et al., 2011; Moore et al., 2012; Perera et al., 2011). Using integrin bait proteins incorporated into bicelles, we identified four, 43 and 156 proteins that were pulled down with α5, αM and αIIb cytoplasmic tails, respectively, and between 55 and 107 proteins with the different β integrin subunits. The number of potential interactors is slightly higher but still in the range of previously published proteomics interaction studies with α or β integrin peptides (Raab et al., 2010; Schiller et al., 2013). The increased number of interactors could be the result of less stringent, detergent-free washing conditions to preserve bicelle integrity or the presence of a membrane to facilitate and stabilize potential interactions. Indeed, the binding of talin and kindlin-2 to the β1 TMcyto domain increases after incorporation of the β1 TMcyto bait into bicelles. The charged membrane either presents an additional binding motif that adds to the overall affinity or favors optimal alignment of the two interactors for high-affinity binding, as has been shown for interaction of the talin FERM domain with the β3 cytoplasmic tail in the presence of negatively charged PtdIns(4,5)P2 (Moore et al., 2012). Despite the different pull-down approaches, we observed similar trends to those previously published regarding the number of interactors with the individual α or β cytoplasmic domains. For example, only a few proteins bind to α5 tails as compared to αIIb (Raab et al., 2010). Likewise, there are a higher number of β3 integrin interactors compared to β1 integrins (Schiller et al., 2013). Further modification of the bicelle composition, for example by adding specific phosphatidyl-inosites, would allow the analysis of protein recruitment to integrin cytosolic domains in specific membrane compartments.
An important property of integrins, usually not taken into account in proteomic-based screening approaches for interactors, is their ability to switch between conformational states. In their inactive state the TMcyto domains of the α and β integrin subunits are associated, while they become separated upon integrin activation. Cells distinguish between the two conformations during integrin activation on the cell surface to control ligand binding and the conformation-specific trafficking of integrins through the endosomal system (reviewed in De Franceschi et al., 2015). Evidence suggests that the inactive conformation of integrins may not be a default state but is dynamically regulated by the binding of intracellular proteins, referred to as integrin inactivators (Bouvard et al., 2013). Intracellular integrin inactivators trigger integrin inactivation and/or maintain integrins in an inactive state, thereby establishing a regulatory mechanism to efficiently prevent unwanted integrin activation. This mode of regulation is particularly important for αIIbβ3 integrin and β2-class integrins expressed on hematopoietic cells to prevent blood clotting or unwanted attachment of leukocytes to the vessel wall, with dramatic consequences for immune functions (Schmidt et al., 2013). With the exception of filamin-2 and moesin, however, we were unable to detect other prominent integrin tail-binding inactivators including sharpin, ICAP, etc. in our pull-down experiments.
To mimic the inactive heterodimeric integrin cytoplasmic tail conformation we developed a bait for pull-down experiments based on the Jun–Fos dimerization domains, which have been used to maintain heterodimeric association of recombinantly expressed α/β integrin ectodomains (Eble et al., 1998; Raynal et al., 2006). Unexpectedly, we observed a large number of proteins pulled down with the Jun–Fos-dimerized αMβ2 TMcyto tails compared to the single αM or β2 TMcyto domain. This could be due to the low stringency binding and washing conditions that pulled down low-affinity binding proteins and complexes as well as unspecific interactors with the Jun–Fos domains. Among the dimer-specific interactors, we identified filamin-2 and moesin, which have been shown to promote integrin inactivation (Das et al., 2011; Vitorino et al., 2015). Filamins have been established as negative regulators of integrin activation by competing with talin-1 for integrin β tails (Das et al., 2011; Ithychanda et al., 2009; Kiema et al., 2006). A recent NMR study showed that filamin-1 forms a ternary complex with the αIIb and β3 cytoplasmic tails to stabilize the integrin inner membrane clasp and thereby retains the integrin in a resting state (Liu et al., 2015). Moesin, a member of the ezrin, radixin and moesin (ERM) protein family, is phosphorylated by MAP4K4 and displaces talin-1 from integrin β tails, leading to integrin inactivation (Vitorino et al., 2015). Our proteomics data also suggest that moesin has binding sites for α and β tails and therefore binds associated αMβ2 integrins. However, further structural and biochemical analyses are necessary to support this hypothesis.
In addition to filamin-2 and moesin, we identified LCP1 as an interactor of clasped, inactive αMβ2 TMcyto tails. LCP1 is a member of the α-actinin family of actin crosslinking proteins, with a restricted expression pattern in hematopoietic cells and cancer tissue (reviewed in Delanote et al., 2005; Morley, 2012). Several members of the α-actinin family of proteins, particularly α-actinin-1, as well as filamin-1, link the actin cytoskeleton to the plasma membrane via interaction with integrins (Otey et al., 1990; Pavalko and LaRoche, 1993; Sharma et al., 1995). Previous studies have shown that LCP1 binds to the cytoplasmic domains of β1 and β2 integrins via its actin-binding domain (Le Goff et al., 2010). However, a structural characterization of how LCP1 engages the heterodimeric integrin cytoplasmic face has not been reported. Our pull-down and immunoprecipitation experiments suggest that LCP1 preferentially binds to clasped αMβ2 TMcyto tails and exhibits a lower affinity for the isolated αM and β2 cytoplasmic domains. A similar increased affinity has been observed for the ternary binding of filamin-1 to the cytoplasmic domains of αIIb and β3 integrin under clasped conditions (Liu et al., 2015).
To address the role of the LCP1–αMβ2 integrin complex, we analyzed the consequences of altering LCP1 protein levels on integrin activation in leukocytes. We found that LCP1 depletion leads to increased adhesion of macrophages and differentiated neutrophil-like PLB985 cells under resting conditions while LCP1 overexpression reduced the levels of active αMβ2 integrins on differentiated PLB985 cells. The increased adhesion of LCP1-depleted Raw264.7 cells to fibrinogen and ICAM-1 was lost upon treatment with PMA but not TNFα, confirming that PMA is the stronger integrin activator (Harokopakis and Hajishengallis, 2005). These findings suggest that the LCP1–αMβ2 complex stabilizes inactive integrins to prevent spontaneous and/or excess integrin activation in resting hematopoietic cells and the pathological consequences such as thrombosis or inflammation. Interestingly, neutrophils isolated from LCP1 knockout mice have previously been reported to show diminished respiratory burst generation and increased adhesion under non-stimulatory conditions, while adhesion was normal after PMA, fMLP and TNFα stimulation (Chen et al., 2003). In addition, in previous studies LCP1 expression in CV-1 fibroblasts showed fewer and smaller focal contacts, often leading to the rounding-up of the cells (Arpin et al., 1994; Timmers et al., 2002). These observations would be in agreement with our findings that LCP1 stabilizes the inactive state of integrins. However, it has to be noted that LCP1 overexpression or knockdown could impact integrin function independent of its direct interaction with integrin cytosolic domains and that it might have additional functions affecting cell adhesion, including cytoskeletal alterations through its actin-bundling activity. We observed reduced binding of LCP1 SS/EE to clasped αMβ2 TMcyto domains. However, these mutations also increase the F-actin-binding activity of LCP1. With more structural information available, it will be possible to design LCP1 mutants that abolish integrin binding without affecting their interaction with actin to resolve the role of LCP1 in integrin activity regulation.
Finally, we found that depletion of LCP1 increased β2 integrin stability and cell-surface levels of αMβ2 integrin without changing β2 integrin mRNA levels in Raw264.7 cells, suggesting an additional role of LCP1 in reducing surface trafficking of inactive αMβ2 integrin. In contrast, depletion of LCP1 in differentiated PLB985 cells had only a small effect on cell-surface levels of αMβ2 integrin, and no discernable effect on integrin stability. Since cell-surface levels of β2 integrin were previously shown to remain unchanged in LCP1-depleted PMNs (Chen et al., 2003), it is possible that the effect of LCP1 on cell-surface levels of integrins is cell type-dependent. Several actin-regulatory proteins, including α-actinin-1 and LCP1, are involved in transmembrane protein trafficking (Burgueño et al., 2003; Foran et al., 2006; Schulz et al., 2004). Previous studies reported a role of Sac6P, the yeast LCP1 homolog, in the organization of the actin cytoskeleton and endocytosis of the maltose transporter (Adams et al., 1995; Penalver et al., 1997; Skau et al., 2011). Other studies have shown a role for LCP1 in E-cadherin endocytosis in colon cancer cells (Foran et al., 2006). Finally, there is evidence that LCP1 may also regulate the trafficking of transmembrane proteins by binding Rab5, a critical GTPase of the endocytic pathway (Hagiwara et al., 2011).
In conclusion, our study reveals that LCP1 regulates integrin-mediated cell adhesion by stabilizing the clasped conformation of αMβ2 integrin. Further investigations will be required to show how and to what extent α/β tail-binding integrin inactivators regulate dynamic adhesion and migration across a variety of cell types.
MATERIALS AND METHODS
Antibodies
The following antibodies, raised against the listed proteins, were used: Talin (8d4, Sigma; 1:1000), Kindlin-2 (MAB2617, Millipore; 1:1000), His (2365, Cell Signaling Technology; 1:1000), FLAG-M2-HRP (A8592, Sigma; 1:10,000), GAPDH (CB1001, Calbiochem; 1:2000), LCP1 (GTX 114524, Genetex; 1:1000), GFP (A11122, Invitrogen; 1:1000), β2 integrin (gift from Dr Melanie Laschinger, Technical University of Munich, Germany; 1:200), β2 integrin (sc-8420, Santa Cruz Biotechnology; 2-4 µg), activated αM integrin (301402, BioLegend; 1:100), αM integrin (301302, BioLegend; 1:100), MAC1-PE (12-0112, eBioscience; 1:200), MAC1-biotin (557395, PharMingen; 1:200), MBP-HRP (E8038, New England Biolabs; 1:1000).
The following secondary antibodies were used: goat anti-mouse Alexa Fluor 546 (A-11003, Life Technologies; 1:200), goat anti-mouse Alexa Fluor 488 (A-11029, Life Technologies; 1:200) donkey anti-mouse Alexa Fluor 647 (A-31571, Life Technologies; 1:200), goat anti-rat HRP (712035150, Dianova; 1:10,000), goat anti-mouse HRP (172-1011, Bio-Rad; 1:10,000) and goat anti-rabbit HRP (172-1019, Bio-Rad; 1:10,000), streptavidin-Cy5 (016160084, Dianova; 1:200).
Recombinant human ICAM-1/CD54 (ADP4-200) and mouse TNFα (410-MT) were obtained from R&D Systems.
Plasmids
For recombinant bacterial expression, integrin β1–TMcyto (residues from 719 to 799 of the full-length mouse β1 integrin subunit), integrin β2–TMcyto (residues from 698 to 771 of the full-length mouse β2 integrin subunit), β3–TMcyto (residues from 714 to 787 of the full-length mouse β3 integrin subunit), α5–TMcyto (residues from 993 to 1054 of the full-length mouse α5 integrin subunit), αIIb–TMcyto (residues from 986 to 1033 of the full-length mouse αIIb integrin subunit), αM–TMcyto (residues from 1106 to 1153 of the full-length mouse αM integrin subunit) were cloned in-frame with the Fos dimerization domain (residues from 161 to 200 of full-length mouse Fos protein) (αIIb–TMcyto, αM–TMcyto and α5–TMcyto) or Jun dimerization domain (residues from 277 to 318 of full-length mouse Jun protein) (β1–TMcyto, β2–TMcyto and β3–TMcyto) and subcloned into the pET15b vector (Clontech) to generate His–Fos–α5–TMcyto, His–Fos–αIIb–TMcyto, His–Fos–αM–TMcyto, His–Jun–β1–TMcyto, His–Jun–β2–TMcyto, and His–Jun–β3–TMcyto constructs. β2–cyto (residues from 726 to 771 of the full-length mouse β2 integrin subunit) was cloned into pCoofy35 and αM–cyto (residues from 1130 to 1153 of the full-length mouse αM integrin subunit) was cloned into pCoofy17 to generate MBP–β2–cyto and His–αM–cyto, respectively. The 6×His tag sequence was replaced with a 3×FLAG tag within pET16b vector to generate 3×FLAG–Jun–β2–TMcyto.
To stably knock down LCP1 in PLB985 cells, shRNA targeting the human LCP1 sequences were introduced into the pSUPER.retro vector (OligoEngine) to produce retroviral particles: 5′-AGTAGCCTCTCCTGTATTT-3′ (shLCP1-1), 5′-AGAAGCTGCAGTGGTATTA-3′ (shLCP1-2). In Raw264.7 cells, shRNA target sequences directed against the mouse LCP1 sequence were introduced into the pSUPER.retro vector to produce retroviral particles: 5′-GATGGCATAGTTCTTTGTA-3′ (shLCP1-1), 5′-CAAGTAGCTTCTGCTATAA-3′ (shLCP1-2). The open reading frame of LCP1 was amplified through the use of PCR from the OCAA human library clone OCAAo5051D1183D (imaGenes) and cloned into pJET1.2 cloning vector (ThermoFisher). LCP1 SS/AA and SS/EE in which the serine 5 and serine 7 were substituted with alanine or glutamic acid were cloned using site-directed mutagenesis. The different LCP1 variants were cloned into pEGFP-N1 (Clontech), and RRL-CMV-GFP lentiviral vector (provided by Dr Alexander Pfeifer, University of Bonn, Germany). Wild-type LCP1 was subcloned into pGEX-6P-1 (GE Healthcare) to express GST-tagged LCP1.
Cell culture
Raw264.7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) and PLB985 cells in RPMI (Gibco), both supplemented with 10% (v/v) FBS at 37°C and 10% CO2, and free of mycoplasma contamination. Differentiation of PLB985 cells into neutrophil-like cells was carried out as described (Pivot-Pajot et al., 2010). Briefly, differentiation was induced through incubation of PLB985 cells in RPMI medium supplemented with 5% (v/v) FBS and 1.25% (v/v) DMSO for five days. The medium was changed once on day three of the differentiation period.
Bone marrow was isolated from C57BL/6 mice and passed through a 70 µm cell strainer (BD Biosciences) to obtain single cell suspensions. 4×106 cells were seeded in 15 cm petri dishes (non-treated) and cultured at 37°C and 5% CO2 in RPMI medium supplemented with 10% (v/v) FBS and 10% macrophage-colony stimulating factor (M-CSF) as described before (Schmidt et al., 2011). Non-adherent cells were removed after 24 h and adherent cells were cultured for an additional 6 days. The cells were lysed in hypotonic lysis buffer [10 mM Tris-HCl, pH 7.6; 5 mM KCl; 1.5 mM MgCl2; 1 mM dithiothreitol (DTT), protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail 2 and 3 (Sigma)] and cleared by centrifugation.
Transient and stable transfection and transduction
Cells were transiently transfected using Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol. To generate stable cell lines, vesicular stomatitis virus G glycoprotein (VSV-G) pseudotyped retroviral and lentiviral vectors were produced through transient transfection of 293T (human embryonic kidney) cells. Viral particles were concentrated from cell culture supernatant as previously described (Pfeifer et al., 2000) and used for infection.
Flow cytometry
Flow cytometry was carried out with a FACSCantoTMII cytometer (BD Biosciences) equipped with FACS DiVa software (BD Biosciences) using standard procedures. Data analysis was carried out with the FlowJo program (version 9.4.10). Cells were incubated with primary antibodies diluted in FACS buffer [1% bovine serum albumin (BSA) in PBS (Sigma)] for 1 h on ice, washed twice with cold FACS buffer and finally incubated with the secondary antibody for 1 h on ice.
Adhesion assay
To measure cell adhesion, Raw264.7 and PLB985 cells were washed, resuspended in FBS-free growth medium and either left untreated or treated for 30 min with 100 ng/ml PMA or TNFα. Adhesion assays were performed in 96-well flat-bottom plates coated for 2 h at room temperature with 10 µg/ml fibrinogen (Sigma) or 4 µg/ml ICAM-1 (R&D systems). PLB985 cells were left to adhere on 96-well flat-bottom plates coated with 100 µg/ml fibrinogen (Sigma). Unspecific binding to the plates was blocked through incubating the wells with 1% BSA/PBS. The wells were washed once with PBS to remove excess BSA before seeding the cells (1×105 cells/well). After adhesion at 37°C, the wells were washed by means of immersion into a plastic tray containing PBS. Adhered cells were fixed with methanol and stained with Crystal Violet overnight (20% methanol, 0.1% Crystal Violet in H2O). After intense washings, cells were solubilized in 0.5% Triton X-100, and the number of cells was determined by measuring the absorbance at 595 nm using an ELISA reader.
Flow chamber assay
A detachment flow chamber assay was performed as described (Schymeinsky et al., 2009). Briefly, microflow chambers (2×0.2 mm; VitroCom) were coated with human fibrinogen (100 µg/ml in 0.1% BSA/PBS, Sigma) for 3 h at room temperature and blocked with 5% casein (Sigma-Aldrich) overnight at 4°C. Before the experiment, chambers were washed with 0.9% NaCl solution (Fresenius Kabi) and then 106 cells/ml (shLCP1, shCtrl, GFP or GFP–LCP1 PLB985 cells) suspended in perfusion medium (HBSS buffer containing 1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 0.25% BSA, 0.1% glucose, pH 7.4) were introduced into the flow chamber. Flow was stopped for 30 min to allow cells to attach. Using a high-precision syringe pump (Harvard Apparatus), flow was started at very low shear stress (<0.5 dyn/cm2) for 60 s to remove debris, and then flow rates were increased every 30 s up to a maximum of 160 dyn/cm2. Experiments were performed with a Zeiss Axioskop2 (equipped with a 20× water objective, 0.5 NA and a Hitachi KP-M1AP camera) and recorded with VirtualDub (Version 1.9.11). The number of adherent cells/FOV was analyzed off-line for baseline conditions and indicated in percentage of baseline adhesion for every shear stress level in the generated movies.
Bicelle preparation
6-cyclohexyl-1-hexylphosphocholine (Cyclofos-6) was purchased from Anatrace. 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) was purchased from Avanti Polar Lipids and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) was purchased from CordenPharma. Lipid stock solutions were prepared by weighing out 500 mg of DMPC in 5 ml of cyto buffer (139 mM K2HPO4, 8.8 mM NaH2PO4, 0.4 mM MgCl2, 3.2 mM NaCl, pH 7.0), 200 mg of Cyclofos-6 in 4 ml of cyto buffer and 75 mg of POPS in 3.75 ml of cyto buffer. For 1 ml 3% bicelle solution, q=4 bicelles, 236 µl DMPC stock, 152 µl POPS stock, and 68 µl Cyclofos-6 stock plus 544 µl cyto buffer were mixed and repeatedly subjected to a temperature cycle: 45°C for 2 min, 5°C (or ice water) for 5 min. In between, the solution was carefully mixed to avoid foaming. The temperature cycle was repeated 10 times until the solution became clear. For 1 ml 3% q=0.25 bicelles, 72 µl DMPC stock, 56 µl POPS stock, and 400 µl Cyclofos-6 stock plus 472 µl of cyto buffer were mixed and incubated as described above. The stocks were snap frozen in liquid nitrogen and stored at −80°C.
LC-MS to determine lipid composition of bicelles
Bicelle samples (1%) were diluted 1:100 with 0.05% trifluoroacetic acid in H2O for determination of their lipid compositions by liquid chromatography-mass spectrometry (LC-MS) using a micrOTOF (Bruker Daltonik) connected to an 1100 HPLC (Agilent). Analyses were performed on a YMC-Pack Butyl 30 mm column (YMC) with a water:acetonitrile (0.05% trifluoroacetic acid) gradient from 30–80% in 15 min at a mass range from 200–2000 m/z in positive mode. Extracted ion chromatograms at 350.2, 678.5 and 762.5 m/z were used to compare the relative ratio of the individual lipids.
Dynamic light scattering
Bicelle samples with different ratios of DMPC, POPS and Cyclofos-6 were freshly prepared, and measured with or without integrin TMcyto domain incorporation. The samples were measured using DLS (DynaPro NanoStar from Wyatt Technology). Each sample was measured at 4°C, the acquisition time was set to 5 s (×10) and each measurement was repeated three times. The effective hydrodynamic radius was calculated with DYNAMICS V7.1.7.16 (Wyatt Technology).
Expression and purification and incorporation of recombinant proteins into bicelles
His–αM–cyto, maltose binding protein (MBP)–β2–cyto, glutathione S-transferase (GST)–LCP1 were transformed into BL21(DE3) Arctic Express E. coli, and protein expression was induced with 1 mM IPTG at 18°C for 24 h before purification of the tagged proteins using Ni-NTA beads (Qiagen, 30210), Amylose resin (NEB, E8021), or GST•Bind Resin (Millipore, 70541) according to manufacturer's instruction. Plasmids encoding His–Fos–α5–TMcyto, His–Fos–αIIb–TMcyto, His–Fos–αM–TMcyto, 3×FLAG–Jun–β2–TMcyto, His–Jun–β1–TMcyto, His–Jun–β2–TMcyto and His–Jun–β3–TMcyto were transformed into BL21 (DE3) Arctic Express E. coli, and protein expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) (30°C, 3 h for α integrin subunits TMcyto domains; 18°C, 24 h for β integrin subunits TMcyto domains). Afterwards, bacteria were pelleted using centrifugation, resuspended in TBS buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl) containing 100 µg/ml lysozyme and 50 µg/ml DNase (ThermoFisher) and rotated at 4°C for 2 h. After the addition of Empigen (Sigma) (30% solution; 1 ml per 10 ml of lysate) the bacterial lysates were rotated at 4°C for 1 h and centrifuged 1500 g at 4°C for 1 h. To purify His- and FLAG-tagged proteins, supernatants were incubated with Ni-NTA agarose beads (Qiagen, 36113) and anti-FLAG M2 affinity gel (Sigma, A2220) for 2 h at 4°C followed by extensive washing of the beads three times with TBS buffer and twice with pre-equilibration buffer [20 mM imidazole; 50 mM Tris-HCl, pH 6.5; 150 mM NaCl in 1% bicelles, q=0.25, as described (Lu et al., 2012)]. The 6×His-tagged proteins were eluted with elution buffer (250 mM imidazole; 50 mM Tris-HCl, pH 7.4; 150 mM NaCl in 1% bicelles). The proteins were concentrated using Amicon Ultra Centrifugal filters (10 kDa molecular weight cutoff, Millipore) by a factor of 10 to obtain a final bicelle concentration of 10%.
Bicelle-incorporated integrin TMcyto pull-down assay
For pull-down experiments, 9× volume of cyto buffer (139 mM K2HPO4, 8.8 mM NaH2PO4, 0.4 mM MgCl2, 3.2 mM NaCl, pH 7.0) was added to 10% TMcyto–bicelle solution to obtain a 1% bicelle solution. To increase the bicelle size after protein incorporation from q=0.25 bicelles into q=2 bicelles, 5.2× volume of 1% q=4 bicelles was added. Integrin TMcyto domains incorporated into q=2* bicelles were incubated with the soluble fraction of bone marrow-derived macrophage (BMDM) lysates generated with hypotonic buffer [10 mM Tris-HCl, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT, protease inhibitor cocktail (Roche)] overnight at 4°C. Importantly, maximal 1.6× the volume of concentrated lysate was added to the TMcyto bicelle solution, which kept the bicelle concentration (above 0.35%) and the Cyclofos-6 concentration (above 2.7 mM) high enough to support bicelle formation. Afterwards, the bicelle–lysate mixture was incubated with Ni-NTA magnetic agarose beads (Qiagen) for 2 h at 4°C. After three washes with q=2* bicelle solution in cyto buffer, proteins were eluted from the beads by boiling with 80 µl 4× Laemmli sample buffer for 5 min, separated using SDS-PAGE and analyzed by means of LC-MS/MS or western blotting.
Mass spectrometry analysis of the pull-down samples
The samples were eluted from Ni-NTA magnetic agarose beads by boiling with Laemmli sample buffer and separated on NuPAGE Novex 4–12% Bis-Tris protein gels (ThermoFisher). The gel was fixed in 50% methanol, 10% acetic acid, 40% H2O for 30 min at room temperature and stained with GelCode Blue Safe Protein Stain reagent (ThermoFisher). Each lane was cut into three bands and digested using the standard in-gel digestion protocol (Shevchenko et al., 2006). Briefly, the gel bands were cut into roughly 1 mm cubes and de-stained in ethanol solution before incubation with 20 mM DTT and 40 mM chloroacetamide to reduce and alkylate the proteins. The gel pieces were then rehydrated in trypsin solution (12.5 ng/µl in 50 mM ammonium bicarbonate) and incubated overnight at 37°C. After overnight digestion, the peptides were extracted in 30% acetonitrile and 3% trifluoroacetic acid solution followed by 100% acetonitrile solution. The extracted peptides were then desalted and concentrated using C18 StageTips (Rappsilber et al., 2003) prior to LC-MS/MS analysis. The peptides were separated on a 120 min gradient in a 15 cm reversed phase column [75 µm inner diameter columns (New Objective) packed in-house with 3 µm Reprosil C18 beads (Dr. Maisch HPLC) using EASY-nLC II (ThermoFisher)] and sprayed directly into a LTQ Orbitrap XL mass spectrometer via a nano-electrospray ion source (ThermoFisher).
Peptides were analyzed using a top five data-dependent acquisition method. Survey scans were acquired in the Orbitrap at a resolution of 60,000 (400 m/z) after accumulating a target of 1×106 ions within a maximum injection time of 1000 ms. From the survey scan, up to the top five most abundant precursors were selected and fragmented in the linear ion trap by means of collisional-induced dissociation with automatic gain control target value of 10,000 within a maximum injection time of 150 ms, and the fragmentation spectra were recorded in the ion trap. The peptide precursors selected for fragmentation were dynamically excluded for 90 s after a repeat count of one in order to minimize the repeat sequencing.
For each set of integrin heterodimer pull-downs, the raw files were processed using the MaxQuant computational platform (Cox and Mann, 2008) (version 1.5.1.8). The peak lists generated were searched against the Uniprot mouse proteome sequence database (59375 entries) using the Andromeda search engine (Cox et al., 2011). The peptide precursors were searched with an initial mass tolerance of 7 ppm and the fragment ions were searched with a tolerance of 0.5 Da. Carbamidomethylation of cysteine was used as a fixed modification and oxidation of methionine and amino terminal protein acetylation were set as variable modification for the database search. The minimum peptide length was set to six amino acids and the identifications were filtered at 1% for the peptide level and 5% for the protein level. The q-value (defined as local false discovery rate based on a target-decoy search with forward and reversed protein sequences) was used for assessing the confidence in the identification of individual proteins. The match between the runs feature was enabled and label-free protein quantitation was performed using the MaxLFQ algorithm (Cox et al., 2014).
All statistical analysis was performed using the Perseus bioinformatics platform (http://www.perseus-framework.org). Student’s t-tests were used to compare two samples with permutation-based FDR (4%) for multiple hypothesis testing.
Solid-phase ligand-binding assays
To determine GST–LCP1 binding to the integrin TMcyto domains, 96-well MaxiSorp plates (Nunc) were coated overnight at 4°C with purified His–Fos–αM–TMcyto, His–Jun–β2–TMcyto and the dimer composed of His–Fos–αM–TMcyto/3×FLAG–Jun–β2–TMcyto at 1.25 mmol/ml in 50 mM sodium bicarbonate buffer, pH 9.6. Plates were blocked with SuperBlock (TBS) blocking buffer (ThermoFisher) for several hours at room temperature, washed twice with washing buffer (50 mM Tris-HCl, pH 7.5, containing 200 mM NaCl and 0.05% Tween) before adding GST or GST–LCP1 in M-PER buffer (ThermoFisher) and incubating overnight at 4°C. After extensive washing with washing buffer, bound proteins were detected with an anti-LCP1 antibody, followed by an anti-rabbit HRP-conjugated secondary antibody. HRP substrate (ATBS, Vector) was added to the wells, and absorbance was monitored at 405 nm. GST was used as control for non-specific binding.
Pull-down experiments using recombinant proteins
To analyze the ternary complex between the cytosolic domains of αM integrin, β2 integrin, and LCP1, His–αM–cyto was incubated with Ni-NTA beads for 3 h at 4°C followed by washing of the beads three times with TBS buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl). Beads were incubated with GST, MBP–β2–cyto or GST–LCP1 and MBP–β2–cyto, and incubated for 2 h at 4°C. The beads were washed three times with TBS to remove unbound proteins. The proteins were eluted from the beads by boiling with 80 µl 4× Laemmli buffer for 5 min, separated by means of SDS-PAGE and analyzed through the use of western blotting with the anti-MBP antibody.
For the talin-1 head competition assays, recombinant His–Fos–αM–TMcyto and dimeric His–Fos–αM–TMcyto/3×FLAG–Jun–β2–TMcyto (5–10 µg) were incubated with 25 µl pre-cleared Talon Metal Affinity Resin (Clontech) in 0.5 ml TBS (50 mM Tris-HCl, pH 7.4; 150 mM NaCl) for 2 h at 4°C under rotation. The beads were washed three times with TBS and recombinant talin-1 head (80 µg), GST–LCP1 or a combination of both proteins were added to the beads in 0.5 ml binding buffer (50 mM Tris-HCl pH 7.5; 200 mM NaCl; 1 mM TCEP) supplemented with BSA 100 μg/ml. After incubation at 4°C overnight, the beads were washed extensively with binding buffer and the proteins were eluted twice with 50 μl elution buffer I (binding buffer plus 250 mM imidazol) using Spin Cups columns (ThermoFisher). Eluted samples were boiled with Laemmli loading buffer and analyzed by means of SDS-PAGE.
For the pull-down of GFP-tagged LCP1, PLB985 cells were lysed for 10 min on ice [lysis buffer: 150 mM NaCl; 1% Triton X-100; 50 mM Tris-HCl, pH 8.0; 2 mM EDTA; 0.05% Na-deoxycholate; Phosphatase Inhibitor Cocktail 2 and 3 (Sigma); cOmplete Mini Protease Inhibitor Cocktail Tablets (Roche)] and the lysate sonicated and cleared using centrifugation at 16,000 g for 10 min at 4°C. 1 mg of protein lysates were incubated with His–Fos–αM–TMcyto or dimeric His–Fos–αM–TMcyto/3×FLAG–Jun–β2–TMcyto (20 µg) bound to 50 µl pre-cleared Talon Metal Affinity Resin (Clontech) and incubated overnight at 4°C. Following repeated washes with lysis buffer, proteins were eluted from the beads using Laemmli buffer and analyzed by means of western blotting using antibodies against GFP. Binding of recombinant integrin tails to the resin was verified using Coomassie Blue staining.
Immunoprecipitation of β2 integrin
For the immunoprecipitation of endogenous β2 integrin, differentiated PLB985 cells expressing GFP–LCP1 were treated with DMSO or 100 ng/ml PMA for 30 min at 37°C before lysis in lysis buffer for 10 min on ice. After sonification and centrifugation at 16,000 g for 10 min at 4°C, 1 mg of protein lysates were incubated with 2–4 µg of the monoclonal mouse anti-β2 integrin antibody (Santa Cruz Biotechnology) for 2 h at 4°C under rotation. Washed Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology) (50 μl/sample) were added to the lysates and incubated overnight at 4°C under rotation. After three washes with lysis buffer and one with PBS, Laemmli loading buffer was added to the beads and boiled for 7 min at 95°C to elute bound proteins.
Stability of cell-surface integrins
The half-life of cell-surface proteins was determined as described previously by means of biotinylation (Böttcher et al., 2012). Briefly, cells were washed twice in cold PBS and surface-biotinylated with 0.2 mg/ml sulfo-NHS-LC-biotin (ThermoFisher) in PBS for 45 min at 4°C. Following washes with cold PBS the cells were incubated in regular growth medium for 0 h and 24 h at 37°C. Cells were lysed in IP buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% Triton X-100; 0.1% sodium deoxycholate; 1 mM EDTA; protease inhibitors) and biotinylated proteins were pulled down with streptavidin-sepharose (GE Healthcare). After three washes with lysis buffer, samples were analyzed using SDS-PAGE and western blotting.
Quantitative real-time polymerase chain reaction
Total RNA was isolated from cultured cells using the RNeasy mini kit (Qiagen), of which 1000 ng was transcribed into cDNA using the iScript cDNA Synthesis kit (Bio-Rad). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed with the LightCycler 480 (Roche) using SYBR green (Bio-Rad) and the following primers: β2 forward-1, 5′-CAGGAATGCACCAAGTACAAAGT-3′; β2 reverse-1, 5′-CCTGGTCCAGTGAAGTTCAGC-3′; β2 forward-2, 5′-AACGGAAACAGCTATCTCCAC-3′; β2 reverse-2, 5′-GAGTAGGAGAGATCCATGAG-3′; αM forward-1, 5′-ATGGACGCTGATGGCAATACC-3′; αM reverse-1, 5′-TCCCCATTCACGTCTCCCA-3′; αM forward-2, 5′-CCATGACCTTCCAAGAGAATGC-3′; αM reverse-2, 5′-ACCGGCTTGTGCTGTAGTC-3′; GAPDH forward, 5′-TCCTGCACCACCAACTGCTTAGC-3′; GAPDH reverse, 5′-TGGATGCAGGGATGATGTTCTGG-3′. Each sample was measured in triplicate and values were normalized to GAPDH.
Statistical analysis
Statistical significance was determined using two-tailed unpaired Student's t-test. Results are expressed as the mean±standard error of the mean (s.e.m.), unless indicated otherwise. Bar graphs throughout the study were generated in Microsoft Office.
Supplementary Material
Acknowledgements
We thank Dr Karim Dib (Queen's University Belfast, UK) for providing PLB985 cells, Dr Nagarjuna Nagaraj (MPI Biochemistry, Martinsried, Germany) for help with the proteomic analysis, Dr Raphael Ruppert (MPI Biochemistry, Martinsried, Germany) for his support with the FACS analysis, Dr Jonas Aretz (MPI Biochemistry, Martinsried, Germany) for the recombinant talin-1 head protein, Dr Sijo Mathew (Vanderbilt University, Nashville, USA) for help advice during bicelle preparation, Elisabeth Weyher for LC-MS analysis, and Hildegard Reiter for expert technical assistance. Some data in the paper forms part of the PhD thesis of Hui-yuan Tseng (2015, Ludwig-Maximilians-University, Munich, Germany).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.F., R.T.B.; Methodology: H.T., P.K., R.Z., C.R.S., R.F., R.T.B.; Validation: H.T., R.F., R.T.B.; Investigation: H.T., A.V.S., S.S., T.Z., R.I., M.S., R.T.B.; Writing - original draft: H.T., R.F., R.T.B.; Writing - review & editing: A.V.S., R.I., R.Z., M.S., C.R.S., R.F., R.T.B.; Supervision: R.F., R.T.B.; Funding acquisition: R.F., R.T.B.
Funding
The work was supported by U.S. Department of Veterans Affairs Merit Review (grant 1I01BX002196 to R.Z.), National Institutes of Health (grants R01-DK075594 and R01-DK383069221 to R.Z.), the Deutsche Forschungsgemeinschaft (SFB914, project A05 to R.T.B. and R.F.; project B01 to M.S.), Deutsches Zentrum für Herz-Kreislaufforschung (grant 81Z1600313 to R.F.), the European Research Council (grant agreement 322652 to R.F.) and the Max-Planck-Gesellschaft. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.218214.supplemental
Reference
- Adams A. E., Shen W., Lin C. S., Leavitt J. and Matsudaira P. (1995). Isoform-specific complementation of the yeast sac6 null mutation by human fimbrin. Mol. Cell. Biol. 15, 69-75. 10.1128/MCB.15.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthis N. J., Wegener K. L., Ye F., Kim C., Goult B. T., Lowe E. D., Vakonakis I., Bate N., Critchley D. R., Ginsberg M. H. et al. (2009). The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 28, 3623-3632. 10.1038/emboj.2009.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpin M., Friederich E., Algrain M., Vernel F. and Louvard D. (1994). Functional differences between L- and T-plastin isoforms. J. Cell Biol. 127, 1995-2008. 10.1083/jcb.127.6.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher R. T., Stremmel C., Meves A., Meyer H., Widmaier M., Tseng H.-Y. and Fässler R. (2012). Sorting nexin 17 prevents lysosomal degradation of beta1 integrins by binding to the beta1-integrin tail. Nat. Cell Biol. 14, 584-592. 10.1038/ncb2501 [DOI] [PubMed] [Google Scholar]
- Bouvard D., Pouwels J., De Franceschi N. and Ivaska J. (2013). Integrin inactivators: balancing cellular functions in vitro and in vivo. Nat. Rev. Mol. Cell Biol. 14, 430-442. 10.1038/nrm3599 [DOI] [PubMed] [Google Scholar]
- Burgueño J., Blake D. J., Benson M. A., Tinsley C. L., Esapa C. T., Canela E. I., Penela P., Mallol J., Mayor F. Jr, Lluis C. et al. (2003). The adenosine A2A receptor interacts with the actin-binding protein alpha-actinin. J. Biol. Chem. 278, 37545-37552. 10.1074/jbc.M302809200 [DOI] [PubMed] [Google Scholar]
- Calderwood D. A., Huttenlocher A., Kiosses W. B., Rose D. M., Woodside D. G., Schwartz M. A. and Ginsberg M. H. (2001). Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat. Cell Biol. 3, 1060-1068. 10.1038/ncb1201-1060 [DOI] [PubMed] [Google Scholar]
- Campbell I. D. and Humphries M. J. (2011). Integrin structure, activation, and interactions. Cold Spring Harb. Perspect Biol. 3, a004994 10.1101/cshperspect.a004994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Mocsai A., Zhang H., Ding R.-X., Morisaki J. H., White M., Rothfork J. M., Heiser P., Colucci-Guyon E., Lowell C. A. et al. (2003). Role for plastin in host defense distinguishes integrin signaling from cell adhesion and spreading. Immunity 19, 95-104. 10.1016/S1074-7613(03)00172-9 [DOI] [PubMed] [Google Scholar]
- Cox J. and Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367-1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V. and Mann M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794-1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N. and Mann M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513-2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M., Ithychanda S. S., Qin J. and Plow E. F. (2011). Migfilin and filamin as regulators of integrin activation in endothelial cells and neutrophils. PLoS ONE 6, e26355 10.1371/journal.pone.0026355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Franceschi N., Hamidi H., Alanko J., Sahgal P. and Ivaska J. (2015). Integrin traffic - the update. J. Cell Sci. 128, 839-852. 10.1242/jcs.161653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanote V., Vandekerckhove J. and Gettemans J. (2005). Plastins: versatile modulators of actin organization in (patho)physiological cellular processes. Acta Pharmacol. Sin. 26, 769-779. 10.1111/j.1745-7254.2005.00145.x [DOI] [PubMed] [Google Scholar]
- Eble J. A., Wucherpfennig K. W., Gauthier L., Dersch P., Krukonis E., Isberg R. R. and Hemler M. E. (1998). Recombinant soluble human alpha 3 beta 1 integrin: purification, processing, regulation, and specific binding to laminin-5 and invasin in a mutually exclusive manner. Biochemistry 37, 10945-10955. 10.1021/bi980175+ [DOI] [PubMed] [Google Scholar]
- Foran E., Mcwilliam P., Kelleher D., Croke D. T. and Long A. (2006). The leukocyte protein L-plastin induces proliferation, invasion and loss of E-cadherin expression in colon cancer cells. Int. J. Cancer 118, 2098-2104. 10.1002/ijc.21593 [DOI] [PubMed] [Google Scholar]
- Goult B. T., Bouaouina M., Harburger D. S., Bate N., Patel B., Anthis N. J., Campbell I. D., Calderwood D. A., Barsukov I. L., Roberts G. C. et al. (2009). The structure of the N-terminus of kindlin-1: a domain important for alphaiibbeta3 integrin activation. J. Mol. Biol. 394, 944-956. 10.1016/j.jmb.2009.09.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult B. T., Bouaouina M., Elliott P. R., Bate N., Patel B., Gingras A. R., Grossmann J. G., Roberts G. C. K., Calderwood D. A., Critchley D. R. et al. (2010). Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. EMBO J. 29, 1069-1080. 10.1038/emboj.2010.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara M., Shinomiya H., Kashihara M., Kobayashi K.-I., Tadokoro T. and Yamamoto Y. (2011). Interaction of activated Rab5 with actin-bundling proteins, L- and T-plastin and its relevance to endocytic functions in mammalian cells. Biochem. Biophys. Res. Commun. 407, 615-619. 10.1016/j.bbrc.2011.03.082 [DOI] [PubMed] [Google Scholar]
- Harokopakis E. and Hajishengallis G. (2005). Integrin activation by bacterial fimbriae through a pathway involving CD14, Toll-like receptor 2, and phosphatidylinositol-3-kinase. Eur. J. Immunol. 35, 1201-1210. 10.1002/eji.200425883 [DOI] [PubMed] [Google Scholar]
- Ithychanda S. S., Das M., Ma Y.-Q., Ding K., Wang X., Gupta S., Wu C., Plow E. F. and Qin J. (2009). Migfilin, a molecular switch in regulation of integrin activation. J. Biol. Chem. 284, 4713-4722. 10.1074/jbc.M807719200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. L., Wang J., Turck C. W. and Brown E. J. (1998). A role for the actin-bundling protein L-plastin in the regulation of leukocyte integrin function. Proc. Natl. Acad. Sci. USA 95, 9331-9336. 10.1073/pnas.95.16.9331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiema T., Lad Y., Jiang P., Oxley C. L., Baldassarre M., Wegener K. L., Campbell I. D., Ylanne J. and Calderwood D. A. (2006). The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell 21, 337-347. 10.1016/j.molcel.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Kim C., Ye F. and Ginsberg M. H. (2011). Regulation of integrin activation. Annu. Rev. Cell Dev. Biol. 27, 321-345. 10.1146/annurev-cellbio-100109-104104 [DOI] [PubMed] [Google Scholar]
- Lau T.-L., Dua V. and Ulmer T. S. (2008a). Structure of the integrin alphaIIb transmembrane segment. J. Biol. Chem. 283, 16162-16168. 10.1074/jbc.M801748200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau T.-L., Partridge A. W., Ginsberg M. H. and Ulmer T. S. (2008b). Structure of the integrin beta3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry 47, 4008-4016. 10.1021/bi800107a [DOI] [PubMed] [Google Scholar]
- Lau T.-L., Kim C., Ginsberg M. H. and Ulmer T. S. (2009). The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 28, 1351-1361. 10.1038/emboj.2009.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebart M.-C., Hubert F., Boiteau C., Ventéo S., Roustan C. and Benyamin Y. (2004). Biochemical characterization of the L-plastin-actin interaction shows a resemblance with that of alpha-actinin and allows a distinction to be made between the two actin-binding domains of the molecule. Biochemistry 43, 2428-2437. 10.1021/bi030151p [DOI] [PubMed] [Google Scholar]
- Lefort C. T., Hyun Y.-M., Schultz J. B., Law F.-Y., Waugh R. E., Knauf P. A. and Kim M. (2009). Outside-in signal transmission by conformational changes in integrin Mac-1. J. Immunol. 183, 6460-6468. 10.4049/jimmunol.0900983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate K. R. and Fassler R. (2009). Mechanisms that regulate adaptor binding to beta-integrin cytoplasmic tails. J. Cell Sci. 122, 187-198. 10.1242/jcs.041624 [DOI] [PubMed] [Google Scholar]
- Le Goff E., Vallentin A., Harmand P. O., Aldrian-Herrada G., Rebiere B., Roy C., Benyamin Y. and Lebart M. C. (2010). Characterization of L-plastin interaction with beta integrin and its regulation by micro-calpain. Cytoskeleton (Hoboken) 67, 286-296. 10.1002/cm.20442 [DOI] [PubMed] [Google Scholar]
- Leventis P. A. and Grinstein S. (2010). The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407-427. 10.1146/annurev.biophys.093008.131234 [DOI] [PubMed] [Google Scholar]
- Lin C. S., Aebersold R. H., Kent S. B., Varma M. and Leavitt J. (1988). Molecular cloning and characterization of plastin, a human leukocyte protein expressed in transformed human fibroblasts. Mol. Cell. Biol. 8, 4659-4668. 10.1128/MCB.8.11.4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. S., Park T., Chen Z. P. and Leavitt J. (1993). Human plastin genes. Comparative gene structure, chromosome location, and differential expression in normal and neoplastic cells. J. Biol. Chem. 268, 2781-2792. [PubMed] [Google Scholar]
- Lin C. S., Lau A. and Lue T. F. (1998). Analysis and mapping of plastin phosphorylation. DNA Cell Biol. 17, 1041-1046. 10.1089/dna.1998.17.1041 [DOI] [PubMed] [Google Scholar]
- Liu J., Fukuda K., Xu Z., Ma Y.-Q., Hirbawi J., Mao X., Wu C., Plow E. F. and Qin J. (2011). Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation. J. Biol. Chem. 286, 43334-43342. 10.1074/jbc.M111.295352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Das M., Yang J., Ithychanda S. S., Yakubenko V. P., Plow E. F. and Qin J. (2015). Structural mechanism of integrin inactivation by filamin. Nat. Struct. Mol. Biol. 22, 383-389. 10.1038/nsmb.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Van Horn W. D., Chen J., Mathew S., Zent R. and Sanders C. R. (2012). Bicelles at low concentrations. Mol. Pharm. 9, 752-761. 10.1021/mp2004687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margadant C., Monsuur H. N., Norman J. C. and Sonnenberg A. (2011). Mechanisms of integrin activation and trafficking. Curr. Opin. Cell Biol. 23, 607-614. 10.1016/j.ceb.2011.08.005 [DOI] [PubMed] [Google Scholar]
- Moore D. T., Nygren P., Jo H., Boesze-Battaglia K., Bennett J. S. and Degrado W. F. (2012). Affinity of talin-1 for the beta3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc. Natl. Acad. Sci. USA 109, 793-798. 10.1073/pnas.1117220108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley S. C. (2012). The actin-bundling protein L-plastin: a critical regulator of immune cell function. Int. J. Cell Biol. 2012, 935173 10.1155/2012/935173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse E. M., Brahme N. N. and Calderwood D. A. (2014). Integrin cytoplasmic tail interactions. Biochemistry 53, 810-820. 10.1021/bi401596q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otey C. A., Pavalko F. M. and Burridge K. (1990). An interaction between alpha-actinin and the beta 1 integrin subunit in vitro. J. Cell Biol. 111, 721-729. 10.1083/jcb.111.2.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavalko F. M. and Laroche S. M. (1993). Activation of human neutrophils induces an interaction between the integrin beta 2-subunit (CD18) and the actin binding protein alpha-actinin. J. Immunol. 151, 3795-3807. [PubMed] [Google Scholar]
- Penalver E., Ojeda L., Moreno E. and Lagunas R. (1997). Role of the cytoskeleton in endocytosis of the yeast maltose transporter. Yeast 13, 541-549. [DOI] [PubMed] [Google Scholar]
- Perera H. D., Ma Y.-Q., Yang J., Hirbawi J., Plow E. F. and Qin J. (2011). Membrane binding of the N-terminal ubiquitin-like domain of kindlin-2 is crucial for its regulation of integrin activation. Structure 19, 1664-1671. 10.1016/j.str.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer A., Kessler T., Silletti S., Cheresh D. A. and Verma I. M. (2000). Suppression of angiogenesis by lentiviral delivery of PEX, a noncatalytic fragment of matrix metalloproteinase 2. Proc. Natl. Acad. Sci. USA 97, 12227-12232. 10.1073/pnas.220399597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pivot-Pajot C., Chouinard F. C., El Azreq M. A., Harbour D. and Bourgoin S. G. (2010). Characterisation of degranulation and phagocytic capacity of a human neutrophilic cellular model, PLB-985 cells. Immunobiology 215, 38-52. 10.1016/j.imbio.2009.01.007 [DOI] [PubMed] [Google Scholar]
- Raab M., Daxecker H., Edwards R. J., Treumann A., Murphy D. and Moran N. (2010). Protein interactions with the platelet integrin alpha(IIb) regulatory motif. Proteomics 10, 2790-2800. 10.1002/pmic.200900621 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y. and Mann M. (2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663-670. 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- Raynal N., Hamaia S. W., Siljander P. R., Maddox B., Peachey A. R., Fernandez R., Foley L. J., Slatter D. A., Jarvis G. E. and Farndale R. W. (2006). Use of synthetic peptides to locate novel integrin alpha2beta1-binding motifs in human collagen III. J. Biol. Chem. 281, 3821-3831. 10.1074/jbc.M509818200 [DOI] [PubMed] [Google Scholar]
- Saltel F., Mortier E., Hytönen V. P., Jacquier M.-C., Zimmermann P., Vogel V., Liu W. and Wehrle-Haller B. (2009). New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J. Cell Biol. 187, 715-731. 10.1083/jcb.200908134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H. B., Hermann M.-R., Polleux J., Vignaud T., Zanivan S., Friedel C. C., Sun Z., Raducanu A., Gottschalk K.-E., Théry M. et al. (2013). beta1- and alphav-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 15, 625-636. 10.1038/ncb2747 [DOI] [PubMed] [Google Scholar]
- Schmidt S., Nakchbandi I., Ruppert R., Kawelke N., Hess M. W., Pfaller K., Jurdic P., Fassler R. and Moser M. (2011). Kindlin-3-mediated signaling from multiple integrin classes is required for osteoclast-mediated bone resorption. J. Cell Biol. 192, 883-897. 10.1083/jcb.201007141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S., Moser M. and Sperandio M. (2013). The molecular basis of leukocyte recruitment and its deficiencies. Mol. Immunol. 55, 49-58. 10.1016/j.molimm.2012.11.006 [DOI] [PubMed] [Google Scholar]
- Schulz T. W., Nakagawa T., Licznerski P., Pawlak V., Kolleker A., Rozov A., Kim J., Dittgen T., Kohr G., Sheng M. et al. (2004). Actin/alpha-actinin-dependent transport of AMPA receptors in dendritic spines: role of the PDZ-LIM protein RIL. J. Neurosci. 24, 8584-8594. 10.1523/JNEUROSCI.2100-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymeinsky J., Gerstl R., Mannigel I., Niedung K., Frommhold D., Panthel K., Heesemann J., Sixt M., Quast T., Kolanus W. et al. (2009). A fundamental role of mAbp1 in neutrophils: impact on beta(2) integrin-mediated phagocytosis and adhesion in vivo. Blood 114, 4209-4220. 10.1182/blood-2009-02-206169 [DOI] [PubMed] [Google Scholar]
- Sharma C. P., Ezzell R. M. and Arnaout M. A. (1995). Direct interaction of filamin (ABP-280) with the beta 2-integrin subunit CD18. J. Immunol. 154, 3461-3470. [PubMed] [Google Scholar]
- Shattil S. J., Kim C. and Ginsberg M. H. (2010). The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 11, 288-300. 10.1038/nrm2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J. V. and Mann M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856-2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Shinomiya H., Hagi A., Fukuzumi M., Mizobuchi M., Hirata H. and Utsumi S. (1995). Complete primary structure and phosphorylation site of the 65-kDa macrophage protein phosphorylated by stimulation with bacterial lipopolysaccharide. J. Immunol. 154, 3471-3478. [PubMed] [Google Scholar]
- Skau C. T., Courson D. S., Bestul A. J., Winkelman J. D., Rock R. S., Sirotkin V. and Kovar D. R. (2011). Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J. Biol. Chem. 286, 26964-26977. 10.1074/jbc.M111.239004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surya W., Li Y., Millet O., Diercks T. and Torres J. (2013). Transmembrane and Juxtamembrane Structure of alphaL Integrin in Bicelles. PLoS ONE 8, e74281 10.1371/journal.pone.0074281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers A. C., Niebel A., Balague C. and Dagkesamanskaya A. (2002). Differential localisation of GFP fusions to cytoskeleton-binding proteins in animal, plant, and yeast cells. Green-fluorescent protein. Protoplasma 220, 69-78. 10.1007/s00709-002-0026-7 [DOI] [PubMed] [Google Scholar]
- Van Dam L., Karlsson G. and Edwards K. (2004). Direct observation and characterization of DMPC/DHPC aggregates under conditions relevant for biological solution NMR. Biochim. Biophys. Acta 1664, 241-256. 10.1016/j.bbamem.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Vitorino P., Yeung S., Crow A., Bakke J., Smyczek T., West K., Mcnamara E., Eastham-Anderson J., Gould S., Harris S. F. et al. (2015). MAP4K4 regulates integrin-FERM binding to control endothelial cell motility. Nature 519, 425-430. 10.1038/nature14323 [DOI] [PubMed] [Google Scholar]
- Wang J., Chen H. and Brown E. J. (2001). L-plastin peptide activation of alpha(v)beta(3)-mediated adhesion requires integrin conformational change and actin filament disassembly. J. Biol. Chem. 276, 14474-14481. 10.1074/jbc.M007324200 [DOI] [PubMed] [Google Scholar]
- Yang J., Ma Y.-Q., Page R. C., Misra S., Plow E. F. and Qin J. (2009). Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc. Natl. Acad. Sci. USA 106, 17729-17734. 10.1073/pnas.0909589106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.