Abstract
Protein phosphatases play a crucial role in cell cycle progression, cell survival, cellular signaling, and genomic integrity. The protein phosphatase 1 (PP1) regulatory subunit SDS22 plays a significant role in cell cycle progression. A recent study showed that SDS22 plays a vital role in epithelial integrity and tumor suppression in Drosophila. However, its tumor suppressive activity remains obscure in the mammalian system. Here, for the first time, we show that SDS22 inhibits the growth of breast cancer cells through induction of apoptosis. SDS22 negatively regulates the AKT kinase signaling pathway through PP1. SDS22 associates predominantly with AKT and dephosphorylates the phospho Thr308 and phospho Ser473 through PP1 and hence abrogates the cell migration, invasion, and tumor growth. Thus, our study deciphers the long-standing question of how PP1 negatively regulates the AKT signaling pathway. Further, we observed a significant converse correlation in the expression levels of SDS22 and phospho form of AKT with reduced levels of SDS22 in the higher grades of cancer. Overall, our results suggest that SDS22 could be a putative tumor suppressor and replenishment of SDS22 would be an important strategy to restrict the tumor progression.
Abbreviations: AKT, AKT serine/threonine kinase/ Protein kinase B; MAPK, Mitogen activated protein kinase; SDS22, Protein phosphatise regulatory subunit 7 or PPP1R7; PP1, Protein phosphatase 1; PP2, Protein phosphatase 2; qRT-PCR, quantitative real-time PCR; TNBC, Triple negative breast cancer; BAX, BCL2-associated X protein; BAD, BCL2 associated agonist of cell death; PUMA, BCL2 binding component 3; GSK3β, Glycogen synthase kinase 3 beta; FOXO1, Forkhead box O1; Bcl2, B cell leukemia/lymphoma 2; IKK, inhibitor of nuclear factor kappa B kinase; PDK1, pyruvate dehydrogenase kinase 1; mTOR, mechanistic target of rapamycin kinase; PTEN, phosphatase and tensin homolog; SHIP, inositol polyphosphate-5-phosphatase D; eIF4E, eukaryotic translation initiation factor 4E; PHLPP1, PH domain and leucine rich repeat protein phosphatase 1; JNK, c-Jun NH2-terminal kinase; MEK, Mitogen-activated protein kinase; ERK, Mitogen-activated protein kinase 1; EMT, Epithelial to mesenchymal transition; FBS, Foetal bovine serum; GFP, Green fluroscent protein; PARP1, poly(ADP-ribose) polymerase 1; APAF1, apoptotic peptidase activating factor 1; SDS, Sodium dodecyl sulphate; PAGE, Polyacrylamide gel electrophoresis; PVDF, Polyvinylidenedifluoride; PBS, Phosphate buffer saline; BSA, Bovine serum albumin; NOD-SCID mice, Non obese diabetic -everely compromised immune deficient mice; FACS, Fluorescence activated cell sorting; DMEM, Dulbecco's modified Eagle's medium; RPMI, Roswell Park Memorial Institute; USA, United states of America; UK, United kingdom
Introduction
Breaching the precise cell cycle progression checkpoints result in uncontrolled cell proliferation, which is closely associated with the aberrant activation of cellular signaling pathways with the initiation, progression, and development of cancer. Therefore, these pathways are important targets to prevent the malignancy. Previous studies have shown that the AKT/protein kinase B (PKB) is most frequently hyperactivated in many cancers and promotes the initiation, progression, and metastasis of cancer [1], [2]. AKT is a serine threonine kinase, and it controls numerous downstream signaling involved in the cell proliferation, metabolism, cell cycle, cellular growth, and cell death [3]. It facilitates the malignancy by inactivating proapoptotic proteins such as PUMA, BAX, GSK3β, FOXO1/3a, BAD, and Caspase 9. On the other hand, it also induces the expression of prosurvival proteins such as BCL2 and Xiap [4].
The kinase activity of AKT is closely associated with its phosphorylation status. It is known that phosphorylation at Thr308 (by PDK1) and Ser473 (by mTORC2) in AKT1 is important for its optimum kinase activity and that phosphorylation at Thr450 is important for its stability [5], [6], [7]. PI3K plays a major role in phosphorylation-dependent activation of AKT. PI3K mediates phosphatidylinositol (3,4,5)-trisphosphate (PIP3)–dependent recruitment of AKT in the membrane wherein it is phosphorylated at Thr308 by PDK1. Increasing evidences suggest that the PI3K-AKT pathway majorly contributes to the development as well as the establishment of higher grades of breast cancer [8]. For instance, hyperactivation of AKT is observed in 40%-70% of the breast cancers [9], and constitutively active AKT signaling was reported in multiple breast cancer cell lines with either HER2 overexpression or PTEN (negative regulator of PI3K-AKT pathway) mutation [10]. Interestingly, PTEN is inactivated only in 38% of invasive breast cancer [7], suggesting that inactivation of PTEN might not be the sole reason for constitutive activation of AKT in 70% breast cancer incidences. This strongly suggests the involvement of other phosphatases in the regulation of AKT pathway.
Protein phosphatase-mediated dephosphorylation is the main pathway of deactivation of AKT. In the human genome, the protein phosphatase 1 (PP1) is the most abundant phosphatase pool in the cell. It accounts for almost 80% of phosphatase activity in the cells. Previous studies have identified different lipid phosphatases that can control the PI3K-AKT pathway which includes PTEN, SHIP, INNPP4B, and eIF4E [11], [12], [13], [14]. These lipid phosphatases mainly dephosphorylate PIP3 and hence abrogate the activation of AKT. In addition to lipid phosphatases, phosphatases such as PPI, PP2A [14], [15], and PHLPP1 [16] are also involved in inactivation of AKT pathways. Association of PP1 and AKT has been reported to mediate dephosphorylation of AKT and induction of apoptosis [17], [18]. However, the regulatory subunit involved in PP1-mediated inactivation of AKT is yet to be discovered.
SDS22 (also called PPP1R7) is a highly conserved regulatory subunit of PP1 [19]. Previous studies have shown that SDS22 functions as a critical regulator of the G2/M cell cycle progression [20], [21]. It is located in the chromosome 2q37.3 and reported to be the regions frequently deleted in many cancers and mainly inactivated due to loss of heterozygosity [22]. Using Drosophila as a model system, Jiang et al. have shown that SDS22/PP1 regulates the epithelial integrity and tumor suppression via regulating myosin II and JNK signaling [22]. It has also been reported to be involved in cell polarity [23] and also associated with the chemoresistance in ovarian cancer [24]. In spite of its involvement in different cellular processes and development of cancer, the exact role of SDS22 in cancer progression needs to be explored.
In this study, we for the first time showed that the PP1 regulatory subunit SDS22 negatively regulates the AKT kinase signaling pathways. Co-immunoprecipitation results demonstrated that SDS22 physically interacts with the AKT and dephosphorylates it on both the Thr308 and Ser473. Further, we show that SDS22-mediated inactivation of AKT signaling pathway is distinct from the earlier known phosphatases. SDS22 inhibits the cancer cell invasion, migration, and survival through attenuation of the AKT signaling pathway. Our study thus suggested that SDS22 might function as a putative tumor suppressor in breast cancer.
Materials and Methods
Cell Culture
Human MCF10A, MCF7, MDA-MB-231, T47D, and HEK293T cell lines were kind gift from Prof. Michael R. Green (University of Massachusetts Medical School, USA). MCF7 and HEK-293 T cells were cultured at 37°C in Dulbecco's modified Eagles medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS). MDA-MB-231 and T47D were maintained in Roswell Park Memorial Institute medium (RPMI 1640) with 10% FBS supplement. MCF10A cells were grown in DMEM-F12 (Gibco) media supplemented with 20 ng/ml epidermal growth factor (Sigma-Aldrich), 10 μg/ml insulin (Sigma-Aldrich), 0.5 μg/ml hydrocortisone (Sigma-Aldrich), 100 ng/ml choleratoxin (Sigma-Aldrich), and 10% horse serum. Cells were cultured in 5% CO2 with humidified atmosphere at 37°C.
Plasmids and Transfection
The expression plasmid of GFP-SDS22 was a kind gift from Dr. Jason Swedlow (UK). FLAG-tagged full length SDS22 was cloned using the forward primer 5′AAGCTTATGGCGGCGGAACGCGGCGCG3′ and the reverse primer 5′GGATCCAGCGTAATCTGGAACATCGTATGGGTAGAACCTGACGAACGTGGCATCGATCTGCCGCACGGAGGGGA3′. psPAX2 (Addgene plasmid # 12260) and pMD2.G (Addgene plasmid # 12259) were obtained from Addgene. MDA-MB-231 cells were transfected with the plasmids using Lipofectamine 3000 according to the manufacturer's protocol. After 12 hours of transfection, transfection media were replaced with the complete media.
Treatment of Cells
Cells were treated with either the vehicle or 5 μM MG132 (Calbiochem, USA), 5 μMAKT inhibitor (Calbiochem; catalog no. 124005), 125 nM U0126 (Calbiochem, Cat#662005, USA), 10 μM KU55933 (TOCRIS, Cat#3544), or 40 μg/ml cycloheximide (Sigma Cat #C104450, USA) for the indicated time periods. The cells were harvested after treatments, and protein extracts were prepared.
Cell Lysate Preparation and Immunoblotting
Protein extraction and Western blotting were performed as described by Santra et al. [25]. Briefly, cells were washed twice with ice-cold PBS, harvested, and collected by centrifugation. Protein extracts were prepared by lysing the cells in lysis buffer containing 50 mM Tris pH 7.4, 200 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 0.5% Triton X-100, and protease inhibitor cocktail on ice for 20 minutes [25]. Equal amounts of total protein were separated using SDS-PAGE and transferred onto the PVDF membranes (Millipore). Blots were developed by chemiluminescence method using LAS 4000 Images (GE). Densitometry analysis of the immunoblot was performed using the Image J software.
Antibody against the α-tubulin, β-actin, and anti-FLAG antibody was obtained from Sigma. Antibodies against the SDS22, GFP, PARP1, phospho AKT(S473), phospho AKT(T308), AKT, PARP1, MEK, ERK, p-cJUN, Twist, Snail, p53, APAF-1, Bcl2, BAX ATM, and AKT were obtained from Santa Cruz Biotechnology (Santacruz, CA). E-cadherin was obtained from BD Biosciences (USA).c-Myc, phospho MEK (Ser217/221), phospho ERK (Thr202/Tyr204), PUMA, BCL-XL, BAX, Caspase 9, and anti-mouse secondary antibody were obtained from Cell Signaling Technology, USA. Phospho-ATM Ser1981 was obtained from Rockland, USA.
Cycloheximide Chase Experiment
Cells were incubated with 40 μg/ml cycloheximide for the indicated time periods. Cells were then harvested, and the whole cell protein extracts were prepared and immunoblotted for the indicated proteins. Band intensity was measured using the Image J software and was normalized with loading control. The protein levels at 0-hour time point was taken as 100%, and the percentage of the remaining protein was calculated with respect to 0 hour.
Co-immunoprecipitation
Co-immunoprecipitation assay was performed as described previously [25]. In brief, 2 μg of antibody was added to 600-700 μg of the whole cell lysate. Protein and antibody mixture was then kept at 4°C with gentle rocking for 12-16 hours. The antibody and cell lysate mixture was then allowed to bind to protein-G-agarose beads for 2 hours at 4°C with gentle rocking. Immune complexes were eluted from the beads using 1× Laemmli buffer for 3-5 minutes and boiled for 5 minutes prior to resolving using SDS-PAGE. Three percent of the protein extracts were used as input in immunoprecipitation experiment.
Generation of Stable Knockdown Cells
Lentiviral shRNAs (For SDS22, shRNA 1: V2LHS_170323, shRNA 2: V3LHS_340369, and V3LHS_340373) were a kind gift from Prof. Michael R. Green, University of Massachusetts Medical School, USA. HEK-293 T cells were co-transfected with the indicated shRNAs along with packaging vectors (pPAX2 and pMD2.G) using polyethylenimine (Polyscience, USA) as reported earlier [26].
Colony Formation Assay
Five thousand cells (transduced/treated) were seeded in 35-mm culture dish and allowed to grow for 12-15 days to form the colonies. Cells were then fixed with 3.7% formaldehyde, washed with PBS, and finally stained with 0.05% crystal violet. Plates were photographed, and the representative images were shown.
Soft Agar Colony Formation Assay
Soft agar anchorage independent growth assay was performed as described previously [27]. Briefly, 35-mm dishes were filled with 0.6% base agar (Invitrogen, Cat # 75510-019) and 2× RPMI 1640 (MDA-MB-231 cells)/2× DMEM (MCF7 cells) with 20% FBS and allowed to solidify. Thereafter, 5000 cells were dissolved in 0.3% of agar containing 2× RPMI 1640 (MDA-MB-231 cells)/2× DMEM (MCF7 cells) 20% FBS were placed on the top of base agar. Twenty days later, cells were observed under microscope and photographed.
Cell Migration and Invasion Assay
Scratch wound healing assay was performed as described previously [28]. Confluent monolayer cells were scratched using a pipette tip in the presence of 5 ng/ml Actinomycin D (Sigma, Cat. #A1410). The migration of the cells was tracked continuously using phase contrast microscope (Olympus IX71). We performed the experiments in triplicate, and a minimum of three independent experiments was performed.
Invasion experiment was performed as described previously with minor modification [29]. Briefly, the cells were serum starved for 24 hours, and 50,000 cells were then suspended in 200 μl of media supplemented with 0.5% FBS in the upper chamber and 10% FBS containing media (1 ml) was added to the lower chamber of the Transwell. Cells were allowed to grow for 16 hours in CO2 incubator under the humid condition at 37°C. Media and nonmigrated cells on the upper chamber were removed, and invaded cells were then fixed with 3.7% formaldehyde followed by washing with PBS and stained by 0.5% crystal violet stain and photographed. Different fields were captured, and the number of invading cells was expressed as the average number of cells per microscopic field. The experiment was repeated three times.
Quantitative Real-Time RT-PCR
Quantitative real time RT-PCR was performed as described previously [26]. The expression of GAPDH was used to normalize the data. SDS22 primers (forward: 5′GGTACCGAATCAAGCAGAGC3′ and reverse: 5′GCAGTAGGAGGACGAGTTGG3′) and GAPDH primers (forward: 5′AATCCCATCACCATCTTCCA3′ and reverse: 5′TGGACTCCACGACGTACTCA3′) were used in this study. The experiment was repeated three times, and the fold change was calculated using the ∆∆CT method.
Fluorescence-Activated Cell Sorting (FACS)
FACS was used to analyze the cell cycle profile as well as the dead cell populations [30]. MDA-MB-231 cells transfected with either the vector or SDS22 were harvested after 48 hours of transfection and fixed in 95% ethanol. On the day of cell acquisition in FACS, cells were washed with ice-cold 1× PBS followed by staining with propidium iodide (DNA staining dye) and immediately acquired in BD FACS Calibur. FACS data were analyzed using the BD Cell Quest Rro Software (BD Bioscience). The percentage of cells at the sub-G1 populations was considered as apoptotic/dead cells. The experiment was repeated three times.
Cell Viability Assay by Dye Exclusion Method
The cell viability was measured by the Trypan blue dye exclusion method as described previously [31]. In brief, equal number of cells was seeded 1 day prior to the transfection. Next day, cells were transfected with either the SDS22 plasmid or vector control. On the day of transfection, cell number was taken as the zero time point. Transfected cells were counted at every 24-hour interval using the Trypan blue dye under the microscope using a hemocytometer. The cell number at zero time point was taken as 100% viable cells. The experiment was repeated three times.
JC1 Dye Staining
MDA-MB-231 cells were placed in the six-well plates and cultured overnight. Cells were transfected with either the vector or SDS22 plasmid. Transfection media were replaced with the complete media after 24 hours of transfection. After 48 hours of transfection, MDA-MB-231 cells were stained with the JC-1 dye (5 mg/ml) for 20 minutes at 37°C in the dark; coverslips were mounted on slides and observed using the Zeiss meta confocal microscope.
Immunofluroscence (IF)
Cells grown on coverslips were fixed with 3.7% formaldehyde (Sigma-Aldrich, USA). Next, permeabilization of cells was achieved by incubating them with 0.1% Triton X-100 (Sigma-Aldrich, USA) in PBS at room temperature for 10 minutes. Blocking of these cells was achieved using 1% BSA for 1 hour followed by incubation with the respective primary antibody for 2 hours at room temperature. Cells were then washed thrice with 1× PBS followed by incubation with the respective secondary antibody for 2 hours. Then, cells were again washed with PBS for three times, and finally, the coverslips were mounted with Vectashield mounting medium. The images were acquired in the Zeiss LSM meta 500 confocal microscope. Alexa Fluor 594 anti-rabbit IgG andAlexa Fluor 488 anti-goat IgG were purchased from Invitrogen (USA).
Immunohistochemistry (IHC)
The tissue samples were obtained from SDM College of Medical Sciences, as per the established core procedures and Institutional Ethical Board approval. Tissue samples were stained with hematoxylin-0eosin to determine the histological type and grade of tumors. Phospho AKT Ser473 and Thr308 and SDS22 protein levels in breast cancer patients, including malignant tissue and adjacent nonmalignant epithelium, were detected using immunohistochemical staining according to the method previously described [32]. The protein expression was scored based on the percentage of positive cells: 0 = 0% of stained positive cells; 1 = weakly stained tissue or 1%-25% of positive cells; score 2 = moderate stained tissue or 26%-50% of positive stained cells; and 3 = strongly stained tissue or more than 50% of stained cells.
Xenograft Assay
Animal housing and all procedures were conducted with protocols approved by the Institutional Animal Ethics Committees of the National Centre for Cell Science, Pune. In brief, 5- to 6-week-old female NOD-SCID mice were used in this study, and MDA-MB-231 cells (5 × 106/mouse) expressing either the vector or SDS22 were injected subcutaneously in the flank of the mice. The tumor volume was observed regularly up to the time palpable tumor has appeared. The length (L) and width (W) of tumors were measured with digital Vernier Caliper, and the tumor volume was calculated using the following formula: V = (L × W2)/2, with W being smaller than L.
Statistical Analysis
All the experiments were performed in triplicate. Values are shown as standard deviation (SD) except otherwise mentioned. P values ≤ .05were considered significant.
Results
SDS22 suppresses growth of breast cancer
SDS22 gene is frequently deleted in six different cancer subtypes and the second most deleted gene in breast cancer with deletion frequency of 28.8% [22].This observation was corroborated by TCGA analysis of breast cancer samples where we observed attenuated expression of SDS22 in majority of the samples (Supplementary Figure 1A). Our first empirical study in a panel of breast cancer cell lines also showed the reduced levels of SDS22 as compared to the normal epithelial cell line MCF10A (Figure 1A). Among the cell lines used, expression levels of SDS22 were significantly declined in most of the aggressive triple-negative breast cancer (TNBC) cell lines. Collectively, data from breast cancer TCGA analysis and cell line raise the possibility of involvement of SDS22 in restricting the progression of breast cancer.
Supplementary Figure 1.
(A) Analysis of SDS22 expression from TCGA database. (B) Tumor growth profile of the vector and SDS22 expressing MDA-MB-231 cells subcutaneously injected in NOD-SCID mice.
Figure 1.
SDS22 is a potential growth suppressor. (A) Immunoblotting was performed to analyze the expression levels of SDS22 in different breast cell lines. β-Actin was used as a loading control. Experiment was repeated three times, and blots represent one of the three independent experiments. (B) Trypan blue cell viable assay. MDA-MB-231 cells were expressing either vector control or SDS22 plasmid for indicated days, and the viable cells were counted using Trypan blue exclusion principle. The experiment was repeated three times. (C) Growth suppression activity of SDS22 was examined by colony formation assay. (D and E) Soft agar assay was performed to examine the anchorage independent growth of MDA-MB-231 cells ectopically expressing either vector or SDS22 (D). Number of colonies was counted and presented as percentage (E). (F and G) Scratch wound healing assay reveals that ectopically expressed SDS22 suppresses cell migration (F). Graphical representation of percentage wound healed (G). Wound healed (in graph) in MDA-MB-231 cells expressing the vector was taken as 100%. (H and I) Boyden chamber assay reveals that SDS22 suppresses cell invasion. Invasion activity of MDA-MB-231 cells ectopically expressing either the vector or SDS22 was measured using Boyden chamber assay (H). (J) SDS22 suppresses the tumor formation in NOD-SCID mice. MDA-MB-231 cells stably expressing either the vector or SDS22 were injected in flank of NOD-SCID mice. After the appearance of tumor, volume was measured at regular interval. All the experiments were repeated three times and data presented as mean ± SD. **P < .005, *P < .05 by Student's t test.
To explore this possibility, we performed a series of experiments. First, the proliferation of MDA-MB-231 TNBC cells was examined following ectopic expression of SDS22 using the Trypan blue exclusion cell count assay. We found that SDS22 significantly suppressed the cell proliferation as compared to the vector infected cells (Figure 1B). Next, we performed the colony formation assay to examine the role of SDS22 on long-term survival of MDA-MB-231, and our results showed that ectopic expression of SDS22 significantly suppressed the growth (Figure 1C). Likewise, soft-agar colony formation assay also revealed that it can prevent anchorage-independent growth of MDA-MB-231 cells (Figure 1D and E). Additionally, we found that the size of colonies of MDA-MB-231 cells expressing SDS22 is much smaller than the vector-expressing cells (Figure 1D).
TNBCs are characterized by their strong metastatic propensity, and the cell migration is required for invasion and metastasis. We therefore performed the scratch wound healing assay (Figure 1F) and Boyden Chamber assay (Figure 1H) to assess the impact of SDS22 using MDA-MB-231 cells. We discovered that wound healing cell migration (Figure 1F& G) and invasiveness were significantly suppressed upon ectopic expression of SDS22 (Figure 1H & I). To consolidate the growth-repressive role of SDS22 in TNBCs in vivo, we made xenografts of MDA-MB-231 cells expressing either the SDS22 or vector control in NOD-SCID mice. Once palpable tumor appeared, tumor growth was measured. Results revealed that the tumor growth was potently suppressed upon ectopic expression of SDS22 (Figure 1J and Supplementary Figure 1B). Collectively, our findings showed that SDS22 significantly inhibits the malignant potential of MDA-MB-231 cells in vitro and in vivo.
SDS22 Facilitates Induction of Apoptosis
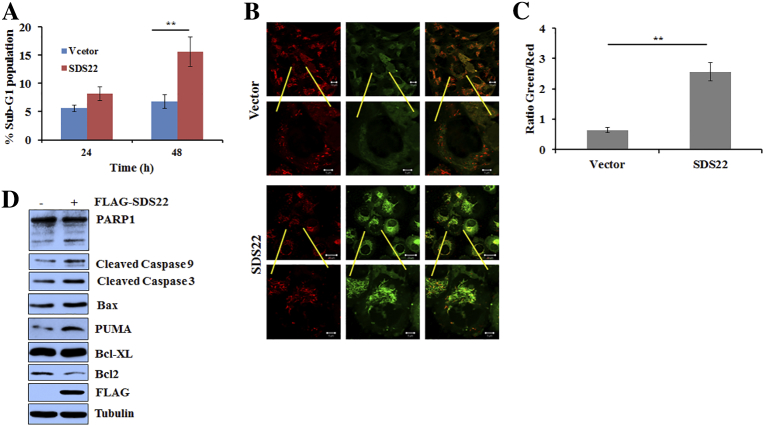
To understand the mode of growth inhibition, first we examined whether it induces the cell death. FACS analysis revealed that ectopic expression of SDS22 significantly increases the sub-G1 population of MDA-MB-231 cells, indicating that SDS22 might induce apoptotic cell death (Figure 2A). To further confirm the mode of growth inhibition, cells were stained with JC1 dye, and fluorescence data revealed that mitochondrial membrane is depolarized following SDS22 expression (Figure 2B and C). Collectively, these results demonstrated that SDS22 may promote apoptosis mediated cell death. We therefore examined the expression levels of multiple apoptotic markers upon ectopic expression of SDS22. Figure 2D showed an increased level of cleaved PARP1, cleaved caspase 9, and Bax and decreased levels of antiapoptotic protein Bcl2 following SDS22 overexpression, suggesting that SDS22 induces apoptotic cell death.
Figure 2.
SDS22 induces apoptosis through intrinsic pathway. (A) FACS analysis reveals that ectopically expressed SDS22 enhances the sub-G1 population of MDA-MB-231 cells. MDA-MB-231 cells were transfected with either the vector or SDS22 plasmid, cells were harvested at the indicated time points, and FACS was performed to find out the sub-G1 population. (B) JC1 dye staining demonstrated that ectopically expressed SDS22 induces the apoptosis. MDA-MB-231 cells were expressing either the vector control or SDS22 for 48 hours, and cells were then grown in the presence of JC1 dye for additional 20 minutes at 37°C in the dark. (C) Quantification of JC1-stained apoptotic cells. (D) SDS22 induces apoptosis. Whole cell lysates of MDA-MB-231 cells ectopically expressing either the vector control or SDS22 for 48 hours were immunoblotted for the indicated proteins, and tubulin was used as a loading control. **P < .005, *P < .05 by Student's t test.
SDS22 Negatively Regulates the Growth-Promoting AKT and MAPK-ERK Signaling Pathways
Being assured by the aforementioned results of soft agar and colony formation, we posited that SDS22 might have impaired two most crucial paradigmatic growth-promoting pathways, AKT and MAPK, as their deregulation is invariably linked with progression of almost every cancer types including breast cancer. In addition, previous study reported that SDS22 enhances chemosensitivity of ovarian cancer through controlling ERK/JNK signaling [24]. Further, it has been reported that activated AKT and MAPK pathways are potential prognostic markers of TNBC [33]. Furthermore, it has been shown that the AKT signaling pathway promotes cancer cell growth, proliferation, glucose metabolism, and metastasis, whereas MEK/ERK is critical for cell survival [34]. We therefore investigated the activation of these two pathways following ectopic expression of SDS22. In line with our supposition, we found that the terminal kinase of MAPK pathway was markedly repressed but no change was observed in JNK's activation status. In agreement with the previous study, we also observe reduced phospho levels of ERK following expression of SDS22 but did not find any change in p-JNK (Figure 3A) [24]. Surprisingly, we found that ectopically expressed SDS22 significantly decreased the phospho levels of MEK, indicating that reduced phospho levels of ERK might be due to the inactivation of upstream kinase MEK (Figure 3A).On the other hand, the major cellular prosurvival pathway was decimated by SDS22 as evident by significant reduction of the phosphorylation on the Thr308 and Ser473 of AKT without change in the total level of AKT (Figure 3A). Next, we studied the physical interaction of SDS22 with these kinases to understand whether SDS22 directly or indirectly regulates these kinases. Immunoblotting of SDS22 immunoprecipitates revealed that SDS22 physically interacts with AKT, MEK, and ERK (Figure 3B). In a reciprocal immunoprecipitation assay, SDS22 was observed in the immunoprecipitates of AKT, MEK, and ERK (Figure 3C), indicating that AKT, MEK, and ERK are the cellular substrates of SDS22. Astonishingly, we observed that AKT has highest affinity and ERK has lowest affinity for SDS22 (Figure 3C).
Figure 3.
SDS22 inactivates AKT and MAPK–ERK signaling pathway through their dephosphorylation. (A) Whole cell lysates of MDA-MB-231 cells ectopically expressing either vector control or SDS22 for 48 hours were immunoblotted for the indicated proteins. (B) Immunoblotting of immunoprecipitates and input whole cell lysates of MCF7 cells for the indicated proteins. Whole cell lysates were immunoprecipitated with either the IgG control or SDS22 antibody. (C) Immunoblotting of immunoprecipitates and input whole cell lysates of MCF7 cells. Whole cell lysates were immunoprecipitated with either the IgG control or AKT/MEK/ERK antibody. Ratio of SDS22 and ERK/MEK/AKT was quantified by the Image J software. (D) Immunoblotting of whole cell lysates of MCF7 cells expressing either the vector control or wild-type SDS22 or mutants SDS22 for 48 hours. (E) Immunoblotting of immunoprecipitates and input whole cell lysates of MCF7 cells ectopically expressing the indicated plasmids for 48 hours. Whole cell lysates were immunoprecipitated with FLAG antibody. The immunoprecipitates and input protein extracts were immunoblotted for the indicated proteins.
SDS22 Inactivates AKT and MAPK-ERK Signaling Pathways Through PP1
SDS22 functions as a regulatory subunit of the PP1 complex [35]. We therefore checked whether it dephosphorylates these kinases through PP1 or not using PP1 interaction defective SDS22 mutants (SDS22E192A and SDS22W302A) [36]. We observed that the wild-type SDS22 as expected significantly dephosphorylates these kinase substrates, but both the mutants failed to do so (Figure 3D). To further understand the reason of inability of the mutants, we performed co-immunoprecipitation assay and immunoblotting. Results of immunoprecipitates showed that these mutants could not form PP1-SDS22 complex despite their potent interaction with the substrates (Figure 3E). Altogether, these results suggested that SDS22 dephosphorylates AKT, MEK, and ERK via PP1.
SDS22 Restricts the Cellular Activity of AKT and MEK-ERK Pathway
Our results revealed that ectopic expression of SDS22 resulted in inactivation of AKT and MAPK signaling pathways. We therefore examined whether SDS22 restricts the basal level activity of AKT and MAPK signaling pathways. Immunoblotting results showed a notable elevation of phospho levels of AKT, MEK, and ERK following depletion of SDS22, indicating that they are the cellular substrates of SDS22 (Figure 4A). Further, increased phospho levels of AKT, MEK, and ERK were also observed in multiple unrelated shRNAs-mediated SDS22 stable knockdown cells, suggesting that elevated phospho levels of these kinases are not due to off-target effect of RNAi (Figure 4A). Collectively, these results demonstrated that SDS22 has role in controlling the activity of these kinases at the physiological level.
Figure 4.
SDS22 regulates AKT and MAPK pathway at physiological level to inhibit malignancy. (A) Whole cell lysates of MCF7 cells stably expressing either scramble (NS) or unrelated SDS22 shRNAs were immunoblotted for the indicated proteins. MCF7 cells were stably knocked down using three unrelated shRNAs against SDS22 through lentivirus transduction. (B) Soft agar colony formation assay of MCF7 cells expressing either scramble (NS) or SDS22 shRNAs in the absence and presence of AKT/MEK inhibitor. (C) Scratch wound healing assay of MCF7 cells stably expressing either NS or SDS22 shRNA. Cells were grown in the presence or absence of AKT/MEK inhibitor as indicated. (D) Quantification of cell migration from scratch wound healing data of panel (C). Cell migration of NS cells at 24 hours was taken as 100%. (E) Cell invasion of MCF7 cells stably expressing either NS or SDS22 shRNA by Boyden chamber assay. Cells were grown in the presence or absence of AKT/MEK inhibitor as indicated. (F) Quantification of cell invasion from data of panel E. Cell invasion of NS cells was taken as 100%. **P < .005, *P < .05 by Student's t test.
SDS22 Retards Cell Migration Through Preferential Inactivation of AKT Signaling Pathway
We showed that SDS22 inhibits the kinase activity of AKT and MEK-ERK through their dephosphorylation (Figures 3A and 4A). AKT and MEK-ERK pathways are associated with initiation and progression of malignancy by promoting the growth, migration, and invasion. We therefore examined the potential of SDS22 depleted cells in regulating these phenotypes. The soft-agar colony formation assay revealed that the depletion of SDS22 in MCF7 cells leads to increase in number as well as size of colonies which was preferentially abrogated following exposure with the AKT inhibitor (Figure 4B). The scratch wound healing assay showed that knockdown of SDS22 significantly increased the migration potential of the MCF7 cells by 42.5% (Figure 4C and D). The increased migratory potential of SDS22-depleted MCF7 cells might be due to increased activity of either AKT or MAPK or in combination of both these kinase pathway. To discern these possibilities, we further measured the migration of cells following inactivation of these kinases (Figure 4C and D). We observed that migration potential of SDS22-depleted cells significantly blocked the following inactivation of AKT than MEK, suggesting that SDS22 controls the cell migration predominantly through inactivation of the AKT pathway (Figure 4C and D). Similarly, increased invasive potential of SDS22-depleted cells markedly declined following exposure to the AKT inhibitor (Figure 4E and F). Taken together, these results suggest that cell growth, migratory potential, and invasion of malignant cells are inhibited by SDS22 predominantly through inactivation of the AKT pathway.
Depletion of SDS22 Promotes EMT
Our study showed that SDS22 noticeably inhibits cell migration, invasion, and anchorage-independent growth (Figures 1 and 4), which are considered to be the key features of malignant mesenchymal cells. Microscopic observations revealed that SDS22-depleted cells have the mesenchymal morphology as compared to wild-type cells (Figure 5A). Further, immunoblotting data revealed that the levels of EMT promoters were augmented following depletion of SDS22, suggesting that SDS22 might be suppressing the EMT process (Figure 5B). Next, we checked whether elevated AKT activity in SDS22 depleted cells is associated with increased levels of EMT promoters. SDS22 knockdown cells were treated with AKT inhibitor, and immunoblotting results revealed that levels of Snail, Twist, and c-Myc were significantly attenuated with increase of E-cadherin upon AKT inactivation (Figure 5B). Typically, the expression levels of EMT players are regulated at the transcriptional level; thus, we next determined the relative transcript levels of these EMT markers in the absence of SDS22. Figure 5C showed that the relative mRNA levels of EMT regulators were augmented following depletion of SDS22. Interestingly, inhibition of AKT leads to restoration of the mRNA levels of the EMT regulators. Converse results were also obtained following ectopic expression of FLAG-SDS22 (Figure 5D). Further, immunofluorescence results suggested that membrane localization of E-cadherin was augmented following overexpression of SDS22 (Figure 5E and F), supporting the immunoblotting data (Figure 5D).
Figure 5.
SDS22 inhibits EMT process by limiting the activity of AKT kinase. (A) Phase contrast image of morphology of MCF7 cells stable expressing either NS or SDS22 shRNA. (B) Whole cell lysates of MCF7 cells expressing either NS or SDS22 shRNA were immunoblotted for the indicated proteins. SDS22 knockdown cells were grown in the absence or presence of 5 μM AKT inhibitor for 12 hours. (C) Real-time RT-PCR demonstrated the relative mRNA levels of EMT regulators. Expression levels of regulators in NS cells were normalized with loading control GAPDH and taken as 1. (D) Whole cell lysates of MDA-MB-231 cells expressing either vector or SDS22 were immunoblotted for the indicated proteins. (E) Immunofluorescence staining was performed to detect the expression levels of E-cadherin. Red color represents E-cadherin, and blue color represents DAPI staining. (F) Quantification of expression levels of E-cadherin following overexpression of SDS22 as in panel E.*P < .05, **P < .005 by Student's t test.
AKT is known to facilitate oncogenic potential of c-Myc which is a key player in EMT [37]. Next, we checked whether SDS22-mediated retardation of EMT is due to alteration of c-Myc via AKT inactivation. Depletion of SDS22 elevated the levels of c-Myc which was revoked following inactivation of AKT (Figure 5B). Conversely, co-expression of c-Myc in SDS22 expressing cells favors elevated levels of EMT promoters (Figure 5D).
Attenuated Expression of SDS22 in Breast Cancer Samples Positively Correlates with Overall Malignancy
We have seen that the depletion of SDS22 facilitates the cell growth and survival of breast cancer cells due to the hyper activation of AKT signaling. To further understand this converse relationship, we examined the correlation of SDS22 expression and activation of AKT (expression levels of pS473 and pT308) using human breast cancer patient samples. Expression levels of SDS22 significantly declined with concomitant increase of phospho AKT in higher grades of breast cancer patient samples, supporting our in vitro data (Figure 6A and B). In addition, Kaplan-Meier analysis of patient samples also demonstrated a nice converse correlation of SDS22 expression with the distance-free metastasis survival of patients (Figure 6C). It was also observed that patients having lower expression levels of SDS22 are associated with poor overall survival (Figure 6D). Taken together, these results suggested that SDS22 negatively regulates the activity of AKT signaling.
Figure 6.
SDS22 expression is conversely correlated with active form of AKT in breast cancer patient samples. (A) Immunohistochemical analysis of SDS22, pAKT473, and pAKT308 in different grades of human breast cancer samples. (B) Statistical analysis of the average score of SDS22 and active form of AKT staining between cancer tissues and corresponding nontumor tissues, P < .0001 (one-way ANOVA). (C and D) Kaplan-Meier analysis of multiple gene expression studies via public database revealed that low expression of SDS22 was associated with poor distance-free metastasis survival (C) and poor overall survival (D). (E) Model depicts the SDS22-mediated suppression of malignancy via inactivation of AKT signaling.
Discussion
Initially, SDS22 was identified as a crucial player in the progression of mitosis and maintaining the genomic stability [20], [21], [36]. Recent studies demonstrated that SDS22 is frequently deleted in many cancers including breast, melanoma, ovarian, oral, and cervical [38], [39], [40], indicating that SDS22 might function as a tumor suppressor. The novel findings in this study suggest that SDS22 function as a putative tumor suppressor through inactivation of the AKT signaling (Figure 6E).
Jiang et al., for the first time, showed that the PP1 regulatory subunit of SDS22 is a putative tumor suppressor in the Drosophila model system and regulates the function of myosin II and JNK activity [22]. Using breast cancer as a model system, we, for the first time, showed that SDS22 functions as a putative tumor suppressor by inactivating the AKT and MAPK signaling pathways. However, we did not find any change in JNK following either ectopic expression or depletion of SDS22, indicating that JNK might not be its putative substrate (Figures 3A and 4A).
AKT is the most frequently hyperactivated kinase in human cancer. It facilitates the tumor growth, proliferation, metastasis, and resistance to anticancer therapy [2]. Therefore, inactivation of AKT is important for controlling the progression of tumor growth and metastasis. Previous studies showed that protein phosphatase PTEN inactivates the AKT signaling pathway by dephosphorylating it. Clinical data revealed that PTEN is inactivated in 24% of breast cancers [41], [42]. However, AKT is hyperactivated in 70% of breast cancers, indicating the presence of other negative regulators. In addition, AKT plays important roles in cell cycle progression, and its activity fluctuates across the cell cycle [43]. We found that SDS22 is underexpressed in most of the breast cancer cells, and overexpression of SDS22 significantly inhibits their growth (Figure 1A-D). We therefore presumed that SDS22 might be involved in inactivating the growth promoting kinases and discovered that SDS22 dephosphorylates crucial growth promoting kinases AKT, MEK, and ERK (Figure 3A). Our study revealed that AKT and MEK are the new substrates of SDS22, which is completely novel. A previous study suggested that SDS22 suppresses the ovarian cancer growth through inactivation of ERK signaling [24], but we here suggest that SDS22-mediated inactivation of ERK signaling observed in ovarian cancer might be partly due to inactivation of upstream kinase MEK. Further, affinity analysis revealed that AKT has more affinity for SDS22 than MEK or ERK (Figure 3C). Our study predicted that reduced chemosensitivity in SDS22-depleted cancer might be due to the hyperactivation of MAPK and AKT signaling.
Protein phosphatases PTEN, PP1, PP2A, and PHLPP inactivate AKT by dephosphorylation [16], [17], [44]. However, PTEN does not dephosphorylate AKT directly. PTEN predominantly dephosphorylates lipid like phosphatidylinositol (3,4,5) triphosphate (PIP3) and phosphatidylinositol (3,4) to inactivate AKT [44]. However, PP1, PP2A, and PHLPP inactivate AKT by direct dephosphorylation [16], [17]. Interestingly, PHLPP dephosphorylates phospho-Ser473, while PP2A and PP1 dephosphorylate at both Thr308 and Ser473 positions. Here, we discovered that SDS22 efficiently dephosphorylated the AKT at Thr308 and Ser473, indicating that PP1-SDS22 might function as AKT phosphatase. Collectively, our study revealed that SDS22 acts as the crucial regulatory subunit of PP1, which is involved in the inactivation of AKT via dephosphorylation. Thus, our study elucidates the molecular mechanism of the long-standing question in the field of how PP1 negatively regulates the AKT signaling pathway.
We observed that ectopically expressed SDS22 can potently induce apoptosis through intrinsic pathway (Figure 2D). Proapoptotic protein PUMA plays a crucial role in induction of apoptosis through activation of caspases, and we found augmentation of PUMA levels following SDS22 expression. Previously, it was demonstrated that AKT functions as antiapoptotic protein by limiting the expression of many proapoptotic proteins including PUMA [45]. Thus, increased levels of PUMA following SDS22 expression might be due to inactivation of AKT. This suggested that SDS22 might function as a proapoptotic by inactivating antiapoptotic AKT.
Ninety percent of cancer deaths are reported to be due to incidences of metastasis [46]. It is associated with cell migration, invasion, and induction of epithelial-mesenchymal transition [47]. Approximately 80% of metastatic tumors have been reported to have hyperactive AKT [48]. AKT has been shown to contribute in each step of metastasis and known to enhance the metastatic potentials of tumors [47], [49]. Notably, our results revealed that SDS22 can abrogate each step of metastasis by predominantly inactivating AKT.
In conclusion, our findings revealed that protein phosphatase regulatory subunit SDS22 functions as a putative tumor suppressor by dephosphorylation-mediated inactivation of AKT and MAPK signaling pathways. It dephosphorylates the AKT at Thr308 and Ser473 and thereby restricts its oncogenic activity. Levels of SDS22 and active form of AKT are in converse relation in cancer with less expression of SDS22 in higher grades of breast cancer, which is correlating the overall survival of the patients. These findings suggest that SDS22 functions as a dual phosphatase of AKT/MAPK. A previous study demonstrated that dual inhibition of AKT and MAPK pathways might be a better therapeutic strategy for breast cancer treatment [50]. Therefore, restoration of SDS22 expression in breast cancer might appear as a potential approach to prevent malignancy through inactivation of AKT and MAPK signaling.
The following are the supplementary data related to this article.
Acknowledgement
We thank Prof. Michael R. Green (University of Massachusetts Medical School, USA) for providing cell lines and shRNAs for this study. We acknowledge Dr. Jayati Mullick (National Institute of Virology, India) and Dr. Biswanath Maity for editing the manuscript. We thank Dr. Ramanamurthy Boppana for assisting in mice experiments.
Footnotes
Financial support: This work was supported by the National Centre for Cell Science, Department of Biotechnology, Ministry of Science and Technology, Government of India (to M.K.S.). D. P. is a Senior UGC research fellow.
Conflict of interest: We declare that we do not have any competing financial interests in relation to the work described here.
Funding: This work was supported in part by the National Centre for Cell Science, Department of Biotechnology, Ministry of Science and Technology, Government of India (to M.K.S.). D. P. is a Senior UGC research fellow.
References
- 1.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 2.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franke TF. PI3K/Akt: getting it right matters. Oncogene. 2008;50:6473–6488. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- 4.Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 5.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice PN, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 6.Kohn AD, Summers SA, Birnbaum MJ, Roth RA. Expression of a constitutively active AktSer/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 7.Aoki M, Batista O, Bellacosa A, Tsichlis P, Vogt PK. The akt kinase: molecular determinants of oncogenicity. Proc Natl Acad Sci U S A. 1998;95:14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Testa JR, Bellacosa A. AKT plays a central role in tumorigenesis. Proc Natl Acad Sci U S A. 2001;98:10983–10985. doi: 10.1073/pnas.211430998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun M, Paciga JE, Feldman RI, Yuan ZQ, Coppola D, Lu YY, Shelley SA, Nicosia SV, Cheng JQ. Phosphatidylinositol-3-OH kinase (PI3K)/AKT2, activated in breast cancer, regulates and is induced by estrogen receptor α (ERα) via interaction between ERα and PI3K. Cancer Res. 2001;61:5985–5991. [PubMed] [Google Scholar]
- 10.Clark AS, West K, Streicher S, Dennis PA. Constitutive and inducible Akt activity promotes resistance to chemotherapy, trastuzumab, or tamoxifen in breast cancer cells. Mol Cancer Ther. 2002;1:707–717. [PubMed] [Google Scholar]
- 11.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 12.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 13.Wendel HG, de Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 14.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 15.Millward TA, Zolnierowicz S, Hemmings BA. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem Sci. 1999;24:186–191. doi: 10.1016/s0968-0004(99)01375-4. [DOI] [PubMed] [Google Scholar]
- 16.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Ren CH, Tahir SA, Ren C, Thompson TC. Caveolin-1 maintains activated Akt in prostate cancer cells through scaffolding domain binding site interactions with and inhibition of serine/threonine protein phosphatases PP1 and PP2A. Mol Cell Biol. 2003;24:9389–9404. doi: 10.1128/MCB.23.24.9389-9404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thayyullathil F, Chathoth S, Shahin A, Kizhakkayil J, Hago A, Patel M, Galadari S. Protein phosphatase 1-dependent dephosphorylation of Akt is the prime signaling event in sphingosine-induced apoptosis in Jurkat cells. J Cell Biochem. 2011;112:1138–1153. doi: 10.1002/jcb.23033. [DOI] [PubMed] [Google Scholar]
- 19.Dinischiotu A, Beullens M, Stalmans W, Bollen M. Identification of sds22 as an inhibitory subunit of protein phosphatase-1 in rat liver nuclei. FEBS Lett. 1997;402:141–144. doi: 10.1016/s0014-5793(96)01514-1. [DOI] [PubMed] [Google Scholar]
- 20.Ohkura H, Yanagida M. S. pombe gene sds22+ essential for a midmitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-1. Cell. 1991;64:149–157. doi: 10.1016/0092-8674(91)90216-l. [DOI] [PubMed] [Google Scholar]
- 21.Posch M, Khoudoli GA, Swift S, King EM, DeLuca JG, Swedlow JR. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J Cell Boil. 2010;191:61–74. doi: 10.1083/jcb.200912046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Scott KL, Kwak SJ, Chen R, Mardon G. Sds22/PP1 links epithelial integrity and tumor suppression via regulation of myosin II and JNK signaling. Oncogene. 2011;30:3248–3260. doi: 10.1038/onc.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grusche FA, Hidalgo C, Fletcher G, Sung HH, Sahai E, Thompson BJ. Sds22, a PP1 phosphatase regulatory subunit, regulates epithelial cell polarity and shape. BMC Dev Biol. 2009;9:14–23. doi: 10.1186/1471-213X-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, Sun Y, Zhang PY, Qian M, Zhang H, Chen X, Ma D, Xu Y, Chen X, Tang KF. The Fra-1–miR-134–SDS22 feedback loop amplifies ERK/JNK signaling and reduces chemosensitivity in ovarian cancer cells. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meshram SN, Paul D, Manne R, Choppara S, Sankaran G, Agrawal Y, Santra MK. FBXO32 activates NF-κB through IκBα degradation in inflammatory and genotoxic stress. Int J Biochem Cell Biol. 2017;92:134–140. doi: 10.1016/j.biocel.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 27.Courtenay VD. A soft agar colony assay for Lewis lung tumour and B16 melanoma taken directly from the mouse. Br J Cancer. 1976;34:39–45. doi: 10.1038/bjc.1976.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yarrow JC, Perlman ZE, Westwood NJ, Mitchison TJ. A high-throughput cell migration assay using scratch wound healing, a comparison of image-based readout methods. BMC Biotechnol. 2004;9:21–29. doi: 10.1186/1472-6750-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, McEwan RN. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239–3245. [PubMed] [Google Scholar]
- 30.Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tennant JR. Evaluation of the trypan blue technique for determination of cell viability. Transplantation. 1964;2:685–694. doi: 10.1097/00007890-196411000-00001. [DOI] [PubMed] [Google Scholar]
- 32.Paul D, Ghorai S, Dinesh US, Shetty P, Chattopadhyay S, Santra MK. Cdc20 directs proteasome-mediated degradation of the tumor suppressor SMAR1 in higher grades of cancer through the anaphase promoting complex. Cell Death Dis. 2017;8:e2882. doi: 10.1038/cddis.2017.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashimoto K, Tsuda H, Koizumi F, Shimizu C, Yonemori K, Ando M, Kodaira M, Yunokawa M, Fujiwara Y, Tamura K. Activated PI3K/AKT and MAPK pathways are potential good prognostic markers in node-positive, triple-negative breast cancer. Ann Oncol. 2014;25:1973–1979. doi: 10.1093/annonc/mdu247. [DOI] [PubMed] [Google Scholar]
- 34.Ripple MO, Kalmadi S, Eastman A. Inhibition of either phosphatidylinositol 3-kinase/Akt or the mitogen/extracellular-regulated kinase, MEK/ERK, signaling pathways suppress growth of breast cancer cell lines, but MEK/ERK signaling is critical for cell survival. Breast Cancer Res Treat. 2005;93:177–188. doi: 10.1007/s10549-005-4794-6. [DOI] [PubMed] [Google Scholar]
- 35.Renouf S, Beullens M, Wera S, Van Eynde A, Sikela J, Stalmans W, Bollen M. Molecular cloning of a human polypeptide related to yeast sds22, a regulator of protein phosphatase-1. FEBS Lett. 1995;375:75–78. doi: 10.1016/0014-5793(95)01180-m. [DOI] [PubMed] [Google Scholar]
- 36.Eiteneuer A, Seiler J, Weith M, Beullens M, Lesage B, Krenn V, Musacchio A, Bollen M, Meyer H. Inhibitor-3 ensures bipolar mitotic spindle attachment by limiting association of SDS22 with kinetochore-bound protein phosphatase-1. EMBO J. 2014;33:2704–2720. doi: 10.15252/embj.201489054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai WB, Aiba I, Long Y, Lin HK, Feun L, Savaraj N, Kuo MT. Activation of Ras/PI3K/ERK pathway induces c-Myc stabilization to upregulate argininosuccinate synthetase, leading to arginine deiminase resistance in melanoma cells. Cancer Res. 2012;72:2622–2633. doi: 10.1158/0008-5472.CAN-11-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narayan G, Pulido HA, Koul S, Lu XY, Harris CP, Yeh YA, Vargas H, Posso H, Terry MB, Gissmann L. Genetic analysis identifies putative tumor suppressor sites at 2q35-q36.1 and 2q36.3-q37.1 involved in cervical cancer progression. Oncogene. 2003;22:3489–3499. doi: 10.1038/sj.onc.1206432. [DOI] [PubMed] [Google Scholar]
- 40.Cengiz B, Gunduz M, Nagatsuka H, Beder L, Gunduz E, Tamamura R, Mahmut N, Fukushima K, Ali MA, Naomoto Y. Fine deletion mapping of chromosome 2q21–37 shows three preferentially deleted regions in oral cancer. Oral Oncol. 2007;43:241–247. doi: 10.1016/j.oraloncology.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Lerma E, Catasus L, Gallardo A, Peiro G, Alonso C, Aranda I, Barnadas A, Prat J. Exon 20 PIK3CA mutations decreases survival in aggressive (HER-2 positive) breast carcinomas. Virchows Arch. 2008;453:133–139. doi: 10.1007/s00428-008-0643-4. [DOI] [PubMed] [Google Scholar]
- 42.Bose S, Chandran S, Mirocha JM, Bose N. The Akt pathway in human breast cancer: a tissue-array-based analysis. Mod Pathol. 2006;19:238–245. doi: 10.1038/modpathol.3800525. [DOI] [PubMed] [Google Scholar]
- 43.Liu P, Begley PM, Michowski W, Inuzuka H, Ginzberg M, Gao D, Tsou P, Gan W, Papa A, Kim BM. Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature. 2014;508:541–544. doi: 10.1038/nature13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stambolic V, Suzuki A, De La Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 45.Green BD, Jabbour AM, Sandow JJ, Riffkin CD, Masouras D, Daunt CP, Salmanidis M, Brumatti G, Hemmings BA, Guthridge MA. Akt1 is the principal Akt isoform regulating apoptosis in limiting cytokine concentrations. Cell Death Differ. 2013;20:1341–1349. doi: 10.1038/cdd.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiao M, Sheng S, Pardee AB. Metastasis and AKT activation. Cell Cycle. 2008;7:2991–2996. doi: 10.4161/cc.7.19.6784. [DOI] [PubMed] [Google Scholar]
- 47.Gupta PB, Mani S, Yang J, Hartwell K, Weinberg RA. The evolving portrait of cancer metastasis. Cold Spring Harb Symp Quant Biol. 2005;70:291–297. doi: 10.1101/sqb.2005.70.033. [DOI] [PubMed] [Google Scholar]
- 48.Sun M, Wang G, Paciga JE, Feldman RI, Yuan ZQ, Ma XL, Shelley SA, Jove R, Tsichlis PN, Nicosia SV. AKT1/PKBalpha kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 cells. Am J Pathol. 2001;159:431–437. doi: 10.1016/s0002-9440(10)61714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grille SJ, Bellacosa A, Upson J, Klein-Szanto AJ, Van Roy F, Lee-Kwon W, Donowitz M, Tsichlis PN, Larue L. The protein kinase Akt induces epithelial mesenchymal transition and promotes enhanced motility and invasiveness of squamous cell carcinoma lines. Cancer Res. 2003;63:2172–2178. [PubMed] [Google Scholar]
- 50.Saini KS, Loi S, de Azambuja E, Metzger-Filho O, Saini ML, Ignatiadis M, Dancey JE, Piccart-Gebhart MJ. Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat Rev. 2013;39:935–946. doi: 10.1016/j.ctrv.2013.03.009. [DOI] [PubMed] [Google Scholar]