Abstract
Angiogenesis is a highly regulated process orchestrated, in large part, by the vascular endothelial growth factor-A (VEGF-A) system of ligands and receptors. Considerable effort has been invested in finding optimal ways to modulate VEGF-A activity to treat disease, however, the mechanisms by which the various components interact remain poorly understood. This is in part because of the difficulty of analyzing the various interactions in an intercomparable manner. In the present study, we established conditions to allow for the detailed characterization of the molecular interactions between VEGF and its receptors and the co-receptor NRP-1 using surface plasmon resonance (SPR). We found that VEGF dissociated 25-times faster from its major signaling receptor, VEGF receptor-2 (VEGFR-2) than from its “decoy” receptor, VEGF receptor-1 (VEGFR-1). Using a systematic approach, we obtained kinetic parameters for each individual interaction under a consistent set of experimental conditions allowing for comparison between various receptors. The set of quantitative kinetic parameters and experimental conditions reported herein will provide valuable tools for developing comprehensive models of the VEGF system.
Keywords: VEGF, SPR, Receptors, Neuropilin, Angiogenesis
INTRODUCTION
The vascular endothelial growth factor (VEGF) family of growth factors includes 5 members expressed in mammals: VEGF-A, also referred to as vascular permeability factor (VFP), VEGF-B, VEGF-C, VEGF-D and placental growth factor (PlGF) 1-3. The most studied member is VEGF-A (referred to hereon as VEGF for simplicity) due to its significant role in vascular homeostasis. Deletion of a single allele of VEGF causes early embryonic death due to failure to develop blood vessels 4 Moreover, moderate overexpression of VEGF also results in lethality 5, highlighting the importance of a very tighly regulated system for proper vascular development. VEGF also plays critical roles in adult organisms where it is required for vascular homeostasis and for the growth of new blood vessels from pre-exisiting vessels (angiogenesis) 6 While angiogenesis is important in a number of normal physiological processes such as during the ovarian cycle and in wound healing, loss of control of angiogenesis contributes to many pathological conditions. Indeed, the ability of cancerous tumors to hijack this process leads to sustained growth and metastasis 7, 8. In addition, uncontrolled growth of blood vessels in other tissues contributes to wet age-related macular degeneration, ulcerative diseases, diabetic retinopathy, and rheumatoid arthritis, among others 9-12. Thus, there remains a need to better understand the mechanisms controlling VEGF activity in order to develop approaches to effectively promote or inhibit angiogenesis.
The potential for the development of highly efficient therapies to target VEGF under various cellular contexts has been limited by the lack of information regarding the complexities of the system. Mathematical models such as those described by the Popel group 13-15 are powerful tools for predicting in vivo behaviors of the VEGF system. These models rely on accurate kinetic and affinity data to simulate cellular responses to changes in normal and pathological conditions. The purpose of this study is to identify new mechanisms for the regulation of VEGF at the cell surface and within the extracellular matrix by conducting a detailed analysis of the various VEGF-receptor interactions. We have previously shown that heparin/heparan sulfate can modulate VEGF interactions with VEGF receptors and neuropilin 16 and that VEGF-VEGF Receptor 2 complexes can interact with binding sites on fibronectin within the extracellular matrix 17. However, a more complete model of this complex system of interacting macromolecules requires that the kinetic parameters for each VEGF binding event be defined. Toward this end, we have undertaken a systematic characterization of the binding interactions that occur in the system using a surface plasmon resonance (SPR)-based approach.
Quantitative analysis of ligand binding kinetics can be used to better understand how signaling is activated and regulated. Thus, we have produced a set of kinetic parameters for the VEGF system that can be used in conjunction with information regarding the composition of the pericellular environment to predict biological responses to VEGF under a range of cellular and extracellular contexts.
MATERIALS AND METHODS
Recombinant human VEGF165 (#293-VE), VEGF121 (#4644-VS), VEGFR-1 and VEGFR-2 Fc chimeras (#321-FL and #357-KD), recombinant rat neuropilin-1 Fc chimera (#566-N1), recombinant human and mouse neuropilin-1 (#s 3870-N1 and 5994-N1) and recombinant human FGFR-1 β (IIIc) Fc chimera (#661-FR) were from R&D Systems (Minneapolis, MN). GLC and NLC SPR sensor chips and other SPR-related reagents were from BIO-RAD (Hercules, CA).
Surface Plasmon Resonance
All interactions were characterized using the ProteOn XPR36 protein interaction array system from BIO-RAD at 25°C in binding buffer (phosphate buffered saline (PBS) with 0.05% Tween 20 and 0.1% bovine serum albumin (BSA)) at a flow rate of 25-100 μL/min unless stated otherwise.
Receptor immobilization on GLC and GLM SPR chips
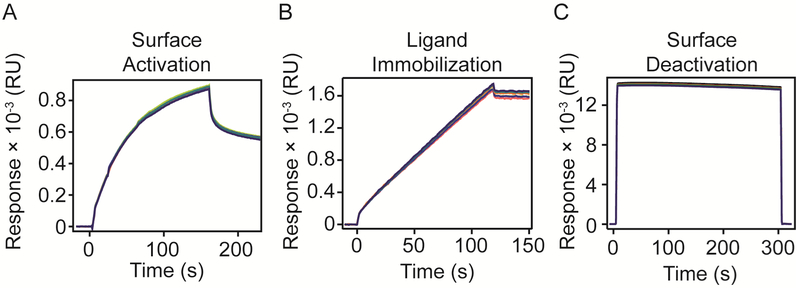
Chip surfaces were pre-conditioned following the manufacturer’s instructions. Briefly, three injections were performed at 30 μL/min flow rate for 60 s in both orientations: 1) 0.5% SDS, 2) 50 mM NaOH, and 3) 100 mM HCl. Immediately after preconditioning the chip, surfaces were exposed to a mixture of 20 mM EDAC (N-(3-Dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride) and 5 mM sulfo-NHS (N-Hydroxysulfosuccinimide) at 30 μL/min for 120-160 s to activate carboxyl groups. Receptors dissolved in running buffer were flowed over the surface at the appropriate concentrations and pH values determined previously 16. After reaching the desired immobilization levels, one step of 1 M ethanolamine HCl at pH 8.5 at 30 μL/min for 300 s was performed to block any activated carboxyl groups left unreacted on the surface. Sensograms recording the responses obtained in each step of immobilization are shown in Figure 1.
Figure 1. VEGFR-1 Fc chimera immobilization on SPR chip.
A) Surface was activated with EDAC – sNHS for 160 s in the horizontal orientation. B) Ligand was immobilized up to 1600 RU. C) Surface was deactivated with ethanolamine for 300 s.
Evaluation of theoretical Rmax
For each immobilization level, a theoretical maximal response (Rmax) was calculated for the analytes to be tested before performing kinetic analyses. The theoretical Rmax is the estimated maximal response expected for an analyte at saturating concentrations over
| (Equation 1) |
a specific immobilization level of ligand (RL). The Rmax value is determined by equation 1, where MWA and MWL are the molecular weight of the analyte and ligand, respectively, and n is the stoichiometry of the interaction (i.e. 1:1 interaction n=1).tRmax = theoretical maximal response; MWA = molecular weight of analyte; MWL = molecular weight of ligand; RL = immobilization level; n = reaction stoichiometry
SPR kinetic studies and sensogram analysis
Increasing concentrations of binding partners in running buffer were injected over immobilized receptors until equilibrium was reached, then the chip surface was washed with running buffer alone to measure dissociation. Injections of 2 M NaCl and 5-10 mM NaOH were used to regenerate surfaces after each binding event.
All sensograms were double-referenced by subtracting buffer injection and a surface containing immobilized reference protein (BSA or FGFR-1 Fc chimera for receptor studies). Interspots were monitored as an additional control of non-specific binding. Association and dissociation rate constants (ka and kd, respectively) were obtained by non-linear regression of data to a 1:1 Langmuir model (Equations 2-4) using OriginLab, Northampton, MA. In Equation 2, Rt represents the response (RU) at time t, Rmax is the maximal response at saturating analyte concentration and [A] is the ligand concentration in M. Equilibrium affinity constants (KD) were derived from kinetic parameters (KD=kd/ka) or determined from equilibrium analysis (Equation 4).
| (Equation 2) |
| (Equation 3) |
| (Equation 4) |
Where [A] = analyte concentration; Rt = measured response over time; Rmax = maximal response; Req = response at equilibrium; Rmin = permanently bound analyte; t0 = start of dissociation phase.
RESULTS
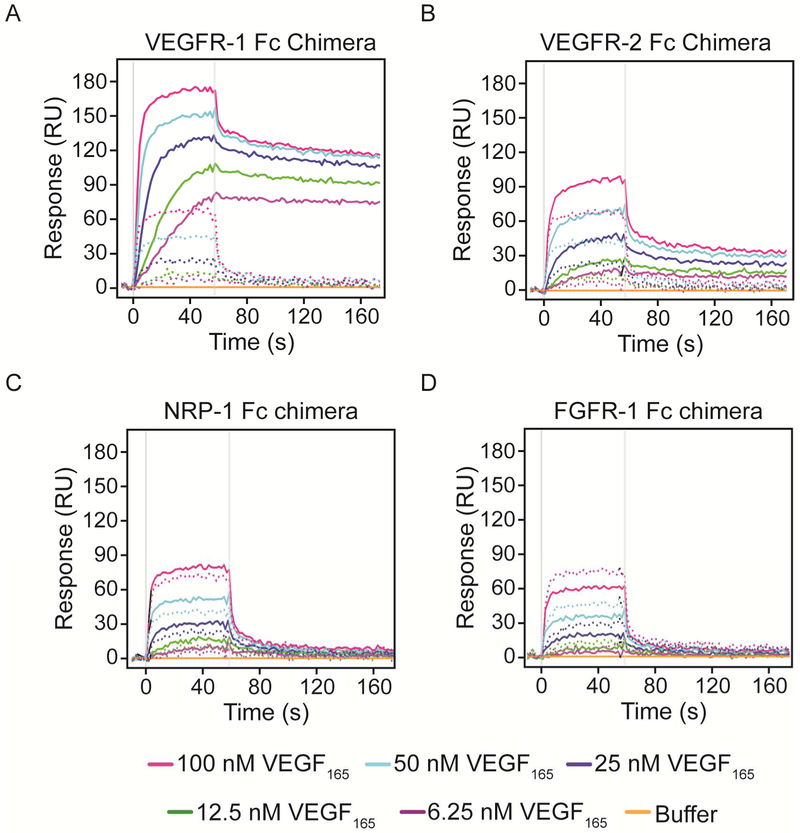
We used SPR and VEGFR Fc chimeras, which are pre-dimerized fusion proteins consisting of the extracellular domains of the VEGFRs and the constant Fc region of human IgG1, to systematically analyze and compare the interactions between VEGF and its different receptors and co-receptors. As a reference, we used bovine serum albumin (BSA) or a non-VEGF binding receptor chimera, FGFR-1 Fc, to subtract non-specific binding. VEGFR-1 Fc, VEGFR-2 Fc, NRP-1 Fc, NRP-1 monomer and FGFR-1 Fc were covalently immobilized to the surfaces of activated SPR chips by amine coupling. Ligand immobilization levels and conditions were optimized to reduce non-specific binding and mass transport effects. In order to determine the best reference surface for the analysis, we compared the extent of VEGF binding non-specifically to the naked surface versus a surface containing immobilized FGFR-1 Fc chimera (Figure 2). Each panel shows initial uncorrected VEGF165 injections at 5 concentrations (6.25, 12.5, 25, 50 and 100 nM) over surfaces with the indicated protein immobilized (solid lines) overlaid with the non-specific binding to the naked chip on adjacent interspots (dotted lines). VEGF165 interacts strongly with the negatively charged unmodified surface; however, the responses are flat and return to baseline after the end of the injection. We concluded that FGFR-1 Fc chimera is a more relevant reference since the surface undergoes the same activation, immobilization and deactivation process of the surface containing VEGFRs. The responses observed on the FGFR surface, although lower than those observed on the interspots, were also flat and returned to baseline. The lower response observed on the FGF surface can be explained by a reduction in the electrostatic charges on the surface after the chemical modifications that occur during immobilization.
Figure 2. VEGF165 flowed over SPR surfaces before correction.
VEGFR-1 Fc chimera (A), VEGFR-2 Fc chimera (B), NRP-1 Fc chimera (C), and FGFR-1 Fc chimera (D) were immobilized onto an SPR chip. Recombinant human VEGF165 diluted at the indicated concentrations in running buffer was flowed over the surfaces after stabilization. Solid lines represent VEGF binding to the surface with the indicated protein immobilized, while dotted lines represent the response observed on interspots adjacent to each surface.
One common artifact that occurs within the SPR experimental setting is mass transport limitation. Mass transport refers to an interaction being limited by the rate of diffusion of the soluble binding partner from the bulk buffer to the chip surface. When analyzing data, it is assumed that the transport rate (time the analyte takes to reach the biosensor surface) is significantly faster than the binding rate, and therefore it can be disregarded. However, in some experimental settings, this assumption is not met and the reaction kinetics are governed by the rate of transport and not the intrinsic molecular binding kinetics 18. Mass transport can skew data and result in false kinetic constant determination or may necessitate the use of more complex binding models that do not necessarily describe true kinetics of the interaction 19. Under appropriate experimental conditions (fast flow rate and low immobilization) mass transport does not have a significant effect on binding kinetics 20.
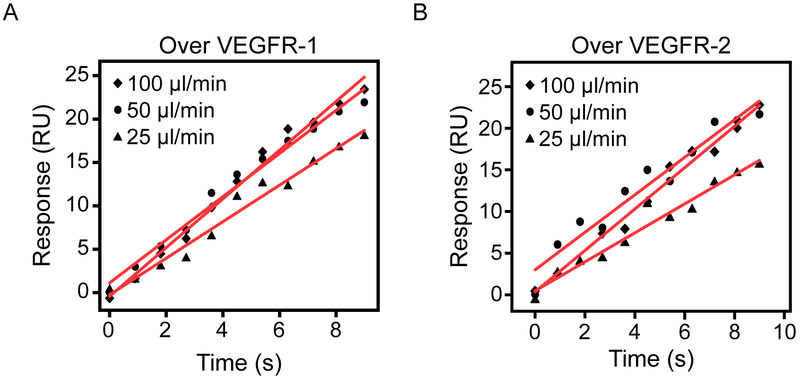
To test for the possibility of mass transport effects in our system, we flowed one VEGF concentration over VEGFR-1 and VEGFR-2 at different flow rates 21 (Figure 3). By comparing the initial linear association phase we did note a small increase in the initial slope, particularly when increasing the flow rate from 25 to 50 μL/min, nevertheless, this analysis was able to rule out significant mass transport effects under these conditions. Slope values are shown on Table 1.
Figure 3. Variations in flow rate to detect mass transport effects.
Symbols represent SPR response over the first 9 s of a VEGF165 injection at different flow rates (25, 50, 100 μL/min) over VEGFR-1 Fc (A) and VEGFR-2 Fc (B) chimeras. Red lines are linear fits and slope values are shown on Table 1.
Table 1.
Initial rate analyses to detect mass transport.
| VEGFR-1 | VEGFR-2 | |
|---|---|---|
| Flow rate (µL/min) | Slope ± SE | Slope ± SE |
| 100 | 5.6 ± 0.2 | 2.5 ± 0.1 |
| 50 | 5.0 ± 0.2 | 2.3 ± 0.2 |
| 25 | 4.2 ± 0.2 | 1.7 ± 0.1 |
Slopes of the initial 10 s binding response of a single VEGF concentration over a surface containing immobilized VEGFR-1 or VEGFR-2 (Figure 3).
VEGF165 binding kinetics to VEGFRs
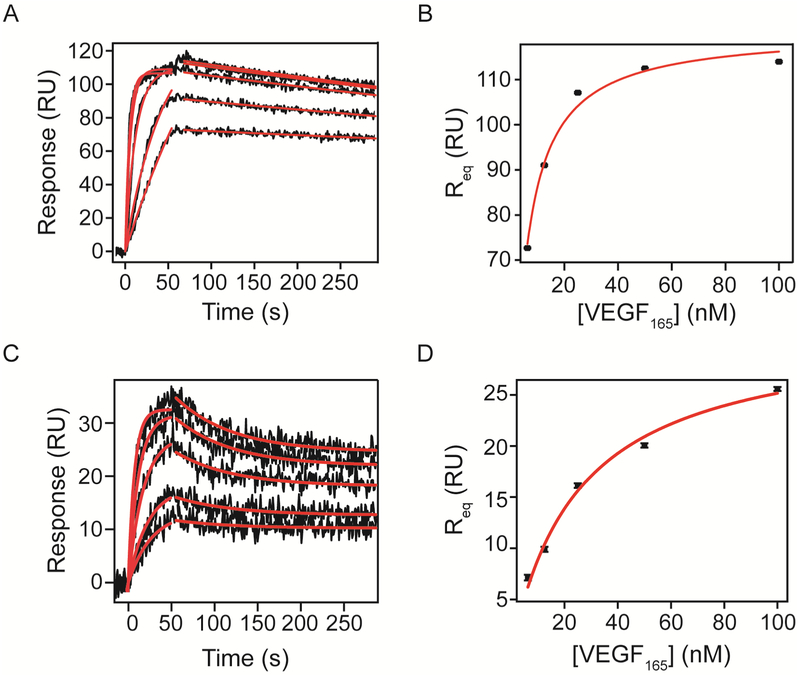
After defining and optimizing the system, we analyzed VEGF165 and VEGF121 binding to VEGFRs and NRP-1 Fc chimeras. Figure 4 shows increasing concentrations of VEGF165 binding to (A) VEGFR-1 and (C) VEGFR-2 Fc chimeras immobilized by amine coupling to an SPR chip (black undulating lines represent binding at concentrations increasing from bottom to top). To obtain kinetic parameters, we performed global non-linear regression (red lines) of the dissociation phase sensograms to equation 3 using OriginLab (Materials and Methods). The dissociation rate constant parameter was shared for all data sets, while equilibrium binding response (Req) was kept local for each concentration. The start time of dissociation (to) was fixed to the time in seconds that the system switched the flow to buffer. For the interaction between VEGF165 and VEGFR-1 Fc chimera, we observed a dissociation rate constant of 5.92 ± 0.07 × 10−4 s−1 (Table 2). Using this dissociation rate constant, we fit the association phase curves to equation 2, with VEGF165 concentration values kept constant. The maximal response at saturation (Rmax) was shared for all data sets since it represents the total number of binding sites on the surface. We obtained an association rate constant of 2.91 ± 0.04 × 106 M−1 s−1 (Table 2). Using the association and dissociation rate constants we calculated the equilibrium dissociation constant (KD = kd/ka) of the interaction to be 196 ± 4 pM.
Figure 4. Kinetic analysis of VEGF165 binding to VEGFR-1 and VEGFR-2 Fc chimeras on an SPR chip.
VEGFR-1 and VEGFR-2 Fc chimeras were immobilized onto an activated chip surface until reaching 728 and 407 RU of immobilization respectively. Increasing concentrations of recombinant human VEGF165 in running buffer (6.25, 12.5, 25, 50 and 100 nM) were flowed over the surface and the response was measured for 60s at 50 μL/min. Dissociation was measured after switching the flow to running buffer. All sensograms were referenced to a surface containing immobilized FGFR-1 Fc and a blank injection to correct for bulk shifts. Association and dissociation phases were analyzed by non-linear curve fitting (red lines). Parameters obtained by fitting are shown in Tables 2 and 3. Req values were plotted against VEGF concentration to implement equilibrium analysis and results are shown in Table 5.
Table 2.
Analysis of kinetics of VEGF165 binding to VEGFR-1.
| A. | |||||
|---|---|---|---|---|---|
| [VEGF165] (nM) |
to (s) Fixed |
Req ± SE (RU) |
kd ± SE (s−1) |
Red. X2 |
Adj. R2 |
| 100 | 61.2 | 113.9 ± 0.1 | 5.69 ± 0.07 × 10 −4 | 2.5 | 0.99 |
| 50 | 112.5 ± 0.1 | ||||
| 25 | 107.4 ± 0.1 | ||||
| 12.5 | 92.1 ± 0.1 | ||||
| 6.25 | 75.0 ± 0.1 | ||||
| B. | ||||||
|---|---|---|---|---|---|---|
| [VEGF165] (M) Fixed |
ka ± SE (M−1s−1) |
Rmax ± SE (RU) |
kd -Fixed (s−1) |
Red. X2 |
Adj. R2 |
KD ± SE (pM) |
| 1.00 × 10−7 | 2.91 ± 0.04 × 106 | 108.7 ± 0.5 | 5.92 × 10 −4 | 24.6 | 0.98 | 196 ± 4 |
| 5.00 × 10−8 | ||||||
| 2.50 × 10−8 | ||||||
| 1.25 × 10−8 | ||||||
| 6.25 × 10−9 | ||||||
A. Dissociation phase analysis VEGF165 from VEGFR-1. Parameters obtained from global curve fitting (red lines Figure 4A) of dissociation data from VEGFR-1 Fc chimera (Equation 3 Rmin = 0). B. Association phase analysis of VEGF165 binding to VEGFR-1. Parameters from fitting association phase data (red lines Figure 4A) and calculated affinity (KD = kd / ka) from - VEGFR-1 interaction.
We applied the same process to all SPR sensograms analyzed. When we examined the interaction of VEGF165 with its the main signaling receptor, VEGFR-2, we observed a dissociation rate constant of 1.51 ± 0.07 × 10−2 s−1 approximately 25 times faster than VEGFR-1 Fc chimera (Figure 2 and Table 3). The association rate constant was 1.76 ± 0.04 × 106 M−1 s−1, comparable to that for VEGFR-1 (Table 3). The calculated KD was 8.6 ± 0.5 nM, about 40 times higher (lower affinity) than that for VEGFR-1, consistent with the differences reported in the literature between the two receptors 22.
Table 3.
Analysis of kinetics of VEGF165 binding to VEGFR-2.
| A. | ||||||
|---|---|---|---|---|---|---|
| [VEGF165] (nM) |
to (s) Fixed |
Req ± SE (RU) |
Rmin ± SE (RU) |
kd ± SE (s−1) |
Red. X2 |
Adj. R2 |
| 100 | 55.8 | 10.2 ± 0.3 | 24.6 ± 0.2 | 1.51 ± 0.07 × 10 −2 | 3.29 | 0.97 |
| 50 | 8.8 ± 0.3 | 21.9 ± 0.1 | ||||
| 25 | 6.4 ± 0.3 | 18.1 ± 0.1 | ||||
| 12.5 | 3.3 ± 0.3 | 12.7 ± 0.1 | ||||
| 6.25 | 1.5 ± 0.3 | 10.2 ± 0.1 | ||||
| B. | ||||||
|---|---|---|---|---|---|---|
| [VEGF165] (M) Fixed |
ka ± SE (M−1s−1) |
Rmax ± SE (RU) |
kd – Fixed (s−1) |
Red. X2 |
Adj. R2 |
KD ± SE (nM) |
| 1.00 × 10−7 | 1.76 ± 0.04 × 106 | 34.9 ± 0.3 | 1.51 × 10 −2 | 1.76 | 0.96 | 8.6 ± 0.5 |
| 5.00 × 10−8 | ||||||
| 2.50 × 10−8 | ||||||
| 1.25 × 10−8 | ||||||
| 6.25 × 10−9 | ||||||
A. Dissociation phase analysis of VEGF165 from VEGFR-2. Parameters obtained from global curve fitting (red lines Figure 4C) of dissociation data from VEGFR-2 Fc chimera (Equation 3). B. Association phase analysis of VEGF165 binding to VEGFR-2. Parameters from fitting association phase data (red lines Figure 4C) and calculated. affinity (KD = kd / ka) from - VEGFR-1 interaction.
The equilibrium values obtained by the association curve fit were plotted as a function of VEGF concentration to perform equilibrium analysis. The equilibrium dissociation constant obtained by fitting equilibrium responses of VEGF165 binding to VEGFR-1 Fc to equation 4 (Figure 4B) was 3.7 ± 0.3 nM, higher than the calculated KD from kinetic values. This discrepancy could be due to limitations of the system when measuring slow dissociation rates 23. On the other hand, we obtained a KD of 18 ± 2 nM for VEGF165 binding to VEGFR-2 Fc chimera, consistent with our kinetic analysis. As expected, VEGF165 bound VEGFR-2 Fc chimera with a lower affinity than to VEGFR-1, evaluated by equilibrium binding.
VEGF binding kinetics to NRP-1: dimeric and monomeric
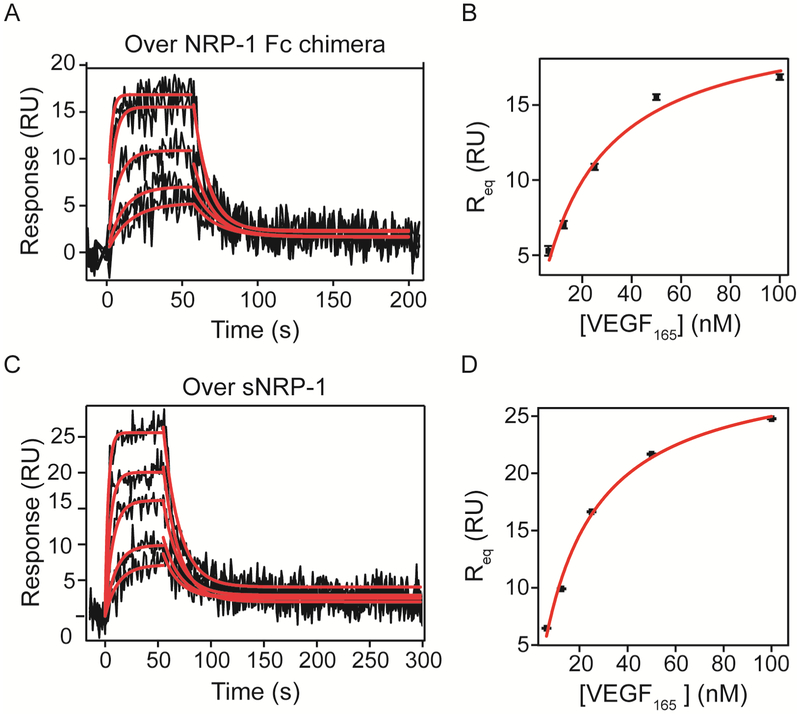
In addition to binding to its receptor tyrosine kinase receptors, VEGF has been demonstrated to interact with the co-receptor, NRP1. Thus, we analyzed the kinetics of VEGF165 binding to NRP-1 Fc chimera and a monomeric NRP-1 protein (Figure 5). NRP-1 Fc chimera contains the extracellular domains of the rat sequence (shares approximately 93% homology with the human sequence) fused to the Fc region of human IgG1. Hence, the protein is expressed as a dimer through formation of disulfide bonds between the Fc domains of each construct. On the other hand, sNRP-1 is a construct containing only the extracellular domains of the mouse sequence (also shares ~93% homology with human NRP-1) expressed as a monomer. Both constructs contain residues important for dimerization and oligomerization located on the c1 domain 24.
Figure 5. Kinetic analysis of VEGF165 binding to NRP-1.
NRP-1 Fc chimera (A and B) or NRP-1 monomer (C and D) were immobilized onto an SPR chip reaching 295 and 166 RU of immobilization, respectively. VEGF165 (6.25, 12.5, 25, 50 and 100 nM) binding responses were measured for 60 s at 50 μL/min. Association and dissociation phases were analyzed by non-linear curve fitting after correction (red lines). Parameters obtained by fitting are shown on Tables 4 and 5.
VEGF165 bound NRP-1 Fc chimera with a kd of 9.0 ± 0.4 ×10−2 s−1 and a ka of 3.6 ± 0.2 × 106 M−1 s−1 (Table 4). The calculated affinity (KD) was 25 ± 2 nM. VEGF165 binding to NRP-1 monomer showed very similar kinetics, with an association rate constant of 2.64 ± 0.08 × 106 M−1 s−1 and a kd of 6.5 ± 0.2 × 10−2 s−1 and a calculated KD of 25 ± 1 nM from kinetic parameters. Equilibrium analysis of both interactions resulted in a KD of 22 ± 7 nM for the Fc chimera and 28 ± 4 nM for the monomeric form of NRP-1 (Table 5). It is interesting that we found such small variation in VEGF165 binding kinetics between the dimeric NRP-1 and the monomer. It is possible that immobilization of NRP-1 to the surface of the chip places each NRP-1 monomer in close proximity to another, and the observed kinetics resemble binding to the “pre-dimerized” NRP-1.
Table 4.
Analysis of kinetics of VEGF165 binding to NRP-1.
| A. | ||||||
|---|---|---|---|---|---|---|
| [VEGF165] (nM) |
to (s) Fixed |
Req ± SE (RU) |
Rmin ± SE (RU) |
kd ± SE (s−1) |
Red. X2 |
Adj. R2 |
| 100 | 57.6 | 13.9 ± 0.7 | 2.3 ± 0.1 | 9.0 ± 0.4 × 10 −2 | 1.75 | 0.66 |
| 50 | 14.2 ± 0.7 | 2.2 ± 0.1 | ||||
| 25 | 8.8 ± 0.6 | 1.7 ± 0.1 | ||||
| 12.5 | 6.1 ± 0.6 | 1.8 ± 0.1 | ||||
| 6.25 | 4.3 ± 0.6 | 1.7 ± 0.1 | ||||
| B. | ||||||
|---|---|---|---|---|---|---|
| [VEGF165] (M) Fixed |
ka ± SE (M−1s−1) |
Rmax ± SE (RU) |
kd – Fixed (s−1) |
Red. X2 |
Adj. R2 |
KD ± SE (nM) |
| 1.00 × 10−7 | 3.6 ± 0.2 × 106 | 21.8 ± 0.4 | 9.0 × 10 −2 | 2.27 | 0.92 | 25 ± 2 |
| 5.00 × 10−8 | ||||||
| 2.50 × 10−8 | ||||||
| 1.25 × 10−8 | ||||||
| 6.25 × 10−9 | ||||||
| C. | ||||||
|---|---|---|---|---|---|---|
| [VEGF165] (nM) |
to (s) Fixed |
Req ± SE (RU) |
Rmin ± SE (RU) |
kd ± SE (s−1) |
Red. X2 |
Adj. R2 |
| 100 | 54.9 | 22.3 ± 0.6 | 4.1 ± 0.1 | 6.5 ± 0.2 × 10−2 | 1.89 | 0.80 |
| 50 | 18.8 ± 0.5 | 2.0 ± 0.1 | ||||
| 25 | 13.1 ± 0.5 | 2.9 ± 0.1 | ||||
| 12.5 | 8.4 ± 0.5 | 2.6 ± 0.1 | ||||
| 6.25 | 6.2 ± 0.5 | 2.5 ± 0.1 | ||||
| D. | |||||
|---|---|---|---|---|---|
| [VEGF165] (M) Fixed |
ka ± SE (M−1s−1) |
Rmax ± SE (RU) |
kd – Fixed (s−1) |
Red. X2 |
KD ± SE (nM) |
| 1.00 × 10−7 | 2.64 ± 0.08 × 106 | 31.4 ± 0.4 | 6.5 × 10−2 | 2.38 | 25 ± 1 |
| 5.00 × 10−8 | |||||
| 2.50 × 10−8 | |||||
| 1.25 × 10−8 | |||||
| 6.25 × 10−9 | |||||
A. Dissociation phase analysis of VEGF165 from NRP-1 Fc chimera. Parameters obtained from global curve fitting (red lines Figure 5A) of dissociation data from NRP-1 Fc chimera (Equation 3). B. Association phase analysis of VEGF165 binding to NRP-1 Fc chimera. Parameters from global fitting of association phase data to equation 2 (red lines Figure 3.4A) and calculated affinity (KD = kd / ka). C. Dissociation phase analysis of VEGF165 from NRP-1 monomer. Parameters obtained from global curve fitting (red lines Figure 5C) of dissociation data from NRP-1 monomer (Equation 3). D. Association phase analysis of VEGF165 binding to NRP-1 monomer. Parameters from global fitting of association phase data to equation 2 (red lines Figure 3.4C) and calculated affinity (KD = kd / ka).
Table 5.
VEGF165 binding affinities to immobilized VEGFRs and NRP-1.
|
KD ± SE (nM) |
Rmax ± SE (RU) |
Red. X2 |
Adj. R2 |
|
|---|---|---|---|---|
| VEGFR-1 Fc Chimera | 3.7 ± 0.3 | 120 ± 2 | 4.2 | 0.98 |
| VEGFR-2 Fc Chimera | 18 ± 2 | 41 ± 1 | 281 | 0.99 |
| NRP-1 Fc Chimera | 22 ± 7 | 18 ± 2 | 3.21 | 0.93 |
| NRP-1 monomer | 28 ± 4 | 28 ± 2 | 2.09 | 0.99 |
Equilibrium curve fit results (red lines in Figures 4B, D, 5B and D) for the indicated receptors binding to VEGF165 (Equation 4).
Using Rmax values and immobilization levels for each surface, we calculated the percentage of protein on the surface that was available for binding VEGF using equation 1. The surface of VEGFR-1 Fc was 86% active while surfaces containing VEGFR-2 Fc chimera, NRP-1 Fc chimera or NRP-1 were closer to 50% active (Table 6).
Table 6.
Theoretical Rmax binding response of VEGF165.
| MW (kDa) |
RL
(RU) |
tRmax
(RU) |
Rmax from Equilibrium Fit (RU) |
Percent Active |
|
|---|---|---|---|---|---|
| VEGFR-1 Fc | 200 | 728.9 | 140 | 109 | 86 |
| VEGFR-2 Fc | 220 | 407.1 | 71.1 | 35 | 58 |
| NRP-1 Fc | 238 | 295.0 | 47.6 | 22 | 38 |
| NRP-1 mon | 94.5 | 166.3 | 67.6 | 31 | 41 |
Theoretical maximal responses were calculated using equation 1 and compared to maximal binding response observed on chips.
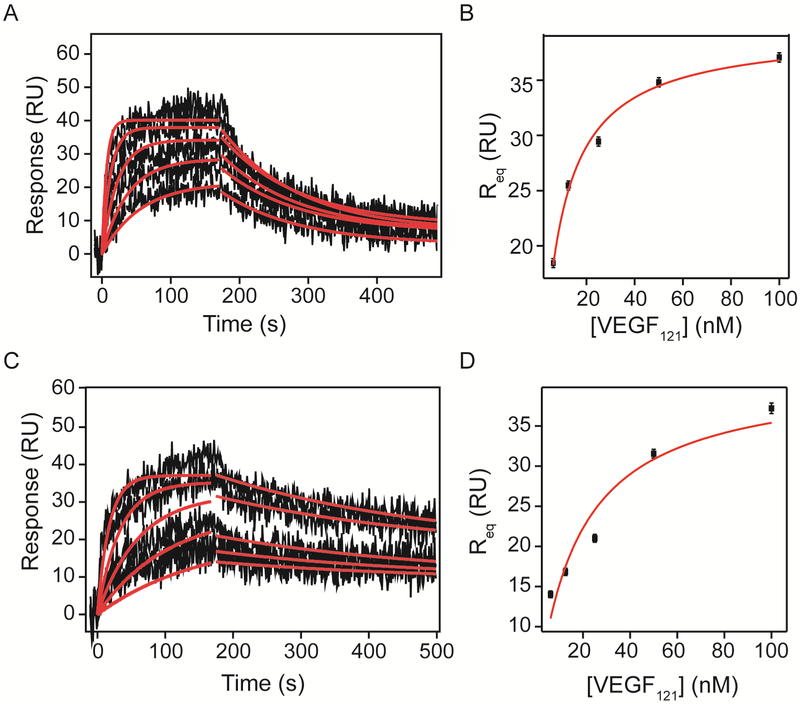
VEGF121 binding kinetics to VEGFRs
VEGF121 analysis revealed fewer differences between binding kinetics to VEGFR-1 and VEGFR-2 Fc chimeras, compared to VEGF165. VEGFR-1 bound VEGF121 with a dissociation rate constant of 8.8 ± 0.2 × 10−3 s−1, 15-times greater than VEGF165 and with a similar association rate of 1.43 ± 0.03 × 106 M−1 s−1, resulting in a KD of 6.2 ± 0.2 nM (Figure 6 and Tables 7). VEGF121 binding to VEGFR-2 showed a kd of 3.0 ± 0.2 ×10−3 s−1 and a ka of 5.47 ± 0.01 × 105 M−1 s−1 with a KD of 5.5 ± 0.3 nM (Table 8). Equilibrium analysis resulted in an affinity of 7.2 ± 0.6 nM for VEGFR-1, consistent with the calculated affinity from kinetic parameters, and an affinity of 17 ± 6 nM for VEGFR-2, which was slightly higher than that determined by kinetic analysis (Table 9).
Figure 6. Kinetic analysis of VEGF121 binding to VEGFR-1 and VEGFR-2 Fc chimera on a SPR chip.
Increasing concentrations (6.25, 12.5, 25, 50 and 100 nM) of recombinant human VEGF121 in running buffer were flowed over the surfaces containing VEGFR-1 and VEGFR-2 Fc chimeras (1610 and 1450 RU immobilized respectively) and response was measured for 180 s at 30 μL/min. Dissociation was measured after switching the flow to running buffer. Association and dissociation phases were analyzed by non-linear curve fitting (red lines). Results of the curve fit are shown in Tables 7–9.
Table 7.
Analysis of kinetics of VEGF121 binding to VEGFR-1.
| A. | |||||
|---|---|---|---|---|---|
| [VEGF121] (nM) |
to (s) Fixed |
Req ± SE (RU) |
kd ± SE (s−1) |
Red. X2 |
Adj. R2 |
| 100 | 174.6 | 30.1 ± 0.4 | 8.8 ± 0.2 × 10 −3 | 6.12 | 0.86 |
| 50 | 26.2 ± 0.4 | ||||
| 25 | 22.8 ± 0.4 | ||||
| 12.5 | 19.0 ± 0.4 | ||||
| 6.25 | 15.6 ± 0.4 | ||||
| B. | ||||||
|---|---|---|---|---|---|---|
| [VEGF121] (M) Fixed |
ka ± SE (M−1s−1) |
Rmax ± SE (RU) |
kd – Fixed (s−1) |
Red. X2 |
Adj. R2 |
KD ± SE (nM) |
| 1.00 × 10−7 | 1.43 ± 0.03 × 106 | 42.6 ± 0.2 | 8.8 × 10 −3 | 11.1 | 0.90 | 6.2 ± 0.2 |
| 5.00 × 10−8 | ||||||
| 2.50 × 10−8 | ||||||
| 1.25 × 10−8 | ||||||
| 6.25 × 10−9 | ||||||
A. Dissociation phase analysis of VEGF121 from VEGFR-1. Parameters obtained from global curve fitting (red lines Figure 6A) of dissociation data from VEGFR-1 (Equation 3). B. Association phase analysis of VEGF121 binding to VEGFR-1. Parameters from global fitting of association phase data to equation 2 (red lines Figure 6A) and calculated affinity (KD = kd / ka).
Table 8.
Analysis of kinetics of VEGF121 binding to VEGFR-2.
| A. | |||||
|---|---|---|---|---|---|
| [VEGF121] (nM) |
to (s) Fixed |
Req ± SE (RU) |
kd ± SE (s−1) |
Red. X2 |
Adj. R2 |
| 100 | 174.6 | 19.6 ± 0.5 | 3.0 ± 0.2 × 10 −3 | 5.63 | 0.89 |
| 50 | 14.6 ± 0.4 | ||||
| 25 | 11.0 ± 0.4 | ||||
| 12.5 | 7.5 ± 0.4 | ||||
| 6.25 | 5.1 ± 0.4 | ||||
| B. | ||||||
|---|---|---|---|---|---|---|
| [VEGF121] (M) Fixed |
ka ± SE (M−1s−1) |
Rmax ± SE (RU) |
kd – Fixed (s−1) |
Red. X2 |
Adj. R2 |
KD ± SE (nM) |
| 1.00 × 10−7 | 5.47 ± 0.01 × 106 | 39.1 ± 0.4 | 3.0 × 10 −3 | 16.3 | 0.86 | 5.5 ± 0.3 |
| 5.00 × 10−8 | ||||||
| 2.50 × 10−8 | ||||||
| 1.25 × 10−8 | ||||||
| 6.25 × 10−9 | ||||||
A. Dissociation phase analysis of VEGF121 from VEGFR-2. Parameters obtained from global curve fitting (red lines Figure 6C) of dissociation data from VEGFR-2 (Equation 3). B. Association phase analysis of VEGF121 binding to VEGFR-2. Parameters from global fitting of association phase data to equation 2 (red lines Figure 6C) and calculated affinity (KD = kd / ka).
Table 9.
VEGF121 binding affinities to immobilized VEGFRs.
|
KD ± SE (nM) |
Rmax ± SE (RU) |
Red. X2 |
Adj. R2 | |
|---|---|---|---|---|
| VEGFR-1 Fc Chimera | 7.2 ± 0.6 | 39.5 ± 0.8 | 3.38 | 0.99 |
| VEGFR-2 Fc Chimera | 17 ± 6 | 42 ± 5 | 37.8 | 0.89 |
Equilibrium curve fit results (red lines in Figures 6B and 6D) for the indicated receptors binding to VEGF121 (Equation 4).
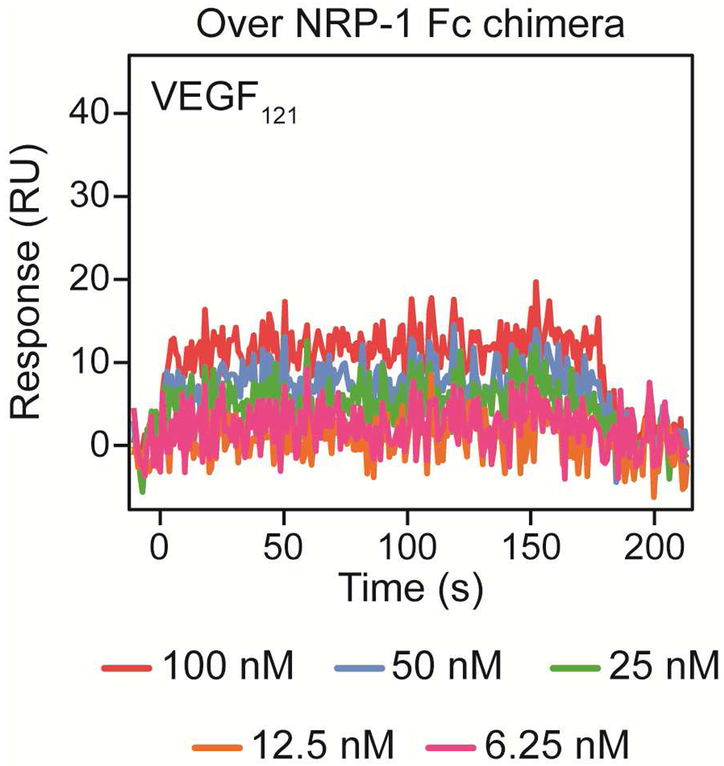
The ability of NRP-1 to bind VEGF121 has been debated. Given that NRP-1 binds the HBD of VEGF located on exon 7, it has been assumed that it is unable to interact with shorter VEGF isoforms. However, some groups have reported VEGF121 interaction with NRP-1 25, 26. We observed no detectable binding of VEGF121 to NRP-1 Fc chimera (Figure 7) or monomer (not shown).
Figure 7. VEGF121 binding to NRP-1 Fc chimera immobilized on SPR chip.
NRP-1 Fc chimera was covalently attached to an activated SPR sensor chip by amine coupling until 1460 RU of immobilization were reached. Increasing concentrations (6.25, 12.5, 25, 50 and 100 nM) of recombinant human VEGF121 in running buffer were flowed over the surface and response was measured for 180 s at 30 μL/min. We observed no significant binding to NRP-1 Fc under these conditions.
DISCUSSION
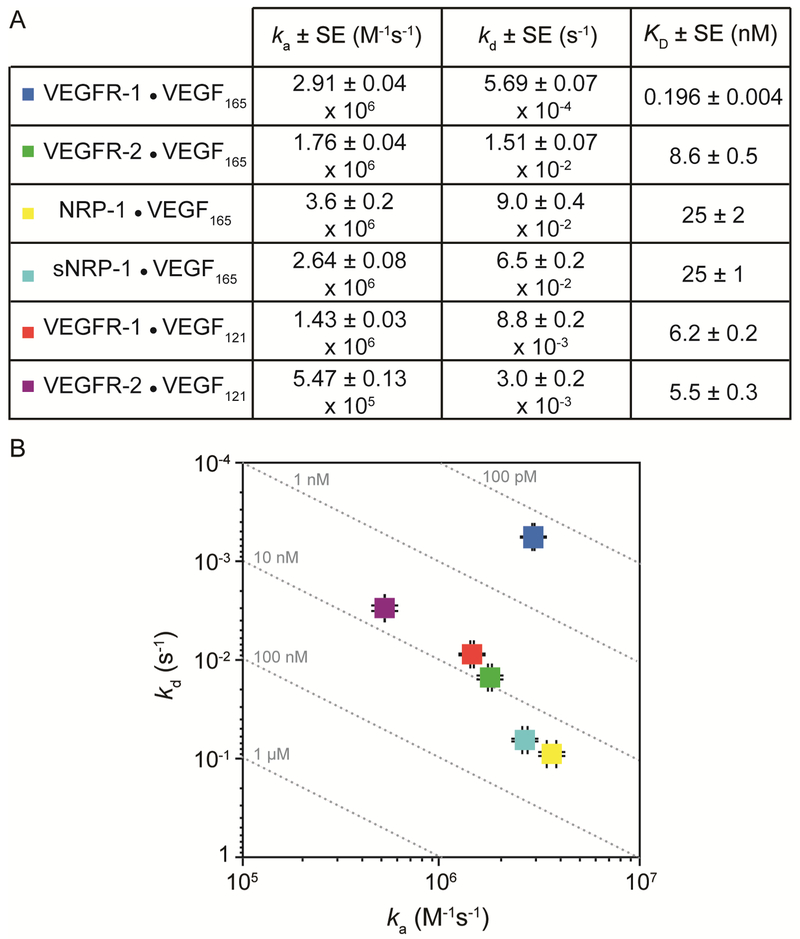
VEGF165 affinities for its receptors reported in the literature are inconsistent and range between 1 and 30 pM 27-31 for VEGFR-1, and between 50 pM and 1 nM 27-29, 32-34 for VEGFR-2. Additionally, nearly all published studies fail to report binding rate constants. Here, we provide additional data on the interactions between VEGF and the different receptors and co-receptors all measured under one set of consistent conditions. The kinetic parameters and calculated affinities from all analyses are summarized in Figure 8.
Figure 8. Iso-affinity plot.
(A) Kinetic and equilibrium rate constants derived from binding data. (B) Rate map summarizing binding affinity and kinetics. Dashed lines indicate different combinations of ka and kd that result in the same KD. Data point colors in the graph correspond to the interaction indicated in the Table above.
We found that the affinities of VEGFR-1 and VEGFR-2 for VEGF165 was lower than reported values 27-31. The discrepancies we observed could be explained by the different experimental settings. For example, most measurements reported in the literature were performed on intact cell surfaces in the presents of components of the ECM and cell surface heparan sulfate proteoglycans (HSPGs), which are known to enhance VEGF binding to its receptors. It is also surprising that the affinities observed in this study using pre-dimerized forms of the receptor are lower than those measured on intact cells, since, in the plasma membrane, receptor dimerization has been shown to be the rate limiting step 35. It is possible that the discrepancies may be related to the nature of the Fc-receptor chimeras used and/or the chemical conjugation to the SPR chip surface. For instance, the orientation of the disulfide linked dimeric proteins might not accommodate the optimal positioning of the dimeric VEGF165 with the two receptor proteins. In an attempt to determine if this is the case, we reduced the disulfide bond linkages within the Fc-receptor chimeras and isolated monomeric receptors. However, the monomeric receptors showed no detectible VEGF165 binding under the conditions used in this study, which could indicate that the receptors were damaged during the chemical reduction or that the dimeric receptors are the active binding forms. Nevertheless, the set of kinetic binding constants reported here, collected under a set of standard conditions, are internally consistent and intercomparable. Understanding how the kinetics of ligand-receptor binding and activation translate to biological output is an area of great interest. The information presented here provides a set of experimental conditions that can be used to analyze the mechanisms controlling VEGF activation of its signaling receptors
The two main VEGF receptors expressed in endothelial cells are VEGFR-1 and VEGFR-2, which share similar structures but different affinities for VEGF165 22. They also have different signaling properties, VEGFR-2 being responsible for activating the specific signaling cascades in endothelial cells that lead to angiogenesis 36. On the other hand, the role of VEGFR-1 in regulation of angiogenesis remains controversial. VEGFR-1−/− mice show early embryonic lethality due to endothelial-cell overgrowth 37 and, unlike VEGFR-2, VEGFR-1 is more widely expressed in tissues apart from the endothelium, suggesting a more diversified function 38. VEGFR-1 has a low intrinsic kinase activity in response to VEGF165, and mice that express a truncated form of VEGFR-1 lacking the entire intracellular kinase domain develop without major vascular defects 39. Hence, VEGFR-1 has been considered to play a negative role in VEGF-induced signaling. The differential functions of VEGFR-1 and VEGFR-2 are crucial for vascular homeostasis; however, how these receptors coordinate to regulate receptor activation remains poorly understood.
The differences in affinities for VEGF165 observed with VEGFR-1 versus VEGFR-2 are largely due to the difference in dissociation rate constants. This finding has profound significance when considering the intact biological system. The hypothesis that, in most cellular contexts, VEGFR-1 functions as a decoy receptor is supported by the observation of a slower VEGF165 dissociation rate compared to VEGFR-2. It is possible that under certain stimuli (i.e. changes in ECM composition and co-receptor availability), the interaction of VEGF165 with VEGFR-2 may be stabilized (slower off rate) such that it becomes the predominant complex in comparison to VEGF165-VEGFR-1. This would require that co-receptors function to decrease the dissociation rate of the ligand from the complex. Thus, modulation of co-receptor levels or pericellular location could act as a switch to shift cells from a VEGF non-responsive to responsive state.
An additional “optimal dwell” model could be relevant in the VEGF system. Ligand-induced receptor activation might require an optimal interaction time for activation, and those ligands that dissociate too fast or remain bound too long may result in reduced activity. This has been demonstrated in other receptor systems 40, and could serve as possible mechanism for VEGFR-1 to “trap” VEGF165 and decrease VEGFR-2 mediated angiogenesis signaling. On the other hand, VEGF121 binds indiscriminately to both receptors, and does not bind NRP-1 directly. This observation correlates with the observed decreased biological activity of VEGF121 compared to VEGF165, since VEGF121 activity is not enhanced by the presence of co-receptors.
NRP-1 is expressed in some cancer cell lines, including PC3, prostate cancer cell line, and MD-MB-231, a breast cancer line 41. PC3 cells bind VEGF165 with an apparent KD of 0.28 nM 42. We determined a KD of 25 nM for the interaction between NRP-1 and VEGF165. However, we have previously found that heparin can enhance VEGF165 binding to NRP-1 16, thus, endogenous HSPGs on PC3 cells might be participating in VEGF binding to NRP-1 leading to the high observed affinity. Understanding how NRP-1 regulates VEGF in disease systems will be important when determining best therapies to treat aberrant or deficient angiogenesis. Here we have determined the kinetics of VEGF binding to NRP-1, in both dimeric and monomeric forms in the absence of HSPGs. This will allow for the quantification of the contribution that different co-receptors have on VEGF binding to VEGFRs and a better understanding of how they regulate VEGF activity.
Another VEGF isoform, VEGF121, lacks the ability to be regulated by coreceptors. Mice expressing only the VEGF120 isoform (the murine version of human VEGF121) display defects in myocardial angiogenesis, and defects in vascular growth and patterning 43, 44, suggesting the roles are not redundant with those of VEGF165. We observed that VEGF121 binds VEGFR-1 and VEGFR-2 with very similar kinetics, suggesting VEGF121 is less sensitive to regulation by co-receptors. Additionally, this finding implies that the residues responsible for differential VEGF binding to VEGFR-1 and VEGFR-2 are located on exon 7, which is absent in isoform VEGF121. Site-directed mutagenesis could provide more information of the specific residues that enhance binding affinity to VEGFR-1 and not VEGFR-2. A VEGF mutant with decreased affinity for VEGFR-1 could render it more efficient at activating VEGFR-2-mediated signaling as a means to induce revascularization of ischemic tissues.
CONCLUSIONS
In the present study, we report a set of consistent conditions for quantitative and robust analysis of VEGF binding to its receptors and the co-receptor NRP-1 using SPR. The set of kinetic parameters that we report has revealed interesting distinctions between VEGF isoforms and binding partners that may be used to develop a more complete understanding of this system.
Table 10.
Theoretical Rmax binding response of VEGF121.
| MW (kDa) |
RL (RU) |
tRmax (RU) |
Rmax from Association Fit (RU) |
Percent Active |
|
|---|---|---|---|---|---|
| VEGFR-1 Fc | 200 | 1610 | 225 | 43 | 19 |
| VEGFR-2 Fc | 220 | 1450 | 185 | 39 | 21 |
Theoretical maximal responses were calculated using equation 1 and compared to maximal binding response observed on chips.
Highlights.
Experimental conditions are provided for analysis of VEGF receptor binding by SPR.
Kinetics of VEGF binding to VEGF-receptor 1 and 2 show alterations in off rate.
A set of quantitative kinetic parameters for VEGF receptor binding are presented.
Conjugation of FGF receptor to SPR surfaces controls VEGF non-specific binding.
VEGF121 binds VEGF receptors 1 and 2 with similar affinity.
Acknowledgments
Funding: This work was supported by the National Institutes of Health grant GM120702.
Footnotes
Declaration of interest: NONE
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Ferrara N (2009) VEGF-A: a critical regulator of blood vessel growth, Eur Cytokine Netw 20, 158–163. [DOI] [PubMed] [Google Scholar]
- [2].Simons M, Gordon E, and Claesson-Welsh L (2016) Mechanisms and regulation of endothelial VEGF receptor signalling, Nat Rev Mol Cell Biol 17, 611–625. [DOI] [PubMed] [Google Scholar]
- [3].Peach CJ, Mignone VW, Arruda MA, Alcobia DC, Hill SJ, Kilpatrick LE, and Woolard J (2018) Molecular Pharmacology of VEGF-A Isoforms: Binding and Signalling at VEGFR2, Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, and Nagy A (1996) Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele, Nature. 380, 435–439. [DOI] [PubMed] [Google Scholar]
- [5].Miquerol L, Langille BL, and Nagy A (2000) Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression, Development 127, 3941–3946. [DOI] [PubMed] [Google Scholar]
- [6].Maharaj AS, and D'Amore PA (2007) Roles for VEGF in the adult, Microvasc Res 74, 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Carmeliet P (2005) VEGF as a key mediator of angiogenesis in cancer, Oncology 69 Suppl 3, 4–10. [DOI] [PubMed] [Google Scholar]
- [8].Rapisarda A, and Melillo G (2012) Role of the VEGF/VEGFR axis in cancer biology and therapy, Adv Cancer Res 114, 237–267. [DOI] [PubMed] [Google Scholar]
- [9].Folkman J (1995) Angiogenesis in cancer, vascular, rheumatoid and other disease, Nat. Med 1, 27–30. [DOI] [PubMed] [Google Scholar]
- [10].Nagy JA, Dvorak AM, and Dvorak HF (2007) VEGF-A and the induction of pathological angiogenesis, In Annual Review of Pathology-Mechanisms of Disease, pp 251–275, Annual Reviews, Palo Alto. [DOI] [PubMed] [Google Scholar]
- [11].Kerbel RS (2008) Tumor Angiogenesis, N. Engl. J. Med 358, 2039–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carmeliet P, and Jain RK (2000) Angiogenesis in cancer and other diseases, Nature. 407, 249–257. [DOI] [PubMed] [Google Scholar]
- [13].Waters SL, Alastruey J, Beard DA, Bovendeerd PH, Davies PF, Jayaraman G, Jensen OE, Lee J, Parker KH, Popel AS, Secomb TW, Siebes M, Sherwin SJ, Shipley RJ, Smith NP, and van de Vosse FN (2011) Theoretical models for coronary vascular biomechanics: progress & challenges, Prog. Biophys. Mol. Biol 104, 49–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mac Gabhann F, Ji JW, and Popel AS (2006) Computational model of vascular endothelial growth factor spatial distribution in muscle and pro-angiogenic cell therapy, PLoS Comput. Biol 2, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mac Gabhann F, and Popel AS (2004) Model of competitive binding of vascular endothelial growth factor and placental growth factor to VEGF receptors on endothelial cells, Am J Physiol Heart Circ Physiol 286, H153–164. [DOI] [PubMed] [Google Scholar]
- [16].Teran M, and Nugent MA (2015) Synergistic Binding of Vascular Endothelial Growth Factor-A and Its Receptors to Heparin Selectively Modulates Complex Affinity, The Journal of biological chemistry 290, 16451–16462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sack KD, Teran M, and Nugent MA (2016) Extracellular Matrix Stiffness Controls VEGF Signaling and Processing in Endothelial Cells, Journal Of Cellular Physiology. 231, 2026–2039. [DOI] [PubMed] [Google Scholar]
- [18].Karlsson R, and Falt A (1997) Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors, J. Immunol. Methods 200, 121–133. [DOI] [PubMed] [Google Scholar]
- [19].Goldstein B, Coombs D, He X, Pineda AR, and Wofsy C (1999) The influence of transport on the kinetics of binding to surface receptors: application to cells and BIAcore, J. Mol. Recognit 12, 293–299. [DOI] [PubMed] [Google Scholar]
- [20].Myszka DG, He X, Dembo M, Morton TA, and Goldstein B (1998) Extending the range of rate constants available from BIACORE: interpreting mass transport-influenced binding data, Biophys. J 75, 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Morton TA, and Myszka DG (1998) Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors, Methods Enzymol. 295, 268–294. [DOI] [PubMed] [Google Scholar]
- [22].Grünewald FS, Prota AE, Giese A, and Ballmer-Hofer K (2010) Structure-function analysis of VEGF receptor activation and the role of coreceptors in angiogenic signaling, Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics 1804, 567–580. [DOI] [PubMed] [Google Scholar]
- [23].O'Shannessy DJ, and Winzor DJ (1996) Interpretation of deviations from pseudo-first-order kinetic behavior in the characterization of ligand binding by biosensor technology, Anal. Biochem 236, 275–283. [DOI] [PubMed] [Google Scholar]
- [24].Nakamura F, Tanaka M, Takahashi T, Kalb RG, and Strittmatter SM (1998) Neuropilin-1 extracellular domains mediate semaphorin D/III-induced growth cone collapse, Neuron 21, 1093–1100. [DOI] [PubMed] [Google Scholar]
- [25].Pan Q, Chanthery Y, Liang WC, Stawicki S, Mak J, Rathore N, Tong RK, Kowalski J, Yee SF, Pacheco G, Ross S, Cheng Z, Le Couter J, Plowman G, Peale F, Koch AW, Wu Y, Bagri A, Tessier-Lavigne M, and Watts RJ (2007) Blocking neuropilin-1 function has an additive effect with anti-VEGF to inhibit tumor growth, Cancer Cell 11, 53–67. [DOI] [PubMed] [Google Scholar]
- [26].Parker MW, Xu P, Li X, and Vander Kooi CW (2012) Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1, J. Biol. Chem 287, 11082–11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shinkai A, Ito M, Anazawa H, Yamaguchi S, Shitara K, and Shibuya M (1998) Mapping of the sites involved in ligand association and dissociation at the extracellular domain of the kinase insert domain-containing receptor for vascular endothelial growth factor, J. Biol. Chem 273, 31283–31288. [DOI] [PubMed] [Google Scholar]
- [28].Quinn TP, Peters KG, De Vries C, Ferrara N, and Williams LT (1993) Fetal liver kinase 1 is a receptor for vascular endothelial growth factor and is selectively expressed in vascular endothelium, Proceedings of the National Academy of Sciences 90, 7533–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Waltenberger J, Claesson-Welsh L, Siegbahn A, Shibuya M, and Heldin CH (1994) Different signal transduction properties of KDR and Flt1, two receptors for vascular endothelial growth factor, J. Biol. Chem 269, 26988–26995. [PubMed] [Google Scholar]
- [30].de Vries C, Escobedo JA, Ueno H, Houck K, Ferrara N, and Williams LT (1992) The fms-like tyrosine kinase, a receptor for vascular endothelial growth factor, Science 255, 989–991. [DOI] [PubMed] [Google Scholar]
- [31].Seetharam L, Gotoh N, Maru Y, Neufeld G, Yamaguchi S, and Shibuya M (1995) A unique signal transduction from FLT tyrosine kinase, a receptor for vascular endothelial growth factor VEGF, Oncogene 10, 135–147. [PubMed] [Google Scholar]
- [32].Fuh G, Li B, Crowley C, Cunningham B, and Wells JA (1998) Requirements for binding and signaling of the kinase domain receptor for vascular endothelial growth factor, J. Biol. Chem 273, 11197–11204. [DOI] [PubMed] [Google Scholar]
- [33].Terman BI, Carrion ME, Kovacs E, Rasmussen BA, Eddy RL, and Shows TB (1991) Identification of a new endothelial cell growth factor receptor tyrosine kinase, Oncogene 6, 1677–1683. [PubMed] [Google Scholar]
- [34].Millauer B, Wizigmann-Voos S, Schnurch H, Martinez R, Moller NP, Risau W, and Ullrich A (1993) High affinity VEGF binding and developmental expression suggest Flk-1 as a major regulator of vasculogenesis and angiogenesis, Cell 72, 835–846. [DOI] [PubMed] [Google Scholar]
- [35].Canals F (1992) Signal transmission by epidermal growth factor receptor: coincidence of activation and dimerization, Biochemistry 31, 4493–4501. [DOI] [PubMed] [Google Scholar]
- [36].Gerber HP, McMurtrey A, Kowalski J, Yan M, Keyt BA, Dixit V, and Ferrara N (1998) Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3'-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation, Journal Of Biological Chemistry. 273, 30336–30343. [DOI] [PubMed] [Google Scholar]
- [37].Fong G-H, Rossant J, Gertsenstein M, and Breitman ML (1995) Role of the Flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium, Nature 376, 66–70. [DOI] [PubMed] [Google Scholar]
- [38].Cao Y (2009) Positive and Negative Modulation of Angiogenesis by VEGFR1 Ligands, Sci. Signal 2, re1. [DOI] [PubMed] [Google Scholar]
- [39].Hiratsuka S, Minowa O, Kuno J, Noda T, and Shibuya M (1998) Flt-1 lacking the tyrosine kinase domain is sufficient for normal development and angiogenesis in mice, Proceedings of the National Academy of Sciences 95, 9349–9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kalergis AM, Boucheron N, Doucey MA, Palmieri E, Goyarts EC, Vegh Z, Luescher IF, and Nathenson SG (2001) Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex, Nat. Immunol 2, 229–234. [DOI] [PubMed] [Google Scholar]
- [41].Miao HQ, Lee P, Lin H, Soker S, and Klagsbrun M (2000) Neuropilin-1 expression by tumor cells promotes tumor angiogenesis and progression, FASEB J. 14, 2532–2539. [DOI] [PubMed] [Google Scholar]
- [42].Soker S, Takashima S, Miao HQ, Neufeld G, and Klagsbrun M (1998) Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor, Cell 92, 735–745. [DOI] [PubMed] [Google Scholar]
- [43].Carmeliet P, Ng YS, Nuyens D, Theilmeier G, Brusselmans K, Cornelissen I, Ehler E, Kakkar VV, Stalmans I, Mattot V, Perriard JC, Dewerchin M, Flameng W, Nagy A, Lupu F, Moons L, Collen D, D'Amore PA, and Shima DT (1999) Impaired myocardial angiogenesis and ischemic cardiomyopathy in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188, Nat. Med 5, 495–502. [DOI] [PubMed] [Google Scholar]
- [44].Maes C, Carmeliet P, Moermans K, Stockmans I, Smets N, Collen D, Bouillon R, and Carmeliet G (2002) Impaired angiogenesis and endochondral bone formation in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188, Mech. Dev 111, 61–73. [DOI] [PubMed] [Google Scholar]