Abstract
Primary bile acids are synthesized from cholesterol in the liver, conjugated to either glycine or taurine and secreted into bile. Bile salts undergo enterohepatic circulation several times each day. During this process, they are biotransformed into a variety of metabolites by gut bacteria. The major biotransformation is the 7α-dehydroxylation of cholic acid and chenodeoxycholic acid yielding deoxycholic acid and lithocholic acid, respectively. 7α-Dehydroxylation is a multi-step pathway. The genes encoding enzymes in this pathway have been identified in two species of “high” activity strains of clostridia. Here, we report the isolation and characterization of a bile acid inducible (bai) operon in Clostridium hylemonae, a “low” activity 7α-dehydroxylating strain. The gene organization and sequence of the baiBCDEFGHI operon was highly conserved between C. hylemonae and “high” activity strains. Surprisingly, the baiA gene was missing from the bai operon of C. hylemonae. The baiA gene was isolated using PCR and degenerate oligonucleotide primers. The mRNA start site for the large bai operon was determined and shown to be only 11 bp from the initiation codon of the first gene. It was also discovered that allocholic acid (5α) induced the bai operon and stimulated the formation of allodeoxycholic acid. Finally, it was discovered that the addition of testosterone to the growth medium markedly increased 7α-dehydroxylation of cholic acid in C. scindens and C. hylemonae. We hypothesize that testosterone may be a gratuitous inducer of genes involved in the reductive arm of the bile acid 7α-dehydroxylation pathway.
Introduction
Bile acids are amphipathic molecules synthesized in the liver from cholesterol which function primarily in absorption of dietary lipids and lipid soluble vitamins. The primary bile acids synthesized in the human liver include cholic acid and chenodeoxycholic acid. After being conjugated to either taurine or glycine, bile salts are concentrated in the gallbladder during the interdigestive period, and released into the lumen of the small intestine after cholecystokinin induced contraction following a meal. Once bile salts reach the terminal ileum, they are actively transported from the ileal lumen, into the portal blood and returned to the liver. This process is known as the enterohepatic circulation and is highly efficient (>95%). However, roughly 400–600 mg of bile salts escape the enterohepatic circulation where they encounter the colonic microbiota consisting of about 2–5 × 1011 bacteria/gram wet weight (1, 2).
Bile salts are first deconjugated by a wide variety of intestinal microbes with bile salt hydrolase activity (3). Hydroxy groups can also be reversibly epimerized by the concerted effort of stereospecific hydroxysteroid dehydrogenases (3). A particularly important microbial biotransformation of primary bile acids is 7α/β-dehydroxylation, forming secondary bile acids. The secondary bile acids in humans include deoxycholic acid and lithocholic acid, formed from the 7α-dehydroxylation of cholic acid and chenodeoxycholic acid, respectively. Secondary bile acids are more hydrophobic than primary bile acids, and are able to passively diffuse across the colonic membrane and can thus reenter the biliary pool. The human liver is unable to 7α-hydroxylate secondary bile acids, and as a result, secondary bile acids can accumulate to high levels in some individuals (3). Numerous lines of evidence suggest that secondary bile acids promote colon cancer (4, 5), and possibly cholesterol gallstone disease, in some patients (6). Recent research has also shown that bile acids are potent signaling molecules regulating a variety of physiological processes (5, 7, 8). Therefore, understanding the molecular biology and ecology of bile acid metabolism in the colon is of importance to human health and disease as the gut flora can control the bile acid pool composition.
Removal of the 7-hydroxy group proceeds through a multi-step biochemical pathway (3). Primary bile acids enter the cell through an H+-dependent primary bile acid transportor encoded by the baiG gene (9). Once inside the cell, the bile acid is ligated to coenzyme A in an ATP-dependent manner (baiB gene) (10). The bile acid-CoA thioester is then oxidized at the 3-hydroxy group by a NAD+(P) dependent 3α-hydroxysteroid dehydrogenase encoded by the baiA gene(s) (11). The baiA gene was shown to be specific for bile acid CoA conjugates (11). The next step in the pathway is formation of a either 3-dehydro-4-CDCA/3-dehydro-4-CA or 3-dehydro-4-UDCA by the NAD-dependent flavoproteins encoded by the baiCD or baiH genes, respectively (12). Following the 7α-hydroxy dehydration, the 3-oxo-4,6-intermediate is sequentially reduced in a 3-step manner. The secondary bile acid is hypothesized to be transported from the cell. The pathway is predicted to provide energy to the bacterium as there is a net 2 electron reduction, and recent evidence suggests that the baiF gene may conserve energy through transfer of coenzyme A in an ATP-independent manner (13). Thus, with a small population of bacteria capable of 7α/β-dehydroxylation of bile acids (~0.0001% of total colonic flora), and a large amount of substrate (400–600 mg/day), these bacteria are likely to occupy a unique niche in the gastrointestinal tract (14–16).
Recent data by Narushima et al. (2006) demonstrated, for the first time, that human intestinal 7α-dehydroxylating clostridia, when inoculated in germ-free mice (GFM) in combination with bile acid deconjugating strains are able to generate deoxycholic acid in vivo (17). This GFM study suggested that strains of Clostridium hylemonae may play an important role in bile acid transformation in the human colon. However, the genes involved in bile acid metabolism in C. hylemonae have yet to be identified and characterized. The current study reports isolation and analysis of bile acid-inducible (bai) genes from C. hylemonae- including conserved putative upstream regulatory elements and comparison with other known bile acid 7α/β-dehydroxylating bacteria. In addition, we have identified bile acid and steroid inducers which may lead to discovery of novel enzymes involved in bile acid 7α-dehydroxylation.
1. Materials and methods
2.1. Bacterial strains and culture conditions
Clostridium hylemonae TN271 was a generous gift of Dr. Fusae Takamine, University Ryukyus, Okinawa. C. hylemonae and C. scindens VPI 12708 were cultivated under anaerobic conditions (hungate tubes; N2) at 37 °C in Brain Heart Infusion broth (BHI) (BD, Sparks, MD, USA). One Shot © Chemically Competent E. coli was purchased from Invitrogen (Carlsbad, CA, USA) and cultivated at 37 °C in LB medium (Fischer, Fair Lawn, NJ, USA) containing 100 μg/ml spectinomycin.
2.2. DNA extraction and general PCR methods
Genomic DNA was extracted from C. hylemonae TN271 and Clostridium scindens VPI 12708 as previously described (18). Plasmid DNA was isolated from E. coli using the QIAprep Spin Miniprep kit (Qiagen, MD, USA). PCR products ≤ 2 kb were amplified using the TITANIUM Taq PCR kit (Clontech) as recommended by the manufacturer. Reactions contained 500 nmole each primer and 50–100 ng genomic DNA template. PCR was performed on an Eppendorf Mastercylcer Gradient PCR machine in 50 μl volumes with 29 cycles of 95°C for 20 s, annealing temperatures and times varied based on primer/template, 68°C extension for 30–90s. PCR products were separated on a 1% agarose gel containing 0.75 μg/ml ethidium bromide.
Genome-walking by polymerase chain reaction
Genome-walking DNA libraries were prepared using the Universal Genome-Walker Kit (Clontech) according to the manufacturer. The DNA extraction method used in this application (18) was slightly modified to obtain high molecular weight DNA (>50 kB). Pipetting steps during phenol/phenol:chloroform:iso-amyl alcohol were changed to decanting the aqueous phase, which prevents substantial DNA sheering. An 18 gauge needle was used to pierce the very bottom of 50 ml falcon conical tubes following centrifugation and the organic phase was emptied. DNA was then precipitated by addition of 10% 3M sodium acetate and 3 volumes of absolute ethanol. DNA pellets were washed with 70% ethanol and resuspended in TE buffer, pH 7.6, overnight with agitation at 45 °C. DNA quality was determined on a 0.5% agarose gel containing 75 μg/ml ethidium bromide. Prior to electrophoresis, the DNA sample was heated to 65°C for 15 min. The gel was run 18 hours at 25 V overnight along with a High Molecular Weight DNA Marker (Invitrogen). DNA (>48 kb) was restriction digested and adapter ligated using GenomeWalker kit. Genomic DNA libraries were stored at −20°C. Oligonucleotides were designed using Primer3 software with Tm values between 60°C and 70°C with ≥ 45% G+C and ≥ 25 mer in length.
PCR reactions were prepared using Advantage cDNA Polymerase Kit or Advantage 2 Polymerase Kit (Clontech) as follows: 40 μl ddH2O, 5 μl 10x Advantage PCR buffer, 1 μl dNTP mix (10 mM each), 1 μl adaptor primer AP1 (10 μM), 1 μl gene-specific primer (10 μM), 1 μl Advantage Polymerase mix (50X) and 1 μl adaptor-ligated DNA restriction library template. Reaction mixtures were mixed briefly and tubes centrifuged. “Touchdown” PCR amplification was performed on an Eppendorf Mastercylcer Gradient PCR machine using the following parameters: 95°C for 25 s, 72°C for 3 min (7 cycles); 94°C for 25 s, 67°C for 3 min (32 cycles); 67°C for 7 min, hold 4°C. Products (5 μl) were separated by electrophoresis on a 1% agarose/EtBr gel and visualized on a Fotodyne analyzer. PCR products of interest were GENECLEAN purified from the gel and ligated into a pCR8 GW TOPO TA Cloning vector (Invitrogen) and transformed into TOP 10 Chemically Competent E. coli according to the manufacturer (Invitrogen). Colonies were cultivated in LB broth containing 100 μg/ml spectinomycin. Plasmids were purified using Qiagen Miniprep Spin Kit according to the manufacturer. Sequencing was performed at the Nucleic Acid Research Facility, School of Medicine, Virginia Commonwealth University on ABI 3700 Sequencer using oligonucleotides synthesized by Integrated DNA Technologies (Coralville, IA, USA).
Isolation and analysis of RNA
C. scindens VPI 12708 as well as C. hylemonae were cultivated in 400 ml BHI containing 0.02% fructose to an O.D.600nm 0.3 (C. scindens) or 0.2 (C. hylemonae) at which point the cultures were induced 3 times at hourly intervals with 50 μM CA. One hour after the third induction, 1 ml culture was transferred to a serum bottle containing 1 ml BHI. [24-14C] CA (0.5 μCi/500 nmoles) was added to initiate the reaction which was then incubated at 37°C for 10 minutes. A non-induced control was also tested for 7α-dehydroxylation activity. The remaining culture was centrifuged at 13,000 × g for 30 min at 4°C. The cell pellet was suspended in 1 ml TE buffer and cells transferred to 1.5 ml microcentrifuge tube and spun 1 min at 13,600 × g. The supernatant was decanted and bacterial cells were stored in RNAlater solution (Ambion, Austin, TX, USA) overnight, centrifuged and the pellet was stored at −70°C until further processing.
Once 7α-dehydroxylating activity was confirmed by presence of [24-14C] DCA in activity assay, total RNA was extracted using the Ribopure Bacteria RNA extraction kit (Ambion) essentially according to the manufacturer with the exception that cells were broken on a Mini-bead beater (Biospec Products Inc, Bartlesville, OK, USA) at 3,000 rpm for two cycles of 1 min. DNase treatment was performed for 1 hour at 37°C. cDNA was generated using the SMART-RACE cDNA synthesis kit (Clontech) according to the manufacturer with some modifications. First strand synthesis was generated using gene-specific primers (Table 1) rather than 5′ CDS primer. cDNA was synthesized starting with 1 μg total RNA. SMART RACE PCR of 5′ ends was performed using the Advantage-GC 2 PCR Kit (Clontech) according to the manufacturer’s instruction. 0.5 μl GC-Melt was added to each 50 μl reaction volume along with 2.5 μl cDNA sample. PCR products were excised from agarose gel and purified using the GENECLEAN Spin Kit (MP Biomedicals, Solon, Ohio). The purified PCR product was then cloned into a pCR8 GW TOPO TA cloning vector (Invitrogen), transformed into TOP 10 Chemically Competent E. coli (Invitrogen) and selected on LB agar containing 100 μg/ml spectinomycin. Plasmids containing insert were sequenced. cDNA for reverse transcriptase PCR was generated using the Advantage RT-for-PCR kit (Clontech) according to the manufacturer using random hexamer primers and 0.5 μg RNA.
Table 1.
Oligonucleotides and plasmids used in this study
| Primer name | Sequence | Reference |
|---|---|---|
| Redundant oligonucleotides | ||
| baiCD-F | GGWTTCAGCCCTCAGATGTTCTTTG | 21 |
| baiCD-R | GAATTCCGGGTTCATGAACATTCTKCKAAG | 21 |
| baiA271F | CAGGYGGMACMMGWGGWATNGG | TS* |
| baiA271R | GGRCTRTAAGCMCCATCWTCGC | TS |
| gyrAF1 | GGVAARTAYCAYCCGCAYGGVGAY | TS |
| gyrAR4 | TCKATRCAKATSCGCATKCCYTCTCT | TS |
| Genome-walking oligonucleotides | ||
| AP1 | GTAATACGACTCACTATAGGGC | Invitrogen |
| AP2 | ACTATAGGGCACGCGTGGT | Invitrogen |
| WALKUP-1 | GAAGTCCTTTATACTCCAGACAGAGC | TS |
| WALKUPNEST-1 | TTGGATCGATGACTGCGTCATAACAGC | TS |
| WALKUP2 | AACCAGAAGCTGTCTCTGCTCAAAGTCC | TS |
| WALKUP2NEST | AGCTGTCTCTGCTCAAAGTCCATGCC | TS |
| WALKDS-1 | GTATACCCAGTATGAGGTGCTAAGAGG | TS |
| WALKDSNEST-1 | CAGTATGAGGTGCTAAGAGGAGAAGC | TS |
| WALKDS2 | TACGAGGAACATTTTATGCGCGACC | TS |
| WALKDSNEST-2 | CGAAGATCAAGATCACGATGAACATGC | TS |
| WALKDS3 | CAATTCCCTGGAGGATATGCTTTATCTGG | TS |
| WALKDSNEST-3 | CCTAAGTTTGAAGCGGCAGGGGTGG | TS |
| baiAWALKDS1 | GAATACGACTCACTATAGGGC | TS |
| baiAWALKUP1 | ATACCCTATTCCGGACAGAGAACC | TS |
| baiAUP1NEST | ACAACCGTAGATGCCTGTCACAGATGC | TS |
| baiADS1NEST | GTGTGATCGGTCTTACACAAGGTCTCG | TS |
| AUP2TN271 | GCACGTATAGAAACTGAACAGAGCA | TS |
| AUP2TN271NEST | CCTGGGAGTCCTTATTACCTGTGC | TS |
| AUP3TN271 | GGCAGTGTTGTATATGATCTCACC | TS |
| AUP3TN271NEST | TCCACGTCCTGTACTTTGTATCC | TS |
| UP4A | TTAACCTCCGTGATCTCATCTTCC | TS |
| UP4ANEST | CATCTTCCGTAACTGTTACATTCACG | TS |
| Primer-walking oligonucleotides | ||
| 271-100-F | ACATCGGGTATGTAGACGAGGAAGG | TS |
| 271-001-F | GTTCTGGGATGAACTGGAGACTACACTCTCC | TS |
| 271-100-R | GCTGATGACTTCAAAGCAGAATCTGCAGC | TS |
| CGAPR1 | CTCTTCCTCTGTCATGTTCTGC | TS |
| RGAPF1 | TCTTACACGGTTGAGAAACTGG | TS |
| BGAPUP | GCTTCCATGTGATCTCCGATAC | TS |
| BGAPF1 | GTTCCCACACTTATGAATCG | TS |
| BGAPR1 | TCTACCACTGCATGCAGC | TS |
| EGAPF1 | TGATGAGACGAACACACTGG | TS |
| 271baifF | ACATGTCACTGAACGGAACG | TS |
| 271baihR | AGATAAAGCATATCCTCCAGG | TS |
| 271baihR2 | CTTAAGTGTCAGGGATCC | TS |
| HGAP1 | ACTTTCCTTTCTCCGCTTCG | TS |
| 271baifF2 | GATATACAGACGAAGAGATCGACG | TS |
| EDS2271baiFf2 | ACTGGATTGCGAGAGAGACG | TS |
| EDS2271BAIHR | GTCAACCGTAACAGCGTCTATG | TS |
| EDS2HGAP2 | GGTCGGTAAGATCAACGATACC | TS |
| EDS2HR2F | TCAGAAGCAGAGAAAACGTAAGC | TS |
| EDS2HR2R | CTTTATGAAAGTGGCGTTGTCC | TS |
| EDS2F2R | CTATTCATCTTTCATGTGAGG | TS |
| EDS2F2F | GTTCATAGTAGGTATCGGAACG | TS |
| DS3BAIHF2 | CATATATGTACCCGCACACACC | TS |
| DS3GLUSYNF1 | CAGAAATTATGATCCGCTCTCG | TS |
| DS3GLUSYNR2 | AGAAAATCGTGGGCAGTTAGC | TS |
| baiHFTN271 | AGGGACGGGATATGTAGTCATAGACG | TS |
| baiIR | ACTCCTCCCCTTTCATATCCATAAGC | TS |
| baiAR1 | CTGATTGGTTCGCGTTCTTCC | TS |
| baiAUPAR2 | ATGTCTCGTCAATGGGAAGC | TS |
| baiAUP2AR2 | CAGGTGTTCCCTGTTGAGG | TS |
| baiAUPARR | GAATATCGGTATCCTGGACTGG | TS |
| baiAUPDUPF2 | GAAGTTTGTGTCGGAGCATACC | TS |
| baiAUPDUPFR | CAGCATTATCTTTTCCACTTCG | TS |
| AUP2GW2R | GTCTTCATATCTGCAACCATGG | TS |
| AUP2GW1F | CTCACAGATGAAAATGGTGG | TS |
| UP4F1 | GAAATACCGGTAGACACTGATCG | TS |
| UP4R1 | CACAATACGTATGTTGGCAAGC | TS |
| UP4F2 | GTCATCCGCTTTAGAAGAAGC | TS |
| UP4AR2 | CATATTCACTGTTGCCTGTTCC | TS |
| UP4F3 | GGAAGTCATCGACGAGATCCTAGAGG | TS |
| UP4AR3 | AAGTCCGTTTGCCTCTGCAGC | TS |
| RT-PCR oligonucleotides | ||
| baiBTN271RTF | CTGCAGGCTCATAAAGGAATATAA | TS |
| BCDInterF | GATAGAAAGAGGAGACAACGGGAAG | TS |
| BCDInterR | GATGGTCGTTGTACAGACCCATATAC | TS |
| CDEInterF | ATTCCGGTACAGGTGATAGGAGAC | TS |
| CDEInterR | GTAGTCTCCAGTTCATCCCAGAACTT | TS |
| EFInterF | GAGACCGGGTATCTTCGTGTTTAC | TS |
| EFInterR | AGGGCTCCGAAACTTGGAAAATCT | TS |
| FGInterF | CTCAGAGACCTTGGATATACAGACGA | TS |
| FGInterR | GGGAAATCCATGTCGTATTGCTGT | TS |
| GHInterF | AGTGTCGTCCATCAAAACACTGAC | TS |
| GHInterR | GTCTTCCGCCTCATAGTCCATACT | TS |
| HIInterF | AGGATTTGAAGCTGCCTACAGTCT | TS |
| HIInterR | CATTATCAGTTCCCGGTCTTTCTC | TS |
| IGLUInterF | GAGACATGGATGGTCTACTGTGATG | TS |
| IGLUInterR | CATATAACGGTAACCGCACTTCTG | TS |
| AgeneF | GAGTTAAAAGAGCTTTATCCCGAAG | TS |
| AgeneR | GCTTCCTCTGTCACTCTTGAAAATA | TS |
| AdsinterF | TCTCGAAGACTACCTGAAATCATTC | TS |
| AdsinterR | TCATGACGCTGAAGGATAAAGATAC | TS |
| gyrATN271F | ACTTTGACGAGACAGAAAAAGAACC | TS |
| gyrATN271R | CAGTATATGTTCGATCGTCGTCTCT | TS |
| SMART-RACE oligonucleotides | ||
| RACEB325 | GTCTTGCTCGATATGGGCATATAACAGGC | TS |
| TNRACEA253 | TCCCGTATTTTTCCGCCACTGCTCTTACC | TS |
TS (This study)
Sequence analysis and Assembly
Sequence ABI files were edited and assembled using Vector NTI software (Invitrogen, Carlsbad, CA, USA), as well as CAP3 Sequence Assembly Program. Pairwise alignments were performed to determine percent identity/similarity using the EMBOSS Needle program. Homology searches were made against the UniProt database. Multiple sequence alignments were prepared using T-COFFEE (19) or CLUSTALW2 with some manual editing based on structural data. Sequence conservation within multiple alignments was emphasized using the BOXSHADE utility. Protein Mr calculations were performed using the Compute pI/MW tool on the Expasy Proteomics Server. Open reading frames (ORF) were detected using the ORF Finder Program at the NCBI website and searched against Genbank by BLAST analysis.
Nucleotide Accession Numbers
Genbank accession numbers for the baiBCDEFGHI operon from C. hylemonae TN271 is (EU675332). The baiA gene from C. hylemonae TN271 can be found using accession number (EU675329).
Induction Experiments
Allocholic acid was tested as a potential inducer of bile acid 7α-dehydroxylating genes by addition of 50 μM allocholic acid (Toronto Research Chemicals) to BHI prior to inoculation with either C. scindens VPI 12708 or C. hylemonae TN271. Cells were grown until mid-log phase at which point one ml culture was added to a 5 ml anaerobic (N2) serum bottle containing 1 ml BHI and 0.1 μ Ci [24-14C] CA. Assay was performed at 37°C for 10 minutes and stopped by addition of 0.1 volume 1N HCl, and bile acids extracted with ethyl acetate and dried under nitrogen gas.
Testosterone was tested as a possible enhancer of bile acid 7α-dehydroxylating activity by addition of 50 μM testosterone (in ethanol) either alone or in conjunction with 50 μM CA. Qualitative assays were performed in a similar manner as the allocholic acid studies. Quantitative assays required addition of 0.1 μCi/ml [24-14C] CA to the culture medium and removal of 1 ml aliquots at various intervals throughout logarithmic and stationary phase. Bile acids were separated via TLC and regions of the TLC containing CA and DCA were extracted and counted via scintillation spectrometry.
3. Results
3.1. Identification and characterization of the baiBCDEFHGI operon
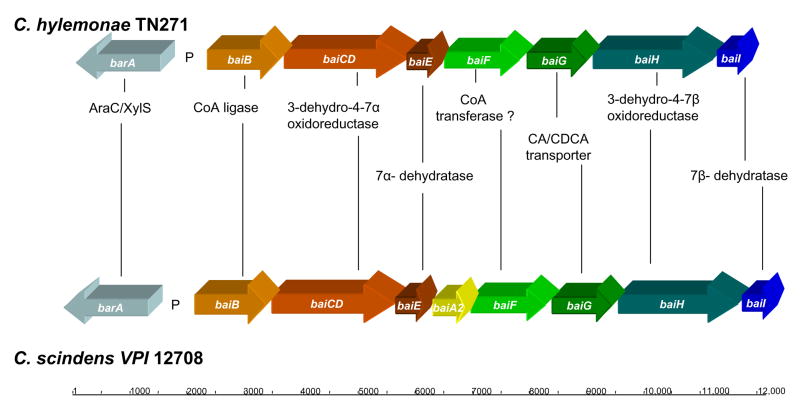
Previously, our laboratory performed activity assays on cultured 7α-dehydroxylating bacteria and found that these organisms clustered into two groups based on relative rate of conversion of [24-14C] CA to [24-14C] DCA (20). Wells et al. (2000) designed redundant PCR primers targeting the baiCD gene which are aimed at detecting 7α-dehydroxylating bacteria in the human colon (21). In that study, the baiCD gene of C. hylemonae TN271 was detected using these oligonucleotides. Therefore, these primers were used to amplify a 1.2 kB segment of the baiCD gene from C. hylemonae which was subsequently cloned into a pCR8 GW TOPO TA vector and sequenced. BLAST analysis of the sequence data and CLUSTALW alignment of the nucleotide and translated amino acid sequence verified the identity of a baiCD gene in C. hylemonae. We hypothesized that the bai genes involved in the oxidative arm of the bile acid 7α-dehydroxylation pathway (baiA – baiI) would be located on a polycistronic operon flanking the baiCD gene. Therefore, a bidirectional genome-walking by PCR strategy was used to isolate the remaining bai genes.
A slight modification of the method of Ridlon et al. (2005) was utilized to prepare genomic DNA (>50 kb) from both C. scindens VPI 12708 and C. hylemonae TN271 for genome-walking by PCR (see Materials and Methods) (18). Restriction libraries derived from this DNA sample yielded PCR products averaging 2 kb with a range of 0.7 kb to 4 kb. The limit of this technique, according to the manufacturer (Clontech GenomeWalker Universal Kit) is around 6 kb. Table 1 lists oligonucleotides used in this study. Upstream genome-walking PCR using primer WALKUP1 and subsequent nested amplification using primer WALKUP1Nest resulted in 2,139 bp of sequence data which overlapped the baiCD gene data by 673 bp resulting in a 2,690 bp contig. Analysis of this data revealed the 5′ end of the baiCD gene. A second open reading frame (ORF) was detected 19 bp upstream of the baiCD gene. BLAST results suggest this ORF encodes an AMP-dependent coenzyme A ligase with highest identity to the baiB gene from C. hiranonis. Continued upstream genome-walking resulted in a PCR product of 1,964 bp. Assembly of this data resulted in a 4,586 bp contig containing the complete baiB gene. The 1.5 kb baiB gene is predicted to encode a 57.7 kDa polypeptide similar in size to the reported 58 kDa subunit recombinant baiB polypeptide from C. scindens VPI 12708 (10).
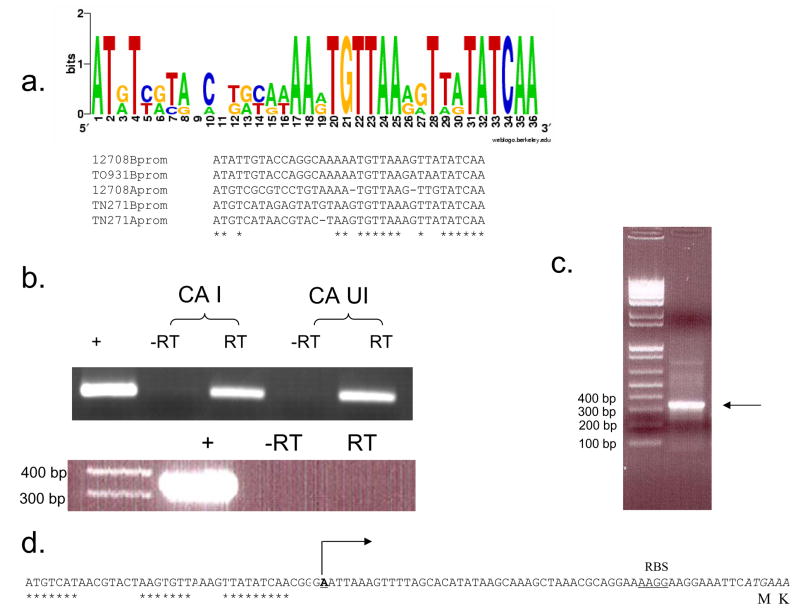
Two additional interesting features were identified within this sequence data. First, a conserved bile acid inducible promoter was located on the sense strand 49 bp upstream of the baiB gene (Figure 1). The second notable feature is a conserved 47 kDa AraC/XylS transcription factor located on the anti-sense strand upstream of the bai promoter which is conserved in C. scindens VPI 12708 and C. hiranonis TO931. Additional genome-walking upstream of the barA did not result in ORFs predicted to be involved in bile acid metabolism. Downstream genome-walking from the baiCD fragment yielded a PCR product of 1,131 bp which increased contig length to 5,625 bp. This stretch of sequence resulted in completion of the 1.9 kb baiCD gene predicted to encode a 70 kDa polypeptide. We recently reported overexpression of the recombinant baiCD gene from C. scindens VPI 12708 in E. coli resulted in a 70 kDa polypeptide as estimated by SDS-PAGE (12). In addition, a 510 bp ORF was located 46 bp downstream of the baiCD gene which was predicted to encode a 19.8 kDa bile acid 7α-dehydratase (baiE gene). Dawson et al (1996) reported that the19 kDa baiE gene from C. scindens VPI 12708 encodes a bile acid 7α-dehydratase (22). Multiple sequence alignment of this ORF with the baiE genes from C. scindens VPI 12708 and C. hiranonis sp. strain TO931 revealed >80% amino acid sequence identity and conservation of active site amino acids predicted through computer modeling and site-directed mutagenesis data (3) (Table 2). Continued downstream GW-PCR resulted in a 3,856 bp PCR product which when added to the previous alignment results in a contig of 9,291 bp. An ORF was located 25 bp downstream of the baiE gene 1,275 bp in length predicted to encode a 47 kDa polypeptide in the Type III CoA transferase family of proteins. BLAST analysis suggests this gene encodes the baiF gene product. Ye et al. (1999) previously determined the Mr of the recombinant baiF from C. scindens VPI 12708 to be 47.5 kDa and to have bile acid CoA hydrolase activity (13). A second ORF, 1.4 kb in length, was located 53 bp downstream of the baiF gene whose deduced amino acid sequence suggests this gene encodes a 49.5 kDa AraJ-like efflux permease. BLAST analysis suggests this ORF encodes the baiG gene product. Mallonee et al (1996) overexpressed the 50 kDa baiG gene product in E. coli and showed that it is involved in transporting primary bile acids across the bacterial cytoplasmic membrane (9). Further analysis revealed a partial ORF 19 bp downstream of the baiG gene encoding an “Old Yellow Enzyme” like FMN oxidoreductase with the baiH gene from C. hiranonis sharing the highest amino acid sequence identity. The third downstream GW-PCR resulted in completion of the bai oxidative operon. The 2,019 bp baiH gene was completed and the deduced amino acid sequence suggested this gene encodes a 71.7 kDa polypeptide. The baiH gene from C. scindens VPI 12708 has been shown to encode a 72 kDa polypeptide with 3-oxo-4-steroid oxidoreductase activity (12). Finally, the 549 bp baiI gene was located 38 bp downstream of the baiH gene and predicted to encode a 21.8 kDa polypeptide.
Figure 1.
SMART RACE PCR analysis to determine TIS for the baiB transcript in Clostridium hylemonae TN271. (a.) RT-PCR analysis of C. hylemonae TN271 cDNA generated from CA induced (I) and uninduced (UI) cultures. Primers against baiB gene (above) and gyrA gene (below) were used to determine whether baiB transcript was expressed. Below RT-PCR is the corresponding [24-14C] CA activity assay performed prior to RNA isolation of I and UI cultures of C. hylemonae. [24-14C] DCA is detected only in CA induced (50 μM) culture. (b.) 5′ SMART RACE PCR of baiB transcript using RACEB325 oligonucleotide against cDNA from induced culture of C. hylemonae TN271. PCR product was purified by GENECLEAN Spin Kit and cloned into a pCR8 GW TOPO TA cloning vector and sequenced. (c.) TIS for baiB transcript (underlined; direction of transcription designated by arrow). “*” designates conserved bai promoter elements (See Figure 4), putative ribosome binding site in bold, “†” designates mRNA start site determined for baiB in C. scindens, “‡” designates mRNA start site for baiB in C. hiranonis sp. strain TO931, italics indicates anticodons with corresponding amino acids (below).
Table 2.
Comparison of bai gene sequences between Clostridium hylemonae TN721 and Clostridium scindens VPI 12708
| Gene or region | Length (bp) C. scindens vs. C. hylemonae | DNA sequence identity (%) | Enzyme function | Amino acid sequence identity (%) | Amino acid sequence similarity(%) | Reference(s) |
|---|---|---|---|---|---|---|
| baiB | 1,563 vs. 1,545 | 65.7 | Bile acid-CoA ligase | 64.4 | 78.9 | 10 |
| baiB-CD(intergene) | 45 vs. 44 | 52 | 23 | |||
| baiCD | 1,920 vs. 1,923 | 75 | 3-dehydro-4-steroid oxidoreductase | 80.6 | 89.2 | 23 |
| baiCD-baiE(intergene) | 43 vs. 46 | 51.1 | 23 | |||
| baiE | 501 vs. 510 | 76.6 | Bile acid 7α-dehydratase | 82.8 | 92.3 | 22 |
| baiA | 747 vs. 750 | 76.2 | 3α-hydroxysteroid dehydrogenase | 85.5 | 92.4 | 11, 25 |
| baiF | 1,281 vs. 1,275 | 78.7 | Bile acid-CoA hydrolase/transferase* | 88 | 94.8 | 13 |
| baiF-baiG(intergene) | 5 vs. 25 | 5 | † | |||
| baiG | 1,434 vs. 1,428 | 71.1 | Bile acid transporter | 80.3 | 88.9 | 9 |
| baiG-baiH(intergene) | 28 vs. 19 | 55.6 | † | |||
| baiH | 1,986 vs. 2,019 | 74.2 | 3-dehydro-4-steroid oxidoreductase | 80.8 | 90.0 | 12, 33 |
| baiH-baiI(intergene) | 17 vs. 38 | 41.9 | † | |||
| baiI | 555 vs. 549 | 68.2 | Bile acid 7β-dehydratase* | 61.4 | 76.6 | ‡ |
Previous studies have shown that bai genes involved in the oxidative arm of the 7α/β-dehydroxylating operon are co-expressed as a polycistronic message in Clostridium scindens VPI 12708 (23) and Clostridium hiranonis TO931 (24). RT-PCR analysis of intergenic regions indicated that the baiBCDEFGHI genes located in Clostridium hylemonae TN271 are also expressed as a polycistronic message (Figure 2).
Figure 2.
RT-PCR of intergenic regions of the baiBCDEFGHI operon from C. hylemonae TN271. RNA was isolated from CA induced cells. “+” represents positive control which is PCR of C. hylemonae TN271 genomic DNA. –RT is the negative control reaction in which cDNA synthesis lacks reverse transcriptase. Organization of genes is shown above for reference. See table 1 for primer sequences.
The complete bai oxidative operon from C. hylemonae is represented schematically in Figure 3 and compared to the gene organization in C. scindens. Table 2 compares amino acid sequence identity/similarity between C. hylemonae and C. scindens. Clearly, there is a high degree of conservation both between the genes and several of the intergenic regions; not surprising given the close phylogenetic relation between these species as determined by neighbor-joining of 16s rDNA sequences of 7α-dehydroxylating bacteria and other gut microbes (21). One of the most striking observations is the fact that the baiA gene is not found within the operon of C. hylemonae as compared to C. scindens and C. hiranonis. Taken together, these data indicate that genes located on the 9.5 kb bai oxidative operon of the “low activity strain” C. hylemonae TN271 are highly conserved both in organization as well as amino acid sequence with the homologous bai polycistronic operons identified in the “high activity” species C. scindens and C. hiranonis with the exception of the baiA gene.
Figure 3.
Schematic representation and comparison of the bai oxidative operons of C. hylemonae TN721 and C. scindens VPI 12708. See Table 2 for comparison of amino acid sequence identity and other parameters. Notice the high degree of conservation in gene organization, with the exception of the baiA gene, present in both C. scindens and C. hiranonis though conspicuously missing from the bai oxidative operon of C. hylemonae TN271.
SMART-RACE PCR analysis of the cDNA from a CA induced culture of C. hylemonae led to the identification of the mRNA start site which appears in a short purine rich stretch corresponding to the predicted ribosome binding site (Figure 1). Interestingly, the distance between the mRNA start site in the baiB polycistronic operon in C. hylemonae is only 11 bp from the initiation codon while in the “high activity strains” there is a distance of 108 bp in C. hiranonis (24) and 68 bp in C. scindens (23). As proof of principle, SMART RACE PCR was also performed on the baiB gene of C. scindens VPI 12708, whose mRNA initiation site was previously determined by primer extension (23). The results obtained in the present study agree with those of Mallonee et al. (1990) providing additional evidence for the TIS of the baiB operon in C. scindens VPI 12708 (Data not shown) (23).
Isolation and characterization of a baiA gene in Clostridium hylemonae TN271
The 27 kDa baiA gene product from C. scindens VPI 12708 catalyzes the first oxidative step in the bile acid 7α-dehydroxylating pathway following ATP-dependent CoA ligation (11). As noted in the previous section, and illustrated in Figure 3, the baiA gene was not found in the oxidative polycistronic bai operon in C. hylemonae. We predicted that the baiA gene may be found as a single cistron with a bai promoter region as observed in C. scindens with the baiA1 and baiA3 genes (25). Therefore, redundant primers were designed based on a multiple sequence alignment of baiA genes from C. scindens and C. hiranonis (Table 1). Using these primers and C. hylemonae genomic DNA, a PCR product of 722 bp was obtained, sequenced and predicted to encode the baiA gene by BLAST analysis. The complete 789 bp gene encoding a predicted 27.9 kDa protein was obtained following bidirectional genome-walking from the initial sequence data. Sequence analysis upstream of the baiA gene revealed a conserved bai promoter and RBS (Figure 4). Regions of the C. hylemonae baiB and baiA upstream regions, previously identified as highly conserved within bai promoters between species showed 100% identity. 5′ SMART RACE analysis suggests the mRNA initiation site is 56 bp upstream of the baiA gene (Figure 4).
Figure 4.
Bioinformatic and transcriptional chacterization of baiA gene and upstream regions. (a.) Conserved upstream regulatory elements from C. scindens VPI 12708 baiBCDEFGHI operon (12708Bprom) and baiA gene (12708Aprom), C. hiranonis sp. TO931 baiBCDEAFG operon (TO931Bprom), and C. hylemonae TN271 baiBCDEFGHI (TN271Bprom) and baiA gene (TN271Aprom) were aligned via ClustalW, and represented by Weblogo. (b.) RT PCR of CA induced vs. uninduced cultures of C. hylemonae (above). “+” represents C. hylemonae TN271 genomic DNA. Intergenic PCR spanning end of baiA gene and start of next predicted ORF (below). “+” represents C. hylemonae TN271 genomic DNA. Reactions performed with same cDNA preparation. (C.) SMART RACE PCR determination of baiA TIS using TNRACEA253 primer (Table 1). (d.) Upstream region of baiA gene. Asterisks denote conserved region (represented in “a.”) common to bai operons. TIS is shown in bold/underline with arrow denoting direction of transcription. Putative ribosome binding site (RBS) and coding sequence (italics) are shown.
Isolation of the gyrA gene from C. hylemonae
At the initiation of this study, the only sequence data available for C. hylemonae was a 16s rDNA sequence (26). We therefore wanted to isolate a gene which we used as a “housekeeping gene” for RT-PCR analysis of bai genes during cholic acid induction studies. The strategy for isolating the gyrA subunit from Clostridium hylemonae TN271 was to generate a multiple sequence alignment of several gyrA genes from various species of Clostridia in addition to two gyrA genes from Alkaliphilus oremlandii OhILAs. After a multiple alignment was generated and searched for regions of low codon degeneracy based on a codon usage table generated for C. hylemonae TN271 (not shown) two regions of high sequence identity and low codon degeneracy were identified. PCR amplification using sixteen combinations of forward and reverse redundant oligonucleotides resulted in five positive reactions. The primer pair used for cloning this gene are listed in table 1. Cloning of PCR product derived from primer pair gyrAF1/gyrAR4, sequencing of this product and alignment of the deduced amino acid sequence confirm that the gyrA gene has been isolated. A set of RT-PCR primers were designed to detect the gyrA gene of C. hylemonae TN271 (gyrATN271F 5′-ACTTTGACGAGACAGAAAAAGAACC-3′; gyrATN271R 5′-CAGTATATGTTCGATCGTCGT) which showed high specificity against genomic DNA. Induction with bile acids did not influence gyrA expression levels (Figure 1).
Identification of inducers of bile acid 7α-dehydroxylating activity
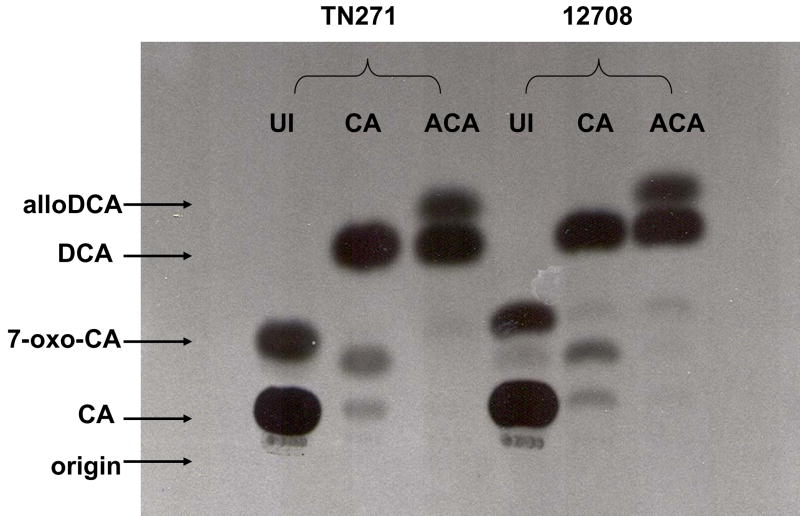
C. hylemonae TN271 and C. scindens VPI 12708 were tested for induction of bile acid 7α-dehydroxylating genes by allocholic acid (ACA), the 5α epimer of cholic acid. Addition of 50 μM ACA to culture medium during early log phase growth in BHI resulted in induction of 7α-dehydroxylating activity as demonstrated by [24-14C] cholic acid activity assay (Figure 5). Interestingly, both species produced [24-14C] DCA when 50 μM CA was added to the culture medium, but when ACA was added as the inducer, both [24-14C] DCA and [24-14C] alloDCA were produced in significant amounts (Figure 5). This data suggests that ACA induces the expression of a 5α-Δ4 bile acid oxidoreductase.
Figure 5.
TLC audoradiograph representing production of [24-14C] allodeoxycholic acid from [24-14C] CA following induction with allocholic acid in C. hylemonae TN271 and C. scindens VPI 12708. Cultures were grown in the presence of either 50 μM cholic acid (“CA”) or 50 μM allocholic acid (“ACA”) and compared to an uninduced control (“UI”). During mid-log phase, 1 ml was taken from each culture and incubated in the presence of 0.1 μCi [24-14C] CA for 15 minutes under anaerobic conditions and 37°C. Reactions were terminated by addition of 75 μl 1 N HCl.
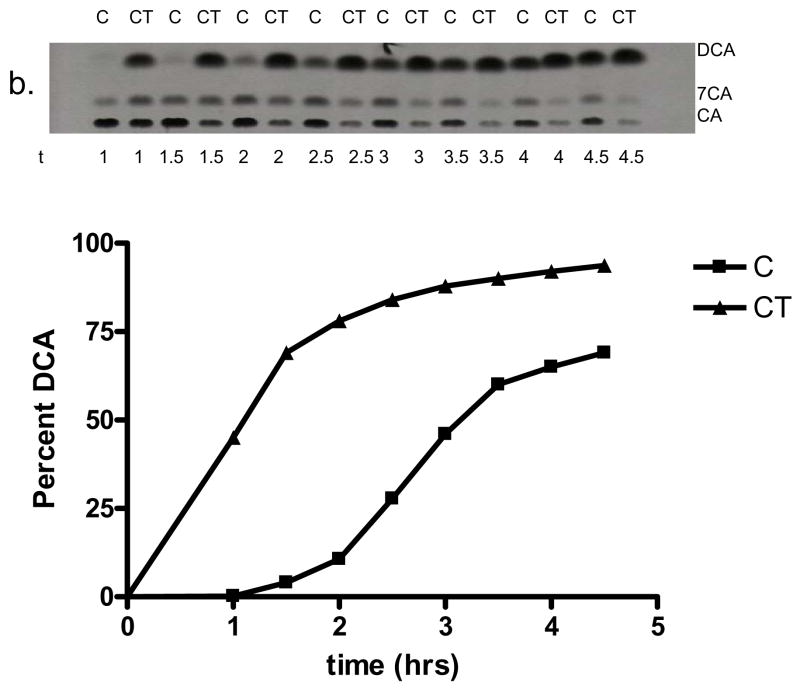
Testosterone shares a common 3-oxo-Δ4 structure with 3-oxo-Δ4-DCA, an intermediate following the 7α-dehydration step. While genes in the oxidative arm of the 7α-dehydroxylating pathway are induced about 100 fold over uninduced control, Δ6-reductase activity appeared to be induced only 6-fold over uninduced (Baron SF and Hylemon PB unpublished data). This suggests that the two arms of the pathway are under different regulation. We hypothesize that 3-oxo-Δ4-DCA or 3-oxo-Δ4,6-DCA may be intermediates that induce genes involved in reducing these intermediates to DCA. Because these intermediates are not commercially available, we tested the hypothesis that a Δ4- 3-oxo-steroid structure will increase 7α-dehydroxylating activity by adding testosterone in the presence of CA in the culture medium. Increased expression of these reductive genes is expected to increase the rate of 7α-dehydroxylation. Addition of 50 μM testosterone to the culture medium in the presence of 50 μM CA resulted in a significant increase in the rate of conversion of [24-14C] CA to [24-14C] DCA relative to cultures induced with CA alone. Figure 6 shows that the rate of [24-14C] DCA production is enhanced significantly in both species of clostridia. Indeed, after 48 hrs of growth, conversion of [24-14C] CA to [24-14C] DCA approached completion only in cultures that contained testosterone. The enhanced activity appears to only act in concert with CA, as testosterone alone is not sufficient to induce 7α-dehydroxylation, as shown in Figure 6. The addition of testosterone appears to “pull” the equilibrium toward secondary bile acid production as the percent conversion of CA to DCA in testosterone induced cultures during stationary phase was nearly double the value of CA induced cultures. These data suggest that ACA and testosterone may induce novel genes involved in bile acid 7α-dehydroxylation.
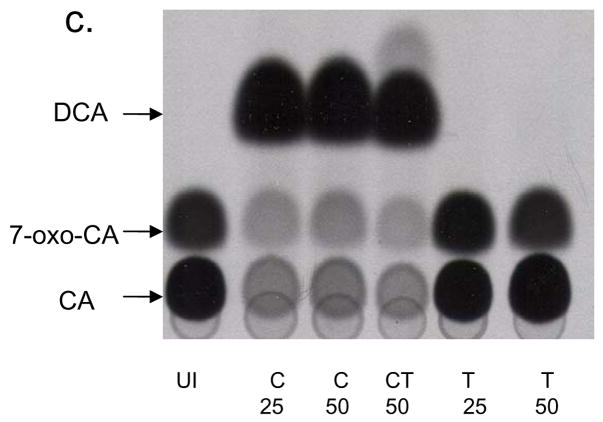
Figure 6.
Effect of testosterone on bile acid 7α-dehydroxylation in C. hylemonae TN271 and C. scindens VPI 12708. Cultures of each species were incubated with either 50 μM cholic acid (“C”) or 50 μM CA and 50 μM testosterone (“CT”). Each contained 0.1 μCi [24-14C] CA ml−1. Samples (1 ml) culture was taken at various intervals and bile acids were extracted and separated by TLC and visualized by audoradiography. (a.) Representative TLC audoradiograph and graphical representation of percent [24-14C] DCA produced by cultures of C. hylemonae TN271. (b.) TLC audoradiograph and graphical representation of percent [24-14C] DCA produced by cultures of C. scindens VPI 12708. (c.) Representative TLC audoradiograph of fixed point [24-14C] CA activity assay of uninduced (“UI”), induction with 25 or 50 μM C, 50 μM CT, and 25 or 50 μM testosterone (“T”).
Discussion
In the current work, we have isolated and characterized a polycistronic bile acid inducible operon and conserved upstream regulatory elements from the human intestinal bacterium Clostridium hylemonae TN271. This is the first report of identification of bai genes from a “low activity” bile acid 7α-dehydroxylating strain (20). The organization and sequence of bai genes is highly conserved between C. hylemonae TN271 and C. scindens VPI 12708 (Figure 3; Table 2). The only exception is the baiA gene, present in the polycistronic operon from C. scindens VPI 12708 (23) and C. hiranonis TO931 (24), but absent from the polycistronic operon in C. hylemonae (Figure 3). We located the baiA gene using redundant primers designed from sequence data from baiA genes in C. scindens VPI 12708 (11, 23) and C. hiranonis TO931 (24). The gene was located as a monocistron with a conserved upstream promoter (Figure 4). Additional monocistronic copies of the baiA gene with conserved bai promoter sequences have been located in C. scindens VPI 12708 (25).
5′ RACE PCR analysis of the baiBCDEFGHI and baiA operons, combined with comparison between previously identified bai promoter regions from two other 7α-dehydroxylating clostridia have allowed identification of several conserved residues upstream of the TIS in bai genes (23, 24). These putative promoter regions are composed of the consensus sequence ATxTxxtaxcxxxxxxAAxTGTTAAxxTtaTATCAA. This consensus sequence may allow identification of other genes regulated by bile acids in closely related clostridia whose genomes have been sequenced in part or in whole-such as C. scindens 35704 (Refseq ID number NZ_ABFY00000000), Clostridium hylemonae DSM 15053 (Refseq ID number NZ_ABYI00000000), and Clostridium hiranonis DSM 13275 (NZ_ABWP00000000).
Previous work identified primary 5β-bile acids as inducers of 7α-dehydroxylating activity. However, in the present study, we have shown that a primary 5α-bile acid, ACA, induces bile acid 7α-dehydroxylating activity (Figure 5). To our knowledge, this is the first report of a primary allo-bile acid inducing bile acid 7α-dehydroxylating genes. This observation is important for two reasons. First, this data provides further information about the regions of the bile acid that are recognized by proteins involved in regulating expression of the genes involved in the oxidative branch of the bile acid 7α-dehydroxylating pathway. Previous work has demonstrated that side-chain conjugation blocks 7α-dehydroxylation (27, 28), and 7α-hydroxy bile acids are required for induction of the baiBCDEAFGHI operon from C. scindens VPI 12708 (29). The presence of a 12-hydroxyl-present in cholic acid, absent in chenodeoxycholic acid-increases 7α-dehydroxylation activity 5-fold relative to the saturated C-12 (27). The current data suggests that substrates with either the cis (5β-bile acids) or trans (5α-bile acids) orientations about the A/B ring junction are able to induce baiBCDEAFGHI and baiBCDEFGHI operons from C. scindens VPI 12708 and C. hylemonae TN271. Secondly, this data suggests that the Δ4-3-oxo-5β-steroid reductase and Δ4-3-oxo-5α-steroid reductase are differentially regulated. Induction with ACA results in conversion of [24-14C] CA to [24-14C] DCA, as well as [24-14C] alloDCA in appreciable amounts. This suggests that ACA is inducing the expression of a 5α-Δ4-bile acid oxidoreductase leading to the production of alloDCA. Previous work with CA induced cell extracts of C. scindens VPI 12708 showed that [24-14C] alloDCA was produced from [24-14C] CA; however, production of this metabolite is sporadic and the conditions favoring production of alloDCA have not been determined when CA is the inducer. Induction with alloCA results in repeatable and consistent production of alloDCA. Identification of alloCA as an inducer of 3-oxo-4-5α-reductase activity may allow isolation of the gene encoding this activity.
In addition, we tested the hypothesis that a 3-oxo-4-steroid structure- in the form of testosterone- would act as a gratuitous inducer of the reductive genes involved in reducing 3-oxo-4,6-DCA → 3-oxo-4-DCA → 3-oxo-DCA → DCA (Figure 7). Testosterone by itself is not sufficient to induce bile acid 7α-dehydroxylating activity, as [24-14C] CA activity assays have shown (Figure 6). However, when added in combination with CA the activity increases significantly in both C. scindens VPI 12708 and C. hylemonae TN271 both in the rate of conversion over time and the equilibrium is also shifted to the right (Figure 6). In the presence of testosterone, C. hylemonae converted 50% more CA to DCA and C. scindens VPI 12708 converted 25% more CA to DCA compared to CA induction alone after 48 hrs. Taken together, this data suggests that testosterone mimics an intermediate in the bile acid 7α-dehydroxylating pathway, probably 3-oxo-4-DCA, which induces genes involved in the reductive arm of the pathway. Reduction of the A-ring of steroids by human intestinal flora has been well documented and appears to be carried out mainly by members of the genus Clostridium under anaerobic conditions (30). Reduction of the A-ring in steroid hormones under anaerobic conditions is a two step process beginning with reduction of the Δ4 bond to either the 5α- or 5β configuration by stereospecific 5α- or 5β-steroid reductases, followed by reduction of the 3-oxo-group to either the 3α- or 3β-hydroxy orientation by stereospecific 3α- or 3β-hydroxysteroid dehydrogenases, respectively (31, 32). Induction of a Δ4- 3-oxo-5β-bile acid reductase and potentially other gene products would be expected to increase the rate of conversion of CA to DCA and also to “pull” the equilibrium toward secondary bile acid production. The fact that induction with testosterone alone was not sufficient to yield [24-14C] DCA in [24-14C] CA activity assays suggests that the baiBCDE(A)FGHI operon is not induced by testosterone simply because the rate limiting step (7α-dehydration) is catalyzed by the baiE gene in this operon and would have yielded some secondary bile acids (figure 6). These data suggest that testosterone induction may allow isolation of inducible polypeptides involved in the reductive arm of the bile acid 7α-dehydroxylating pathway resulting in DCA.
Figure 7.
Comparison of structure between testosterone and 3-oxo-4-DCA. Note the 3-oxo-4 conjugated double bond common to both molecules.
Toward the question of what makes C. hylemonae TN271 a “low activity” 7α-dehydroxylating bacterium, there are a few possibilities. First, the distance between the TIS and the start codon is shorter between C. hylemonae TN271 and the “high activity” strains C. scindens VPI 12708 and C. hiranonis sp. TO931. Second, there may be some residue changes in the C. hylemonae TN271 putative promoter region which is responsible for the reduced activity seen in vitro. However, the testosterone data suggests an additional scenario. Testosterone appears to be acting as a gratuitous inducer acting on genes involved in reducing the 3-oxo-4,6-cholenoic structure to secondary bile acids. Perhaps regulation of genes involved in the reductive arm of the pathway is responsible for controlling the relative activity between the “high activity” and “low activity” strains. Identification of novel genes involved in the reductive arm of the bile acid 7α-dehydroxylating pathway and their regulation will likely be important in our understanding of the differences in relative CA 7α-dehydroxylating activity observed in static culture.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human gastrointestinal tract. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Nutr. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 3.Ridlon JM, Kang D, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Bayerdörffer E, Mannes GA, Richter WO, Ochsenkühn T, Wiebecke B, Kopcke W, Paumgartner G. Increased serum deoxycholic acid levels in men with colorectal adenomas. Gastroenterology. 1993;104:145–151. doi: 10.1016/0016-5085(93)90846-5. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005 Jan;589(1):47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Berr F, Kullak-Ublick GA, Paumgartner G, Munzig W, Hylemon PB. 7 alpha-dehydroxylating bacteria enhance deoxycholic acid input and cholesterol saturation of bile in patients with gallstones. Gastroenterology. 1996;111:1611–1620. doi: 10.1016/s0016-5085(96)70024-0. [DOI] [PubMed] [Google Scholar]
- 7.Pai R, Tarnawski AS, Tran T. Deoxycholic Acid Activates β-Catenin Signaling Pathway and Increases Colon Cell Cancer Growth and Invasiveness. Mol Biol Cell. 2004;15(5):2156–2163. doi: 10.1091/mbc.E03-12-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25(7):1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallonee DH, Hylemon PB. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J Bacteriol. 1996;178:7053–7058. doi: 10.1128/jb.178.24.7053-7058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallonee DH, Adams JL, Hylemon PB. The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A ligase. J Bacteriol. 1992;174:2065–2071. doi: 10.1128/jb.174.7.2065-2071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mallonee DH, Lijewski MA, Hylemon PB. Expression in Escherichia coli and characterization of a bile acid-inducible 3α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Curr Microbiol. 1995;30:259–263. doi: 10.1007/BF00295498. [DOI] [PubMed] [Google Scholar]
- 12.Kang D, Ridlon JM, Moore DR, Barnes S, Hylemon PB. Clostridium scindens baiCD and baiH genes encode stereo-specific 7α/7β-hydroxy-3-oxo-Δ4-cholenoic acid oxidoreductases. Biochim Biophys Act. 2008;1781:16–25. doi: 10.1016/j.bbalip.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye HQ, Mallonee DH, Wells JE, Björkhem I, Hylemon PB. The bile acid-inducible baiF gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A hydrolase. J Lipid Res. 1999;40:17–23. [PubMed] [Google Scholar]
- 14.Stellwag EJ, Hylemon PB. Characterization of 7α-dehydroxylase in Clostridium leptum. Am J Clin Nutr. 1978;31 (Suppl 10):243–247. doi: 10.1093/ajcn/31.10.S243. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari A, Scolastino C, Baretta L. Activity on bile salts of a Clostridium bifermentans cell-free extract. FEBS Lett. 1977;75:166–168. doi: 10.1016/0014-5793(77)80076-8. [DOI] [PubMed] [Google Scholar]
- 16.Wells JE, Berr F, Thomas LA, Dowling RH, Hylemon PB. Isolation and characterization of cholic acid 7adehydroxylating fecal bacteria from cholesterol gallstone patients. J Hepatol. 2000;32:4–10. doi: 10.1016/s0168-8278(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 17.Narushima S, Itoh K, Miyamoto Y, Park S, Nagata K, Kuruma K, Uchida K. Deoxycholic acid formation in gnotobiotic mice associated with human intestinal bacteria. Lipids. 2006;41(9):835–843. doi: 10.1007/s11745-006-5038-1. [DOI] [PubMed] [Google Scholar]
- 18.Ridlon JM, McGarr SE, Hylemon PB. Development of methods for the detection and quantification of 7alpha-dehydroxylating clostridia, Desulfovibrio vulgaris, Methanobrevibacter smithii, and Lactobacillus plantarum in human feces. Clin Chim Acta. 2005;357(1):55–64. doi: 10.1016/j.cccn.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Notredame C, Higgins D, Heringa J. T-Coffee: a novel method for multiple sequence alignments. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 20.Doerner KC, Takamine F, LaVoie CP, Mallonee DH, Hylemon PB. Assessment of fecal bacteria with bile acid 7α-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol. 1997;63(3):1185–1188. doi: 10.1128/aem.63.3.1185-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wells JE, Williams KB, Whitehead TR, Heuman DM, Hylemon PB. Development and application of a polymerase chain reaction assay for the detection and enumeration of bile acid 7α-dehydroxylating bacteria in human feces. Clin Chim Act. 2003;331:127–134. doi: 10.1016/s0009-8981(03)00115-3. [DOI] [PubMed] [Google Scholar]
- 22.Dawson JA, Mallonee DH, Björkhem I, Hylemon PB. Expression and characterization of a C24 bile acid 7α-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J Lipid Res. 1996;37:1258–1267. [PubMed] [Google Scholar]
- 23.Mallonee DH, White WB, Hylemon PB. Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain VPI 12708. J Bacteriol. 1990;172:7011–7019. doi: 10.1128/jb.172.12.7011-7019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells JE, Hylemon PB. Identification and characterization of a bile acid 7α-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl Environ Microbiol. 2000;66(3):1107–1113. doi: 10.1128/aem.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopal-Srivastava R, Mallonee DH, White WB, Hylemon PB. Multiple copies of a bile acid-inducible gene in Eubacterium sp. strain VPI 12708. J Bacteriol. 1990;172(8):4420–4426. doi: 10.1128/jb.172.8.4420-4426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitahara M, Takamine F, Imamura T, Benno Y. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7α-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov. isolated from human faeces. Int J Syst Evol Microbiol. 2000;50:971–978. doi: 10.1099/00207713-50-3-971. [DOI] [PubMed] [Google Scholar]
- 27.White BA, Lipsky RL, Fricke RJ, Hylemon PB. Bile acid induction specificity of 7α-dehydroxylase activity in an intestinal Eubacterium species. Steroids. 1980;35(1):103–109. doi: 10.1016/0039-128x(80)90115-4. [DOI] [PubMed] [Google Scholar]
- 28.Batta AK, Salen G, Arora R, Shefer S, Batta M, Person A. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J Biol Chem. 1990;265(19):10925–10928. [PubMed] [Google Scholar]
- 29.White BA, Fricke RJ, Hylemon PB. 7β-dehydroxylation of ursodeoxycholic acid by whole cells and cell extracts of the intestinal anaerobic bacterium, Eubacterium species VPI 12708. J Lipid Res. 1982;23:145–153. [PubMed] [Google Scholar]
- 30.Glass TL, Wheeler LA, Sutter VL, Finegold SM. Transformation of 4-androsten-3,17-dione by growing cultures and cell extracts of Clostridium paraputrifium. Biochim Biophys Act. 1979;573:332–342. doi: 10.1016/0005-2760(79)90066-3. [DOI] [PubMed] [Google Scholar]
- 31.Bokkenheuser VD, Winter J, Dehazya P, Deleon O, Kelley WG. Formation and metabolism of tetrahydrodeoxycorticosterone by human fecal flora. J Steroid Biochem. 1976;7:837–843. doi: 10.1016/0022-4731(76)90187-4. [DOI] [PubMed] [Google Scholar]
- 32.Baron SF, Hylemon PB. Biotransformation of bile acids, cholesterol, and steroid hormones. In: Mackie RI, White BA, editors. Gastrointestinal Microbiology. New York: Chapman & Hall; 1997. pp. 470–510. [Google Scholar]
- 33.Franklund CV, Baron SF, Hylemon PB. Characterization of the baiH gene encoding a bile acid-inducible NADH:flavin oxidoreductase from Eubacterium sp. strain VPI 12708. J Bacteriol. 1993;175(10):3002–3012. doi: 10.1128/jb.175.10.3002-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]