Abstract
The combination of DNA alkylating agents busulfan (Bu) and cyclophosphamide is the most commonly used myeloablative pre-transplant conditioning therapy for myeloid leukemias. However, it is associated with significant non-relapse mortality, which prohibits dose-escalation to control relapse. We hypothesized that combining these two drugs with an epigenetic modifier would increase anti-leukemic efficacy without jeopardizing patient safety. A pre-clinical study was performed to determine the synergistic cytotoxicity of Bu, 4-hydroperoxycyclophosphamide (4HC) and the hypomethylating agent decitabine (DAC) in human AML cell lines. Exposure of KBM3/Bu2506 (P53-null) and OCI-AML3 (P53-wild-type) cells to [Bu+4HC] inhibited cell proliferation by ~35-39%; addition of DAC increased the inhibition to ~60-62%. The observed synergistic interactions correlated with DNA-damage response activation, increased production of reactive oxygen species, decreased mitochondrial membrane potential, release of mitochondrial proapoptotic proteins into the cytoplasm, and induction of caspase-dependent programmed cell death. The [Bu+4HC+DAC] combination further caused chromatin trapping of DNMT1 with a concomitant increase in DNA damage. In contrast, FLT3-ITD-positive AML cell lines were not sensitized to [Bu+4HC] by inclusion of DAC; addition of the FLT3 kinase inhibitor sorafenib (Sor) sensitized the FLT3-ITD-positive MV4-11 and MOLM13 cell lines to the triple drug combination by inhibiting the FLT3 signal transduction pathway. Our results therefore provide a rationale for the development of personalized conditioning therapy for patients with P53-mutated and FLT3-ITD-positive AML.
Keywords: busulfan, cyclophosphamide, decitabine, acute myeloid leukemia, pre-transplant regimen
INTRODUCTION
Busulfan (Bu) is a bifunctional DNA alkylating agent commonly used in combination with other agents for high-dose pre-transplant conditioning therapy for hematopoietic stem cell transplantation (HSCT) [1]. Its combination with cyclophosphamide (Cy) has been used as an alternative to a Cy-total body irradiation (TBI)-containing myeloablative regimen and found to be effective for acute leukemias [2–7]. The “BuCy2” preparative regimen is more effective than Cy+TBI in patients ≤ 40 years old [6,8]. However, the “BuCy2” regimen is commonly associated with high non-relapse mortality [9]. To reduce its high toxicity, possibly due to adverse interactions of Cy metabolites, Cy has been replaced with nucleoside analog(s) such as fludarabine (Flu) and/or clofarabine (Clo) and found to be a safe and effective preparative regimen for HSCT [10–14]. Open-label, multicenter, randomized studies in acute myeloid leukemia (AML) patients comparing Bu+Cy with Bu+Flu showed lower transplant-related mortality in the Bu+Flu group but similar results in 5-year leukemia-free survival [15,16].
We hypothesized that the combination of Bu+Cy with a DNA hypomethylating nucleoside analog such as decitabine (DAC) would increase the antileukemic efficacy without jeopardizing patient safety. The ability of DAC to induce leukemic differentiation and re-expression of epigenetically silenced putative tumor suppressor genes [17] and its synergistic cytotoxicity with Bu in AML cell lines [18] suggest that the Bu+Cy+DAC combination could be a more efficacious regimen. Moreover, DAC has been used for epigenetic priming prior to induction therapy in AML patients [19, 20].
The cytotoxicity of Cy is attributed to its active metabolites. Upon absorption, Cy is converted to its main metabolite 4-hydroxycyclophosphamide (HCy) by the cytochrome P450 system in the liver [21, 22]. 4-Hydroxycyclophosphamide exists in equilibrium with aldophosphamide, which is converted to phosphoramide mustard and acrolein [23]. Since Cy is a stable pro-drug requiring activation, 4-hydroperoxycyclophosphamide (4HC) is commonly used to determine its synergism with other chemotherapeutic drugs in in vitro studies; 4HC is converted to HCy, which is further converted to active metabolites. In this regard, we performed a pharmacological study to determine the anti-leukemic synergism of Bu, 4HC and DAC in established AML cell lines. Strong synergistic interactions were observed regardless of P53 status. AML cells positive for FMS-like tyrosine kinase 3 internal tandem duplications (FLT3-ITD) were found to be less sensitive to [Bu+4HC+DAC] but were sensitized when sorafenib (Sor) was added to the combination. The results from this study provide a rationale for the development of personalized anti-leukemic therapy specifically as a pre-transplant conditioning regimen for patients undergoing HSCT for P53-mutated or FLT3-ITD-positive AML.
MATERIALS AND METHODS
Cell lines and drugs
KBM3/Bu2506 is an AML cell line established from one of our patients and made resistant to Bu as described previously [24]. The OCI-AML3, THP1 and MOLM13 AML cell lines were kindly provided by Dr. Michael Andreeff’s laboratory (University of Texas MD Anderson Cancer Center, Houston, TX). The OCI-AML3/shP53 cell line [25] was obtained from Dr. Paul Corn (University of Texas MD Anderson Cancer Center, Houston, TX). The MV4-11 AML cell line was obtained from the American Type Culture Collection (Manassas, VA). Cells were grown in RPMI-1640 medium (Mediatech, Manassas, VA) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Inc., Flowery Branch, GA) and 100 IU/mL penicillin and 100 μg/mL streptomycin (Mediatech) at 37°C in a humidified atmosphere of 5% CO2 in air. Busulfan was obtained from Sigma-Aldrich (St. Louis, MO), and DAC (10 mM solution in dimethyl sulfoxide (DMSO)) and Sor were purchased from Selleck Chemicals LLC (Houston, TX). 4-Hydroperoxycylophosphamide was a generous gift from Dr. Scott Rowley (Hackensack University Medical Center, Hackensack, NJ). Busulfan and 4HC were dissolved in DMSO immediately prior to each experiment.
Cytotoxicity and apoptosis assays
Cells (6 ml of 0.5 × 106 cells/ml) in T25 flasks were exposed to drugs, alone or in combination, for 48 hrs, aliquoted (100 μl) into 96-well plates and analyzed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay [26]. Briefly, 50 μl of 2 mg/ml MTT reagent (Sigma-Aldrich) in phosphate-buffered saline (PBS) was added per well and incubated for 4 hours at 37°C. The solid reaction product was dissolved by adding 100 μl of solubilization solution (0.1 N HCl in isopropanol containing 10% Triton X-100) to each well, mixing, and incubating at 37°C overnight. Absorbance at 570 nm was measured using a Victor X3 (Perkin Elmer Life and Analytical Sciences, Shelton, CT) plate reader. The number of metabolically-active (MTT-positive) cells was determined relative to the control cells exposed to solvent alone.
Apoptosis was determined by flow-cytometric measurements of phosphatidylserine externalization [27] with Annexin-V-FLUOS (Roche Diagnostics, Indianapolis, IN) and 7-aminoactinomycin D (BD Biosciences, San Jose, CA) using a Muse Cell Analyzer (EMD Millipore, Billerica, MA). Drug combination effects were estimated based on the combination index (CI) values [28] calculated using the CalcuSyn software (Biosoft, Ferguson, MO).
Western blot analysis
Cells exposed to solvent or drug(s) were collected by centrifugation, washed with cold PBS, and lysed with cell lysis buffer (Cell Signaling Technology, Danvers, MA). The protein concentrations were determined using a BCA Protein Assay kit (ThermoFisher Scientific, Rockford, IL). Proteins were resolved on polyacrylamide-SDS gels and blotted onto nitrocellulose membranes (Bio-Rad, Hercules, CA). Western blot analyses were done by chemiluminescence using the Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore). The sources of the antibodies and their optimum dilutions are provided in Supplementary Table 1.
Real-time polymerase chain reaction
Real-time polymerase chain reaction (PCR) was performed to determine the extent of DNA demethylation of the P16/INK4A gene promoter and its level of expression as we previously described [29].
Flow cytometric analysis of H2AX phosphorylation
Cells were collected after 48-hr drug exposure and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature with occasional mixing. Cells were resuspended in 0.3 ml PBS, 0.7 ml ethanol was added and the suspension kept at least overnight at −20°C. Cells were then pelleted and washed with PBS containing 3% goat serum. After centrifugation, cells were resuspended in 0.5 ml PBS with 5% goat serum and incubated at room temperature for 1 hr with occasional mixing. Cells were again centrifuged and resuspended in 100 μl PBS with 3% goat serum containing 1:500 diluted anti-γ-H2AX antibody (mouse) and gently mixed for 2 hrs at room temperature. Cells were washed with PBS/3% goat serum and resuspended in 200 μl PBS/3% goat serum containing 1:200 diluted Alexa Fluor 488-conjugated anti-mouse IgG antibody and mixed for 1 hr in the dark. Cells were washed twice with PBS/3% goat serum and kept in the same wash buffer overnight at 4°C. Cells were centrifuged and resuspended in 0.5 ml PBS/3% goat serum containing 15 μg/ml propidium iodide (PI) and 2.5 μg/ml RNAse A for 30 min at 4°C prior to flow cytometry using 488 nm excitation and 518 nm emission for Alexa Fluor 488 and 625 nm for PI.
Preparation of cytoplasmic and mitochondrial extracts
Cells were exposed to solvent or drugs for 48 hrs, collected and washed with ice-cold PBS. The cytoplasmic extract was a by-product supernatant from the mitochondrial isolation using the Mitochondria Isolation Kit for Cultured Cells - Reagent-based method (ThermoFisher Scientific). Mitochondria were resuspended in cell lysis buffer and boiled with loading buffer and dithiothreitol (DTT) (Cell Signaling Technology). Cytoplasmic extracts were boiled with the same loading buffer and DTT. Denatured protein samples were analyzed by Western blotting. β-ACTIN and COX IV proteins were used as loading controls for cytoplasmic and mitochondrial extracts, respectively.
Analysis of production of reactive oxygen species (ROS) and changes in mitochondrial membrane potential (MMP)
Cells exposed to drug(s) for 48 hrs were analyzed for production of ROS using CM-H2DCFDA (5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester), an ROS indicator that diffuses into cells where it is oxidized to a fluorescent product (Life Technologies, Grand Island, NY). Briefly, cells were aliquoted (0.5 ml) into 5-ml tubes and 2 μl of 1.5 mM CM-H2DCFDA (freshly dissolved in DMSO) was added. Cells were incubated at 37°C for 1 hr and immediately analyzed with a Gallios Flow Cytometer (Beckman Coulter, Inc., Brea, CA) using excitation/emission wavelengths of 492/520 nm. Geometric means of the fluorescence intensities were used in the analysis and fold increase relative to the control was reported.
Changes in the MMP were measured using a JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbo cyanine iodide) Mitochondrial Membrane Potential Detection kit (Cayman Chemical, Ann Arbor, MI). Cells were exposed to drugs for 48 hrs and 0.5 ml of cell suspension was aliquoted into 5-ml tubes. Diluted (1:10 with cell growth medium, 40 μl) MMP-sensitive fluorescent dye JC-1 reagent was added to each tube, incubated at 37°C for 20 min, and immediately analyzed by flow cytometry (λex = 488 nm) using the 530-nm (FL-1 channel, green) and 585-nm (FL-2 channel, red) band-pass filters simultaneously. Healthy cells with functional mitochondria and high MMP exhibit red fluorescence (aggregated JC-1), whereas cells with disrupted mitochondria and low MMP show green fluorescence (monomeric JC-1). The ratio of monomer/aggregate JC-1 was calculated.
Isolation of soluble and chromatin-bound nuclear extracts
Cells were exposed to drugs for 24 hrs, collected by centrifugation, and washed with cold PBS. Soluble and chromatin-bound nuclear extracts were prepared using the Subcellular Protein Fractionation Kit for Cultured Cells (Pierce Biotechnology, Rockford, IL). Protein concentrations were determined using the BCA protein assay kit and Western blot analysis was performed as described above. Changes in the level of chromatin-bound DNMT1 were calculated as described previously [30].
Analysis of FLT3-ITD mutations by PCR
The presence of FLT3-ITD mutations was detected as described [31]. Briefly, genomic DNA from AML cells was isolated using the Wizard Genomic DNA Purification kit (Promega, Madison, WI). DNA amplification by PCR was performed using primers specific for FLT3 exons 14 and 15 (forward primer: 5’-GCAATTTAGGTATGAAAGCCAGC-3’ and reverse primer: 5′-TTTCAGCATTTTGACGGCAACC-3′), where duplications usually occur [32]. DNA amplification was performed by mixing 1 μg genomic DNA, 10 pM of each primer, 10 μM dNTPs, and 1.25 units Taq DNA polymerase in a 50-μ1 reaction mixture. Denaturation, annealing, and extension steps were done at 94°C for 30 sec, 60°C for 30 sec, and 72°C for 40 sec, respectively, for 30 cycles. An initial 2-min denaturation step at 94°C and a final extension step at 72°C for 10 min were included in the protocol. PCR products were resolved on a 2% agarose gel and analyzed after staining with ethidium bromide. Samples that showed PCR products longer than 330 bp were considered positive for FLT3-ITD mutations.
Statistical analysis
Results are presented as the mean ± standard deviation of at least three independent experiments and statistical significance of the difference between two groups was determined by paired, two-tailed, non-parametric Wilcoxon test using Graph Pad Prism 7.03. P values ≤ 0.05 were considered statistically significant.
RESULTS
Combination of Bu, 4HC and DAC provides synergistic cytotoxicity towards acute myeloid leukemia cell lines
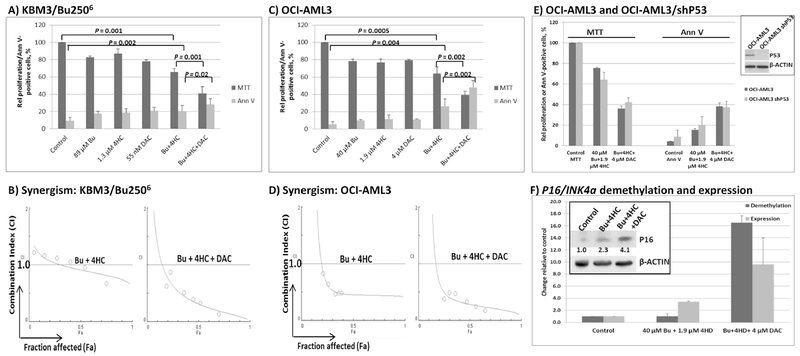
Exposure of KBM3/Bu2506 and OCI-AML3 cell lines to concentrations close to the IC20 values (the concentration of drug required for 20% growth inhibition) of Bu, 4HC and DAC inhibited cell proliferation by ~20%, as expected, when administered individually, as measured by the MTT assay (Figure 1 A, C). Using the same drug concentrations, the two-drug combination [Bu+4HC] decreased KBM3/Bu2506 and OCI-AML3 proliferation to ~65% and ~61%, respectively, versus untreated control cultures (Figure 1A and C). Addition of DAC to this two-drug combination significantly decreased proliferation of KBM3/Bu2506 and OCI-AML3 cultures to ~40% and ~38% of control, respectively (P = 0.001 and 0.002). Analysis of activation of apoptosis by Annexin V (Ann V) assay showed similar results. Combination of 89 μM Bu and 1.3 μM 4HC resulted in ~20% Ann V-positive cells (P = 0.002, compared with control) in KBM3/Bu2506 cells; addition of 55 nM DAC resulted in ~29% positivity (P = 0.02, when the 2- and 3-drug combinations were compared). Even more significant differences were observed in OCI-AML3 cells. Exposure to [40 μM Bu + 1.9 μM 4HC] resulted in ~25% Ann V-positive cells (P = 0.004) and addition of 4 μM DAC further increased this effect to ~47% positivity (P = 0.002, Figure 1C).
Figure 1.
Synergistic anti-proliferative and cytotoxic effects of the various drug combinations in KBM3/Bu2506 (A) and OCI-AML3 (C) cell lines. Cells were exposed to drugs, alone or in combination, for 48 hrs then analyzed for cell proliferation by MTT assay and for apoptosis by Annexin V (Ann V) assay. The relationships between combination index (CI; y-axis) and fraction affected (Fa; x-axis) in each cell line are shown in panels B and D (representatives of two independent experiments). CI < 1 indicates synergism. (E) Comparison of the effects of the indicated drug combinations on OCI-AML3 (P53-wild type) and OCI-AML3/shP53 clone with downregulated P53 expression (inset shows Western blot for P53 protein level). (F) DNA demethylation of P16/INK4A gene promoter and its expression. OCI-AML3 cells were exposed to solvent (Control) or the indicated drug combinations for 48 hrs. Genomic DNA and total RNA were isolated and used for real-time PCR. The inset shows Western blot for the level of P16/INK4A protein. Signals were scanned and normalized relative to β-ACTIN. The numbers refer to fold difference in P16/INK4A relative to Control. Results (bar graphs) are the average ± SD of at least three independent experiments. Statistically significant differences are indicated by P values. Bu, busulfan; 4HC, 4-hydroperoxycyclophosphamide; DAC, decitabine.
To test for possible synergistic interactions, KBM3/Bu2506 and OCI-AML3 cells were exposed to different concentrations of individual drugs, or to the drug combinations at a constant concentration ratio, and the MTT assay was performed after 48 hrs. The combination index (CI) values at increasing drug effects were graphically analyzed according to the Chou-Talalay method [28] as shown in Figure 1B and D. The calculated CI values less than 1 suggest significant synergism in these cell lines. The combination of Bu and 4HC in KBM3/Bu2506 cells showed a modest synergistic effect at higher fraction affected (Fa) and addition of DAC significantly increased the synergistic interactions as suggested by a further decrease in the CI values (Figure 1B). The synergism of Bu and 4HC was more pronounced in OCI-AML3 cells and addition of DAC further increased the synergism (Figure 1D).
Since KBM3/Bu2506 is P53-null, we sought to further determine if the effects of [Bu+4HC+DAC] were P53-independent. OCI-AML3 parental cell line and its clone with down-regulated P53 (OCI-AML3/shP53) were exposed to the same drug concentrations and analyzed by MTT and Annexin V assays. Figure 1E shows similar effects of the indicated drug combinations on these two cell lines, suggesting lack of P53-dependency.
Addition of DAC significantly enhanced the inhibition of proliferation mediated by [Bu+4HC] in KBM3/Bu2506 and OCI-AML3 cells (Figure 1A and C). To determine if DAC exposure resulted in DNA hypomethylation, genomic DNA was isolated from OCI-AML3 cells exposed to solvent control, [Bu+4HC] or [Bu+4HC+DAC]. Unmethylated, but not methylated, cytosines were converted to uracil by bisulfite modification. Since the expression of cyclin-dependent kinase inhibitor 4A (P16/INK4A) depends on the demethylation of its gene promoter [33], we analyzed its methylation status by methyl-specific real-time PCR. Figure 1F shows ~16-fold increase in the level of unmethylated P16/INK4A promoter when DAC was added to [Bu+4HC]. The level of P16/INK4A mRNA and expressed protein concomitantly increased by ~3- and ~2-fold, respectively (Figure 1F). These results suggest that the efficacy of [Bu+4HC+DAC] was partly due to the hypomethylating activity of DAC and the up regulation of expression of P16/INK4A, a known tumor suppressor gene.
The three-drug combination strongly activates the DNA-damage response
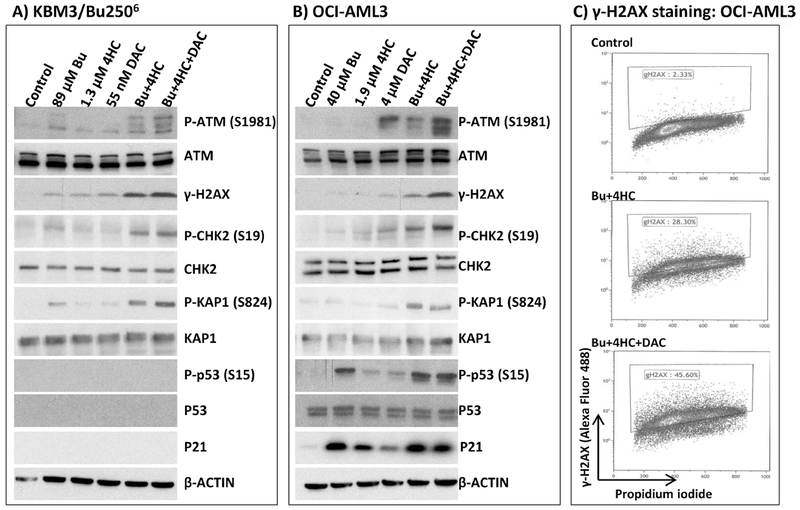
To determine possible mechanisms of the observed synergistic cytotoxicity, we examined changes in the level of biomarkers of the DNA-damage response (DDR). One of the earliest events in DDR is the activation of the ATM kinase by self-phosphorylation [34]; analysis by Western blotting shows a marked increase in the level of P-ATM (S1981) when KBM3/Bu2506 and OCI-AML3 cells were exposed to [Bu+4HC] that was even greater with the 3-drug combination (Figure 2A, 2B). Increased phosphorylation of known ATM substrates including histone 2AX (H2AX), CHK2 and KAP1 was observed in the 2-drug combination, and again this effect was further enhanced in cells exposed to [Bu+4HC+DAC] (Figure 2A, 2B).
Figure 2.
Effects of drug combinations on biomarkers of the DNA-damage response. Cells were exposed to the indicated drugs for 48 hrs and analyzed by Western blotting for changes in protein levels and status of their phosphorylation (A and B). Similar cells were double-stained with anti-γ-H2AX antibody-Alexa Fluor 488 conjugate and propidium iodide and analyzed by flow cytometry (C). The inside windows in panel C show the extent of H2AX phosphorylation. Results are representatives of two independent experiments. Bu, busulfan; 4HC, 4-hydroperoxycyclophosphamide; DAC, decitabine.
KBM3/Bu2506 cells are P53-null, as confirmed by Western blotting, and expression of P21 was also not observed (Figure 2A). The drug-dependent phosphorylation of P53 correlates with the induction of P21 in OCI-AML3 (P53-wild-type) cells (Figure 2B). The increased level of γ-H2AX in OCI-AML3 cells exposed to 2-drug and 3-drug combinations versus solvent-treated control cells was further indicated by flow cytometric analysis. Exposure to [Bu+4HC] or [Bu+4HC+DAC] increased the phosphorylation of H2AX from ~2% to ~28% or ~46%, respectively, relative to the control cells (Figure 2C), consistent with the Western blotting data (Figure 2B). Overall, these results suggest that [Bu+4HC+DAC] combination activates DDR in a P53-independent manner in AML cell lines, consistent with the results shown in Figure 1.
The three-drug combination activates the apoptosis pathway
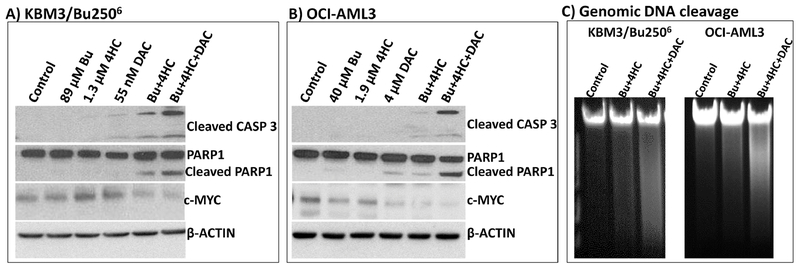
Whether the observed DDR activation would lead to cell death was assessed by analyzing the cleavage of caspase 3 and PARP1. Significant protein cleavage was observed in KBM3/Bu2506 and OCI-AML3 cells exposed to [Bu+4HC+DAC] (Figure 3A, 3B), suggesting activation of apoptosis consistent with the observed increase in Annexin V-positive cells (Figure 1). This activation of programmed cell death correlates with the decrease in the level of pro-survival c-MYC (Figures 3A, 3B). Since apoptosis is known to be accompanied by genomic DNA cleavage/degradation [35], we isolated the DNA from drug-treated cells and analyzed it by agarose gel electrophoresis. Ethidium bromide staining showed extensive cleavage of genomic DNA in cells exposed to [Bu+4HC+DAC] (Figure 3C). These results suggest that the synergistic cytotoxicity of [Bu+4HC+DAC] in AML cell lines is due to activation of apoptosis.
Figure 3.
Effect of drug combinations on biomarkers of apoptosis. Cells were exposed to the indicated drugs for 48 hrs and extracted proteins were analyzed by Western blotting (A and B). Genomic DNA was isolated from similar cells and analyzed by agarose gel electrophoresis with ethidium bromide staining (C). Results are representatives of two independent experiments. Bu, busulfan; 4HC, 4-hydroperoxycyclophosphamide; DAC, decitabine.
Possible mechanisms of apoptosis induction mediated by [Bu+4HC+DAC]
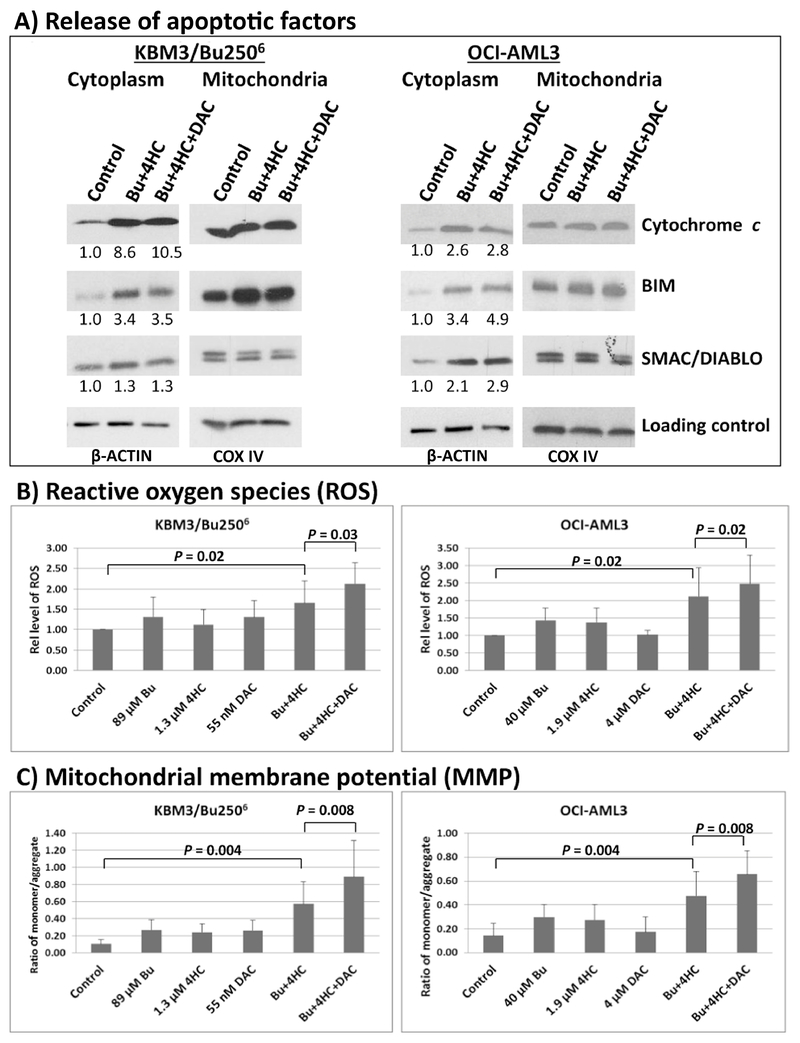
Induction of intrinsic apoptosis involves release of pro-apoptotic factors from mitochondria into the cytoplasm, which initiates a cascade of events leading to the activation of caspases and programmed cell death [36]. We, therefore, compared the levels of cytochrome c, BIM, and SMAC/DIABLO in cytoplasmic extracts from cells exposed to solvent control or drug combinations. A significant increase in the level of these pro-apoptotic proteins in the cytoplasm was observed in KBM3/Bu2506 and OCI-AML3 cells exposed to [Bu+4HC] or [Bu+4HC+ DAC] (Figure 4A). Quantitative analysis showed 2.6- to 10.5-fold increase in the level of cytochrome c, 3.4- to 4.9-fold increase in BIM, and a slight increase in SMAC/DIABLO in the cytoplasmic extracts from cells exposed to drugs (Figure 4A).
Figure 4.
Analysis of factors contributing to the observed drug-induced apoptosis. Cells were exposed to drug(s) for 48 hrs and cytoplasmic and mitochondrial protein extracts were analyzed by Western blotting (A). Western blot signals for cytoplasmic extracts were normalized relative to the β-ACTIN loading control and the calculated values were compared with untreated control cells (set to 1.0) as shown. Production of reactive oxygen species (B) and changes in mitochondrial membrane potential (C) were determined by flow cytometry. Results are representatives of two (A) or average ± SD of five (B, C) independent experiments. Statistically significant differences are indicated by P values. Bu, busulfan; 4HC, 4-hydroperoxycyclophosphamide; DAC, decitabine.
The observed release of pro-apoptotic proteins from the mitochondria might be due to an increase in the level of reactive oxygen species (ROS) and decrease in mitochondrial membrane potential (MMP) as reported previously [37,38]. We were thus prompted to determine changes in these parameters by flow cytometry. Figure 4B shows 2.1- and 2.0-fold increase in the level of ROS in KBM3/Bu2506 and OCI-AML3 cells exposed to [Bu+ 4HC+DAC], respectively, which are statistically significant (P = 0.02-0.03). The ratio of monomer: aggregate JC-1 increased by as much as 8-fold in triple drug-treated cells versus control, suggesting increased monomeric JC-1 in the cytoplasm relative to its aggregated form in the mitochondria and implied a decrease in MMP (Figure 4C). These results suggest that leakage of pro-apoptotic factors possibly due to increased level of ROS and decreased MMP, contribute to the [Bu+4HC+DAC]-mediated induction of apoptosis in AML cells.
Addition of DAC increases chromatin trapping of DNMT1
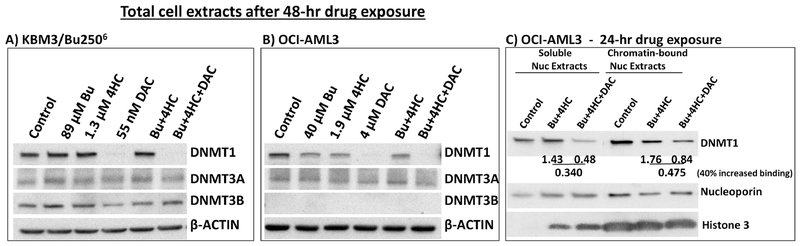
The hypomethylating activity of DAC is partly due to degradation of DNA methyl transferases [39]. We, therefore, examined the expression of DNMT1, DNMT3A and DNMT3B in cells exposed to DAC alone, or in combination with [Bu+4HC], for 48 hrs. Figures 5A and 5B show abrogation of DNMT1 in KBM3/Bu2506 cells exposed to 55 nM DAC and in OCI-AML3 cells exposed to 4 μM DAC. Addition of [Bu+4HC] did not inhibit the DAC-mediated degradation of DNMT1. DAC did not significantly affect the levels of DNMT3A or DNMT3B in KBM3/Bu2506 cells. Similarly the level of DNMT3A was not significantly affected by the drug(s) in OCI-AML3 cells and no expression of DNMT3B was observed in this cell line (Figure 5B).
Figure 5.
Effect of DAC on the level of DNMT1 and its chromatin trapping. Cells were exposed to drug(s) for 48 hrs (A and B) or 24 hrs (C) and analyzed by Western blotting for changes in the level of DNA methyltransferases DNMT1 and DNMT3A/3B (A and B) and chromatin-bound DNMT1 (C) as described under Materials and Methods. Results are representatives of two independent experiments. Bu, busulfan; 4HC, 4-hydroperoxycyclophosphamide; DAC, decitabine.
The cytotoxicity of DAC is mediated primarily by covalent trapping of DNMT1 to DNA [40]. Since we anticipated that DNMT1 trapping would be a relatively early event, we analyzed the level of DNMT1 in soluble and chromatin-bound nuclear extracts from cells exposed to [Bu+4HC±DAC] for 24 hrs (Figure 5c). DNMT1 signals were first normalized to nucleoporin; the ratio of these normalized values for [Bu+4HC+DAC] and [Bu+4HC] was then calculated. The calculated ratios for the soluble and chromatin-bound DNMT1 in OCI-AML3 cells were 0.34 and 0.475, respectively, showing an ~40% increase in DAC-mediated binding of DNMT1 to chromatin (Figure 5C). The quality of the chromatin preparations was confirmed by the presence of histone 3 in the chromatin-bound extracts and its absence in the soluble nuclear extract from control cells. The presence of histone 3 in the soluble nuclear extracts from drug-treated cells (Figure 5C) suggests its release from the chromatin, possibly due to changes in chromatin structure and DNA degradation (Figure 3C). Overall, these results suggest that addition of DAC to [Bu+4HC] leads to chromatin trapping and degradation of DNMT1, which may have implications for their synergism.
FLT3-ITD-positive AML cells are relatively resistant to [Bu+4HC+DAC] but they are sensitized with Sor
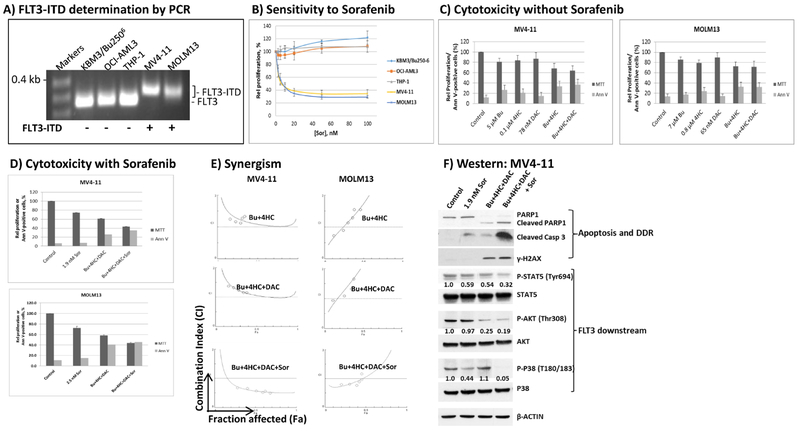
Figure 1 suggests that the sensitivity of AML cells to [Bu+4HC+DAC] is P53-independent. We next determined the sensitivity of two FLT3-ITD-positive AML cell lines to this drug combination since almost 30% of AML patients are FLT3-ITD-positive and have a poor prognosis [32,41,42]. The presence of FLT3-ITD mutations in MV4-11 and MOLM13 AML cells is suggested by their longer PCR products compared with the FLT3 wild type cell lines KBU/Bu2506, OCI-AML3 and THP1 (Figure 6A). These findings are consistent with the high resistance of KBU/Bu2506, OCI-AML3 and THP1 cells and profound sensitivity of MV4-11 and MOLM13 cells to Sor (Figure 6B). Exposure of MV4-11 and MOLM13 cells to [Bu+4HC+DAC] for 48 hrs inhibited cell proliferation by ~37% and ~30%, respectively, using individual drug concentrations equivalent to their IC20 values (Figure 6C). Addition of Sor to the triple-drug combination inhibited cell proliferation by ~60% in both cell lines with the levels of apoptosis, as indicated by Annexin-V staining, broadly tracking with the effects on proliferation (Figure 6D). Whereas synergism between Bu, 4HC and DAC was not observed in MV4-11 and M0LM13 cells, synergistic effects were obtained when Sor was added to the three-drug combination (Figure 6E). Western blot analysis showed increased cleavage of PARP1 and caspase 3 and phosphorylation of H2AX when FLT4-ITD-positive cells were exposed to [Bu+4HC+DAC+Sor] (Figure 6F). The observed sensitivity correlated with decreased phosphorylation of proteins downstream of the pro-survival FLT3 kinase signaling pathway [43], including STAT5, AKT and p38 (Figure 6F). Taken all together, these results strongly suggest that the resistance of FLT3-ITD-positive AML cells to [Bu+4HC+DAC] can be reversed by inclusion of Sor to inhibit the FLT3-ITD-activated tyrosine kinase and commit the cells to apoptosis.
Figure 6.
Effect of FLT3-ITD status on the response of AML cell lines to cytotoxic drug treatments. Genomic DNA was isolated from FLT3 wild type (KBM3/Bu2506, OCI-AML3 and THP1) and FLT3-ITD-positive AML cell lines (MV4-11 and MOLM13) and used as template for PCR analysis. PCR products were resolved on 2% agarose gel with ethidium bromide staining (A). The sensitivity of the five AML cell lines to Sor was compared by analyzing cell proliferation using MTT assay (B). The cytotoxicity of [Bu+4HC+DAC] in the absence (C) or presence (D) of Sor was determined in FLT3-ITD cells. Drug synergism (E) was analyzed as in Figure 1. Western blot analysis (F) shows increased cleavage of PARP1 and caspase 3 and phosphorylation of H2AX, and decreased levels of FLT3 downstream targets, when Sor was combined with [Bu+4HC+DAC]. The levels of phosphorylated proteins were first normalized to their non-phosphorylated forms, and the ratio of the normalized phosphoprotein level relative to the untreated control was calculated. All cells were exposed to drug(s) for 48 hrs (B-F). Results are representatives of two (A, E, F) or average±SD of three (B-D) independent experiments. Bu, busulfan; 4HC, 4-hydroperoxycyclophosphamide; DAC, decitabine; Sor, sorafenib.
DISCUSSION
We report in this study the synergistic cytotoxicity of a double alkylating agent combination (Bu and 4HC) with an epigenetic modifier (DAC). The efficacy of the triple-drug combination in AML cell lines is seen in both P53-wild type and –mutant cell lines but it is greatly decreased in cells with FLT3-ITD mutations. However, AML cell lines positive for FLT3-ITD are sensitized to [Bu+4HC+DAC] in the presence of Sor, where synergistic interactions are again observed. The efficacy of these drug combinations are broadly paralleled by the levels of DDR activation and apoptosis seen in the various exposure groups across different AML cell lines.
The cytotoxicity of the [Bu+4HC+DAC] combination might be mediated at the level of the initial DNA damage invoked by the two alkylating agents as suggested by the activation of the ATM kinase pathway, which is further enhanced by the addition of DAC (Figure 2), a DNA hypomethylating agent (Figure 1F) known to induce genome-wide DNA damage [44]. Moreover, the ability of DAC to trap DNMT1 to chromatin [40], as also suggested by our results with AML cell lines shown in Figure 5C, might prevent repair of the damaged DNA and exacerbate the generation of double-strand DNA breaks. In addition, DAC-mediated epigenetic changes (ie. hypomethylaton of DNA) may upregulate the expression of cell cycle-related/tumor suppressor proteins such as P16/INK4A (Figure 1F) and downregulate pro-survival proteins (Figure 3). The drug-mediated exacerbation of DNA damage and decreased expression of pro-survival proteins appears to transmit molecular signals to mitochondria and increase ROS production, decreasing the mitochondrial membrane potential and causing leakage of pro-apoptotic mitochondrial factors into the cytoplasm, initiating the cascade of events leading to activation of caspases resulting in apoptosis (Figures 3 and 4).
We previously suggested that chromatin remodeling is an important intermediary in the efficacy and synergism of nucleoside analog-alkylating agent combinations; in particular, nucleoside analogs likely induce changes in the chromatin structure that confer a greater susceptibility of the DNA to alkylating agents [45,46]. By extension, a similar mechanism might underlie the synergistic cytotoxicity observed in this study. Addition of the hypomethylating agent DAC might have sensitized AML cells to [Bu+4HC] in part by inducing chromatin remodeling and increasing DNA alkylation.
These mechanisms appear to be circumvented by the FLT3-mediated signal transduction pathway. Although a modest cell killing effect was observed in FLT3-ITD-positive cell lines MV4-11 and MOLM13 (Figure 6) exposed to [Bu+4HC+DAC], no synergistic interaction was obtained (Figure 6E). Inhibition of the FLT3 kinase with Sor, however, sensitized these cell lines resulting in synergistic cytotoxicity of the four drugs (Figure 6E). Such a mechanism is similar to our previous report on the synergistic cytotoxicity of Sor with clofarabine, fludarabine and Bu in FLT3-ITD-positive AML cells [32]. Our present and previous results provide a basis for designing personalized therapy in the treatment of FLT3-ITD-positive AML patients.
In conclusion, the present study suggests that the P53-independent synergistic cytotoxicity of [Bu+4HC+DAC] in AML cells might be due to the molecular interactions between DNA alkylation and epigenetic modification events that result in the trapping of DNMT1 to chromatin, induction of the DDR, ROS production, and decreased mitochondrial membrane potential, which ultimately commit tumor cells to apoptosis. Furthermore, inhibition of the activated FLT3 signal transduction pathway with Sor sensitizes FLT3-ITD-positive cells to [Bu+4HC+DAC], suggesting that patients with abnormal expression of FLT3 might benefit more from this four-drug combination. Our results provide a basis to evaluate these drug combinations as a cytoreductive treatment for AML patients with P53 and FLT3 mutations. Specifically, such drug combinations may be used as part of the pre-transplant conditioning regimen for AML patients undergoing hematopoietic stem cell transplantation.
Supplementary Material
Highlights.
Bu+Cy is an effective pre-transplant conditioning therapy but toxic.
Epigenetic modifiers enhance the efficacy of DNA alkylators.
Bu+Cy+DAC is synergistically cytotoxic to AML cells regardless of P53 status.
FLT3-ITD-positive AML cells are sensitized to Bu+Cy+DAC with sorafenib.
Bu+Cy+DAC±Sor may be used to personalize pre-transplant conditioning regimen.
ACKNOWLEDGMENTS
Part of this research was performed in the Flow Cytometry & Cellular Imaging Facility, which is supported in part by the National Institutes of Health through M.D. Anderson’s Cancer Center Support Grant CA016672. This work was also supported by the Stephen L. and Lavinia Boyd Fund for Leukemia Research, funds donated by grateful patients, and research grants to X. Tang from the National Natural Science Foundation of China (81270645), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Frontier Clinical Technical Project of the Science and Technology Department of Jiangsu Province (BE2017655), and Jiangsu Provincial Medical Talent (ZDRCA2016045).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Ciurea SO, Andersson BS. Busulfan in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2009;15:523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos GW, Tutschka PJ, Brookmeyer R, et al. Marrow transplantation for acute nonlymphocytic leukemia after treatment with busulfan and cyclophosphamide. N Engl J Med. 1983;309:1347–1353. [DOI] [PubMed] [Google Scholar]
- 3.Andersson BS, Gajewski J, Donato M, et al. Allogeneic stem cell transplantation (BMT) for AML and MDS following i.v. busulfan and cyclophosphamide (i.v. BuCy). Bone Marrow Transplant. 2000. May;25 Suppl 2:S35–S38. [DOI] [PubMed] [Google Scholar]
- 4.Andersson BS, Kashyap A, Gian V, et al. Conditioning therapy with intravenous busulfan and cyclophosphamide (IV BuCy2) for hematologic malignancies prior to allogeneic stem cell transplantation: a phase II study. Biol Blood Marrow Transplant. 2002;8:145–154. [DOI] [PubMed] [Google Scholar]
- 5.Bredeson C, LeRademacher J, Kato K, et al. Prospective cohort study comparing intravenous busulfan to total body irradiation in hematopoietic cell transplantation. Blood. 2013;122:3871–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copelan EA, Hamilton BK, Avalos B, et al. Better leukemia-free and overall survival in AML in first remission following cyclophosphamide in combination with busulfan compared with TBI. Blood. 2013;122:3863–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Barouch S, Cohen O, Vidal L, Avivi I, Ram R. Busulfan fludarabine vs busulfan cyclophosphamide as a preparative regimen before allogeneic hematopoietic cell transplantation: systematic review and meta-analysis. Bone Marrow Transplant. 2016;51:232–240. [DOI] [PubMed] [Google Scholar]
- 8.Nagler A, Rocha V, Labopin M, et al. Allogeneic hematopoietic stem-cell transplantation for acute myeloid leukemia in remission: comparison of intravenous busulfan plus cyclophosphamide (Cy) versus total-body irradiation plus Cy as conditioning regimen--a report from the acute leukemia working party of the European group for blood and marrow transplantation. J Clin Oncol. 2013;31:3549–3556. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen JJ, van Putten WL, Verdonck LF, et al. Results of a HOVON/SAKK donor versus no-donor analysis of myeloablative HLA-identical sibling stem cell transplantation in first remission acute myeloid leukemia in young and middle-aged adults: benefits for whom? Blood. 2007;109:3658–3666. [DOI] [PubMed] [Google Scholar]
- 10.Russell JA, Tran HT, Quinlan D, et al. Once-daily intravenous busulfan given with fludarabine as conditioning for allogeneic stem cell transplantation: study of pharmacokinetics and early clinical outcomes. Biol Blood Marrow Transplant. 2002;8:468–476. [DOI] [PubMed] [Google Scholar]
- 11.de Lima M, Couriel D, Thall PF, et al. Once-daily intravenous busulfan and fludarabine: clinical and pharmacokinetic results of a myeloablative, reduced-toxicity conditioning regimen for allogeneic stem cell transplantation in AML and MDS. Blood. 2004;104:857–864. [DOI] [PubMed] [Google Scholar]
- 12.Andersson BS, Valdez BC. Pretransplant conditioning with fludarabine and IV busulfan, reduced toxicity and increased safety without compromising antitumor efficacy and overall treatment effect? Bone Marrow Transplant. 2016;51:919–920. [DOI] [PubMed] [Google Scholar]
- 13.Kebriaei P, Bassett R, Lyons G, et al. Clofarabine plus busulfan is an effective conditioning regimen for allogeneic hematopoietic stem cell transplantation in patients with acute lymphoblastic leukemia: Long-term study results. Biol Blood Marrow Transplant. 2017;23:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersson BS, Thall PF, Valdez BC, et al. Fludarabine with pharmacokinetically guided IV busulfan is superior to fixed-dose delivery in pretransplant conditioning of AML/MDS patients. Bone Marrow Transplant. 2017;52:580–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Zhai X, Song Z, et al. Busulfan plus fludarabine as a myeloablative conditioning regimen compared with busulfan plus cyclophosphamide for acute myeloid leukemia in first complete remission undergoing allogeneic hematopoietic stem cell transplantation: a prospective and multicenter study. J Hematol Oncol. 2013;6:15 doi: 10.1186/1756-8722-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rambaldi A, Grassi A, Masciulli A, et al. Busulfan plus cyclophosphamide versus busulfan plus fludarabine as a preparative regimen for allogeneic haemopoietic stem-cell transplantation in patients with acute myeloid leukaemia: an open-label, multicentre, randomised, phase 3 trial. Lancet Oncol 2015;16:1525–1536. [DOI] [PubMed] [Google Scholar]
- 17.Momparler RL. Molecular, cellular and animal pharmacology of 5-aza-2’-deoxycytidine. Pharmacol Ther. 1985;30:287–299. [DOI] [PubMed] [Google Scholar]
- 18.Valdez BC, Li Y, Murray D, Corn P, Champlin RE, Andersson BS. 5-Aza-2’-deoxycytidine sensitizes busulfan-resistant myeloid leukemia cells by regulating expression of genes involved in cell cycle checkpoint and apoptosis. Leuk Res. 2010;34:364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scandura JM, Roboz GJ, Moh M, et al. Phase 1 study of epigenetic priming with decitabine prior to standard induction chemotherapy for patients with AML. Blood. 2011;118:1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gore L, Triche TJ Jr, Farrar JE, et al. A multicenter, randomized study of decitabine as epigenetic priming with induction chemotherapy in children with AML. Clin Epigenetics. 2017;9:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen JL, Jao JY. Enzymatic basis of cyclophosphamide activation by hepatic microsomes of the rat. J Pharmacol Exp Ther. 1970;174: 206–210. [PubMed] [Google Scholar]
- 22.McDonald GB, Slattery JT, Bouvier ME, et al. Cyclophosphamide metabolism, liver toxicity, and mortality following hematopoietic stem cell transplantation. Blood. 2003; 101:2043–2048. [DOI] [PubMed] [Google Scholar]
- 23.Boddy AV, Yule SM. Metabolism and pharmacokinetics of oxazaphosphorines. Clin Pharmacokinet. 2000;38:291–304. [DOI] [PubMed] [Google Scholar]
- 24.Valdez BC, Murray D, Ramdas L, et al. Altered gene expression in busulfan-resistant human myeloid leukemia. LeukRes. 2008;32:1684–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojima K, Shimanuki M, Shikami M, et al. The dual PI3 kinase/mTOR inhibitor PI-103 prevents p53 induction by Mdm2 inhibition but enhances p53-mediated mitochondrial apoptosis inp53 wild-type AML.Leukemia.2008;2:1728–1736. [DOI] [PubMed] [Google Scholar]
- 26.Mosmann T Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. [DOI] [PubMed] [Google Scholar]
- 27.Martin SJ, Reutelingsperger CP, McGahon AJ, et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exptl Med. 1995;182:1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. [DOI] [PubMed] [Google Scholar]
- 29.Valdez BC, Li Y, Murray D, et al. Comparison of the cytotoxicity of cladribine and clofarabine when combined with fludarabine and busulfan in AML cells: Enhancement of cytotoxicity with epigenetic modulators. Exp Hematol. 2015;43:448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valdez BC, Li Y, Murray D, Liu Y, Nieto Y, Champlin RE, Andersson BS. Combination of a hypomethylating agent and inhibitors of PARP and HD AC traps PARP1 and DNMT1 to chromatin, acetylates DNA repair proteins, down-regulates NuRD and induces apoptosis in human leukemia and lymphoma cells. Oncotarget. 2018;9:3908–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song G, Valdez BC, Li Y, Liu Y, Champlin RE, Andersson BS. Synergistic cytotoxicity of sorafenib with busulfan and nucleoside analogs in human FMS-like tyrosine kinase 3 internal tandem duplications-positive acute myeloid leukemia cells. Biol Blood Marrow Transplant. 2014;20:1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiyoi H, Naoe T, Nakano Y, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93:3074–3080. [PubMed] [Google Scholar]
- 33.Merlo A, Herman JG, Mao L, et al. 5’ CpG island methylation is associated with transcriptional silencing of the tumour suppressor pl6/CDKN2/MTSl in human cancers. Nat Med. 1995;1:686–692. [DOI] [PubMed] [Google Scholar]
- 34.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. [DOI] [PubMed] [Google Scholar]
- 35.Nagata S, Nagase H, Kawane K, Mukae N, Fukuyama H. Degradation of chromosomal DNA during apoptosis. Cell Death Differ. 2003;10:108–116. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996; 86:147–157. [DOI] [PubMed] [Google Scholar]
- 37.Ly JD, Grubb DR, Lawen A. The mitochondrial membrane potential (deltapsi(m)) in apoptosis; an update. Apoptosis. 2003;8:115–128. [DOI] [PubMed] [Google Scholar]
- 38.Marchi S, Giorgi C, Suski JM, et al. Mitochondria-ros crosstalk in the control of cell death and aging. J Signal Transduct. 2012;2012:329635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA 1984;81:6993–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jüttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2’-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A. 1994;91:11797–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakao M, Yokota S, Iwai T, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- 42.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. [DOI] [PubMed] [Google Scholar]
- 43.Cauchy P, James SR, Zacarias-Cabeza J, et al. Chronic FLT3-ITD Signaling in acute myeloid leukemia is connected to a specific chromatin signature. Cell Rep. 2015;12:821–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2’-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28:752–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valdez BC, Andersson BS. Interstrand crosslink inducing agents in pretransplant conditioning therapy for hematologic malignancies. Environ Mol Mutagen. 2010;51:659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valdez BC, Li Y, Murray D, Champlin RE, Andersson BS. The synergistic cytotoxicity of clofarabine, fludarabine and busulfan in AML cells involves ATM pathway activation and chromatin remodeling. Biochem Pharmacol. 2011;81:222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.