Abstract
MoaA is one of the founding members of the radical S-adenosyl-L-methionine (SAM) superfamily, and together with the second enzyme, MoaC, catalyzes the construction of the pyranopterin backbone structure of the molybdenum cofactor (Moco). However, the exact functions of both MoaA and MoaC had remained ambiguous for more than 2 decades. Recently, their functions were finally elucidated through successful characterization of the MoaA product as 3′,8-cyclo-7,8-dihydro-GTP (3′,8-cH2GTP), which was shown to be converted to cyclic pyranopterin monophosphate (cPMP) by MoaC. 3′,8-cH2GTP was produced in a small quantity and was highly oxygen sensitive, which explains why this compound had previously eluded characterization. This chapter describes the methodologies for the characterization of MoaA, MoaC, and 3′,8-cH2GTP, which together significantly altered the view of the mechanism of the pyranopterin backbone construction during the Moco biosynthesis. Through this chapter, we hope to share not only the protocols to study the first step of Moco biosynthesis but also the lessons we learned from the characterization of the chemically labile bio-synthetic intermediate, which would be informative for the study of many other metabolic pathways and enzymes.
1. INTRODUCTION
Molybdenum cofactor (Moco) is an essential organometallic cofactor found in nearly all organisms (Mendel & Schwarz, 2011). Moco-dependent enzymes take advantage of the redox ability of Moco and catalyze biologically important reactions. Unlike many other organic cofactors, Moco cannot be directly taken up as a nutrient, thus requiring de novo biosynthesis. Therefore, Moco biosynthesis is essential for the survival of many organisms (Leimkuhler, Wuebbens, & Rajagopalan, 2011). In human, Moco deficiency (MoCD) caused by genetic mutations in one of the Moco biosynthesis enzymes can lead to activity loss of all Moco-dependent enzymes, resulting in fatal neurodegeneration and early childhood death (Johnson et al., 1980). Moco biosynthesis is also found essential for virulence of several clinically important bacterial pathogens, such as Mycobacterium tuberculosis (Boshoff & Barry, 2005) and Pseudomonas aeruginosa (Folkesson et al., 2012). These organisms rely on several Moco-dependent enzymes in the anaerobic respiration to survive the hypoxic conditions of the host body (Hurdle, O’Neill, Chopra, & Lee, 2011). More recently, Moco biosynthesis in enterobacteria in gut microbiome was shown to be essential for these organisms to cause inflammation, and small molecule inhibitors of Moco biosynthesis was shown effective to prevent such inflammation (Zhu et al., 2018).
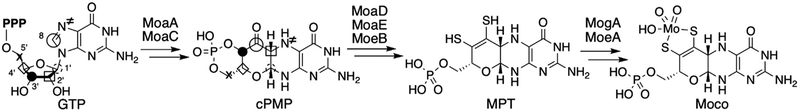
Moco biosynthetic pathway is conserved among most organisms and can be divided into three major steps (Fig. 1) (Leimkuhler et al., 2011; Mendel & Schwarz, 2011). The biosynthesis is initiated by the rearrangement of guanosine 5′-triphosphate (GTP) into cyclic pyranopterin monophosphate (cPMP), followed by insertion of two sulfur atoms to form molybdopterin (MPT). Finally, molybdate, the only absorptive form of molybdenum, is ligated to the dithiolene moiety of MPT to form Moco. The conversion of GTP into cPMP is of particular mechanistic interests, as it establishes the pyranopterin backbone of Moco from GTP through highly complex and unique rearrangement reaction. This transformation has been affected in more than 60% of the human MoCD patients (Reiss, 2000). Early isotope-labeling experiments had demonstrated that the transformation involves a unique ring rearrangement where guanine C-8 is inserted between C-2′ and C-3′ of ribose (Fig. 1; see symbols on GTP and cPMP) (Rieder et al., 1998; Wuebbens & Rajagopalan, 1995). This transformation is distinct from other pterin biosynthetic pathways, in which C-8 is hydrolyzed as formate and does not retain in the pterin product (Burg & Brown, 1968; Shiota, Palumbo, & Tsai, 1967). Thus, the mechanism of pyranopterin ring formation during the Moco biosynthesis was considered as a critical mystery in cofactor biosynthesis and enzymology fields.
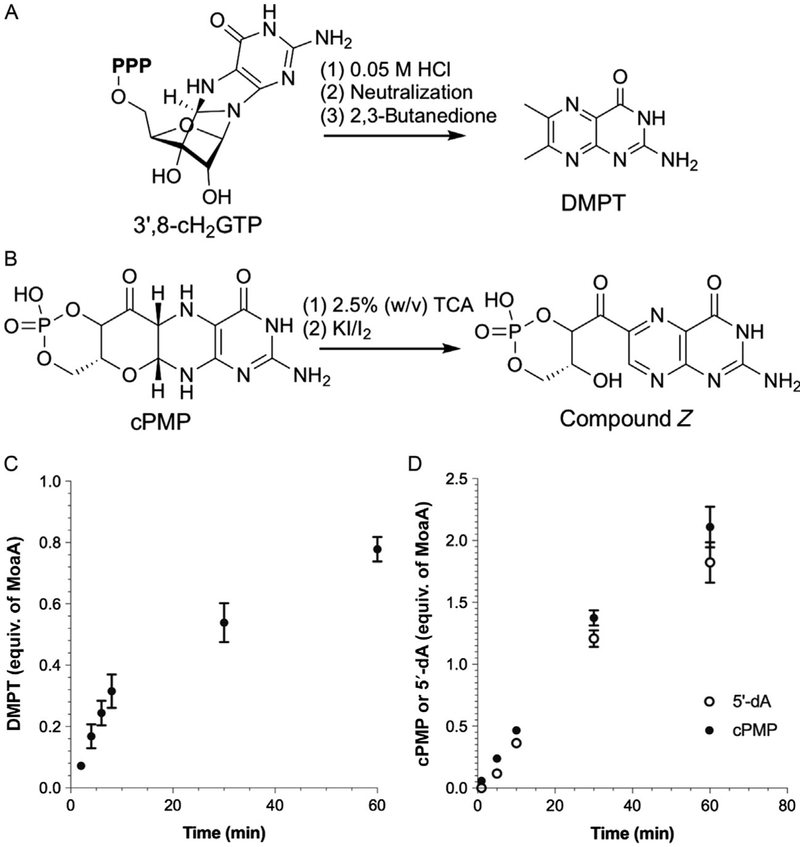
Fig. 1.
Moco biosynthetic pathway in bacteria. The symbols on GTP and cPMP show the fates of carbon and nitrogen atoms during the construction of the pyranopterin backbone as determined by isotope-labeling studies (Rieder et al., 1998; Wuebbens & Rajagopalan, 1995).
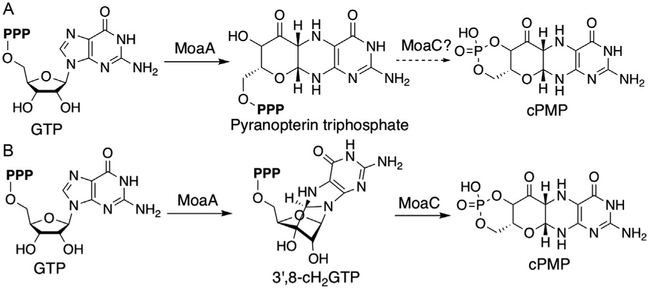
In bacteria, the transformation from GTP to cPMP is catalyzed by two enzymes, MoaA and MoaC. MoaA belongs to the radical S-adenosyl-L-methionine (SAM) superfamily (Sofia, Chen, Hetzler, Reyes-Spindola, & Miller, 2001), while MoaC shows no obvious sequence or structural similarities to other characterized enzymes (Wuebbens, Liu, Rajagopalan, & Schindelin, 2000). Because of the general awareness of radical SAM enzymes catalyzing novel chemical reactions, MoaA was thought to catalyze the complex reactions to rearrange GTP to cPMP (Hänzelmann & Schindelin, 2004; Leimkuhler et al., 2011). MoaC was thought to be either a regulatory protein (Hänzelmann & Schindelin, 2004) or an enzyme responsible only for the cyclic phosphate formation (Leimkuhler et al., 2011). In 2006, Hänzelmann and Schindelin mentioned in their publication that when MoaA was assayed with GTP and SAM in the absence of MoaC, they observed a compound that can be chemically derivatized to dimethylpterin (DMPT) (Hänzelmann & Schindelin, 2006). Although no data were published, the statement implied the presence of a potential intermediate during pyranopterin ring formation. By the early 2010s, the predominant view in the field was that MoaA catalyzes the transformation of GTP into pyranopterin triphosphate, while MoaC catalyzes the cyclic phosphate formation (Fig. 2A) (Kanaujia et al., 2010; Leimkuhler et al., 2011). However, pyranopterin triphosphate would not be converted to DMPT (Wuebbens & Rajagopalan, 1993). Thus, such model is inconsistent with the statement by Hänzelmann and Schindelin (2006), and the function of MoaA remained ambiguous.
Fig. 2.
Proposed models for the pyranopterin ring formation catalyzed by MoaA and MoaC during the Moco biosynthesis. (A) Pyranopterin triphosphate as the MoaA product and (B) 3′,8-cH2GTP as the MoaA product.
When we initiated the project in 2011, no experimental data were available to support even the presence of the putative product of MoaA. Therefore, we initially focused on detection of the putative MoaA product using a stepwise assay (Hover, Loksztejn, Ribeiro, & Yokoyama, 2013). In this assay, MoaA was first incubated with GTP and SAM in the absence of MoaC, followed by separation of MoaA and its small molecule products by ultrafiltration. Incubation of the small molecule fraction with MoaC resulted in the formation of cPMP. The putative MoaA product was also derivatized to DMPT (Hover et al., 2013), consistent with the statement by Hänzelmann and Schindelin (2006). The quantitative agreement between cPMP and DMPT suggested that the MoaA product was quantitatively converted into cPMP (Hover et al., 2013). These observations were the first quantitative evidence for the presence of a small molecule that is diffusible from MoaA and converted to cPMP by MoaC.
The isolation and structural characterization of this small molecule, however, were challenging due to the substoichiometric turnover of MoaA and the limited chemical stability of the MoaA product. In particular, the MoaA product was quickly decomposed in the presence of oxygen (t1/2 70min) or at acidic pH (t1/2<10min) (Hover et al., 2013). These challenges were overcome by the development of large-scale preparation of MoaA and the purification of the MoaA product under strictly anaerobic conditions at mild basic pH and low temperature. The isolated MoaA product was characterized using comprehensive approaches including chemical and enzymo-logical derivatizations, UV–vis absorption spectroscopy, mass spectrometry (MS), and nuclear magnetic resonance (NMR) spectroscopy, based on which the structure was proposed as 3′,8-cyclo-7,8-dihydro-GTP (3′,8-cH2GTP, Fig. 2B) (Hover et al., 2013). 3′,8-cH2GTP has an acid-labile 6-hydroxy-2,4,5-triaminopyrimidine partial structure that can be converted to DMPT, which is consistent with the Hänzelmann and Schindelin’s statement (Hänzelmann & Schindelin, 2006). In parallel to our efforts, Mehta et al. reported an liquid chromatography–mass spectrometry (LC–MS) analysis of a MoaA product that absorbs light at 320nm and shows a low-resolution mass signal at m/z=524 [M+H]+ (Mehta et al., 2013). While the authors assigned this compound to pyranopterin triphosphate, the compound was never isolated. Since 3′,8-cH2GTP absorbs light at 322nm and has a molecular weight of 523, the compound observed by Mehta et al. was most likely 3′,8-cH2GTP as well. Thus, 3′,8-cH2GTP was observed by multiple groups, but its structure was assigned incorrectly due to the lack of its isolation.
The identification of 3′,8-cH2GTP as the MoaA product revealed that MoaA is responsible for the CdC bond formation between guanine C-8 and ribose C-3′, while MoaC, in contrast to the previous consensus, catalyzes the complicated rearrangement reaction during the conversion of 3′,8-cH2GTP into cPMP (Fig. 2B). Therefore, the successful characterization of the MoaA product revealed novel enzyme functions and mechanisms during the construction of the pyranopterin ring of Moco. This success was made possible through quantitative and comprehensive characterization of MoaA, MoaC, and 3′,8-cH2GTP. Therefore, in this chapter, we will describe the protocols for preparation and characterization of MoaA and MoaC, and for the isolation and characterization of 3′,8-cH2GTP. These protocols can be used to study MoaA and MoaC from other organisms, including humans, to investigate the generality of their functions, and to elucidate detailed mechanisms of these enzymes.
Also, the characterization of 3′,8-cH2GTP highlights important lessons for characterization of enzymes and biosynthetic pathways in general. As will be detailed in this chapter, the study emphasizes the importance of quantitative characterization of enzymes and their products for accurate determination of enzyme functions. The study also demonstrated the importance of understanding the chemical properties of small molecules for their isolation and structural characterization. We believe these are important lessons applicable to many other biosynthetic enzymes and pathways, especially those with unstable intermediates.
2. PREPARATION AND CHARACTERIZATION OF MoaA
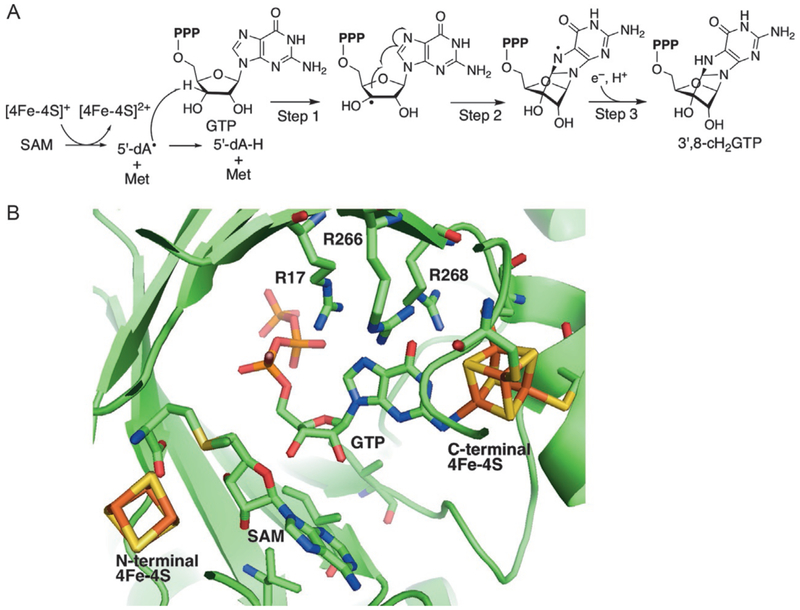
MoaA catalyzes the transformation of GTP into 3′,8-cH2GTP. Fig. 3A shows the current model for the catalytic mechanism (Hover et al., 2013). The reactionisinitiatedbyreductivecleavageofSAMtogenerate5′-deoxyadenosyl radical(5′-dA•)and L-methionine. Theresulting5′-dA• abstracts theH-atomat the 3′-position of GTP and produces C3′-radical intermediate (step 1), which then attacks C-8 of guanine base (step 2). The resulting aminyl radical is reduced by transfers of a proton and an electron to produce the final product 3′,8-cH2GTP (step 3). The C-terminal cluster was proposed to be the electron donor (Hover et al., 2013; Mehta et al., 2013), but no evidence is currently available.
Fig. 3.
Proposed mechanism and crystal structure of MoaA. (A) Current model for the catalytic mechanism of MoaA and (B) model of the MoaA active site. The position of SAM is modeled into the crystal structure of MoaA in complex with GTP (PDB ID: 2FB3) (Hänzelmann & Schindelin, 2006) by structural alignment with the crystal structure of MoaA in complex with SAM (PDB ID: 1TV8) (Hänzelmann & Schindelin, 2004). Reprinted with permission from Hover, B. M., Loksztejn, A., Ribeiro, A. A., & Yokoyama, K. (2013). Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. Journal of the American Chemical Society, 135(18), 7019–7032. doi:10.1021/ja401781t. Copyright 2013 American Chemical Society.
MoaA is a representative member of the twitch subfamily (Goldman, Grove, Booker, & Drennan, 2013), one of the subclasses in the radical SAM superfamily. Members of this subfamily are characterized by the presence of two [4Fe–4S] clusters; one is the canonical radical SAM cluster and the other is an auxiliary [4Fe–4S] cluster in an extended C-terminal domain (Goldman et al., 2013). The crystal structure of Staphylococcus aureus MoaA in complex with SAM (Fig. 3B) showed that the canonical radical SAM cluster was bound at the N-terminus of MoaA, and ligated by three Cys residues while the fourth Fe ligated by SAM (Hänzelmann & Schindelin, 2004). The auxiliary cluster at the C-terminus of MoaA is also ligated by three Cys. The crystal structure of S. aureus MoaA in complex with GTP (Hänzelmann & Schindelin, 2006), together with an electron nuclear double resonance (ENDOR) spectroscopy analysis (Lees et al., 2009), revealed that the fourth Fe interacts with N-1 of the guanine base of GTP. This interaction was proposed to change the tautomerization of guanine base to generally unfavorable enol tautomer (Lees et al., 2009), although the role of tautomerization on the catalytic mechanism of MoaA is not known. The C-terminal cluster is also critical for the stability of MoaA, and muta-genesis of any of the Cys ligands results in expression of insoluble proteins (Hänzelmann & Schindelin, 2006). Both [4Fe–4S] clusters are essential for the catalytic function of MoaA and are both oxygen sensitive. Therefore, the preparation and characterization of MoaA require anaerobic methods. In this section, we will describe our protocols for expression, purification, and characterization of MoaA using the enzyme from S. aureus as a model.
2.1. Expression and Purification of MoaA
In general, purification of radical SAM enzymes is performed under strictly anaerobic conditions because of the oxygen-sensitive nature of the [4Fe–4S] clusters. The previous protocol for MoaA purification described by Hänzelmann and Schindelin also used anaerobic conditions (Hänzelmann & Schindelin, 2004). However, anaerobic purification limits the amount of protein that can be purified in a single experiment. In our current protocol, ~1.6g of MoaA is required for isolation of 3–5 mg of the MoaA product (see Section 4.2). To facilitate such isolation effort, we developed a protocol for MoaA purification under aerobic conditions. In this procedure, coexpression of the suf operon encoding Fe–S cluster assembly enzymes (SufA, SufB, SufC, SufD, SufS, and SufE) (Hänzelmann et al., 2004) was critical. Also, to maintain the reduced form of all the Cys residues, thiol-reducing reagent, either β-mercaptoethanol (βME) or dithiothreitol (DTT), was always included in the buffer throughout the purification.
With these modifications, MoaA can be overexpressed and purified under aerobic conditions. Our typical purification of MoaA is performed from Escherichia coli harvested from over 50L of culture (usually around 200g of cell paste) using Ni-NTA agarose resin in the presence of βME as a Cys protection reagent, followed by Sephadex G-25 size exclusion chromatography for exchange into buffer containing DTT. The aerobically purified MoaA harbors only 1.5–2 equiv. of iron and sulfide bound to each monomer, suggesting that even the structurally essential C-terminal cluster is only partially loaded. Therefore, to avoid inactivation of MoaA in the absence of the fully loaded C-terminal cluster, we usually reconstitute the cluster immediately after the purification. The cluster reconstitution and all the subsequent characterizations are carried out under strict anaerobic conditions (<0.1ppm O2).
Subclone S. aureus moaA gene into the NdeI and BamHI sites of the pET15b vector to give pET15b-HisMoaA (Hover et al., 2013).
Transform E. coli BL21(DE3) cells with the pET15b-HisMoaA and SUF plasmids (Hänzelmann et al., 2004) and culture the transformant overnight on an Lysogeny broth (LB) agar plate with 100mg/L ampicillin (Amp) and 35mg/L chloramphenicol (Cam) at 37°C.
Pick up a single colony and culture overnight in 5mL of LB medium (100mg/L Amp and 35mg/L Cam) at 37°C.
Inoculate 250mL of LB medium (100mg/L Amp and 35mg/L Cam) with the 5mL overnight culture and incubate at 37°C and 200rpm until saturation (~18 h).
Inoculate each of 6 × 1.5L of LB medium (100mg/L Amp and 35mg/L Cam) with 40mL of saturated overnight culture and incubate at 37°C, 200rpm.
When OD600nm reaches 0.9–1.1, cool down the culture to 30°C and induce the overexpression by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to the final concentration of 0.5mM. Continue the culture at 30°C and 200rpm for 4–5h.
Harvest the cells by centrifugation at 5000 × g, 20min, 4°C. The 9L culture typically yields ~30g of cell paste. The cell paste can be stored at −20°C.
Repeat steps 2–7 to accumulate ~200g cell paste (usually requires over 50L of LB culture).
Resuspend the cell paste (~200g) with 500mL of buffer A (50mM Tris–HCl, pH 7.6, 0.3M NaCl, 10% glycerol) supplemented with 40mM imidazole, 3mM βME, and 10μL benzonase nuclease (EMD Millipore) and homogenize the cell suspension.
Lyse the cells by two passages through French Press at 14,000psi.
Remove the cell debris by centrifugation at 25,000 × g for 30min at 4°C. The supernatant should be dark brown.
Load the supernatant onto a Ni-NTA column (100–150mL resin).
Wash the column with buffer A supplemented with 40mM imidazole and 3mM βME until no protein is eluted based on Bradford assay (typically need ~1L buffer).
Elute MoaA with buffer A supplemented with 400mM imidazole and 3mM βME until no protein is eluted based on Bradford assay (typically need ~200mL buffer).
Concentrate the resultant solution to ~100mL by ultrafiltration (30kDa MWCO).
Load the eluted protein onto 1L Sephadex G-25 column equilibrated in buffer A supplemented with 5mM DTT.
Elute the protein with buffer A supplemented with 5mM DTT. Check the fractions by UV–vis absorption spectra and pool the fractions containing MoaA.
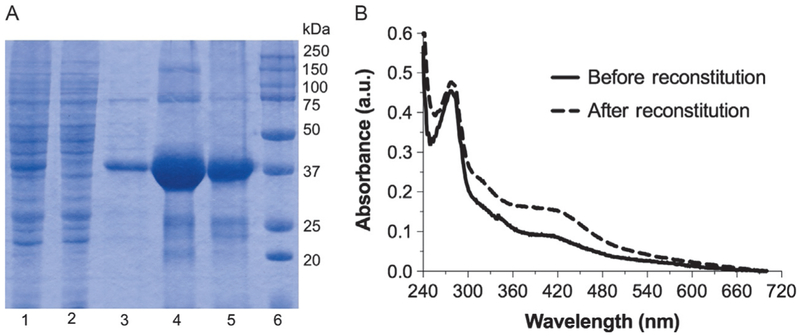
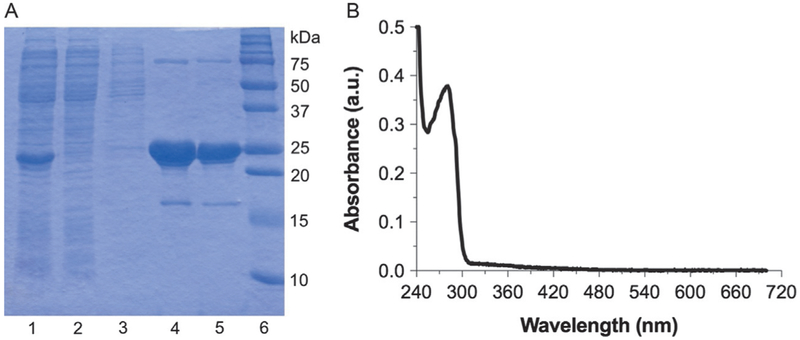
Analyze each step of purification by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) to check the purity and recovery (Fig. 4A).
Determine UV–vis spectra of as-isolated MoaA from 700 to 200nm (Hitachi U-3900 UV–vis spectrometer). There should be absorption band from protein at 280nm and that from the [4Fe–4S] clusters at 300–500nm with a shoulder at ~420nm (Fig. 4B, solid line).
Fig. 4.
Purification and reconstitution of S. aureus MoaA. (A) SDS-PAGE of MoaA at different purification steps. Lanes: 1, crude cell lysate; 2, Ni-NTA flow through; 3, Ni-NTA wash with 40mM imidazole; 4, Ni-NTA elution with 400mM imidazole; 5, after G-25; and 6, molecular weight marker. (B) UV–vis absorption spectra of MoaA (11.4μM) before (solid line) and after reconstitution (dashed line).
2.2. MoaA Concentration Determination
Determination of protein concentrations for radical SAM enzymes is not trivial. The residual Fe–S cluster in the as-isolated MoaA significantly contributes to the absorption at 280nm, and thus the concentration of MoaA cannot be determined directly by the UV absorbance of proteins at 280nm. Instead, these enzymes are the best quantified by colorimetric protein assays, such as Bradford assay, compatible with Fe and other reducing agents. Since bovine serum albumin (BSA) standards give variable errors depending on the protein of interests (for MoaA, BSA gave >10% errors), it is necessary to prepare MoaA standards with known concentrations. The MoaA standards can be prepared by first removing all the copurified iron and sulfur under a denaturing condition, followed by refolding by exchanging the protein back into the native condition. The concentration of the apo-MoaA is determined using the UV absorption at 280nm (Gill & Von Hippel, 1989). The refolded apo-MoaA is then used as a concentration standard for the Bradford assay of as-isolated and reconstituted MoaA.
Mix 0.5mL of as-isolated MoaA with 1.5mL of 8M guanidine HCl, 13.3mM ethylenediaminetetraacetic acid (EDTA), pH 8. Incubate at 4°C for 1h.
Load onto a G-25 column (20mL) equilibrated in buffer A supplemented with 5mM DTT and elute with the same buffer.
Collect fractions every 0.5mL of elution. Identify the fractions containing MoaA judged by Bradford assay and combine.
Centrifuge at 16,000 × g for 5min to remove precipitants.
Dilute one part of the resulting apo-MoaA in three parts of 8 M guanidine HCl. The final concentration of guanidine HCl should be 6M.
Determine the absorption at 280nm by UV–vis spectrometer and calculate the protein concentration using ε280nm =26,200M−1 cm−1. The extinction coefficient of the protein in 6 M guanidine HCl is calculated based on the following equation (Pace, Vajdos, Fee, Grimsley, & Gray, 1995): ε280nm(M−1cm−1)=(NTrp)(5500)+(NTyr)(1490)+(NCys)(125).
Use the apo-MoaA to make concentration standards. Determine the concentration of as-isolated MoaA by the Bradford assay. Typically, each purification yields 6–7mg of as-isolated MoaA per gram of wet cell paste.
2.3. Reconstitution of the [4Fe–4S] Clusters
The two [4Fe–4S] clusters of MoaA can be chemically reconstituted. This reconstitution and all the following steps are performed under strictly anaerobic conditions in a glove box (M. Braun) that is maintained at <0.1ppm O2 and 10°C. While there have been several different ways to reconstitute the [4Fe–4S] clusters of radical SAM enzymes, such as using cysteine desulfurase as a sulfide source (Mahanta, Zhang, Hudson, van der Donk, & Mitchell, 2017), we found that chemical reconstitution using (NH4)2Fe(SO4)2 and Na2S as iron and sulfide source, respectively, resulted in reproducible reconstitution.
For the chemical reconstitution, it is critical to minimize the formation of insoluble ferrous sulfide precipitant. This can be achieved by slow and careful titration of the solutions of (NH4)2Fe(SO4)2 and Na2S to give enough time for self-assembly of the [4Fe–4S] clusters in the active site of MoaA. Also, to avoid an unnecessary excess of Fe2+ and S2−, the amount of Fe–S cluster copurified in the as-isolated MoaA is quantified to adjust the amount of (NH4)2Fe(SO4)2 and Na2S added during the reconstitution. The successful reconstitution typically gives slightly yellowish dark brown color while ferrous sulfide forms black precipitation or colloid. The quality of the reconstituted [4Fe–4S] clusters can also be assessed by UV–vis absorption spectra, which should show strong absorption of the [4Fe–4S] clusters in the 300–500nm region with a characteristic shoulder at ~420nm (Fig. 4B). Contamination of ferrous sulfide colloid results in very broad and featureless absorption from 200 to 800nm.
Degas 1L of buffer A and 1L of Sephadex G-25 resin equilibrated and suspended in ~2L of buffer A by 15min evacuation followed by 10min Ar refill.
Evacuate the flasks containing the degassed buffer and resin after the last cycle in step 1, and bring them into the glove box. Equilibrate the buffer and resin in the glove box atmosphere for at least 15h.
Bring two portions of preweighted DTT powder (5 and 10mmol) into the glove box and add 5mmol to the degassed buffer A and 10mmol to the degassed Sephadex G-25 resin immediately before use. Pack the Sephadex G-25 resin to a column (5 × 50 cm2).
Remove oxygen from the as-isolated MoaA (~0.2mM) solution by exchanging the atmosphere to Ar using a Schlenk line by brief evacuation (<30s) followed by Ar refill and equilibration for 5min. Repeat this cycle four times. During the evacuation, it is critical to minimize formation of bubbles to avoid unfolding of MoaA.
Evacuate the flask after the last cycle in step 4 and bring the sample into the glove box.
Determine the protein concentration by the Bradford assay as described in Section 2.2.
Quantitate the amount of Fe2+/3+ and S2− by following the reported protocols for the ferrozine (Wayne, 1988) and sulfide assays (Beinert, 1983), respectively. Typically, there is around 1.5–2equiv. of Fe2+/3+ and S2− per monomer of MoaA.
Based on the concentration of MoaA and the amounts of Fe2+/3+ and S2− determined in steps 6 and 7, respectively, determine the amounts of (NH4)2Fe(SO4)2 and Na2S needed for the reconstitution. The sum of the copurified Fe2+/3+ and S2− and those added should be 9equiv. per monomer of MoaA.
Introduce the preweighted (NH4)2Fe(SO4)2 and Na2S crystals into the glove box and dissolve with degassed H2O to make the final concentration 25mM. Calculate the volume needed for the reconstitution. Typically 9–10mL each of this solution is required.
Dropwise and gently add 1mL of the 25mM (NH4)2Fe(SO4)2 solution followed by 1mL of the 25mM Na2S solution and repeat this cycle until the required volumes are added. Perform the addition over the course of 1h and incubate the mixture for another 45min.
Load the protein solution onto the 1L Sephadex G-25 column prepared in step 3. Elute the protein using the same buffer.
Identify and combine the fractions containing the reconstituted MoaA based on UV–vis absorption spectra.
Aliquot the reconstituted MoaA solution in appropriate volumes, flash–freeze in liquid nitrogen and store at −80°C. Store under aerobic conditions at this temperature does not influence the activity. Do not thaw under aerobic conditions.
Determine the UV spectra of reconstituted MoaA using a septum-sealed screw-cap quartz cuvette (Starna Cells Inc.). As shown in Fig. 4B (dashed line), the absorption intensity at 300–500nm derived from the [4Fe–4S] clusters should be higher for the successfully reconstituted MoaA compared to the as-isolated MoaA.
Determine the concentration of reconstituted MoaA by Bradford assay as described in Section 2.2.
Determine the amount of Fe2+/3+ and S2− bound to each monomer by ferrozine (Wayne, 1988) and sulfide assays (Beinert, 1983), respectively. Typically the yield is around 4–4.5mg of reconstituted MoaA per gram of wet cell paste, with 6–7equiv. of Fe2+/3+ and S2− per monomer of MoaA.
2.4. Activity Assays of MoaA
To characterize the MoaA catalytic activity, we established three activity assay methods, all of which are important to verify the quality of the reconstituted MoaA. In the first method, the MoaA product is chemically derivatized to DMPT through acid hydrolysis and incubation with 2,3-butanedione (Fig. 5A). DMPT is fluorescent and thus can be quantified by high-performance liquid chromatography (HPLC). This assay quantifies 3′,8-cH2GTP and therefore can be used to characterize the function of MoaA independent of MoaC. However, many compounds can be converted to DMPT under the same conditions (Bracher et al., 1999; Hover, Tonthat, Schumacher, & Yokoyama, 2015). Therefore, it is advisable to confirm the observation by the coupled assay, in which the MoaA assays are performed in the presence of MoaC. In this assay, 3′,8-cH2GTP is converted to cPMP in situ and the resulting cPMP is subsequently derivatized to compound Z (Fig. 5B) and quantified by HPLC. Compound Z authentic standard can be isolated from the ΔmoeB E. coli strain by following the published protocol (Wuebbens & Rajagopalan, 1993). This assay allows characterization of only the MoaA activity relevant to the cPMP formation. The third assay is the quantitation of 5′-deoxyadenosine (5′-dA) produced during the MoaA reaction, which can be combined with the coupled assay to determine the reaction stoichiometry. This stoichiometry is important for evaluating the quality of enzymes. MoaA with good quality exhibits excellent coupling of SAM cleavage to 3′,8-cH2GTP formation. No more than 5% of abortive SAM cleavage should be observed (Hover & Yokoyama, 2015).
Fig. 5.
Activity assays of MoaA. (A and B) Chemical derivatizations of 3′,8-cH2GTP to DMPT (A) and cPMP to compound Z (B). (C) Time course of the MoaA reaction in the absence of MoaC. The amount of 3′,8-cH2GTP formation was quantified by HPLC after its chemical derivatization to DMPT. (D) Time course of the formation of 5′-dA and cPMP during the catalysis in the coupled assay. cPMP was quantified by HPLC after its conversion to compound Z. Panels (C) and (D) are prepared using the previously published data (Hover et al., 2013).
2.4.1. Quantitative Detection of the MoaA Product by Chemical Derivatization
3′,8-cH2GTP has a 6-hydroxy-2,4,5-triaminopyrimidine partial structure connected to the other parts of the molecule through acid-labile bonds (aminal and glycosidic bonds; see Fig. 5A). Thus, 3′,8-cH2GTP can be derivatized to DMPT by acid hydrolysis that releases 6-hydroxy-2,4,5-triaminopyrimidine from 3′,8-cH2GTP, followed by neutralization and incubation with 2,3-butanedione, which derivatizes 6-hydroxy-2,4,5-triaminopyrimidine into DMPT. The resulting DMPT is fluorescent with excitation wavelength at 365nm, and emission wavelength at 445nm, and thus can be quantified by HPLC with excellent sensitivity. The lower limit of detection in typical MoaA assays is ~1pmol.
Preincubate MoaA (10μM) and sodium dithionite (SDT, 2mM) in buffer A supplemented with 5mM DTT at 25°C in the glove box for 5min.
Preincubate SAM (2mM), GTP (2mM) in buffer A supplemented with 5mM DTT at 25°C in the glove box for 5min.
Initiate the reaction by mixing equal volumes of solutions from steps 1 and 2, and incubate the reaction at 25°C.
Quench the reaction by mixing 40μL of the reaction mixture with 5μL of 0.5M HCl at different time points (e.g., 2, 4, 6, 8, 10, 30, and 60min). Usually, the product formation is in linear correlation with time over the first 10min, after which it slows down significantly (Fig. 5C).
Transfer the quenched samples out of the glove box and incubate the samples at 95°C for 5min.
Cool down and neutralize the sample by adding 3μL of 1M NaOH.
Add 12.5μL of 0.66% (v/v) 2,3-butanedione in 0.9M Tris–HCl (pH 8.5) and incubate at 95°C for 45min.
Centrifuge at 16,000 × g for 10min to remove precipitants.
Analyze the supernatant by HPLC (Hitachi L-2139 pump, L-2455 diode array detector, L-2485 fluorescence detector, L-2200 auto-sampler, and L-2300 column oven) equipped with an ODS hypersil column (250 × 4.6mm2, 3μm) at 40°C. Chromatography is performed by isocratic elution with 92.5% 30mM ammonium formate, pH 4.5, and 7.5% methanol at a flow rate of 1.5mL/min and monitored by fluorescence (excitation 365nm and emission 445nm). Under this condition, DMPT elutes out at ~5.5min.
To determine a standard curve for DMPT quantitation, inject different amounts of DMPT standards (e.g., 5, 10, 15, 20, and 25pmol) to the HPLC system and analyze under the conditions described in step 9. From the resulting HPLC traces, determine the DMPT peak areas and plot against the amount of injection.
Determine the DMPT peak areas in the HPLC traces of the samples and quantify the amount of DMPT using the standard curve determined in step 10. As shown in Fig. 5C, the amount of DMPT increases with prolonged incubation time with a sign of product inhibition after ~10min.
2.4.2. Coupled Assay of MoaA with MoaC
The DMPT assay quantifies the amounts of compounds with 6-hydroxy-2,4,5-triaminopyrimidine partial structure, which is present in not only 3′,8-cH2GTP but many other compounds (Bracher et al., 1999; Hover et al., 2015). Also, as will be detailed in Section 4, 3′,8-cH2GTP is not chemically stable. Therefore, it is preferable to assay MoaA in the presence of MoaC and detect cPMP. This assay is more relevant to the actual Moco biosynthesis inside the cells because MoaA and MoaC are always present in healthy cells. MoaC has nM-order Km values toward 3′,8-cH2GTP with the kcat value 4–13 folds higher than that of MoaA (see Section 3, for details), and does not exhibit apparent product inhibition (compare Fig. 5C and D). Thus, the inclusion of MoaC in the MoaA assay allows selective and quantitative conversion of 3′,8-cH2GTP to cPMP. The amount of cPMP can be quantified after its oxidative derivatization to compound Z, which fluoresces with the excitation and the emission wavelengths of 367 and 450nm, respectively. In the protocol described below, the typical lower limit of detection for compound Z is ~1pmol.
Preincubate MoaA (2μM) and SDT (2mM) in buffer A supplemented with 5mM DTT at 25°C in a glove box for 5min.
Preincubate SAM (2mM), GTP (2mM), MoaC (20μM), and MgCl2 (2mM) in buffer A supplemented with 5mM DTT at 25°C in a glove box for 5min.
Initiate the reaction by mixing equal amounts of solutions from steps 1 to 2, and incubate the reaction at 25°C.
Quench the reaction by mixing 45μL of the reaction mixture with 5μL of 25% (w/v) trichloroacetic acid (TCA) at different time points (e.g., 2, 4, 6, 8, 10, 30, and 60min).
Transfer the samples out of the glove box and add 5μL of a solution containing 2% (w/v) KI and 1% (w/v) I2 to each sample. Incubate at room temperature for 20min.
Centrifuge at 16,000 × g for 10min to remove precipitants.
Analyze the supernatant by the HPLC system equipped with an ODS hypersil column (250 × 4.6mm2, 3μm). Perform chromatography by isocratic elution with 92.5% of 0.1% trifluoroacetic acid (TFA)/H2O buffer, and 7.5% methanol with a flow rate of 1mL/min and monitor the elution by fluorescence (excitation 367nm and emission 450nm). Under this condition, compound Z elutes at ~2.5min.
To determine a standard curve for compound Z quantitation, inject different amounts of compound Z standards (e.g., 5, 10, 15, 20, and 25pmol) to the HPLC system and analyze under the conditions described in step 7. From the resulting HPLC traces, determine the compound Z peak areas and plot against the amount of injection.
Determine the compound Z peak areas on the HPLC traces of the samples and quantify the amount of compound Z using the standard curve determined in step 8.
2.4.3. Determination of the Reaction Stoichiometry
MoaA initiates the radical reaction by reductive cleavage of SAM and uses a stoichiometric amount of SAM to produce 3′,8-cH2GTP. In general, many radical SAM enzymes catalyze the abortive cleavage of SAM. In some cases, the abortive cleavage is caused by the inappropriate position of affinity tag (Grove, Ahlum, Sharma, Krebs, & Booker, 2010), or the use of non-physiological substrate (Yang et al., 2012), or reductant (Barr et al., 2016). Therefore, the extent of the abortive cleavage can be used for quality control of radical SAM enzyme preparation. For MoaA, under our assay conditions, we observed less than 5% abortive cleavage, suggesting an excellent coupling of SAM cleavage to 3′,8-cH2GTP formation (Hover & Yokoyama, 2015). Therefore, any increase in the abortive cleavage from this level is a sign for inappropriate enzyme preparation or assay conditions. The reaction stoichiometry can be determined by quantifying the amount of 5′-dA produced during the catalysis, and its comparison with the amount of 3′,8-cH2GTP. The amount of 5′-dA can be quantified by HPLC while the amount of 3′,8-cH2GTP can be determined by the coupled assay as described in Section 2.4.2. The lower limit of 5′-dA detection is ~50pmol.
Preincubate MoaA (40μM) and SDT (2mM) in buffer A supplemented with 5mM DTT at 25°C in the glove box for 5min.
Preincubate SAM (2mM), GTP (2mM), MoaC (80μM), and MgCl2 (2mM) in buffer A supplemented with 5mM DTT at 25°C in the glove box for 5min.
Initiate the reaction by mixing equal volumes of solutions from steps 1 to 2 and incubate the reaction at 25°C.
Quench the reaction by mixing 90μL reaction system with 10μL 25% (w/v) TCA at different time points during the course of 60min. The typical time points are 2, 4, 6, 8, 10, 30, and 60min.
For quantification of cPMP, use 50μL of the quenched samples and follow steps 4–8 in Section 2.4.2.
For quantification of 5′-dA, centrifuge the remaining quenched samples at 16,000 × g for 10min to remove precipitants.
Analyze the supernatant from step 6 by the HPLC system equipped with an ODS hypersil column, 250 × 4.6mm2, 3μm. Chromatography is performed by a linear gradient of 0%–15% methanol in 27mM KH2PO4, pH 4.5 in 25min with a flow rate of 2mL/min and monitored by UV absorption (256nm). Under this condition, 5′-dA elutes out at ~3.8min.
To determine a standard curve for 5′-dA quantitation, inject different amounts of 5′-dA standards (e.g., 50, 100, 150, 200, and 250pmol) to the HPLC system and analyze under the conditions described in step 7. From the resulting HPLC traces, determine the 5′-dA peak areas and plot against the amount of injection.
Determine the amounts of 5′-dA and cPMP formed at each time point using the standard curves. Stoichiometric formation of 5′-dA and 3′,8-cH2GTP is expected (Fig. 5D).
3. PREPARATION AND CHARACTERIZATION OF MoaC
MoaC catalyzes the complicated rearrangement of 3′,8-cH2GTP into cPMP (see Fig. 2B) (Hover et al., 2013). MoaC from two bacteria E. coli (Hover et al., 2015) and S. aureus (Hover et al., 2013), and MOCS1B, a human homolog of MoaC (Hover et al., 2013), have been functionally characterized. As shown in Table 1, the kcat of His-tagged S. aureus MoaC with purified 3′,8-cH2GTP is ~4 folds greater than that for S. aureus MoaA (Hover et al., 2013). The kcat of His-tagged E. coli MoaC is even greater, although the rate for E. coli MoaA is unknown because E. coli MoaA has not been successfully expressed as a soluble protein (Hover et al., 2015). In addition, the Km values toward 3′,8-cH2GTP is <0.060, 0.21–0.25, and0.79μM for His-tagged S. aureus, E. coli MoaC, and human MOCS1B, respectively (Hover et al., 2013), suggesting the recognition of 3′,8-cH2GTP at nM concentrations regardless of the organismal origins. These observations suggest that the rate-determining step of cPMP formation is likely the MoaA catalysis and explain the absence of accumulation of 3′,8-cH2GTP when both enzymes are present in vitro or in vivo.
Table 1.
Steady-State Kinetic Parameters of MoaA and MoaC From Different Organisms (Hover et al., 2013, 2015)
| Enzyme | Organism | Tag | Substrate | Km (μM) | Kcat (min−1) |
|---|---|---|---|---|---|
| MoaA | S. aureus | His-tagged | GTP | 1.4 ± 0.2 | 0.045 ± 0.003 |
| SAM | 4.1 ± 1.3 | 0.043 ± 0.004 | |||
| MoaC | S. aureus | His-tagged | 3′,8-cH2GTP | <0.060 | 0.17 ± 0.026 |
| E. coli | His-tagged | 3′,8-cH2GTP | 0.21 ± 0.058 | 0.56 ± 0.027 | |
| Nontagged | 3′,8-cH2GTP | 0.25 ± 0.040 | 0.53 ± 0.061 | ||
| MOCS1B | Homo sapiens | His-tagged | 3′,8-cH2GTP | 0.79 ± 0.24 | 0.092 ± 0.020 |
The crystal structures have been reported for MoaC from E. coli (Hover et al., 2015; Wuebbens et al., 2000), M. tuberculosis (Srivastava, Srivastava, Arora, & Pratap, 2012), and an archaea Sulfolobus tokodaii (Yoshida, Yamada, Kuramitsu, & Kamitori, 2008). Based on these crystal structures, all MoaC adopts a hexameric structure, organized as a trimer of homodimers. The active site is located at the interface of two monomers in a dimer unit and consisted of six strictly conserved and catalytically essential amino acid residues (Hover et al., 2015).
Since MoaC is not sensitive to oxygen, the enzyme can be purified under aerobic conditions. The activity assays, on the other hand, should be performed under anaerobic conditions because 3′,8-cH2GTP is oxygen sensitive. Therefore, the purified MoaC is degassed and assayed in the anaerobic glove box. The ability to prepare and assay MoaC is critical for elucidation of cPMP biosynthesis, as this enzyme is responsible for the complicated rearrangement reaction of 3′,8-cH2GTP into cPMP. Also, MoaC is used for quantitative detection of 3′,8-cH2GTP (Section 4.1.2), as well as the coupled assay of MoaA (Section 2.4.2). Therefore, in this section, we will describe the preparation and characterization of E. coli MoaC as a model.
3.1. Expression and Purification of MoaC
The recombinant E. coli MoaC can be homologously expressed in E. coli with robust expression level and solubility. While E. coli MoaC was initially purified without an affinity tag by taking advantage of its uniquely hydro-phobic nature (Wuebbens et al., 2000), we have demonstrated that N-terminal His-tag provides even shorter purification without significantly affecting the activity or crystallization (Hover et al., 2015). The His-tagged and nontagged E. coli MoaC exhibited essentially indistinguishable kinetic parameters (Table 1) (Hover et al., 2015). Therefore, here we describe His-tag affinity purification of E. coli MoaC.
Subclone E. coli moaC gene into the NcoI and BamHI sites of the pET30b vector to give pET30b-HisMoaC (Hover et al., 2015).
Transform E. coli BL21(DE3) with pET30b-HisMoaC plasmid and culture the transformant overnight on LB agar plate with 50mg/L kanamycin (Kan) at 37°C.
Pick up a single colony and culture overnight in 5mL of LB medium (50mg/L Kan) at 37°C.
Inoculate 250mL of LB medium (50mg/L Kan) with the 5mL of overnight culture and incubate at 37°C, 200rpm until saturation (~18 h).
Inoculate each of 6 × 1.5L of LB medium (50mg/L Kan) with 40mL of saturated overnight culture and incubate at 37°C, 200rpm.
When OD600nm reaches 0.9–1.1, induce the overexpression by adding IPTG to the final concentration of 0.5mM. Continue the culture at 37°C, 200rpm for 4–5 h.
Harvest the cells by centrifugation at 5000 × g, 20min, 4°C. The 9L culture typically yields ~30g of cell paste. The cell paste can be stored at −20°C.
Resuspend the cell pellet with 75mL of buffer A supplemented with 40mM imidazole and 3mM βME.
Lyse the cells by two passages through French Press at 14,000psi.
Remove the cell debris by centrifugation at 25,000 × g for 30min at 4°C.
To remove DNA, add 0.2 volumes of 6% (w/v) streptomycin sulfate in buffer A dropwise over 15min at 4°C and stir the solution at 4°C for another 15min.
Remove the precipitation by centrifugation at 25,000 × g for 20min at 4°C.
Load the supernatant onto a Ni-NTA column (~15mL resin).
Wash the column with buffer A supplemented with 40mM imidazole and 3mM βME until no protein is eluted based on Bradford assay (typically need ~200mL buffer).
Elute MoaC with buffer A supplemented with 400mM imidazole and 3mM βME until no protein is eluted based on Bradford assay (typically need ~30mL buffer).
Concentrate the purified MoaC to less than 15mL by ultrafiltration (3kDa MWCO).
To remove imidazole, load the protein solution onto a ~150mL Sephadex G-25 column equilibrated in buffer A and elute MoaC with buffer A. Check the fractions by Bradford assay and pool the fractions containing MoaC.
Analyze each step of purification by SDS-PAGE to make sure the purity and recovery (Fig. 6A).
Determine the UV–vis absorption spectra of purified MoaC by scanning from 700 to 200nm (Fig. 6B). Pure MoaC should not absorb above 300nm.
Concentrate MoaC to 0.8–1mM by ultrafiltration (3kDa MWCO). MoaC is stable for storage at these concentrations.
Remove oxygen from the MoaC solution by exchanging the atmosphere to Ar using a Schlenk line by brief evacuation (<30s) followed by equilibration for 5min. Repeat this cycle four times. During the evacuation, it is critical to minimize formation of bubbles to avoid unfolding of MoaC. Repeat this cycle for four times.
Evacuate the flask after the last cycle of step 21 and bring the sample into the glove box.
Determine MoaC concentration by the Edelhoch’s method (ε280nm = 7370M−1 cm−1) (Gill & Von Hippel, 1989; Pace et al., 1995). Typically, 9–10mg of MoaC is purified from each gram of wet cell paste.
Aliquot into appropriate volume, flash–freeze in liquid nitrogen and store at −80°C.
Fig. 6.
Purification of E. coli MoaC. (A) SDS-PAGE of MoaC at different purification steps. Lanes: 1, crude cell lysate; 2, Ni-NTA flow through; 3, Ni-NTA wash with 40mM imidazole; 4, Ni-NTA elution with 400mM imidazole; 5, after G-25; and 6, molecular weight marker.(B) UV–vis absorption spectrum of MoaC.
3.2. Activity Assay of MoaC Using a Crude MoaA Product
As described earlier, the kcat value for MoaC is much greater than that for MoaA. As a result, in the presence of both enzymes, the rate of MoaC catalysis is masked by the rate-determining MoaA reaction. Therefore, the coupled assay described in Section 2.4.2 cannot be used to determine the activity of MoaC. Thus, we designed a stepwise assay which uses a crude MoaA product as MoaC substrate. In this assay, MoaA reaction is performed in the absence of MoaC and the resulting solution is passed through ultra-filtration. The resulting flow through fraction contains 3′,8-cH2GTP. While this fraction also contains SAM, 5′-dA, L-methionine, GTP, and SDT, these molecules do not affect MoaC at the concentrations used for the MoaA assay. Therefore, this crude 3′,8-cH2GTP can be used for MoaC characterization in the absence of purified 3′,8-cH2GTP. The concentration of 3′,8-cH2GTP can be determined by the DMPT derivatization, and the MoaC product, cPMP, can be quantified by HPLC after its chemical derivatization to compound Z.
Perform 5mL MoaA reaction using 200μM MoaA, 200μM GTP, 1mM SAM, and 1mM SDT in buffer A with 5mM DTT. Incubate the reaction at 25°C in a glove box for 1h.
Remove MoaA by ultrafiltration (3kDa MWCO).
Collect the small molecule fraction (typically around 4mL). This crude MoaA product can be aliquoted, frozen in liquid nitrogen, and stored at −80°C.
Take out 40μL of the small molecule fraction and mix with 5μL 0.5M HCl. Quantify 3′,8-cH2GTP by DMPT assay as described in Section 2.4.1 (typically contains 60–100μM 3′,8-cH2GTP).
Initiate MoaC reaction using 0.1μM MoaC, 10μM crude MoaA product in buffer A with 5mM DTT and incubate the reaction at 25°C in a glove box.
Quench the reaction by mixing 45μL of reaction mixture with 5μL of 25% (w/v) TCA at different time points (e.g., 0.5, 1, 1.5, 2, 3, 5, 8, and 10min).
Quantify the amount of cPMP by chemical derivatization as described in Section 2.4.2 (steps 4–8).
4. ISOLATION AND CHARACTERIZATION OF 3′,8-cH2GTP
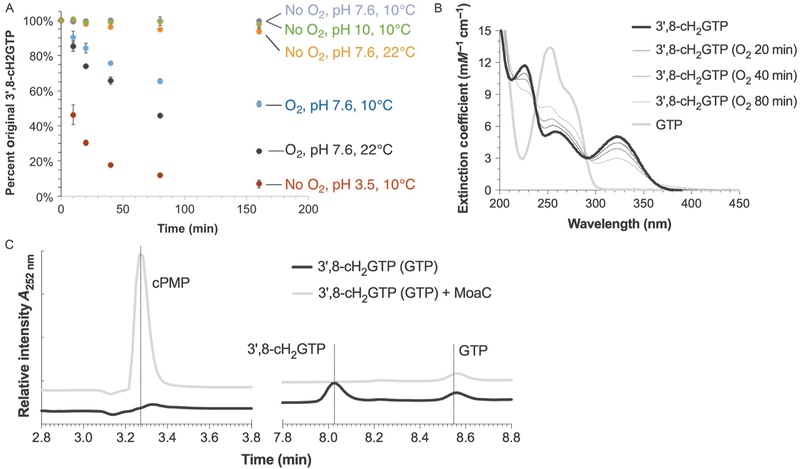
Although the presence of the MoaA product was implied in 2006 (Hänzelmann & Schindelin, 2006), the compound was not isolated and structurally characterized until our report in 2013 (Hover et al., 2013). In our study, it quickly became apparent that the MoaA product is not chemically stable. To identify the conditions in which the MoaA product can be handled, it was critical to understand its chemical properties. Therefore, we performed a stability test (Fig. 7A) (Hover et al., 2013), in which the MoaA product was incubated under various conditions. Such investigation revealed that the compound was particularly sensitive to the presence of oxygen or at acidic pH. Temperature also played some role because the degradation was significantly slower at lower temperatures. In contrast, no degradation was detectable for at least 160min under an anaerobic condition at pH 9 and 10°C. These initial experiments formed the basis for the conditions to handle the MoaA product.
Fig. 7.
Detection and quantitation of 3′,8-cH2GTP. (A) Stability test of 3′,8-cH2GTP. Crude MoaA product was incubated under different conditions and quantified after its conversion to cPMP by MoaC. (B) UV–vis absorption spectra of 3′,8-cH2GTP (solid black line) and GTP (solid gray line). The thin lines represent the spectra of 3′,8-cH2GTP after exposure to O2 for the specified period. (C) Anaerobic HPAEC analysis of 3′,8-cH2GTP, cPMP, and GTP monitored at 252nm. Panel (A) was reprinted with permission from Hover, B. M., Loksztejn, A., Ribeiro, A. A., & Yokoyama, K. (2013). Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. Journal of the American Chemical Society, 135(18), 7019–7032. doi:10.1021/ja401781t. Copyright 2013 American Chemical Society.
For the successful purification, facile approaches were required to quantitatively detect the MoaA product to identify the fractions after each chromatography step. Thus, we developed three approaches: UV–vis absorption spectra, MoaC assay, and anaerobic high-performance anion exchange chromatography (HPAEC). As detailed later, these assays have complementary benefits, and therefore, a combination of these assays allowed us to identify fractions containing the MoaA product after each chromatography.
Even after the above-described developments, when we attempted isolation of 3′,8-cH2GTP, we encountered two major problems. First, the rate of MoaA product formation was slow and did not exceed the amount of MoaA even after 60min of incubation (see Fig. 5C). Prolonged incubation resulted in the competition between the formation of 3′,8-cH2GTP and its decomposition. Second, the separation of the MoaA product with some of the components in the regular assay conditions, such as DTT, glycerol, and GTP, was challenging. As detailed in Section 4.2, these challenges were overcome by optimizing the MoaA reaction conditions.
In this section, we will describe protocols for the quantitative detection of 3′,8-cH2GTP, the optimized protocol for the 3′,8-cH2GTP isolation and the structural characterization of 3′,8-cH2GTP. We also describe our newly developed in situ NMR characterization, which allows structural characterization of the MoaA product without its isolation. These protocols will allow comprehensive characterization of the long-sought MoaA product, 3′,8-cH2GTP, and can be used to characterize MoaA and MoaC homologs from other organisms.
4.1. Detection and Quantitation of 3′,8-cH2GTP
We established three methods for the detection and quantitation informative during the isolation of 3′,8-cH2GTP. The first method uses UV–vis spectroscopy. 3′,8-cH2GTP absorbs light at 322nm, which is well outside the envelope of GTP, and thus detectable even in samples dominated by GTP. The second method incubates 3′,8-cH2GTP with MoaC and detects cPMP after its conversion to compound Z. The third approach is an anaerobic HPAEC for quantitative detection of the intact 3′,8-cH2GTP without derivatization.
4.1.1. Detection by UV–Vis Absorption Spectroscopy
Based on the unique UV absorption feature of 3′,8-cH2GTP at 322nm, UV–vis spectroscopy is an easy way to detect the compound. Fig. 7B shows the UV–vis absorption spectrum of 3′,8-cH2GTP, together with the UV spectra of GTP which does not have UV absorbance above 300nm. In the presence of oxygen, 3′,8-cH2GTP has a relatively short half-life (~70min at 10°C, Fig. 7A), and thus the detection has to be either quick or under anaerobic conditions. We found Nanodrop 1000 (Thermo Fisher Scientific) to be a convenient instrument to test multiple fractions quickly and it consumes only 1μL of solution for each measurement. 3′,8-cH2GTP can be quantified using the reported extinction coefficient (5.1 1.1mM−1 cm−1) at its λmax (322nm) (Hover et al., 2013). For a more accurate determination, we recommend the use of wavelength scanning UV–vis absorption spectrometer (U-3900H, Hitachi High Technologies America Inc., TX) and a septum-sealed screw-cap quartz cuvette (Starna Cells Inc., CA) to protect the anaerobic samples from the atmosphere.
The UV–vis absorption spectra can also be used to roughly estimate the purity of the compound. The isolated MoaA product has two major absorption bands at 252 and 322nm with the extinction coefficients of 5.8 and 5.1mM−1 cm−1, respectively. Since GTP absorbs at 252nm but not at 322nm, the contamination of GTP can be measured based on the ratio of A252nm/A322nm. An increase of this ratio from 1.0 ± 0.1 indicates a GTP contamination.
It is important to note that the 322nm absorption feature is derived from the base moiety, and thus this analysis does not distinguish di- and monophosphates (3′,8-cH2GDP and 3′,8-cH2GMP, respectively), which we frequently detected in our purification (see Section 4.2, for details). Therefore, complementary assays are required to distinguish different phosphor-ylation states.
4.1.2. Detection by MoaC Assay
More accurate amount of 3′,8-cH2GTP can be determined based on MoaC assays. In a typical analysis, 10μL of a sample is incubated with 80μL of 20μM MoaC in buffer A at 25°C for 30min to make sure complete conversion to cPMP. The reaction is then quenched by TCA and chemically derivatized using the method described in Section 2.4.2 (steps 4–8). Since the HPLC analysis is time consuming (15–20min per sample), this assay is usually performed during lyophilization of the pooled fractions (see Section 4.2). 3′,8-cH2GDP and 3′,8-cH2GMP are not the substrate of MoaC, and thus this MoaC assay distinguishes these decomposed compounds from 3′,8-cH2GTP and allows accurate quantitation of 3′,8-cH2GTP in each fraction.
4.1.3. Detection by Anaerobic Anion Exchange HPLC (HPAEC)
HPLC under anaerobic conditions can be used to characterize the intact 3′,8-cH2GTP. While we previously reported an ion-pairing HPLC under anaerobic conditions to detect 3′,8-cH2GTP without derivatization (Hover et al., 2015), this approach gives a significant fluctuation in the retention times as well as relatively broad peak shapes, which limits the sensitivity. Therefore, we recently developed an anaerobic anion exchange HPLC method. In particular, we used Dionex ICS-5000+ HPAEC system equipped with DNAPac PA-100 column (4 × 250mm2, 2.7μm), a column specialized for separation of nucleotides. This system features the ability to place the solvents under the inert gas atmosphere, allowing chromatography under anaerobic conditions.
Fig. 7C shows our representative analysis of 3′,8-cH2GTP, GTP, and cPMP. Using GTP as a concentration standard, the amount of 3′,8-cH2GTP could also be estimated even in the absence of purified 3′,8-cH2GTP (note that accurate quantitation still requires purified 3′,8-cH2GTP as the standard). Based on such quantitation, the purity of 3′,8-cH2GTP can be determined. Thus, this HPAEC approach is complementary to the two methods described earlier.
Inside the glove box, place aliquots (~30μL) of purification fractions into septum-sealed screw-cap HPLC vials with inserts for small volume samples.
Take out the sample vials and place into the autosampler rack maintained at 4°C.
Using an autosampler, inject an aliquot (5–25μL) of the sample into an HPAEC system (Dionex ICS-5000+) equipped with a DNAPac PA100 column. The system and the column should be equilibrated in anaerobic H2O for at least 30min at 1.5mL/min.
Perform chromatography by a gradient of 0–50% anaerobic 1 M NH4OAc, pH 6 at 1mL/min over 11min, and monitor the elution by an ICS-3000 PDA diode array. Under these conditions, 3′,8-cH2GTP elutes at ~8.0min, GTP at ~8.6min, and cPMP at ~3.2min (Fig. 7C).
Using the diode array data, identify peaks with UV–vis absorption spectra that agree with 3′,8-cH2GTP (λmax=322nm, Fig. 7B).
Using the chromatograms at 252nm, determine the areas for the 3′,8-cH2GTP peaks.
Analyze GTP standards with a series of injection amounts (e.g., 10, 30, 50, 70, and 90pmol) by HPAEC under the conditions identical to step 1.
Using the chromatograms at 252nm, determine the areas for the GTP peaks and compute the standard curve.
Using the extinction coefficient of GTP (13.6mM−1 cm−1) and 3′,8-cH2GTP (5.3 ± 1.1mM−1 cm−1) at 252nm (Hover et al., 2013), and the GTP standard curve determined in step 8, quantify the peak areas for 3′,8-cH2GTP.
4.2. Isolation of 3′,8-cH2GTP
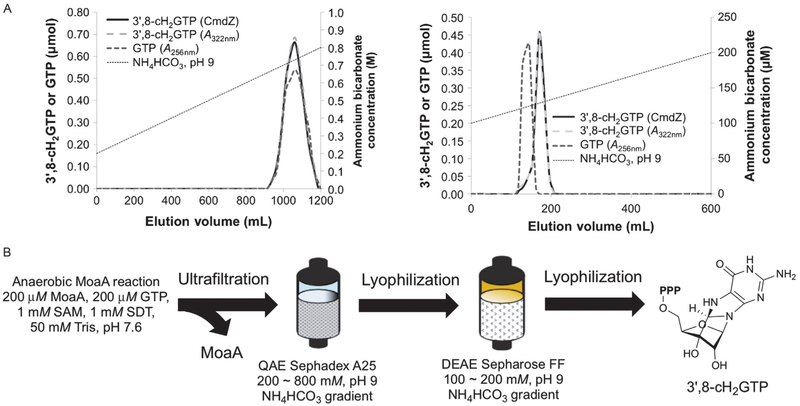
As described earlier, we met two major challenges during the isolation of 3′,8-cH2GTP, the low turnover number of MoaA and the difficulty in the separation of 3′,8-cH2GTP from other components in the assays. These problems were solved based on following optimizations. First, the concentration of GTP for the MoaA reaction was optimized to minimize the residual GTP after the reaction. Based on our investigation, the use of a stoichiometric amount of GTP and MoaA (200μM) gave the maximum (~50%) conversion of GTP to 3′,8-cH2GTP. Second, the assay buffer was modified by eliminating DTT and glycerol that were difficult to separate from 3′,8-cH2GTP. Glycerol, in particular, binds to anion exchange resin, elutes broadly, and contaminates the sample. We found that MoaA remains active at least for ~1h without the stabilizing reagents (note that longer-term storage of MoaA requires DTT and glycerol). Therefore, we exchanged MoaA into 50mM Tris–HCl, pH 7.6 immediately before the MoaA reaction. When MoaA reaction was performed in this buffer, no molecules other than GTP migrated close to 3′,8-cH2GTP during the QAE Sephadex A25 anion exchange chromatography (Fig. 8A, left). The separation of GTP and 3′,8-cH2GTP is still close, but they are separable on DEAE Sepharose FF column when GTP consumption is maximized by limiting the initial concentration of GTP (Fig. 8A, right) (Hover et al., 2013).
Fig. 8.
Isolation of 3′,8-cH2GTP. (A) Chromatograms of 3′,8-cH2GTP purification on QAE Sephadex A25 (left) and DEAE Sepharose FF (right) columns. The gradient of NH4HCO3 (pH 9) buffer for each chromatography is indicated by dotted lines. The amount of 3′,8-cH2GTP is quantified by either UV absorbance (gray dashed lines) at 322nm or MoaC assay (black solid lines). The amount of GTP is quantified by UV absorbance at 256nm after subtraction of the contribution from 3′,8-cH2GTP based on its extinction coefficient at 256nm (black dashed lines). (B) Overall scheme for the 3′,8-cH2GTP isolation. Panel (A) is reprinted with permission from Hover, B. M., Loksztejn, A., Ribeiro, A. A., & Yokoyama, K. (2013). Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. Journal of the American Chemical Society, 135(18), 7019–7032. doi:10.1021/ja401781t. Copyright 2013 American Chemical Society.
Fig. 8B shows the overview of the purification procedure, which typically provides >90% pure 3′,8-cH2GTP in a 7.5–12.5% overall yield from GTP. The current major sources of loss are (1) the hydrolysis of 3′,8-cH2GTP to 3′,8-cH2GDP and 3′,8-cH2GMP, (2) ultrafiltration, in which some of 3′,8-cH2GTP binds MoaA and does not come through the membrane, and (3) the DEAE chromatography, where 3′,8-cH2GTP elution partially overlaps with GTP. The hydrolysis is particularly problematic, and its cause is still unknown. Our current hypothesis is that it is caused by a minor contamination of E. coli endogenous phosphatase activity because the extent of hydrolysis varies by the MoaA preparation. The use of purer MoaA in the future may solve the problem. The protocol described below is our current optimized conditions, in which all the problems discussed earlier have been taken into consideration.
Prepare the following degassed buffer: 1.5L of 1M NH4HCO3 (pH9.0), 3L of H2O, 1L of 50mM Tris–HCl buffer (pH 7.6).
Prepare the following degassed resin: Sephadex G-25 resin (~1L, equilibrated in 50mM Tris–HCl buffer, pH 7.6), QAE Sephadex A25 resin (~300mL, equilibrated in 200mM NH4HCO3, pH 9.0), and DEAE Sepharose FF resin (~20mL, equilibrated in 100mM NH4HCO3, pH 9.0).
Exchange MoaA into 50mM Tris–HCl buffer (pH 7.6) by the degassed Sephadex G-25 column.
Perform a large-scale MoaA reaction in the glove box. 200μM MoaA (~1.6g) is incubated with 200μM GTP, 1mM SAM, and 1mM SDT in 200mL of 50mM Tris–HCl buffer (pH 7.6) at 25°C in a metal beads bath for 60min. (This scale of reaction requires MoaA from two large-scale preparation of MoaA with 200g of E. coli cell paste as described earlier.)
Pass the reaction solution through an Amicon 30kDa MWCO filtration membrane (Millipore) and collect the flow through.
Wash with 30mL of ddH2O for four times and combine the flow through.
Save an aliquot (10μL) from steps 4–6 to perform MoaC assay.
Apply the combined flow through fractions onto the QAE Sephadex A25 column (300mL) and wash with the column 750mL of H2O.
Elute the bound nucleotides by a 1200mL linear gradient of 200–800mM NH4HCO3 buffer and fractionate the elution every 14mL using a fraction collector (AKTA frac-920).
Identify fractions containing 3′,8-cH2GTP by Nanodrop. 3′,8-cH2GTP usually comigrates with GTP. Save an aliquot (10μL) from each fraction to perform MoaC assay.
Combine the fractions containing 3′,8-cH2GTP and freeze in liquid nitrogen.
Take out from the glove box and lyophilize.
While lyophilizing, perform MoaC assays for the 10μL aliquots saved in steps 7 and 10 by following the protocol described in Section 4.1.2 to make sure the detected 322nm absorption in these fractions indeed derives from 3′,8-cH2GTP and quantify its amount.
Bring the lyophilized fractions back to the glove box and dissolve the powder in 3–5mL of 100mM NH4HCO3 buffer in the glove box.
Apply the solution to a 15mL DEAE Sepharose FF column and wash the column with 50mL of 100mM NH4HCO3 buffer.
Elute 3′,8-cH2GTP with a 600mL linear gradient of 100–200mM NH4HCO3 buffer and fractionate the elution every 9mL using a fraction collector (AKTA Frac-920).
Check the UV–vis absorption spectra for each fraction. Fractions with pure 3′,8-cH2GTP should exhibit the A252nm/A322nm ratio of 1.0 ± 0.1. GTP contamination increases this ratio. Save an aliquot (10μL) from each fraction to perform MoaC assay.
Freeze individual fractions containing pure 3′,8-cH2GTP in liquid nitrogen.
Take out from the glove box and lyophilize.
While lyophilizing, perform MoaC assays for the 10μL aliquots saved in step 17 by following the protocol described in Section 4.1.2 to quantify the amount of 3′,8-cH2GTP. Alternatively, this step can be replaced by the anaerobic HPAEC analysis (Section 4.1.3), for which 40μL aliquots should be saved in step 17.
Dissolve the lyophilized fractions with pure 3′,8-cH2GTP in H2O.
Measure the final concentration by the MoaC assay and UV absorption at 322nm (ε322nm =5.1 ± 1.1mM−1cm−1).
Determine the purity based on the anaerobic HPAEC analysis (Section 4.1.3). Typically, this protocol provides 3–5μmol of 3′,8-cH2GTP (7.5%–12.5% yield) with a purity >90%.
Aliquot, flash–freeze in liquid nitrogen, and store the purified 3′,8-cH2GTP at −80°C.
4.3. Structural Characterization of 3′,8-cH2GTP
The structural characterization of 3′,8-cH2GTP can be performed using MS and NMR spectroscopy. For NMR, it is recommended to use uniformly 13C-labeled GTP ([U-13C10]GTP), whereas the MS analysis should be performed with natural isotope abundance to avoid any complication from partial labeling.
4.3.1. Mass Spectroscopy
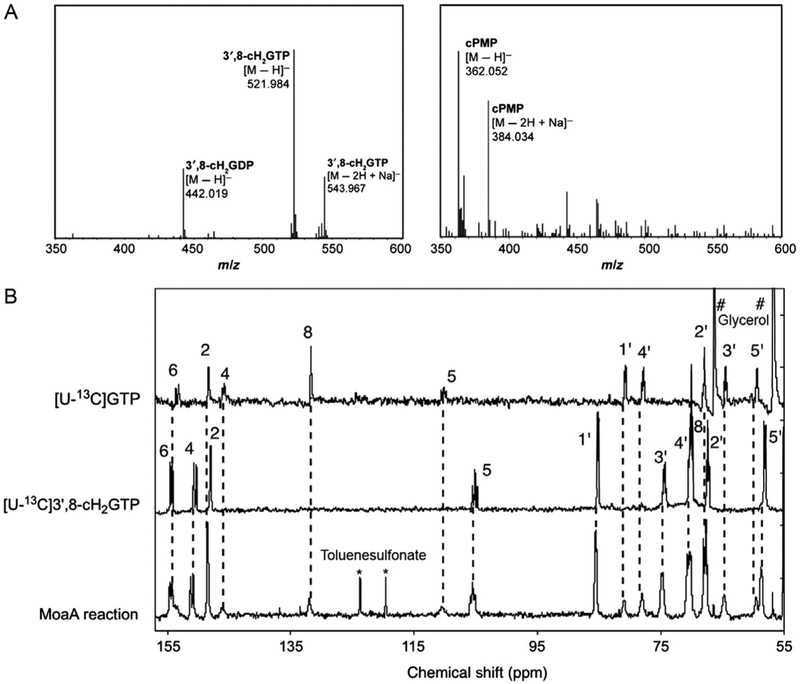
The molecular weight of purified 3′,8-cH2GTP can be determined by a high-resolution electrospray ionization time-of-flight mass spectrometry (ESI-TOF-MS), which revealed m/z=521.984 [M H]− (Fig. 9A, left). This mass signal is not only consistent with the molecular weight of 3′,8-cH2GTP but also with that of GTP which migrates very close to 3′,8-cH2GTP during the purification. To rule out the possibility that the signal is derived from contaminated GTP, incubate the sample with MoaC, remove MoaC by passing through 3kDa MWCO membrane, and then analyze by MS. The original peak at 521.984 should disappear, with a new peak forming at m/z=362.052 [M H]− (Fig. 9A, right), consistent with cPMP. These analyses will confirm the identity of the original mass signal indeed comes from 3′,8-cH2GTP.
Fig. 9.
Characterization of the MoaA product, 3′,8-cH2GTP. (A) ESI-TOF-MS spectra of isolated 3′,8-cH2GTP (left) and the same sample incubated with MoaC (right). (B) In situ 13C NMR spectra. Shown are the spectra for the MoaA reaction with [U-13C]GTP (bottom), [U-13C]3′,8-cH2GTP (middle), and [U-13C]GTP (top) standards. * and # are signals derived from toluenesulfonate and glycerol, respectively. Panel (A) is reprinted with permission from Hover, B. M., Loksztejn, A., Ribeiro, A. A., & Yokoyama, K. (2013). Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. Journal of the American Chemical Society, 135(18), 7019–7032. doi:10.1021/ja401781t. Copyright 2013 American Chemical Society. Panel (B) was adapted with permission from Hover, B. M., Tonthat, N. K., Schumacher, M. A., & Yokoyama, K. (2015). Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis. Proceedings of the National Academy of Sciences of the United States of America, 112(20), 6347–6352. doi:10.1073/pnas.1500697112.
4.3.2. NMR
Detailed structural characterization is mainly accomplished using NMR. To facilitate the analysis, uniformly 13C-labeled 3′,8-cH2GTP ([U-13C]3′,8-cH2GTP) is prepared from [U-13C]GTP (Sigma-Aldrich) by following the protocol identical to that for the nonlabeled compound (Section 4.2). In our analysis, 13C NMR was a surprise as it demonstrated the presence of a tertiary carbon (C-3′) that was absent in any of the previously proposed MoaA product, including pyranopterin triphosphate. The most informative experiment was 13Cd13C homonuclear correlation spectroscopy (COSY), which demonstrated the covalent bridge between C-3′ and C-8, and allowed annotation of all carbon signals. Together with 1H NMR, heteronuclear multiple quantum coherence (HMQC), heteronuclear multiple bond correlation (HMBC), and nuclear Overhauser effect spectroscopy (NOESY), these experiments unambiguously determined the structure of 3′,8-cH2GTP. The details of the NMR data and analysis can be found in our previous publication (Hover et al., 2013).
4.4. In Situ NMR Characterization
The isolation and NMR characterization of the MoaA product are powerful ways to determine its structure. However, the current protocol requires a lot of materials and efforts, which may not be affordable to many researchers. Although the chemical derivatization and UV–vis spectroscopy are simple and easy to perform, they give limited information about the structure of the molecule. Therefore, we developed the in situ 13C NMR method that allows 13C NMR detection of the MoaA product without isolation or derivatization (Hover et al., 2015). In this approach, the MoaA reaction is performed with [U-13C]GTP (1mM) as the substrate and 13C NMR spectrum of the reaction mixture is determined.
Incubate MoaA (0.4mM) with [U-13C]GTP (1mM), SAM (1mM), and SDT (5mM) in buffer A supplemented with 1mM MgCl2 and 2mM DTT at 25°C in a glove box for 120min.
Add 5% (v/v) anaerobic D2O, transfer the solution to an NMR tube, and seal the tube with a rubber septum.
Perform a control reaction without MoaA and add 5% (v/v) anaerobic D2O.
Collect 13C NMR data for the two samples at 6°C for overnight (~16h).
Fig. 9B shows our representative in situ 13C NMR data. By comparing the signals from the MoaA reaction mixture (bottom) with the spectra of authentic [U-13C]GTP (top) and [U-13C]3′,8-cH2GTP (middle), all the observed signals were assigned to either GTP or 3′,8-cH2GTP except for some known impurities. In practice, [U-13C]3′,8-cH2GTP standard is not required because under these conditions, the chemical shifts of [U-13C] 3′,8-cH2GTP are highly reproducible. A comparison with the published spectra (Hover et al., 2013, 2015) is sufficient to assign all the signals derived from [U-13C]3′,8-cH2GTP. This in situ NMR method provides structural information about the MoaA product without any workup, and therefore avoids potential artifacts during isolation or chemical derivatization. Owing to the simplicity of the methodology, it can be used to test the generality of 3′,8-cH2GTP as the product of MoaA from various organisms.
5. SUMMARY AND CONCLUSION
The mechanism of biosynthesis of the pterin ring of Moco was the long-standing mystery in the enzymology and cofactor biosynthesis fields. This transformation requires two enzymes, MoaA and MoaC, but their functions had remained ambiguous since the 1990s. Recently, successful isolation and structural characterization of the long-sought MoaA product, 3′,8-cH2GTP, has finally elucidated the functions of the two enzymes and provided breakthrough insights into the early steps in the Moco biosyn-thesis. In this chapter, we described protocols for characterization of MoaA and MoaC, and the isolation of 3′,8-cH2GTP.
The discovery of 3′,8-cH2GTP was made possible based on the quantitative analyses. The quantitative activity assay of MoaA revealed its substoichiometric turnover, and the necessity to perform large-scale preparation of MoaA to isolate 3′,8-cH2GTP. The quantitative assays of MoaC allowed identification of 3′,8-cH2GTP during its purification. The quantitative analysis of 3′,8-cH2GTP was critical to identify the conditions to handle this highly oxygen-sensitive compound. Thus, the quantitative characterization protocols described in this chapter all contributed to the successful identification of 3′,8-cH2GTP.
Importantly, many of the protocols described in this chapter are based on the rich history of the biochemistry of Moco and other pterin cofactors. For example, the MoaA/MoaC coupled assay was based on the report that cPMP can be selectively and oxidatively derivatized to compound Z (Hänzelmann et al., 2004; Wuebbens & Rajagopalan, 1993). The chemical derivatization of MoaA product to DMPT was first mentioned in 2006 (Hänzelmann & Schindelin, 2006), but has its origin in the characterization of an intermediate in the GTP cyclohydrolase I catalysis (Bracher et al., 1999). Therefore, the discovery of 3′,8-cH2GTP is a result of decades of studies on Moco and other pterin cofactor biosynthesis by numbers of labs in the field. We hope that the protocols described in this chapter will be useful for the future research in the Moco biosynthesis or other related fields.
The characterization of 3′,8-cH2GTP emphasizes the importance of structural characterization of unstable intermediates existing in biosynthetic pathways. To this end, it is critical to be able to characterize the biosynthetic enzymes and the intermediates quantitatively. Therefore, while the protocols described here are specific to Moco biosynthesis, the fundamental concepts of quantitative biochemistry apply to other biosynthetic pathways and enzymes.
ABBREVIATIONS
- 3′,8-cH2GTP
3′,8-cyclo-7,8-dihydro-GTP
- 5′-dA
5′-deoxyadenosine
- 5′-dA•
5′-deoxyadenosyl radical
- Amp
ampicillin
- BSA
bovine serum albumin
- Cam
chloramphenicol
- COSY
homonuclear correlation spectroscopy
- cPMP
cyclic pyranopterin monophosphate
- DMPT
dimethylpterin
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- ENDOR
electron nuclear double resonance
- ESI-TOF-MS
electrospray ionization time-of-flight mass spectrometry
- GTP
guanosine 5′-triphosphate
- HMBC
heteronuclear multiple bond correlation
- HMQC
heteronuclear multiple quantum coherence
- HPAEC
high-performance anion exchange chromatography
- HPLC
high-performance liquid chromatography
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- Kan
kanamycin
- LB
Lysogeny broth
- LC–MS
liquid chromatography–mass spectrometry
- MoCD
molybdenum cofactor deficiency
- Moco
molybdenum cofactor
- MPT
molybdopterin
- MS
mass spectrometry
- NMR
nuclear magnetic resonance
- NOESY
nuclear Overhauser effect spectroscopy
- SAM
S-adenosyl-L-methionine
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCA
trichloroacetic acid
- TFA
trifluoroacetic acid
- βME
β-mercaptoethanol
REFERENCES
- Barr I, Latham JA, Iavarone AT, Chantarojsiri T, Hwang JD, & Klinman JP (2016). Demonstration that the radical S-adenosylmethionine (SAM) enzyme PqqE catalyzes de novo carbon–carbon cross-linking within a peptide substrate PqqA in the presence of the peptide chaperone PqqD. Journal of Biological Chemistry, 291(17), 8877–8884. https://doi.org/10.1074/jbc.C115.699918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H (1983). Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron–sulfur proteins. Analytical Biochemistry, 131(2), 373–378. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, & Barry CE 3rd. (2005). Tuberculosis—Metabolism and respiration in the absence of growth. Nature Reviews. Microbiology, 3(1), 70–80. https://doi.org/10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- Bracher A, Fischer M, Eisenreich W, Ritz H, Schramek N, Boyle P, et al. (1999). Histidine 179 mutants of GTP cyclohydrolase I catalyze the formation of 2-amino-5-formylamino-6-ribofuranosylamino-4 (3H)-pyrimidinone triphosphate. Journal of Biological Chemistry, 274(24), 16727–16735. [DOI] [PubMed] [Google Scholar]
- Burg AW, & Brown GM (1968). The biosynthesis of folic acid VIII. Purification and properties of the enzyme that catalyzes the production of formate from carbon atom 8 of guanosine triphosphate. Journal of Biological Chemistry, 243(9), 2349–2358. [PubMed] [Google Scholar]
- Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, et al. (2012). Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nature Reviews. Microbiology, 10(12), 841–851. https://doi.org/10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- Gill SC, & Von Hippel PH (1989). Calculation of protein extinction coefficients from amino acid sequence data. Analytical Biochemistry, 182(2), 319–326. [DOI] [PubMed] [Google Scholar]
- Goldman PJ, Grove TL, Booker SJ, & Drennan CL (2013). X-ray analysis of butirosin biosynthetic enzyme BtrN redefines structural motifs for AdoMet radical chemistry. Proceedings of the National Academy of Sciences of the United States of America, 110(40), 15949–15954. https://doi.org/10.1073/Pnas.1312228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove TL, Ahlum JH, Sharma P, Krebs C, & Booker SJ (2010). A consensus mechanism for radical SAM-dependent dehydrogenation? BtrN contains two [4Fe-4S] clusters. Biochemistry, 49(18), 3783–3785. https://doi.org/10.1021/bi9022126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann P, Hernandez HL, Menzel C, Garcia-Serres R, Huynh BH, Johnson MK, et al. (2004). Characterization of MOCS1A, an oxygen-sensitive iron–sulfur protein involved in human molybdenum cofactor biosynthesis. Journal of Biological Chemistry, 279(33), 34721–34732. https://doi.org/10.1074/Jbc.M313398200. [DOI] [PubMed] [Google Scholar]
- Hänzelmann P, & Schindelin H (2004). Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proceedings of the National Academy of Sciences of the United States of America, 101(35), 12870–12875. https://doi.org/10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann P, & Schindelin H (2006). Binding of 5′-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proceedings of the National Academy of Sciences of the United States of America, 103(18), 6829–6834. https://doi.org/10.1073/pnas.0510711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hover BM, Loksztejn A, Ribeiro AA, & Yokoyama K (2013). Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. Journal of the American Chemical Society, 135(18), 7019–7032. https://doi.org/10.1021/ja401781t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hover BM, Tonthat NK, Schumacher MA, & Yokoyama K (2015). Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis. Proceedings of the National Academy of Sciences of the United States of America, 112(20), 6347–6352. https://doi.org/10.1073/pnas.1500697112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hover BM, & Yokoyama K (2015). C-terminal glycine-gated radical initiation by GTP 3′,8-cyclase in the molybdenum cofactor biosynthesis. Journal of the American Chemical Society, 137(9), 3352–3359. https://doi.org/10.1021/ja512997j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurdle JG, O’Neill AJ, Chopra I, & Lee RE (2011). Targeting bacterial membrane function: An underexploited mechanism for treating persistent infections. Nature Reviews. Microbiology, 9(1), 62–75. https://doi.org/10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Waud WR, Rajagopalan KV, Duran M, Beemer FA, & Wadman SK (1980). Inborn errors of molybdenum metabolism: Combined deficiencies of sulfite oxidase and xanthine dehydrogenase in a patient lacking the molybdenum cofactor. Proceedings of the National Academy of Sciences of the United States of America, 77(6), 3715–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaujia SP, Jeyakanthan J, Nakagawa N, Balasubramaniam S, Shinkai A, Kuramitsu S, et al. (2010). Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8. Acta Crystallographica. Section D, Biological Crystallography, 66(Pt 7), 821–833. https://doi.org/10.1107/S0907444910019074. [DOI] [PubMed] [Google Scholar]
- Lees NS, Hänzelmann P, Hernandez HL, Subramanian S, Schindelin H, Johnson MK, et al. (2009). ENDOR spectroscopy shows that guanine N1 binds to [4Fe-4S] cluster II of the S-adenosylmethionine-dependent enzyme MoaA: Mechanistic implications. Journal of the American Chemical Society, 131(26), 9184–9185. https://doi.org/10.1021/ja903978u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimkuhler S, Wuebbens MM, & Rajagopalan KV (2011). The history of the discovery of the molybdenum cofactor and novel aspects of its biosynthesis in bacteria. Coordination Chemistry Reviews, 255(9–10), 1129–1144. https://doi.org/10.1016/j.ccr.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanta N, Zhang Z, Hudson GA, van der Donk WA, & Mitchell DA (2017). Reconstitution and substrate specificity of the radical S-adenosyl-methionine thiazole C-methyltransferase in thiomuracin biosynthesis. Journal of the American Chemical Society, 139(12), 4310–4313. https://doi.org/10.1021/jacs.7b00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AP, Hanes JW, Abdelwahed SH, Hilmey DG, Hänzelmann P, & Begley TP (2013). Catalysis of a new ribose carbon-insertion reaction by the molybdenum cofactor biosynthetic enzyme MoaA. Biochemistry, 52(7), 1134–1136. https://doi.org/10.1021/bi3016026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel RR, & Schwarz G (2011). Molybdenum cofactor biosynthesis in plants and humans. Coordination Chemistry Reviews, 255, 1145–1158. [Google Scholar]
- Pace CN, Vajdos F, Fee L, Grimsley G, & Gray T (1995). How to measure and predict the molar absorption coefficient of a protein. Protein Science, 4(11), 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss J (2000). Genetics of molybdenum cofactor deficiency. Human Genetics, 106(2),157–163. https://doi.org/10.1007/s004399900223. [DOI] [PubMed] [Google Scholar]
- Rieder C, Eisenreich W, O’Brien J, Richter G, Gotze E, Boyle P, et al. (1998). Rearrangement reactions in the biosynthesis of molybdopterin—An NMR study with multiply 13C/15N labelled precursors. European Journal of Biochemistry, 255(1), 24–36. [DOI] [PubMed] [Google Scholar]
- Shiota T, Palumbo MP, & Tsai L (1967). A chemically prepared formamidopyrimidine derivative of guanosine triphosphate as a possible intermediate in pteridine biosynthesis. Journal of Biological Chemistry, 242(8), 1961–1969. [PubMed] [Google Scholar]
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, & Miller NE (2001). Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms-functional characterization using new analysis and information visualization methods. Nucleic Acids Research, 29(5), 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Srivastava VK, Arora A, & Pratap JV (2012). Overexpression, purification, crystallization and preliminary X-ray analysis of putative molybdenum cofactor biosynthesis protein C (MoaC2) from Mycobacterium tuberculosis H37Rv. Acta Crystallographica. Section F: Structural Biology and Crystallization Communications, 68(Pt 6), 687–691. https://doi.org/10.1107/S174430911201665X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne W (1988). Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods in Enzymology, 158, 357–364. [DOI] [PubMed] [Google Scholar]
- Wuebbens MM, Liu MT, Rajagopalan K, & Schindelin H (2000). Insights into molybdenum cofactor deficiency provided by the crystal structure of the molybdenum cofactor biosynthesis protein MoaC. Structure, 8(7), 709–718. [DOI] [PubMed] [Google Scholar]
- Wuebbens MM, & Rajagopalan K (1993). Structural characterization of a molybdopterin precursor. Journal of Biological Chemistry, 268(18), 13493–13498. [PubMed] [Google Scholar]
- Wuebbens MM, & Rajagopalan KV (1995). Investigation of the early steps of molybdopterin biosynthesis in Escherichia coli through the use of in vivo labeling studies. The Journal of Biological Chemistry, 270(3), 1082–1087. [DOI] [PubMed] [Google Scholar]
- Yang L, Lin G, Nelson RS, Jian Y, Telser J, & Li L (2012). Mechanistic studies of the spore photoproduct lyase via a single cysteine mutation. Biochemistry, 51(36), 7173–7188. https://doi.org/10.1021/bi3010945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Yamada M, Kuramitsu S, & Kamitori S (2008). Structure of a putative molybdenum-cofactor biosynthesis protein C (MoaC) from Sulfolobus tokodaii (ST0472). Acta Crystallographica. Section F: Structural Biology and Crystallization Communications, 64(Pt 7), 589–592. https://doi.org/10.1107/S174430910801590X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, et al. (2018). Precision editing of the gut microbiota ameliorates colitis. Nature, 553(7687), 208–211. https://doi.org/10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]