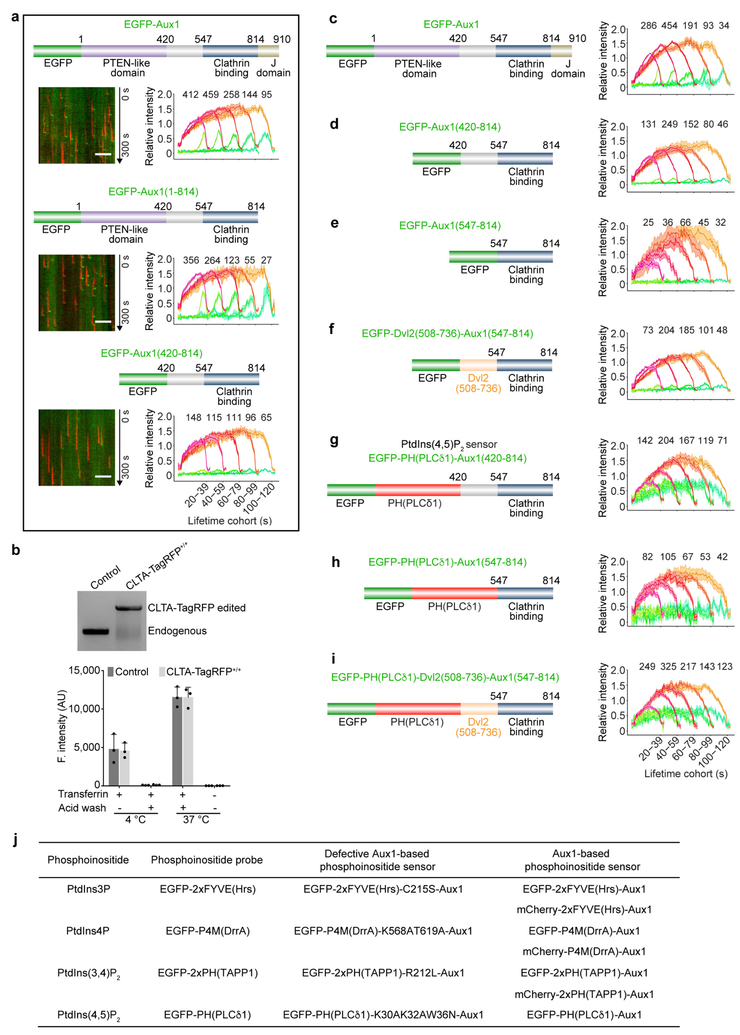
Extended Data Figure 1 |. Characterization of gene-edited SUM159 cells expressing clathrin light chain A and minimal requirements for the targeting specificity of the Aux1-based PtdIns(4,5)P2 sensor to endocytic coated pits and vesicles.
a, Schematic representation of the constructs EGFP-Auxl, EGFP-Aux1(1–814), and EGFP-Aux1(420–814) used for transient expression in CLTA-TagRFP+/+ cells. The representative kymographs are from 300-s time series imaged every 1 s by TIRF microscopy at the bottom surface of the cells. The kymographs were shifted laterally by six pixels. Scale bars, 5 μm. The plots show averaged fluorescence intensity traces (mean ± s.e.m.) of CLTA-TagRFP (red) and the Aux1 constructs (green) from endocytic clathrin-coated pits and vesicles automatically identified using the 2D tracking software in 11, 6 and 12 cells, respectively, and then grouped in cohorts according to lifetimes. Note that removal of the PTEN-like domain but not the J domain largely inhibited the burst-like recruitment of Aux1. The numbers of analysed traces are shown above each cohort. Data are representative of at least two independent experiments. b, Genomic PCR analysis showing biallelic integration of the TagRFP sequence into the CLTA genomic locus in the SUM159 clonal gene-edited CLTA-TagRFP+/+ cell line (top), representative of three independent experiments. Transferrin uptake comparing parental, unedited SUM159 control cells and the gene-edited CTLA-TagRFP+/+ cells determined by flow cytometry (n = 3 independent experiments, mean ± s.d.) (bottom). c-e, Schematic representation of the constructs EGFP-Aux1, EGFP-Aux1(420–814) and EGFP-Aux1(547–814) used for transient expression in gene-edited AP2-TagRFP+/+ SUM159 cells and then imaged by TIRF microscopy. The plots show averaged fluorescence intensity traces (mean ± s.e.m.) of AP2-TagRFP+/+ (red) and of the Aux1 constructs (green) from endocytic clathrin-coated pits and vesicles identified in 5, 6 and 6 cells, respectively, and then grouped in cohorts according to lifetimes. Note that removal of residues 420–546 from EGFP-Aux1(420–814) has largely inhibited its small, burst-like recruitment. The numbers of analysed traces are shown above each cohort. Data are representative of two independent experiments. f, A linker derived from the unstructured region (residues 508–736) of Dishevelled2 (Dvl2) was inserted between EGFP and Aux1(547–814) to make the construct EGFP-Dvl2(508–736)-Aux1(547–814), which showed very small burst-like recruitment, similar to that of EGFP-Aux1(420–814), which also lacked the PTEN-like domain (n = 5 cells). Data are representative of two independent experiments. g, h, The constructs EGFP-PH(PLCδ1)-Aux1(420–814) or EGFP-PH(PLCδ1)-Aux1(547–814) were used for transient expression in AP2-TagRFP+/+ cells and then imaged by TIRF microscopy. The plots show averaged fluorescence intensity traces (mean ± s.e.m.) of AP2-TagRFP+/+ (red) and of the Aux1-based PtdIns(4,5)P2 sensors (green) associated with endocytic clathrin-coated pits and vesicles identified in 9 and 6 cells, and then grouped in cohorts according to their lifetimes. Note that the removal of the linker Aux1 residues 420–546 from EGFP-PH(PLCδ1)-Aux1 (420–814) largely inhibited its recruitment to coated pits (h). The numbers of analysed traces are shown above each cohort. Data are representative of two independent experiments. i, The Dvl2(508–736) linker was inserted between EGFP-PH(PLCδ1) and Aux1(547–814) to make the PtdIns(4,5)P2 sensor EGFP-PH(PLCδ1)-Dvl2(508–736)-Aux1(547–814). This chimaera is recruited to coated pits with an efficiency similar to that of EGFP-PH(PLCδ1)-Aux1(420–814) (n = 12 cells). Data are representative of two independent experiments. j, Table summarizing general phosphoinositide probes and Aux1-based phosphoinositide sensors used in this study. The results obtained with sensors specific for PtdIns3P, PtdIns4P, PtdIns(4,5)P2, and PtdIns(3,4)P2, were obtained using the tandem FYVE domains of Hrs, the P4M domain of DrrA, the PH domain of PLCδ1 and the tandem PH domains of TAPP1, respectively.