Abstract
Despite recent advance of therapeutic development, coronary artery disease (CAD) remains one of the major issues to public health. The use of genomics and systems biology approaches to inform drug discovery and development have offered the possibilities for new target identification and in silico drug repurposing. In this study, we propose a network-based, systems pharmacology framework for target identification and drug repurposing in pharmacologic treatment and chemoprevention of CAD. Specifically, we build in silico models by integrating known drug-target interactions, CAD genes derived from the genetic and genomic studies, and the human protein-protein interactome. We demonstrate that the proposed in silico models can successfully uncover approved drugs and novel natural products in potentially treating and preventing CAD. In case studies, we highlight several approved drugs (e.g., fasudil, parecoxib, and dexamethasone) or natural products (e.g., resveratrol, luteolin, daidzein and caffeic acid) with new mechanism-of-action in chemical intervention of CAD by network analysis. In summary, this study offers a powerful systems pharmacology approach for target identification and in silico drug repurposing on CAD.
Keywords: Drug repurposing, drug-target network, systems pharmacology, genomics, protein-protein interaction, coronary artery disease, natural product
Graphical Abstract
1. INTRODUCTION
Coronary artery disease (CAD) is the leading cause of morbidity and mortality worldwide among multiple types of cardiovascular diseases [1, 2]. Although technological advances have been successfully used to develop new treatment, the mortality rate of CAD has remained unimpressive owing to the heterogeneous factors of causing disease, including genetic factors, environmental factors, and life styles [3]. According to the data from National Health and Nutrition Examination Survey (NHANES) from 2011 to 2014, an estimated 16.5 million adults (age≥20) in America suffer from CAD with the prevalence of 7.4% for males and 5.3% for females [4]. Meanwhile, the estimated direct and indirect cost of myocardial disease in United States (U.S.) is approximate 200 billion annually, placing a heavy burden for economic society [4, 5]. Currently, antihypertensive drugs, lipid reducing drugs, and antiplatelet drugs are the most common medications for treatment or prevention of myocardial diseases (e.g., CAD). However, due to heterogeneous population on CAD, current approved agents are not efficient enough to reduce the disease burden as well as mortality. Thus, it is a pressing need to develop novel treatment and chemoprevention strategies for CAD.
Drug repurposing or repositioning has been recognized as powerful approaches [6, 7] toward treatment of various complex diseases, including CAD [8]. Drug repurposing focuses on the detection and development of new clinical indication from approved drugs, which significantly shortens the time and reduces the cost from random clinical trials and drug development. In addition to approved drugs with ideal pharmacokinetics and well-known pharmacodynamics profile, natural products have been demonstrated another abundant medical resources for developing new treatment strategies [9, 10]. Chinese medicine, including various medicinal natural products, has achieved great success to the health and well-being of the people in Asia [11]. Natural products derived from Chinese medicine have been used to treat or prevent a group of diseases including cardiovascular disease [12]. Accumulating evidence has suggested that Chinese medicine is effective for stable CAD patients, which might serve as a complementary and alternative strategy to the primary prevention of CAD [13, 14]. Altogether, identification of effective treatment and chemoprevention strategies from approved drugs and natural products offer possible strategies to reduce morbidity and mortality of CAD. However, effective identification of approved drugs or natural products with novel mechanism-of-action for treatment of CAD is very challenging for traditional experimental approaches owing to lack of ideal in vitro and in vivo models.
The traditional drug discovery approach focusing on “one drug, one target, one disease” is suboptimal for leading to off-target toxicity or unintended beneficial effects. Quantitative and systems pharmacology combines computational and experimental tools toward discovering novel therapeutic agents and understanding of the therapeutic mechanisms of complex diseases [10, 15]. Recent development of systems pharmacology approaches have offered new insights into drug discovery and development, especially focusing on approved drugs (e.g., drug repurposing) and natural products [9, 16–18], offering the possibilities for development of new therapeutic agents and prevention strategies in CAD. For example, recent studies have applied systems pharmacology approaches for discovering new indications for natural products (e.g., resveratrol and quercetin) via unique integration of drug-target interaction (DTI) network and disease-associated genes, in potentially treating cancer and aging disorders [16, 17].
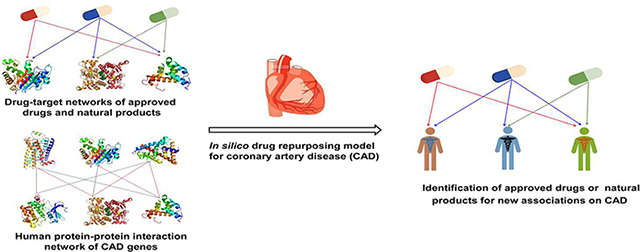
In this study, we presented an integrated systems pharmacology framework (Fig. 1) for development of new therapeutic and prevention strategies in CAD by focusing on approved drugs and natural products. Specifically, we built two drug-target networks for both approved drugs and natural products respectively via assembling experimentally validated drug-target interactions from 11 chemoinformatics and bioinformatics sources (see Methods). We then built in silico models to predict new potential associations of approved drugs or natural products on CAD by incorporating drug-target networks and known CAD gene products (proteins) from genetic and genomic studies into the human protein-protein interaction (PPI) network. We demonstrated that our systems pharmacology approaches can identify new therapeutic indications of approved drugs or uncover natural products in treatment and chemoprevention of CAD potentially. If broadly applied, the systems pharmacology-based framework presented here offers useful in silico tools for development of new treatment and chemoprevention strategies for CAD and other complex diseases.
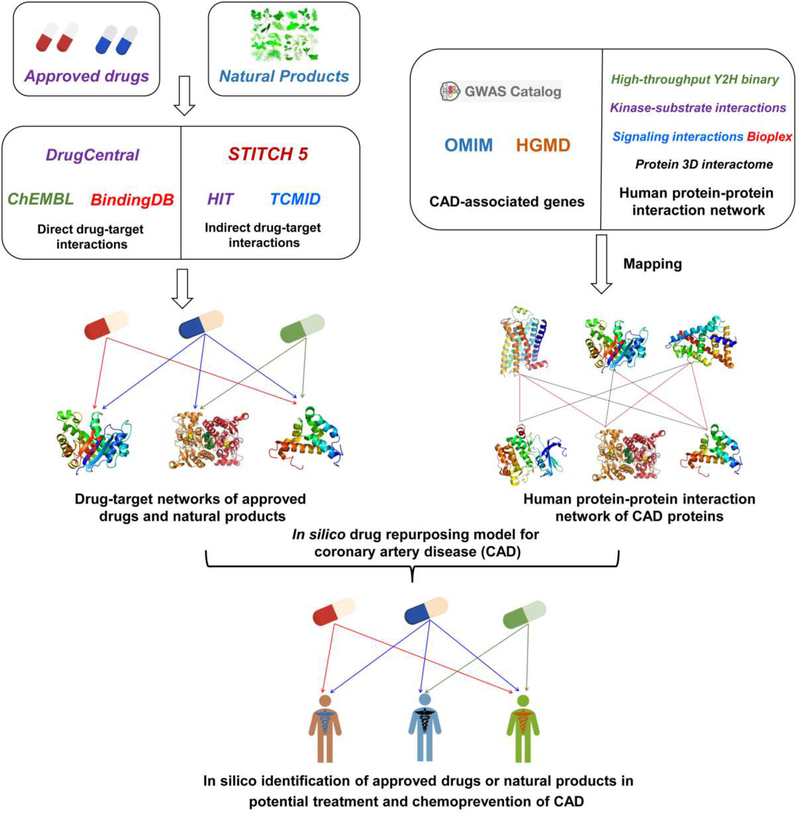
Fig. 1. Schematic diagram illustrating an integrated systems pharmacology framework for predicting new associations of approved drugs and natural products on coronary artery disease (CAD).
(A) Construction of drug-target (compound-protein) networks for both approved drugs and natural products. (B) Building a protein-protein interaction (PPI) network for CAD proteins. (C) Predicting new associations of approved drugs and natural products on CAD via in silico models (see Methods).
2. RESULTS
2.1. Drug–target network analysis in CAD
We constructed the drug-target network for approved drugs (DTnet) and the compound-protein interaction network for natural products (NPnet) respectively by uniquely integrating multiple types of experimental data (see Methods). The DTnet contains 11,333 drug-target interactions connecting 1,812 approved drugs and 1,335 human targets (proteins), and NPnet contains 6,229 compound-protein interactions connecting 500 unique natural products and 2,020 human targets (Table 1). Interestingly, the average degree (connectivity) of natural products (12.46±1.45, P = 3.069×10−6, Fig. S1) in NPnet is significantly stronger than that of approved drugs (6.25±0.33) based on data from DrugCentral database [19], suggesting a significant ‘promiscuity’ of natural products. The detailed information of two drug-target networks are provided in Table S1.
Table 1.
The statistics comparison of four drug-target (compound-protein) networks.
| Data set | ND | NT | NDTI | Sparsity (%) |
|---|---|---|---|---|
| DTnet | 1,812 | 1,335 | 11,333 | 0.468 |
| CADDTnet | 634 | 58 | 931 | 2.535 |
| NPnet | 500 | 2,020 | 6,229 | 0.617 |
| CADNPnet | 170 | 71 | 395 | 3.273 |
ND: the number of drugs in network, NT: the number of targets in network, NDTI: the number of drug-target interactions (DTIs) or compound-protein interactions, Sparsity: the ratio of NDTI to the number of all possible DTIs. CADDTnet represents the specific network connecting approved drugs with coronary artery disease (CAD) proteins derived from DTnet, while CADNPnet denotes to the specific network connecting natural products with CAD proteins extracted from NPnet.
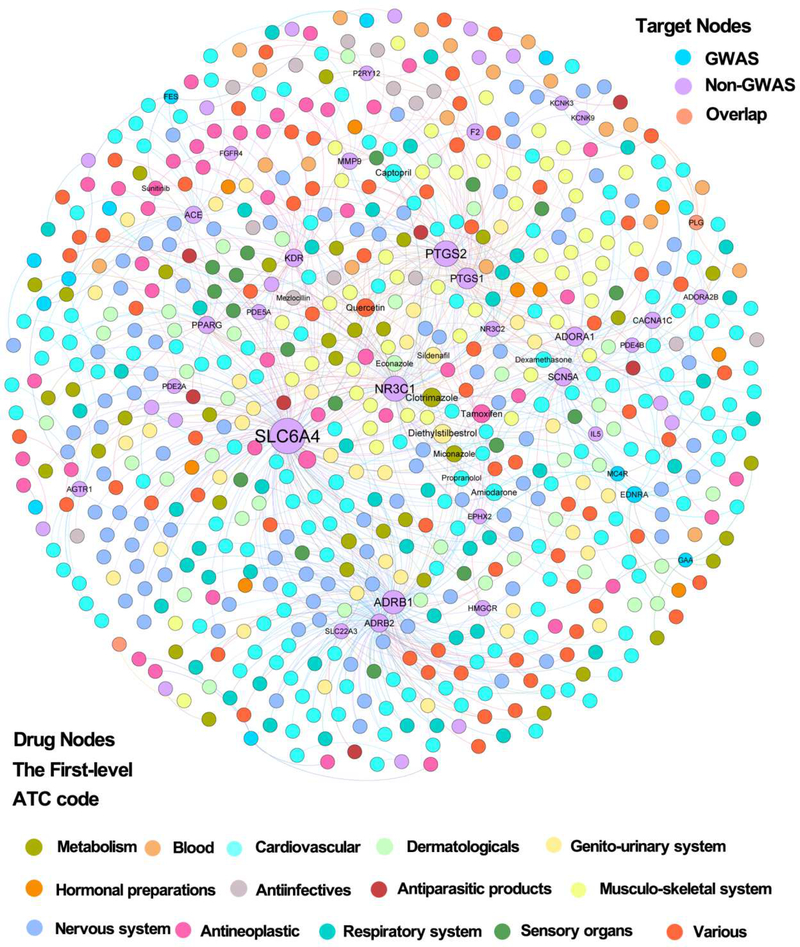
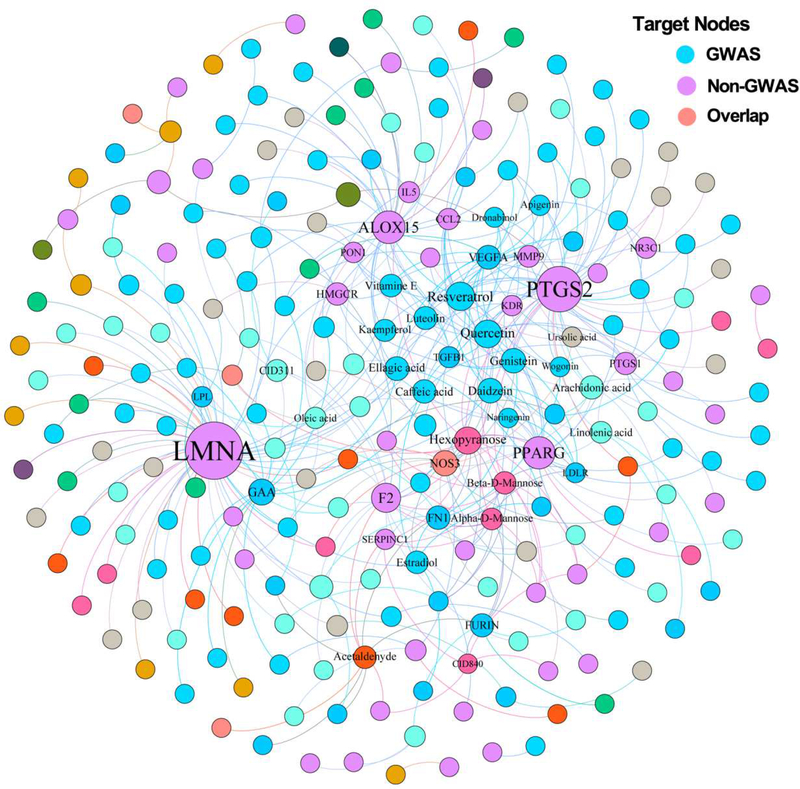
We further reconstructed two disease-centered drug-target networks (Table S1) by focusing on approved drugs or natural products that specifically target proteins encoded by CAD genes: CADDTnet and CADNPnet (see Methods). Figure 2 displays a bipartite drug-target network for CADDTnet that contains 931 drug-target interactions connecting 634 approved drugs and 58 known CAD proteins (e.g., angiotensin-converting enzyme [ACE] and HMG-CoA Reductase [HMGCR]). Figure 3 displays a bipartite drug-target network for CADNPnet that contains 395 compound-protein interactions connecting 170 unique natural products and 71 CAD proteins. We examined the protein overlap between CADDTnet and CADNPnet. In total, 37 CAD proteins targeted by both approved drugs and natural products, including ACE, nitric oxide synthase (NOS3), phosphodiesterase 5A (PDE5A), and HMGCR. Interestingly, we found that 24.1% (14/58) of CAD proteins in CADDTnet and 38.0% (27/71) of that in CADNPnet are derived from genome-wide association studies (GWAS). This implies that GWAS may contribute to the identification of CAD targets for approved drugs and natural products, consistent with a previous study [20].
Fig. 2. A bipartite drug–target interaction network (CADDTnet) for coronary artery disease (CAD)-associated drugs from DrugCentral database.
[19]. CADDTnet contains 931 interactions connecting 634 approved drugs and 58 CAD proteins (including 12 proteins from GWAS, 44 from non-GWAS, and 2 overlapping genes shared by multiple data sources [see Methods]). The CADDTnet was drawn using Gephi (v0.9.2). The label font size and node size are proportional to degree (connectivity). The label name is shown only when its degree is higher than 5. The color of drug node is colored by their first-level Anatomical Therapeutic Chemical Classification (ATC) codes from the DrugBank database [29].
Fig. 3. A bipartite natural product–protein interaction network (CADNPnet) for coronary artery disease (CAD)-associated natural products from 6 traditional Chinese medicine (TCM) formulae.
CADNPnet contains 395 interactions connecting 170 natural products and 71 CAD proteins (including 23 proteins from GWAS, 44 from non-GWAS, and 4 overlapping proteins shared by multiple sources). The CADNPnet was drawn using Gephi (v0.9.2). The label font size and node size are proportional to degree (connectivity). The label name is shown only when its degree is higher than 5. The color of drug nodes is colored by the chemical clustering analysis (see Fig. 4).
We found that the average degree (2.32±0.20, P = 0.0027 [one-side Wilcoxon test], Fig. S2) of natural products in CADNPnet is slightly stronger than that of approved drugs (1.47±0.03) in CADDTnet, further suggesting the ‘promiscuity’ of natural products (Fig. S1). Among 170 natural products (Fig. 3), 9 natural products targeting CAD proteins have degree (K) greater than 8: quercetin (K = 14), resveratrol (K = 14); hexopyranose (K = 14), genistein (K = 11), beta-D-Mannose (K = 11), arachidonic acid (K = 9), daidzein (K = 9), acetaldehyde (K = 9), and alpha-D-Mannose (K = 9). Of note, several natural products (e.g., quercetin and genistein) have been reported to exert cardio-protective effects [21, 22]. For example, quercetin supplementation can reduce the risk of cardiovascular disease in an epidemiologic study [21]. Figure 3 shows that quercetin binds with 14 known CAD proteins, including 3 CAD proteins (NOS3, TGFB1 and VEGFA) identified by GWAS. Quercetin was reported to attenuate endothelial oxidative damage by modulating the endothelial NO synthase (NOS)-related signaling pathway in human umbilical vein endothelial cells (HUVECs) [22]. Genistein interacts with 11 CAD proteins, including 5 CAD proteins (FN1, LDLR, NOS3, TGFB1 and VEGFA) derived from GWAS (Fig. 3). Genistein was reported to regulate low-density lipoprotein receptor (LDLR) expression in vitro [23].
Altogether, drug-target network analysis suggests that genetic studies (e.g., GWAS) may contribute to the target identification in CAD. In addition to approved drugs, natural products further offer novel candidates in development of new chemoprevention strategies on CAD. These network analyses promote us develop in silico models to further uncover potential therapeutic strategies for CAD by exploiting both approved drugs and natural products.
2.2. Chemical diversity of approved drugs and natural products on CAD
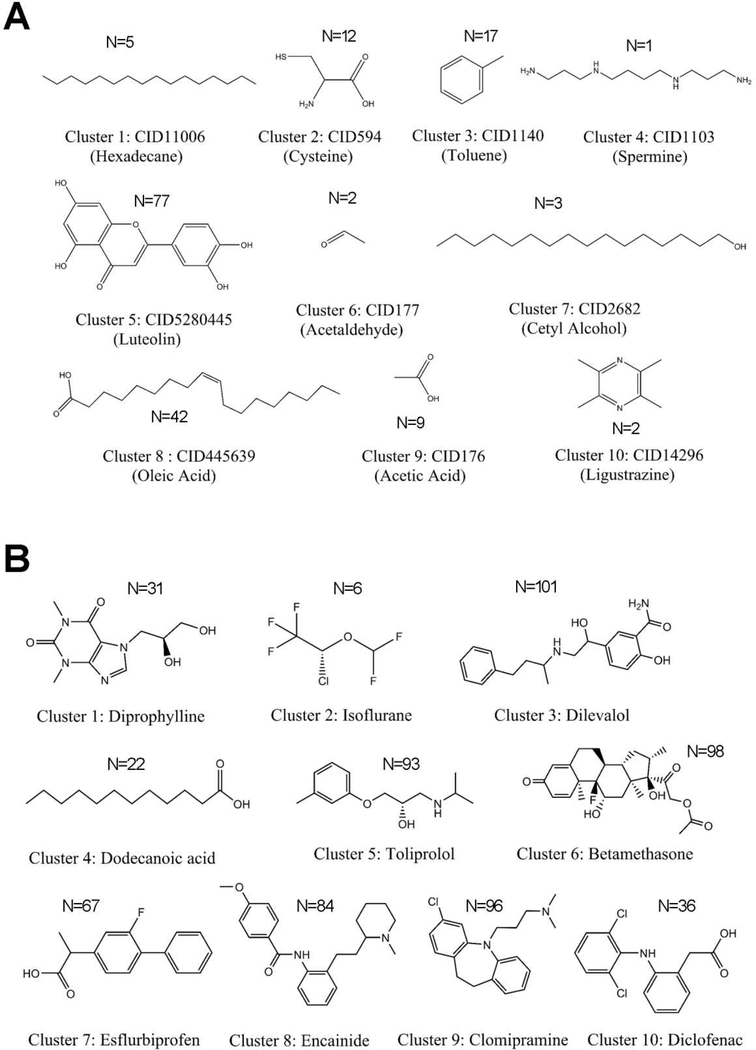
To investigate the chemical feature of known therapeutic agents on CAD, we performed clustering analysis (Fig. 4) for 634 approved drugs and 170 natural products that target at least one CAD protein. The chemical clustering analysis was performed by computing the root-mean-square-difference (RMSD) of the Tanimoto distance of pairwise compounds using FCFP_4 fingerprint implemented in Discovery Studio 4.0 (version 4.0, Accelrys Inc.). In total, 170 natural products are clustered into 10 chemical groups with cluster centers: hexadecane, cysteine, toluene, spermine, luteolin, acetaldehyde, cetyl alcohol, oleic acid, acetic acid, and ligustrazine, respectively (Fig. 4A). Among them, cluster 5 (Cluster center: luteolin) are represented as flavonoids, with the largest number (N=77) of natural products. For example, kaempferol [24] and quercetin [25] belonging to flavonoids, have been reported to reduce the risk of CAD [26]. In addition, 634 approved drugs in CADDTnet are clustered into 10 groups with cluster centers: diprophylline, isoflurane, dilevalol, dodecanoic acid, toliprolol, betamethasone, esflurbiprofen, encainide, clomipramine, and diclofenac. Compared with natural products, the second largest cluster (cluster 6) of approved drugs is steroids, including 96 approved drugs (Fig. 4B). For example, digoxin, a glycoside with steroid scaffold, is an FDA-approved drug for congestive cardiac insufficiency, arrhythmias and heart failure [27]. However, a recent study has revealed that long-term anabolic-androgenic steroid use may cause adverse cardiovascular complications, such as myocardial dysfunction and accelerated coronary atherosclerosis [28]. Taken together, natural products (i.e., flavonoid) and approved drugs (i.e., steroids) show unique chemical features (Fig. 4), offering diverse pharmacologic mechanisms-of-action for deep understanding of cardiovascular therapeutics versus side effects of approved drugs and natural products.
Fig. 4. Chemical diversity analysis for natural products (A) from 6 traditional Chinese medicine (TCM) formulae (see Methods) and for approved drugs (B) from DrugCentral database.
[19]. The structures of 10 cluster centers during chemical structural clustering analysis for 171 natural products (A) and 634 approved drugs (B) were illustrated. N denotes the number of drugs or natural products in each cluster.
2.3. Uncovering new association of CAD with approved drugs or natural products
We next turn to build in silico models for predicting new associations of CAD with approved drugs or natural products through integrating experimentally validated drug-target networks, known CAD proteins, and the human protein-protein interactions. By applying the threshold of adjusted p-value (q) < 0.05, we computationally identified potential associations for 61 approved drugs (Fig. 5A) and 46 natural products (Fig. 6A) on CAD. In total, 51 approved drugs as well as 27 natural products are predicted to have significant associations on CAD (q<10−5), including several well-known anti-CAD natural products (e.g., quercetin and luteolin) and known cardiovascular drugs (e.g., eplerenone and metoprolol).
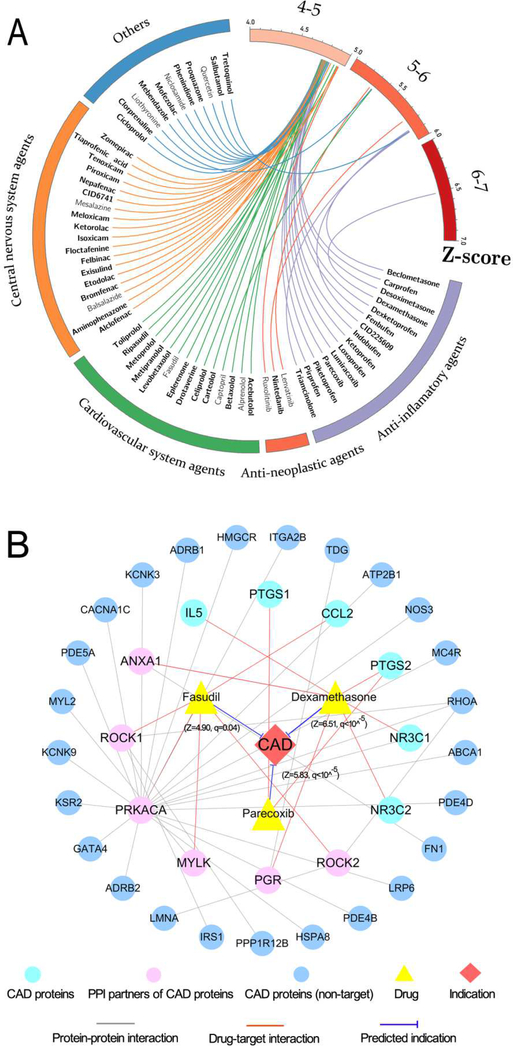
Fig. 5. The predicted associations of approved drugs on coronary artery disease (CAD).
(A) Circos plot representing the predicted associations connecting 61 approved drugs with CAD. The predicted q values with corresponding Z-score are exhibited as connected lines (edges). The drugs with significant q values (q<10−5) are highlighted in bold font. Drugs are grouped by their first-level Anatomical Therapeutic Chemical Classification (ATC) codes. The circos plot was drawn using Circos (v0.69). (B) A discovered drug-target-disease network connecting three selected drugs (fasudil, parecoxib, and dexamethasone) as well as their corresponding targets of CAD proteins and their protein-protein interaction neighbors. The thickness of blue line is proportional to the predicted Z-score.
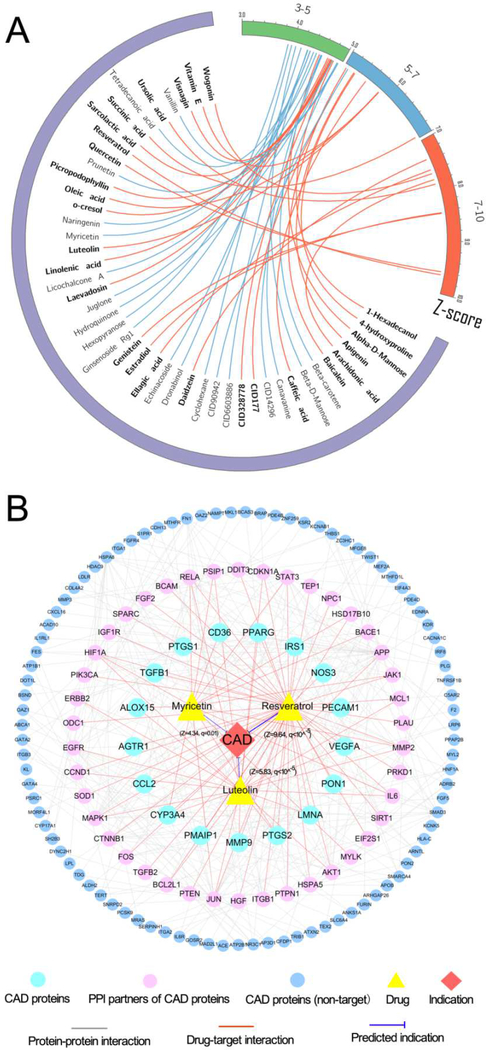
Fig. 6. The predicted associations of natural products on coronary artery disease (CAD).
(A) Circos plot representing the predicted associations connecting 46 natural products with CAD. The predicted q values with corresponding Z-score are exhibited as connected lines (edges). Natural products with significant q values (q<10−5) are highlighted in bold font. The circos plot was drawn using Circos (v0.69). (B) A discovered drug-target-disease network connecting three typical natural products (resveratrol, luteolin and myricetin) as well as their corresponding targets of CAD proteins and their protein-protein interaction neighbors. The thickness of blue line is proportional to the predicted Z-score.
Based on 19 approved CAD drugs defined by first-level Anatomical Therapeutic Chemical (ATC) classification codes [29], we found that the area under the ROC curve (AUC) was 72 % (Fig. S3), which is comparable to a previous network proximity approach [30]. We further retrieved previously reported literature data of potential associations for the 61 significantly predicted, approved drugs and 46 natural products on CAD (Table S2). In total, we found 24 approved drugs (a success rate of 39.3 % [24/61]) and 19 natural products (a success rate of 41.3 % [19/46]) that have the reported experimental data to support the predicted anti-CAD effects, suggesting a high hit rate compared to traditional virtual screening approaches [31, 32]. After excluding the compounds with reported experimental evidences, the remaining 37 approved drugs and 27 natural products offer novel, potential anti-CAD candidates.
2.4. Mechanism-of-action to approved drugs on CAD: a network-based analysis
We next selected three typical categories of approved drugs (Those three drugs have the most reported experimental evidences), including Rho-kinase (ROCK) inhibitors, nonsteroidal anti-inflammatory drugs (NSAIDs), and glucocorticoids, to illustrate the mechanism-of-action on CAD.
Rho-kinases has been identified as potentially therapeutic targets for developing anti-cardiovascular agents [33]. Fasudil, known as ROCK inhibitors, is under investigating to treat cardiovascular disease in several clinical trials, including atherosclerosis (NCT00120718) and carotid stenosis (NCT00670202). As shown in Fig. 5B, fasudil interacts with only one CAD protein (encoded by CCL2) as well as 4 PPI partners (encoded by PRKACA, MYLK, ROCK1 and ROCK2). Obviously, fasudil cannot be predicted to have significant indication on CAD based on the curated CAD protein (CCL2) only. After adding 4 PPI partners, fasudil is predicted to have a significant association on CAD (Z=4.90, q=0.04). Inhibition of ROCK was reported to offer an alternative strategy to treat CAD in a preclinical study [34]. Fasudil has been reported to be a strong inhibitor on both ROCK1 (IC50=0.26 μM) and ROCK2 (IC50=0.32 μM) [35]. Interestingly, fasudil has been reported to ameliorate myocardial ischemia in patients with coronary microvascular spasm [36].
Cardiovascular complications account for the withdrawal of multitudinous post-marketed drugs [37]. NSAIDs have been widely reported to induce various adverse effects, especially in patients with cardiovascular complications [38]. For example, parecoxib, a cyclooxygenase-2 (COX2) selective inhibitor, is approved for short term perioperative pain control in the European Union [39]. Parecoxib has been reported to increase the risk of cardiovascular events [40]. Fig. 5B shows that parecoxib binds with both COX1 and COX2 (encoding by PTGS1 and PTGS2) involving CAD. Herein parecoxib was predicted to have a significant association with CAD (Z= 4.75, q<10−5, Fig. 5B), consistent with previous experimental reports [40]. Parecoxib has been reported to be a strong inhibitor of COX2 (IC50=5 nM) [41], while inhibition of COX2 contributes to several cardiovascular complications, such as coronary atherothrombosis and heart injury due to the decrease of prostaglandin E2 (PGE2) and prostacyclin (PGI2) [42].
Cardiovascular adverse effects are essential concerns for glucocorticoids as well [43, 44]. The understanding of the molecular mechanism of cardiotoxicities of glucocorticoids might offer potential prevention strategies for reducing cardiovascular events, including CAD. Dexamethasone, known as a glucocorticoid agonist, was approved for treatment of multiple types of human diseases [45]. As displayed in Fig. 5B, dexamethasone binds with 4 CAD proteins (NR3C1, NR3C2, IL5, and PTGS2) and two PPI partners (ANXA1 and PGR).
Here dexamethasone was predicted to have potential association with CAD (Z=6.51, q<10−5), consistent with the clinically reported cardiotoxicities [46]. Activation of glucocorticoid receptor (NR3C1), has been reported to induce cardiovascular complications such as obesity and hypertension [43]. Dexamethasone is a high effective glucocorticoid receptor agonist with half maximal effective concentration (EC50) of 0.1 nM [47], which may help explain the molecular mechanism of its cardiovascular complications.
2.5. Mechanism-of-action to natural products on CAD: a network-based analysis
Previous studies have reported that several Chinese medicines have potential protective effects on CAD [13, 48]. However, the detailed molecular mechanism of protective effects of Chinese medicine on CAD remain unclear. We next turned to illustrate mechanism-of-action of resveratrol and luteolin on cardiovascular systems via a network-based analysis
Resveratrol, a naturally occurring phenol abundant in the skin of grapes and in red wine, shows cardiovascular protective effects in multiple preclinical studies [49–51]. There are over 12 clinical trials (http://clinicaltrials.gov/) being conducted or completed to test resveratrol’s therapeutic effects on CAD (e.g., NCT02137421) or other cardiovascular diseases (e.g., NCT01449110 and NCT01564381). Fig. 6B shows that resveratrol binds with 15 CAD proteins and 35 PPI partners (e.g., SIRT1). Herein resveratrol was predicted to show a significant association on CAD (Z = 9.64, q<10−5). Activation of sirtuins 1 (SIRT1) has been reported to offer therapeutic effect on CAD [52]. Fig. 6B shows that resveratrol not only targets two CAD proteins (IRS1 and NOS3) but also a PPI partner (SIRT1, EC50=23.6 μM [53]), suggesting a protective mechanism of resveratrol on CAD.
Luteolin, a flavonoid rich in fruits and herbs, has been reported to show cardio-protective effects in vitro and in vivo, including CAD, heart failure, and atherosclerosis [54, 55]. The exact molecular mechanism of cardio-protective effects (e.g., CAD) by luteolin remains unclear. In Fig. 6B, luteolin interacts with 7 CAD proteins and 16 PPI partners (e.g. AKT1 and MAPK1), and is predicted to have potential association on CAD (Z= 5.83, q<10−5). AKT regulates the apoptotic pathological process of myocardial ischemia/reperfusion (I/R) injury, while mitogen-activated protein kinases (MAPKs) has been reported to regulate cardiomyocyte function after I/R injury [56, 57]. Recent studies have demonstrated that luteolin can suppress apoptosis by activation of AKT in a simulated I/R model and inhibit MAPK pathway in myocardial I/R injury [58, 59], suggesting a protective mechanism by luteolin on CAD. In summary, quantitative network analysis offers potential mechanism for deep understanding of cardio-protective effects by natural products on CAD and other cardiovascular systems if broadly applied (Fig. 6B).
2.6. Elucidating protective effects of natural products on coronary artery disease via targeting inflammatory-associated proteins.
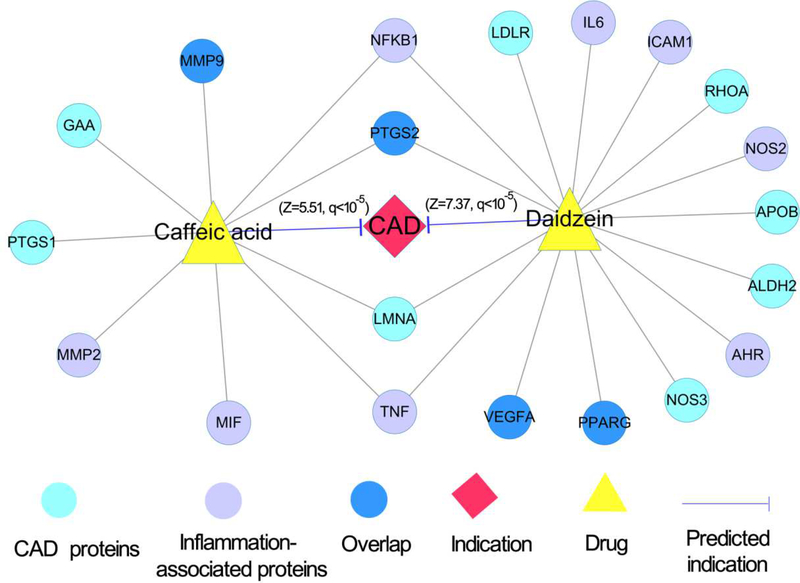
Recent experimental and clinical studies have suggested that reducing inflammation reduce the risk of cardiovascular disease, including CAD [60]. We next inspected whether natural products target inflammatory response pathways on CAD, by showcasing two typical natural compounds: daidzein and caffeic acid. Daidzein, a soybean isoflavone, has been reported to show cardiovascular protective and anti-inflammatory effects in several preclinical studies [61, 62]. Here daidzein was computationally predicted to have significant association on CAD (Z= 7.37, q<10−5). Figure 7 reveals that daidzein binds with 9 inflammatory-associated proteins (e.g., NFKB1 and PPARG) and 9 CAD proteins. Daidzein was reported to attenuate I/R-induced myocardial damage via inhibiting NF-kappa B activation [61]. In addition, daidzein regulated pro-inflammatory adipokines by activation of PPARG [62]. Caffeic acid, a phenolic acid, is effectively used as a natural antioxidant. Figure 7 shows that caffeic acid interacts with 6 inflammation-associated proteins (e.g., NFKB1 and TNF) and 5 CAD proteins (e.g., MMP9 and LMNA). We found that caffeic acid was predicted to have a significant association on CAD (Z= 5.51, q<10−5). Specifically, caffeic acid has been reported to improve cardiac mitochondrial dysfunction in Wistar rats [63] and to inhibit tumor necrosis factor alpha (TNFA)-induced vascular inflammation in HUVECs by inhibiting NF-kappa B activation [64]. Collectively, in silico prediction and network analysis reveal that daidzein and caffeic acid have potential protective effects on CAD by targeting inflammatory response pathways. However, further pre-clinical and clinical studies are warranted to inspect these network-predicted anti-inflammatory pathways on CAD further.
Fig. 7. A discovered drug-target-disease network for two typical natural products by targeting inflammatory response pathways.
The network displays the predicted associations of two natural products (daidzein and caffeic acid) on coronary artery disease (CAD) as well as their corresponding CAD proteins (targets) and genes on inflammatory response pathways (see Methods).
3. DISSCUSSION AND CONCLUSION
Network-based systems pharmacology approaches offer potential tools for developing chemical intervention strategies on CAD and help use better characterization of pharmacological mechanisms of approved drugs and natural products on cardiovascular systems. In this study, we computationally investigated approved drugs and natural products on CAD using an integrated systems pharmacology framework: (i) constructing two global drug-target networks for both approved drugs and natural products, (ii) building in silico models for predicting potential associations of approved drugs and natural products on CAD through integrating drug-target networks, known CAD proteins, and the protein-protein interactions, (iii) identifying potential mechanism-of-action of the computationally predicted therapeutic profiles or adverse effects of approved drugs and natural products on CAD by network analysis. In summary, we demonstrated a powerful systems pharmacology framework for the development of potential therapeutic and prevention strategies for CAD by exploiting the wealth of approved drugs and natural products.
We highlighted several significant improvements compared to previous studies [9, 16, 17]. First, we integrated the human protein-protein interaction of known CAD proteins, which can complement the incompleteness of known disease genes of CAD and increase the statistical power of in silico models [65]. We thereby predicted potential associations on CAD for those approved drugs or natural products targeting only one or none of known CAD proteins, such as fasudil. Second, we extended the systems pharmacology framework by searching cardio-protective natural products, offering potential natural products in treating and chemoprevention of CAD comparing to our recent network proximity-based approach that focused on the approved drugs only [30].
However, several potential limitations should be acknowledged. First, although we assembled large-scale, experimentally reported drug-target interactions for both approved drugs and natural products from publicly available databases based on our sizeable efforts, the incompleteness of drug-target networks may exist as well. In addition, some drug-target or compound-protein interactions on natural products come from functional assays, not physical binding studies, which could cause the risk of false positive rate. An integration of the computational predicted drug-target network may help overcome the incompleteness of the known drug-target networks based on our recent studies [66, 67]. In addition, adding large-scale drug-induced transcriptome [68] or proteome [69] data may help improve the accuracy of in silico models further, such as Connectivity Map. Second, our current in silico models cannot separate the therapeutic effects against side effects owing to lack of the detailed functional effects of drug targets and disease proteins. Drug targets representing nodes within cellular networks are often intrinsically coupled in both therapeutic and adverse profiles [70]. Drugs can inhibit or activate protein functions (including antagonists vs. agonists), while disease alleles from genetic or genomic studies contain loss-of-function or gain-of-function. For example, an inhibitor that targets loss-of-function disease proteins often causes adverse effects. Hence, integration of functional genomic assays or large-scale disease gene expression profiles (upregulation or downregulation), along with patient data (e.g., health insurance claims data) validation and in vitro or in vivo mechanistic studies will improve in silico drug repurposing models further [30, 71].
In summary, we suggested that a network-based, systems pharmacology framework offered potential strategies for in silico drug repurposing in treating and chemoprevention of CAD by exploiting promiscuity of approved drugs and natural products. If broadly applied, the systems pharmacology approaches presented here can be applied for development of new chemical intervention strategies in other diseases as well.
4. EXPERIMENTAL SECTION
4.1. Construction of drug–target networks for approved drugs
To construct a drug-target network for approved drugs, we downloaded the latest data from DrugCentral database (accessed in Dec 2017) [19], and only kept data items by the following criteria: (i) the target organism is homo sapiens; (ii) the target can be transformed into a symbol gene; (iii) the drug can be transformed to canonical SMILES format.
4.2. Construction of compound–protein networks for natural products
We manually collected 6 traditional Chinese medicine (TCM) formulae with clinically-proven efficacy on CAD, including ShengMaiSan (SMS) [72], ZhiGanCao Decoction (ZGC), SuHeXiang Pill (SHX), BuYangHuanWu Decoction (BYHW) [73], ShiXiaoSan (SXS), and XiaoXianXiong Decoction (XXD), from references [72, 73] and website [http://cnda.cfda.gov.cn] of China Food and Drug Administration (CFDA).
We next collected the constituting herbs for each TCM formula, and extracted related herb ingredients for 6 TCM formulae from six publicly available TCM data sources: Traditional Chinese Medicine database (TCMDb) [74], Traditional Chinese Medicine integrated database (TCMID) [75], Traditional Chinese Medicine Systems Pharmacology (TCMSP) [76], Traditional Chinese Medicine database@Taiwan (TCM@Taiwan) [77], TCM-MESH [78] and TM-MC [79]. In total, we obtained 6,077 herb-ingredient pairs connecting 30 herbs to 4,297 unique natural products (Table S3).
We further integrated a compound-protein interaction network of natural products from two types of data sources: 1) direct DTI databases (including ChEMBL [80] and BindingDB [81]), and 2) indirect DTI databases (including STITCH 5 [82], Herbal Ingredients’ Targets Database (HIT) [83], and TCMID [75]. We downloaded the latest data as below: ChEMBL (v21), and BindingDB (accessed in September 2017). We filtered data items that met the following criteria: (i) inhibitory constant (Ki), dissociation constant (Kd), half maximal inhibitory concentration (IC50) or half maximal effective concentration (EC50) ≤ 10 μM; (ii) the target is a human protein; (iii) the target has a unique UniProt accession number; (iv) each drug can be transformed to canonical SMILES format. Subsequently all drug structures were carefully standardized by removing salt ions and standardizing dative bonds using Open Babel toolkit (v2.3.2) [84].
We collected indirect DTIs by following the below steps. For STITCH source (accessed in Sep 2017), the thickness of each interaction pair represents the confidence score of the association. Only DTIs from homo sapiens were retained, and compound-protein interactions with experimental evidence score over 0.7 were used in this work. We further extracted compound-protein interactions from HIT and TCMID using a web crawler approach, and deleted the duplicated DTIs. Finally, we constructed a global compound-protein network for natural products after integrating direct and indirect interactions (edges) via mapping 4,297 unique natural products into the five databases using the “InChIKey” derived from chemical structures (SMILES format).
4.3. Manual curation of disease genes for coronary artery disease and inflammatory response pathways.
In this study, CAD disease-associated genes (CAD genes) were collected from Online Mendelian Inheritance in Man (OMIM) [85], The Human Gene Mutation Database (HGMD) [86], and GWAS Catalog [87], while inflammation-associated genes were integrated from Comparative Toxicogenomics Database (CTD) [88], DisGeNet [89], HGMD [86] and GWAS Catalog [87]. OMIM is a comprehensive bioinformatics resource of curated descriptions of human genes and phenotypes as well as the relationships between them [85], while HGMD is a repository of inherited mutation data manually curated from published references [86]. Only single-nucleotide polymorphism (SNPs) with Genome-wide significance (P < 5.0×10−8) on CAD were extracted from GWAS Catalog. Finally, 243 CAD disease-associated genes and 112 inflammation-associated genes were obtained. In this study, we grouped 243 CAD genes into three source types: GWAS, non-GWAS, and their overlapped gene sets. The details are provided in Table S4.
4.4. Construction of a specific PPI network of CAD genes
We integrated a large-scale, high-quality human protein interactome from 5 types of PPIs: high-throughput Y2H binary, kinase-substrate interactions, signaling interactions, protein three-dimensional (3D) interactome, protein complexes (Bioplex 1&2), and literatures, as described in our recent studies [30]. In addition, we mapped 243 CAD genes to the interactome to extract a specific PPI network of CAD genes.
We further filtered PPI interactor genes (neighbors of CAD proteins) that are not significantly expressed in CAD tissue (blood vessel). Firstly, the RNA-seq data (RPKM value) of 32 tissues were downloaded from GTEx V6 release (accessed on April 01, 2016, https://gtexportal.org/home/). Those genes with RPKM ≥ 1 in over 80% of samples were defined as tissue-expressed genes and the remaining genes as tissue-unexpressed. Secondly, to quantify the expression significance of tissue-expressed gene i in tissue t, we computed the average expression E(i) and the standard deviation δ_E (i) of a gene’s expression across all considered tissues. The significance of gene expression in tissue t is defined as below:
| (1) |
In this study, only the PPI interactor genes with Z-score higher than 0 were kept. Finally, we compiled a specific PPI network of CAD proteins, which consisted of 3,176 PPIs connecting 243 CAD proteins and 1,433 PPI partners (Table S5).
4.5. In silico prediction
Here, we built in silico models to predict new associations of approved drugs (or natural products) on CAD by incorporating drug-target (compound-protein) interactions, the known CAD genes, and the human PPI network. We first excluded the approved drugs or natural products that had less than two targets as well as those without any known proteins encoded by CAD genes or the first neighbors (direct interacting proteins) of CAD proteins in the human PPI network.
The hypothesis of our framework asserts that an approved drug (or a natural product) with high promiscuity shows a higher possibility to treat CAD if its targets are more likely to be CAD proteins or partners of CAD PPIs. A permutation testing was used to calculate the statistical significance of an approved drug (or a natural product) to be prioritized for potential association on CAD. The null hypothesis supposes that the targets of an approved drug (or a natural product) randomly locate at CAD proteins or their neighbors of CAD PPIs across the human proteome. We performed the permutation testing as below:
| (2) |
A nominal P was computed for each drug (or natural product) by counting the number of observed CAD or PPI interactor genes greater (Sm (p)) than the permutations (Sm).
Here we repeated 100,000 permutations by randomly selecting 1,638 proteins (the same number of CAD proteins and their neighbors of CAD PPIs) from the genome-wide simulation (20,462 human protein-coding genes from the NCBI database [https://www.ncbi.nlm.nih.gov/gene], Table S6). Then, the nominal P-values from the permutation tests were corrected as adjusted P-values (q) using R based on Benjamini-Hochberg approach [90].
Subsequently, a Z-score was calculated for each drug (or natural product) to be prioritized for potential association on CAD during permutation testing as previously described [9, 16], where “ is the real number of CAD or PPI interactor genes targeted by a given natural product (or drug), μ is the mean number of CAD or their PPI partners targeted by a given natural product (or drug) during 100,000 permutations, and σ is the standard deviation.
| (3) |
4.6. Network Visualization and Statistical Analysis
The statistical analysis in this study was carried out using the Python (v3.2, http://www.python.org/) and R platforms (v3.01, http://www.r-project.org/). Networks were visualized by Cytoscape (v3.2.0, http://www.cytoscape.org/) and Gephi (v0.9.2, https://gephi.org/).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 81603318), Research Fund for Characteristic Innovation Projects of Guangdong Province (2016KTSCX013) and Open Tending Project for the Construction of High-Level University (A1-AFD018171Z11027) and the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number K99HL138272 and R00HL138272 to F.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Gaziano TA, Bitton A, Anand S, Abrahams-Gessel S, Murphy A, Growing Epidemic of Coronary Heart Disease in Low- and Middle-income Countries, Curr. Probl. Cardiol 35 (2010) 72–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dalen JE, Alpert JS, Goldberg RJ, Weinstein RS, The Epidemic of the 20(th) century: Coronary Heart Disease, Am J Med. 127 (2014) 807–812. [DOI] [PubMed] [Google Scholar]
- [3].Antman EM, Loscalzo J, Precision Medicine in Cardiology, Nat. Rev. Cardiol 13 (2016) 591–602. [DOI] [PubMed] [Google Scholar]
- [4].Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P, Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association, Circulation 135 (2017) e146–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ford ES, Roger VL, Dunlay SM, Go AS, Rosamond WD, Challenges of Ascertaining National Trends in the Incidence of Coronary Heart Disease in the United States, J. Am. Heart Assoc 3 (2014) e001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li J, Zheng S, Chen B, Butte AJ, Swamidass SJ, Lu Z, A Survey of Current Trends in Computational Drug Repositioning, Brief. Bioinform. 17 (2016) 2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Novac N, Challenges and Opportunities of Drug Repositioning, Trends Pharmacol. Sci. 34 (2013) 267–272. [DOI] [PubMed] [Google Scholar]
- [8].Katare PB, Banerjee SK, Repositioning of Drugs in Cardiometabolic Disorders: Importance and Current Scenario, Curr. Top. Med. Chem 16 (2016) 2189–2200. [DOI] [PubMed] [Google Scholar]
- [9].Fang J, Wu Z, Cai C, Wang Q, Tang Y, Cheng F, Quantitative and Systems Pharmacology. 1. In Silico Prediction of Drug-Target Interactions of Natural Products Enables New Targeted Cancer Therapy, J. Chem. Inf. Model 57 (2017) 2657–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fang J, Liu C, Wang Q, Lin P, Cheng F, In Silico Polypharmacology of Natural Products, Brief. Bioinform (2017). doi: 10.1093/bib/bbx045 [DOI] [PubMed] [Google Scholar]
- [11].Wang J, Wong YK, Liao F, What Has Traditional Chinese Medicine Delivered for Modern Medicine?, Expert Rev. Mol. Med 20 (2018) e4. [DOI] [PubMed] [Google Scholar]
- [12].Hao P, Jiang F, Cheng J, Ma L, Zhang Y, Zhao Y, Traditional Chinese Medicine for Cardiovascular Disease: Evidence and Potential Mechanisms, J. Am. Coll. Cardiol 69 (2017) 2952–2966. [DOI] [PubMed] [Google Scholar]
- [13].Gao ZY, Xu H, Shi DZ, Wen C, Liu BY, Analysis on Outcome of 5284 Patients with Coronary Artery Disease: the Role of Integrative Medicine, J. Ethnopharmacol. 141 (2012) 578–583. [DOI] [PubMed] [Google Scholar]
- [14].Zhang J, Meng H, Zhang Y, Zhang X, Shao M, Li C, Tu P, The Therapeutical Effect of Chinese Medicine for the Treatment of Atherosclerotic Coronary Heart Disease, Curr. Pharm. Des 23 (2017) 5086–5096. [DOI] [PubMed] [Google Scholar]
- [15].Vicini P, van der Graaf PH, Systems Pharmacology for Drug Discovery and Development: Paradigm Shift or Flash in the Pan?, Clin. Pharmacol. Ther 93 (2013) 379–381. [DOI] [PubMed] [Google Scholar]
- [16].Fang J, Cai C, Wang Q, Lin P, Zhao Z, Cheng F, Systems Pharmacology-Based Discovery of Natural Products for Precision Oncology Through Targeting Cancer Mutated Genes, CPT Pharmacometrics Syst. Pharmacol 6 (2017) 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fang J, Gao L, Ma H, Wu Q, Wu T, Wu J, Wang Q, Cheng F, Quantitative and Systems Pharmacology 3. Network-Based Identification of New Targets for Natural Products Enables Potential Uses in Aging-Associated Disorders, Front. Pharmacol 8 (2017) 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cai H, Luo Y, Yan X, Ding P, Huang Y, Fang S, Zhang R, Chen Y, Guo Z, Fang J, Wang Q, Xu J, The Mechanisms of Bushen-Yizhi Formula as a Therapeutic Agent against Alzheimer’s Disease, Sci. Rep 8 (2018) 3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ursu O, Holmes J, Knockel J, Bologa CG, Yang JJ, Mathias SL, Nelson SJ, Oprea TI, DrugCentral: Online Drug Compendium, Nucleic Acids Res. 45 (2017) D932–d939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, Cardon LR, Whittaker JC, Sanseau P, The Support of Human Genetic Evidence for Approved Drug Indications, Nat. Genet 47 (2015) 856–860. [DOI] [PubMed] [Google Scholar]
- [21].Egert S, Bosy-Westphal A, Seiberl J, Kurbitz C, Settler U, Plachta-Danielzik S, Wagner AE, Frank J, Schrezenmeir J, Rimbach G, Wolffram S, Muller MJ, Quercetin Reduces Systolic Blood Pressure and Plasma Oxidised Low-density Lipoprotein Concentrations in Overweight Subjects with a High-cardiovascular Disease Risk Phenotype: a Double-blinded, Placebo-controlled Cross-over Study, Br. J. Nutr 102 (2009) 1065–1074. [DOI] [PubMed] [Google Scholar]
- [22].Hung CH, Chan SH, Chu PM, Tsai KL, Quercetin is a Potent Anti-atherosclerotic Compound by Activation of SIRT1 Signaling under OxLDL Stimulation, Mol. Nutr. Food Res 59 (2015) 1905–1917. [DOI] [PubMed] [Google Scholar]
- [23].Caruso MG, Messa C, Orlando A, D’Attoma B, Notarnicola M, Early Induction of LDL Receptor Gene Expression by Genistein in DLD-1 Colon Cancer Cell Line, Fitoterapia 79 (2008) 524–528. [DOI] [PubMed] [Google Scholar]
- [24].Xu YC, Leung SW, Leung GP, Man RY, Kaempferol Enhances Endothelium-dependent Relaxation in the Porcine Coronary Artery through Activation of Large-conductance Ca(2+) -activated K(+) Channels, Br. J. Pharmacol 172 (2015) 3003–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Suri S, Liu XH, Rayment S, Hughes DA, Kroon PA, Needs PW, Taylor MA, Tribolo S, Wilson VG, Quercetin and its Major Metabolites Selectively Modulate Cyclic GMP-dependent Relaxations and Associated Tolerance in Pig Isolated Coronary Artery, Br. J. Pharmacol 159 (2010) 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jiang W, Wei H, He B, Dietary Flavonoids Intake and the Risk of Coronary Heart Disease: a Dose-response Meta-analysis of 15 Prospective Studies, Thromb. Res 135 (2015) 459–463. [DOI] [PubMed] [Google Scholar]
- [27].Grzesk G, Stolarek W, Kasprzak M, Krzyzanowski M, Szadujkis-Szadurska K, Wicinski M, Grzesk E, Therapeutic Drug Monitoring of Digoxin-20 Years of Experience, Pharmacol. Rep 70 (2018) 184–189. [DOI] [PubMed] [Google Scholar]
- [28].Baggish AL, Weiner RB, Kanayama G, Hudson JI, Lu MT, Hoffmann U, Pope HG Jr., Cardiovascular Toxicity of Illicit Anabolic-Androgenic Steroid Use, Circulation 135 (2017) 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, Tang A, Gabriel G, Ly C, Adamjee S, Dame ZT, Han B, Zhou Y, Wishart DS, DrugBank 4.0: Shedding New Light on Drug Metabolism, Nucleic Acids Res. 42 (2014) D1091–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cheng F, Desai JR, Handy ED, Wang R, Schneeweiss S, Barabasi AL, Loscalzo J, Network-based Approach to Prediction and Population-based Validation of In Silico Drug Repurposing, Nat. Commun 9 (2018) 5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Truchon JF, Bayly CI, Evaluating Virtual Screening Methods: Good and Bad Metrics for the “Early Recognition” Problem, J. Chem. Inf. Model 47 (2007) 488–508. [DOI] [PubMed] [Google Scholar]
- [32].Zhu T, Cao S, Su PC, Patel R, Shah D, Chokshi HB, Szukala R, Johnson ME, Hevener KE, Hit Identification and Optimization in Virtual Screening: Practical Recommendations Based on a Critical Literature Analysis, J. Med. Chem 56 (2013) 6560–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shimokawa H, Sunamura S, Satoh K, RhoA/Rho-Kinase in the Cardiovascular System, Circ. Res 118 (2016) 352–366. [DOI] [PubMed] [Google Scholar]
- [34].Nohria A, Grunert ME, Rikitake Y, Noma K, Prsic A, Ganz P, Liao JK, Creager MA, Rho Kinase Inhibition Improves Endothelial Function in Human Subjects with Coronary Artery Disease, Circ. Res 99 (2006) 1426–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pireddu R, Forinash KD, Sun NN, Martin MP, Sung SS, Alexander B, Zhu JY, Guida WC, Schonbrunn E, Sebti SM, Lawrence NJ, Pyridylthiazole-based Ureas as Inhibitors of Rho Associated Protein Kinases (ROCK1 and 2), MedChemComm 3 (2012) 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mohri M, Shimokawa H, Hirakawa Y, Masumoto A, Takeshita A, Rho-kinase Inhibition with Intracoronary Fasudil Prevents Myocardial Ischemia in Patients with Coronary Microvascular Spasm, J. Am. Coll. Cardiol 41 (2003) 15–19. [DOI] [PubMed] [Google Scholar]
- [37].Cai C, Fang J, Guo P, Wang Q, Hong H, Moslehi J, Cheng F, In Silico Pharmacoepidemiologic Evaluation of Drug-Induced Cardiovascular Complications Using Combined Classifiers, J. Chem. Inf. Model 58 (2018) 943–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Danelich IM, Wright SS, Lose JM, Tefft BJ, Cicci JD, Reed BN, Safety of Nonsteroidal Antiinflammatory Drugs in Patients with Cardiovascular Disease, Pharmacotherapy 35 (2015) 520–535. [DOI] [PubMed] [Google Scholar]
- [39].Lloyd R, Derry S, Moore RA, McQuay HJ, Intravenous or Intramuscular Parecoxib for Acute Postoperative Pain in Adults, Cochrane Database Syst. Rev (2009) Cd004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Aldington S, Shirtcliffe P, Weatherall M, Beasley R, Increased Risk of Cardiovascular Events with Parecoxib/valdecoxib: a systematic review and meta-analysis, N. Z. Med. J 118 (2005) U1755. [PubMed] [Google Scholar]
- [41].Talley JJ, Bertenshaw SR, Brown DL, Carter JS, Graneto MJ, Kellogg MS, Koboldt CM, Yuan J, Zhang YY, Seibert K, N-[[(5-methyl-3-phenylisoxazol-4-yl)-phenyl]sulfonyl]propanamide, sodium salt, parecoxib sodium: A potent and selective inhibitor of COX-2 for parenteral administration, J. Med. Chem 43 (2000) 1661–1663. [DOI] [PubMed] [Google Scholar]
- [42].Patrono C, Cardiovascular Effects of Cyclooxygenase-2 Inhibitors: a Mechanistic and Clinical Perspective, Br. J. Clin. Pharmacol 82 (2016) 957–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Walker BR, Glucocorticoids and Cardiovascular Disease, Eur. J. Endocrinol 157 (2007) 545–559. [DOI] [PubMed] [Google Scholar]
- [44].Fardet L, Feve B, Systemic Glucocorticoid Therapy: a Review of its Metabolic and Cardiovascular Adverse Events, Drugs 74 (2014) 1731–1745. [DOI] [PubMed] [Google Scholar]
- [45].Tang E, Chen Y, Luo Y, Dexamethasone for the Prevention of Acute Mountain Sickness: Systematic Review and Meta-analysis, Int. J. Cardiol 173 (2014) 133–138. [DOI] [PubMed] [Google Scholar]
- [46].Zecca E, Papacci P, Maggio L, Gallini F, Elia S, De Rosa G, Romagnoli C, Cardiac Adverse Effects of Early Dexamethasone Treatment in Preterm Infants: a Randomized Clinical Trial, J. Clin. Pharmacol 41 (2001) 1075–1081. [DOI] [PubMed] [Google Scholar]
- [47].Miner JN, Ardecky B, Benbatoul K, Griffiths K, Larson CJ, Mais DE, Marschke K, Rosen J, Vajda E, Zhi L, Negro-Vilar A, Antiinflammatory Glucocorticoid Receptor Ligand with Reduced Side Effects Exhibits an Altered Protein-protein Interaction Profile, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 19244–19249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Qiu Y, Xu H, Shi D, Traditional Chinese Herbal Products for Coronary Heart Disease: an Overview of Cochrane Reviews, Evid. Based Complement. Alternat. Med 2012 (2012) 417387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xia N, Daiber A, Forstermann U, Li H, Antioxidant Effects of Resveratrol in the Cardiovascular System, Br. J. Pharmacol 174 (2017) 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zordoky BN, Robertson IM, Dyck JR, Preclinical and Clinical Evidence for the Role of Resveratrol in the Treatment of Cardiovascular Diseases, Biochim. Biophys. Acta 1852 (2015) 1155–1177. [DOI] [PubMed] [Google Scholar]
- [51].Penumathsa SV, Maulik N, Resveratrol: a Promising Agent in Promoting Cardioprotection against Coronary Heart Disease, Can. J. Physiol. Pharmacol 87 (2009) 275–286. [DOI] [PubMed] [Google Scholar]
- [52].Chan SH, Hung CH, Shih JY, Chu PM, Cheng YH, Lin HC, Tsai KL, SIRT1 Inhibition Causes Oxidative Stress and Inflammation in Patients with Coronary Artery Disease, Redox Biol. 13 (2017) 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wu J, Zhang D, Chen L, Li J, Wang J, Ning C, Yu N, Zhao F, Chen D, Chen X, Chen K, Jiang H, Liu H, Liu D, Discovery and Mechanism Study of SIRT1 Activators that Promote the Deacetylation of Fluorophore-labeled Substrate, J. Med. Chem 56 (2013) 761–780. [DOI] [PubMed] [Google Scholar]
- [54].Hu W, Xu T, Wu P, Pan D, Chen J, Chen J, Zhang B, Zhu H, Li D, Luteolin Improves Cardiac Dysfunction in Heart Failure Rats by Regulating Sarcoplasmic Reticulum Ca(2+)-ATPase 2A, Sci. Rep 7 (2017) 41017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Luo Y, Shang P, Li D, Luteolin: A Flavonoid that Has Multiple Cardio-Protective Effects and Its Molecular Mechanisms, Front. Pharmacol 8 (2017) 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Clark JE, Sarafraz N, Marber MS, Potential of p38-MAPK Inhibitors in the Treatment of Ischaemic Heart Disease, Pharmacol. Ther 116 (2007) 192–206. [DOI] [PubMed] [Google Scholar]
- [57].Ha T, Hua F, Liu X, Ma J, McMullen JR, Shioi T, Izumo S, Kelley J, Gao X, Browder W, Williams DL, Kao RL, Li C, Lipopolysaccharide-induced Myocardial Protection against Ischaemia/Reperfusion Injury is Mediated through a PI3K/Akt-dependent Mechanism, Cardiovasc. Res 78 (2008) 546–553. [DOI] [PubMed] [Google Scholar]
- [58].Fang F, Li D, Pan H, Chen D, Qi L, Zhang R, Sun H, Luteolin Inhibits Apoptosis and Improves Cardiomyocyte Contractile Function through the PI3K/Akt Pathway in Simulated Ischemia/Reperfusion, Pharmacology 88 (2011) 149–158. [DOI] [PubMed] [Google Scholar]
- [59].Yu D, Li M, Tian Y, Liu J, Shang J, Luteolin Inhibits ROS-activated MAPK Pathway in Myocardial Ischemia/Reperfusion Injury, Life Sci. 122 (2015) 15–25. [DOI] [PubMed] [Google Scholar]
- [60].Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ, Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease, N. Z. Med. J 377 (2017) 1119–1131. [DOI] [PubMed] [Google Scholar]
- [61].Kim JW, Jin YC, Kim YM, Rhie S, Kim HJ, Seo HG, Lee JH, Ha YL, Chang KC, Daidzein Administration in Vivo Reduces Myocardial Injury in a Rat Ischemia/Reperfusion Model by Inhibiting NF-kappaB Activation, Life Sci. 84 (2009) 227–234. [DOI] [PubMed] [Google Scholar]
- [62].Sakamoto Y, Naka A, Ohara N, Kondo K, Iida K, Daidzein Regulates Proinflammatory Adipokines Thereby Improving Obesity-related Inflammation through PPARgamma, Mol. Nutr. Food Res 58 (2014) 718–726. [DOI] [PubMed] [Google Scholar]
- [63].Kumaran KS, Prince PS, Caffeic Acid Protects Rat Heart Mitochondria against Isoproterenol-induced Oxidative Damage, Cell Stress Chaperones 15 (2010) 791–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Moon MK, Lee YJ, Kim JS, Kang DG, Lee HS, Effect of Caffeic Acid on Tumor Necrosis Factor-alpha-induced Vascular Inflammation in Human Umbilical Vein Endothelial Cells, Biol. Pharm. Bull 32 (2009) 1371–1377. [DOI] [PubMed] [Google Scholar]
- [65].Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Yoshida S, Graham RR, Manoharan A, Ortmann W, Bhangale T, Denny JC, Carroll RJ, Eyler AE, Greenberg JD, Kremer JM, Pappas DA, Jiang L, Yin J, Ye L, Su DF, Yang J, Xie G, Keystone E, Westra HJ, Esko T, Metspalu A, Zhou X, Gupta N, Mirel D, Stahl EA, Diogo D, Cui J, Liao K, Guo MH, Myouzen K, Kawaguchi T, Coenen MJ, van Riel PL, van de Laar MA, Guchelaar HJ, Huizinga TW, Dieude P, Mariette X, Bridges SL Jr., Zhernakova A, Toes RE, Tak PP, Miceli-Richard C, Bang SY, Lee HS, Martin J, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Rantapaa-Dahlqvist S, Arlestig L, Choi HK, Kamatani Y, Galan P, Lathrop M, Eyre S, Bowes J, Barton A, de Vries N, Moreland LW, Criswell LA, Karlson EW, Taniguchi A, Yamada R, Kubo M, Liu JS, Bae SC, Worthington J, Padyukov L, Klareskog L, Gregersen PK, Raychaudhuri S, Stranger BE, De Jager PL, Franke L, Visscher PM, Brown MA, Yamanaka H, Mimori T, Takahashi A, Xu H, Behrens TW, Siminovitch KA, Momohara S, Matsuda F, Yamamoto K, Plenge RM, Genetics of Rheumatoid Arthritis Contributes to Biology and Drug Discovery, Nature 506 (2014) 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wu Z, Cheng F, Li J, Li W, Liu G, Tang Y, SDTNBI: an Integrated Network and Chemoinformatics Tool for Systematic Prediction of Drug-target Interactions and Drug Repositioning, Brief. Bioinform 18 (2017) 333–347. [DOI] [PubMed] [Google Scholar]
- [67].Wu Z, Lu W, Wu D, Luo A, Bian H, Li J, Li W, Liu G, Huang J, Cheng F, Tang Y, In Silico Prediction of Chemical Mechanism of Action via an Improved Network-based Inference Method, Br. J. Pharmacol 173 (2016) 3372–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, Lahr DL, Hirschman JE, Liu Z, Donahue M, Julian B, Khan M, Wadden D, Smith IC, Lam D, Liberzon A, Toder C, Bagul M, Orzechowski M, Enache OM, Piccioni F, Johnson SA, Lyons NJ, Berger AH, Shamji AF, Brooks AN, Vrcic A, Flynn C, Rosains J, Takeda DY, Hu R, Davison D, Lamb J, Ardlie K, Hogstrom L, Greenside P, Gray NS, Clemons PA, Silver S, Wu X, Zhao WN, Read-Button W, Wu X, Haggarty SJ, Ronco LV, Boehm JS, Schreiber SL, Doench JG, Bittker JA, Root DE, Wong B, Golub TR, A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles, Cell 171 (2017) 1437–1452.e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Litichevskiy L, Peckner R, Abelin JG, Asiedu JK, Creech AL, Davis JF, Davison D, Dunning CM, Egertson JD, Egri S, Gould J, Ko T, Johnson SA, Lahr DL, Lam D, Liu Z, Lyons NJ, Lu X, MacLean BX, Mungenast AE, Officer A, Natoli TE, Papanastasiou M, Patel J, Sharma V, Toder C, Tubelli AA, Young JZ, Carr SA, Golub TR, Subramanian A, MacCoss MJ, Tsai LH, Jaffe JD, A Library of Phosphoproteomic and Chromatin Signatures for Characterizing Cellular Responses to Drug Perturbations, Cell Syst. 6 (2018) 424–443.e427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bedi O, Dhawan V, Sharma PL, Kumar P, Pleiotropic Effects of Statins: New Therapeutic Targets in Drug Design, Naunyn Schmiedebergs Arch. Pharmacol. 389 (2016) 695–712. [DOI] [PubMed] [Google Scholar]
- [71].Cheng F, Zhao J, Fooksa M, Zhao Z, A Network-based Drug Repositioning Infrastructure for Precision Cancer Medicine through Targeting Significantly Mutated Genes in the Human Cancer Genomes, J. Am. Med. Inform. Assoc 23 (2016) 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhang YC, Lu BJ, Zhao MH, Rong YZ, Chen RM, Effect of Shengmai Injection on Vascular Endothelial and Heart Functions in Patients with Coronary Heart Disease Complicated with Diabetes Mellitus, Chin. J. Integr. Med 14 (2008) 281–285. [DOI] [PubMed] [Google Scholar]
- [73].Zhang H, Wang WR, Lin R, Zhang JY, Ji QL, Lin QQ, Yang LN, Buyang Huanwu Decoction Ameliorates Coronary Heart Disease with Qi Deficiency and Blood Stasis Syndrome by Reducing CRP and CD40 in Rats, J. Ethnopharmacol 130 (2010) 98–102. [DOI] [PubMed] [Google Scholar]
- [74].He M, Yan X, Zhou J, Xie G, Traditional Chinese Medicine Database and Application on the Web, J. Chem. Inf. Comput. Sci 41 (2001) 273–277. [DOI] [PubMed] [Google Scholar]
- [75].Xue R, Fang Z, Zhang M, Yi Z, Wen C, Shi T, TCMID: Traditional Chinese Medicine Integrative Database for Herb Molecular Mechanism Analysis, Nucleic Acids Res. 41 (2013) D1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y, Xu X, Li Y, Wang Y, Yang L, TCMSP: a Database of Systems Pharmacology for Drug Discovery from Herbal Medicines, J. Cheminform 6 (2014) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chen CY, TCM Database@Taiwan: the World’s Largest Traditional Chinese Medicine Database for Drug Screening in Silico, PLoS One 6 (2011) e15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhang RZ, Yu SJ, Bai H, Ning K, TCM-Mesh: The Database and Analytical System for Network Pharmacology Analysis for TCM Preparations, Sci Rep. 7 (2017) 2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kim SK, Nam S, Jang H, Kim A, Lee JJ, TM-MC: a Database of Medicinal Materials and Chemical Compounds in Northeast Asian Traditional Medicine, BMC Complement. Altern. Med. 15 (2015) 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrian-Uhalte E, Davies M, Dedman N, Karlsson A, Magarinos MP, Overington JP, Papadatos G, The ChEMBL Database in 2017, Nucleic Acids Res. 45 (2017) D945–d954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J, BindingDB in 2015: A Public Database for Medicinal Chemistry, Computational Chemistry and Systems Pharmacology, Nucleic Acids Res. 44 (2016) D1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M, STITCH 5: Augmenting Protein-chemical Interaction Networks with Tissue and Affinity Data, Nucleic Acids Res. 44 (2016) D380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ye H, Ye L, Kang H, Zhang D, Tao L, Tang K, Liu X, Zhu R, Liu Q, Chen YZ, Li Y, Cao Z, HIT: Linking Herbal Active Ingredients to Targets, Nucleic Acids Res. 39 (2011) D1055–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR, Open Babel: An Open Chemical Toolbox, J. Cheminform 3 (2011) 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A, OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an Online Catalog of Human Genes and Genetic Disorders, Nucleic Acids Res. 43 (2015) D789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN, The Human Gene Mutation Database: towards a Comprehensive Repository of Inherited Mutation Data for Medical Research, Genetic Diagnosis and Next-generation Sequencing Studies, Hum. Genet 136 (2017) 665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H, The NHGRI GWAS Catalog, A Curated Resource of SNP-trait Associations, Nucleic Acids Res. 42 (2014) D1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ, The Comparative Toxicogenomics Database: Update 2017, Nucleic Acids Res. 45 (2017) D972–d978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Pinero J, Bravo A, Queralt-Rosinach N, Gutierrez-Sacristan A, Deu-Pons J, Centeno E, Garcia-Garcia J, Sanz F, Furlong LI, DisGeNET: A Comprehensive Platform Integrating Information on Human Disease-associated Genes and Variants, Nucleic Acids Res. 45 (2017) D833–d839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Benjamini Y, Hochberg Y, Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing, Journal of the royal statistical society. Series B (Methodological), (1995) 289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.