Abstract
Most bacterial genomes display contrasting strand asymmetry in a variety of features, such as nucleotide composition and gene orientation, of the two replichores separated by the replication origin and terminus. The cause for the polarization is often attributed to mutations arising from the asymmetric replication machinery. Notably, a base compositional bias known as a GC skew is focused on as a footprint of the bacterial genome evolution driven by DNA replication. Previously, although a replication driven mutation pattern responsible for the GC skew formation or the related mathematical models have been well reported, an exact impact of the replication-related elements on the genomic structure is yet actively debated, and not confirmed experimentally. However, the GC skew formation is very time consuming and challenging in the laboratory. We, therefore, used cytosine deaminase as a DNA mutator, and by monitoring the mutations during an accelerated laboratory evolution procedure with Illumina sequencing, we enabled the trial and error of the GC skew formation in high resolution. Using this technology, we succeeded in reconfirming the influence of bacterial replication machinery on the genomic structure at high resolution.
Keywords: circular chromosome, ultra-sensitive quantification of heterogeneous mutations, replication-directed genomic asymmetry, genomic polarity
Introduction
Nucleotide composition bias between the leading and lagging strands was discovered in the earliest bacterial genome sequencing projects (Lobry 1996; Casjens 1998; Frank and Lobry 1999; Bentley and Parkhill 2004), and its analysis is now an indispensable means to computationally locate the replication origin in order to define the first base position of circular genomes (Frank and Lobry 2000). The biological cause of such bias, including multifactorial causes for the mutations and selection pressures, has been actively debated, with controversial views on the extent of the contribution of replication and transcription (Francino et al. 1996; Rocha et al. 2006; Chen et al. 2016). However, Bhagwat et al. (2016) recently experimentally demonstrated replication-driven GC skew by observing ssDNA deamination. As replication involvement is now evident, the investigation of the specific impact of the replication machinery on the bacterial genome structure is required. For example, the degree to which replication directly processes the genomic structure can be observed by deleting the replication-related element. Previously, we simulated the impact of replication-related elements on the alteration of the genomic structure and demonstrated that termination-related factors have a much greater impact on the GC skew than do transcription-related factors (Kono et al. 2012). Then, these computationally predicted results should be confirmed experimentally. However, because their method requires bottleneck passage culturing to collect numerous mutation sites, it is time-consuming, and verification under multiple conditions is not easy. Furthermore, although the 0.01% mutation rate can demonstrate the outline of the GC skew, it does not detect subtle changes, because this mutational resolution will be indistinguishable at a single-gene level.
To address the above technological problem, a novel approach combining accelerated laboratory evolution with cytosine deamination mutation (Harris et al. 2002) and ultrasensitive quantification of heterogeneous mutations is required. Cytosine deamination of ssDNA is a spontaneous mutation and is thought to be the most direct cause of the deviation of nucleotide composition in proteobacterial replichores (Coulondre et al. 1978; Reyes et al. 1998; Frank and Lobry 1999; Rocha et al. 2006; Khrustalev and Barkovsky 2010). Thus, a laboratory evolution experiment designed to control cytosine deamination is required to test this hypothesis and observe the transition of bacterial genome polarization. Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (APOBEC3G) is a human enzyme that deaminates cytidine to uridine in an ssDNA-specific manner (Conticello et al. 2007; Chiu and Greene 2008). A3G-CTD is the carboxyl terminal domain (CTD) (Carpenter et al. 2010) of human APOBEC3G and shows activity in heterologous hosts such as E. coli (Harris et al. 2002; Petersen-Mahrt et al. 2002) and yeast (Mayorov et al. 2005). Therefore, A3G-CTD mutation analogously promotes the spontaneous deamination of ssDNA for the efficient induction of genotypic diversity (Bhagwat et al. 2016). Additionally, the deletion of a uracil DNA glycosylase (ung) gene, which excises the deaminated cytosine, enhances the effectiveness of the mutation (Bhagwat et al. 2016). However, even in the presence of these mutations, many generations are required to verify the mutational bias with existing methodologies that employ single colony bottlenecking. Moreover, single colony bottlenecking is a selection step using single colony isolation, and determining whether the substitutions result from mutation or selection is difficult.
Materials and Methods
Escherichia coli Strains and Plasmids
The E. coli strains and plasmids used in this study are listed in supplementary table S1, Supplementary Material online. All E. coli mutants were based on JW strains from the Keio collection (Baba et al. 2006). The E. coli BANK12035 (Δung) and BANK12034 (Δtus) strains are JW2564 (Δung::Km) and JW1602 (Δtus::Km) strains, respectively, in which markers were eliminated by an l-arabinose-induced flippase (FLP) recombinase expressed in pKD322. Gene knockout was performed using the protocol for constructing Keio collection mutants (Baba et al. 2006). The BANK12037 (Δtus Δung::Km) strain was isolated by deleting the ung gene using an l-arabinose-induced λ-red recombinase (Datsenko and Wanner 2000) expressed in pKD46 and an appropriate flippase recognition target (FRT)-flanked kanamycin (Km) resistance gene fragment from the BANK12034 strain. The BANK12049 (Δung Δdif::Km) strain was isolated by deleting the dif sequence using an l-arabinose-induced λ-red recombinase (Datsenko and Wanner 2000) expressed in pKD46 and an appropriate FRT-flanked Km resistance gene fragment from the BANK12035 strain. Each FRT-flanked Km resistance gene fragment and the target gene or sequence was amplified from pKD13 using the appropriate primers (Baba et al. 2006). The pGST-A3G-CTD plasmid was constructed as described in previous studies (Carpenter et al. 2010; Bhagwat et al. 2016). The artificially synthesized A3G-CTD sequence (Eurofins Genomics) was cloned into the SmaI and XhoI sites of the pGST-6p-2 expression vector (GE Healthcare Life Sciences). The pGST-A3G-CTD vector was transformed into the E. coli DH5α strain, and the transformant was selected after culture on a carbenicillin (Carb)-treated plate.
Culture Conditions
Escherichia coli strains were grown in Luria–Bertani (LB) broth or agar (1.5% w/v) supplemented with 100 μg/ml Carb or 30 μg/ml Km for selection, and 100 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) was used to induce A3G-CTD expression from pGST-A3G-CTD. Overnight cultures were prepared in 2 ml of LB broth in a 14-ml round-bottom tube and incubated at 37 °C for 16 h with rotation. Strains harboring pKD46 for λ-recombination or pKD322 for FLP recombination were grown at 30 °C to induce repA101ts expression.
Computational Analysis and Databases
All bioinformatics analyses were conducted using custom Perl scripts in G-language Genome Analysis Environment (v1.9.1) (Arakawa et al. 2003). The cumulative GC skews were calculated using the “gcskew” function with the cumulative parameter, and the generalized GC skew indexes (GCSIs) (Arakawa et al. 2009) were calculated using the “gcsi” function in G-language GAE. The statistical analyses and visualizations were performed using the R statistics package, version 3.2.1. The genomic sequence (CP009273.1: October 30, 2014) of the E. coli parent strain (BW25113) was obtained from the National Center for Biotechnology Information (NCBI) FTP Repository, and the A3G-CTD sequence was obtained from a previous study (Carpenter et al. 2010). RNA-seq data for E. coli strain K-12 substrain MG1655 were obtained from the NCBI Gene Expression Omnibus (GEO) database (GSM1104387-9, containing data for three biological replicates; McClure et al. 2013). The E. coli gene expression profile was calculated using Kallisto (v0.42.2.1) with the default parameters. The randomized genomes used for the GC skew calculation were computationally constructed based on randomly shuffled substitution sites with 100 replications. The average scores at each position were used as the randomized genome GC skew score. The sequenced reads from the ultrasensitive quantification of heterogeneous substitutions were assessed with FastQC (v0.10.1) and mapped on each parent genome sequence using BWA-MEM (0.7.11-r1034) (Li and Durbin 2009). We extracted only 1-bp mismatch reads from the mapped reads using custom Perl scripts. Using the extracted mismatch reads, various de novo substitutions were collected, and the coverage was calculated for each position based on the alignment results. The collected substitutions were based on an appropriate coverage threshold (fig. 1c). The sequenced reads in the strand-specific ssDNA sequencing (4S-seq) data contained a molecular tag sequence in addition to the adapter, and this tag sequence was used to determine whether the read originated from the Watson or the Crick strand. The calculated strand bias was represented as the strand skew: (Watson−Crick)/(Watson + Crick). A sequence logo was generated using WebLogo (Crooks et al. 2004).
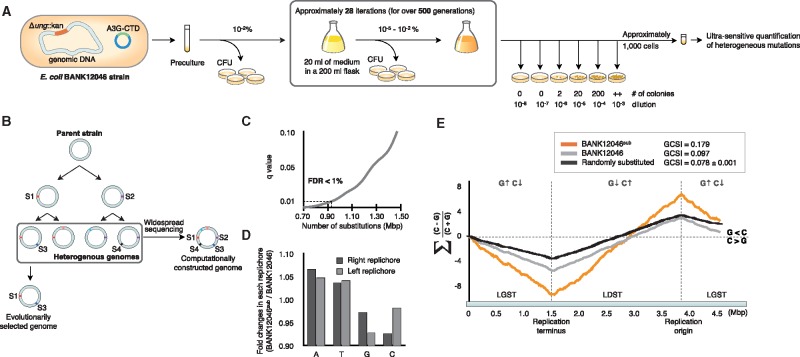
Fig. 1.
—Laboratory evolution approach without single colony bottlenecking and with ultrasensitive quantification of heterogeneous substitutions. (a and b) Overview of the serial transfer culture method and ultrasensitive quantification of heterogeneous substitutions. Our laboratory evolution strategy uses a culture passaging experiment that calculates the colony-forming units (CFUs) for each transfer without the isolation step. The method used for the ultrasensitive quantification of heterogeneous substitutions combines a sequencing method with the accumulation of heterogeneous genomes by serial culture. The conventional sequencing method (vertical arrow) sequences the homogeneous genome, and thus only two substitution sites (S1 and S2) are obtained. However, because our ultrasensitive quantification of heterogeneous substitutions method (horizontal arrow) uses heterogeneous genomes, it comprehensively collects all single nucleotide variations (see the Materials and Methods section for details). (d) Ratios of each nucleotide in both replichores. (e) Cumulative GC skew after laboratory evolution. The x axis shows the genome position (Mb), and the y axis shows the cumulative GC skew score. The black graph shows the BANK12046 genome (before laboratory evolution), the orange graph represents the BANK12046sub genome (after laboratory evolution), and the gray graph shows the GC skew in the mutated positions of the shuffled genome with error bars (SD).
Serial Transfer Culture Experiment
Escherichia coli strains were revived from frozen stocks by streaking on LB plates and culturing overnight at 37 °C. Isolated single colonies were picked, placed in 2 ml of LB in 14-ml round-bottom tubes and incubated overnight at 37 °C with rotation. We transferred 2–0.02 µl of culture in medium to 20 ml of fresh LB supplemented with Carb-IPTG in a 200-ml flask. This transfer was conducted when the culture growth reached the late exponential phase (approximate OD600 = 0.2) to maintain the fastest doubling time. The incubation was implemented using a T-2S thermostatic water bath (Thomas Scientific) with a shaker (110 rpm). At transfer, spread plating was also performed to enumerate the colony-forming units (CFUs)/ml. The number of generations was calculated based on the CFUs. The serial transfer culture experiments were performed until the number of generations exceeded 500.
Ultrasensitive Quantification of Heterogeneous Substitutions
Ultrasensitive quantification of heterogeneous substitutions is a technique used to collect the various substitutions from low-coverage sequencing of heterogeneous genomes. Therefore, the serial transfer culture was diluted to ∼1,000 cells, according to the CFU, to obtain the minimum coverage based on the sequencing instrument. A sequence library was prepared from each sample using the standard protocol from a KAPA HyperPlus Kit (KAPA Biosystems). Genomic DNA was purified using a DNeasy Blood & Tissue Kit (Qiagen). The DNA was eluted in 10-mM Tris–HCl (pH 8.0), dissociated into 350-bp fragments using a Covaris M220 instrument (Covaris), end repaired, poly (A)-tailed, and ligated to Illumina TruSeq adapters. The library amplification step was performed in seven cycles using KAPA HiFi HotStart ReadyMix (KAPA Biosystems). Each sample was sequenced with a NextSeq 500 instrument (Illumina, Inc.) using a 150-bp paired-end read chemistry with a NextSeq 500 High Output Kit v.2 (300 cycles). Control sequencing reactions lacking plasmid were performed on strains BANK12035 (Δung), BANK12037 (Δung, Δtus), and BANK12049 (Δung, Δdif) to investigate the relationship between the error rate and sequence coverage and to curate the substitutions collected from the ultrasensitive quantification of heterogeneous substitutions. The methods used to purify the genomic DNA, prepare the library, and perform sequencing are described earlier.
Error Correction for the Ultrasensitive Quantification of Heterogeneous Substitutions
Ultrasensitive quantification of heterogeneous substitutions is a method to comprehensively collect substitutions from a heterogeneous genome pool using a sequencer. Therefore, the collection system should remove error-derived substitutions from the obtained substitution candidates using minimum coverage. The probability distributions of the sequencing error for 12 substitution patterns in each of three E. coli strains, BANK12046 (Δung, pGST-A3G-CTD), BANK12040 (Δtus, Δung, pGST-A3G-CTD), and BANK12050 (Δdif, Δung, pGST-A3G-CTD), were calculated based on the Poisson distribution. The actual measured sequencing error rate for these three E. coli strains was obtained from the sequencing the control strain lacking each plasmid (BANK12035, BANK12037, BANK12049) as described earlier. Using the calculated probability distribution, the false discovery rate (FDR) of the frequency of alleles obtained from the ultrasensitive quantification of heterogeneous substitutions was estimated, and a 1% FDR threshold was used for validating the substitution.
Strand-Specific ssDNA Sequencing
The detailed 4S-seq protocol is described in the supplementary methods, Supplementary Material online. The overall 4S-seq protocol is broadly divided into ssDNA enrichment and strand-specific sequencing steps. The ssDNA was enriched by cleaving the dsDNA contained in the fragmented genomic DNA using a duplex-specific nuclease (DSN; Evrogen). Strand-specific sequencing was implemented using a biotinylated adapter with an exclusive uniquely designed tag sequence (5′-GGGAANNNNNNNNTAGGGATAACAGGGTAATAGGAGGA-3′). Adapter-ligated ssDNAs were immobilized on streptavidin-coated beads, and complementary strands were synthesized with a polymerase. Because the designed adapter contains an I-SceI site (5′-TAGGGATAACAGGGTAAT-3′), I-SceI digestion releases the dsDNA fragments from the streptavidin-coated beads. Because a low amount of released fragments was expected, the library was prepared using a ThruPLEX DNA-seq Kit (RUBICON GENOMICS). Sequencing was performed on the NextSeq 500 instrument (Illumina, Inc.) using 75-bp single-end read chemistry with a NextSeq 500 High Output Kit (75 cycles). To validate this 4S-seq approach, a model DNA substrate (a mixture of known dsDNA and ssDNA) was used. We used a pUC19 plasmid (2,686 bp) as the dsDNA, and the ssDNA was obtained by denaturing the amplified region of the pUC19 plasmid. Using the pUC19 plasmid as a template, we amplified a multiple cloning region (98 bp) with an M13 primer set (5′-AGTCACGACGTTGTA-3′/5′-CAGGAAACAGCTATGAC-3′). The PCR cycling parameters were as follows: one cycle at 94 °C for 3 min; 30 cycles at 94 °C for 20 s, 55 °C for 20 s, and 72 °C for 30 s; and one cycle at 72 °C for 1 min. The amplified DNA was denatured at 95 °C for 5 min and was rapidly cooled on ice. The dsDNA and ssDNA were mixed in equal molar amounts for the 4S-seq validation test. The sequence data are available from the DNA Data Bank of Japan (DDBJ) under bioproject no. PRJDB6239 (supplementary table S2, Supplementary Material online).
Results
Ultrasensitive Quantification of Heterogeneous Substitutions
We accelerated the laboratory evolution by combining Illumina sequencing with A3G-CTD mutation and ung gene deletion. We conducted serial transfer culture of E. coli without the single colony isolation step. By removing the isolation step, substitutions produced under lower selection pressure are generated, and the accumulated substitutions produce heterogeneous genome pools with nonuniform substitutions (fig. 1a). Using the isolated E. coli BANK12046 strain (Δung, A3G-CTD plasmid), we performed serial culturing for >500 generations (supplementary fig. S1, Supplementary Material online) and produced heterogeneous genomes. No change in the doubling time occurred over the 500 generations of culture passaging (supplementary fig. S1, Supplementary Material online). The obtained heterogeneous genomes were sequenced en masse with the Illumina sequencer (fig. 1b). Here, the innate heterogeneity was extremely high; therefore, the coverage of each of the lineages was sparse, even with the large number of reads. Sequencing errors and substitutions were determined according to the probability distribution of the sequencing error for each minor allele (12 patterns). The probability distribution was estimated by a control sequence of an E. coli strain lacking the A3G-CTD plasmid (see Materials and Methods for details). Consequently, ∼960,000 substitution sites were classified as derived from the E. coli genome, not as a result of a sequencing error (P ≤ 0.01, fig. 1c). Based on these protocols for the ultrasensitive quantification of heterogeneous substitutions, the substitution ratio was increased by 250-fold compared with that in the previous study (Bhagwat et al. 2016). Furthermore, as we confirmed the sequence context of the C: G to T: A substitution, an appropriate sequence context (5′-CCCR-3′; supplementary fig. S2, Supplementary Material online) was preferred by the A3G-CTD mutation, as previously reported (Bhagwat et al. 2016).
Asymmetrically Accumulated Substitutions in Each Replichore
Using the collected substitution data, we reconstructed an artificial genome data set in silico that included all identified de novo substitutions (hereinafter called the BANK12046sub genome) and was based on the BANK12046 genome. As expected, the genomic GC content decreased, consistent with the decrease in the C content caused by C deamination (table 1). Moreover, the compositions of each nucleotide were asymmetric in both replichores, suggesting a replication-related strand bias in the substitutions (fig. 1d). The asymmetric substitution patterns in the BANK12046sub genome were confirmed using GC skew visualization (fig. 1e). The GC skew is a measure of the base distribution (Lobry 1996; Frank and Lobry 1999) and is calculated as (C−G)/(C + G). The cumulative GC skew in the BANK12046 genome sharply decreased in the lagging strand template (LGST), indicating that compared with the C content, the G content in the LGST is enriched. In contrast, the leading strand template (LDST) is more enriched in C than in G. Although the GC skew in the BANK12046sub genome showed a similar trend as that in the BANK12046 genome, the skew was more evident, and the generalized GCSI (Arakawa et al. 2009), an index used to quantify the bias intensity, increased from 0.097 to 0.179 (fig. 1e). On the other hand, the GC skew in the randomized genomes constructed by randomly shuffling the substitution sites was more equilibrated, and the generalized GCSI decreased to 0.078 ± 0.001. Hence, the combination of serial transfer culture and ultrasensitive quantification of heterogeneous substitutions revealed the progression of the base composition bias through laboratory evolution on a realistic time scale and showed that C deamination accumulates asymmetrically in the two replichores. This result of increasing the GCSI by enhancing the GC skew bias is due to the combination of the A3G-CTD plasmid and ung deletion, as was shown in the previous study (Bhagwat et al. 2016). According to Bhagwat and colleagues, the mutation rate in the strain combining the A3G-CTD plasmid and ung deletion is also high in ordinary passage culture. The mutation rate decreases by 42% if the A3G-CTD plasmid is absent or catalytically inactive, and it drops to 12% in the wild-type (WT) strain. Furthermore, the frequency of the accumulation of asymmetric mutations has been clearly proven to be significantly higher in the strain harboring the A3G-CTD plasmid and ung deletion.
Table 1.
Genome substitution information
| Strains | Genome Size (bp) | Generationsa | Substitutions (P < 1%) | Cellsb | Mutation Rate Substitutions/(Generations×Cells×Genome size) | AT Content (%) | GC Content (%) | GCSI (GC Skew Index) |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli wild type | 4,641,652 | — | — | — | — | 49.22 | 50.78 | 0.097 |
| BANK12046sub | 4,630,884 | 547.98 | 961,509 | 900 | 4.21E-07 | 51.59 | 48.41 | 0.173 |
| BANK12040sub | 4,631,281 | 537.26 | 810,696 | 1,100 | 2.96E-07 | 49.05 | 50.95 | 0.157 |
| BANK12050sub | 4,632,159 | 552.22 | 1,020,558 | 1,000 | 3.99E-07 | 51.94 | 48.06 | 0.201 |
The number of generation in serial-transfer culture experiment.
The number of cells used for widespread sequencing.
Contributions of the Replication and Transcription Machinery
Mutagenesis by C deamination by A3G-CTD is based on ssDNA substrates, but the DNA duplex frequently separates within bacterial cells during both replication and transcription. The directions of replication and transcription are collinear in most genes, and the mutational bias may be due to transcription-coupled machinery. Therefore, we verified the contributions of replication and transcription to mutational bias by determining the variances of the substitution types and the correlations between the expression levels and substitution ratios. As a result, unlike the substitution patterns observed in the two replichores, the variance of the substitution types is nonsignificant in the leading/lagging strand gene regions (fig. 2a, F test P value < 0.001). Additionally, no significant correlations were observed between the gene expression levels and the collective substitution frequency in all gene regions or 4-fold synonymous sites (ρ = −0.051 or 0.051, supplementary fig. S3, Supplementary Material online). Furthermore, even if the mutation rate was calculated by grouping for each gene size, no significant correlations were found (ρ = 0.311, supplementary fig. S3, Supplementary Material online).
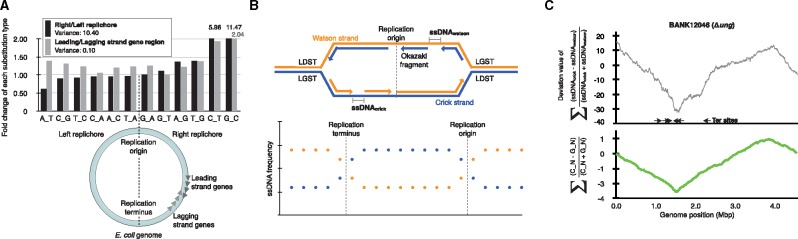
Fig. 2.
—Relationship between genome-associated events and substitution frequency. (a) The fold change in the number of collected substitutions between replichores (black bars) or gene strands (gray bars). The substitution types show the asymmetric deviation in the right and left replichores (variance = 10.40). On the other hand, the substitution types observed in the transcribed regions were independent of the strand (variance = 0.10). (b) Overview of the 4S-seq method. The enriched ssDNA region was sequenced and quantified according to the strand (Watson or Crick) data. (c) The upper panel shows the 4S-seq plot. The lower panel graphs the skew of C to N and G to N substitutions.
Substituted positions must be verified to be located in single-stranded regions in order to investigate the contribution of the DNA replication process to mutational strand bias. Many strand-specific sequencing technologies are available for eukaryotic genomes, with various selection protocols for nascent DNA (Hyrien 2015). For example, nascent DNA is commonly purified by immunoprecipitation with BrdU (Karnani et al. 2010) or a combination of antibodies against strand-specific binding proteins (Yu et al. 2014). Other protocols utilize an agarose gel trap (Mesner et al. 2011) or a customized label (Smith and Whitehouse 2012) to select nascent DNA of a certain size. However, these strand-specific sequencing technologies for eukaryotic organisms are not readily applicable to bacterial genomes because the BrdU incorporation efficiency is overwhelmingly low in bacteria. Therefore, we developed a novel 4S-seq method for the bacterial genome. The 4S-seq method reveals the ssDNA content in each strand (Watson/Crick) at each position of the genome (fig. 2b). The developed 4S-seq method was validated with a model DNA substrate (supplementary fig. S4a, Supplementary Material online). As a result, the target ssDNA region was robustly enriched and sequenced (supplementary fig. S4b, Supplementary Material online), and the 4S-seq protocol was experimentally validated. ssDNA was enriched in the genomic DNA with a dsDNA-specific nuclease, and a strand-specific library was prepared using a biotin label with a strand-specific sequence tag (see the supplementary methods and supplementary fig. S5, Supplementary Material online, for details). As a result, ssDNA was clearly enriched in the LGST and correlated with the substitution bias (fig. 2c).
Relationship between the Substitution Tendency and Replication-Related Factors
We examined how the DNA replication style affects the distribution of de novo substitutions to further evaluate the relationship between the DNA replication process and biased substitution. For this purpose, we selected two mutants lacking either the terminus utilization sequence (tus) gene or the deletion-induced filamentation (dif) sequence (fig. 3a). The tus gene encodes a DNA-binding protein (Tus) and forms a unidirectional barrier to replication fork progression that terminates the DNA replication process (Kamada et al. 1996). In the absence of the tus gene, DNA replication terminates at the site opposite the replication origin in Bacillus subtilis (Kono et al. 2014). The dif sequence is a widely conserved cis element in bacteria (Kono et al. 2011) that plays a central role in chromosome dimer resolution (CDR) as the binding site for tyrosine recombinases (Lesterlin et al. 2004). Owing to inefficient CDR and cell division (Hendricks et al. 2000), the dif deletion mutant exhibits slow growth, and the replication cycle is lengthened.
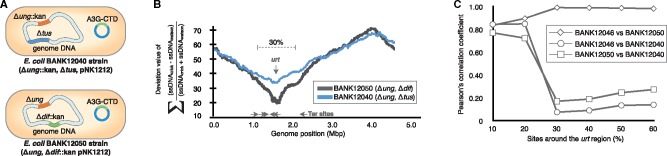
Fig. 3.
—Use of strand-specific ssDNA sequencing (4S-seq) to elucidate the effects of the replication termination machinery on replication-associated substitutions. (a) Schematic of the mutants isolated for 4S-seq. (b) 4S-seq plot of each mutant. The black and blue lines represent the BANK12050 and BANK12040 strains, respectively. The gray horizontal arrows show Ter sites, and the vertical arrow shows the urt site. (c) Correlation coefficient for the fold change in the collected substitution types between replichores around the urt region (10–60% of the genomic region). The deviations, as shown by a decrease in the correlation coefficient in regions located in 30% of the sites around urt, show that the BANK12040 (Δung, Δtus) strain utilizes replication termination machinery different from that used by the BANK12050 strain.
Substitution Bias around the Replication Terminus Region
The ssDNA behavior was observed in each mutant using 4S-seq, and as expected, the mode of replication reflected a change in the ssDNA regions around the replication terminus (fig. 3b). In the E. coli BANK12040 (tus−) strain, the strand bias of single-stranded regions around the terminus decreased because the fork trap was deleted and replication did not terminate at replication termination (Ter) sites, resulting in a weaker shift in polarity. As shown in our previous study, an undesigned replication terminus (urt) is formed at the site opposite the replication origin in this strain (Kono et al. 2014). Although the doubling time was longer in the E. coli BANK12050 (dif−) strain (supplementary fig. S1, Supplementary Material online), the 4S-seq result for this strain was similar to that for the E. coli BANK12046 strain (fig. 2c), supporting the hypothesis that substitution bias is predominantly related to replication.
The substitutions in each mutant were collected as described earlier using >500 generations of serial culture and the ultrasensitive quantification of heterogeneous substitutions method (supplementary fig. S1, Supplementary Material online). The statistical data (table 1) and base composition bias (the GC skew shown in supplementary fig. S6, Supplementary Material online) were similar to those for the BANK12046 strain. Although the fold changes in the substitution frequencies between the right and left replichores in the BANK12046 and BANK12050 strains always showed similar patterns (Pearson’s correlation coefficient ≥ 0.838, fig. 3b), the BANK12040 strain, lacking tus, resulted in altered substitution patterns in 30% of the sites located around the urt region (Pearson’s correlation coefficient ≤ 0.276, fig. 3b). Again, substitution bias was clearly influenced by the replication machinery rather than by cell division timing and other mechanisms.
Discussion
The contributions of the replication machinery to the base composition polarization have been proposed in numerous previous studies (Fix and Glickman 1987; Rocha et al. 2006; Kono et al. 2012; Bhagwat et al. 2016). Here, based on our laboratory evolution experiments with ultrasensitive quantification of heterogeneous de novo substitutions induced by C deaminase and the use of 4S-seq to identify ssDNA positions within the bacterial chromosome, nascent ssDNA formed during replication is the substrate for C deamination. In E. coli, the experimentally confirmed linkage between substitution strand bias and the DNA replication process suggests that the DNA replication machinery drives the formation of genomic compositional polarity in the two replichores. Furthermore, our technology enabled the investigation of the impact of replication-related factors on the GC skew. As shown in figure 3, the lack of barriers to the replication fork drastically altered the GC skew in the terminal region, which is thought to be due to the reduction in mutation bias in the terminal region; this result supports our previous prediction (Kono et al. 2012).
The precise mechanisms of C deamination in WT E. coli are not yet clear, as an efficient and active cytosine deamination system, such as A3G-CTD, has not yet been identified. In yeast and human tumors using members of the APOBEC family, such mutational bias has been confirmed by whole-genome sequencing (Haradhvala et al. 2016; Hoopes et al. 2016). However, many different DNA glycosylases are widely conserved in bacteria. Thus, a combination of pathways for the induction, inhibition, and repair of deamination may exist, and the diversity of such pathways may result in a variety of substitution directions, as observed in a previous study (Rocha et al. 2006), although the mutations are predominantly related to replication. Moreover, our basic strategy for detecting de novo substitutions coupled with 4S-seq can be applied to other mutators in studies to elucidate the cause of the asymmetry in the genomic composition in other bacteria.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Nozomi Abe and Yuki Takai (Keio University) for DNA sequencing, and Prof. Masakazu Kataoka (Shinshu University) for providing strains. This research was supported by research funds from the Yamagata Prefectural Government and Tsuruoka City, Japan.
Literature Cited
- Arakawa K, et al. 2003. G-language genome analysis environment: a workbench for nucleotide sequence data mining. Bioinformatics 192:305–306. [DOI] [PubMed] [Google Scholar]
- Arakawa K, Suzuki H, Tomita M.. 2009. Quantitative analysis of replication-related mutation and selection pressures in bacterial chromosomes and plasmids using generalised GC skew index. BMC Genomics 101:640.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Parkhill J.. 2004. Comparative genomic structure of prokaryotes. Ann Rev Genet. 38:771–792. [DOI] [PubMed] [Google Scholar]
- Bhagwat AS, et al. 2016. Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli. Proc Natl Acad Sci U S A. 1138:2176–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MA, Rajagurubandara E, Wijesinghe P, Bhagwat AS.. 2010. Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair (Amst) 95:579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. 1998. The diverse and dynamic structure of bacterial genomes. Annu Rev Genet. 321:339–377. [DOI] [PubMed] [Google Scholar]
- Chen WH, Lu G, Bork P, Hu S, Lercher MJ.. 2016. Energy efficiency trade-offs drive nucleotide usage in transcribed regions. Nat Commun. 7:11334.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Greene WC.. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 261:317–353. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Yang Z, Neuberger MS.. 2007. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 94:37–73. [DOI] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W.. 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 2745673:775–780. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 146:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL.. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 9712:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fix DF, Glickman BW.. 1987. Asymmetric cytosine deamination revealed by spontaneous mutational specificity in an Ung-strain of Escherichia coli. Mol Gen Genet. 2091:78–82. [DOI] [PubMed] [Google Scholar]
- Francino MP, Chao L, Riley MA, Ochman H.. 1996. Asymmetries generated by transcription-coupled repair in enterobacterial genes. Science 2725258:107–109. [DOI] [PubMed] [Google Scholar]
- Frank AC, Lobry JR.. 1999. Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene 2381:65–77. [DOI] [PubMed] [Google Scholar]
- Frank AC, Lobry JR.. 2000. Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics 166:560–561. [DOI] [PubMed] [Google Scholar]
- Haradhvala NJ, et al. 2016. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell 1643:538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS.. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 105:1247–1253. [DOI] [PubMed] [Google Scholar]
- Hendricks EC, Szerlong H, Hill T, Kuempel P.. 2000. Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli. Mol Microbiol. 364:973–981. [DOI] [PubMed] [Google Scholar]
- Hoopes JI, et al. 2016. APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep. 146:1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. 2015. Peaks cloaked in the mist: the landscape of mammalian replication origins. J Cell Biol. 2082:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K.. 1996. Structure of a replication-terminator protein complexed with DNA. Nature 3836601:598–603. [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A.. 2010. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell. 213:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrustalev VV, Barkovsky EV.. 2010. The probability of nonsense mutation caused by replication-associated mutational pressure is much higher for bacterial genes from lagging than from leading strands. Genomics 963:173–180. [DOI] [PubMed] [Google Scholar]
- Kono N, et al. 2014. Undesigned selection for replication termination of bacterial chromosomes. J Mol Biol. 42616:2918–2927. [DOI] [PubMed] [Google Scholar]
- Kono N, Arakawa K, Tomita M.. 2011. Comprehensive prediction of chromosome dimer resolution sites in bacterial genomes. BMC Genomics 12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono N, Arakawa K, Tomita M.. 2012. Validation of bacterial replication termination models using simulation of genomic mutations. PLoS One 74:e34526.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesterlin C, Barre FX, Cornet F.. 2004. Genetic recombination and the cell cycle: what we have learned from chromosome dimers. Mol Microbiol. 545:1151–1160. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2514:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR. 1996. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 135:660–665. [DOI] [PubMed] [Google Scholar]
- Mayorov VI, et al. 2005. Expression of human AID in yeast induces mutations in context similar to the context of somatic hypermutation at G-C pairs in immunoglobulin genes. BMC Immunol. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure R, et al. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 4114:e140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, et al. 2011. Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res. 213:377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS.. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 4186893:99–103. [DOI] [PubMed] [Google Scholar]
- Reyes A, Gissi C, Pesole G, Saccone C.. 1998. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol. 158:957–966. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Touchon M, Feil EJ.. 2006. Similar compositional biases are caused by very different mutational effects. Genome Res. 1612:1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Whitehouse I.. 2012. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 4837390:434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, et al. 2014. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 564:551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.