Abstract
Objective.
Advances in genome-wide association studies have made possible the return of genetic risk results for complex diseases. Two concerns about these results are: a) negative psychological consequences; and b) viewing probabilistic results as deterministic, leading to misinterpretation and inappropriate decisions. The present study evaluates these concerns through a meta-analytic review of existing literature.
Methods.
Seventeen genetic testing studies of complex disease, including 1,171 participants and reporting 195 effects, 104 of which were unadjusted for covariates, were meta-analyzed under a random effects model. Diseases included Alzheimer’s, cardiovascular and coronary heart disease, lung cancer, melanoma, thrombophilia, and type II diabetes. Six domains of behavioral/psychological reactions were examined.
Results.
Carriers showed significantly increased self-reported behavior change compared to non-carriers when assessed six months or later after results return (Hedge’s g = .36, p = .019).
Conclusions.
Return of genetic testing results for complex disease does not strongly impact self-reported negative behavior or psychological function of at-risk individuals. Return of results does appear to moderately increase self-reported healthy behavior in carriers, although research on objectively observed behavioral change is needed. This is a growing area of research, with preliminary results suggesting potential positive implications of genetic testing for complex disease on behavior change.
Keywords: genetic testing, return of results, meta-analysis, complex disease, genetic essentialism
The return of genetic test results in the population at large is increasingly feasible as the cost of genome sequencing drops and knowledge about genotype-disease relationships increases (Mardis, 2011; Ritchie et al., 2015). To date, hundreds of thousands of individual whole genome sequences have been generated, including individual research studies that have sequenced over 10,000 individuals each (Telenti et al., 2016). An important question is whether and how such data may be returned to the individuals or patients who provided their DNA and, if returned, how those individuals may interpret and be affected by this information. In the traditional clinical genetic test results return model, doctors and genetic counselors act as conduits and gatekeepers. They order, interpret, and return results from genetic tests of targeted genomic loci for specific medical purposes (Yu et al., 2013). The system is not structured to return results for hundreds of thousands of individuals, each of whom could be tested at hundreds of millions of known varying sites in their genome where each variant is, in principle, a candidate for disease-related interpretation.
What is more, interpretations of genetic variants will change over time as research results continue to accumulate. For example, between 1995 and 2015 there were on average ~80,000 updates per year to the Online Mendelian Inheritance in Man (OMIM) (McKusick-Nathans Institute of Genetic Medicine, 2016a) database. From 2005 to 2015 there were an average of nearly 4,000 new entries each year in the NHGRI-EBI Catalog of published genome-wide association studies (GWAS), a repository of associations between genetic variants and (usually) complex diseases and traits (Burdett et al., 2016; McKusick-Nathans Institute of Genetic Medicine, 2016b). On the other hand, the number of genetic counselors and physicians available to interpret and return genetic test results is growing slowly. There are currently somewhere between two and three thousand licensed practicing genetic counselors in the United States (Bureau of Labor Statistics, 2015a), a number that is not expected to grow rapidly in the coming years (Accreditation Council for Genetic Counseling, 2016; Sarah Lawrence College, 2016). The number of practicing physicians is larger, of course (Bureau of Labor Statistics, 2015b), but only a small fraction believe they can capably and correctly interpret genetic test results. In a survey of 10,000 physicians, only 10% indicated comfort with genetic testing results (Christensen, Vassy, et al., 2016; Patay & Topol, 2012).
Another challenging and central aspect of returning genetic test results are the potential negative emotional and behavioral reactions when an individual learns about their genetic risk for some disease or trait. Individuals who receive genetic test results may overweight or misunderstand disease risk information. Termed “genetic essentialism,” this is the idea that lay-persons view genetic influences as powerful, deterministic, and immutable (Gould & Heine, 2012). In the case of disease risk, lay-persons may see genetic explanations as the main, or only, factor in disease etiology and may be more likely to feel distress or hopelessness when they are presented with genetic risk information (Dar-Nimrod & Heine, 2011). Indeed, the notion that individuals may be harmed by receiving too much information, or may misinterpret the results they are given, has influenced regulatory positions by the Food and Drug Administration (FDA), which has cited the possibility of misinterpretation as a potential source of harm to consumers who engage in direct-to-consumer testing (The Food and Drug Administration, 2013).
Historically, returning results for highly penetrant (often monogenic) disease-related mutations has been a common focus of genetic testing. Extensive reviews over the last few decades have evaluated results return for Huntington’s disease, hereditary breast and ovarian cancer, and Lynch syndrome (Hirschberg, Chan-Smutko, & Pirl, 2015; Leblond et al., 2011; Meiser & Dunn, 2001). These reviews generally contend that receipt of carrier status for highly penetrant mutations is related to a number of negative psychological outcomes including worry (Vansenne, Bossuyt, & de Borgie, 2009), general distress (Vansenne et al., 2009), depression (Leblond et al., 2011; Meiser & Dunn, 2001), anxiety (Hirschberg et al., 2015; Leblond et al., 2011), disease-specific distress (Hirschberg et al., 2015; Leblond et al., 2011), risk perception (Leblond et al., 2011; Vansenne et al., 2009), hopelessness (Meiser & Dunn, 2001), poorer general well-being (Meiser & Dunn, 2001), and decreased quality of life (Leblond et al., 2011), although carrier status is also associated with increases in report of healthy behavior (Leblond et al., 2011).
The largest effects on these outcomes are seen shortly after receipt of results (Leblond et al., 2011; Meiser & Dunn, 2001), with a steady tendency of individuals to return to pre-testing levels within six months of receipt (Meiser & Dunn, 2001). One significant limitation in this area of research is the lack of long-term follow-ups years after testing (Leblond et al., 2011; Meiser & Dunn, 2001), which have suggested incipient negative effects of genetic testing for severe disorders like Huntington’s (Timman et al., 2004). One review also noted positive behavioral aspects of genetic testing, including improved health and screening behavior in carriers as compared to non-carriers (Leblond et al., 2011).
In contrast to the extensive literature on diseases with large and clear genetic influences, there is relatively little research on complex diseases that show polygenic inheritance. This is due in part to the fact that the effects of individual genetic variants on complex traits are often small, and have only in the past few years begun to be discovered through large population-based genetic association studies. It is currently unclear what psychological harms or benefits, if any, can be expected from the return of genetic test results for complex diseases. This is perhaps especially true as the association between carrier status and disease is probabilistic, with most common mutations affecting complex diseases having odds ratios far less than 2 (Manolio et al., 2009). Thus, proper understanding of how a genetic variant affects one’s disease risk is more nuanced than for, say, Huntington’s or other monogenic diseases. Compounding this issue are the concerns about genetic essentialism described above, where individuals who receive genetic test results for mutations with small effects on disease risk may nevertheless disproportionately weight such findings in their life decisions based on the genetic test results.
In recent years, studies have begun to directly examine the psychological and behavioral consequences of returning genetic test results for common, complex diseases. Hollands et al. (2016) recently reviewed and meta-analyzed 18 studies comparing individuals randomly assigned to receive genetic test results for a complex disease (regardless of whether they carried a risk allele or not) versus individuals randomly assigned to not undergo genetic testing. They found no significant differences between these two conditions for outcomes ranging from depression and anxiety to adaptive behavioral changes such as increased sunscreen use to ward off skin cancer. Hollands et al. also conducted sub-analyses comparing outcomes between carriers of a risk allele and non-carriers (k = 10 studies included in the sub-analyses), but found no significant differences across outcomes with one exception. APOE-e4 carriers for Alzheimer’s disease showed significantly increased positive change in dietary supplements compared to non-carriers. Although the carrier vs. non-carrier subgroup analysis in Hollands et al. provides suggestive evidence of behavioral change following genetic testing receipt, conclusions are limited by their study inclusion/exclusion criteria, which required every included study to have a control group that never underwent genetic testing in the first place. Although all studies included in Hollands et al. were randomized control trials (RCTs), it is impossible to randomize carriers and non-carriers of a risk allele. As such, the randomized “intervention” arm in Hollands et al. contained two non-randomized groups (Hollands et al., 2016).
The Hollands et al. (2016) meta-analysis provides an important first step to evaluate the overall impact of return of genetic testing results for complex disease. The present article builds on this work by conducting a systematic review and meta-analysis of risk allele carrier versus non-carrier comparisons, analogous to the approach used to study the impact of test results for monogenic disorders. To do this, we compared the behavioral and psychological reactions of those who learned they are at higher genetic risk for common complex disease (i.e., carried a risk genotype) versus those who learn they are genetically protected from disease (i.e., did not carry a risk genotype). In contrast to Hollands et al. (2016), we included all studies that returned carrier status for common, complex disease, regardless of whether a control group was available, which resulted in a total of 17 relevant studies. This comparison of carriers and non-carriers is a crucial test of the psychological and behavioral effects of receiving positive versus negative test results from genetic testing.
Consistent with the theory of genetic essentialism, we expected that participants would show significant and disproportional behavioral change and psychological distress upon receipt of positive test results compared to negative results, even when those test results are positive for variants with small to moderate effects on disease risk.
Methods
Inclusion and Exclusion Criteria
To ensure that the meta-analysis only contained studies of common variants associated with common, complex diseases, we used the effect size associated with the APOE risk allele epsilon-4 (e4). This risk allele is perhaps the most well-known example of a common variant with a large effect on a common complex disease, thus we used it a benchmark for inclusion. Therefore, any studies returning information for mutations with effect sizes greater than that of a heterozygous APOE e3/e4 carrier (OR vs e3/e3 carrier = 3.6, 95% CI [3.4–3.9] (Genin et al., 2011)) under an additive model were excluded. Conditions where carriers could be already affected and therefore know their carrier status prior to testing, such as Alpha-1 antitrypsin deficiency (Zorzetto et al., 2008), were also excluded. In total, inclusion criteria were (1) comparison of genetic mutation carriers and non-carriers; (2) test results for a complex disease affected by a genetic mutation with an effect size lower than 3.6 under an additive model; (3) measure(s) of psychological or behavioral outcomes (self-reports or objective measures) after return of results; (4) quantitative measures of outcomes; (5) asymptomatic or unaffected subjects; (6) clear established link between genetic mutation and disease; (7) human; (8) English language; (9) study subjects were of adult age (18+ years old); and (10) either published in a peer-reviewed journal or an unpublished study or dissertation. Exclusion criteria were (1) test results for high-effect genetic mutations or monogenic disorders such as Huntington’s, BRCA1/2 mutations, or Lynch Syndrome; (2) hypothetical or vignette-based studies for measuring psychological or behavioral outcomes; (3) lack of comparison between carriers and non-carriers; (4) lack of specific disease or condition caused by or associated with mutation(s) of interest; (5) studies of participants who are already affected or diagnosed participants; (6) lack of quantitative information; (7) non-human; (8) pediatric (under 18 years old); (9) non-English; or (10) meeting abstract, poster presentation, or conference proceeding.
Literature Search
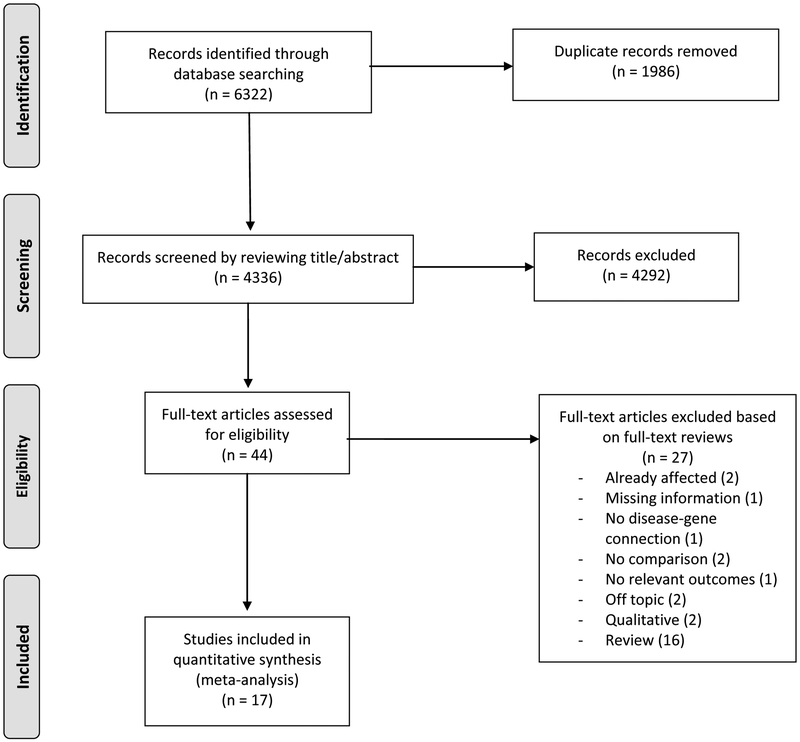
The literature search was conducted in February 2018 using the Web of Science, PsycINFO, and MEDLINE databases. Keywords used in the database searches were combinations of the terms ‘genetic test*’ AND (‘return of result*’ OR ‘psychol* impact*’ OR ‘behav* impact*’ OR ‘distress’). The search was limited to human studies published in English. In addition, references from studies read in full (detailed below) were examined for potential inclusion, as well as references from studies citing those read in full. The database and reference searches resulted in 4,336 unique articles. Titles and abstracts for all 4,336 articles were screened for inclusion, and 4,292 articles that did not fit the inclusion criteria were excluded. Forty-four articles were read in full, and 17 of these were included in the meta-analysis (see Figure 1 for the PRISMA flowchart of the literature review). Two coders independently reviewed all 44 full-text articles, and fully agreed on the inclusion and exclusion of all articles. No unpublished studies, including dissertations, met the inclusion/exclusion criteria. Therefore, all 17 studies included in the meta-analysis were studies published in peer-reviewed journals.
Figure 1. PRISMA flowchart of literature review.
Study Coding
The 17 studies that met inclusion criteria were coded for the following: (1) sample description; (2) disease(s) and genetic mutation(s) of interest; (3) psychological and/or behavioral outcome(s) measured; (4) outcome measurement tools; (5) timepoint(s) of outcome measurement; and (6) statistics for the calculation of effect sizes. An overview of all included studies, including gene and disease of interest, outcome(s) measured and measurement tools, and timepoint(s) can be found in Table 1. Two coders independently coded each of the 17 included studies. Interrater reliability coefficients (Cohen’s kappa for categorical ratings, intraclass correlation coefficients [ICCs] for continuous ratings) for the independent initial coding (prior to discussion and resolution by both coders, as needed) are noted below. By convention, kappas and ICCs ≥ .75 denote excellent agreement beyond chance and kappas and ICCs between .40 and .74 denote fair-to-good agreement (Fleiss, 1981). Any coding disagreements were resolved by consensus and the final consensus ratings were used in analyses.
Table 1.
Meta-Analysis Study Descriptions
| First Author, year | Gene, disease | Measure(s) | Outcome(s) | Timepoint(s) |
|---|---|---|---|---|
| (Aspinwall et al., 2008) | CDKN2A/p16, melanoma | Self-report questionnaire | Behavior Change | Baseline, immediately after, 1 month |
| (Aspinwall et al., 2009) | CDKN2A/p16, melanoma | Self-report questionnaire | Behavior Change, Behavioral Attitude | Baseline, immediately after, 1 month |
| (Aspinwall et al., 2013) | CDKN2A/p16, melanoma | Self-report questionnaire | Behavior Change | Baseline, 24 months |
| (Aspinwall et al., 2014) | CDKN2A/p16, melanoma | Self-report questionnaire | Behavior Change | Baseline, 24 months |
| (Chao et al., 2008)2 | ApoE-4, Alzheimer’s | Self-report questionnaire | Behavior Change | 12 months |
| (Christensen, Roberts, et al., 2016)1 | ApoE-4, Alzheimer’s and cardiovascular disease (CVD) | Self-report questionnaire | Behavior Change | 12 months |
| (Grant et al., 2013) | 21-allele risk score, type II diabetes | Self-report questionnaire | Behavior Change, Behavioral Attitude | Baseline, 12 weeks |
| (Green et al., 2009)1 | ApoE-4, Alzheimer’s | BAI, CES-D, IES | Anxiety, Depression, Disease-Specific | Baseline, 6 weeks, 12 months |
| (Heshka et al., 2008) | Factor V Leiden and prothrombin, thrombophilia | Self-report questionnaire | Behavior Change, Other Psychological | 12 months |
| (Hietaranta-Luoma et al., 2014)1 | ApoE-4, CVD | HTAS, self-report questionnaire | Behavior Change | Baseline, 10 weeks, 6 months, 12 months |
| (Hietaranta-Luoma et al., 2015)1 | ApoE-4, CVD | STAI. RBD, self-report questionnaire | Anxiety, Behavior Change, Other Psychological | 10 weeks, 12 months |
| (Kasparian et al., 2009) | CDKN2a, melanoma | HADS-A, HADS-D, IES, Self-report questionnaire | Anxiety, Behavior Change, Depression, Disease-Specific | Baseline, 2 weeks, 12 months |
| (Kullo et al., 2016) | 28-SNP risk score, coronary heart disease | STAI, PEFS, TAPAQ | Anxiety, Behavior Change | Baseline, 3 months, 6 months |
| (Legnani et al., 2006)1 | Factor V Leiden and G20210A, Thrombophilia | CBA-H, Self-report questionnaire | Other Psychological | Baseline, 20 days |
| (Sanderson et al., 2008)1 | GSTM1, lung cancer | IPQ-R, Self-report questionnaire | Anxiety, Behavior Change, Behavioral Attitude, Depression, Disease-Specific | Baseline, 1 week, 2 months |
| (Sanderson et al., 2009) | GSTM1, lung cancer | Self-report questionnaire | Behavior Change, Behavioral Attitude, Other Psychological | Baseline, immediately after, 6 months |
| (Vernarelli et al., 2010)2 | ApoE-4, Alzheimer’s | Self-report questionnaire | Behavior Change | 6 weeks |
Table 1. Descriptions of the final studies included in the meta-analysis. Descriptions include which gene(s) and/or mutation(s) and disease(s) were the results given in the study, the measures of different outcomes used, the outcomes measured and more specific descriptions, and the timepoints at which measures were taken. For descriptions of the outcomes, see Table S2. BAI= Beck Anxiety Inventory, CBA-H= Cognitive Behavioral Assessment Hospital Form, CES-D= Center for Epidemiologic Studies-Depression, HADS-A= Hospital Anxiety and Depression Scale-Anxiety, HADS-D= Hospital Anxiety and Depression Scale-Depression, HTAS= Health and Taste Screener, IES= Impact of Event Scale, IPQ-R= Revised Illness Perception Questionnaire, MICRA= Multidimensional Impact of Cancer Risk Assessment, PEFS= Percentage Energy from Fat Screener, RBD= Risk Behavior Diagnosis, STAI: State-Trait Anxiety Inventory, TAPAQ= Telephonic Assessment of Physical Activity Questionnaire.
outcomes reported in this study were adjusted, usually for demographics and baseline values.
outcomes reported in this study were adjusted, but non-adjusted values were reported and used.
Study Coding Ratings
Information coded for each study included average age of carriers and non-carriers (ICC = 1.00) and the percentage female of carriers and non-carriers (ICC = 1.00); any sample overlap with another included study was also noted. Information coded for disease and gene included disease name(s) (kappa = 1.00) and specific gene name(s) and/or mutation(s) of interest (kappa = 1.00). The diseases included were Alzheimer’s disease (k = 4), cardiovascular disease (k = 3), coronary heart disease (k = 1), lung cancer (k = 2), melanoma (k = 5), thrombophilia (k = 2), and type II diabetes (k = 1). One study included results for both Alzheimer’s disease and cardiovascular disease. This was the only study that included two disease outcomes, and we treated the Alzheimer’s results return separately from the Alzheimer’s/cardiovascular combined results return. Note that these two conditions (Alzheimer’s only versus Alzheimer’s and cardiovascular disease combined) used non-overlapping sets of participants, so treating them in the meta-analysis as separate samples is justified. The five melanoma studies were all conducted in two unique samples, and overlapping samples were only counted once for analysis. All study samples were >75% Caucasian or of European ancestry.
Psychological and Behavioral Outcomes Ratings
Three composite measures were constructed from individual effects and used as outcomes in the final meta-analysis: Behavior Change, Behavioral Attitudes, and Other Psychological. These outcomes were measured either through study-specific self-report questionnaires or previously-validated self-report questionnaires (See Table 1 for specific measures used in each study). Although either self-reported or objective measures of outcomes would have met our inclusion criteria, ultimately, all of the included studies and outcomes relied on self-reports. Effects for each outcome were synthesized across type of measure. The category ‘Behavior Change’ included self-reported measures of concrete changes in health behaviors, such as increases in exercise, diet change, and enacting preventative measures such as wearing sunscreen. ‘Behavioral Attitudes’ was comprised of measures of motivation, confidence, and intention around changing health behaviors and knowledge about one’s ability to prevent or alleviate disease risk. The category ‘Other Psychological’ included psychological outcomes that were not obviously related to clinical measures of anxiety and depression, such as stress, well-being, and overall emotional changes, both positive and negative. Non-composite measures were Anxiety, Depression, and Disease-Specific Distress. Information coded for each study included the specific psychological (Anxiety, Depression, Disease-Specific Distress, Other Psychological outcomes) and behavioral (Behavior Change, Behavioral Attitudes) outcomes measured (kappa = 1.00). Information coded for the questionnaires used to measure these study outcomes included the names of validated measures used to assess psychological and behavioral outcomes (e.g., the Impact of Event Scale, used to measure Disease-Specific distress) or specification of study-specific self-report questionnaires (kappa = 1.00). Information coded for the effect size calculations included sample sizes of carrier (ICC = 1.00) and non-carrier groups (ICC = 1.00) and statistics for comparisons between the two groups (ICC = 1.00).
Potential Risk of Study Bias
To assess bias in the studies included in this meta-analytic review, all included studies were reviewed by two independent coders for potential sources of bias following Cochrane guidelines for non-randomized studies (all included studies were non-randomized case-control studies, given that participants cannot be randomly assigned by a researcher to carrier versus non-carrier groups) (Becker L.A. & Oxman, 2011). All included studies were rated for bias (“high”, “low or no”, or “unclear”) in the following categories: selection bias (comparability of carrier and non-carrier groups at baseline) (ICC = 1.00), detection bias (blinding of study/outcome assessment personnel to carrier/non-carrier status) (ICC = .85), attrition bias (participant attrition at follow-up assessments) (ICC = .82), reporting bias (completeness and appropriateness of statistical outcome reporting) (ICC = .53), and potential sources of other bias (ICC = 1.00). Any coding disagreements were resolved by consensus.
Data Analysis
Effect sizes for comparing carriers and non-carriers were calculated from each study for each included outcome at each included timepoint. A Hedges g and respective standard error were calculated for every reported result from these studies by either calculating a standardized mean difference or a log odds ratio, which was then converted into Hedges g (Lipsey & Wilson, 2001). When a cell had zero observations (e.g., no controls reported feeling “worried”), 0.5 was added to the cell in order to estimate a standard error and allow the cell to contribute to the analysis (Lipsey & Wilson, 2001). Sample overlap was taken into account such that any sample reported on multiple times was counted only once in analysis. Observations taken after receipt of results clustered into three timepoints: 1) baseline, before testing results were returned; 2) six months or sooner after results; and 3) six months or longer after results. These timepoints are referred to as “Baseline”, “≤ 6 months”, and “> 6 months” for analysis. The average time of follow-up in the ≤ 6 month timepoint was 1.29 months after results return, and the average time of follow-up in the > 6 month timepoint was 14.14 months after results return. Not every study had an observation in each of the three timepoint categories. If a study had two or more assessments after results return but before six months had passed, we used the earliest measure. If a study had two or more assessments after results return and after six months had elapsed, we took the latest assessment possible. This allowed for examination of the most proximal and distal effects on participants. The earliest timepoints give insight into the most immediate impact of results. There is a lack of studies in the literature focusing on the longitudinal impacts of returning genetic testing results (Hirschberg et al., 2015; Landsbergen et al., 2009), making it important to examine the furthest possible outcomes to address this gap.
An overall Hedges g effect size, variance, 95% confidence interval, and a two-tailed p-value were calculated for each result at each timepoint (Lipsey & Wilson, 2001). Effects from similar measures within a study (e.g., depression and anxiety) are likely correlated to some degree, but accounting for such dependencies among all measures, including highly study-specific measures such as disease-specific distress, is not feasible. Hence, we chose to report all outcomes at all timepoints to retain analytical and interpretive simplicity and generate a set of conservative results that can be interpreted with greater confidence.
The effect sizes from each measure were coded in standard meta-analytic format (Lipsey & Wilson, 2001), with effect sizes coded as positive or negative depending on the outcome in question. For Depression, Anxiety, Disease-specific Distress, and Other Psychological distress, a positive effect size (>0) indicates that carriers reported greater depression, anxiety or distress than non-carriers. For Behavioral Change and Behavioral attitudes, a positive effect size (>0) indicates that carriers reported greater healthy change or healthier attitudes than non-carriers. All analyses were conducted in R version 3.2.2 (R Core Team, 2015). The R package ‘MAd’ (Del Re, 2014) was used to compute a Hedges g and variance for each unique effect. Most studies (k = 13) reported multiple measures of one outcome (e.g., two different anxiety questionnaires assessed at < 6 months). All such measures are expected to be highly correlated, and thus were aggregated using ‘MAd’ as though they were correlated at .50 (the default (Wampold et al., 1997)) to create one overall “aggregate” effect per outcome per timepoint, following standard recommendations (Del Re, 2014). Study effects were then combined in a random-effects meta-analysis using the R package ‘metafor’ (Viechtbauer, 2010). Total study heterogeneity was measured with the standard Q statistic and summarized with I2, the percentage of the variability in the effect size due to heterogeneity across studies.
Several studies included in the meta-analysis reported only statistics adjusted for covariates, usually accounting for baseline measures, age, sex, education, or other demographic variables. When both unadjusted and adjusted statistics were reported, we used only the unadjusted statistics. We conducted a primary meta-analysis that included only unadjusted statistics; as the unadjusted results are the most robust, we highlight these in the main text of the paper. Since this is a nascent field, with only 17 studies, we chose to conduct a secondary meta-analysis of both unadjusted and, if unadjusted statistics were not available, adjusted statistics, in order to maximize the number of studies and the total meta-analytic sample size. We consider the combined analysis as exploratory, and so focus almost all attention here on the primary meta-analysis of only unadjusted statistics. (Full results for the combined meta-analysis are presented in the Supplement; for completeness, we also conducted a meta-analysis of adjusted statistics only, also presented in the Supplement. See Peters & Mengersen, 2008; Voils, et al., 2010 for further discussion of including unadjusted and adjusted statistics in meta-analysis (Peters & Mengersen, 2008; Voils et al., 2011).)
Results
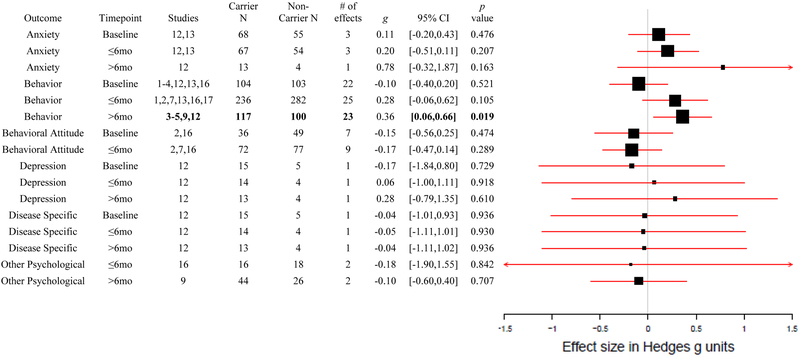
In total, 219 unique effects were identified and computed from a total of 17 studies. All 17 studies compared individuals who received a positive genetic test result for increased risk of disease (carriers), with individuals who received a negative result (non-carriers). After removing all but the most proximal and distal measures from return of results, a total of 195 unique effect sizes from 17 studies remained (number of studies at Baseline, k = 11; ≤ 6 months k = 12; > 6 months k = 9). Of these 195 effects, 104 were unadjusted and 91 had been adjusted for covariates (Baseline, k = 7; ≤ 6 months, k = 7; > 6 months, k = 5). No studies reported both adjusted and unadjusted effects on the same samples. All Baseline measures were taken before return of results, so the Baseline effects represent any systematic carrier and non-carrier differences before the effect of disclosure. An overall meta-analytic effect for each of the six outcomes was computed for each of the three timepoint clusters, resulting in 16 unadjusted meta-analytic effects (no unadjusted data for Baseline Other Psychological Distress or > 6 months Behavioral Attitude). Eleven of the total 17 studies reported unadjusted statistics (see Figure 2 for a forest plot of the unadjusted-only meta-analysis). In the separate combined analysis including adjusted and unadjusted statistics, there were 17 overall meta-analytic effects (please see the Supplement for more details on the combined meta-analysis).
Figure 2. Forest Plot of Only Unadjusted Meta-Analytic Results.
Forest plot of only unadjusted meta-analytic effect sizes for each of the six psychological and behavioral outcomes at the three timepoints; Baseline, ≤6 months, > 6 months, including which studies the effects were calculated from, carrier and non-carrier total N, number of effects included in the overall effect, and a 95% confidence interval and p value. Study number key: 1: Aspinwall, 2008, 2: Aspinwall, 2009, 3: Aspinwall, 2013, 4: Aspinwall, 2014, 5: Chao, 2008, 6: Christensen, 2016, 7: Grant, 2013, 8: Green, 2009, 9: Heshka, 2008, 10: Hietaranta-Luoma, 2014, 11: Hietaranta-Luoma, 2015, 12: Kasparian, 2009, 13: Kullo, 2016, 14: Legnani, 2006, 15: Sanderson, 2008, 16: Sanderson, 2009, 17: Vernarelli, 2010.
The meta-analytic outcomes of the analysis ranged from −.18 to.78. Of the 16 meta-analytic effects computed from unadjusted statistics, one was significant at p < .050. Carriers were significantly more likely to self-report positive Behavior Change than non-carriers at the > 6 months timepoint (k = 5, g = .36, p = .019). The unadjusted-only Behavior Change effects (k = 5) at the > 6 months timepoint were not significantly heterogeneous, Q(4) = 2.06, p =.724, I2 = 0.00%. There was less evidence for significant positive Behavior Change more proximal to return of results at ≤ 6 months (k = 6, g = .28, p = .105). This Behavior Change outcome ≤ 6 months after results also had the smallest non-significant p-value (.105). Unlike the significant > 6 months Behavior Change finding, the ≤ 6 months Behavior Change effects (k = 6) were significantly heterogeneous, Q(5) = 13.76, p = .017, I2 = 61.94%. While the shorter term ≤ 6 months Behavior Change was not significant, the effect size (g=.28) was similar to the longer term > 6 months Behavior Change finding, (g=.36), suggesting the existence of an effect, albeit non-significant at the < 6 months timepoint in the present sample sizes. Indeed, in the more exploratory combined meta-analysis, described in the supplement, which had a larger number of available studies, both Behavior Change effects were significant (≤ 6 months k = 9, g = .25, p = .038; > 6 months, k = 9, g = .37, p < .001). Neither effect was heterogeneous.
No other effect was significant in our meta-analysis of unadjusted effects, at any timepoint. Effect sizes ranged from −.18 to .78 in the non-Behavior Change outcomes, with p-values ranging from .163 to .936. Nearly all effects were in the expected direction, with carriers reporting greater distress/depression/anxiety than non-carriers, and greater healthy behavior change and attitudes, although all these effects were non-significant in the present study. For example, the largest single effect in the analysis was carriers reporting more Anxiety than non-carriers > 6 months after results, but this outcome was both non-significant and based on a single effect from a single study (k = 1, g = .78, p = .163). At the earlier (≤ 6 months) timepoint, carriers also reported more Anxiety, but again non-significantly (k = 3, g = .20, p = .207). Carriers reported less improvement in Behavioral Attitudes than non-carriers > 6 months after results, although not significantly (k = 3, g = −.17, p = .289). There was no strong evidence for any increased Disease Specific Distress or Other Psychological distress at any timepoint. In fact, carriers reported lower increases in these outcomes than non-carriers. However, these outcomes were based off of only one study each, respectively. None of the other meta-analytic effects showed heterogeneity. With one exception, these effects all remained non-significant in the combined meta-analysis (described in the supplement), even though in all cases save for Baseline Depression the sample size and number of studies increased. The one exception was that Disease Specific Distress became significantly associated in the combined analysis (k = 2, g = .38, p = .036), and the result was not heterogeneous.
Bias Ratings
All included studies were rated for bias (“high”, “low or no”, or “unclear”) in the previously-mentioned categories of selection, detection, attrition, reporting, and other bias. Selection bias was high in half of the included articles, meaning there were significant baseline differences between carriers and non-carriers, although no difference was detected for any meta-analysis results at Baseline. Detection bias was generally “unclear”, as only four studies provided information regarding blinding. All four of these studies were not blinded. Half of the included studies also had high attrition bias, losing many participants to follow-up, although six studies had no or very little loss to follow-up. Almost all included studies (k = 14) were coded as “High” risk of reporting bias. Many studies identified in the literature review did not report necessary statistics for inclusion in meta-analysis, instead reporting means without standard deviations, or providing graphs that were poorly labeled and not fully described in the article itself. Several of the included studies also failed to report appropriate information for all their measures, so only some information could be utilized. Fourteen of the 17 included studies were rated as “High” risk for other sources of bias. Often this high risk referred to reliance upon self-reported measures, which are subject to a number of biases themselves. See Table 2 for the full results of bias coding.
Table 2.
Bias Evaluations
| First Author, Year | Selection | Detection | Attrition | Reporting | Other |
|---|---|---|---|---|---|
| Studies Reporting Unadjusted Effects | |||||
| Aspinwall, 2008 | Low | Unclear | High | High | High |
| Aspinwall, 2009 | Low | Unclear | High | High | High |
| Aspinwall, 2013 | Low | Unclear | High | High | High |
| Aspinwall, 2014 | Low | Unclear | High | High | High |
| Chao, 2008 | High | Unclear | Low | Low | High |
| Grant, 2013 | High | Unclear | Low | High | High |
| Heshka, 2008 | High | Unclear | High | Low | High |
| Kasparian, 2009 | Unclear | Unclear | High | High | High |
| Kullo, 2016 | Low | Unclear | Low | High | Low |
| Sanderson, 2009 | High | High | Unclear | Low | High |
| Vernarelli, 2010 | Unclear | Unclear | Low | High | High |
| Studies Reporting Adjusted Statistics | |||||
| Christensen, 2016 | High | Unclear | High | High | High |
| Green, 2009 | High | Unclear | Low | High | Low |
| Hietaranta-Luoma, 2015 | High | High | High | High | High |
| Hietaranta-Luoma, 2014 | High | High | High | High | High |
| Legnani, 2006 | Unclear | Unclear | Low | High | Low |
| Sanderson, 2008 | High | High | Unclear | High | High |
Discussion
We conducted a meta-analysis of self-reported behavioral and psychological reactions to receipt of carrier/non-carrier status for complex diseases. This systematic review of the literature included comprehensive searches of several databases combined with a recursive citation search.
The search of published and unpublished articles identified 17 articles that met inclusion/exclusion criteria, all of which in peer-reviewed publications. All included studies compared carriers who received a positive (i.e., who carried a risk genotype) genetic testing result to non-carriers (i.e., who did not carry a risk genotype), and evaluated self-reported psychological or behavioral changes that stemmed from receipt of these results. While either self-reported or objective measures were considered for inclusion in the meta-analysis, all 17 studies relied exclusively on self-report. Ten of the 17 studies included genetic testing for either Alzheimer’s or melanoma, and eight came from two research groups, limiting generalizability of our findings. Many studies only reported statistics adjusted for covariates, posing an analytical challenge. We chose to conduct meta-analysis of unadjusted statistics (11 studies) as our primary analysis but to maximize sample size also reported in the supplement a meta-analysis of all results, regardless of whether the statistics were adjusted or unadjusted. Sample overlap was taken into account, such that any study sample reported on in multiple studies was only counted once.
There was evidence of significant positive health-related behavioral change self-reported more frequently by carriers than non-carriers six months or longer after receiving genetic test results. In our primary meta-analysis, which included only unadjusted statistics, there was no strong evidence that receipt of genetic test results affected any of the other 16 outcome categories, including distress or short-term behavioral change. This is perhaps surprising, given the many concerns over how individuals will react to genetic risk information. The theory of genetic essentialism would suggest that individuals who receive genetic test results would treat their risk information as immutable and unchangeable, and therefore be more likely to have adverse psychological reactions and/or fail to engage in preventative behaviors when presented with genetic information (Dar-Nimrod & Heine, 2011). We found no evidence that the individuals in these studies operated under an essentialist perspective. Further longitudinal work is needed to examine if the changes in behavior that carriers self-reported does last past two years after return, or if this effect as well will return to baseline levels.
Given the lack of significant adverse psychological reactions coupled with the significant self-reported Behavior Change in carriers, we wanted to examine the Behavior Change effect more closely using Alzheimer’s, a disease that currently cannot be prevented or effectively treated, and for which the APOE-e4 allele is a large genetic risk factor. Only one of the five studies with unadjusted effects included in the Behavior Change analysis was an Alzheimer’s disease study. Under a genetic essentialism hypothesis, one would expect APOE-e4 carriers to show the same behavior as non-carriers, because neither group can modify their genetic risk (or genetic protection). We did not observe substantial heterogeneity of the significant result in the analysis of unadjusted statistics, indicating that the Alzheimer’s result was not distinguishably larger or smaller as compared to the other diseases. That is, individuals who carried APOE-e4 reported more engagement with healthy behaviors than non-carriers. This result is interesting in part because return of results for APOE e4 carrier status and Alzheimer’s disease is not typically recommended (Burke et al., 2013), as it is not clinically actionable (Green et al., 2013; Hegde et al., 2015; National Society of Genetic Counselors, 2013), despite its strong effect (Corder et al., 1993). If our results hold, the participants in these research studies report engaging in healthy behaviors with respect to Alzheimer’s, despite the expected ineffectiveness of those behaviors.
The present results complement and extend a recent meta-analysis by Hollands et al. (2016), in which the authors compared self-reported health behaviors from individuals randomly assigned to receive genetic test results (grouping together carriers and non-carriers) and those randomly assigned to not receive results. Hollands et al. did not find any significant effect of receiving genetic test results on behavioral change. However, they did find in subgroup analyses directly comparing carriers versus non-carriers that receiving positive APOE-e4 results led to self-reported increased use of dietary supplements (Hollands et al. did not distinguish between unadjusted and adjusted statistics). The present study comprehensively expands on this preliminary result, and expands the finding of significant positive changes in risk carriers to diseases other than Alzheimer’s, such as melanoma and lung cancer. Taken together, the results of Hollands et al. and the present meta-analysis suggest that the simple act of obtaining genetic testing (regardless of carrier status) for complex disease does not impact behavior or psychological outcomes in substantial, or statistically significant ways. The receipt of carrier status, however, does appear to influence self-reported behavioral change, even for conditions for which there are no effective preventative measures or treatments available.
The present meta-analysis has several limitations that highlight areas of needed future research. Although we conducted a comprehensive, systematic review of the existing literature, including searching multiple databases, reference sections, and citing articles, only a small number of studies were available for inclusion (k = 17). Because these were not RCTs, many of them did not include a control group that did not receive genetic testing. Control groups allow one to evaluate whether information about risk or protective alleles (or both) result in change, and allow evaluation of unmeasured factors and secular trends that may not be apparent in comparisons of carriers and non-carriers. Sample sizes within each study tended to be small, and some of the included studies were different reports based on the same sample. The largest study consisted of 111 carriers and 161 non-carriers, perhaps indicating the difficulty in conducting these sorts of studies. Ultimately, 1,171 unique individuals were represented in the included studies (500 carriers and 671 non-carriers). In our primary analysis of only unadjusted statistics, 719 unique individuals were represented (346 carriers and 373 non-carriers).
Another limitation is the small number of individual effects within a given domain at a given timepoint. Some outcomes were only measured for one, or just a few, effects. In some cases, the effects were correlated to an unknown degree, making adjustment for correlated statistical significance tests difficult or impossible. As we note above, for almost half of the overall outcomes, we did not have access to unadjusted statistics from each included study. This is important, as many studies reported effects adjusted for demographics or other idiosyncratic confounders. Not all outcomes using unadjusted statistics could be properly evaluated due to lack of availability of unadjusted statistics. The majority of outcomes included in the unadjusted-only meta-analysis were based on three or fewer unique effects each, and many were based off of only one reported effect. However, results the combined meta-analysis of adjusted and unadjusted effects mirrored those from the unadjusted-only meta-analysis.
Although either self-reported or objective measures satisfied the inclusion criteria, all included studies ultimately relied on self-report questionnaires to measure outcomes. Recall can be biased or influenced by demand characteristics (Christensen, Roberts, et al., 2016). As a result, the effect of genetic information on true behavior changes, as opposed to self-reported changes, may be different than that reported here. The majority of studies included >75% white individuals, likely of European ancestry, although no study indicated this was due to exclusion of other ancestries. Given the differences in allele frequencies between ethnicities, the results of this meta-analysis may be confounded in unknown ways by ancestry. However, this concern is mitigated by largely ancestrally homogeneous samples, as well as the observation that the driving effect of APOE (the most commonly studied mutation) does not show gross allele frequency differences across major ancestral groups.
The present review highlights the lack of research on the consequences of genetic testing for complex disease. Indeed, the long-term effects, spanning > 1 year, of receiving genetic testing results are essentially unknown. To properly assess the impact of genetic information, future research with larger samples, conducted by independent research groups, that assess both proximal and more distal psychological and behavior outcomes, using both subjective self-reports and more objective methods, is needed. It is also critical that such research studies fully report relevant statistics, including unadjusted statistics. Despite the limitations, the results of this meta-analysis suggest that potential benefits from health behavior change are an important factor to consider in discussions about the return of genetic test results for complex disease. This also matches findings from the more-robust literature around returning Mendelian risk information, where carriers indicate improved health behavior after receipt of results (Leblond et al., 2011).
This conclusion may have implications for the process of returning results from genetic testing. If in fact individuals do not report or experience significant lasting harm from return of results, then strict regulation of the return of results for complex disease may be unwarranted. However, the true potential long-term effects of these returns remain unknown, especially in terms of disease-specific distress, and some individuals may respond in extreme ways to genetic risk information, which is not captured in tests of mean differences reported herein. On the other hand, the results suggest that individuals may benefit from the return of results, for example through increasing or adopting healthy behaviors, even if they are not mitigating specific genetic risks. If this is the case, then strict regulation of return of results for genetic tests of complex disease may do more harm than good.
Supplementary Material
Acknowledgments
This research was supported through National Institutes of Health grants R01AA023974, R01DA037904, R21DA040177, U01DA141120, and K01DA037280.
References
- Accreditation Council for Genetic Counseling. (2016). Accredited Programs. Retrieved from http://gceducation.org/Pages/Accredited-Programs.aspx
- Aspinwall LG, Leaf SL, Dola ER, Kohlmann W, & Leachman SA (2008). CDKN2A/p16 genetic test reporting improves early detection intentions and practices in high-risk melanoma families. Cancer Epidemiology Biomarkers & Prevention, 17(6), 1510–1519. doi:10.1158/1055-9965.epi-08-0010 [DOI] [PubMed] [Google Scholar]
- Aspinwall LG, Leaf SL, Kohlmann W, Dola ER, & Leachman SA (2009). Patterns of photoprotection following CDKN2A/p16 genetic test reporting and counseling. J Am Acad Dermatol, 60(5), 745–757. doi:10.1016/j.jaad.2008.12.034 [DOI] [PubMed] [Google Scholar]
- Aspinwall LG, Taber JM, Kohlmann W, Leaf SL, & Leachman SA (2014). Unaffected family members report improvements in daily routine sun protection 2 years following melanoma genetic testing. Genetics in Medicine, 16(11), 846–853. doi:10.1038/gim.2014.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinwall LG, Taber JM, Leaf SL, Kohlmann W, & Leachman SA (2013). Melanoma Genetic Counseling and Test Reporting Improve Screening Adherence Among Unaffected Carriers 2 Years Later. Cancer Epidemiology Biomarkers & Prevention, 22(10), 1687–1697. doi:10.1158/1055-9965.epi-13-0422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker LA, & Oxman AD (2011). Chapter 22: Overviews of reviews . In Higgins JPT & Green S (Eds.), Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0: The Cochrane Collaboration. [Google Scholar]
- Burdett T, Hall PN, Hasting E, Hindoff LA, Junkins HA, Klemm AK, . . . Welter D (2016). The NHGRI-EBI Catalog of published genome-wide association studies. Retrieved from www.ebi.ac.uk/gwas
- Bureau of Labor Statistics. (2015a). Occupational Outlook Handbook, 2016–17 Edition, Genetic Counselors; Retrieved from http://www.bls.gov/ooh/healthcare/genetic-counselors.htm [Google Scholar]
- Bureau of Labor Statistics. (2015b). Occupational Outlook Handbook, 2016–17 Edition, Physicians and Surgeons; Retrieved from http://www.bls.gov/ooh/healthcare/physicians-and-surgeons.htm [Google Scholar]
- Burke W, Antommaria AHM, Bennett R, Botkin J, Clayton EW, Henderson GE, . . . Zimmern R (2013). Recommendations for returning genomic incidental findings? We need to talk! Genetics in Medicine: Official Journal of the American College of Medical Genetics, 15(11), 854–859. doi:10.1038/gim.2013.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao S, Roberts JS, Marteau TM, Silliman R, Cupples LA, & Green RC (2008). Health Behavior Changes After Genetic Risk Assessment for Alzheimer Disease: The REVEAL Study. Alzheimer disease and associated disorders, 22(1), 94–97. doi:10.1097/WAD.0b013e31815a9dcc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen KD, Roberts JS, Whitehouse PJ, Royal CD, Obisesan TO, Cupples LA, . . . Green RC (2016). Disclosing Pleiotropic Effects During Genetic Risk Assessment for Alzheimer Disease: A Randomized Trial. Ann Intern Med, 164(3), 155–163. doi:10.7326/m15-0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen KD, Vassy JL, Jamal L, Lehmann LS, Slashinski MJ, Perry DL, . . . McGuire AL (2016). Are physicians prepared for whole genome sequencing? a qualitative analysis. Clin Genet, 89(2), 228–234. doi:10.1111/cge.12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, . . . Pericak-Vance MA (1993). Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science, 261(5123), 921–923. [DOI] [PubMed] [Google Scholar]
- Dar-Nimrod I, & Heine SJ (2011). Genetic Essentialism: On the Deceptive Determinism of DNA. Psychological Bulletin, 137(5), 800–818. doi:10.1037/a0021860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Re AC, Hoyt WT (2014). MAd: Meta-Analysis with Mean Differences. R package version 0.8–2 . Retrieved from http://cran.r-project.org/web/packages/MAd
- Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, . . . Campion D (2011). APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry, 16(9), 903–907. doi:10.1038/mp.2011.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould WA, & Heine SJ (2012). Implicit Essentialism: Genetic Concepts Are Implicitly Associated with Fate Concepts. PLoS ONE, 7(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RW, O’Brien KE, Waxler JL, Vassy JL, Delahanty LM, Bissett LG, . . . Meigs JB (2013). Personalized Genetic Risk Counseling to Motivate Diabetes Prevention: A randomized trial. Diabetes Care, 36(1), 13–19. doi:10.2337/dc12-0884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, . . . Biesecker LG (2013). ACMG Recommendations for Reporting of Incidental Findings in Clinical Exome and Genome Sequencing. Genetics in medicine : official journal of the American College of Medical Genetics, 15(7), 565–574. doi:10.1038/gim.2013.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Roberts JS, Cupples LA, Relkin NR, Whitehouse PJ, Brown T, . . . Farrer LA (2009). Disclosure of APOE Genotype for Risk of Alzheimer’s Disease. New England Journal of Medicine, 361(3), 245–254. doi:doi:10.1056/NEJMoa0809578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde M, Bale S, Bayrak-Toydemir P, Gibson J, Bone Jeng LJ, Joseph L, . . . Mao R (2015). Reporting Incidental Findings in Genomic Scale Clinical Sequencing—A Clinical Laboratory Perspective. The Journal of Molecular Diagnostics, 17(2), 107–117. doi:10.1016/j.jmoldx.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heshka J, Palleschi C, Wilson B, Brehaut J, Rutberg J, Etchegary H, . . . Wells PS (2008). Cognitive and behavioural effects of genetic testing for thrombophilia. J Genet Couns, 17(3), 288–296. doi:10.1007/s10897-008-9148-1 [DOI] [PubMed] [Google Scholar]
- Hietaranta-Luoma HL, Luomala HT, Puolijoki H, & Hopia A (2015). Using ApoE Genotyping to Promote Healthy Lifestyles in Finland – Psychological Impacts: Randomized Controlled Trial. Journal of Genetic Counseling, 24(6), 908–921. doi:10.1007/s10897-015-9826-8 [DOI] [PubMed] [Google Scholar]
- Hietaranta-Luoma HL, Tahvonen R, Iso-Touru T, Puolijoki H, & Hopia A (2014). An intervention study of individual, apoE genotype-based dietary and physical-activity advice: impact on health behavior. J Nutrigenet Nutrigenomics, 7(3), 161–174. doi:10.1159/000371743 [DOI] [PubMed] [Google Scholar]
- Hirschberg AM, Chan-Smutko G, & Pirl WF (2015). Psychiatric implications of cancer genetic testing. Cancer, 121(3), 341–360. doi:10.1002/cncr.28879 [DOI] [PubMed] [Google Scholar]
- Hollands GJ, French DP, Griffin SJ, Prevost AT, Sutton S, King S, & Marteau TM (2016). The impact of communicating genetic risks of disease on risk-reducing health behaviour: systematic review with meta-analysis. Bmj, 352, i1102. doi:10.1136/bmj.i1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasparian NA, Meiser B, Butow PN, Simpson JM, & Mann GJ (2009). Genetic testing for melanoma risk: a prospective cohort study of uptake and outcomes among Australian families. Genetics in Medicine, 11(4), 265–278. Retrieved from http://dx.doi.org/10.1097/GIM.0b013e3181993175 [DOI] [PubMed] [Google Scholar]
- Kullo IJ, Jouni H, Austin EE, Brown S-A, Kruisselbrink TM, Isseh IN, . . . Bailey KR (2016). Incorporating a Genetic Risk Score into Coronary Heart Disease Risk Estimates: Effect on LDL Cholesterol Levels (the MIGENES Clinical Trial). Circulation. doi:10.1161/circulationaha.115.020109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsbergen K, Prins J, Brunner H, Kraaimaat F, & Hoogerbrugge N (2009). Genetic testing for Lynch syndrome in the first year of colorectal cancer: a review of the psychological impact. Familial Cancer, 8(4), 325–337. doi:10.1007/s10689-009-9239-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond D, Bredart A, Dolbeault S, De Pauw A, Lyonnet DS, Flahault C, & Sultan S (2011). Cognitive, emotional and behavioral impact of an uncertain outcome after study of BRCA1/2: review of the literature. Bulletin Du Cancer, 98(2), 184–198. doi:10.1684/bdc.2011.1309 [DOI] [PubMed] [Google Scholar]
- Legnani C, Razzaboni E, Gremigni P, Ricci Bitti PE, Favaretto E, & Palareti G (2006). Psychological impact of testing for thrombophilic alterations. Thromb Haemost, 96(3), 348–355. doi:10.1160/th06-01-0015 [DOI] [PubMed] [Google Scholar]
- Lipsey MW, & Wilson DB (2001). Practical meta-analysis. Thousand Oaks, CA: Sage Publications. [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, . . . Visscher PM (2009). Finding the missing heritability of complex diseases. Nature, 461(7265), 747–753. Retrieved from http://dx.doi.org/10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER (2011). A decade’s perspective on DNA sequencing technology. Nature, 470(7333), 198–203. doi:10.1038/nature09796 [DOI] [PubMed] [Google Scholar]
- McKusick-Nathans Institute of Genetic Medicine. (2016a). OMIM-Online Mendelian Inheritance in Man. Retrieved from http://www.omim.org/about
- McKusick-Nathans Institute of Genetic Medicine. (2016b). OMIM Update List. Retrieved from http://www.omim.org/statistics/update
- Meiser B, & Dunn S (2001). Psychological effect of genetic testing for Huntington’s disease: an update of the literature. Western Journal of Medicine, 174(5), 336–340. Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1071392/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Society of Genetic Counselors. (2013). Incidental Findings in Genetic Testing. Retrieved from http://nsgc.org/p/bl/et/blogid=47&blogaid=30
- Patay BA, & Topol EJ (2012). The Unmet Need of Education in Genomic Medicine. The American Journal of Medicine, 125(1), 5–6. doi:10.1016/j.amjmed.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Peters J, & Mengersen K (2008). Selective Reporting of Adjusted Estimates in Observational Epidemiology Studies: Reasons and Implications for Meta-analyses. Evaluation & the Health Professions, 31(4), 370–389. doi:10.1177/0163278708324438 [DOI] [PubMed] [Google Scholar]
- R Core Team. (2015). R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing; Retrieved from https://www.R-project.org/ [Google Scholar]
- Ritchie MD, Holzinger ER, Li R, Pendergrass SA, & Kim D (2015). Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet, 16(2), 85–97. doi:10.1038/nrg3868 [DOI] [PubMed] [Google Scholar]
- Sanderson SC, Humphries SE, Hubbart C, Hughes E, Jarvis MJ, & Wardle J (2008). Psychological and behavioural impact of genetic testing smokers for lung cancer risk: a phase II exploratory trial. J Health Psychol, 13(4), 481–494. doi:10.1177/1359105308088519 [DOI] [PubMed] [Google Scholar]
- Sanderson SC, O’Neill SC, White DB, Bepler G, Bastian L, Lipkus IM, & McBride CM (2009). Responses to online GSTM1 genetic test results among smokers related to patients with lung cancer: a pilot study. Cancer Epidemiology Biomarkers & Prevention, 18(7), 1953–1961. doi:10.1158/1055-9965.epi-08-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarah Lawrence College. (2016). Application Process, Tuition & Financial Aid: Master of Science in Human Genetics. Retrieved from https://www.sarahlawrence.edu/human-genetics/application/
- Telenti A, Pierce LCT, Biggs WH, di Iulio J, Wong EHM, Fabani MM, . . . Venter JC (2016). Deep sequencing of 10,000 human genomes. Proceedings of the National Academy of Sciences, 113(42), 11901–11906. doi:10.1073/pnas.1613365113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Food and Drug Administration. (2013). 23andMe, Inc. 11/23/13. Retrieved from http://www.fda.gov/ICECI/EnforcementActions/WarningLetters/2013/ucm376296.htm.
- Timman R, Roos R, Maat-Kievit A, & Tibben A (2004). Adverse effects of predictive testing for Huntington disease underestimated: long-term effects 7–10 years after the test. Health Psychol, 23(2), 189–197. doi:10.1037/0278-6133.23.2.189 [DOI] [PubMed] [Google Scholar]
- Vansenne F, Bossuyt PMM, & de Borgie C (2009). Evaluating the Psychological Effects of Genetic Testing in Symptomatic Patients: A Systematic Review. Genetic Testing and Molecular Biomarkers, 13(5), 555–563. doi:10.1089/gtmb.2009.0029 [DOI] [PubMed] [Google Scholar]
- Vernarelli JA, Roberts JS, Hiraki S, Chen CA, Cupples LA, & Green RC (2010). Effect of Alzheimer disease genetic risk disclosure on dietary supplement use. Am J Clin Nutr, 91(5), 1402–1407. doi:10.3945/ajcn.2009.28981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viechtbauer W (2010). Conducting meta-analyses in R with the metafor package. Journal of Statistical Software 36(3), 1–48. Retrieved from http://www.jstatsoft.org/v36/io3 [Google Scholar]
- Voils CI, Crandell JL, Chang Y, Leeman J, & Sandelowski M (2011). Combining adjusted and unadjusted findings in mixed research synthesis. Journal of evaluation in clinical practice, 17(3), 429–434. doi:10.1111/j.1365-2753.2010.01444.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wampold BE, Mondin GW, Moody M, Stich F, Benson K,, & Ahn H (1997). A meta-analysis of outcome studies comparing bona fide psychotherapies: Empiricially, “all must have prizes.”. Psychological Bulletin, 122(3). [Google Scholar]
- Yu J-H, Jamal SM, Tabor HK, & Bamshad MJ (2013). Self-guided management of exome and whole-genome sequencing results: changing the results return model. Genetics in Medicine, 15(9), 684–690. doi:10.1038/gim.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzetto M, Russi E, Senn O, Imboden M, Ferrarotti I, Tinelli C, . . . Probst-Hensch N (2008). SERPINA1 gene variants in individuals from the general population with reduced alpha1-antitrypsin concentrations. Clin Chem, 54(8), 1331–1338. doi:10.1373/clinchem.2007.102798 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.