Abstract
Following a year of valganciclovir prophylaxis, a lung transplant recipient developed cytomegalovirus (CMV) infection that became resistant to ganciclovir, as confirmed by detection of UL97 kinase mutation M460V and a previously uncharacterized UL54 DNA polymerase mutation L516P. The latter mutation is now shown to confer ganciclovir and cidofovir resistance. As predicted from the viral genotype, foscarnet therapy was effective, but resumption of valganciclovir as secondary prophylaxis resulted in a plasma viral load rebound to 3.6 log10 copies/mL several weeks later. Valganciclovir was then replaced by letermovir, resulting in gradual viral load reduction in the first 5 weeks to below the quantitation limit (2.7 log10 copies/mL) for one week, followed by 10 weeks of rising viral loads reaching 4.3 log10 copies/mL while on letermovir. At this point, CMV genotypic testing revealed UL56 mutation C325Y, which confers absolute resistance to letermovir. Retreatment with foscarnet was successful. This case adds to the considerable list of proven ganciclovir resistance mutations, and provides an early experience with letermovir resistance after off-label therapeutic use. This experience is consistent with in vitro observations of rapid emergence of letermovir-resistant CMV after drug exposure.
1. INTRODUCTION
The recent approval of the human cytomegalovirus (CMV) terminase inhibitor letermovir for prophylaxis in stem cell transplantation represents a significant advance in antiviral options for this major viral opportunistic pathogen.1 Because of a drug target different from the standard DNA polymerase inhibitors, oral bioavailability, and a good safety profile to date, letermovir is being considered for off-label use as therapy for CMV infection that has become resistant to approved agents or as an alternative to poorly tolerated intravenous options. However, clinical experience with letermovir as treatment for active CMV infection is limited. A Phase 2a trial of letermovir as preemptive therapy at suboptimal doses was inconclusive.2 There is one case report of successful use of letermovir for drug-resistant CMV in a lung transplant recipient who became intolerant of foscarnet.3 However, the doses used were a quarter to half of the FDA-approved prophylaxis dose, and immunosuppression was substantially reduced, leaving doubt about the specific role of letermovir. Here, we report a case with letermovir as treatment of ganciclovir-resistant CMV infection, which resulted in the development of letermovir resistance.
2. CASE REPORT
A 57 year-old woman received a bilateral lung transplant for fibrotic nonspecific interstitial pneumonitis and pulmonary hypertension. She was CMV-seronegative and her donor was CMV-seropositive. Accordingly, CMV prophylaxis was started, initially with CMV immune globulin and intravenous ganciclovir, followed by oral valganciclovir for a total of 51 weeks and monthly CMV immunoglobulin. The patient received basiliximab at the time of transplant, followed by cyclosporine (goal trough 300–350 ng/mL), mycophenolate 1000 mg twice daily, and prednisone for maintenance immunosuppression. Mycophenolate was discontinued and valganciclovir dosing was reduced from 450 mg twice a day to 450 mg daily because of leukopenia at week 47; renal function was not impaired at that time.
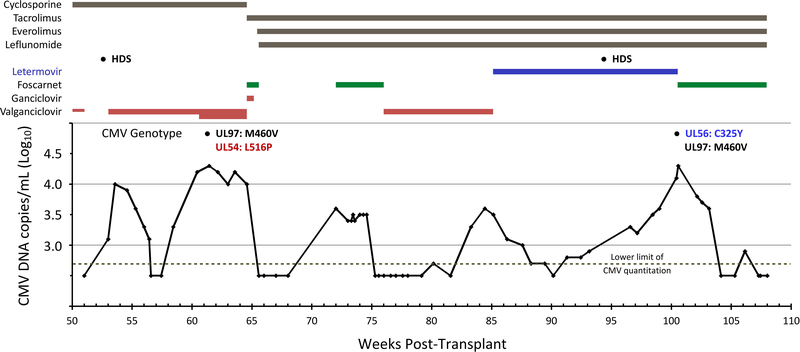
A timeline of medications, plasma CMV DNA loads, and viral genotypes from week 50 post-transplant is shown in Figure 1. Following discontinuation of valganciclovir (week 51), high dose corticosteroids were administered for reduced spirometric flows and suspected rejection. Plasma CMV DNA was noted at week 53 and CMV immune globulin and valganciclovir were initiated with a decrease in viral load from a peak of 4 log10 copies/mL to < 2.7 log10 copies/mL at week 57. Adverse effects of leukopenia, nausea, and vomiting led to valganciclovir dose reduction (from 900mg twice a day to 450 mg twice a day). The viral load then began to rise, reaching 4.3 log10 copies/mL at week 61. The valganciclovir dose was increased to 900 mg twice daily and CMV immune globulin was given. At week 65, the patient was hospitalized and started on ganciclovir (2.5 mg/kg twice daily) and foscarnet (30 mg/kg twice daily) reflecting dose reduction for estimated creatinine clearance of 40–50 mL/min. Viral genotypic testing sent at week 61 returned with mutations in genes UL97 (M460V) and UL54 (L516P) and ganciclovir was therefore discontinued, while foscarnet dosing was increased to 70 mg/kg twice daily after renal function improvement. The viral DNA load decreased to < 2.7 log10 copies/mL one week later (week 66). Several changes were made to the patient’s immunosuppressive regimen during this admission. The patient was converted from cyclosporine to tacrolimus (goal level 4–6 ng/mL). Leflunomide 20 mg daily and everolimus (goal level 4–6 ng/mL) were initiated for their anticipated anti-CMV properties. Five weeks after stopping foscarnet treatment, a plasma viral DNA rebound to 3.6 log10 copies/mL was detected, and foscarnet therapy was resumed for 4 weeks, with decrease in viral load to below < 2.7 log10 copies/mL (week 75). Valganciclovir 450 mg twice daily as secondary prophylaxis was resumed, but viral rebound was noted after 8 weeks, reaching 3.6 log10 copies/mL. At this point (week 85), valganciclovir was stopped and oral letermovir was initiated at a dose of 480 mg daily. The plasma viral DNA progressively decreased to below the quantitation limit 5 weeks later (week 90) but rose to 2.8 log10 copies/mL the following week and increased over the next 9 weeks to a level of 4.1 log copies/mL (week 100). The viral DNA rebound began before high dose steroid therapy was given at week 94. At week 100, viral genotypic analysis revealed a CMV UL56 mutation C325Y, which is known to confer absolute resistance to letermovir (50% inhibitory concentration increased by >8000-fold over wild type).4,5 The previously detected UL97 mutation M460V was again detected, but the UL54 mutation L516P was no longer detected. We attempted to recover residual plasma specimens from the weeks prior to 100 (Fig. 1) to assay for UL56 mutation, but all had been discarded. Foscarnet therapy was initiated with decreasing plasma viral DNA over the next 4 weeks. During the extended period of fluctuating CMV reactivation as shown in Fig. 1, there was no symptomatic disease attributed to CMV.
Figure 1. Timeline of medications, CMV loads and genotypes.
CMV DNA copies/mL were determined on whole blood specimens prior to 70 weeks at an outside hospital (copies/mL = IU/mL), and on plasma specimens using a locally developed assay (not calibrated to IU/mL) thereafter. Loads below the lower limit of quantitation (500 or 2.7 log10 for each assay) are not to scale. Thicknesses of bars for valganciclovir denote doses of 450 mg daily (end of prophylaxis, week 51), 450 mg twice daily, and 900 mg twice daily. HDS = high-dose steroids at weeks 52 and 94.
Because the UL54 mutation resulting in the L516P amino acid substitution has not been characterized previously, its relationship to drug resistance was determined by recombinant phenotyping.6 The mutation was transferred by recombination into a cloned derivative of laboratory strain AD169 (BD1) that contained a secreted alkaline phosphatase (SEAP) reporter gene for quantitation of viral growth. After reconstitution and genetic verification of live recombinant virus containing UL54 mutation L516P in modified retinal epithelial cells (ARPEp), drug concentrations required to reduce SEAP growth by 50% (EC50) were determined for ganciclovir, cidofovir and foscarnet.7 Results of multiple rounds of phenotypic testing with accompanying control strains are shown in Table 1. The L516P mutant was resistant to ganciclovir and cidofovir but remained susceptible to foscarnet. This phenotype is qualitatively similar to the D515Y mutant4,8 involving the adjacent amino acid of the UL54 DNA polymerase gene product, though the L516P mutant conferred a slightly higher level of resistance to ganciclovir and cidofovir. The UL54 mutant with codons 981–982 deleted was included as a known multi-drug resistant control and showed the expected degree of resistance to all three polymerase inhibitors.4,7
Table 1.
Genotypes and phenotypes of recombinant CMV strains
| Strain | UL54 pol Genotypea | Cidofovir | Ganciclovir | Foscarnet | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC50b | SDc | Nd | Folde | EC50b | SDc | Nd | Folde | EC50b | SDc | Nd | Folde | ||
| 4198 | wild type | 0.24 | 0.04 | 25 | 1.1 | 0.21 | 31 | 42 | 5.3 | 24 | |||
| 4376 | 981del2 | 0.77 | 0.16 | 26 | 3.2 | 8.0 | 1.5 | 29 | 7.0 | 114 | 22 | 24 | 2.7 |
| 4439 | D515Y | 0.62 | 0.13 | 22 | 2.6 | 3.7 | 0.59 | 32 | 3.2 | 44 | 5.4 | 22 | 1.1 |
| 4446 | L516P | 1.42 | 0.24 | 14 | 5.9 | 5.0 | 0.66 | 16 | 4.4 | 38 | 5.0 | 14 | 0.9 |
Amino acid substitutions where different from wild type. 981del2 represents an in frame deletion of codons 981 and 982, used as a multi-drug resistant control strain
Drug concentration in μM required to reduce viral SEAP growth by 50%
Standard deviation of EC50 values
Number of replicates of testing performed over at least 5 setup dates
Fold change in EC50 compared with wild type control strain (4198). Values in bold indicate drug resistance
3. DISCUSSION
This case illustrates a typical evolution of ganciclovir resistance in a lung transplant recipient, with two notable exceptions. First, the patient developed a newly phenotyped UL54 DNA polymerase exonuclease domain mutation, which increased the level of ganciclovir resistance conferred by the UL97 kinase mutation. Second, treatment with letermovir led to the rapid emergence of a UL56 terminase gene mutation that conferred absolute letermovir resistance.
Risk factors for ganciclovir resistance included a lung transplant, a CMV seropositive organ transplanted into a seronegative recipient (D+R-), reduced valganciclovir dosing for leukopenia possibly resulting in subtherapeutic drug levels, and one year duration of valganciclovir prophylaxis before re-use for treatment of active CMV infection in an immunosuppressed patient.9
Genotypic testing at 61 weeks confirmed the presence of UL97 kinase mutation M460V, which by itself confers a 5- to 10-fold increase in ganciclovir EC50 for the drug in multiple reports.9 It could be suspected that the previously uncharacterized UL54 exonuclease domain mutation L516P might confer ganciclovir and cidofovir resistance,9–11 although the level of resistance was difficult to predict from the conflicting phenotypes published for mutations in the immediate vicinity. For example, UL54 mutations D515G and L516M were reported to confer no drug resistance,10,12 while D515Y was reported to confer ganciclovir and foscarnet resistance without cidofovir resistance.8 Because the foscarnet-resistant phenotype was unexpected for D515Y, we independently repeated its recombinant phenotyping and discovered that both D515Y and L516P (Table 1) have the ganciclovir- and cidofovir-resistant, foscarnet-susceptible phenotype. In addition, the UL54 mutation L516P is predicted to combine with the UL97 mutation M460V to give an overall ganciclovir EC50 increase of about 30-fold,7 a level of resistance that puts it out of reach of ganciclovir dose escalation as a realistic management option.9 According to recently published clinical guidelines,9 foscarnet is the antiviral therapy of choice in this setting.
Separate courses of foscarnet successfully treated the ganciclovir-resistant virus at weeks 64, 72–76 and 100–108. Secondary prophylaxis with valganciclovir would not be expected to be effective in view of the level of drug resistance predicted above, and it did not prevent viral rebound at weeks 83–85, nor did the leflunomide or everolimus being given at the time. At week 85, guidelines again suggest retreatment with foscarnet,9 but given the relatively low viral load of 3.6 log10 or 4000 copies/mL and no CMV disease it seemed reasonable to attempt oral letermovir as a simpler alternative. The letermovir treatment appeared effective in gradually reducing the plasma CMV DNA load in the first 5 weeks, reaching below the limit of quantitation for 1 week but rebounding thereafter (Fig. 1). Within the following 10 weeks, or 15 weeks after starting letermovir therapy, the terminase gene UL56 mutation C325Y emerged to confer absolute letermovir resistance, thus explaining the viral rebound. Without intermediate specimens for genotypic testing, we cannot specify more precisely at what point resistance would be detectable after the viral rebound started at 6 weeks. No issues with adherence to the 480 mg daily letermovir dose were identified. It is unclear if higher doses of letermovir would be more effective or safe for prolonged use, in the absence of adequate data.
It is difficult from this one case to compare the in vivo evolution of ganciclovir and letermovir resistance. In the literature, the median duration of ganciclovir therapy prior to detection of resistance is about 22 weeks, although 3 of 17 lung recipients were reported to develop ganciclovir resistance much sooner.13 Extensive in vitro studies have reported the rapid evolution of letermovir resistance after drug exposure, much sooner than in parallel experiments with ganciclovir4 and foscarnet.14 Unlike ganciclovir and foscarnet, letermovir is affected by a single step mutation at UL56 codon 325 (C325) that confers absolute resistance with minimal to low impact on viral growth fitness.4,5,14 In vitro, multiple independent selections of C325 mutations have been observed in the approximate order of frequency of C325F > C325R ~ C325Y > C325W,4,5,14 probably reflecting the relative growth fitness of the mutants. Each of these mutations increases the EC50 by >3000-fold and effectively eliminates the antiviral activity of letermovir at any nontoxic dose. In treated individuals, C325W was found once in the Phase III letermovir prophylaxis trial,4 while the C325Y detected in this case is another early instance. Apart from these important C325 mutations, various mutations conferring lower levels of letermovir resistance have been selected in vitro in gene UL56 (clustered at codons 229–369 and codon 25), and occasionally in other terminase genes UL51 and UL89.4,5,14–16 The UL56 mutation V236M has been reported twice in letermovir prophylaxis trials.1,17 It is too early to determine how long it takes for letermovir resistance mutations to clear after drug discontinuation, as observed with the UL54 mutation L516P in this case.
In conclusion, the relatively low genetic barrier to high-grade letermovir resistance observed in vitro, combined with case experience as documented here, suggests a cautious approach in off-label use of letermovir for salvage therapy of ganciclovir-resistant CMV infection, even at low starting viral loads. Viral load suppression using alternative therapy such as foscarnet to below the starting level observed in this case, or to an undetectable level, may be advisable prior to use of letermovir as treatment or secondary prophylaxis. Care should be taken to ensure consistent drug delivery to avoid subtherapeutic drug levels. Monitoring for letermovir resistance is indicated soon after observation of any viral rebound while on therapy. More information is needed about the optimal use of letermovir as therapy, including dosing regimens, incidence of drug resistance at various starting viral loads and trajectories, and the possible benefit of combining letermovir with other antivirals.
ACKNOWLEDGMENTS
We thank Ronald J. Ercolani and L. Elizabeth Satterwhite for technical assistance with recombinant phenotyping. The phenotyping work was supported by NIH grant AI116635 and use of Department of Veterans Affairs facilities and resources.
Abbreviations:
- ARPEp
modified retinal epithelial cells
- CMV
cytomegalovirus
- SEAP
secreted alkaline phosphatase
Footnotes
DISCLOSURE
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. SC is principal investigator on Department of Veterans Affairs cooperative research and development agreements with Merck and Shire (all payments to institution only). The other authors have no conflicts of interest to disclose.
REFERENCES
- 1.Marty FM, Ljungman P, Chemaly RF, et al. Letermovir prophylaxis for cytomegalovirus in hematopoietic-cell transplantation. N Engl J Med. 2017;377:2433–44. [DOI] [PubMed] [Google Scholar]
- 2.Stoelben S, Arns W, Renders L, et al. Preemptive treatment of cytomegalovirus infection in kidney transplant recipients with letermovir: results of a Phase 2a study. Transpl Int. 2014;72(1):77–86. [DOI] [PubMed] [Google Scholar]
- 3.Kaul DR, Stoelben S, Cober E, et al. First report of successful treatment of multidrug-resistant cytomegalovirus disease with the novel anti-CMV compound AIC246. Am J Transplant. 2011;11(5):1079–84. [DOI] [PubMed] [Google Scholar]
- 4.Chou S, Satterwhite LE, Ercolani RJ. New locus of drug resistance in the human cytomegalovirus UL56 gene revealed by in vitro exposure to letermovir and ganciclovir. Antimicrob Agents Chemother. 2018;62(9):e00922–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldner T, Hempel C, Ruebsamen-Schaeff H, Zimmermann H, Lischka P. Geno- and phenotypic characterization of human cytomegalovirus mutants selected in vitro after letermovir (AIC246) exposure. Antimicrob Agents Chemother. 2014;58:610–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou S Foscarnet resistance mutations mapping to atypical domains of the cytomegalovirus DNA polymerase gene. Antiviral Res. 2017;138:57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou S, Van Wechel LC, Lichy HM, Marousek GI. Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene. Antimicrob Agents Chemother. 2005;49(7):2710–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andouard D, Mazeron MC, Ligat G, et al. Contrasting effect of new HCMV pUL54 mutations on antiviral drug susceptibility: Benefits and limits of 3D analysis. Antiviral Res. 2016;129:115–9. [DOI] [PubMed] [Google Scholar]
- 9.Kotton CN, Kumar D, Caliendo AM, et al. The third international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation. 2018;102(6):900–31. [DOI] [PubMed] [Google Scholar]
- 10.Chou S, Lurain NS, Thompson KD, Miner RC, Drew WL. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J Infect Dis. 2003;188(1):32–9. [DOI] [PubMed] [Google Scholar]
- 11.Fischer L, Imrich E, Sampaio KL, et al. Identification of resistance-associated HCMV UL97- and UL54-mutations and a UL97-polymorphism with impact on phenotypic drug-resistance. Antiviral Res. 2016;131:1–8. [DOI] [PubMed] [Google Scholar]
- 12.Fischer L, Laib Sampaio K, Jahn G, Hamprecht K, Gohring K. Generation and characterization of a GCV resistant HCMV UL97-mutation and a drug sensitive UL54-mutation. Antiviral Res. 2013;100(3):575–7. [DOI] [PubMed] [Google Scholar]
- 13.Fisher CE, Knudsen JL, Lease ED, et al. Risk factors and outcomes of ganciclovir-resistant cytomegalovirus infection in solid organ transplant recipients. Clin Infect Dis. 2017;65(1):57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou S Rapid in vitro evolution of human cytomegalovirus UL56 mutations that confer letermovir resistance. Antimicrob Agents Chemother. 2015;59:6588–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou S Comparison of cytomegalovirus terminase gene mutations selected after exposure to three distinct inhibitor compounds. Antimicrob Agents Chemother. 2017;61(11):e01325–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou S A third component of the human cytomegalovirus terminase complex is involved in letermovir resistance. Antiviral Res. 2017;148:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lischka P, Michel D, Zimmermann H. Characterization of cytomegalovirus breakthrough events in a Phase 2 prophylaxis trial of letermovir (AIC246, MK 8228). J Infect Dis. 2016;213(1):23–30. [DOI] [PubMed] [Google Scholar]