Abstract
Despite significant improvements in pediatric brain tumor therapy and outcome, too many children still die from disease, and too many survivors suffer significant sequelae as a result of conventional therapies. Molecular characterization of pediatric brain tumors has afforded tremendous insight into the basic biology and clinical management of these deadly childhood diseases. Genomic, epigenomic, and transcriptional profiling have facilitated the identification of significant heterogeneity among previously uniform disease entities. In particular, DNA methylation profiling has emerged as a robust tool for identifying key disease-specific subgroups that can exhibit distinct clinical outcomes. These approaches, which also complement classical histologic techniques, can suggest key mechanistic underpinnings of tumorigenesis and open the door for better informed and more tailored therapy. By leveraging the results of large-scale classifications of disease cohorts, novel driver mutations and pathways can be uncovered, enabling generation of faithful animal models, promoting targeted drug design, informing development biology, and ultimately translating to improved clinical management. In this review, the progress in epigenetic classification of common malignant pediatric brain tumors – medulloblastoma, ependymoma, high-grade glioma, atypical teratoid/rhabdoid tumor, and CNS embryonal tumors – will be discussed, and the potential role of DNA methylation profiling as a frontline diagnostic modality will be emphasized.
Keywords: Molecular pathology, epigenetics, methylation, pediatric, neuro oncology
Introduction
As the most common solid tumors in children, pediatric central nervous system (CNS) tumors represent an array of pathologically and molecularly diverse entities. [1, 2] Treatment of malignant CNS tumors in children presents a unique challenge given the exquisite susceptibility of the developing nervous system to damage from conventional treatment modalities of neurosurgery, craniospinal irradiation, and cytotoxic chemotherapy. Though refinement of these conventional modalities has improved survival in many cases, neurodevelopmental sequelae continue to confer significant morbidity to many children. [3, 4] Moreover, many patients still succumb to their disease despite these incremental advances. As such, improving clinical management and the ultimate long-term outcomes of these patients relies on uncovering subtle differences within disease categories that can refine the risk stratifications that dictate treatments and their intensities.
Given the vast diversity of pediatric CNS tumors, disease classification ultimately dictates and defines treatment. Traditional risk stratification schemes have relied on factors such as patient age, tumor histology, and metastatic status at diagnosis to define treatment. While these factors represent surrogate markers of complex underlying tumor biology and the subsequent natural disease history, they fail to recognize subtle molecular differences that now define prognosis. Similarly, while inclusion of immunohistochemical (IHC) and cytogenetic markers have greatly refined prognostication, these techniques are limited in their capacity to identify the numerous unique biologic subgroups that more advanced molecular techniques are uncovering. Driven by unbiased molecular profiling, the therapeutic implications of such subgroupings are immediate and major. By tailoring therapy based on risk of relapse or responsiveness to conventional treatments, treatment escalation or dose reductions can be implemented to mitigate treatment failure or neurodevelopmental sequelae, respectively. Furthermore, those patients predicted to relapse despite maximal conventional therapy highlight the necessity for sensible alternative therapies.
Molecular approaches to study pediatric CNS tumors have flourished in the age of next-generation sequencing (NGS), providing a wealth of clinically and biologically significant insights into these deadly childhood diseases. By interrogating the molecular signatures of tumors, these methods have helped reconcile disparate outcomes among previously uniform disease classifications, driving refinements of risk stratifications and therapeutic decision making. Among methods for molecular subgrouping, genome-wide methylation profiling has emerged as a powerful molecular tool to classify tumors. With the development of high-density arrays and improved bioinformatics pipelines, methylation classification of pediatric CNS tumors provides a robust, reproducible modality for classification with high concordance to subgroups initially identified by other molecular methods, such as gene expression profiling and genome sequencing. With its reasonable cost and favorable turnaround time, DNA methylation profiling represents a feasible implementation of molecular methods for diagnostic use and clinical protocol stratification.
In this review, we highlight the utility of genome-wide DNA methylation profiling in the subgrouping of pediatric brain tumors. After an overview of methodologies, discussion of recently characterized entities with clinical implications are presented for medulloblastoma, ependymoma, high-grade glioma, atypical teratoid/rhabdoid tumor, and CNS-embryonal tumors (formerly known as CNS primitive neuroectodermal tumor). Finally, the rationale and proposal for integration of methylation profiling in the clinical diagnostic pipeline is set forth.
Molecular characterization by genetic, transcriptomic, and, now, epigenetic analyses
Criteria for classification of CNS tumors has historically been based on histology that relied on the anatomic location and morphologic similarity of tumors under light microscopy to specific cell types in the normal or developing brain. The advent of IHC and cytogenetic methods allowed for further refinement of classification schemes by substratifying tumors based on expression patterns of lineage markers and the presence of specific structural abnormalities. Modern advances in molecular methods to interrogate tumors, including genomic, transcriptomic, and epigenomic characterizations, have provided deeper insights and highlighted tumor substructure even among tumors that are indistinguishable by conventional methods. A more refined classification scheme layered with rich molecular data provides a significant step toward improving diagnostic reproducibility, improving risk stratification, informing the underlying mechanism of tumorigenesis, and facilitating the development of targeted therapeutic approaches for precision medicines.
Genomic analyses
Our understanding of the genetic landscape of pediatric CNS tumors has improved significantly by comprehensive genomic analyses, which have identified a significant number of recurrent, disease-specific abnormalities. These analyses have also highlighted key differences between pediatric CNS tumors and their adult counterparts while informing prognostication. Yet even though genomic analysis of tumors by NGS is a powerful tool to interrogate the cancer genome, these methods are still costly and time-consuming, precluding routine implementation into diagnostic and clinical pipelines. Moreover, a large proportion of tumors, particularly pediatric tumors, have been found to be devoid of pathognomonic genetic lesions or specific drivers that NGS uncovers. Rather, for many pediatric entities, epigenetic mechanisms are believed to be contributing to tumor etiology. [5]
Transcriptomic profiling
Transcriptomic analyses have contributed significantly to the sub-classification of pediatric CNS tumors by identifying molecular derangements that are not evident at the genetic level. Disruptions of key developmental pathways and cellular processes have been identified by gene expression signatures allowing for insights into cell of origin and development of therapies targeted to combat specifically deranged pathways. [6, 7] Profiling of large disease cohorts has uncovered the convergence of some diseases on altered canonical signaling pathways that may represent avenues of novel therapeutic intervention.
Though an extremely powerful tool in the sub-classification of pediatric CNS tumors, several features have prevented transcriptomic analyses from widespread adoption into clinical workflows. While most anatomic pathology laboratories routinely collect formalin-fixed, paraffin-embedded (FFPE) tissue for diagnostic purposes, historically, transcriptomic analysis largely relied on the availability of frozen tissue to gather high-quality RNA for array or next-generation sequencing methods. Utilization of fresh frozen tissue rather than FFPE invokes significant demands in terms of resources and added cost for proper tissue cryopreservation and storage. Furthermore, reliance on fresh frozen tissue prevented utilization of the large retrospective repositories of CNS tumors available in FFPE blocks or slides. More modern techniques have emerged for recovery and restoration of RNA from FFPE material. [8] Despite these advances, fresh frozen tissue remains the gold standard for transcriptome analysis, and the relative stability and versatility of DNA compared to RNA make DNA-based platforms more attractive to guide clinical decision making.
DNA methylation profiling
In addition to genomic and transcriptomic approaches, epigenomics has emerged as central focus in tumor biology. Epigenetic mechanisms regulate gene activity in precise spatiotemporal programs during development. Of these mechanisms, DNA methylation and histone tail modification have been extensively characterized. Distributed in a biased manner throughout the human genome, CpG islands are often located in gene promoters and function as key regulators of gene expression. [9] The cytosine bases in CpG islands undergo methylation to 5-methylcytosine through the action of DNA methyltransferases. Such covalent modification not only prevents accessibility of transcription factors to DNA but also leads to recruitment of other chromatin modifiers that can lead to the formation of heterochromatin through changes in histone modifications. [10]
In cancer, aberrant patterns of DNA methylation can lead to transcriptional inactivation of tumor suppressors. [11] Conversely, decreased levels of methylation at oncogenes can drive tumorigenesis. [12] The epigenetic crosstalk between DNA methylation and histone modification also implicates the role of aberrant chromatin modulation in the development of cancer. Failure of transcriptional programs for proper cellular differentiation during development can thus lead to the inappropriate persistence of a proliferative progenitor-like cell population. Furthermore, patterns of DNA methylation and histone post-translational modification may provide indirect assessment of developmental origins.
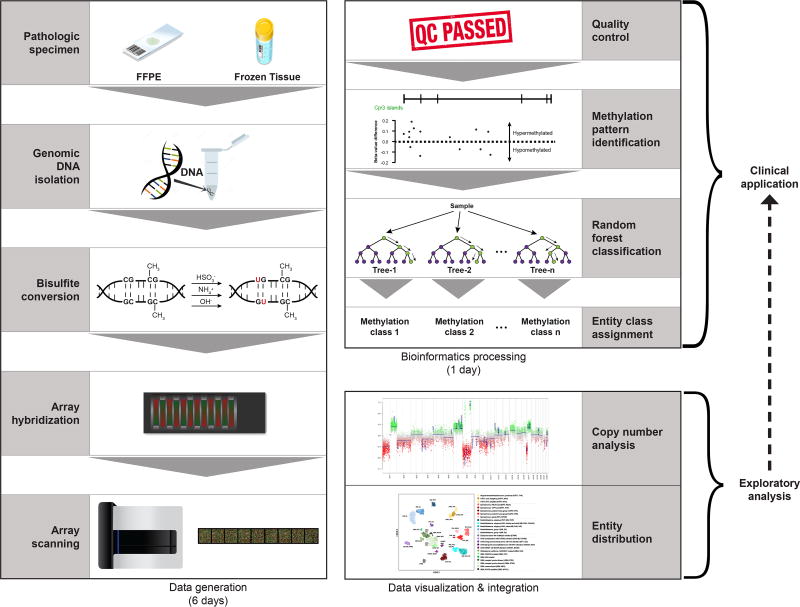
Methods for detecting cytosine modifications hinge on treatment of DNA with bisulfite, which converts cytosine to uracil while not affecting 5-methylcytosine. [13] Upon bisulfite treatment, DNA thus retains only methylated cytosine allowing single-nucleotide resolution of methylation status (Figure 1). With a priori targets, methylation specific PCR can be used to interrogate specific genomic loci while next-generation sequencing methods can cover much larger regions of the genome without prior knowledge of specific targets. Despite its cost, whole-genome bisulfite sequencing can provide genome-wide, single-nucleotide methylation patterns. Array-based methods have evolved to include over 850,000 CpG dinucleotides throughout the human genome. [14] As rapid, cost-effective methods for genome-wide coverage of methylation patterns mature, these data provide crucial insights into the epigenetic landscape of cancer.
Figure 1. Schematic workflow for DNA methylation analysis.
With the ability to utilize FFPE or frozen tissue as input, generation of DNA methylation data using array-based technologies can be accomplished in a timeline feasible for clinical implementation. In addition to class prediction outputs, DNA methylation data can provide robust copy number information while also facilitating powerful exploratory analyses that could eventually feedback into the core clinical pipeline.
Array-based assays for DNA methylation, such as the Illumina Infinium Human BeadChip arrays (450K and EPIC), have certain characteristics rendering them amenable to utilization in the clinical environment. First, since these assays utilize DNA and query a relatively stable epigenetic mark, issues of degradation and sample quality that plague transcript-based assays are avoided. Additionally, comparable performance on FFPE and frozen tumor samples facilitates utilization of archived tumor samples. The relatively low DNA input required for these assays (approximately 250 nanograms) can typically be obtained from less than five unstained FFPE sections. Since fixed tissue can be utilized, isolation of tumor from surrounding normal tissue can be accomplished by macrodissection of unstained sections or core biopsy of tissue blocks. Importantly, these assays can also be performed and analyzed within a week, a reasonable timeframe for diagnostic implementation and use in the clinic (Figure 1).
In parallel with methodological advances in interrogating the methylation landscape in biological samples, bioinformatics pipelines and classifiers have been developed to process the raw data into biologically and clinically applicable outputs. [15, 16] These computational methods utilize statistical tests and algorithms to make class predictions. Ensemble methods, such as random forest analyses, utilize the combinatorial power of many decision trees to improve predictive accuracy. [17] With the generation of larger and more numerous training reference data sets with well annotated metadata, many of these classifiers have potential to exploit advanced machine learning techniques to optimize predictions of not only tumor class assignment, but also clinical behavior or therapeutic response. The following discussion exemplifies how DNA methylation profiling can and will benefit the management of pediatric CNS tumors.
Medulloblastoma
Medulloblastoma is the most common CNS embryonal tumor with a cure rate of about 70%. [18–20] Standard treatment consists of maximal surgical resection, craniospinal irradiation, and adjuvant chemotherapy. Traditional risk stratification is based on age, extent of resection, evidence of metastasis, and histological variants. [21] Prognosis for high risk disease remains suboptimal, and survivors from all risk groups often experience debilitating long-term morbidities. [22] Research in the past decade has revealed clinically relevant molecular heterogeneity within medulloblastoma, setting the stage for similar work to be done in other pediatric and adult CNS tumors. [23, 24] Molecular subgrouping of medulloblastoma was integrated into the 2016 update to the WHO classification of tumors of the central nervous system and is currently being used to stratify patients in prospective clinical trials. [25, 26]
Data from transcriptomic analysis on multi-institutional medulloblastoma cohorts led to the consensus classification. Unsupervised hierarchical clustering on expression arrays saw the disease split into 4 subgroups (named WNT, sonic hedgehog (SHH), Group 3, and Group 4), each with distinct patient-related characteristics, somatic genetic alterations, upregulated signaling pathways, and clinical course. [23, 24, 27–31] Patients with WNT-subgroup medulloblastoma were found to respond exceptionally well to conventional treatment modalities with nearly uniform overall survival even across treatment regimens of varying intensity. On the other hand, patients with Group 3 medulloblastoma tended to do poorly with overall survival of approximately 50–60% while SHH and Group 4 tumors had intermediate survival outcomes (70–80%). Thus, by subgrouping the disease, it became conceivable to propose dose de-escalation strategies for certain patients carrying disease with good prognosis (i.e. WNT) in order to reduce treatment-related morbidity without compromising outcome. Furthermore, subgroup stratification facilitated the identification of certain patients with very poor outcome, particularly Group 3 tumors with metastasis at diagnosis.
Despite these groundbreaking discoveries, routine application of transcriptomic methods carries practical issues as mentioned previously. Hence, the immediate translation of these findings into the clinic was stunted, leading to a search for alternative platforms. [32] An RNA-based NanoString method and use of IHC stains emerged as alternative strategies to translate the transcriptomic findings. NanoString has been employed at a few centers but has failed to secure wide-spread acceptance. Meanwhile, the use of IHC stains to beta-catenin, YAP1 and GAB1, has enabled pathologists to rapidly recognize and distinguish WNT and SHH medulloblastomas from non-WNT/non-SHH medulloblastoma using standard histopathological techniques. Nonetheless, this method has been difficult to standardize across all centers and does not distinguish Group 3 from Group 4 tumors. [33] On the other hand, methylation profiling faithfully recapitulates the four transcriptomic medulloblastoma subgroups even with limited quantity of DNA from readily available FFPE material. [34] Comparison between the RNA-based NanoString method and methylation profiling demonstrated superiority of the latter both in terms of subgroup assignment and the ability to identify tumors with alternative diagnosis.[35]
Furthermore, methylation profiling also allows for sub-classification or “subtyping” within the four subgroups, a characteristic that has proven to be particularly important for describing clinical and molecular heterogeneity within each of the non-WNT subgroups. (Figure 2, 3) [36–38] Recent publications have shown that SHH exhibits significant clinical heterogeneity according to the age of diagnosis (infants, children, adults) as well as somatic genetic alterations carried. [39] While PTCH1 mutations are evenly distributed among the three age groups; infants often carry SUFU mutations; tumors from children are more likely to be TP53 mutated and GLI2 amplified; and SHH-subgroup medulloblastomas in adults are characteristically enriched with SMO, DDX3X and TERT promotor mutations. Importantly, survival outcome is different for each of these SHH subtypes. Fittingly, methylation profiling on large reference datasets, such as Molecular-Neuropathology 2.0 developed at the German Cancer Research Center (DKFZ), can reproducibly and systematically recognize differences within the methylomes of SHH tumors that distinguish infant SHH MB from childhood or adult SHH MB (see MB_SHH_INF vs MB_ SHH_CHL/AD in Figure 2). Furthermore, new evidence shows methylation profiling of infant SHH MB can detect low and high risk groups within infants in this tumor subgroup (Robinson GW et al. In press. Lancet Oncology. 2018). [37]
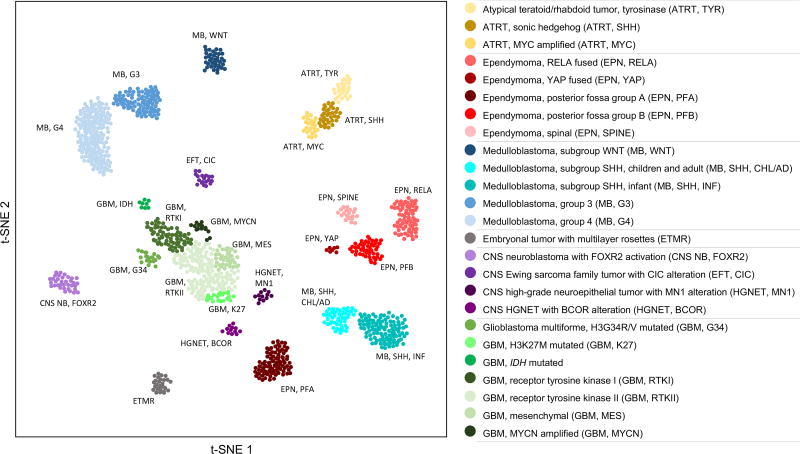
Figure 2. Global methylome (t-SNE structure) of discussed pediatric tumors.
Using the t-distributed stochastic neighbor embedding (t-SNE) algorithm to reduce high dimensional methylation data, the landscape of selected pediatric CNS tumors and their molecularly defined subgroups/subtypes can be readily visualized. In simple terms, the Euclidean distances between objects or clusters can be conceptualized as a metric of relatedness. Adapted by permission from Springer Nature: DNA methylation-based classification of central nervous system tumours by David Capper et al. 2018 [15].
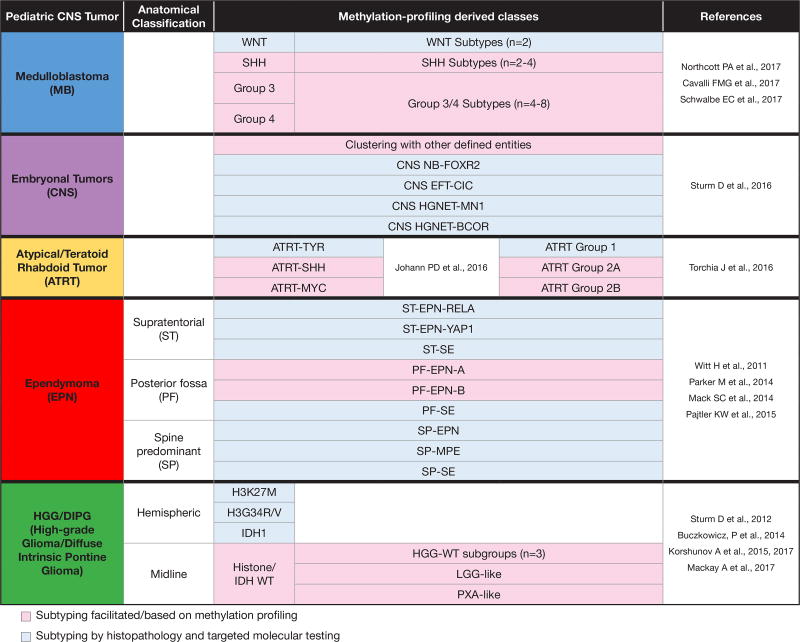
Figure 3. Summary of various sub-classifications defined by methylation.
The methylation-profile derived classes of various pediatric CNS tumors are outlined based on the referenced studies. The necessity for reconciliation of subtypes and consensus definition is evident, particularly for medulloblastoma and ATRT. When incidences of the various entities are considered, methylation profiling facilitates subtyping in approximately two-thirds of cases (pink).
Akin to the intertumoral heterogeneity observed within SHH medulloblastoma, similar observations have been described among Group 3 and Group 4 medulloblastomas. Definitively distinguishing Group 3 from Group 4 medulloblastoma represents a challenge because of the now accepted inherent molecular overlap between the two groups that precludes complete separation. However, methylation profiling has facilitated the subtyping of consensus Group 3 and Group 4 tumors, confirming their close molecular relationship (Figure 2). Such identification of molecular overlap among consensus Group 3 and Group 4 tumors has led to its recent restructuring into at least four and as many as eight subtypes in a series of recently published manuscripts. [36–38] Despite a current lack of subtype consensus among these studies, common themes are emerging. The existence of a Group 3 signature enriched with tumors harboring MYC amplification is irrefutable. Similarly, Group 4 methylome signatures identify a set of medulloblatomas that contain recurrent chromatin remodeling aberrations. Finally a mixed group 3/4 signature seems to be emerging that captures both Group 3 and Group 4 medulloblastomas harboring GFI1 overexpression with OTX2 copy number gain. Overlay of cytogenetic data onto these analyses has highlighted the broad distribution of abnormalities, such as 7q gain, 8p loss, and isochromosome 17q, spanning the Group 3/4 continuum. Nonetheless, the next step is to determine how applicable and relevant these new categories are from a clinical perspective.
Methylation profiling has thus augmented the discovery of clinically informative heterogeneity. The application of this test to readily available samples with minimal processing along with the rapid turnaround time makes this an ideal platform for the development of molecularly-driven, risk-stratified clinical trial design with an eye toward targeted therapeutics and therapy de-escalation. Methylation profiling therefore appeals as a practical adjunct for subgroup assignment in medulloblastoma for prognostication and treatment stratification. Nonetheless, consensus subtype assignments based on methylation profiling must be established.
Atypical teratoid/rhabdoid tumor
Atypical teratoid/rhabdoid tumor (ATRT) is an aggressive embryonal tumor of infancy and young childhood, characterized by abnormalities in SWI/SNF complex genes leading to loss of INI1 (SMARCB1) or, rarely, BRG1 (SMARCA4) protein expression. [40, 41] Prognosis is exceptionally poor and toxicities resulting from conventional therapy in very young patients are significant. [42–45] Attempts to mitigate such a malignant disease course will depend on our ability to gain a deeper understanding of ATRT biology and the molecular basis of the disease. SMARCB1 is a core component of the SWI/SNF chromatin-remodeling complex, which carries a crucial role in tumor suppression and has been implicated in the pathogenesis of numerous cancers. [46, 47] Whole exome sequencing studies of rhabdoid tumors has shown that recurrent genetic alterations other than that those in SMARCB1/SMARCA4 are surprisingly scarce, prompting evaluation of non-coding regions and the epigenome. [48, 49]
Two recent studies of ATRT based on methylation profiling have uncovered heterogeneity within what had previously been considered a single entity. Methylation profiling of 192 ATRTs and unsupervised clustering yielded three distinct subgroups (TYR, SHH, MYC), supported by gene expression data. [49] (Figures 2, 3) ATRT-TYR occurred predominantly in patients of age < 1 year and were more likely infratentorial. Expression data featured overexpression of melanosomal markers and genes involved in ciliogenesis. TYR, encoding tyrosinase, appeared to be a sensitive and specific biomarker for this subgroup. The ATRT-SHH subgroup comprised of tumors located both supratentorially and infratentorially in patients < 6 years of age with upregulation of SHH and NOTCH signaling. In ATRT-MYC tumors where MYC overexpression was prevalent, disease often developed supratentorially and in patients ≥ 6 years of age. Torchia and colleagues performed integrated genomic, transcriptomic and methylomic analysis on another cohort of 191 primary ATRTs. [50] Three epigenetic subgroups, Group 1, 2A and 2B, were again defined with DNA methylation arrays. Group 1 tumors were predominantly supratentorial, had a median age of diagnosis of 2 years, exhibited recurrent chromosome 14 gains and chromosome 19 losses, and had SMARCB1 loss due to focal or subgenic alterations.Group 2A tumors were predominantly infratentorial, had a median age of diagnosis of 1 year and were genomically bland. Group 2B included tumors arising from various locations, comprised of most patients > 3 years of age, had more focal genomic alterations across multiple chromosomes and SMARCB1 loss resulting from large deletions that encompassed additional genes on chromosome 22. These subgroups exhibited distinct methylation and expression patterns associated with lineage and developmental signaling genes.
With the lack of obvious concordance between the two subgrouping schemes of ATRT, a dire need to reach consensus in subgrouping and nomenclature exists. However, once again the utility of methylation profiling in identifying heterogeneity that corresponds with other characteristics such as tumor location, age at diagnosis, and patterns of molecular aberrations is apparent. Nonetheless, uncovering correlations between ATRT epigenetic subgroups and patient survival along with translations into direct clinical applicability are outstanding tasks. It is possible that patient response in on-going trials incorporating novel agents (i.e. alisertib and tazemetostat) will be divergent among ATRT epigenetic subgroups. [51, 52] Therefore we predict that targeted therapy will be applied in future studies according to molecular stratification supported by data from subgroup-specific pathway dysregulation and in vitro treatment models.
CNS-embryonal tumors [formerly known as primitive neuroectodermal tumor (CNS-PNET)]
A keen example of how DNA-methylation profiling has revolutionized our understanding and approach to pediatric brain tumors has come through the study of what was previously known as CNS-PNET. [53] CNS-PNET has long represented a diagnostic challenge due to the lack of specific morphological features and molecular markers. Distinguishing CNS-PNET from histologic mimics such as high grade glioma carries important clinical implications, as patients with CNS-PNET are often prescribed craniospinal rather than local radiation. Historically, this embryonal tumor of childhood has been managed with medulloblastoma therapy based on the assumption that CNS-PNETs represent a supratentorial equivalent of the infratentorially-located medulloblastoma. [54]
Recent work has suggested such a framing of CNS-PNET is incorrect. In a large collaborative study, 323 tumors institutionally diagnosed as CNS-PNET were compared to 211 reference samples by unsupervised clustering of their DNA methylation profiles. [53] Unexpectedly, 61% of cases diagnosed as CNS-PNET clustered with other well- defined entities including high-grade glioma (MYCN-amplified, RTK-amplified, IDH-mutant, H3G34R/V-mutant), H3K27M-mutant diffuse midline glioma, embryonal tumor with multilayered rossettes (ETMR), ependymoma, ATRT, medulloblastoma, pineal tumors, Ewing sarcoma, and choroid plexus carcinoma. Such re-classification was confirmed by orthogonal validation in most instances. Of the remaining tumors diagnosed as CNS-PNET, 24% formed four distinct clusters. These newly described clusters were characterized by distinct genomic drivers as well as clinicopathologic findings and were designated – CNS neuroblastoma with FOXR2 activation (CNS NB-FOXR2), CNS Ewing sarcoma family tumor with CIC alteration (CNS EFT-CIC), CNS high-grade neuroepithelial tumor with MN1 alteration (CNS HGNET-MN1), and CNS high-grade neuroepithelial tumor with BCOR alteration (CNS HGNET-BCOR). (Figure 2, 3) Of the new groups, only the CNS NB-FOXR2 group met classic histologic criteria for CNS-PNET. Methylation profiling thus defined unknown subgroups within CNS-PNET and facilitated the recognition of histological features within these novel entities. Acknowledging the difficulty in diagnosis and frequency of misclassification, CNS-PNET was eliminated as a diagnostic entity from the WHO 2016 classification of CNS tumors. [25]
Future studies must address the clinical behavior of these new tumor entities in addition to identifying appropriate treatment modalities and regimens for these novel groups. The traditional treatment paradigms for high grade pediatric brain tumors may also require re-evaluation. Early work has begun to address these questions. The CNS-PNET arm of Children’s Oncology Group (COG) ACNS0332 closed prematurely with the expectation that such newly introduced heterogeneity would now undermine the statistical power assumed in the study. Indeed of the 31 non-pineal CNS-PNETs with tissue available 22 (71%) were re-classified to high grade glioma (n=18), ATRT (n=2) and ependymoma (n=2). [55] Molecularly-classified high grade glioma had a 5-year event free and overall survival of 5.6% and 12%, respectively, compared with 62.8% and 78.5% respectively in those with molecularly-classified embryonal tumors. Data on the clinical course of some newly defined entities are emerging but remains anecdotal. [56–58]
Ependymoma
Ependymoma is the third most common CNS tumor of childhood after glioma and medulloblastoma. [59] Standard of care consists of maximal surgical resection and adjuvant radiotherapy, while the role of chemotherapy is still being investigated in ongoing trials. [60] Predictors for inferior outcome include infratentorial location of primary tumor, residual disease after resection, young age at diagnosis, and management without adjuvant radiation. Tumor grading (WHO grade II versus WHO grade III) has been shown to display high inter-observer variability along with questionable association with patient outcome and is no longer recommended to dictate treatment options. [61–63] Tumors from different anatomical compartments have indistinguishable histological features, and it was not until a decade ago that the application of gene expression arrays illustrated distinct profiles among them. [64] Subsequent whole genome and epigenome analyses have characterized recurrent fusion between RELA and C11orf95 in > 70% of supratentorial ependymoma, a new entity in the latest WHO classification, while also identifying two molecular subgroups among posterior fossa EPN with prognostic significance. [65–68]
Methylation profiling of 500 ependymomas from various anatomical locations and patient ages have consolidated earlier findings, allocating ependymoma into nine subgroups. [69] (Figure 2, 3) Three of the molecular subgroups encompass WHO Grade I subependymoma each separated by the three anatomical compartments in which these are found: supratentorial subependymoma (ST_SE), posterior fossa subependymoma (PF_SE), and spinal subependymoma (SP_SE). Non-subependymoma supratentorial tumors were classified into supratentorial ependymoma with RELA fusion (ST-EPN-RELA) and supratentorial ependymoma with YAP1 fusions (ST-EPN-YAP1). Likewise, non-subependymoma posterior fossa tumors were classified into posterior fossa ependymoma group A (PF-EPN-A) and posterior fossa ependymoma group B (PF-EPN-B). Finally spinal non-subependymomas were classified into groups enriched in WHO Grade I myxopapillary ependymoma (SP-MPE) and WHO Grade II ependymoma (SP-EPN). [69] ST-EPN-RELA represented two-thirds of the non-subependymoma tumors in the supratentorial compartment with a median age at diagnosis of 8 years and three-quarter of cases being assigned WHO Grade III. The less common ST-EPN-YAP1 was characterized younger patients (median age at diagnosis 1.4 years) and better outcome. In the posterior fossa, PF-EPN-A was associated with younger age at diagnosis (median 3 years), laterally located tumors, a balanced genome, and worse prognosis, particularly with an increased risk of metastatic recurrence. [66, 67, 69] Patients with PF-EPN-B were older children and adults (median 30 years), often with centrally located tumors and higher degree of genomic instability. Gain of chromosome 1q, despite being observed in PF-EPN-A, PF-EPN-B, and ST-EPN-RELA tumors, only manifested itself as poor prognostic marker in PF-EPN-A tumors. [69]
The clinical value of molecular subgrouping in posterior fossa ependymoma was subsequently validated in a large retrospective study consisting of 820 patients from four independent cohorts. [70] Multivariate analysis accounting for molecular subgrouping, age, gender, extent of surgery, and adjuvant therapy revealed molecular subgrouping as the most powerful predictor of progression free and overall survival. [70] In addition to confirming the poor prognosis in PF-EPN-A patients, it was shown that a proportion of PF-EPN-B patients could be cured by gross total resection alone. These findings are thus prompting the development of new strategies in upcoming clinical trials, where therapy is expected to be risk-stratified by molecular classification. [61, 63]
High Grade Glioma [including Diffuse Intrinsic Pontine Glioma (DIPG) and Glioblastoma multiforme (GBM)]
High grade glioma accounts for 15–20% of pediatric CNS tumors and is largely incurable with median survival between 10 and 18 months. [71] Current standard of care for high grade glioma involves maximal safe surgical resection, which is often incomplete due to the tumor location and its infiltrative nature, followed by adjuvant focal radiotherapy. Therapeutic interventions in pediatric high grade glioma, including those extrapolated from adult studies, have been disappointing to date. [72, 73]
Clinical and molecular studies in high grade glioma across age groups suggest that pediatric and adult tumors have distinct developmental origins and disease biology. [74] Driver mutations in genes encoding histones H3.1 and H3.3 now characterize pediatric GBM and DIPG. [75, 76] Sturm and colleagues have since identified six epigenetic subgroups (K27, G34, IDH, RTK I, RTK II, mesenchymal). [77]. These findings were validated and expanded upon by methylation profiling in larger cohorts of pediatric high grade glioma. [78, 79] Hierarchical clustering of 202 pediatric cases resulted in 4 subsets: H3.3/H3.1K27M-mutated, H3.3G34R-mutated, IDH1-mutated and H3/IDH-wild type (wt), each characterized by unique clinical and molecular profiles in line with the previous study (Figure 2, 3).
H3.3/H3.1K27M-mutated tumors were the commonest and had dismal prognosis. They occurred in younger children, were predominantly midline, metastasized in half of the cases, and rarely displayed O6-methylguanine-DNA methyltransferase (MGMT) promotor methylation. The devastating DIPG (also known as brainstem glioma) fell into this category. The H3.3G34R and IDH1-mutated tumors had better outcome, occurred in older patients, were often located in the hemispheres, and were enriched with MGMT promotor methylation. Unlike their IDH1-mutated adult counterparts, there was no evidence that these pediatric GBMs transformed from precursor lesions. [80]
Within H3/IDH-wt, three clearly defined epigenetic clusters with respective oncogene amplification, namely pedGBM_MYCN, pedGBM_RTK1 (enriched for PDGRFA amplification), and pedGBM_RTK2 (enriched for EGFR amplification), were recognized. [77, 79] Multivariate analysis identified methylation profile-derived molecular subtype as the only significant prognostic factor. Additional analysis of cases that fell into this category, prompted by identification of extended survival in some patients, revealed samples that molecularly resembled low grade glioma (LGG) and pleomorphic xanthoastrocytoma (PXA). The LGG-like tumors were diagnosed mostly in young children < 3 years and had a better prognosis (3-year overall survival of 91%), supporting the long-held observation that high grade gliomas in infants respond better to treatment. [81, 82] PXA-like tumors had the highest frequency BRAF V600E and intermediate prognosis. Notably, histological review of LGG-like and PXA-like tumors confirmed typical high grade glioma appearances in > 80% of the cases, highlighting the utility of methylation profiling in accurately identifying these subgroups in the clinical setting.
The molecular landscape of pediatric high grade glioma was most recently consolidated by integrated molecular analysis of more than 1000 cases. [71] Even though most of these entities are universally fatal, it has become apparent that molecular characteristics are important. Methylation profiling gives valuable prognostic information by subgroup definition, providing an opportunity to better counsel families and to focus efforts against unique molecular aberrations.
Integration of methylation profiling with first-line diagnostics in pediatric CNS tumors
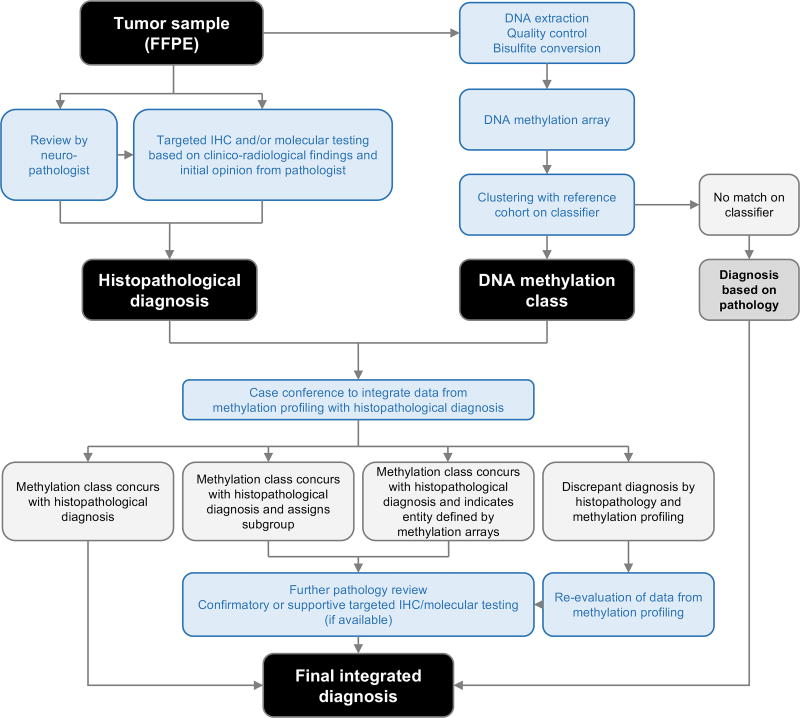
Despite greatly enhancing our understanding of and ability to classify pediatric CNS tumors, DNA methylation profiling still remains primarily a research rather than clinical tool. However, given the profound effect this modality has had on expanding and defining the molecular landscape of pediatric CNS tumors and its relative ease of application, more widespread adoption in the clinical setting seems inevitable. The potential as an aid to clinical diagnosis, treatment decision-making, and study trial design appear boundless (Figure 4).
Figure 4. Proposed workflow for utilization of methylation profiling in clinical diagnosis.
DNA methylation profiling of CNS tumors must be utilized alongside established histopathologic and clinico-radiologic approaches to arrive at a confident, integrated diagnosis.
Nonetheless, certain limitations and caveats must be acknowledged before blindly accepting methylation profiling into the clinic. The introduction of methylation-based methods does not mean more traditional approaches will or should be completely supplanted. Moreover, similar to other tools utilized in neuro-pathology, such as IHC, fluorescence in-situ hybridization, and NGS, methylation profiling needs to remain an instrument that assists in diagnosis rather than declaring it. Classification by methylation profiling may be impossible in cases with insufficient biopsy tissue or poor DNA yields. Hypocellular low grade tumors or those with stromal contamination may not contain sufficient tumoral DNA to yield an accurate classification, as is the case for some diffuse astrocytoma. [83] Even in cases with adequate material, some tumors may still remain unclassifiable by methylation, as observed in a recent comprehensive cohort with 82 methylation classes where 12% of samples scored too poorly for confident classification. [15] The reference cohort used to train the classification model may not contain sufficient samples from a particular rare entity. Since classifiers are often designed to generate a best-fit for a given input, incorrect grouping may occur in such cases. Continued expansion of reference cohorts and the inclusion of uncommon as well as novel entities would be required to improve the versatility and accuracy in classification over time. Further work will also be needed to validate DNA methylation profiling for classifying LGG, due to frequent difficulty in class assignment. Furthermore, intracranial (non-germinomatous) germ cell tumors will need careful evaluation by this method because of inherent variability in the contribution and composition of teratomatous elements, the often substantial stromal contribution to the tumor, and often focal nature of malignant germ cell tumor elements. [83, 84]
For rare tumor types or tumors that present with challenging histomorphology, methylation based approaches will allow for better standardization of diagnoses across centers. As discussed, tumor subgrouping and subtyping based on methylation profiling are expected to improve prognostication, risk assignment and treatment stratification. Medulloblastoma, ependymoma and infant GBM represent key examples where molecular subgrouping has shown remarkable proficiency at predicting treatment outcome in large retrospective molecular cohorts. [61, 78, 85]
Methylation profiling must not supplant clinical acumen. Clinico-radiological and histopathological features should remain core to the classification of pediatric CNS tumors, while methylation analysis should serve to augment current practices. Figure 4 depicts our vision of how DNA methylation profiling may be applied to clinical diagnosis. It is our expectation that classification by methylation-based methods will improve histomorphologic methods of classification by empowering pathologists to recognize previously unappreciated patterns that associate with specific molecular subgroups and to better develop orthogonal biomarkers for novel entities or cases in which tissue is limiting. Even in the case of widespread adoption of methylation profiling, the necessity to produce and assess histologic sections remains paramount, particularly but not limited to quality control, assessing tumor grade, and clinical stage.
Conclusion
DNA methylation profiling brings a highly robust and accurate molecular characterization system to abundant and accessible pathology samples. As a clinically feasible tool, the integration of DNA methylation profiling in the diagnosis and management of children with brain tumors represents an obtainable goal to assist diagnosis and improve clinical trial design. Coupled with histopathologic, genetic, and transcriptomic analyses, epigenetic profiling will also provide fundamental advances in understanding basic tumor biology.
Acknowledgments
Funding: This work was supported in part by American Lebanese Syrian Associated Charities, St Jude Children’s Research Hospital, NCI Cancer Center Grant (P30CA021765).
Footnotes
Conflicts of Interest: None
References
- 1.Linabery AM, Ross JA. Trends in childhood cancer incidence in the U.S. (1992–2004) Cancer. 2008;112(2):416–32. doi: 10.1002/cncr.23169. [DOI] [PubMed] [Google Scholar]
- 2.Gajjar A, et al. Molecular insights into pediatric brain tumors have the potential to transform therapy. Clin Cancer Res. 2014;20(22):5630–40. doi: 10.1158/1078-0432.CCR-14-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinchon M, et al. Morbidity and tumor-related mortality among adult survivors of pediatric brain tumors: a review. Childs Nerv Syst. 2011;27(5):697–704. doi: 10.1007/s00381-010-1385-6. [DOI] [PubMed] [Google Scholar]
- 4.Netson KL, et al. Executive dysfunction is associated with poorer health-related quality of life in pediatric brain tumor survivors. J Neurooncol. 2016;128(2):313–21. doi: 10.1007/s11060-016-2113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li SC, et al. Cancer genomic research at the crossroads: realizing the changing genetic landscape as intratumoral spatial and temporal heterogeneity becomes a confounding factor. Cancer Cell Int. 2014;14(1):115. doi: 10.1186/s12935-014-0115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glod J, et al. Pediatric Brain Tumors: Current Knowledge and Therapeutic Opportunities. J Pediatr Hematol Oncol. 2016;38(4):249–60. doi: 10.1097/MPH.0000000000000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang ZJ, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14(2):135–45. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedegaard J, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One. 2014;9(5):e98187. doi: 10.1371/journal.pone.0098187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 10.Fujita N, et al. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol. 1999;19(9):6415–26. doi: 10.1128/mcb.19.9.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21(35):5427–40. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 12.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21(35):5400–13. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 13.Clark SJ, et al. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22(15):2990–7. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8(3):389–99. doi: 10.2217/epi.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capper D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahamed MT, et al. MethPed: an R package for the identification of pediatric brain tumor subtypes. BMC Bioinformatics. 2016;17(1):262. doi: 10.1186/s12859-016-1144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breiman L. Random Forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 18.Ostrom QT, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro-Oncology. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gajjar A, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. The lancet oncology. 2006;7(10):813–820. doi: 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 20.Packer RJ, et al. Phase III study of craniospinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. Journal of clinical oncology. 2006;24(25):4202–4208. doi: 10.1200/JCO.2006.06.4980. [DOI] [PubMed] [Google Scholar]
- 21.Rutkowski S, et al. Survival and prognostic factors of early childhood medulloblastoma: an international meta-analysis. Journal of Clinical Oncology. 2010;28(33):4961–4968. doi: 10.1200/JCO.2010.30.2299. [DOI] [PubMed] [Google Scholar]
- 22.Armstrong GT, et al. Long-term outcomes among adult survivors of childhood central nervous system malignancies in the Childhood Cancer Survivor Study. JNCI: Journal of the National Cancer Institute. 2009;101(13):946–958. doi: 10.1093/jnci/djp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor MD, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta neuropathologica. 2012;123(4):465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Northcott PA, et al. Molecular subgroups of medulloblastoma. Expert review of neurotherapeutics. 2012;12(7):871–884. doi: 10.1586/ern.12.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louis DN, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 26.Gajjar A. A Clinical and Molecular Risk-Directed Therapy for Newly Diagnosed Medulloblastoma. 2013 NCT01878617. ClinicalTrials.gov.
- 27.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–5. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–10. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148(1–2):59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488(7409):43–8. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Northcott PA, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41(4):465–72. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mack SC, Northcott PA. Genomic Analysis of Childhood Brain Tumors: Methods for Genome-Wide Discovery and Precision Medicine Become Mainstream. J Clin Oncol. 2017;35(21):2346–2354. doi: 10.1200/JCO.2017.72.9921. [DOI] [PubMed] [Google Scholar]
- 33.Ellison DW, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta neuropathologica. 2011;121(3):381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hovestadt V, et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathologica. 2013;125(6):913–916. doi: 10.1007/s00401-013-1126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korshunov A, et al. DNA-methylation profiling discloses significant advantages over NanoString method for molecular classification of medulloblastoma. Acta Neuropathol. 2017;134(6):965–967. doi: 10.1007/s00401-017-1776-9. [DOI] [PubMed] [Google Scholar]
- 36.Schwalbe EC, et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 2017;18(7):958–971. doi: 10.1016/S1470-2045(17)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavalli FMG, et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell. 2017;31(6):737–754. e6. doi: 10.1016/j.ccell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Northcott PA, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kool M, et al. Genome Sequencing of SHH Medulloblastoma Predicts Genotype-Related Response to Smoothened Inhibition. Cancer cell. 2014;25(3):393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biegel JA, et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer research. 1999;59(1):74–79. [PubMed] [Google Scholar]
- 41.Rorke LB, Packer RJ, Biegel JA. Central nervous system atypical teratoid/rhabdoid tumors of infancy and childhood: definition of an entity. Journal of neurosurgery. 1996;85(1):56–65. doi: 10.3171/jns.1996.85.1.0056. [DOI] [PubMed] [Google Scholar]
- 42.Buscariollo DL, et al. Survival outcomes in atypical teratoid rhabdoid tumor for patients undergoing radiotherapy in a Surveillance, Epidemiology, and End Results analysis. Cancer. 2012;118(17):4212–4219. doi: 10.1002/cncr.27373. [DOI] [PubMed] [Google Scholar]
- 43.Chi SN, et al. Intensive multimodality treatment for children with newly diagnosed CNS atypical teratoid rhabdoid tumor. J Clin Oncol. 2009;27(3):385–9. doi: 10.1200/JCO.2008.18.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lafay-Cousin L, et al. Central nervous system atypical teratoid rhabdoid tumours: the Canadian Paediatric Brain Tumour Consortium experience. Eur J Cancer. 2012;48(3):353–9. doi: 10.1016/j.ejca.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Fossey M, et al. Atypical teratoid rhabdoid tumor in the first year of life: the Canadian ATRT registry experience and review of the literature. J Neurooncol. 2017;132(1):155–162. doi: 10.1007/s11060-016-2353-0. [DOI] [PubMed] [Google Scholar]
- 46.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nature Reviews Cancer. 2011;11(7):481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 47.Alver BH, et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nature Communications. 2017;8:14648. doi: 10.1038/ncomms14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kieran MW, et al. Absence of oncogenic canonical pathway mutations in aggressive pediatric rhabdoid tumors. Pediatric blood & cancer. 2012;59(7):1155–1157. doi: 10.1002/pbc.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johann PD, et al. Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell. 2016;29(3):379–93. doi: 10.1016/j.ccell.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 50.Torchia J, et al. Integrated (epi)-Genomic Analyses Identify Subgroup-Specific Therapeutic Targets in CNS Rhabdoid Tumors. Cancer Cell. 2016;30(6):891–908. doi: 10.1016/j.ccell.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Upadhyaya S. Phase 2 Study of Alisertib Therapy for Rhabdoid Tumors (SJATRT) 2014 NCT02114229. ClinicalTrials.gov.
- 52.Robinson GW. A Phase 1 Study of the EZH2 Inhibitor Tazemetostat in Pediatric Subjects With Relapsed or Refractory INI1-Negative Tumors or Synovial Sarcoma. 2015 NCT02601937. ClinicalTrials.gov.
- 53.Sturm D, et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell. 2016;164(5):1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Braganca KC, Packer RJ. Treatment Options for Medulloblastoma and CNS Primitive Neuroectodermal Tumor (PNET) Current treatment options in neurology. 2013;15(5):593–606. doi: 10.1007/s11940-013-0255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang E, et al. Pdct-05. Molecular Diagnostics Reveal 60% Higher Survival For Molecularly-verified Versus Histopathologically-diagnosed Pediatric Supratentorial Central Nervous System Embryonal Tumors And Pineoblastomas; A Report From The Children’s Oncology Group Acns0332 Trial. Neuro-oncology. 2017;19(suppl_6):vi184–vi185. [Google Scholar]
- 56.Yoshida Y, et al. CNS high-grade neuroepithelial tumor with BCOR internal tandem duplication: a comparison with its counterparts in the kidney and soft tissue. Brain Pathology. :n/a–n/a. doi: 10.1111/bpa.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Appay R, et al. HGNET-BCOR Tumors of the Cerebellum: Clinicopathologic and Molecular Characterization of 3 Cases. Am J Surg Pathol. 2017;41(9):1254–1260. doi: 10.1097/PAS.0000000000000866. [DOI] [PubMed] [Google Scholar]
- 58.Wood MD, et al. Multimodal molecular analysis of astroblastoma enables reclassification of most cases into more specific molecular entities. Brain Pathol. 2017 doi: 10.1111/bpa.12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marinoff AE, et al. Rethinking childhood ependymoma: a retrospective, multi-center analysis reveals poor long-term overall survival. J Neurooncol. 2017;135(1):201–211. doi: 10.1007/s11060-017-2568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merchant TE, et al. Conformal radiotherapy after surgery for paediatric ependymoma: a prospective study. Lancet Oncol. 2009;10(3):258–66. doi: 10.1016/S1470-2045(08)70342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pajtler KW, et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 2017;133(1):5–12. doi: 10.1007/s00401-016-1643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ellison DW, et al. Histopathological grading of pediatric ependymoma: reproducibility and clinical relevance in European trial cohorts. J Negat Results Biomed. 2011;10:7. doi: 10.1186/1477-5751-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rudà R, et al. EANO guidelines for the diagnosis and treatment of ependymal tumors. Neuro-oncology. 2017 doi: 10.1093/neuonc/nox166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor MD, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8(4):323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 65.Parker M, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506(7489):451–5. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Witt H, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20(2):143–57. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wani K, et al. A prognostic gene expression signature in infratentorial ependymoma. Acta Neuropathol. 2012;123(5):727–38. doi: 10.1007/s00401-012-0941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mack SC, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506(7489):445–50. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pajtler KW, et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell. 2015;27(5):728–43. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramaswamy V, et al. Therapeutic Impact of Cytoreductive Surgery and Irradiation of Posterior Fossa Ependymoma in the Molecular Era: A Retrospective Multicohort Analysis. J Clin Oncol. 2016;34(21):2468–77. doi: 10.1200/JCO.2015.65.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mackay A, et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell. 2017;32(4):520–537. e5. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stupp R, et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. New England Journal of Medicine. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 73.Cohen KJ, et al. Temozolomide in the treatment of high-grade gliomas in children: a report from the Children's Oncology Group. Neuro Oncol. 2011;13(3):317–23. doi: 10.1093/neuonc/noq191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones C, Perryman L, Hargrave D. Paediatric and adult malignant glioma: close relatives or distant cousins? Nature Reviews Clinical Oncology. 2012;9:400+. doi: 10.1038/nrclinonc.2012.87. [DOI] [PubMed] [Google Scholar]
- 75.Wu G, et al. Somatic Histone H3 Alterations in Paediatric Diffuse Intrinsic Pontine Gliomas and Non-Brainstem Glioblastomas. Nature genetics. 2012;44(3):251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 77.Sturm D, et al. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 78.Korshunov A, et al. Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol. 2015;129(5):669–78. doi: 10.1007/s00401-015-1405-4. [DOI] [PubMed] [Google Scholar]
- 79.Korshunov A, et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathologica. 2017;134(3):507–516. doi: 10.1007/s00401-017-1710-1. [DOI] [PubMed] [Google Scholar]
- 80.Nobusawa S, et al. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clinical Cancer Research. 2009;15(19):6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 81.Davis T, et al. Case Report of Spontaneous Resolution of a Congenital Glioblastoma. Pediatrics. 2016 doi: 10.1542/peds.2015-1241. [DOI] [PubMed] [Google Scholar]
- 82.Kameda M, et al. Congenital Glioblastoma with Distinct Clinical and Molecular Characteristics: Case Reports and a Literature Review. World Neurosurg. 2017;101:817 e5–817 e14. doi: 10.1016/j.wneu.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 83.Jeyapalan JN, et al. DNA methylation analysis of paediatric low-grade astrocytomas identifies a tumour-specific hypomethylation signature in pilocytic astrocytomas. Acta Neuropathologica Communications. 2016;4:54. doi: 10.1186/s40478-016-0323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fukushima S, et al. Genome-wide methylation profiles in primary intracranial germ cell tumors indicate a primordial germ cell origin for germinomas. Acta Neuropathologica. 2017;133(3):445–462. doi: 10.1007/s00401-017-1673-2. [DOI] [PubMed] [Google Scholar]
- 85.Archer TC, Mahoney EL, Pomeroy SL. Medulloblastoma: Molecular Classification-Based Personal Therapeutics. Neurotherapeutics. 2017;14(2):265–273. doi: 10.1007/s13311-017-0526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]