Abstract
The complex interplay between genetic and environmental factors, such as diet and lifestyle, defines the initiation and progression of multifactorial diseases, including cancer, cardiovascular and metabolic diseases, and neurological disorders. Given that most of the studies have been performed in controlled experimental settings to ensure the consistency and reproducibility, the impacts of environmental factors, such as dietary perturbation, on the development of animals with different genotypes and the pathogenesis of these diseases remain poorly understood. By analyzing the cdk8 and cyclin C (cycC) mutant larvae in Drosophila, we have previously reported that the CDK8-CycC complex coordinately regulates lipogenesis by repressing dSREBP (sterol regulatory element-binding protein)-activated transcription and developmental timing by activating EcR (ecdysone receptor)-dependent gene expression. Here we report that dietary nutrients, particularly proteins and carbohydrates, modulate the developmental timing through the CDK8/CycC/EcR pathway. We observed that cdk8 and cycC mutants are sensitive to the levels of dietary proteins and seven amino acids (arginine, glutamine, isoleucine, leucine, methionine, threonine, and valine). Those mutants are also sensitive to dietary carbohydrates, and they are more sensitive to monosaccharides than disaccharides. These results suggest that CDK8-CycC mediates the dietary effects on lipid metabolism and developmental timing in Drosophila larvae.
Keywords: CDK8, dietary effects, ecdysone receptor, developmental transition, Drosophila larvae
Introduction
Deciphering the molecular mechanisms underlying multi-factorial diseases, including cancer, obesity, diabetes, cardiovascular diseases, mental disorders, and neuro-degenerative diseases, has been the major challenge of modern biomedical research. These diseases are determined by genetic, environmental, and chance factors (such as diet, temperature, and stress), as well as the complicated interactions among these factors (Berg, 2016). To mitigate the profound social and economic burdens caused by these multifactorial diseases, it is essential to gain mechanistic insights into the complex gene-environment interactions. Because the effects of genetic and environmental factors are often entangled, thereby obscuring mechanistic insights (compounded by the fact that most of developmental genetic analyses have been carried out in model organisms maintained in standard laboratory settings), the impacts of environmental exposures, with nutrient fluctuations being of major importance, on mutant phenotypes are little understood.
Drosophila represents a powerful model organism to gain insights into the mechanisms underpinning the complex gene-environment interactions. First, the major signaling events and metabolic regulations are conserved but simpler in Drosophila. For example, the underlying biochemical reactions for metabolic processes, such as glycolysis, the tricarboxylic acid cycle, lipogenesis, lipolysis, and fatty acid β-oxidation, are virtually identical to those in mammals. Second, the biological processes from the molecular and cellular to organismal and behavioral levels are well characterized in Drosophila. Third, a rich literature about the function and regulation of most of the genes has been accumulated in the past century. Most of these studies were performed with standard diets, such as the “cornmeal, molasses and yeast medium” (CMY), at standard temperature (25°C), and with the routine day-night cycle. Fourth, there are abundant genetic tools available, as over two-thirds of Drosophila genes have at least one mutant alleles maintained in stock centers or in the research community. Finally, Drosophila has a short life span and the environmental factors, such as diet and temperature, can be manipulated with relative ease. These combined advantages make Drosophila ideally suited to serve as an experimental system to gain insights into our understanding of the aforementioned multi-factorial diseases (Arrese and Soulages, 2010; Cox et al., 2017; Kuhnlein, 2012; Li and Tennessen, 2017; Liu and Huang, 2013; Mattila and Hietakangas, 2017; Schlegel and Stainier, 2007; Sieber and Spradling, 2017).
The developmental timing, particularly the larval-pupal transition in Drosophila, is an ideal phenomenon to analyze the interplay between dietary factors and genetic factors. During the larval stage, larvae keep eating and their body weight increases over 200 times within 4 days largely due to elevated de novo lipogenesis. In contrast, they are immobile during the pupal stage and the energy required for metamorphosis is mainly derived from lipid catabolism. The larval-pupal transition is under the control of two major hormones, juvenile hormone (JH) and steroid hormone ecdysone, and their corresponding receptors (King-Jones and Thummel, 2005; Nakagawa and Henrich, 2009; Riddiford, 1993; Riddiford et al., 2000; Yamanaka et al., 2013). Although hormonal regulation is critical for coordinating larval growth, metabolism, and environmental factors such as dietary nutrients, the underlying molecular mechanism has not yet been fully understood (Andersen et al., 2013; Danielsen et al., 2013; Rewitz et al., 2013; Tennessen and Thummel, 2011; Thummel, 2001).
Our previous studies in analyzing the cdk8 (cyclin-dependent kinase 8) and cycC (cyclin C) mutants suggest that the CDK8-CycC complex may serve as a regulatory node in linking nutrient intake, i.e. feeding and starvation, to developmental timing and fat metabolism during Drosophila development (Xie et al., 2015). CDK8 and CycC, along with MED12 and MED13, form the kinase module of the Mediator complex, which functions as a bridge between different transcription factors and the general transcription machinery, which includes RNA Polymerase II (Clark et al., 2015; Conaway and Conaway, 2011; Kornberg, 2005; Malik and Roeder, 2005; Poss et al., 2013; Xu and Ji, 2011; Yin and Wang, 2014). Our previous work revealed that CDK8-CycC negatively regulates lipogenesis by directly inhibiting the dSREBP (sterol response element binding protein)-dependent gene expression (Zhao et al., 2012), and that CDK8-CycC can regulate the larval-pupal transition by positively regulating EcR (ecdysone receptor)-dependent gene expression (Xie et al., 2015). Interestingly, CDK8 protein levels are increased by nutrient deprivation, but decreased by refeeding during the larval stage (Xie et al., 2015). These results suggest that CDK8-CycC may serve as a regulatory node in linking nutritional perturbations to fat metabolism and developmental transitions in Drosophila larvae.
To further test this model, we analyzed the effects of quality of nutrient intake, i.e. different types of amino acids and sugars (monosaccharides and disaccharides), on the developmental transitions in the cdk8 and cycC mutant larvae. Here we show that the cdk8 and cycC mutants are sensitive to nutritional variations in diets, particularly a few types of dietary amino acids and monosaccharides. These results provide further evidence supporting a critical role of CDK8-CycC in mediating the dietary effects on lipid metabolism and developmental transition in Drosophila larvae.
Results
cdk8 and cycC mutants are sensitive to dietary perturbations
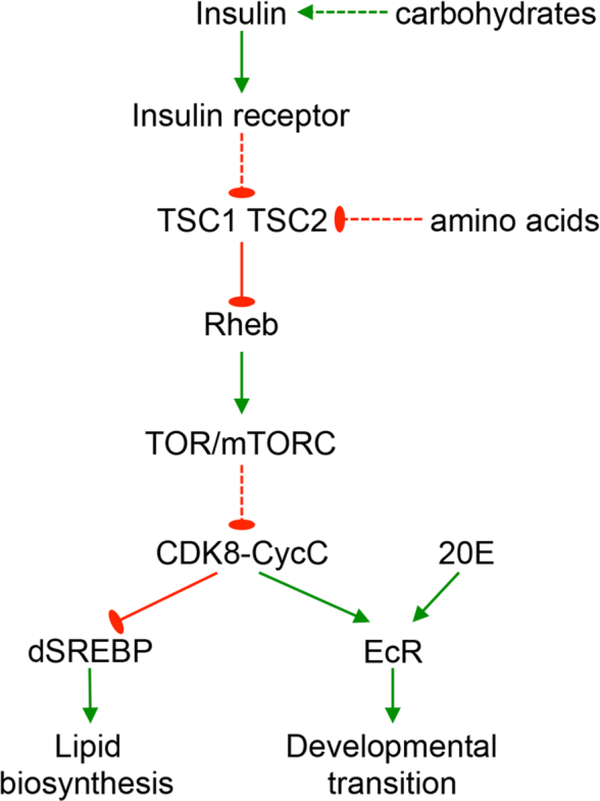
Based on our previous work, regarding the effects of starvation and refeeding on the levels of CDK8 and developmental transition as well as the effects of CDK8-CycC in regulating EcR and dSREBP-dependent gene expression, we have proposed that CDK8-CycC serves as a regulatory node linking nutrient availability and the larval-pupal transition in Drosophila (Xie et al., 2015). In this model (Fig. 1A), the CDK8 activity is inhibited by insulin/TOR (Target of Rapamycin) signaling during the larval stage, which allows dSREBP to activate the expression of lipogenic genes, thereby promoting lipid accumulation. The wandering behavior triggered by a pulse of ecdysteroids allows the accumulation of CDK8-CycC, which inhibits dSREBP but activates EcR-dependent gene expression, thereby blocking de novo lipogenesis while promoting metamorphosis during the pupal stage.
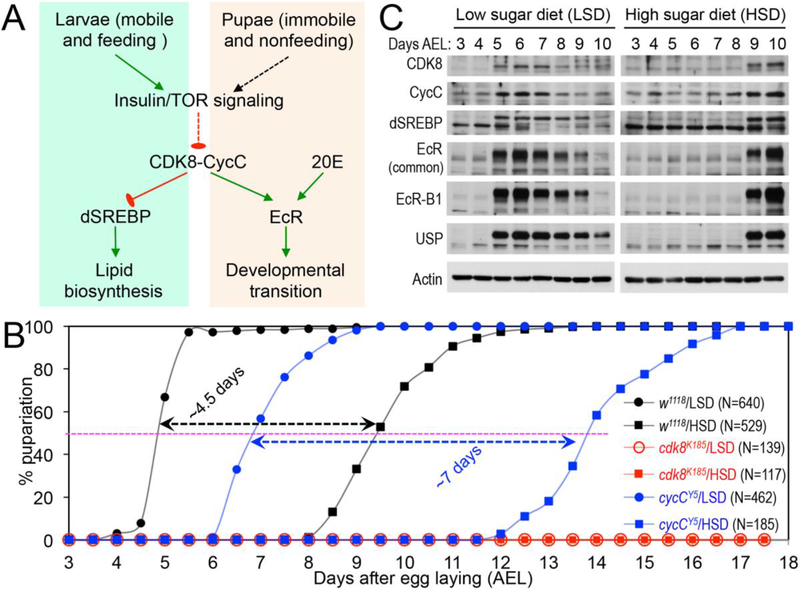
Fig. 1.
CDK8-CycC couples nutrition, lipid biosynthesis, and developmental timing. (A) Model: CDK8-CycC coordinately regulates lipogenesis by directly repressing dSREBP-activated gene expression and developmental timing by activating EcR-activated gene expression during the larval and pupal stages. Arrows represent activation, and blunt arrows represent inhibition. (B) Percentage of pupariated animals raised on either the low sugar diet (LSD) or the high sugar diet (HSD) foods between 3 and 18 days AEL. (C) The protein levels of CDK8, CycC, dSREBP, EcR-B1 and USP (upper band is the 54kDa full-length USP) in wild-type larvae cultured on either the LSD or the HSD between 3 and 10 days AEL. For dSREBP, the lower band (~49kDa) is the mature nuclear form, while the upper band (53~54kDa) is the N-terminal fragment of dSREBP after cleavage by the S1P (Xie et al., 2015).
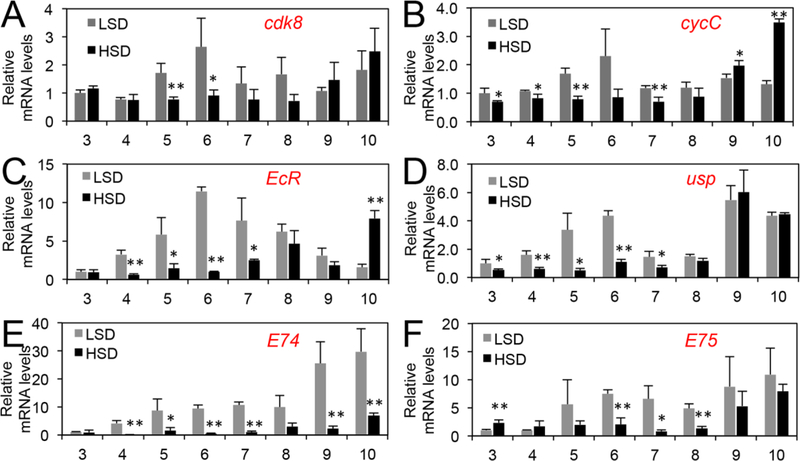
Although the data based on the starvation and re-feeding of early third instar larvae, as well as developmental genetic analyses of the phenotypes of the cdk8 and cycC mutants, support this model (Xie et al., 2015), we sought to further validate this model using dietary perturbations because of their effects on insulin/mTOR signaling and growth. Initially, we analyzed the effects on developmental transition by feeding both the cdk8 or cycC mutants and the control larvae with either a low sugar diet (LSD, see below), or a high sugar diet (HSD, i.e., LSD with 20% of sucrose). The ingredients of LSD are better defined than the CMY recipe, and the HSD has also previously been shown to delay the larval-pupal transition by prolonging the period of activation of insulin signaling (Musselman et al., 2011; Na et al., 2013; Pasco and Leopold, 2012). As expected, HSD dramatically delays the developmental timing in the control larvae (w1118; Fig. 1B). In control animals fed with the LSD, the levels of CDK8, CycC, EcR and USP proteins were increased and the mature nuclear dSREBP level was reduced at the onset of the larval-pupal transition at the end of day 5 after egg laying (AEL, Fig. 1C). In contrast, the levels of CDK8 and CycC remained low in animals fed on a HSD until day 9 AEL, when pupariation was initiated (Fig. 1B, Fig. 1C). The effect of HSD on CDK8 and CycC was evident in steady-state mRNA levels of cdk8 and cycC, which were significantly lower in HSD-fed animals during day 5 and day 6 AEL (Fig. 2A, Fig. 2B). The protein levels of EcR and USP also closely mirrored their mRNA levels (Fig. 1C, Fig. 2C, Fig. 2D) and EcR activity, as monitored by the levels of EcR target genes (Fig. 2E, Fig. 2F). Similarly, reduced CDK8-CycC levels in HSD-fed animals were correlated with increased levels of nuclear dSREBP (Fig. 1C) and dSREBP activity, which were monitored by expression of dSREBP target genes including dFAS (Fig. S1A), dACC (Fig. S1B), and dACS (Fig. S1C). The mRNA level of dSREBP was not significantly affected by HSD (Fig. S1D), indicating that HSD affects dSREBP protein level. Importantly, low CDK8 and CycC levels are correlated with high mature dSREBP and low EcR and USP levels (Fig. 1C). These observations suggest that the regulation of dSREBP and EcR by CDK8 is delayed upon HSD feeding, consistent with the model in which CDK8-CycC integrates diet and feeding with the timing of pupariation (Fig. 1A).
Fig. 2.
The relative mRNA levels of different genes in LSD- or HSD-fed larvae between 3 and 10 days AEL as assayed using qRT-PCR. HSD refers to LSD with addition of 20% of sucrose. * p < 0.05; ** p < 0.01; *** p < 0.001 based on t-tests.
If the CDK8-CycC complex is required for the dietary effects on developmental transitions, then cdk8 (cdk8K185 homozygous) and cycC (cycCY5 homozygous) mutants are expected to be more sensitive in response to HSD than the control. Indeed, cycC mutant larvae fed with HSD took ~7 more days to pupariate than mutant larvae fed with a LSD, which is significantly longer than the ~4.5 day delay in the control (Fig. 1B). Strikingly, when fed with either HSD or LSD, no cdk8 mutant larvae could pupariate and they died either before or during the third instar, even 18 days AEL (Fig. 1B). These observations suggest that both the cdk8 and the cycC mutants are particularly sensitive to nutrient conditions, but it is unexpected and puzzling as to why the cdk8 mutant larvae become hypersensitive when fed with either a HSD or a LSD.
cdk8 mutants are sensitive to dietary protein levels
To understand why cdk8 mutants are hypersensitive to both a HSD and a LSD, we tested a different HSD recipe by analyzing the timing of pupariation using the standard “cornmeal, molasses and yeast” medium (referred to as the “CMY-LSD” diet in this work) to the same medium with additional 20% sucrose (designated as the “CMY-HSD” diet). Compared to larvae fed with CMY-LSD, feeding with CMY-HSD significantly delayed the larval-pupal transition for ~4.5 days and ~3.5 days in cdk8 and cycC mutants, respectively, while the control animals were delayed for ~2.5 days (Fig. 3A). Consistent with this observation, EcR target genes such as E74 and E78 are reduced in wild-type larvae fed by the CMY-HSD, and they are also lower in cdk8 mutants than the wild-type control (Fig. S2A and Fig. S2B). Increasing dietary sucrose has little effects on the mRNA levels of cdk8 and cycC in wild-type larvae (Fig. S2C). These observations differ from the data that the cdk8 larvae fail to pupariate when fed with either a HSD or a LSD (Fig. 1B), indicating that other dietary components may contribute to the larval lethality under both HSD and LSD.
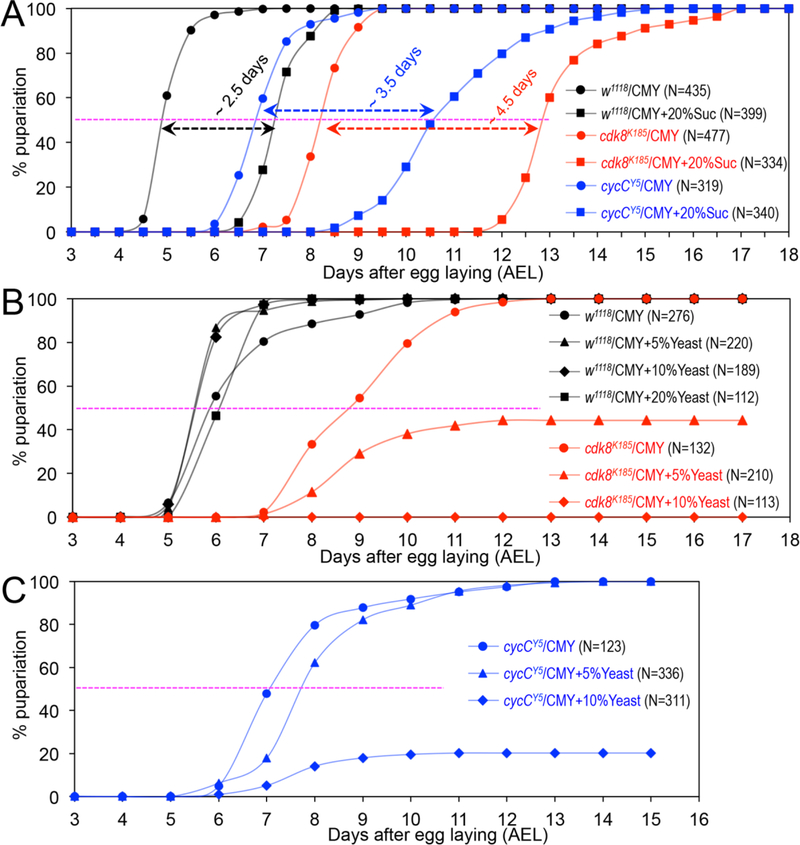
Fig. 3.
The effects of dietary protein levels on the timing for pupariation of the cdk8 and cycC mutants. (A) Percentage of pupariated cdk8 and cycC mutants raised on the CMY (referred to as “CMY-LSD”) or CMY-HSD (CMY with 20% sucrose) foods between 3 and 18 days AEL. Percentage of pupariated cdk8 (B) and cycC (C) mutants raised on the CMY with increasing levels of yeast.
The LSD is also known as the “German food” or sick fly formulation (Backhaus, 1984). Compared to standard CMY-LSD medium, the recipe of LSD is featured with ~5X higher levels of protein from extra yeast (~12% in LSD or “German food” vs. 2.5% yeast in CMY-LSD medium). If a higher level of total protein in a LSD causes the larval lethality of the cdk8 mutants, then increasing protein levels in the CMY-LSD medium is expected to have similar effects on the larval-pupal transition. Inactive dried yeast is the major protein component of the LSD (Table S1), and it also provides low levels of carbohydrates, salts, vitamins, and cholesterols. Indeed, when feeding with the CMY-LSD medium with 5% extra yeast, ~60% of the cdk8 mutants never pupariate, and they die in the third instar larval stage (Fig. 3B). Strikingly, all of the cdk8 mutants are larval lethal when fed with the CMY-LSD medium with 10% extra yeast and they never pupariate (Fig. 3B), similar to animals fed with LSD (Fig. 1B). Similar analyses using cycC mutant larvae showed that CMY-LSD with 5% extra yeast had a weak effect on the pupariation curve, but feeding the cycC mutants with CMY-LSD with 10% extra yeast caused ~80% larval lethality (Fig. 3C). Further increasing the levels of yeast to 20% in the CMY medium caused complete larval lethality in both cdk8 and cycC mutants (data not shown). These observations suggest that the larval lethality of cdk8 and cycC mutants induced by feeding LSD and HSD may be caused by a higher level of proteins in the LSD and HSD recipes compared to the CMY-LSD medium.
cdk8 mutants are sensitive to several specific amino acids
To further validate the effects of dietary protein in lethal stage of the cdk8 mutants, we asked whether reducing proteins levels in LSD can alleviate the effects of LSD on larval lethality and enable them to go through the larval-pupal transition. To test this, we fed both the cdk8 mutant larvae and the control (w1118) in LSD containing reduced the levels of dry yeast by 50% and 75%, without altering other ingredients in the LSD recipe. As shown in Fig. 4A, about 20% of the cdk8 mutant larvae pupariated when fed with LSD containing 50% less dry yeast (“LSD-50%Yeast”), while 60% of cdk8 mutants pupariated when fed with LSD with a 75% reduction of dry yeast (“LSD-75%Yeast”). These observations indicate that a higher level of protein in LSD may cause the larval lethality of the cdk8 mutants. Together with the results showed in Fig. 3, these observations suggest that the cdk8 mutant larvae are particularly sensitive to the levels of dietary proteins.
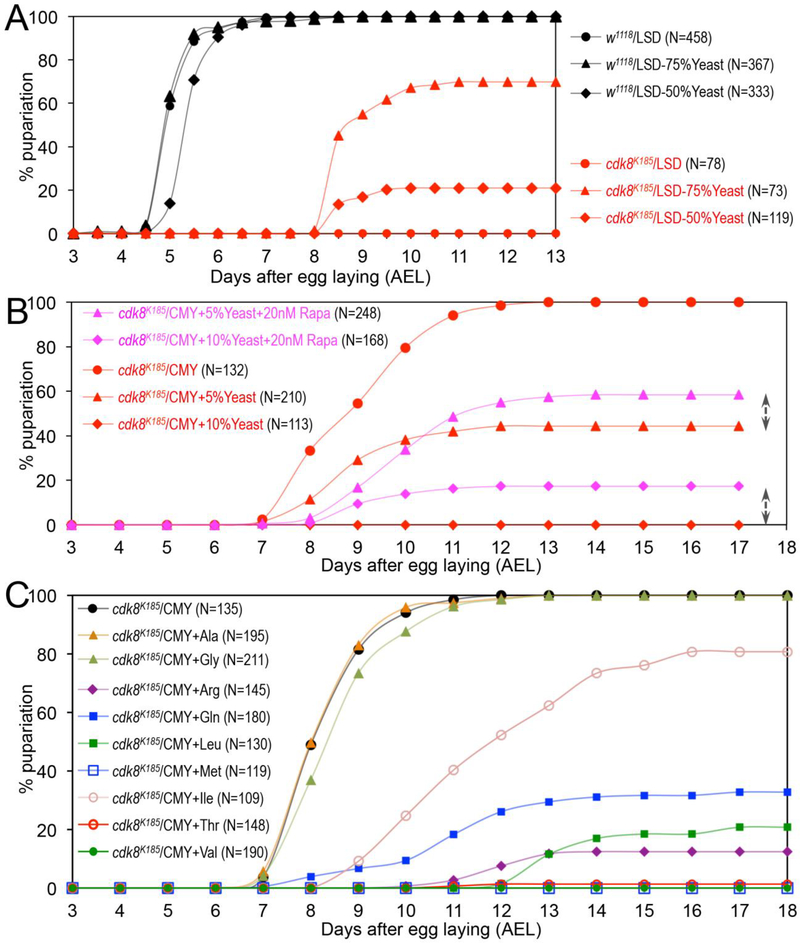
Fig. 4.
The effects of dietary levels of protein, rapamycin, and amino acids on the timing for pupariation of the cdk8 mutants. (A) Percentage of pupariated wild-type and cdk8 mutants raised on the LSD or LSD with reduced levels of yeast: “LSD-50%Yeast” represents the diet with 50% reduction of the yeast in the LSD recipe; “LSD-75%Yeast” is the diet with 75% reduction of yeast in the LSD recipe (see Table S1 for details). (B) Percentage of pupariated cdk8 mutants raised on the CMY diet with 5% or 10% of extra yeast, with or without rapamycin (Rapa). (C) Percentage of pupariated cdk8 mutants raised on the CMY diet with extra amino acids as indicated.
Proteins and amino acids in diets are known to stimulate the activity of TOR, an atypical protein kinase that plays important roles in controlling growth and metabolism (Bar-Peled and Sabatini, 2014; Jewell et al., 2013; Loewith and Hall, 2011; Sabatini, 2017). Activation of TOR signaling in prothoracic gland (PG) by nutrients regulates biosynthesis of ecdysone and the larval-pupal transition (Layalle et al., 2008). Interestingly, mammalian TOR (mTOR) was recently shown to inhibit CDK8-CycC in mammalian cells (Feng et al., 2015). Therefore, we asked whether feeding the cdk8 mutant larvae with Rapamycin, a compound known to inhibit mTOR activity (Ballou and Lin, 2008; Benjamin et al., 2011), could alleviate the effects of extra yeast on the viability of the cdk8 mutant larvae. As shown in Fig. 4B, addition of 20nM of Rapamycin in diets with 5% or 10% extra yeast increased the pupariation rates of the cdk8 mutant larvae by ~20%, indicating that the effects of extra yeast on pupariation rate is in part through mTOR.
Since mTOR is specifically activated by certain amino acids, such as Leu (leucine) and Arg (arginine) (Bar-Peled and Sabatini, 2014; Jewell et al., 2013; Kim, 2009), we asked whether different types of amino acids have the same or different impacts on the development of the cdk8 mutant larvae. Specifically, we tested each of the 20 amino acids for their effects on the pupation course of cdk8 mutants using the concentration of the 20 amino acids as 10 times higher as described previously (Grandison et al., 2009). As shown in Fig. 4C, increasing the concentration of seven amino acids, including Arg, Gln (glutamine), Ile (isoleucine), Leu, Met (methionine), Thr (threonine), and Val (valine), in a diet strongly induced the larval lethality of the cdk8 mutants. In contrast, addition of the other 13 amino acids had no obvious effect on the viability of the cdk8 mutants (Fig. 4C and data not shown). The mRNA levels of EcR target gene such as E74 and E78 are not significantly altered in wild-type larvae fed with CMY diet with increased Val, but their levels are reduced in cdk8 mutants as expected (Fig. S3A, Fig. S3). Consistent with this observation, increasing these amino acids in diet does not affect the growth nor the developmental transitions of the wild-type larvae (Fig. S4 and data not shown). Thus, cdk8 mutants are particularly sensitive to specific types of dietary amino acids. Increased dietary Val also does not affect the mRNA levels of cdk8 and cycC in wild-type larvae (Fig. S3C), suggesting the effect likely occurs at the CDK8-CycC protein level.
The effects of different types of dietary sugars on timing of pupariation of the cdk8 mutants
Although the larval viability of the cdk8 and cycC mutants is sensitive to the levels of dietary protein, the mutant larvae that did pupariate made the larval-pupal transition between day 7 and day 9 AEL when fed with CMY diets with or without extra yeast (Fig. 3B, Fig. 3C). When fed with high protein diets (CMY with 5%, 10% or 20% of yeast), the control (w1118) larvae grow well and pupariate more synchronously at the normal timing for the larval-pupal transition (Fig. 3B). These observations suggest that the extra sucrose in CMY-HSD delays larval-pupal transition in the cdk8 and cycC mutants but does not affect the viability of the mutant larvae, while extra protein from yeast affects the larval viability but has a weaker effect on the timing for pupariation than the levels of dietary sugars.
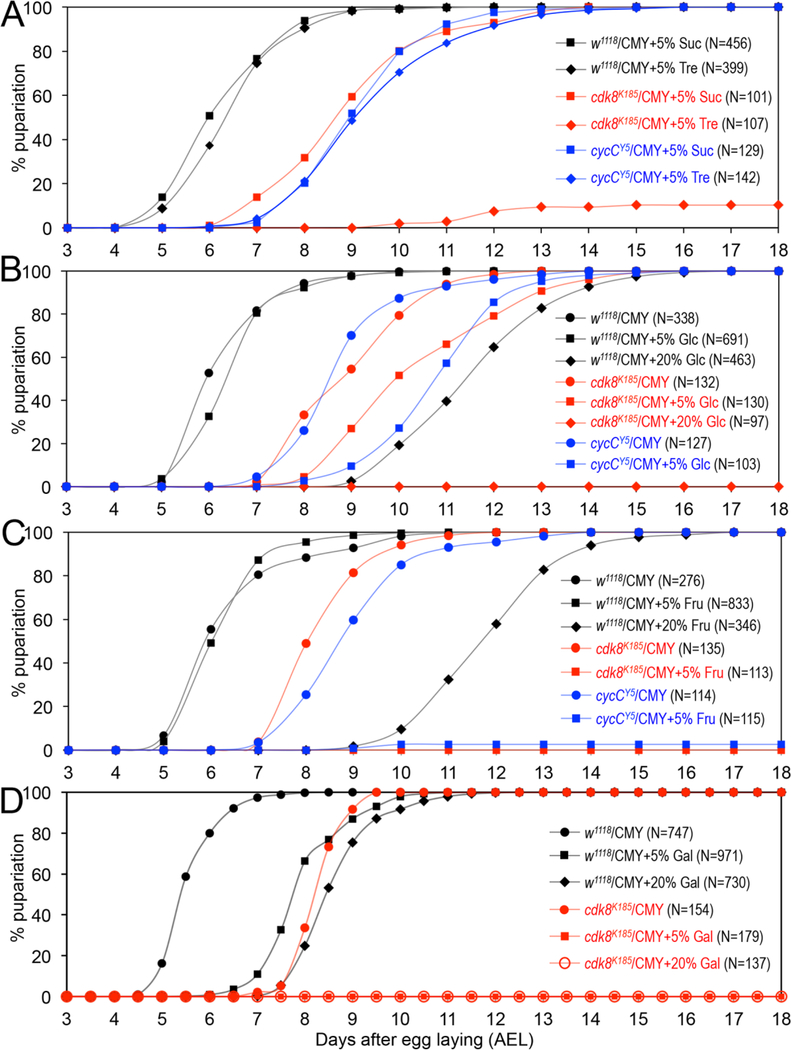
To characterize the effects of different types of sugars on developmental timing for the cdk8 and cycC mutants, we first tested sucrose and trehalose, the two types of disaccharides. As shown in Fig. 5A, adding 5% of sucrose and trehalose in CMY diet does not affect the timing of pupriation for the wild-type animals (the control is shown in Fig. 5B). 5% of sucrose in CMY food does not further delay the timing for pupariation of the cdk8 mutants, but less than 10% the cdk8 mutants can pupariate when fed with 5% of trehalose in CMY food. Most of these mutants die as larvae (Fig. 5A). In contrast, 5% of trehalose or sucrose does not have obvious effects on the timing for pupariation for cycC mutant animals (Fig. 5A), consistent with the notion that cdk8 mutants display more severe defects than cycC mutants (Xie et al., 2015). These observations suggest that the cdk8 mutants display different sensitivity to different dietary disaccharides.
Fig. 5.
The effects of dietary carbohydrate levels on the timing for pupariation of the cdk8 and cycC mutants. (A) Percentage of pupariated wild-type, cdk8, and cycC mutants raised on the CMY with 5% of sucrose (Suc) or trehalose (Tre). (B) Percentage of pupariated wild-type, cdk8, and cycC mutants raised on the CMY with 5% or 20% of glucose (Glc). The data for cycC mutants on 20% of Glc are identical to those of cdk8 mutants on 20% of Glc in CMY diet, thus not shown. (C) Percentage of pupariated wild-type, cdk8, and cycC mutants raised on the CMY with 5% of frucose (Fru). (D) Percentage of pupariated wild-type and cdk8 mutants raised on the CMY with 5% or 20% of galactose (Gla).
Given that dietary polysaccharides need to be digested into monosaccharides to be absorbed, we asked whether different types of monosaccharides could impact the developmental transition in the same or different fashions in wild-type and the cdk8 or cycC mutants. For this reason, we tested three types of monosaccharides, glucose (Fig. 5B), fructose (Fig. 5C), and galactose (Fig. 5D). We have observed that 20% of glucose more severely delayed the timing for pupariation of the wild-type animals than the 20% of sucrose (Fig. 5B vs. Fig. 3A). While 5% of glucose in CMY food does not obviously affect the developmental timing for the control animals (Fig. 5B), 5% of glucose in CMY further delays the timing for pupariation of both cdk8 and cycC mutants, and all these mutants die as larvae when fed with CMY food with 20% glucose (Fig. 5B shows the data for cdk8 mutants; identical data for cycC mutants are not shown). These observations suggest that glucose has a stronger impact on timing of pupariation in the wild-type animals than disaccharides, and both cdk8 and cycC mutants are more sensitive than the control when tested with the same type of dietary sugars.
Similar analyses were performed to test the dietary effects of fructose and galactose. For wild-type animals, both 5% and 20% of fructose (Fig. 5C) had the same effects on the timing for pupariation as glucose (Fig. 5B), while 5% and 20% galactose has stronger effect in delaying this transition (Fig. 5D vs. Fig. 5B/5C). In contrast, 5% (or 20%) of either fructose (Fig. 5C) or galactose (Fig. 5D) caused larval lethality for both cdk8 and cycC mutants, and the mutants never pupariate (data for cycC mutants are not shown). These observations show that different types of monosaccharides have different impacts on the wild-type animals, and fructose and galactose have stronger effects than glucose on developmental transition for both cdk8 and cycC mutants. Taken together, these results show that different types of dietary sugars have different effects on timing of pupariation in Drosophila. Monosaccharides have stronger effects than disaccharides in delaying the timing of pupariation, and both the cdk8 and cycC mutants are more sensitive than the wild-type when fed with the same diets. These observations support the model that CDK8 and CycC play important roles in linking the dietary perturbations and developmental transitions in Drosophila (Fig. 6, see below).
Fig. 6.
CDK8-CycC mediates the dietary effects on lipid metabolism and developmental timing in Drosophila. The model schematizes the effects of dietary factors such as carbohydrates and amino acids through the insulin/TOR pathway and CDK8-CycC complex, which coordinate the activities of transcription factors such as dSREBP and EcR during the larval-pupal transition.
Discussion
Studies in the past two decades have significantly advanced our understanding of how dietary nutrients regulate organismal growth and developmental transitions in Drosophila. At the tissue and organ levels, crosstalks among the fat body, brain, prothoracic gland (PG), imaginal discs, and gut integrate the nutrient conditions with organismal growth and biosynthesis and release of the major hormone ecdysone (Andersen et al., 2013; Danielsen et al., 2013; Edgar, 2006; Nijhout et al., 2014; Tennessen and Thummel, 2011). Coordination of these processes among different tissues depends on hormonal regulation within the larva. Several major signaling pathways have been identified to play critical roles in those processes, including the insulin/insulin-like growth factor signaling (IIS) pathway regulated by carbohydrates, the TOR signaling pathway regulated by amino acids in both fat body and discs, the Upd2-mediated crosstalk between the fat body and the brain in response to the HSD, and the prothoracicotropic hormone (PTTH)-mediated crosstalk between the brain and PG (see below). These elegant studies have established an intellectual framework to understand how larval growth and developmental timing are coordinated with environmental factors such as dietary nutrients and photoperiod, but the exact molecular mechanisms underlying these crosstalks are still not fully understood. It is important to identify additional players and elucidate how they are involved in these processes. In this report, we summarize our characterization of effects of different diets on the timing for pupariation of the cdk8 and cycC mutant larvae, and our results suggest that CDK8-CycC is necessary for mediating the dietary effects on the developmental timing during the larval stage.
This study has raised more questions than it can answer. A key question related to these results is: why do the cdk8 mutant larvae display different sensitivities to excessive levels of different types of carbohydrates and amino acids? This is a complex question, and here we discuss potential models to answer it from the following aspects, all of which are centered on the idea that the cdk8 mutant larvae may be defective in multiple signaling pathways critical in coordinating the inter-organ crosstalks in linking dietary effects on developmental timing.
Given the essential roles of CDK8-CycC and the Mediator complexes in regulating RNA Pol II-dependent gene expression, mutations in CDK8 may disrupt the outputs of multiple signaling pathways involved in coordinating the inter-organ crosstalks. For example, the single insulin-like receptor expressed in various tissues mediates the effects of eight Drosophila insulin-like peptides secreted by insulin-producing cells in the larval brain, fat body, or impaired growth imaginal discs (Kannan and Fridell, 2013). Our previous work suggests that CDK8-CycC is negatively regulated by insulin signaling (Zhao et al., 2012). Consistent with this notion, CDK8-CycC is down-regulated by mTORC1 in mouse liver and cultured HEK293T cells (Feng et al., 2015), and the larval fat body in Drosophila (Tang et al., 2018). In addition, embryos without the maternal cycC do not hatch, suggesting that the pupal lethality of the cdk8 and cycC mutant is due to gradual depletion of maternal CDK8-CycC (Xie et al., 2015). Thus, one possibility is that the residual maternal CDK8 (or CycC) in the cdk8 (or cycC) mutant larvae is further down-regulated by IIS signaling activated by dietary carbohydrates, which may further postpone the regulatory network (Fig. 1A, Fig. 6). This model may explain why the cdk8 and cycC mutant larvae display further delay than the control upon feeding on diets overloaded with carbohydrates. Along this line, why do cdk8 mutants respond differently to different types of carbohydrates? It has been proposed that different tricarboxylic acid intermediates from the fat body, such as fumarate, isocitrate, and malate, can prompt the release of ecdysone from the PG by stimulating the synthesis of PTTH in the brain (Xu et al., 2012). Thus, we speculate that different carbohydrates may be absorbed and metabolized at different rates, thus affecting the dynamics of different tricarboxylic acid intermediates and resulting in different effects on ecdysone release. It will be intriguing to apply metabolomic approaches to analyze the larvae feed with different levels of carbohydrates in the future (Cox et al., 2017).
The crosstalk between the IIS pathway and TOR, the major sensor for amino acids, is conserved between Drosophila and mammals (Fig. 6) (Bar-Peled and Sabatini, 2014; Dennis et al., 2011; Edgar, 2006; Sabatini, 2017). It is interesting to note that cdk8 mutant larvae seem to be particularly sensitive to increased dietary levels of seven amino acids: Arg, Gln, Ile, Leu, Met, Thr, and Val. Of these, Leu, Ile, and Val are branched-chain amino acids, which are known to stimulate mTORC1 activities (Lynch and Adams, 2014; Neishabouri et al., 2015; Yoon, 2016; Zhang et al., 2017). Besides these branched-chain amino acids, the other four amino acids (Arg, Gln, Met, and Thr) can also stimulate TOR in different experimental systems (for examples, (Feng et al., 2013; Gallinetti et al., 2013; Lansard et al., 2011; Sabatini, 2017; Zhai et al., 2015)). Recently, it was reported that the abundance of CDK8-CycC is down-regulated by mTORC1 activation in Drosophila, as well as cultured mammalian cells and mouse liver (Feng et al., 2015; Tang et al., 2018). Therefore, it is likely that increased dietary amino acids may decrease the residual maternal CDK8 and CycC through the activation of TOR, which may explain why cdk8 mutants are particularly sensitive to these amino acids (Fig. 6). However, it is also possible that these amino acids may feed into the CDK8 regulatory network in distinct modes. For example, besides activating TOR, Leu and Ile can also stimulate the secretion of the insulin-like peptides from insulin-producing cells in the larval brain, thereby indirectly activating TOR (Geminard et al., 2009). To understand the different dietary effects of amino acids on cdk8 mutants, further analyses are necessary to distinguish these scenarios.
Besides the dietary effects on CDK8-CycC through the IIS pathway and TOR, defective functions of CDK8 may also disrupt the activities of other signaling pathways involved in coordinating different organs and growth control in response to dietary perturbations. For example, the transforming growth factor β (TGFβ)/Activin signaling pathway activates the expression of Insulin-like receptor and torso in the PG, a process required for inducing biosynthesis of ecdysteroids and the larval-pupal transition (Gibbens et al., 2011). In mammalian systems, CDK8 and CDK9 can positively regulate the activity of SMADs, the key transcription factors downstream of TGFβ signaling (Alarcon et al., 2009). CDK8 and CDK8 play similar role in activating Mad-dependent gene expression in Drosophila (Aleman et al., 2014; Li, 2018), thus the PTTH-regulated synthesis and release of ecdysone may be affected in cdk8 mutants. Consistent with this prediction, the expression of a few Halloween genes, regulated by PTTH and its receptor Torso (Rewitz et al., 2009), are lower in cdk8 and cycC mutants than the control, accompanied with reduced levels of ecdysteroids in these mutant larvae (Xie et al., 2015). Similarly, the JAK-STAT signaling pathway is shown to mediate the crosstalk between the fat body and insulin-producing cells in response to dietary fats and sugars (Rajan and Perrimon, 2012). Interestingly, CDK8 can directly phosphorylate the transactivation domain of mammalian STAT1, STAT3, and STAT5 (Ser727 of STAT1) and modulate the expression of STAT-target genes in IFN-γ-induced antiviral responses (Bancerek et al., 2013). Although the mammalian STAT1-Ser727 is not conserved in Stat92E (not shown), the STAT1 homolog in Drosophila, we could not exclude the possibility that dietary components modulate the activity of JAK-STAT signaling through CDK8 in Drosophila.
Finally, all of our analyses are at the organismal level, and it is unknown whether the status of CDK8 and CycC affects cellular responses to different nutrients. Given that mutations of subunits of the CDK8 module have been identified in a variety of human diseases including cancer and cardiovascular diseases (Clark et al., 2015; Schiano et al., 2014a; Schiano et al., 2014b; Spaeth et al., 2011; Xu and Ji, 2011), future studies are needed to explore the cellular effects of dietary perturbations on the survival and proliferation of cancer cells with mutations in these subunits. This will advance our understanding of how dysregulation of the CDK8 module contributes to tumorigenesis, and impacts of fluctuations of dietary ingredients on this process. These may also aid cancer treatment when considering the dietary factors.
In conclusion, our study reveals that CDK8-CycC plays important roles in mediating the dietary effects on the larval-pupal transition in Drosophila. The cdk8 and cycC mutants display different sensitivities to varied levels of dietary proteins, amino acids, and carbohydrates. Given that both CDK8 and CycC are either mutated or amplified in a variety of human cancers and cardiovascular diseases, understanding the impact and the underpinning mechanisms of dietary perturbations on CDK8-CycC and gene expression may have important clinical implications for the design of strategies to treat these diseases.
Materials and Methods
Drosophila stocks and regents:
The null alleles of cdk8 (cdk8K185) and cycC (cycCY5) alleles were described previously (Loncle et al., 2007; Xie et al., 2015). The flies were kept at 25°C with a 12:12 hour on/off light cycle. Inactivated dry yeast, yeast extract (YE), different amino acids, and sugars were purchased from VWR Life Science. Inactive dry yeast and Drosophila agar were purchased from Genesee Scientific.
Preparation of fly food:
The LSD used in this work is the semi-defined medium (https://bdsc.indiana.edu/information/recipes/germanfood.html) and was prepared according to description by Na et al (Na et al., 2013). HSD food was prepared by adding 20% sucrose into the LSD food. Table S1 includes the detailed recipes for preparation of 500ml of media, which were stored in the fridge at 4°C until use. The standard Cornmeal-Molasses-Yeast (CMY) recipe was used to prepare CMY food (https://bdsc.indiana.edu/information/recipes/molassesfood.html). Additional yeast or different types of sugars were added to the CMY food. For example, “CMY+20% sucrose” food was prepared by mixing 20g sucrose to 100ml of the CMY medium. To increase the concentration of the 20 different amino acids (supplied by Alfa Aesar, purchased from VWR Life Science), we used CMY food with addition of each amino acid at the final concentrations as 10X higher as described previously (Grandison et al., 2009).
Analyses of the timing for the larval-pupal transition:
Flies at the 5–10 days of age, maintained on standard CMY diet, were used to collect embryos. After 1–2 hours of pre-collections on CMY food, the flies were transferred into bottles containing different diets and embryos were collected for another 2–3 hours. The mid-point of this collection time was considered as the beginning point to count the pupation timing. The number of pupariated animals of specific genotypes was scored once every 12 or 24 hours according to respective experiment. The pupariation curves were plotted using Microsoft Excel.
Western blot analysis and antibodies:
Western blot analysis was performed as previously described (Xie et al., 2015). Briefly, larvae were collected and homogenized in a buffer containing NaCl (420 mM), Tris HCl (50 mM, pH 8.0), EDTA (0.1 mM), NP-40 (0.5%), glycerol (10%), dithiothreitol (DTT, 1 mM), phenylmethanesulfonyl fluoride (PMSF, 2.5 mM), the cOmplete protease inhibitor and the PhosSTOP phosphatase inhibitor cocktail tablets (Roche Applied Science). After centrifugation at 2000 ×g for 15 min at 4°C, supernatants were collected and protein concentrations were measured with a Bradford protein assay kit (Bio-Rad), followed by the standard sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Each experiment was repeated at least three times and the representative results were shown.
The following antibodies were used in this work: anti-EcR-common DDA2.7 (mouse, 1:250, Developmental Studies Hybridoma Bank), anti-EcR-common D4.4 (mouse, 1:250, Developmental Studies Hybridoma Bank), anti-USP (guinea pig, 1:2000; (Xie et al., 2015)), anti-CDK8 (guinea pig, 1:500; obtained from Dr. Henri-Marc Bourbon), anti-CycC (rabbit, 1:2000; provided by Dr. Terry Orr-Weaver), anti-dSREBP (rabbit, 1:100; (Xie et al., 2015)), and anti-actin monoclonal antibody (mouse 1:4000, Thermo Scientific MA5–11869).
Analysis of the mRNA levels by quantitative Reverse Transcription PCR (qRT-PCR) Assay and Statistical analyses:
This assay and the specific primer sequences were described in detail previously (Xie et al., 2015; Zhao et al., 2012). Briefly, whole larvae were homogenized in TRIzol reagent (Invitrogen) followed by RNA isolation as described by manufacturer’s instructions. Isolated RNAs were quantified and then the cDNAs were prepared using the the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The quantitative PCR (qPCR) was performed using SYBR Green (Applied Biosystems) as described previously. Three independent biological repeats of each genotype or treatment were conducted. Standard deviation and t-tests were performed using Microsoft Excel. Statistical significance was shown in figures and all error bars indicate standard deviation. Statistical significance was shown in figures: * p < 0.05; ** p < 0.01; *** p < 0.001.
Supplementary Material
Fig S1. The relative mRNA levels of lipogenic genes in LSD- or HSD-fed larvae between 3 and 10 days AEL as assayed using qRT-PCR. * p < 0.05; ** p < 0.01; *** p < 0.001 based on t-tests.
Fig. S2. The relative mRNA levels of EcR target genes in CMY-LSD (i.e., CMY) or CMY-HSD (i.e., CMY+20%Suc) in both wild-type (w1118) and cdk8 (cdk8K185) mutant larvae at day 6 (A) or day 8 (B) AEL as assayed using qRT-PCR. The genotype and treatments are color-coded, shown between (A) and (B). (C) Effects of CMY-HSD on the mRNA levels of cdk8 and cycC in wild-type larvae at day 4, day 6, and day 8 AEL. Pair-wise statistical analyses were performed using one-tailed t-tests. * p < 0.05; ** p < 0.01; *** p < 0.001.
Fig. S3. The relative mRNA levels of EcR target genes in both wild-type (w1118) and cdk8 (cdk8K185) mutant larvae fed with control food (CMY) and food with extra Valine (CMY-Val), analyzed at day 5 (A) or day 6 (B) AEL as assayed using qRT-PCR. The genotype and treatments are color-coded, shown between (A) and (B). (C) Effects of extra Val (CMY-Val) on the mRNA levels of cdk8 and cycC in wild-type larvae at day 4 and day 5 AEL. Pair-wise statistical analyses were performed using one-tailed t-tests. * p < 0.05; ** p < 0.01; *** p < 0.001.
Fig. S4 Effects of increased amino acids on the timing of pupariation of the wild-type (w1118) larvae. Together with the control diet (CMY), data for a few representative amino acids, including Ala, Met, Thr, and Val are shown in the chart.
Table S1. Recipes for preparation of 500 ml of different diets based on the LSD (“German food”) recipe.
Highlights:
The cdk8 and cycC mutant Drosophila larvae are hypersensitive to increased levels of dietary proteins.
Of the 20 kinds of amino acids, the cdk8 and cycC mutant larvae are particularly sensitive to the levels of seven specific amino acids.
Compared to disaccharides, dietary monosaccharides have stronger effects in delaying the timing of the larval-pupal transition in cdk8 and cycC mutant larvae. CDK8-CycC may play important roles in mediating the dietary effects on lipid metabolism and developmental transition in Drosophila larvae.
Acknowledgments
We thank the Bloomington Drosophila Stock Center (NIH P40OD018537) for providing Drosophila strains, Dr. Henri-Marc Bourbon for anti-CDK8 antibody, and Dr. Terry Orr-Weaver for anti-CycC antisera. The monoclonal antibodies against EcR, developed by Drs. Carl Thummel and David Hogness, were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa. We are grateful to Dr. Fajun Yang for helpful discussions, and Stephen Yu and Diana Luna for their careful reading of the manuscript. We would also like to thank the anonymous reviewers for their insightful comments and valuable suggestions. This work was supported by NIH R01 DK095013 to J.Y.J.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, Sapkota G, Pan D, Massague J, 2009. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 139, 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleman A, Rios M, Juarez M, Lee D, Chen A, Eivers E, 2014. Mad linker phosphorylations control the intensity and range of the BMP-activity gradient in developing Drosophila tissues. Scientific reports 4, 6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen DS, Colombani J, Leopold P, 2013. Coordination of organ growth: principles and outstanding questions from the world of insects. Trends in cell biology 23, 336–344. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL, 2010. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol 55, 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhaus B, Sulkowski E, Schlote FW, 1984. A semi-synthetic, general-purpose medium for Drosophila melanogaster. Dros. Inf. Serv. 60, 210–212. [Google Scholar]
- Ballou LM, Lin RZ, 2008. Rapamycin and mTOR kinase inhibitors. J Chem Biol 1, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, Dolken L, Strobl B, Muller M, Taatjes DJ, Kovarik P, 2013. CDK8 Kinase Phosphorylates Transcription Factor STAT1 to Selectively Regulate the Interferon Response. Immunity 38, 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Sabatini DM, 2014. Regulation of mTORC1 by amino acids. Trends in cell biology 24, 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin D, Colombi M, Moroni C, Hall MN, 2011. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov 10, 868–880. [DOI] [PubMed] [Google Scholar]
- Berg J, 2016. Gene-environment interplay. Science 354, 15. [DOI] [PubMed] [Google Scholar]
- Clark AD, Oldenbroek M, Boyer TG, 2015. Mediator kinase module and human tumorigenesis. Critical reviews in biochemistry and molecular biology 50, 393–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW, 2011. Function and regulation of the Mediator complex. Curr Opin Genet Dev 21, 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JE, Thummel CS, Tennessen JM, 2017. Metabolomic Studies in Drosophila. Genetics 206, 1169–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen ET, Moeller ME, Rewitz KF, 2013. Nutrient signaling and developmental timing of maturation. Curr Top Dev Biol 105, 37–67. [DOI] [PubMed] [Google Scholar]
- Dennis MD, Baum JI, Kimball SR, Jefferson LS, 2011. Mechanisms involved in the coordinate regulation of mTORC1 by insulin and amino acids. The Journal of biological chemistry 286, 8287–8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, 2006. How flies get their size: genetics meets physiology. Nature reviews. Genetics 7, 907–916. [DOI] [PubMed] [Google Scholar]
- Feng D, Youn DY, Zhao X, Gao Y, Quinn WJ 3rd, Xiaoli AM, Sun Y, Birnbaum MJ, Pessin JE, Yang F, 2015. mTORC1 Down-Regulates Cyclin-Dependent Kinase 8 (CDK8) and Cyclin C (CycC). PloS one 10, e0126240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Peng Y, Wu P, Hu K, Jiang WD, Liu Y, Jiang J, Li SH, Zhou XQ, 2013. Threonine affects intestinal function, protein synthesis and gene expression of TOR in Jian carp (Cyprinus carpio var. Jian). PloS one 8, e69974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinetti J, Harputlugil E, Mitchell JR, 2013. Amino acid sensing in dietary-restriction-mediated longevity: roles of signal-transducing kinases GCN2 and TOR. Biochem J 449, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geminard C, Rulifson EJ, Leopold P, 2009. Remote control of insulin secretion by fat cells in Drosophila. Cell metabolism 10, 199–207. [DOI] [PubMed] [Google Scholar]
- Gibbens YY, Warren JT, Gilbert LI, O’Connor MB, 2011. Neuroendocrine regulation of Drosophila metamorphosis requires TGFbeta/Activin signaling. Development 138, 2693–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, Partridge L, 2009. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462, 1061–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell JL, Russell RC, Guan KL, 2013. Amino acid signalling upstream of mTOR. Nature reviews. Molecular cell biology 14, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Fridell YW, 2013. Functional implications of Drosophila insulin-like peptides in metabolism, aging, and dietary restriction. Front Physiol 4, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, 2009. Mechanisms of amino acid sensing in mTOR signaling pathway. Nutr Res Pract 3, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS, 2005. Nuclear receptors--a perspective from Drosophila. Nature reviews. Genetics 6, 311–323. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, 2005. Mediator and the mechanism of transcriptional activation. Trends in biochemical sciences 30, 235–239. [DOI] [PubMed] [Google Scholar]
- Kuhnlein RP, 2012. Thematic review series: Lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. J Lipid Res 53, 1430–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansard M, Panserat S, Plagnes-Juan E, Dias K, Seiliez I, Skiba-Cassy S, 2011. L-leucine, L-methionine, and L-lysine are involved in the regulation of intermediary metabolism-related gene expression in rainbow trout hepatocytes. J Nutr 141, 75–80. [DOI] [PubMed] [Google Scholar]
- Layalle S, Arquier N, Leopold P, 2008. The TOR pathway couples nutrition and developmental timing in Drosophila. Developmental cell 15, 568–577. [DOI] [PubMed] [Google Scholar]
- Li H, Tennessen JM, 2017. Methods for studying the metabolic basis of Drosophila development. Wiley interdisciplinary reviews. Developmental biology 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu M, Ren X, Loncle N, Wang Q, Hemba-Waduge R, Boube M, Bourbon H-MG, Ni JQ, and Ji JY, 2018. The Mediator CDK8-Cyclin C complex modulates vein patterning in Drosophila by stimulating Mad-dependent transcription. BioRxiv https://doi.org/10.1101/360628 [DOI] [PMC free article] [PubMed]
- Liu Z, Huang X, 2013. Lipid metabolism in Drosophila: development and disease. Acta biochimica et biophysica Sinica 45, 44–50. [DOI] [PubMed] [Google Scholar]
- Loewith R, Hall MN, 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncle N, Boube M, Joulia L, Boschiero C, Werner M, Cribbs DL, Bourbon HM, 2007. Distinct roles for Mediator Cdk8 module subunits in Drosophila development. The EMBO journal 26, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CJ, Adams SH, 2014. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol 10, 723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG, 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends in biochemical sciences 30, 256–263. [DOI] [PubMed] [Google Scholar]
- Mattila J, Hietakangas V, 2017. Regulation of Carbohydrate Energy Metabolism in Drosophila melanogaster. Genetics 207, 1231–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman LP, Fink JL, Narzinski K, Ramachandran PV, Hathiramani SS, Cagan RL, Baranski TJ, 2011. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis Model Mech 4, 842–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na J, Musselman LP, Pendse J, Baranski TJ, Bodmer R, Ocorr K, Cagan R, 2013. A Drosophila model of high sugar diet-induced cardiomyopathy. PLoS genetics 9, e1003175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, Henrich VC, 2009. Arthropod nuclear receptors and their role in molting. The FEBS journal 276, 6128–6157. [DOI] [PubMed] [Google Scholar]
- Neishabouri SH, Hutson SM, Davoodi J, 2015. Chronic activation of mTOR complex 1 by branched chain amino acids and organ hypertrophy. Amino Acids 47, 1167–1182. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Riddiford LM, Mirth C, Shingleton AW, Suzuki Y, Callier V, 2014. The developmental control of size in insects. Wiley interdisciplinary reviews. Developmental biology 3, 113–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasco MY, Leopold P, 2012. High sugar-induced insulin resistance in Drosophila relies on the lipocalin Neural Lazarillo. PloS one 7, e36583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss ZC, Ebmeier CC, Taatjes DJ, 2013. The Mediator complex and transcription regulation. Critical reviews in biochemistry and molecular biology 48, 575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A, Perrimon N, 2012. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151, 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz KF, Yamanaka N, Gilbert LI, O’Connor MB, 2009. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science 326, 1403–1405. [DOI] [PubMed] [Google Scholar]
- Rewitz KF, Yamanaka N, O’Connor MB, 2013. Developmental checkpoints and feedback circuits time insect maturation. Curr Top Dev Biol 103, 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford LM, 1993. Hormones and Drosophila development, in: Bate M.a.M.A., A. (Ed.), The Development of Drosophila melanogaster, 1993/01/01 ed, pp. 899–939. [Google Scholar]
- Riddiford LM, Cherbas P, Truman JW, 2000. Ecdysone receptors and their biological actions. Vitam Horm 60, 1–73. [DOI] [PubMed] [Google Scholar]
- Sabatini DM, 2017. Twenty-five years of mTOR: Uncovering the link from nutrients to growth. Proceedings of the National Academy of Sciences of the United States of America 114, 11818–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiano C, Casamassimi A, Rienzo M, de Nigris F, Sommese L, Napoli C, 2014a. Involvement of Mediator complex in malignancy. Biochimica et biophysica acta 1845, 66–83. [DOI] [PubMed] [Google Scholar]
- Schiano C, Casamassimi A, Vietri MT, Rienzo M, Napoli C, 2014b. The roles of mediator complex in cardiovascular diseases. Biochimica et biophysica acta 1839, 444–451. [DOI] [PubMed] [Google Scholar]
- Schlegel A, Stainier DY, 2007. Lessons from "lower" organisms: what worms, flies, and zebrafish can teach us about human energy metabolism. PLoS genetics 3, e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Spradling AC, 2017. The role of metabolic states in development and disease. Curr Opin Genet Dev 45, 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaeth JM, Kim NH, Boyer TG, 2011. Mediator and human disease. Seminars in cell & developmental biology 22, 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HW, Hu Y, Chen CL, Xia B, Zirin J, Yuan M, Asara JM, Rabinow L, Perrimon N, 2018. The TORC1-Regulated CPA Complex Rewires an RNA Processing Network to Drive Autophagy and Metabolic Reprogramming. Cell metabolism 27, 1040–1054 e1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JM, Thummel CS, 2011. Coordinating growth and maturation - insights from Drosophila. Current biology : CB 21, R750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS, 2001. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Developmental cell 1, 453–465. [DOI] [PubMed] [Google Scholar]
- Xie XJ, Hsu FN, Gao X, Xu W, Ni JQ, Xing Y, Huang L, Hsiao HC, Zheng H, Wang C, Zheng Y, Xiaoli AM, Yang F, Bondos SE, Ji JY, 2015. CDK8-Cyclin C Mediates Nutritional Regulation of Developmental Transitions through the Ecdysone Receptor in Drosophila. PLoS Biol 13, e1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Ji JY, 2011. Dysregulation of CDK8 and Cyclin C in tumorigenesis. J Genet Genomics 38, 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WH, Lu YX, Denlinger DL, 2012. Cross-talk between the fat body and brain regulates insect developmental arrest. Proceedings of the National Academy of Sciences of the United States of America 109, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N, Rewitz KF, O’Connor MB, 2013. Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol 58, 497–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JW, Wang G, 2014. The Mediator complex: a master coordinator of transcription and cell lineage development. Development 141, 977–987. [DOI] [PubMed] [Google Scholar]
- Yoon MS, 2016. The Emerging Role of Branched-Chain Amino Acids in Insulin Resistance and Metabolism. Nutrients 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Sun Z, Zhang J, Kang K, Chen J, Zhang W, 2015. Activation of the TOR Signalling Pathway by Glutamine Regulates Insect Fecundity. Scientific reports 5, 10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zeng X, Ren M, Mao X, Qiao S, 2017. Novel metabolic and physiological functions of branched chain amino acids: a review. J Anim Sci Biotechnol 8, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Feng D, Wang Q, Abdulla A, Xie XJ, Zhou J, Sun Y, Yang ES, Liu LP, Vaitheesvaran B, Bridges L, Kurland IJ, Strich R, Ni JQ, Wang C, Ericsson J, Pessin JE, Ji JY, Yang F, 2012. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J Clin Invest 122, 2417–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. The relative mRNA levels of lipogenic genes in LSD- or HSD-fed larvae between 3 and 10 days AEL as assayed using qRT-PCR. * p < 0.05; ** p < 0.01; *** p < 0.001 based on t-tests.
Fig. S2. The relative mRNA levels of EcR target genes in CMY-LSD (i.e., CMY) or CMY-HSD (i.e., CMY+20%Suc) in both wild-type (w1118) and cdk8 (cdk8K185) mutant larvae at day 6 (A) or day 8 (B) AEL as assayed using qRT-PCR. The genotype and treatments are color-coded, shown between (A) and (B). (C) Effects of CMY-HSD on the mRNA levels of cdk8 and cycC in wild-type larvae at day 4, day 6, and day 8 AEL. Pair-wise statistical analyses were performed using one-tailed t-tests. * p < 0.05; ** p < 0.01; *** p < 0.001.
Fig. S3. The relative mRNA levels of EcR target genes in both wild-type (w1118) and cdk8 (cdk8K185) mutant larvae fed with control food (CMY) and food with extra Valine (CMY-Val), analyzed at day 5 (A) or day 6 (B) AEL as assayed using qRT-PCR. The genotype and treatments are color-coded, shown between (A) and (B). (C) Effects of extra Val (CMY-Val) on the mRNA levels of cdk8 and cycC in wild-type larvae at day 4 and day 5 AEL. Pair-wise statistical analyses were performed using one-tailed t-tests. * p < 0.05; ** p < 0.01; *** p < 0.001.
Fig. S4 Effects of increased amino acids on the timing of pupariation of the wild-type (w1118) larvae. Together with the control diet (CMY), data for a few representative amino acids, including Ala, Met, Thr, and Val are shown in the chart.
Table S1. Recipes for preparation of 500 ml of different diets based on the LSD (“German food”) recipe.