NMR spectroscopy, one of the two major analytical platforms utilized in the metabolomics field, has witnessed a large number of advances in fundamental studies, methods development, and applications in a broad variety of disciplines. While biomedical science studies have dominated the field, applications to other areas including plant science, food and nutrition, drug development, energy and environmental sciences are on the rise. NMR has contributed very significantly to the metabolomics field despite its lower sensitivity compared to mass spectrometry (MS), the other major analytical platform used in metabolomics. Numerous unsurpassed characteristics of NMR, including its excellent reproducibility, highly quantitative nature, ability to identify unknown metabolites unambiguously, and capacity to detect metabolites using intact biospecimens compensate for its relatively low sensitivity. In addition, and importantly, NMR offers some unique capabilities to reliably identify active metabolic pathways, measure metabolic fluxes and enzyme activity through tracing the flow of nuclei such as 2H, 13C, and 15N from labeled substrates as they are transformed into downstream metabolites. Thus, NMR-based metabolomics applications have exploited the unique strengths of NMR that are largely complementary to mass spectrometry.
In the area of methods development, numerous efforts have been focused on enhancing the sensitivity and resolution of NMR, improving unknown identification and quantifying a larger number of metabolites. The extreme complexity of biological mixtures has also prompted exploitation of the combined strength of the two main analytical platforms, NMR and MS, for direct comparison of the metabolite data, unknown identification and reliable biomarker discovery. The field has already witnessed significant progress in combining NMR and MS methods. An increased number of quantifiable metabolites are now routinely detected, which opens avenues for cross-study comparison and new applications. New databases and software tools have made large scale data analyses and interpretation much easier.
In this review, we discuss recent advances in NMR-based metabolomics with an emphasis on the development of new methods, databases, software tools, and applications during the past 2 years, from mid-2014 to mid-2016. During this period, over 1000 papers have been published (based on PubMed search using the keywords NMR and metabolomics, metabonomics, or metabolic profiling). An excellent, earlier review in Analytical Chemistry on this topic reported the developments from 2011 until mid-2014,1 and numerous aspects of the field have also been highlighted in a number of other recent reviews.2–6 Because of space constraints, the goal of the present review is not to cover all NMR-based metabolomics work exhaustively. Instead, we highlight new developments and some important applications that will help provide a representative overview of the current status and likely future directions of the NMR-based metabolomics field.
FOCUSED REVIEWS
In view of NMR’s favorable analytical characteristics, NMR-based metabolomics has witnessed rapid growth in both applications and methods development. In the following paragraphs, we highlight some recent focused reviews that cover a range of investigations, acknowledging that it is beyond the scope of this Review to cover the full breadth of applications.
Applications that focus on biomarker discovery continue to be a major area driven by the promise of earlier detection and better outcomes, and numerous reviews on this topic highlight recent developments. A description of metabolomics of cardiovascular diseases as viewed from a clinical perspective provides an overview of recent 1H NMR-based findings in the atherosclerosis field7 as well as information on the design and scope of studies conducted to date. In cancer, the detection of biomarkers is of major interest for the metabolomics field in view of the potential impact worldwide. A recent chapter on cancer biomarkers discusses metabolomics approaches and applications,8 and also covers various aspects of sample preparation, analysis methods, and statistical analysis tools for handling large data sets important to the goal of identifying robust cancer biomarkers. More specifically, investigations of renal cancer were described that touch on numerous aspects of NMR-based metabolomics investigations and highlight the current challenges and future directions.9 A recent review on prostate cancer emphasizes the published NMR work analyzing both body fluids and tissue for biomarker identification.10 The potential of metabolomics applications in the area of critical care medicine seems promising. There are a number of approaches using NMR and/or MS for analysis of a variety of biofluids from patients in the intensive care unit with various clinical manifestations including septic shock and community acquired pneumonia.11 Airway diseases have also been investigated based on the NMR analysis of exhaled breadth condensate (EBC).12 The potential of NMR-based metabolomics of EBC, known as breathomics, as well as electronic noses based on other diagnostic platforms in distinguishing patients with respiratory diseases including asthma, chronic obstructive pulmonary disease (COPD), lung cancer, cystic fibrosis, or primary ciliary dyskinesia associated with the respiratory system are discussed. One major hypothesis related to the development of obesity is related to intestinal microbiota, apart from lifestyle habits and genetic factors. Therefore, there has been significant interest to investigate the role of gut microbiota in the pathogenesis of metabolic dysfunctions and increased adiposity. Clavani et al. discuss some important findings on the involvement of gut microbiota in the pathogenesis of obesity and how NMR could be used to understand microbial activity and assess treatments.13
Investigations that involve the use of stable isotope labeled substrates for mechanistic studies continue to attract major attention owing to the ability to answer a number of key mechanistic questions in systems biology and biochemistry. A recent review of this area describes various direct and isotope-edited 1D and 2D NMR methods to profile metabolites and their isotopomer distributions.14 The article also illustrates in detail various in vitro, ex vivo, and in vivo studies that have been made, focusing on gaining systematic and novel metabolic insights in different biological systems, including humans. A number of critical steps involving sample preparation, including rapid cryoquenching, extraction, and chemoselective derivatization are required to achieve robustness in the analysis. The investigation of cancer metabolism is a major topic in the metabolomics field, and over the past 20 years NMR has become a core tool to study altered cellular metabolism specifically using stable isotope tracers. Recent progress made in this direction, along with the current challenges to the field and the benefit of combining MS with NMR, is also described.15
It is increasingly realized that diet has a significant impact on human health and well-being for a variety of reasons. However, detailed knowledge regarding the food metabolome that makes up the human diet as well as food production processes that alter that metabolome is still quite limited. Metabolomics is now widely employed to study various aspects of food products including the impact of agricultural practices, processing, and storage on the global metabolite composition of food; the identification of novel bioactive compounds; and the authentication and region-of-origin classification. Numerous reviews provide an overview of the current application of metabolomics to this broad area.16 The past 5 years of work on food plants, emphasizing NMR-based metabolomics for the quality control of dietary natural products and assessment of their nutritional value has been summarized,17 and current research in the areas of food analysis, dietary biomarker identification, diet-related diseases and nutritional interventions is provided by a number of reviews.18–20 Various steps involved in NMR-based metabolomics studies in nutritional research along with some important applications was also described recently.21
The plant metabolome is widely considered to be the most complex with an estimated number of metabolites in the plant kingdom exceeding well over 200 000 due to the significant presence of secondary metabolites. Research interest in this area is focused on identifying metabolites that correlate with changes in genotype or phenotype. A review on plant metabolomics describes developments over nearly the last 15 years highlighting numerous examples.22 It also describes recent advances in numerous analytical technologies including NMR and MS. Identification of metabolites by integrating NMR and MS platforms is also described in this review. Drug discovery, based on natural sources, is another area that is related to plant metabolomics. An overview of the development of NMR-based plant metabolomics was provided with emphasis on 2D NMR techniques and their applications in phytomedicine quality control analysis and drug discovery from natural sources.23
More broadly, the drug discovery process has been enhanced by the use of metabolomics. The ability to follow dynamic changes in the metabolome resulting not only from pathologies but also from drug treatment and toxicity has been of immense value for the development of new therapeutic approaches. The key features of NMR for applications to drug discovery have been described in detail,24 while the current state of drug discovery with emphasis on the role of NMR-based metabolomics is provided in an article by Powers.4 Notably, inclusion of metabolomics as a routine component of the drug discovery process potentially provides important information on the binding affinity of drug molecules, the selectivity, and the in vivo mechanism of action. In addition, metabolomics facilitates evaluation and validation of lead molecules through high-throughput screening and helps eliminate undesired candidates.
While numerous advances in NMR-based metabolomics have enabled greater insights into biochemical mechanisms in health and disease and provided numerous disease biomarker candidates, the amount of metabolite information currently obtainable from biological specimens is limited owing to the high complexity of such systems. Clearly, such complexity of biological mixtures far outweighs the current capabilities of NMR. Major efforts in the field are focused on addressing this challenge. A recent article presents some perspective on the ability of NMR to alleviate this and other bottlenecks in metabolomics.25 The emerging trends in NMR-based metabolomics in the areas of improved metabolite detection, better unknown identification, and more accurate quantitation of larger numbers of metabolites are discussed in some detail, highlighting both the opportunities and challenges. The complexity of biological specimens demands new approaches for unknown identification. While NMR databases and search tools allow faster and more reliable identification of known metabolites, the identification of unknown metabolites using new strategies that integrate NMR with MS, cheminformatics, and computational methods was reviewed.26,27 One novel approach is the use of certain chemical additives to the NMR tube can permit identification of metabolites with specific physicochemical properties.28 A new method named “blind source separation (BSP)” that is aimed at decomposing the NMR signal into simpler pieces was also described.29 BSP analysis is an alternative to the conventional Fourier transform and uses potentially more appropriate mathematical methods for data processing. It is particularly suitable for analysis of complex mixtures and is aimed at addressing the analytical challenges faced in metabolomics and other fields.
Since each biofluid can potentially present its own set of challenges in terms of preparation and analysis in metabolomics, efforts have been made to start addressing this issue. As an example, urine is one of the most frequently analyzed biofluids (after blood serum/plasma) in metabolomics studies. Metabolic profiling of urine offers numerous benefits as it can be obtained noninvasively and it provides a rich source of information as it generally contains a significantly higher number of NMR detectable metabolites compared to serum/plasma, cells, or tissue. However, contributions from numerous factors including diet, medications, personal habits such as physical activity or smoking, gender, age, gut microbe diversity, as well as genetics, all can affect the urinary metabolome and add to the spectral complexity. Further, varied pH and salt concentrations induce peak shifts for many metabolites, especially, for those with functional groups with pKas near physiological pH. A major effort was made recently by investigators from many institutions to alleviate these challenges by providing standardized protocols for experiments and metabolite quantitation using urine.30,31 To accomplish this task, a large number of published studies in the field were reviewed and key variables that potentially affect results were identified; subsequently, recommendations were made for standardized sample collection protocols and experimental design.30 Separately, a number of issues regarding urinary metabolite analysis with emphasis on quantification for disease biomarker napplications are discussed.31
NEW PROTOCOLS
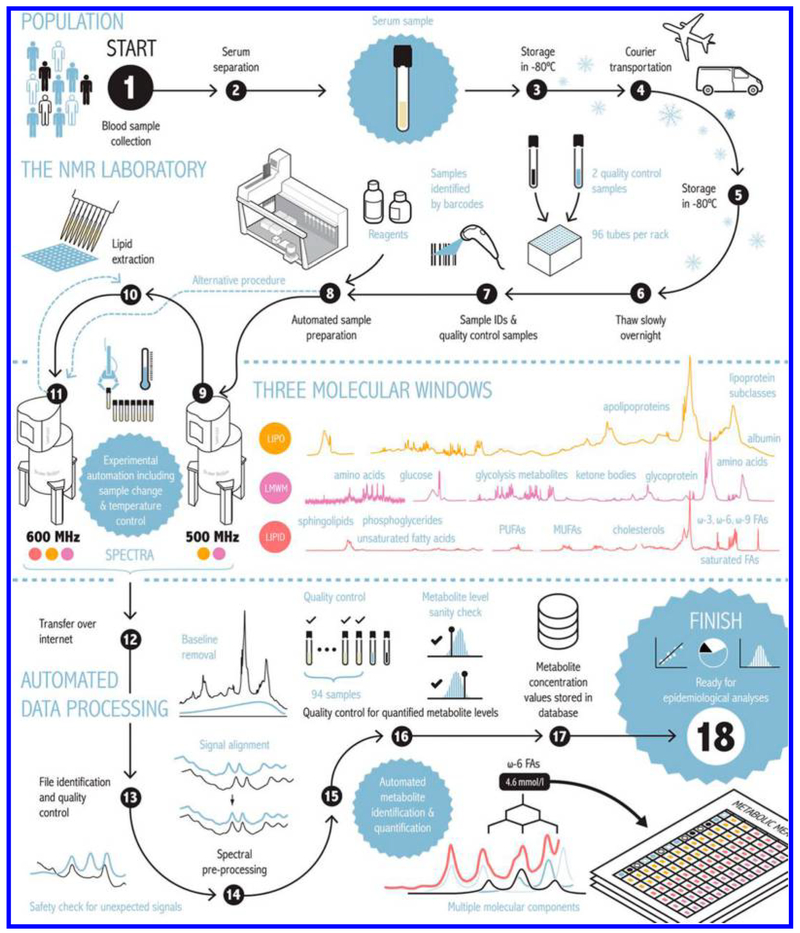
Given the subtle changes in metabolite profiles due to disease combined with numerous confounding effects, the need for large scale studies with high-throughput is increasingly recognized for biomarker discovery and validation. An effort that recognizes such a need is the development of an automated high-throughput serum NMR-based metabolomics platform.32 Impressively, the platform is shown to provide quantitative data for more than 200 metabolic measurements including 14 lipoprotein subclasses, their lipid concentrations and composition, apolipoprotein A-I and B, multiple cholesterol and triglyceride measurements, albumin, various fatty acids, as well as numerous low-molecular-weight metabolites, including amino acids, glycolysis related measures, and ketone bodies (Figure 1). Using this platform, the authors have analyzed nearly 250 000 samples from nearly 100 epidemiological cohorts and biobanks, and the new international setup of multiple platforms in Finland and United Kingdom is anticipated to allow an annual throughput of more than 250 000 samples. A number of new biomarkers for early detection of diseases such as atherosclerosis, type 2 diabetes, diabetic nephropathy, and cardiovascular disease have been identified from such large scale data analysis. Nicholson and coworkers at Imperial College also focused on providing highly reproducible methods for large scale studies and standardized protocols that minimize technical or experimental bias in metabolomics, have provided a detailed set of updated protocols that carefully consider major experimental conditions, including sample preparation, spectrometer parameters, NMR pulse sequences, throughput, reproducibility, quality control, and resolution.33 These results provide an experimental platform that facilitates NMR spectroscopy usage across different large cohorts of biofluid samples, enabling integration of global metabolic profiling.
Figure 1.
Automated high-throughput serum NMR metabolomics platform used to obtain molar concentrations of more than 200 metabolic measures. The samples are handled in 96-well plates, with every plate containing 2 quality control samples, a serum mimic and a mixture of two low-molecular-weight metabolites. All liquid handling steps for serum samples are performed by a PerkinElmer JANUS Automated Workstation equipped with an 8-tip dispensor arm with Varispan. This platform has two NMR spectrometers, each equipped with a SampleJet that holds 480 samples at a cooled (+6 °C) temperature that allow analysis of ≈80 000 samples annually. Reproduced from Soininen, P.; Kangas, A. J.; Würtz, P.; Suna, T.; Ala-Korpela, M.Circ. Cardiovasc. Genet. 2015, 8, 192–206 (ref 32). Copyright 2015 Wolters Kluwer Health, Inc.
Analysis of human blood serum/plasma is of major interest in the metabolomics investigations of virtually all human diseases. However, the large amount of proteins present in serum/plasma presents a major challenge. To alleviate this bottleneck, it has been the norm in MS analysis to physically remove serum/plasma proteins before analysis. For NMR, the samples have typically been used without protein removal; however, it is increasingly realized to be of benefit to remove proteins before analysis. Many approaches including ultra-filtration and protein precipitation using an organic solvent have been the subject of investigation and use. Given the importance for reliable analysis, this topic continues to be of significant interest for further investigations and improvements. Recently, a new method that involves addition of nanoparticles for removal of proteins was explored.34 Treating serum with nanoparticles prior to ultrafiltration offers excellent metabolite recovery and clean NMR experiments. Earlier, the same group presented a charged nanoparticle-based strategy to selectively suppress metabolites with specific charge to simplify NMR spectra.35 By adding electrically charged silica nanoparticles to the solution, metabolites of opposite charge were shown to bind to the nanoparticles and weaken or entirely suppress their NMR signals.
NEW TECHNIQUES AND METHODOLOGIES
The extremely large number of metabolites in biological samples that span a vast range of concentrations requires the development of improved methods for efficient analysis of metabolites. A significant exploration of new techniques and methodologies has occurred over the past 2 years.
1H Techniques.
The reliable quantification of metabolites continues to be a major challenge in metabolomics, and the development of new methods is being sought to overcome this challenge. Recently, a new method focused on accurately quantifying metabolites and proteins in blood plasma as well as other body fluids from one single NMR measurement was presented.36 This method is based on chemical shift changes caused by protein-induced bulk magnetic susceptibility (BMS). BMS shifts were obtained using isolated and exchange-free metabolite resonances such as valine, alanine, glucose, leucine, and lactate and by external referencing with respect to TSP (3-(trimethylsilyl)propionic-2,2,3,3-d4 acid, sodium salt) in a coaxial insert. This method is shown to quantify proteins with an accuracy of 5% or better, which is comparable to the conventional colorimetric method.
NMR provides a unique opportunity to profile metabolites from the intact tissue, nondestructively, using high-resolution magic angle spinning (HRMAS) techniques.37 There are numerous benefits to HRMAS; importantly, it enables obtaining highly resolved, liquid-like spectra without sample preparation. Also, the tissue can be recovered after NMR experiments for use in other studies such as proteomic and genomic analysis. Advances in this area have enabled metabolomics studies that use as little as a few nanograms of tissue and have showed that tissue obtainable from core needle biopsy is sufficient for such analysis. One challenge for HRMAS NMR, however, is the extreme forces exerted on fragile tissue samples due to sample spinning, even at speeds of several kilohertz. To overcome this issue, a protocol for acquiring the spectra at very low spinning speeds of a few hundred hertz was provided.38 In this experiment, the suppression of broad spectral features is achieved using a modified version of the recently introduced PROJECT experiment with water suppression and rotor synchronization.
The lack of advanced analytical tools to investigate metabolism in living cells is still a challenge for metabolomics. To try to improve current analytical capabilities, a miniaturized microslot NMR detector was fabricated with an on-board heater that was integrated with a microfluidic device as an NMR sample holder.39 Using this device, a tumor spheroid of 500 μm diameter consisting of ~9000 cells was studied noninvasively. The production and degradation of 23 intra- and extracellular metabolites could be monitored. The planar geometry of the microslot NMR detector potentially allows its hyphenation with versatile lab-on-a chip (LOC) technology.
Techniques for 13C and Other Nuclei.
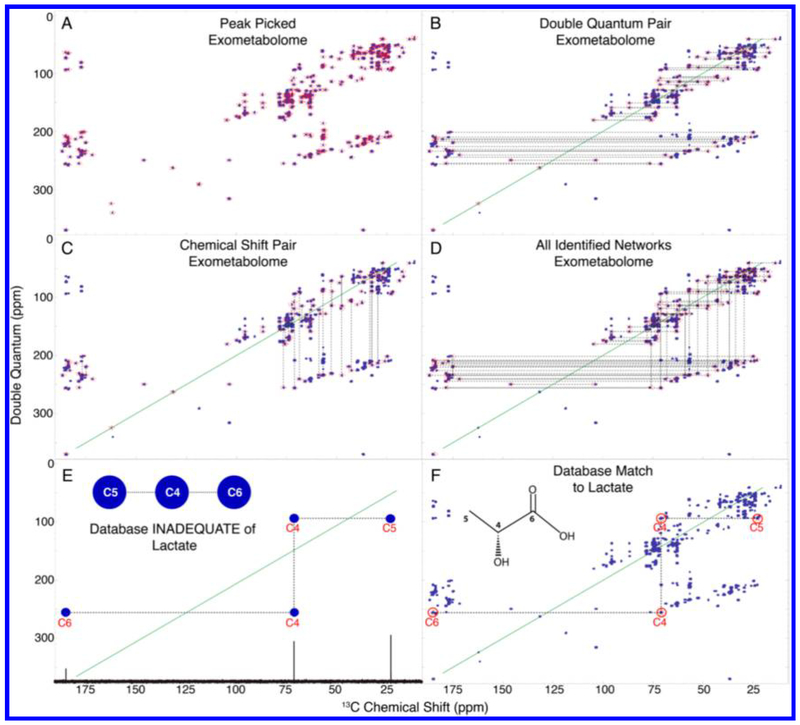
13C NMR, despite its many advantages for metabolomics studies, has not been used widely owing to its relatively low sensitivity and natural abundance. To alleviate this bottleneck, a custom-built 13C-optimized probe was used to obtain high-quality 13C NMR spectra.40 A lower limit of 13C NMR detection of approximately 40 nmol was obtained, and a combined analysis of 13C and 1H data using statistical correlation was shown to improve metabolite identification. Another technique that is often used for small molecule NMR applications is the 13C–13C incredible natural abundance double quantum transfer experiment (INADEQUATE), which is a highly useful 2D NMR method for metabolite identification. However, its potential is largely untapped in metabolomics owing to the low sensitivity of 13C at natural abundance. Recently, using samples derived from isotopically labeled Caenorhabditis, a new INADEQUATE-based, semiautomated approach called INETA (INADEQUATE network analysis) was proposed41 (Figure 2). Using INETA, 53 and 49 networks were identified from the endo- and exometabolomes, respectively, and from these networks, 31 compounds were assigned by combining these data with the BMRB42 database. The authors envisioned that using this approach provides an outstanding way to discover unknown molecules.
Figure 2.
INETA (INADEQUATE network analysis) method for metabolite identification, which uses an INADEQUATE experiment for 13C enriched samples to connect peaks. Peaks are picked (A) and their double quantum signal pair is found (B). Peaks lacking a partner are not considered in the analysis. Vertical pairs are matched on the basis of chemical shifts (C). Vertical and horizontal pairs are connected to form a network. All networks are shown in part D. 1D 13C spectra and carbon connectivity information are downloaded from the BMRB, and a 2D INADEQUATE in silico database is generated by adding the chemical shifts of two directly bonded carbons (E). The INADEQUATE spectra of database matches or candidates can then be plotted onto the peak-picked experimental spectra (denoted by red circles above carbon numbers) to give positive identification (F). Reproduced from Clendinen, C. S.; Pasquel, C.; Ajredini, R.; Edison, A. S. Anal. Chem. 2015, 87, 5698–5706 (ref 41). Copyright 2015 American Chemical Society.
A method for real-time metabolite monitoring in live cancer cells using 13C6-glucose and heteronuclear two-dimensional (2D) NMR was proposed recently.43 This method allowed for metabolic differentiation between cancer and normal cells on the basis of time-dependent changes in metabolite concentrations. The anticancer effects of galloflavin were also monitored, which revealed previously unknown functional targets of galloflavin. This method is potentially applicable to the study of real-time monitoring of metabolic alterations in a wide variety of cellular systems.
Signal Enhancement and Fast Acquisition.
A new methodology, known as signal enhancement by spectral integration (SENSI), was shown to overcome the low signal-to-noise ratio (S/N ratio) problem in 13C NMR by integration of a large number of spectra without additional measurements.44 Here, by the addition of 181 different spectra, a single spectrum was synthesized, which exhibited increased signal-to-noise ratio by about 10-fold and the number of peaks detected by more than 100 compared to a single spectrum. Such an increase in signal-to-noise ratio is equivalent to a 100-fold savings in data acquisition time, and hence this approach is considered highly time efficient. Adoption of the SENSI approach to 2D NMR (SENSI 2D) was also described recently.45 Here, using 1000 2D JRES NMR spectra, it was shown that a signal enhancement of ~14-fold and identification of 80–250 additional peaks could be achieved without any additional measurements.
It has been known for quite some time that data acquisition techniques such as nonuniform sampling (NUS) significantly reduces the measurement time or alternatively enhances spectral resolution. With the incorporation of NUS capabilities in current NMR spectrometers for routine use, NMR-based metabolomics applications are anticipated to benefit significantly. A demonstration that NUS substantially increases the resolution of 2D NMR spectra of complex mixtures of small molecules indicates the potential benefit of NUS to NMR-based metabolomics.46 Another article from the same group has evaluated ultrafast NMR and NUS in the context of metabolomics.47 In this latter paper, model samples mimicking the metabolic profile variations in serum from subjects affected by colorectal cancer and controls were analyzed by 1D 1H NMR along with conventional and fast DQF-COSY and HSQC spectra. It was shown that the fast 2D NMR techniques lead to similar results as conventional 2D NMR, which substantiates their utility for high-throughput metabolomics studies. Another new fast acquisition method that leverages a priori knowledge to tailor polychromatic pulses and customized time delays for an efficient Fourier encoding of the indirect domain of an NMR experiment was explored recently.48 This approach avoids potential artifacts associated with NUS schemes and uses a minimum number of scans equal to the number of resonances present in the indirect dimension. The method uses the conventional Fourier transform to process the generated data and is potentially useful in metabolomics for both in-cell and in in vivo NMR studies.
Hyperpolarization.
Methods that can significantly boost the nuclear polarization can achieve superb sensitivity enhancement to enable even real time monitoring of metabolism in vivo. Among the major approaches, parahydrogen induced polarization (PHIP) and dynamic nuclear polarization (DNP) offer hyperpolarization of nuclei such as 13C or 15N and detection of downstream reaction products, and thereby provide significant promise for metabolomics applications. Recently, for the first time, a PHIP approach was applied for quantitative analysis of biofluid extracts.49 Here, a combination of solid-phase extraction, para-hydrogen induced hyperpolarization, and selective NMR detection quickly enabled the detection of a doping substance, nikethamide, at sub-μM concentrations in urine. Hyperpolarization by dissolution dynamic nuclear polarization (D-DNP) assisted by cross-polarization (CP) was also shown to offer significant sensitivity enhancement suitable for investigations of dilute mixtures, in this case for plant and cancer cell extracts.50 The precision of the 13C D-DNP method was recently evaluated in tomato extract samples to assess its applicability for metabolomics applications.51 It was shown that the average reproducibility of D-DNP NMR is 3.6% for signals above the limit of quantification and 6.4% when all signals above the limit of detection are included.
DATA ANALYSIS
Data Preprocessing.
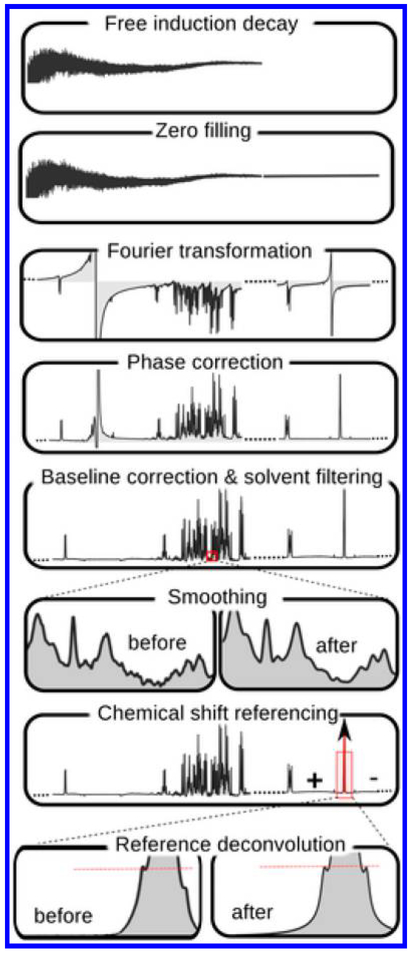
NMR spectra contain up to thousands of signals arising from many hundreds of detected metabolites. Preprocessing of the raw data represents an important and challenging step in NMR-based metabolomics, especially in studies involving large data sets. A number of data pretreatments are generally required before subjecting the data to statistical analysis to obtain meaningful information. Important steps include apodization, Fourier transformation, phasing, baseline correction, chemical shift calibration, and data normalization. For quantitative metabolomics, metabolite identification and quantification represent additional steps. While many previous reports describe each step in detail, large-scale metabolomics data sets obtained from multiple cohorts and analysis platforms, often obtained currently, demand even more robust processing approaches to ensure reliable data for statistical analysis. To meet such needs, a workflow was presented recently for integrated processing of multicohort NMR data.52 In this investigation, NMR data from COMBIBIO project consisting of ~8000 individuals from three different cohorts were used with appropriate quality controls and a pipeline for preprocessing was suggested. Two additional reports published recently describe preprocessing steps for large-scale NMR data in metabolomics.53,54 Data normalization is an essential step and the choice of the appropriate normalization method is also critical. A recent study using NMR of spiked urine samples shows that commonly used normalization and scaling methods fail to retrieve true metabolite concentrations in the presence of increasing amounts of glucose added to simulate unbalanced regulation.55 To address this challenge, Linear Baseline Normalization, Probabilistic Quotient Normalization, and Variance Stabilization Normalization were shown to account well for different dilutions of the samples. Studies focused on interactions between metabolites rely on investigations of correlations between metabolites. Such investigations need proper normalization of metabolomics data. To meet this need, a new normalization approach based on a mixed model that involves simultaneous estimation of a correlation matrix was proposed.56 Data binning is one of the preprocessing steps widely employed in 1D NMR-based metabolomics. With the aim of extending this approach to multidimensional NMR data, a generalization of Adaptive Intelligent binning was described.57 It allows the direct use of nD NMR data in bilinear factorizations such as principal component analysis (PCA) and partial least-squares (PLS) analysis methods.
Spectral and Data Analysis.
A new, simple-to-implement, and quantitative methodology to assess the confidence of NMR-based identification of known metabolites was introduced.58 This method is based on a topological analysis of metabolite identification information available from NMR spectroscopy studies. Biofluids such as urine require improved protocols for analysis as the large variations in pH and ionic concentrations contribute to positional noise and deleteriously affect unknown identification as well as biomarker discovery. To alleviate this challenge, a two-dimensional buffering system using potassium fluoride and phosphate buffer was presented to reduce positional noise.59 It was also shown that recursive segment-wise peak alignment leads to a further improvement in data reliability. A new method, ChemSMP (Chemical Shifts to Metabolic Pathways), which facilitates rapid data analysis by identifying the active metabolic pathways directly from chemical shifts obtained from a single 2D 1H–13C correlation NMR spectrum was described recently.60 The method does not require the identification or assignment of individual metabolites; instead, ChemSMP employs an indexing and scoring system for metabolite identification and uses metabolic pathway data from the Small Molecule Pathway Database (SMPDB; http://smpdb.ca/) and chemical shifts from the Human Metabolome Database (HMDB; http://www.hmdb.ca/).
Focused on efficient identification of metabolites in complex NMR spectra, Dubey and co-workers also proposed a new method that relies on matching the pattern of peaks rather than absolute tolerance thresholds.61 The new approach uses a combination of geometric hashing and similarity scoring techniques and was shown to exhibit 100% accuracy for 719 metabolites tested from the Human Metabolome Database (HMDB). Unlike conventional methods, it was shown to be highly tolerant of alterations in sample pH, temperature, and ionic strength. A new statistical analysis concept that involves classifying a component as “reliable” or “unreliable” depending on the reproducibility of its appearance was proposed.62 Using clustering method for classification, the concept was applied to multivariate curve resolution-alternating least-squares (MCRALS). The new method was shown to detect clusters containing little information but with good reliability, increasing the plausibility of results.
Statistical Data Analysis.
Metabolite data are generally complex and hence multivariate statistical methods are often used to summarize or extract the relevant data, develop classification models, and identify differentiating metabolites. Numerous unsupervised and supervised methods exist currently and the methods have witnessed continuous advances, owing to the need to detect subtle changes in the metabolic profiles and also to alleviate the effects of confounding factors. A recent report describes different steps involved in the metabolomics characterization of biofluids for clinical applications, ranging from study design to biological interpretation with an emphasis on the specific procedures required for processing and interpreting the NMR data.63 Another study presents a new approach to analyze NMR data to find discriminatory variables using minimum classification error rates as test statistics.64 Error rates are transformed into p-values, referred to as ERp, which are potentially useful in addressing many characteristics of metabolomics data. Another study discussed the fact that the now-conventionally used statistical methods such as PLS-DA and OPLS-DA can mislead in the absence of proper validation and provides practical guidelines and cross-validation recommendations for reliable inference from PCA and OPLS-DA models.65
It has also been realized that the conventional metabolomics data analysis approach, in which two or more sample groups are compared, is often unsuitable for complex data analysis, since it does not consider the unpredictable differences between subjects such as responders and nonresponders to treatment. To address this challenge, an approach based on statistical process control (SPC), which is able to monitor the response to a treatment or the development of a pathological condition, was recently proposed.66 It is suggested that this approach can potentially lead to a better understanding of the individual pathological processes and to a personalized diagnosis and therapy. A major challenge to biomarker discovery using metabolomics is the deleterious effects on the metabolite levels from the contributions of confounding factors. Recently, using a powerful statistical approach, seemingly unrelated regression (SUR), the effects of numerous demographic covariates such as gender, BMI, alcohol, and smoking status on serum metabolite levels could be modeled.67 After accounting for the contributions of such confounding factors, the potential of metabolites and pathway-related metabolite groups to differentiate patients with colon polyps and age-matched healthy controls was investigated.
Absolute Quantitation.
As compared to the traditional global data analysis methods that rely on relative comparisons, absolute metabolite quantitation offers numerous benefits, which include reducing the potential errors arising from factors such as broad spectral background, strong solvent signals and peak misalignments. Comparison across multiple studies and even analytical platforms becomes possible when the concentrations of metabolites are recorded. The ability to analyze intact samples with no need for sample preparation or separation represents an important characteristic that has traditionally drawn NMR-based metabolomics to prominence. However, it is increasingly realized that while biospecimens such as urine and extracted metabolite mixtures from cells and tissues enjoy the benefit of analysis using intact samples as they are generally devoid of macromolecules, biospecimens such as human blood serum or plasma, which have high amounts of macromolecules, are much more challenging. Further, numerous challenges including the limited resolution and unknown metabolite identification have deleteriously affected the utility of NMR for absolute metabolite quantitation.
As a remedy to this challenge, removing proteins physically using ultrafiltration, solid phase extraction, or protein precipitation using one or more organic solvents such as methanol, acetonitrile, perchloric acid, and trichloroacetic acid has been proposed. Further studies in this area have shown that protein precipitation using methanol exhibits superior performance over other organic solvents as well as ultrafiltration and allows a dramatic increase in the number of detectable and quantifiable metabolites.68,69 Incidentally, the results also reveal a surprisingly poor performance for protein precipitated samples using acetonitrile, which is often used instead of methanol. This is due to the poor solubility of many metabolites in acetonitrile, and hence it is clearly unsuitable for quantitative analysis of, specifically, hydrophilic blood metabolites.68 Using methanol precipitation and combining with an array of 1D/2D NMR experiments, database searches, and spiking with authentic compounds, quantitation of nearly 70 blood metabolites, a large number (nearly 1/3) of which had not been previously identified in the NMR spectra of blood was reported. Further, experimental protocols and comprehensive peak annotations were provided to take advantage of the high reproducibility and quantitative nature of NMR and to mitigate against the sensitivity of NMR chemical shifts to altered sample conditions. Such details enable easy reproduction of NMR spectra and serve as a visual guide for identification of the enhanced pool of blood metabolites for routine applications. Importantly, this protocol enables even nonexpert NMR users to easily identify and quantify blood metabolites, thereby allowing effective use of this tool.
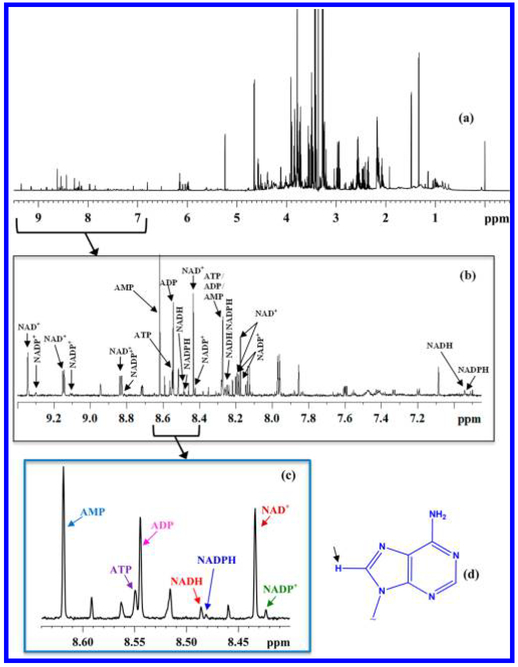
Coenzymes of cellular redox reactions and cellular energy mediate biochemical reactions fundamental to the functioning of all living cells. Recently, the major coenzymes of redox reaction and energy, NAD+ (nicotinamide adenine dinucleo-tide, oxidized), NADH (nicotinamide adenine dinucleotide, reduced), NADP+ (nicotinamide adenine dinucleotide phosphate, oxidized) and NADPH (nicotinamide adenine dinucleo-tide phosphate, reduced), adenosine triphosphate (ATP), adenosine diphosphate (ADP), and adenosine monophosphate (AMP) were detected and their identity established based on the combination of an array of 1D/2D NMR experiments, database searches, and spiking with authentic compounds70 (Figure 3). All the coenzymes were then shown to be quantitated simultaneously. The analysis method was also evaluated by measuring cardiac NADH and NAD+ ratios/pool sizes using mouse models with cardiac specific knockout (cKO) of the Ndufs4 gene (the Ndufs4 protein is critical for the integrity of mitochondrial Complex I) and cardiac-specific overexpression of nicotinamide phosphoribosyl transferase (cNAMPT) as examples. This is the first successful effort to provide the absolute quantitation of major coenzymes of cellular redox reactions and energy, simultaneously, with high fidelity.
Figure 3.
(a) Typical 800 MHz 1H NMR spectrum of a mouse liver tissue; (b) expanded spectral region with annotations for peaks for oxidized nicotinamide adenine dinucleotide (NAD+), oxidized nicotinamide adenine dinucleotide phosphate (NADP+), reduced nicotinamide adenine dinucleotide (NADH), reduced nicotinamide adenine dinucleotide phosphate (NADPH), adenosine triphosphate (ATP), adenosine diphosphate (ADP), and adenosine mono-phosphate (AMP); (c) expanded spectral region showing the characteristic fingerprint of the redox and energy coenzymes; and (d) adenine moiety with the lone hydrogen atom on the five membered ring indicated by an arrow; all peaks in the fingerprint region shown in part c arise from this hydrogen atom. Reproduced from Nagana Gowda, G. A.; Abell, L.; Lee, C. F.; Tian, R.; Raftery, D. Anal. Chem. 2016, 88, 4817–4824 (ref 70). Copyright 2016 American Chemical Society.
One key characteristic of NMR is its ability to provide absolute concentrations for many metabolites based on a single internal reference. Recently, a quantitation method using the synthetic ERETIC (Electronic REference To access In vivo Concentrations) signal was used in combination with the Chenomx NMR software suite to provide absolute quantitation of serum metabolites.71 Internal references such as 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) and trimethylsilyl propionate (TSP) can bind to proteins in the sample and thereby reduce their accuracy as quantitation standards and hence the use of synthetic signal can alleviate this challenge. However, the ERETIC, being a synthetic signal does not account for the sensitivity of NMR signals to sample conditions that affect critical parameters such as pulse width and relaxation parameters. Further, many metabolites that are attenuated due to protein binding are also difficult to quantify even using this approach. Another method that uses diffusion-weighted 1H NMR to separate lipoprotein and metabolite signals based on their large differences in translational diffusion was recently shown to work for blood plasma metabolite quantitation.72 The method, based on the combination of minimal sample preparation and minimal user interaction during processing and quantification, offers an alternative technique for automated quantification of human plasma metabolites. Here, the authors demonstrate an excellent correlation for 28 metabolites between results obtained with diffusion NMR and ultrafiltration.
A new method that permits the sensitive quantification of metabolites in the extracellular medium of cells in cultures was also published recently.73 This method uses the procedure PULCON (pulse length-based concentration determination), which was first applied for protein concentration measurements, requires one reference spectrum for quantitation of metabolites in all samples.
Absolute quantitation of metabolites using 2D NMR such as HSQC offers the significant benefit of improved resolution due to the extra spectral dimension. However, the dependence of peak integrals on a number of parameters such as rf pulse power, interpulse delays, the magnitude of heteronuclear J-couplings, T1 and T2 relaxation times, and the number of protons attached to carbon poses majorz challenges. Numerous efforts have focused on alleviating these challenges, including calibration curves, correction factors calculated from solving the Bloch equations, calculating a “time zero” HSQC spectrum derived by extrapolating the peak intensity back to zero time using a series of HSQC spectra and modulation of coherence transfer delays in the HSQC pulse sequence. Recently, a new quantitative HSQC pulse sequence was developed by introducing a refocusing period after the first INEPT transfer which discards the unwanted excess magnetization from methyl and methylene carbons and is designed to make 2D NMR peak integrals for all types of carbons the same.74 This experiment, which is called Quantitative, Equal Carbon HSQC (QECHSQC), also provides a uniform response over a wide range of 1JCH couplings and potentially promises new avenues for absolute quantification of metabolites in metabolomics.
Unknown Identification.
Unambiguous peak assignment, especially for low concentration metabolites, continues to be a challenge because most of the small metabolite peaks in the NMR spectrum are partially or completely overlapped by large signals from more abundant metabolites. Further, metabolite signals that do not appear in chemical shift databases pose added challenges to their identification.
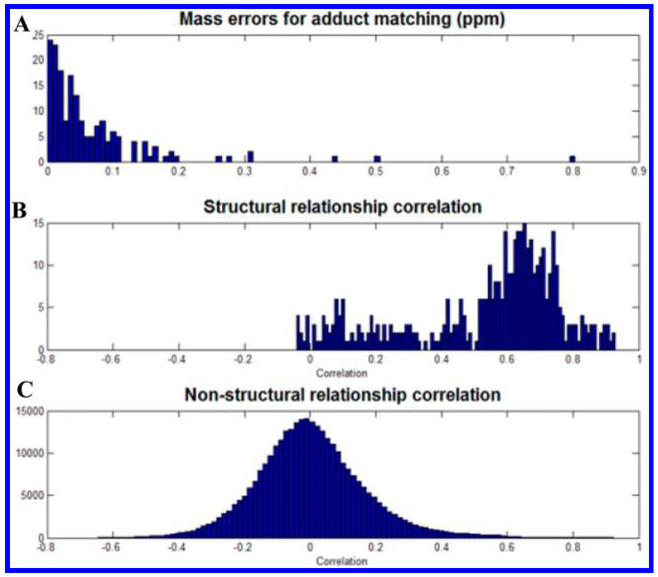
An increasing trend in the area of unknown identification is the utilization of the combined strengths and capabilities of NMR and MS.75 Using statistical correlation analysis of NMR and MS data, it was shown that peaks of the structurally related compounds between the two data sets can be linked. Here, on the basis of the correlation between direct-infusion Fourier transform ion cyclotron resonance mass spectrometry and NMR data of tissue extracts, more than half of the NMR detected signals were shown to exhibit high correlations, which is helpful in prioritizing candidate assignments and unknown identification (Figure 4). Statistical correlation was also applied using a set of 50 1H–13C Heteronuclear Single Quantum Correlation spectra and showed that the approach can be successfully used to identify biofluid metabolites.76 A recent paper describes the combination of NMR and electrospray ionization (ESI) MS for identification of metabolites.77 Here, authors also provide a new software tool for this approach that works with the Chenomx NMR Suite, which is commonly used for metabolite identification and quantitation. Another recent article provides a detailed guide to use different NMR methods and parameters for the identification of metabolites with illustrations using examples from recent studies.78
Figure 4.
Statistical correlation between NMR and MS: (A) distribution of mass errors for MS peaks that matched to at least one assigned NMR bin, (B) structural correlations between NMR and MS, and (C) nonstructural correlations between NMR and MS. Reproduced from Hao, J.; Liebeke, M.; Sommer, U.; Viant, M. R.; Bundy, J. G.; Ebbels, T. M. Anal. Chem. 2016, 88, 2583–2589 (ref 75). Copyright 2016 American Chemical Society.
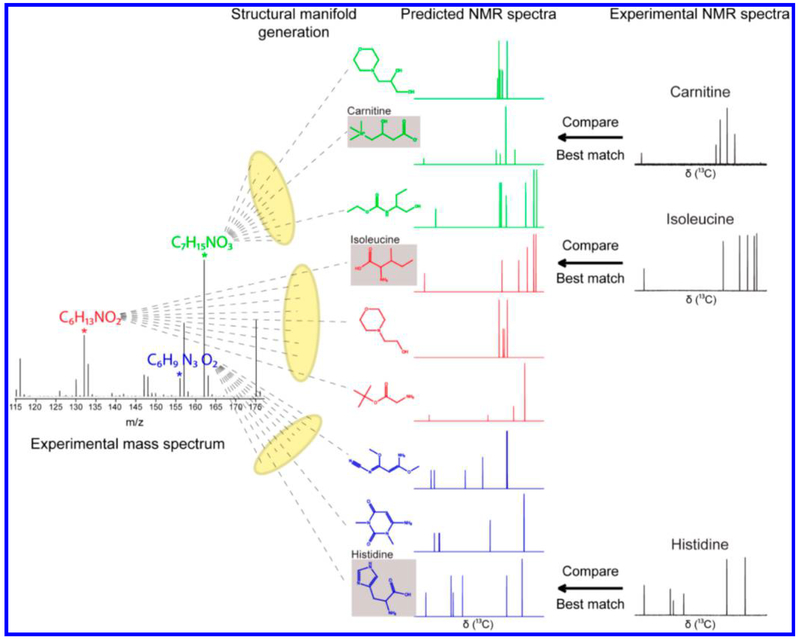
Separately, Brüshweiler and co-workers have developed novel metabolite identification strategies by exploiting the combined strength of NMR and MS methods. One approach identifies metabolite candidates from NMR spectra by database query, followed by the determination of the masses (m/z) of their possible ions, adducts, fragments, and characteristic isotope distributions.79 It allows assignment of mass spectra with very high confidence and, simultaneously, validates NMR-detected metabolites. Using this approach, a number of metabolites in urine were detected that had not been reported in the NMR literature previously. This approach, termed as a NMR/MS Translator, is fast, works in an automated fashion, and offers a new avenue for identifying and validating new metabolites. Another strategy, reported by the same group, combines high-resolution MS with NMR for the identification of unknown metabolites.80 (Figure 5). In this approach, chemical formulas are first identified from accurate mass data and all feasible structures consistent with these chemical formulas are generated. Then, for each structure, the NMR spectrum is predicted and compared with the experimental NMR spectrum. The structure that best matches both the MS and NMR data represents the putative identity of the metabolite. This approach was successfully applied to the identification of a large number of metabolites in an E. coli extract. Interestingly, this approach, termed SUMMIT MS/NMR (Structures of Unknown Metabolomic Mixture components by MS/NMR), does not require metabolite databases and is potentially useful for the discovery of new metabolites in complex biological mixtures.
Figure 5.
Schematic representation of the strategy for identifying metabolites in complex metabolomic mixtures by combining mass spectrometry with data from predicted and experimental 1D NMR spectroscopy. Reproduced from Bingol, K.; Bruschweiler-Li, L.; Yu, C.; Somogyi, A.; Zhang, F.; Brüschweiler, R.Anal. Chem. 2015, 87, 3864–3870 (ref 80). Copyright 2015 American Chemical Society.
A new dimension to the detection of an enhanced pool of metabolites in biological mixtures using NMR is provided by chemical derivatization.25 A selected class of metabolites can be reacted with chemoselective tags containing a stable isotope such as 13C, 15N, or 31P, which is favorable to NMR detection with enhanced resolution and sensitivity. Recently, the chemoselective derivatization approach was extended to the identification of metabolites with free keto and aldehyde groups.81 Here, by incorporating a 15N isotope in the aminooxy functional group, 15N-edited 2D NMR was used to select exclusively metabolites containing a free carbonyl function while suppressing all other metabolites. More than 30 carbonyl-containing compounds were detected in cell extracts and the identity for six was established.
COMBINING NMR AND MS
Interest in combining NMR and MS to exploit their combined strength in areas outside of unknown identification is growing quickly for a number of reasons. The need to improve biomarker discovery, to measure and quantitate many metabolites in complex biological mixtures, and better interpret biological function and mechanisms are all important driving forces. As demonstrated by the metabo-ring initiative,82 untargeted metabolomics data obtained using many NMR and MS instruments was highly convergent between and across different NMR and MS platforms, indicating that the necessary robustness for combining NMR and MS data was present. Recent reports describe some of the advances made so far in combining NMR and MS3,25,80 and indicate that the work is primarily focused on methods development and applications to biomarker discovery.
An optimized protocol for effectively integrating NMR and DI-ESI-MS was described recently and used for a biomarker discovery application.83 To minimize the potential variability between the two methods, a single sample was split into two parts and used for analysis by MS and NMR, simultaneously. Detailed steps to analyze the samples using both platforms and statistical analysis are provided, starting from sample preparation. The effectiveness of this approach in metabolic profiling was demonstrated utilizing multiblock principal component analysis (MB-PCA) and multiblock partial least-squares (MBPLS) to improve the mechanistic understanding of dopaminergic cell death. Another methodology-focused study presented analysis of intracellular metabolites in adherent mammalian cells combining NMR, GC–MS, and LC–MS methods.84 Here, different methods were first evaluated by NMR to quench the metabolism in adherent cells and extract the metabolites. The optimized method, quenching at −40 °C using methanol and extraction with dichloromethane–methanol–water (3:3:2 ratio), was then used for analysis by MS.
In the area of biomarker discovery, a methodology development study presented an algorithm to perform backward variable elimination partial least-squares discriminant analysis embedded with Monte Carlo cross validation (MCCVBVE-PLSDA) and combined NMR and targeted liquid chromatography (LC)–MS data.85 The study focused on differentiating colorectal cancer (CRC) and polyps from healthy controls as an example to demonstrate that variable selection is vitally important in combining NMR and MS data and obtaining improved predictive accuracy. On the basis of the obtained results, the article recommends using such a statistical approach for analysis of data from multiple analytical platforms in biomarker discovery studies focused on improved statistical performance and a more accurate biological interpretation.
A number of recently published studies that combine NMR and MS have focused on a variety of applications, including human diseases, plants, natural products, marine life, environmental effects, and toxicity. One study combined NMR and GC–MS platforms to generate a panel of five urinary metabolite biomarkers for Bipolar Disorder (BD) (four GC–MS-derived and one NMR-derived).86 This composite biomarker panel discriminated BD subjects from healthy controls with impressive area under the receiver operating characteristic curve (AUC) values of 0.974 in a training set and 0.964 in a test set, which is significantly superior to the previous single platform-derived metabolite panels. Urinary metabolic profiles of renal transplant patients with major injury factors at critical stages following transplantation were investigated by combining NMR and GC–MS.87 In this study, markers for medullary injury, tubule cell oxidative metabolism, impaired tubular reabsorption or secretion, and affected metabolic pathways upon transplantation as well as the variation of metabolic profiles with time were identified. Another NMR-and MS-based study investigated the intervention effect of curcumin on hyperlipidemic mice induced by a high-fat diet.88 While the serum biochemical assay indicated that treatment of curcumin improves the lipid profiles, metabolic pathways derived from urine metabolic profiles provided more detail and indicated reversal of the metabolic disorder.
An investigation of two varieties of maize combining NMR and GC–MS methods89 yielded extensive metabolic variation in grain from both varieties and also differentiation in patterns for subpopulations within each variety. Overall, this study provides insights into the extensive metabolic diversity associated with the conventional maize germplasm. In another study, fractions of natural product mixtures were subjected to combined NMR and MS analysis for identification of features related to bioactive compounds in mixtures, without the need for complete isolation.90
In the environmental area, a marine life study combined NMR and UPLC–MS methods to profile metabolite composition of the soft coral genus Sarcophyton with respect to its species and different habitats along the coastal Egyptian Red Sea.91 A total of 120 metabolites including 65 diterpenes, 8 sesquiterpenes, 18 sterols, and 15 oxylipids were identified. A number of these contributed to the differentiation of species and their unique habitats. Another environmental study combined NMR, GC–MS, and LC–MS for analyzing bronchial wash (BW) and bronchoalveolar lavage fluid (BALF) from healthy volunteers following exposure to biodiesel exhaust and filtered air.92 This study detected a total of 82 metabolites, the greatest number of unique metabolites reported so far in BW and BAL fluid. Notably, the GC–MS data revealed an effect of biodiesel exhaust exposure on the levels of 1-monostearylglycerol, sucrose, inosine, nonanoic acid, and ethanolamine (in BAL) and pentadecanoic acid (in BW), whereas LC–MS analysis indicated a shift in the levels of niacinamide (in BAL). The NMR assay only identified lactic acid (in BW) as being responsive to biodiesel exhaust exposure. A toxicity study combined NMR and LC–MS/MS to evaluate the subacute effects of hexabromocyclododecane (HBCD) on the mouse urinary metabolome.93 The results indicated subchronic exposure to HBCD caused a disturbance of mouse metabolism, especially in the TCA cycle, lipid metabolism, gut microbial metabolism, and homeostasis of amino acids. The study demonstrates the ability of the combined NMR and MS approach to provide comprehensive information and aid in predicting the health risks of such pollutants.
APPLICATIONS
Numerous studies including the investigations of human diseases, environmental toxicity, food and nutrition, and plants have been made use of NMR techniques. Among these, specifically, investigations of human diseases continue to be the major focus, and many diseases have been the subject of recent investigations including different types of cancers, heart disease, diabetes, obesity, Huntington’s disease, preeclampsia, and infections.
Cancer.
Mechanistic understanding of the development of cancer and its early detection are the dominant research areas that continue to exploit advances in metabolomics. A number of studies have focused on distinguishing cancer patients from matched controls based on measurements of biofluids and tissue from tumors. Many mechanistic studies have used animal models and cell lines for investigation. These studies have provided a new body of knowledge on altered metabolism in cancer. Notably, investigations of almost all major types of cancers have resulted in the identification of numerous potential metabolite biomarkers, although rigorous validation still remains to be performed in most cases.
Among these studies, the investigation of lung cancer has gained significant attention owing to the fact that it is the most lethal among all cancers. One study focused on diagnostic biomarkers of lung cancer investigated tissue using both ex vivo HRMAS NMR and in vitro NMR.94 Another study combined NMR with targeted rapid resolution liquid chromatography to obtain serum metabolite profiles of Stage I lung cancer (n = 25) and matched healthy controls; 25 metabolites were identified that distinguished the two groups with 100% sensitivity and specificity.95 Notably, this study highlighted the ability of NMR-based metabolomics for early stage detection of lung cancer. In a separate study, 17 metabolites associated with disease progression in serum were detected, in addition to 18 metabolites that distinguished lung cancer from healthy controls.96 Efforts have also been made to distinguish between adenocarcinoma (AdC) and squamous cell carcinoma (SqCC) based on investigations of tumor and tumor adjacent tissue from 56 patients using HRMAS NMR.97 Thirteen metabolites differentiated AdC and SqCC. In another study, NMR-based serum metabolite profiles could distinguish chronic obstructive pulmonary disease and lung cancer.98 And a study focused on the investigation of the effects of tumorigenesis and tumor growth on the noninvolved organs evaluated the metabolic phenotypes of multiple noninvolved mouse organ tissues such as heart, liver, spleen, lung, and kidney using a lung cancer xenograft model.99 It is interesting to note that the noninvolved organs exhibit significant metabolic changes due to the tumor growth.
Gastric cancer is the second most deadly and fourth most common cancer in the world. As with many cancers, gastric cancer exhibits very poor prognosis. Although endoscopy may prove to be useful for its detection, it is impractical to use endoscopy for general screening purposes. Hence, there is significant interest in detecting gastric cancer based on molecular markers, and a number of recent investigations have thus used NMR-based metabolomics approaches to study humans and animal models. One such study used a rat model involving four pathological groups, gastritis, low-grade gastric dysplasia, high-grade gastric dysplasia, and gastric cancer along with a healthy control group, and measured the serum metabolome by NMR.100 This study detected perturbed metabolic networks associated with the four pathological stages along with continually disturbed oxidative stress-related metabolic pathways during gastric carcinogenesis, which provides new information to address the molecular mechanisms underlying the gastric carcinogenesis. Focused on identifying diagnostic markers, an investigation of human urine has resulted in the identification of distinct metabolites for gastric cancer.101 In a separate study, urine as well as tumor tissue from gastric cancer were investigated using solution and HRMAS NMR, respectively; it showed significant differences between patients and healthy controls based on the combination of the two sample types.102 Investigation of tissue specimens from a large cohort of gastric cancer patients showed, apart from discrimination between cancer and controls, discrimination between cancer stages based on changes in 13 metabolites.103 And finally, a study focused on evaluating the effects of different culture media on cellular metabolic profiling has used a human gastric cancer cell line (SGC7901) cultured in two media, RPMI1640 and DMEM.104 This study demonstrated the distinct effect of media on the levels of several metabolites and cautions researchers to account for such media effects in comparative metabolomics analysis of cell lines.
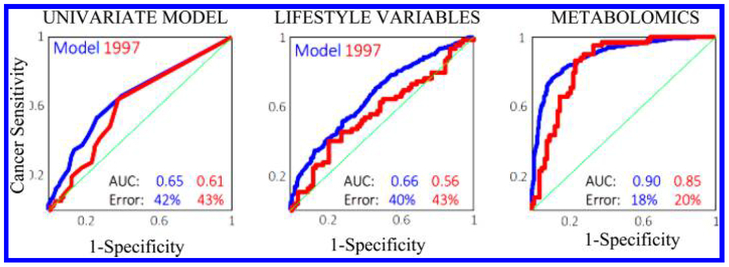
Breast cancer has also been the focus of a large number of metabolomics investigations. An interesting epidemiological study demonstrated accurate prediction of breast cancer several years ahead of its clinical detection based on the combined analysis of serum NMR metabolic profiles and lifestyle variables, referred to as biocontours.105 While mammography can diagnose current breast cancer with a sensitivity and specificity of ~75%, respectively, the new approach can predict the breast cancer risk 2–5 years ahead with sensitivity and specificity above 80%. This study was made using the samples from women enrolled in the Danish Diet, Cancer and Health cohort enrolled in the years 1993–1997. A statistical model was built on data obtained in 1993–1996 and tested on women sampled a year later, in 1997. Clearly, the results of the model are impressive and are far superior to the model obtained from a single molecular marker or even from lifestyle parameters alone (Figure 6). Limitations of this study, however, are that the cases and controls were not matched on age and the identities of many NMR variables that positively contributed to cancer prediction are not known. In another study, HRMAS NMR was used to distinguish between ductal carcinoma in situ (DCIS) lesions with or without an invasive component.106 It showed that the GPC/PC ratio as well as multivariate models discriminated between pure DCIS and DCIS accompanying invasive carcinoma. HRMAS NMR was also used to investigate breast tumor cell lines MCF-7 and MDA-MB-231, along with the nontumor breast cell line MCF-10A, to evaluate cancer metabolism in intact cells.107 Among the metabolites detected, phosphocholine exhibited higher concentrations in both tumor cells relative to controls and the level was higher for MDA-MB-231 than in MCF-7. Phosphocholine is recognized as a tumor marker for breast cancer.
Figure 6.
Comparison of the prediction accuracy for breast cancer incidence several years early obtained from three different models: a univariate model (left), a model of the 47 lifestyle variables (middle) and the model based on the combination of the serum NMR metabolite profile and lifestyle variables (right). Blue curve: Model from the data obtained in 1993–1996. Red curve: validation using independent data obtained in 1997. Reproduced from Bro, R.; Kamstrup-Nielsen, M. H.; Engelsen, S. B.; Savorani, F.; Rasmussen, M. A.; Hansen, L.; Olsen, A.; Tjønneland, A.; Dragsted, L. O. Metabolomics 2015, 11, 1376–1380 (ref 105).
Many other cancer types including prostate cancer, colorectal cancer, pancreatic cancer, ovarian cancer and thyroid cancer have also been the subject of NMR-based metabolomics investigations. For example, a study focused on prostate cancer biomarkers identification investigated serum from low-grade and high-grade prostate cancer patients along with healthy controls.108 It was shown that prostate cancer could be distinguished from controls with 84.4% sensitivity and 92.9% specificity, low-grade cancer could be distinguished from and high-grade cancer with 92.5% sensitivity and 93.3% specificity. In another study by the same group, filtered serum samples from prostate cancer, benign prostatic hyperplasia, and healthy controls were investigated for identifying distinguishing metabolites.109 Here, on the basis of discriminant function analysis, metabolite profiles showed higher discrimination power than the clinical laboratory tests. The identification of robust metabolite biomarkers for prostate cancer has been challenging, and controversy regarding the validated performance of any new biomarkers will likely continue. In the area of colon cancer, some recent investigations have reported fecal metabolite profiles.110,111 These studies have detected a number of short-chain fatty acids as common distinguishing markers. Such metabolites have been the focus of a number of recent gut microbiome studies, which provides some level of biological validation for these studies.112,113 Investigations of pancreatic cancer using animal models have been made based on metabolite profiling of tissue extract and serum.114,115 The disease model included both pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma and the results showed significant differences in metabolite profiles between the two pathological conditions. In the area of ovarian cancer, ovarian cyst fluid samples from benign, borderline, and malignant ovarian tumors were investigated.116 It showed significant metabolic differences among the three groups. Finally, focused on evaluating the potential for NMR to help diagnose preoperative thyroid cancer, percutaneous fine-needle aspiration of tissue specimens were subjected to metabolite profiling.117 The metabolite profiles significantly discriminated between benign and malignant nodules. Similar discrimination was observed in another study that investigated malignant and benign tumors as well as adjacent noncancerous tissue.118
Diabetes.
Several NMR studies have focused on investigations of diabetes using animal models and humans. Many of these studies evaluated the antidiabetic activity of potential drugs or supplements using animal models. For example, a rat model study investigated genipin119 and found that it improves many perturbed pathways including glucose metabolism, the TCA cycle, lipid metabolism, and amino acid metabolism. Investigation of antidiabetic properties of Phyllanthus niruri indicated that it decreased serum glucose levels and improved the lipid profile in diabetic rats.120 The study of antidiabetic effects of metformin and vildagliptin showed altered metabolite profiles in urine of a mouse model.121 Notably, it showed evidence for significant correlation between a number of metabolites including N-carbamoyl-β-alanine and glucose; N-carbamoyl-β-alanine was thus indicated to be a potential diabetes marker. In another study, plasma from a total of 71 patients with diabetes, coronary heart disease, diabetic coronary heart disease, or healthy controls were investigated to evaluate the metabolic risk for diabetic coronary heart disease.122 Interestingly, the diagnostic model constructed based on the metabolites detected diabetic coronary heart disease with 93% sensitivity and 93% specificity. It is known that obesity is a major risk factor for diabetes. Focused on identifying metabolic risk factors, a recent study compared serum metabolite profiles from obese and diabetic subjects with both high and low BMI.123 The study found higher concentrations of five metabolites including fatty acids, valine, isoleucine, phenylalanine, and lactate in nondiabetic obese subjects compared to nondiabetics with low BMI. Further, 19 metabolites including saturated fatty acids, several amino acids, lactic acid, 3-hydroxybutyric acid, choline, 3,7-dimethyluric acid, pantothenic acid, myoinositol, sorbitol, glycerol, and glucose were found to be higher in diabetic subjects compared to controls. Metabolite profiling of saliva offers a noninvasive approach that can be quite beneficial such as in the investigations of small children. Using this approach, an investigation of type I diabetes in small children (<6 years age) was made124 and found that many metabolites associated with glycolysis and the TCA cycle were altered between the diabetic and control groups.
Other Diseases.
Many other diseases including heart disease, Huntington’s disease, and preeclampsia have also been the recent focus of metabolomics investigations using NMR. An investigation of heart failure measured blood metabolites from heart failure patients with severe versus mild to moderate impairment of the left ventricle ejection fraction.125 Levels of four metabolites, 2-hydroxybutyrate, glycine, methylmalonate, and myo-inositol, were found to distinguish the three groups. Another investigation studied serum metabolite profiles from heart failure patients and matched controls focused on evaluating pathophysiological mechanisms of elevated myocardial energy expenditure in heart failure.126 It showed that three metabolites, 3-hydroxybutyrate, acetone, and succinate were associated with myocardial energy expenditure.
Huntington’s disease (HD) represents an autosomal neuro-degenerative disorder and affects one in every 10 000 subjects in the U.S. Metabolomics studies that focus on HD currently are scarce. While a few NMR-based studies have used animal models, investigations of HD using humans had not yet been made. Recently, the first NMR-based metabolomics study focused on HD in humans evaluated brain metabolites in the post-mortem striatum and frontal lobe from Huntington’s disease patients along with matched controls.127 On the basis of 39 quantified metabolites in brain tissue, multivariate discriminant models differentiated between HD and controls. Importantly, the metabolite profiles revealed that both parts of the brain studied were affected differently in HD compared to controls and that the striatum was more perturbed than the frontal lobe, metabolically.
A few NMR-based metabolomics investigations were focused on gaining insights into the pathogenesis of preeclampsia. In one study, late-onset preeclampsia was compared to matched controls based on the measurement of the serum metabolome.128 Altered pathways involving branched-chain amino acids, propanoate, glycolysis, gluconeogenesis, and ketone bodies were detected, which are suggestive of abnormalities including insulin resistance and metabolic syndrome, mitochondrial dysfunction and disturbance of energy metabolism, oxidative stress and lipid dysfunction in the pathogenesis of late-onset preeclampsia. A fairly large study used both urine and serum metabolite profiles to evaluate the potential of metabolomics to predict preeclampsia and gestational hyper-tension in early pregnancy.129 It showed that urinary metabolites provide higher sensitivity than serum metabolite profiles for the prediction of preeclampsia and gestational hypertension. The sensitivity was 51.3% (from urine) vs 15% (serum) for preeclampsia and 40% (urine) vs 33% (serum) for gestational hypertension.
Infection.
For infectious disease diagnosis, identification of the specific microorganism in biological specimens is critical to prescribe appropriate therapy. In principle, the unique metabolism linked to a specific bacterium or virus could be used to establish their identity based on metabolite profiling of infected samples. Many NMR-based metabolomics investigations have thus been focused on this hypothesis. A first metabolomics application to evaluate urinary metabolic profiles to distinguish bacterial and viral causes in children with respiratory tract infections was presented recently.130 While the sample size was small, the interesting result was that the urinary metabolome could differentiate these two causes based on statistical modeling. Another study focused on distinguishing Plasmodium falciparum infection from other symptomatically similar diseases by evaluating plasma metabolite profiles.131 It found an association of the infection with elevated levels of plasma lipoproteins. Focused on rapid discrimination and identification of several bacterial species, culture media of six species were analyzed.132 The results show that the six bacterial species including E. coli, Pseudomonas aeruginosa, Proteus mirabilis, Enterococcus faecalis, Staphylococcus aureus, and Staphylococcus saprophyticus could be discriminated. A study of urinary tract infections (UTI) was focused on the diagnosis of E. coli infection based on the metabolite profiles in urine.133 Trimethylamine was found to be a potential urinary biomarker for UTI involving E. coli. Finally, altered metabolite profiles due to pleural infection in children with Streptococcus pneumoniae pneumonia was investigated using pleural fluid.134 Results of this study indicated the ability of NMR to detect the progress of pneumococcal pneumonia infections by direct visualization of the bacterial metabolism.
Obesity.
Obesity is a major risk factor for many diseases, and many now recognize it as a disease itself. The search for a metabolic basis of obesity-induced diseases continues to be an important topic of many investigations. Several NMR-based metabolomics investigations of obesity have been made recently using both animal models and humans. A study using a rat model investigated metabolic profiles of portal vein and peripheral blood plasma, urine, and fecal water of obese and lean animals with a major focus on circulating host-microbial metabolites.135 This study demonstrated the important association of urinary host-microbial co-metabolites with phenotype. In another study, blood samples collected before and, periodically, after bariatric procedures were investigated to evaluate the metabolic alterations due to bariatric surgery in obese individuals.136 It was shown that surgery reverses the metabolic alterations associated with obesity and the results suggest profound changes in gut microbiome-host interactions after surgery.
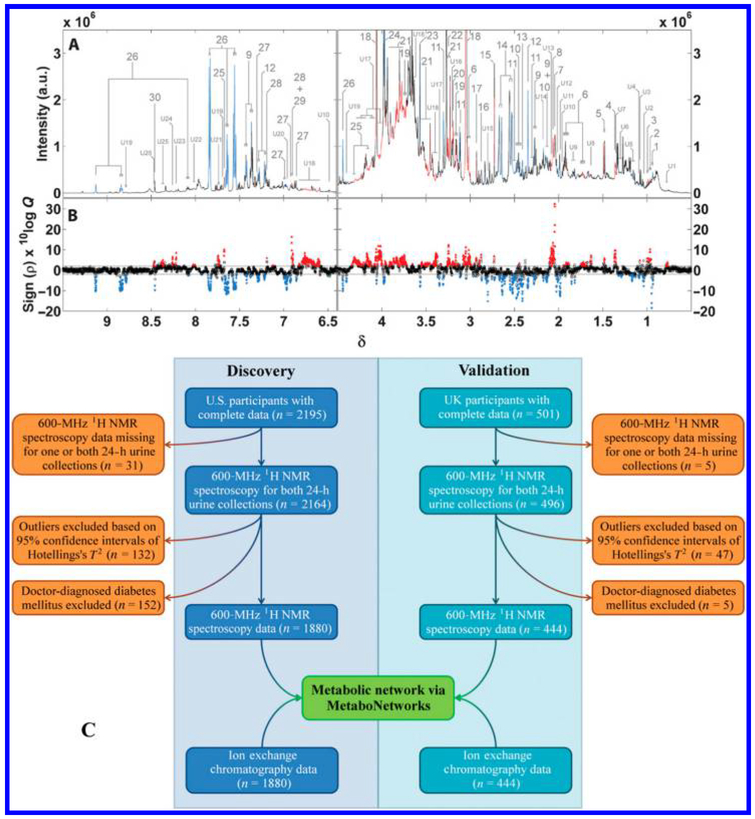
An impressive, international study of over 2 000 patients involving cohorts from U.S. and U.K. investigated urine to characterize metabolic signatures of adiposity.137 This study showed that metabolite profiles of urine collected over two 24-h time periods 3 weeks apart provided reproducible patterns of metabolites associated with adiposity (Figure 7). Further, the cohorts from both U.S. and U.K. showed multiple associations between urinary metabolites and body mass index that importantly could be validated across these two geographically distinct collections.
Figure 7.
An international (U.S. and U.K.) study focused on urinary metabolic signatures of human adiposity. (A) An average spectrum of 1880 urine spectra from the U.S. cohort; (B) associations of BMI with urinary metabolites. Statistically significant peaks are colored red if directly associated with BMI and blue if inversely associated. (C) Overall study design. Reproduced from Elliott, P.; Posma, J. M.; Chan, Q.; Garcia-Perez, I.; Wijeyesekera, A.; Bictash, M.; Ebbels, T. M.; Ueshima, H.; Zhao, L.; van Horn, L.; Daviglus, M.; Stamler, J.; Holmes, E.; Nicholson, J. K. Sci. Transl. Med. 2015, 7, 285ra62 (ref 137). Copyright 2015 American Association for the Advancement of Science.
Environmental Toxicity.
Environmental metabolomics, which deals with the investigation of interactions of the environment with living organisms, is an emerging area within the metabolomics field. Numerous pollutants including toxic chemicals, metals, and their products that have leached into the environment constitute major health hazards for humans as well as animals. Several NMR-based metabolomics studies have focused on investigations of such environmental toxicity. A study using a rat model investigated zinc oxide (ZnO) induced molecular changes on the respiratory system.138 BALF and lung tissue samples obtained after treating rats with different doses of 35 and 250 nm ZnO particles were analyzed. Only moderate and high doses of 250 nm ZnO particles were found to induce altered metabolism. Specifically, metabolites such as isoleucine and valine were increased while acetate, trimethylamine n-oxide, taurine, glycine, formate, ascorbate, and glycerophosphocholine were decreased in the BALF. In addition, lipids were increased, while taurine and glucose were decreased in the lung tissue of ZnO treated rats compared to controls. In another study, Daphnia magna exposed to atrazine, propranolol, and perfluorooctanesulfonic acid (PFOS) at sublethal levels were investigated.139 Depending on the type of contaminant, neonates and adults responded uniquely. Specifically, exposure to propranolol and PFOS altered metabolite profiles significantly in both neonates and adults, whereas exposure to atrazine caused altered metabolism only in adult Daphnia magna. Bioindicators of exposure to pharmaceuticals and personal care products were identified by exposing Daphnia magna to a range of sublethal concentrations of triclosan, carbamazepine, and ibuprofen.140 Separately, investigation of urine from rats exposed to polybrominated diphenyl ethers (PBDE) along with controls using NMR (and MS) indicated altered metabolite levels that are suggestive of endocrine disruption and neurodevelopmental toxicity caused by PBDEs.141 The toxic effects of 3,5,6-trichloro-2-pyridinol (TCP), which is a primary degradation product of chlorpyrifos and chlorpyrifos-methyl, was investigated using a mouse model.142 It was found that the TCP-treated mice exhibited lower body weight relative to controls; a number of metabolites related to hepatotoxicity and nephrotoxicity were detected in the TCP treated mice.
A recent human study evaluated oxidative/nitrosative stress biomarkers and NMR-based metabolite profiles using residents living within a 40-km radius of a petrochemical complex. These subjects were exposed to different levels of environmental pollutants such as vanadium and polycyclic aromatic hydro-carbons.143 The results showed that urine biomarkers, vanadium and 1-hydroxypyrene, were higher by a factor of 40 and 20, respectively, in high-exposure subjects compared to low exposure subjects; on the other hand, results of the metabolomics studies showed lower levels of amino acids and carbohydrates in the high-exposure subjects. Importantly, the combination of exposure biomarkers and metabolomics provide new insights into a number of potentially affected pathways including insulin signaling and oxidative stress. Finally, in the quest for preclinical serum markers of environmental exposure to lead, cadmium, and arsenic, serum metabolic profiles in healthy smelter workers were studied using NMR.144 In this study, the disturbance of energy, lipids, and amino acid metabolism due to exposure to heavy metals was identified.
Food and Nutrition.
Research on food and nutrition has witnessed numerous NMR-based metabolomics studies focused on a variety of issues, including food processing, quality, taste, composition, adulteration, contaminants, and nutritional values. In the area of food processing, a recent study investigated changes in metabolite composition of chicory root during the roasting process.145 The results showed that many metabolites, including most amino acids, degraded while several compounds were newly formed during roasting. Separately, it was shown that metabolic profiles can predict the taste of commercial coffee beans.146 Metabolic profiles in carrot root juices associated with different pedoclimatic conditions were identi-fied based on the analysis of carrot roots cultivated in different geographical areas,147 and metabolomics could also distinguish between monofloral and polyfloral honey.148 Metabolomic analysis provided insights into the metabolic variations of diverse rice cultivars and their associations with environmental conditions and genetic backgrounds.149 Such insights help to facilitate improvement in rice grain quality. Metabolite profiling of different varieties of cherry tomatoes identified variations due to climatic changes, specifically, the association of nutritional metabolites with both the variety as well as climatic conditions.150
Identifying dietary biomarkers is of major interest for numerous reasons including the goal of achieving calorie restriction without malnutrition that is increasingly shown to extend longevity and delay the onset of age-associated disorders in many animal studies. A study focused on detecting the effects of long-term dietary interventions investigated serum and urinary metabolic profiles in mice fed with different diets, with or without reduced caloric intake, over their life span.151 Notable findings of this study include the result that, irrespective of diet, calorie intake exerts a dominant effect on metabolic perturbations. Diet-induced metabolic changes were also investigated using urine metabolic profiles from individuals fed with seafood or nonseafood diets. It was found that seafood intake reduces urinary excretion of metabolites involved in mitochondrial lipid and energy metabolism, possibly facilitating a higher lipid catabolism in healthy subjects.152 It is also known that overeating different dietary fatty acids influence the amount of liver fat stored during weight gain. Focused on understanding the mechanism of such weight gain, a metabolomics study investigated the effect of overfeeding of saturated fatty acids (SFA) and polyunsaturated fatty acids (PUFA).153 Here polar metabolites were evaluated for differentiation between SFA and PUFA diet-based body fat deposition. Finally, a metabolomics study focused on identifying dietary markers for coffee consumption identified 2-furoylglycine as a reliable marker.154
Plants.
Plant metabolomics is another growing area of NMR-based metabolomics investigations. One study focused on biotechnological production of secondary metabolites in plants and presented a biomimetic approach in which a real tissue is simulated by a microfluidic chamber system.155 This study provided proof-of-principle results that a microfluidic system of cultivated plant cells can mimic the metabolic flows in a real plant tissue. Another study investigated the metabolic evolution and cultivar-dependent metabolite variation in soybean leaves using NMR-based metabolomics.156 The results identified distinct metabolism for adaptation to environmental conditions and their intrinsic metabolic phenotype, which provides new insights into the physiology of soybean plants. Another metabolomics study investigated plant species associated with benzylisoquinoline alkaloid metabolism, which accumulates pharmacological agents.157 This study found significant species- and tissue-specific variations including different alkaloids. Separately, metabolomics investigations of lettuce leaves focused on characterizing metabolic adaptations during leaf growth and exposure to the pesticide mancozeb were described.158 Finally, the effect of genotype on the metabolite composition in carrots was investigated and showed that different genotypes were characterized by differences in a number of metabolites, indicating the potential of this approach to map the carrot metabolome and identify genetic differences between varieties.159
NEW RESOURCES
The improved analytical tools in metabolomics generate increasingly large and complex data sets and thus drive the demand for improved processing and analysis software tools to unravel and understand the resulting data. In response, developments in numerous resources including software tools for automated data processing, analysis, and interpretation, as well as databases and data repositories have been made and continue to be developed in the field. One major benefit of these efforts is that virtually all such resources have been made publicly available, which has enabled the capabilities to perform fast data analysis and interpretation of the complex data by even nonexpert users. A recent overview of the freely available and open-source tools, algorithms, and frameworks was published with the goal of making new as well as established metabolomics researchers aware of the developments.160 Important topics included processing, annotation, and visualization of untargeted data from both NMR and MS-based metabolomics investigations.
A new software platform named BAYESIL was provided recently that focused on fast, reliable, and automated analysis of NMR spectra.161 It performs fully automated spectral processing and spectral profiling for 1D 1H NMR spectra collected on standard instruments, at several different frequencies (Figure 8). It was shown that >50 metabolites in biological samples such as serum and CSF can be analyzed in less than 5 min with good quantitative accuracy for ~90% of metabolites. Notably, this is the first fully automatic publicly accessible system that performs quantitative profiling for 1D NMR spectra with an accuracy that meets or exceeds the performance of trained experts. For optimal performance, BAYESIL requires NMR spectra from ultrafiltered serum/plasma. NMRPro, a software tool which enables easy-to-use online interactive processing and visualization of NMR spectra, was also recently developed.162 NMRPro is Web-based and hence is platform independent. This publicly available tool integrates server-side processing with client-side interactive visualization. Another software package, Dolphin, can analyze 2D NMR, in addition to 1D NMR, and automatically quantify a set of target metabolites accurately.163 It takes advantage of the 2D J-resolved NMR spectroscopy signal dispersion to avoid inconsistencies in signal position detection, which enhances the reliability and confidence in metabolite matching.
Figure 8.
NMR spectral processing steps in BAYESIL for the automated identification and quantitation of metabolites. Reproduced from Ravanbakhsh, S.; Liu, P.; Bjorndahl, T. C.; Mandal, R.; Grant, J. R.; Wilson, M.; Eisner, R.; Sinelnikov, I.; Hu, X.; Luchinat, C.; Greiner, R.; Wishart, D. S. PLoS One 2015, 10, e0124219 (ref 161).
A new database, termed 1H–13C-TOCCATA, that contains complete 1H and 13C chemical shifts for individual spin systems and isomeric states of common metabolites was recently developed.164 This database has been shown to help identification of metabolites using 2D 1H–13C TOCSY and 2D 13C–1H HSQC-TOCSY spectra at natural abundance. More recently, the same group introduced another metabolomics database and query algorithm, which unifies NMR spectroscopic information on 555 metabolites from both the Biological Magnetic Resonance Data Bank (BMRB) and the Human Metabolome Database (HMDB), for the analysis of 13C–1H HSQC spectra.165 The database, termed Complex Mixture Analysis by NMR (COLMAR) is specific to isomers and can be queried via an interactive, easy-to-use Web interface that is publicly available. Another software tool, Pathomx, enables processing, analysis, and visualization of NMR data including 1D and 2D spectra.166 This easy-to-use tool should be useful to new, nonexpert users as well as experts and also supports complex analysis.
A number of resources have also been devoted to statistical analysis, data visualization, and interpretation. One important and very successful endeavor among these is MetaboAnalyst, a comprehensive Web-based package for a range of metabolomics applications.167 MetaboAnalyst handles targeted as well as untargeted data from both NMR and MS platforms. It incorporates a number of normalization procedures and a range of univariate and multivariate methods including PCA, PLS-DA, heatmap clustering, and machine learning methods. It also offers a number of data interpretation tools such as MSEA (metabolite set enrichment analysis), MetPA (metabolite pathway analysis), and biomarker selection via ROC (receiver operating characteristic) curve analysis, as well as time series and power analysis. So far, MetaboAnalyst has been used for processing more than 1 million data sets by more than 50 000 users all over the world and currently more than 100 000 jobs are processed per month. Another new open-source platform, MVAPACK, was developed for NMR-based metabolomics applications168 (Figure 9). Developed for the GNU Octave programming environment, this publicly available package represents a complete platform for NMR-based chemometric data handling and was shown to be applicable for both routine NMR-based metabolomics applications as well as the development of new chemometric algorithms.
Figure 9.
Flow diagram showing the steps handled by the MVAPACK software package. Reproduced from Worley, B.; Powers, R. ACS Chem. Biol. 2014, 9, 1138–1144 (ref 168). Copyright 2014 American Chemical Society.
Major repositories for a variety of metabolomics research related tools have also been made available recently. One such repository is the Metabolomics Workbench, an international collection of metabolomics data and metadata, metabolite standards, metabolite structures, protocols, tutorials and training, and analysis tools.169 This public repository is part of the data coordinating effort of the National Institute of Health (NIH) Common Fund’s Metabolomics Program and provides a computational platform to integrate, analyze, track, deposit, and disseminate large volumes of heterogeneous data from NMR and MS platforms from more than 20 different species including humans and other mammals, plants, insects, invertebrates, and microorganisms. Another international open-access database repository, MetaboLights, collects data from cross-platform and cross-species metabolomics research at the European Bioinformatics Institute (EMBL-EBI).170 It provides Metabolomics Standard Initiative compliant metadata and raw experimental data associated with metabolomics experiments. These data repositories and new software tools are already being heavily used and promise to have a large impact on the field.
CONCLUSIONS
The field of metabolomics continues to witness multifaceted growth in methods and tools development as well as a wide range of applications. With its numerous unique characteristics and capabilities, NMR spectroscopy continues to play an important role in the field. While the intrinsically low sensitivity of NMR limits the number of detectable metabolites compared to MS, the unique characteristics of NMR far outweigh the sensitivity limitations. As a result, a large number of successful studies have been completed using NMR-based metabolomics. Also, there have been major efforts to improve the sensitivity and enhance the pool of detected metabolites by NMR; such advances help close the gap between the number of metabolites reliably detected by NMR and MS and enable researchers to exploit the combined strength of the two analytical platforms. Recent advances in NMR methods development have led to the detection of a large number of unknown metabolites, faster data acquisition, as well as automated spectral and data analysis. While the biomedical sciences continues to be the major beneficiary of NMR-based metabolomics investigations, studies in numerous other areas are also increasing. Another major trend over the past few years is the increased use of NMR and MS together. This development highlights the increasingly realized complexity of biological systems and the need for multifaceted and complementary analytical tools for better visualization of metabolite profiles in biological mixtures. We anticipate this trend will continue as more researchers become aware of the improved results from exploitation of the strength of the complementary methods. Further, as highlighted in a recent article,171 a number of technological advances focused on enhancing the resolution and sensitivity of NMR and speed of data collection and analysis will witness major advances in the NMR-based metabolomics applications to the benefit of all in the field.
ACKNOWLEDGMENTS
Financial support from NIH (Grants R01GM085291, P30DK035816, and P30CA015704–40) is gratefully acknowledged.
Biographies
G. A. Nagana Gowda is a Research Assistant Professor at the Northwest Metabolomics Research Center, Department of Anesthesiology and Pain Medicine, University of Washington, Seattle. From 2006 to 2012, he was a Research Scientist in the Chemistry Department, Purdue University, West Lafayette, Indiana. Previously, he was an Assistant Professor at the Centre of Biomedical Magnetic Resonance, Lucknow, India. He did his Postdoctoral work at the State University of New York, Buffalo, NY (1996–1998), and worked at the NMR Research Centre, Indian Institute Science, Bangalore, India (1986–2001). He received his B.S. degree (Physics, Chemistry, and Mathematics) in 1983 and M.S. degree (Analytical Chemistry) in 1985 from Mysore University, India, and Ph.D. degree (NMR Spectroscopy) in 1999 from Bangalore University, India. Broadly, his research is in the area of NMR-based metabolomics, methods development, and applications.
Daniel Raftery is the Founding Director of the Northwest Metabolomics Research Center and holds multiple academic positions at the University of Washington and Fred Hutch Cancer Research Center. He is a Medical Research and Education Endowed Professor in the Department of Anesthesiology and Pain Medicine, Adjunct Professor in the Department of Chemistry, and Director of the NORC Metabolomics Sub-Core Facility at the University of Washington, and Member at the Fred Hutchinson Cancer Research Center, Seattle. Prior to 2012, he was a Professor of Chemistry at Purdue University, West Lafayette, Indiana, where he began his faculty career in 1994 in the Analytical Division. He received his A.B. degree from Harvard and his Ph.D. from U.C. Berkeley. Before joining Purdue, he was an NSF Postdoctoral Fellow at the University of Pennsylvania. The Raftery research group focuses on the development of new methods and applications of metabolite profiling using a combination of NMR and mass spectrometry platforms which provide a broad-based approach for biomarker discovery and systems biology research.
Notes
The authors declare the following competing financial interest(s): Daniel Raftery reports holding equity and an executive role in Matrix-Bio, Inc.
REFERENCES
- (1).Larive CK; Barding GA Jr; Dinges MM Anal. Chem 2015, 87, 133–146. [DOI] [PubMed] [Google Scholar]
- (2).Nagana Gowda GA; Raftery D In Fundamentals of Advanced Omics Technologies: From Genes to Metabolites Comprehensive Analytical Chemistry; Simo C, Cifuentes A, García-Canas V, Eds.; Elsevier: New York, 2014; pp 187–211. [Google Scholar]
- (3).Bingol K; Brüschweiler R Anal. Chem 2014, 86, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Powers RJ J. Med. Chem 2014, 57, 5860–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Wolfender JL; Marti G; Thomas A; Bertrand SJ Chromatogr. A 2015, 1382, 136–164. [DOI] [PubMed] [Google Scholar]
- (6).Holmes E; Wijeyesekera A; Taylor-Robinson SD; Nicholson JK Nat. Rev. Gastroenterol. Hepatol 2015, 12, 458–471. [DOI] [PubMed] [Google Scholar]
- (7).Rankin NJ; Preiss D; Welsh P; Burgess KE; Nelson SM; Lawlor DA; Sattar N Atherosclerosis 2014, 237 (1), 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Trezzi JP; Vlassis N; Hiller K Adv. Exp. Med. Biol 2015, 867, 41–57. [DOI] [PubMed] [Google Scholar]
- (9).Gil AM; de Pinho PG; Monteiro MS; Duarte IF Bioanalysis 2015, 7 (18), 2361–2374. [DOI] [PubMed] [Google Scholar]
- (10).Kumar V; Dwivedi DK; Jagannathan NR NMR Biomed 2014, 27 (1), 80–89. [DOI] [PubMed] [Google Scholar]
- (11).Banoei MM; Donnelly SJ; Mickiewicz B; Weljie A; Vogel HJ; Winston BW Clin. Invest. Med 2014, 37 (6), E363–E376. [DOI] [PubMed] [Google Scholar]
- (12).Santini G; Mores N; Penas A; Capuano R; Mondino C; Trové A; Macagno F; Zini G; Cattani P; Martinelli E; Motta A; Macis G; Ciabattoni G; Montuschi P Curr. Top. Med. Chem 2016, 16 (14), 1610–1630. [DOI] [PubMed] [Google Scholar]
- (13).Calvani R; Brasili E; Praticò G; Sciubba F; Roselli M; Finamore A; Marini F; Marzetti E; Miccheli AJ Clin. Gastroenterol 2014, 48 (Suppl 1), S5–S7. [DOI] [PubMed] [Google Scholar]
- (14).Fan TW; Lane AN Prog. Nucl. Magn. Reson. Spectrosc 2016, 92–93, 18–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Hollinshead KE; Williams DS; Tennant DA; Ludwig C Adv. Exp. Med. Biol 2016, 899, 89–111. [DOI] [PubMed] [Google Scholar]
- (16).Johanningsmeier SD; Harris GK; Klevorn CM Annu. Rev. Food Sci. Technol 2016, 7, 413–438. [DOI] [PubMed] [Google Scholar]
- (17).Simmler C; Kulakowski D; Lankin DC; McAlpine JB; Chen SN; Pauli GF Adv. Nutr 2016, 7 (1), 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).O’Gorman A; Brennan LJ J. Sci. Food Agric 2015, 95 (13), 2567–2570. [DOI] [PubMed] [Google Scholar]
- (19).Ramakrishnan V; Luthria DL J. Sci. Food Agric 2017, 97, 33–42. [DOI] [PubMed] [Google Scholar]
- (20).van Duynhoven JP; Jacobs DM Prog. Nucl. Magn. Reson. Spectrosc 2016, 96, 58–72. [DOI] [PubMed] [Google Scholar]
- (21).Brennan L Prog. Nucl. Magn. Reson. Spectrosc 2014, 83, 42–49. [DOI] [PubMed] [Google Scholar]
- (22).Sumner LW; Lei Z; Nikolau BJ; Saito K Nat. Prod. Rep 2015, 32, 212–229. [DOI] [PubMed] [Google Scholar]
- (23).Mahrous EA; Farag MA J. Adv. Res 2015, 6 (1), 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Leenders J; Frédérich M; de Tullio P Drug Discovery Today: Technol 2015, 13, 39–46. [DOI] [PubMed] [Google Scholar]
- (25).Nagana Gowda GA; Raftery DJ J. Magn. Reson 2015, 260, 144–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Bingol K; Brüschweiler R Curr. Opin. Clin. Nutr. Metab. Care 2015, 18 (5), 471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Bingol K; Brüschweiler R Curr. Opin. Biotechnol 2017, 43, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Bingol K; Bruschweiler-Li L; Li D; Zhang B; Xie M; Brüschweiler R Bioanalysis 2016, 8 (6), 557–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Toumi I; Caldarelli S; Torrésani B Prog. Nucl. Magn. Reson. Spectrosc 2014, 81, 37–64. [DOI] [PubMed] [Google Scholar]
- (30).Emwas AH; Luchinat C; Turano P; Tenori L; Roy R; Salek RM; Ryan D; Merzaban JS; Kaddurah-Daouk R; Zeri AC; Nagana Gowda GA; Raftery D; Wang Y; Brennan L; Wishart DS Metabolomics 2015, 11 (4), 872–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Emwas AH; Roy R; McKay RT; Ryan D; Brennan L; Tenori L; Luchinat C; Gao X; Zeri AC; Nagana Gowda GA; Raftery D; Steinbeck C; Salek RM; Wishart DS J. Proteome Res 2016, 15 (2), 360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Soininen P; Kangas AJ; Würtz P; Suna T; Ala-Korpela M Circ.: Cardiovasc. Genet 2015, 8 (1), 192–206. [DOI] [PubMed] [Google Scholar]
- (33).Dona AC; Jiménez B; Schäfer H; Humpfer E; Spraul M; Lewis MR; Pearce JT; Holmes E; Lindon JC; Nicholson JK Anal. Chem 2014, 86 (19), 9887–9894. [DOI] [PubMed] [Google Scholar]
- (34).Zhang B; Xie M; Bruschweiler-Li L; Brüschweiler R Anal. Chem 2016, 88 (1), 1003–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zhang B; Xie M; Bruschweiler-Li L; Bingol K; Brüschweiler R Anal. Chem 2015, 87 (14), 7211–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jupin M; Michiels PJ; Girard FC; Wijmenga SS Magn. Reson. Med 2015, 73 (2), 459–468. [DOI] [PubMed] [Google Scholar]
- (37).Giskeødegård GF; Cao MD; Bathen TF Methods Mol. Biol 2015, 1277, 37–50. [DOI] [PubMed] [Google Scholar]
- (38).André M; Dumez JN; Rezig L; Shintu L; Piotto M; Caldarelli S Anal. Chem 2014, 86 (21), 10749–10754. [DOI] [PubMed] [Google Scholar]
- (39).Kalfe A; Telfah A; Lambert J; Hergenröder R Anal. Chem 2015, 87 (14), 7402–7410. [DOI] [PubMed] [Google Scholar]
- (40).Clendinen CS; Lee-McMullen B; Williams CM; Stupp GS; Vandenborne K; Hahn DA; Walter GA; Edison AS Anal. Chem 2014, 86 (18), 9242–9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Clendinen CS; Pasquel C; Ajredini R; Edison AS Anal. Chem 2015, 87 (11), 5698–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ulrich EL; Akutsu H; Doreleijers JF; Harano Y; Ioannidis YE; Lin J; Livny M; Mading S; Maziuk D; Miller Z; Nakatani E; Schulte CF; Tolmie DE; Kent Wenger R; Yao H; Markley JL Nucleic Acids Res 2007, 36, D402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wen H; An YJ; Xu WJ; Kang KW; Park S Angew. Chem., Int. Ed 2015, 54 (18), 5374–5377. [DOI] [PubMed] [Google Scholar]
- (44).Misawa T; Komatsu T; Date Y; Kikuchi J Chem. Commun. (Cambridge, U. K.) 2016, 52, 2964–2967. [DOI] [PubMed] [Google Scholar]
- (45).Misawa T; Wei F; Kikuchi J Anal. Chem 2016, 88 (12), 6130–6134. [DOI] [PubMed] [Google Scholar]
- (46).Le Guennec A; Dumez JN; Giraudeau P; Caldarelli S Magn. Reson. Chem 2015, 53 (11), 913–920. [DOI] [PubMed] [Google Scholar]
- (47).Guennec AL; Giraudeau P; Caldarelli S Anal. Chem 2014, 86 (12), 5946–5954. [DOI] [PubMed] [Google Scholar]
- (48).Zhang Z; Smith PE; Frydman LJ J. Chem. Phys 2014, 141 (19), 194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Reile I; Eshuis N; Hermkens NK; van Weerdenburg BJ; Feiters MC; Rutjes FP; Tessari M Analyst 2016, 141 (13), 4001–4005. [DOI] [PubMed] [Google Scholar]
- (50).Dumez JN; Milani J; Vuichoud B; Bornet A; Lalande-Martin J; Tea I; Yon M; Maucourt M; Deborde C; Moing A; Frydman L; Bodenhausen G; Jannin S; Giraudeau P Analyst 2015, 140 (17), 5860–5863. [DOI] [PubMed] [Google Scholar]
- (51).Bornet A; Maucourt M; Deborde C; Jacob D; Milani J; Vuichoud B; Ji X; Dumez JN; Moing A; Bodenhausen G; Jannin S; Giraudeau P Anal. Chem 2016, 88 (12), 6179–6183. [DOI] [PubMed] [Google Scholar]
- (52).Karaman I; Ferreira DL; Boulange CL; Kaluarachchi MR; Herrington D; Dona AC; Castagné R; Moayyeri A; Lehne B; Loh M; de Vries PS; Dehghan A; Franco O; Hofman A; Evangelou E; Tzoulaki I; Elliott P; Lindon JC; Ebbels TM J. Proteome Res 2016, DOI: 10.1021/acs.jproteome.6b00125. [DOI] [PubMed] [Google Scholar]
- (53).Euceda LR; Giskeødegård GF; Bathen TF Scand. J. Clin. Lab. Invest 2015, 75 (3), 193–203. [DOI] [PubMed] [Google Scholar]
- (54).Vettukattil R Methods Mol. Biol 2015, 1277, 123–136. [DOI] [PubMed] [Google Scholar]
- (55).Hochrein J; Zacharias HU; Taruttis F; Samol C; Engelmann JC; Spang R; Oefner PJ; Gronwald WJ Proteome Res 2015, 14 (8), 3217–3228. [DOI] [PubMed] [Google Scholar]
- (56).Jauhiainen A; Madhu B; Narita M; Narita M; Griffiths J; Tavaré S Bioinformatics 2014, 30 (15), 2155–2161. [DOI] [PubMed] [Google Scholar]
- (57).Worley B; Powers R Chemom. Intell. Lab. Syst 2015, 146, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Sanchon-Lopez B; Everett JR J. Proteome Res 2016, 15 (9), 3405–3419. [DOI] [PubMed] [Google Scholar]
- (59).Gil RB; Lehmann R; Schmitt-Kopplin P; Heinzmann SS Anal. Bioanal. Chem 2016, 408 (17), 4683–4691. [DOI] [PubMed] [Google Scholar]
- (60).Dubey A; Rangarajan A; Pal D; Atreya HS Anal. Chem 2015, 87 (24), 12197–12205. [DOI] [PubMed] [Google Scholar]
- (61).Dubey A; Rangarajan A; Pal D; Atreya HS Anal. Chem 2015, 87 (14), 7148–7155. [DOI] [PubMed] [Google Scholar]
- (62).Motegi H; Tsuboi Y; Saga A; Kagami T; Inoue M; Toki H; Minowa O; Noda T; Kikuchi J Sci. Rep 2015, 5, 15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Puchades-Carrasco L; Jantus-Lewintre E; Pérez-Rambla C; García-García F; Lucas R; Calabuig S; Blasco A; Dopazo J; Camps C; Pineda-Lucena A Oncotarget 2016, 7 (11), 12904–12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).van Reenen M; Reinecke CJ; Westerhuis JA; Venter JH BMC Bioinf 2016, 17, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Worley B; Powers R; Curr. Curr. Metabolomics 2016, 4 (2), 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Del Carratore F; Lussu M; Kowalik MA; Perra A; Griffin JL; Atzori L; Grosso M Anal. Chem 2016, 88 (16), 7921–7929. [DOI] [PubMed] [Google Scholar]
- (67).Chen C; Deng L; Wei S; Nagana Gowda GA; Gu H; Chiorean EG; Abu Zaid M; Harrison ML; Pekny JF; Loehrer PJ; Zhang D; Zhang M; Raftery DJ Proteome Res 2015, 14 (6), 2492–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Nagana Gowda GA; Gowda YN; Raftery D Anal. Chem 2015, 87, 706–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Nagana Gowda GA; Raftery D Anal. Chem 2014, 86, 5433–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Nagana Gowda GA; Abell L; Lee CF; Tian R; Raftery D Anal. Chem 2016, 88 (9), 4817–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Jung YS; Hyeon JS; Hwang GS Anal. Chim. Acta 2016, 934, 194–202. [DOI] [PubMed] [Google Scholar]
- (72).de Graaf RA; Prinsen H; Giannini C; Caprio S; Herzog RI Metabolomics 2015, 11 (6), 1702–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Goldoni L; Beringhelli T; Rocchia W; Realini N; Piomelli D Anal. Biochem 2016, 501, 26–34. [DOI] [PubMed] [Google Scholar]
- (74).Mäkelä V; Helminen J; Kilpeläinen I; Heikkinen SJ Magn. Reson 2016, 271, 34–39. [DOI] [PubMed] [Google Scholar]
- (75).Hao J; Liebeke M; Sommer U; Viant MR; Bundy JG; Ebbels TM Anal. Chem 2016, 88 (5), 2583–2589. [DOI] [PubMed] [Google Scholar]
- (76).Öman T; Tessem MB; Bathen TF; Bertilsson H; Angelsen A; Hedenström M; Andreassen T BMC Bioinf 2014, 15, 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Walker LR; Hoyt DW; Walker SM; Ward JK; Nicora CD; Bingol K Magn. Reson. Chem 2016, 54, 998. [DOI] [PubMed] [Google Scholar]
- (78).Dona AC; Kyriakides M; Scott F; Shephard EA; Varshavi D; Veselkov K; Everett JR Comput. Struct. Biotechnol. J 2016, 14, 135–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Bingol K; Brüschweiler RJ Proteome Res 2015, 14 (6), 2642–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Bingol K; Bruschweiler-Li L; Yu C; Somogyi A; Zhang F; Brüschweiler R Anal. Chem 2015, 87 (7), 3864–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Lane AN; Arumugam S; Lorkiewicz PK; Higashi RM; Laulhé S; Nantz MH; Moseley HN; Fan TW Magn. Reson. Chem 2015, 53 (5), 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Martin JC; Maillot M; Mazerolles G; Verdu A; Lyan B; Migné C; Defoort C; Canlet C; Junot C; Guillou C; Manach C; Jabob D; Bouveresse DJ; Paris E; Pujos-Guillot E; Jourdan F; Giacomoni F; Courant F; Favé G; Le Gall G; Chassaigne H; Tabet JC; Martin JF; Antignac JP; Shintu L; Defernez M; Philo M; Alexandre-Gouaubau MC; Amiot-Carlin MJ; Bossis M; Triba MN; Stojilkovic N; Banzet N; Molinié R; Bott R; Goulitquer S; Caldarelli S; Rutledge DN Metabolomics 2015, 11 (4), 807–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Marshall DD; Lei S; Worley B; Huang Y; Garcia-Garcia A; Franco R; Dodds ED; Powers R Metabolomics 2015, 11 (2), 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Madji Hounoum B; Blasco H; Nadal-Desbarats L; Diémé B; Montigny F; Andres CR; Emond P; Mavel S Anal. Bioanal. Chem 2015, 407 (29), 8861–8872. [DOI] [PubMed] [Google Scholar]
- (85).Deng L; Gu H; Zhu J; Nagana Gowda GA; Djukovic D; Chiorean EG; Raftery D Anal. Chem 2016, 88 (16), 7975–7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Chen JJ; Liu Z; Fan SH; Yang DY; Zheng P; Shao WH; Qi ZG; Xu XJ; Li Q; Mu J; Yang YT; Xie P Sci. Rep 2014, 4, 5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Kienana M; Lydie ND; Jean-Michel H; Binta D; Matthias B; Patrick E; Hélène B; Chantal le G Mol. BioSyst 2015, 11 (9), 2493–2510. [DOI] [PubMed] [Google Scholar]
- (88).Li ZY; Ding LL; Li JM; Xu BL; Yang L; Bi KS; Wang ZT PLoS One 2015, 10 (3), e0120950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Venkatesh TV; Chassy AW; Fiehn O; Flint-Garcia S; Zeng Q; Skogerson K; Harrigan GG J. Agric. Food Chem 2016, 64 (10), 2162–2172. [DOI] [PubMed] [Google Scholar]
- (90).Bertrand S; Azzollini A; Nievergelt A; Boccard J; Rudaz S; Cuendet M; Wolfender JL Molecules 2016, 21 (3), 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Farag MA; Porzel A; Al-Hammady MA; Hegazy ME; Meyer A; Mohamed TA; Westphal H; Wessjohann LA J. Proteome Res 2016, 15 (4), 1274–1287. [DOI] [PubMed] [Google Scholar]
- (92).Surowiec I; Karimpour M; Gouveia-Figueira S; Wu J; Unosson J; Bosson JA; Blomberg A; Pourazar J; Sandström T; Behndig AF; Trygg J; Nording ML Anal. Bioanal. Chem 2016, 408 (17), 4751–4764. [DOI] [PubMed] [Google Scholar]
- (93).Wang D; Zhang P; Wang X; Wang Y; Zhou Z; Zhu W Environ. Sci. Pollut. Res 2016, 23 (9), 8500–8507. [DOI] [PubMed] [Google Scholar]
- (94).Chen W; Lu S; Ou J; Wang G; Zu Y; Chen F; Bai C Cancer Biomark 2016, 16 (4), 653–664. [DOI] [PubMed] [Google Scholar]
- (95).Zhang X; Zhu X; Wang C; Zhang H; Cai Z Oncotarget 2016, 7 (39), 63437–63448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Puchades-Carrasco L; Palomino-Schätzlein M; Pérez-Rambla C; Pineda-Lucena A Briefings Bioinf 2016, 17 (3), 541–552. [DOI] [PubMed] [Google Scholar]
- (97).Rocha CM; Barros AS; Goodfellow BJ; Carreira IM; Gomes A; Sousa V; Bernardo J; Carvalho L; Gil AM; Duarte IF Carcinogenesis 2015, 36 (1), 68–75. [DOI] [PubMed] [Google Scholar]
- (98).Deja S; Porebska I; Kowal A; Zabek A; Barg W; Pawelczyk K; Stanimirova I; Daszykowski M; Korzeniewska A; Jankowska R; Mlynarz PJ J. Pharm. Biomed. Anal 2014, 100, 369–380. [DOI] [PubMed] [Google Scholar]
- (99).Xu S; Tian Y; Hu Y; Zhang N; Hu S; Song D; Wu Z; Wang Y; Cui Y; Tang H Sci. Rep 2016, 6, 28057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Gu J; Hu X; Shao W; Ji T; Yang W; Zhuo H; Jin Z; Huang H; Chen J; Huang C; Lin D Oncotarget 2016, 7 (37), 60053–60073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Chan AW; Mercier P; Schiller D; Bailey R; Robbins S; Eurich DT; Sawyer MB; Broadhurst D Br. J. Cancer 2016, 114 (1), 59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Jung J; Jung Y; Bang EJ; Cho SI; Jang YJ; Kwak JM; Ryu DH; Park S; Hwang GS Ann. Surg. Oncol 2014, 21 (Suppl 4), 736–742. [DOI] [PubMed] [Google Scholar]
- (103).Wang H; Zhang H; Deng P; Liu C; Li D; Jie H; Zhang H; Zhou Z; Zhao YL BMC Cancer 2016, 16, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Huang Z; Shao W; Gu J; Hu X; Shi Y; Xu W; Huang C; Lin D Mol. BioSyst 2015, 11 (7), 1832–1840. [DOI] [PubMed] [Google Scholar]
- (105).Bro R; Kamstrup-Nielsen MH; Engelsen SB; Savorani F; Rasmussen MA; Hansen L; Olsen A; Tjønneland A; Dragsted LO Metabolomics 2015, 11 (5), 1376–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Chae EY; Shin HJ; Kim S; Baek HM; Yoon D; Kim S; Shim YE; Kim HH; Cha JH; Choi WJ; Lee JH; Shin JH; Lee HJ; Gong G PLoS One 2016, 11 (8), e0161038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Maria RM; Altei WF; Andricopulo AD; Becceneri AB; Cominetti MR; Venâncio T; Colnago LA Anal. Biochem 2015, 488, 14–18. [DOI] [PubMed] [Google Scholar]
- (108).Kumar D; Gupta A; Mandhani A; Sankhwar SN J. Proteome Res 2015, 14 (3), 1455–1464. [DOI] [PubMed] [Google Scholar]
- (109).Kumar D; Gupta A; Mandhani A; Sankhwar SN Prostate 2016, 76 (12), 1106–1119. [DOI] [PubMed] [Google Scholar]
- (110).Lin Y; Ma C; Liu C; Wang Z; Yang J; Liu X; Shen Z; Wu R Oncotarget 2016, 7 (20), 29454–29464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Amiot A; Dona AC; Wijeyesekera A; Tournigand C; Baumgaertner I; Lebaleur Y; Sobhani I; Holmes EJ Proteome Res 2015, 14 (9), 3871–3881. [DOI] [PubMed] [Google Scholar]
- (112).Rooks MG; Garrett WS Nat. Rev. Immunol 2016, 16, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Ríos-Covián D; Ruas-Madiedo P; Margolles A; Gueimonde M; de Los Reyes-Gavilán CG; Salazar N Front. Microbiol 2016, 7, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Wen S; Li Z; Feng J; Bai J; Lin X; Huang H Cancer Sci 2016, 107 (6), 836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Lin X; Zhan B; Wen S; Li Z; Huang H; Feng J Mol. BioSyst 2016, 12 (9), 2883–2892. [DOI] [PubMed] [Google Scholar]
- (116).Kyriakides M; Rama N; Sidhu J; Gabra H; Keun HC; El-Bahrawy M Oncotarget 2016, 7 (6), 7216–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Ryoo I; Kwon H; Kim SC; Jung SC; Yeom JA; Shin HS; Cho HR; Yun TJ; Choi SH; Sohn CH; Park S; Kim JH Sci. Rep 2016, 6, 30075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Tian Y; Nie X; Xu S; Li Y; Huang T; Tang H; Wang Y Sci. Rep 2015, 5, 14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Shen XL; Liu H; Xiang H; Qin XM; Du GH; Tian JS J. Pharm. Biomed. Anal 2016, 129, 80–89. [DOI] [PubMed] [Google Scholar]
- (120).Mediani A; Abas F; Maulidiani M; Khatib A; Tan CP; Ismail IS; Shaari K; Ismail A; Lajis NH J. Pharm. Biomed. Anal 2016, 128, 302–312. [DOI] [PubMed] [Google Scholar]
- (121).Pelantová H; Bugáňová M; Holubova M; Šedivá B; Zemenová J; Sýkora D; Kaválková P; Haluzík M; Železná B; Maletínská L; Kuneš J; Kuzma M Mol. Cell. Endocrinol 2016, 431, 88–100. [DOI] [PubMed] [Google Scholar]
- (122).Liu X; Gao J; Chen J; Wang Z; Shi Q; Man H; Guo S; Wang Y; Li Z; Wang W Sci. Rep 2016, 6, 30785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Gogna N; Krishna M; Oommen AM; Dorai K Mol. BioSyst 2015, 11 (2), 595–606. [DOI] [PubMed] [Google Scholar]
- (124).de Oliveira LR; Martins C; Fidalgo TK; Freitas-Fernandes LB; de Oliveira Torres R; Soares AL; Almeida FC; Valente AP; de Souza IP J. Proteome Res 2016, 15 (8), 2491–2499. [DOI] [PubMed] [Google Scholar]
- (125).Deidda M; Piras C; Dessalvi CC; Locci E; Barberini L; Torri F; Ascedu F; Atzori L; Mercuro GJ Transl. Med 2015, 13, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Du Z; Shen A; Huang Y; Su L; Lai W; Wang P; Xie Z; Xie Z; Zeng Q; Ren H; Xu D PLoS One 2014, 9 (2), e88102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Graham SF; Kumar PK; Bjorndahl T; Han B; Yilmaz A; Sherman E; Bahado-Singh RO; Wishart D; Mann D; Green BD Biochim. Biophys. Acta, Mol. Basis Dis 2016, 1862 (9), 1675–1684. [DOI] [PubMed] [Google Scholar]
- (128).Bahado-Singh RO; Syngelaki A; Mandal R; Graham SF; Akolekar R; Han B; Bjondahl TC; Dong E; Bauer S; Alpay-Savasan Z; Turkoglu O; Ogunyemi D; Poon LC; Wishart DS; Nicolaides KH J. Matern.-Fetal Neonat. Med 2016, 1–7. [DOI] [PubMed] [Google Scholar]
- (129).Austdal M; Tangerås LH; Skråstad RB; Salvesen K; Austgulen R; Iversen AC; Bathen TF Int. J. Mol. Sci 2015, 16 (9), 21520–21538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Adamko DJ; Saude E; Bear M; Regush S; Robinson JL BMC Infect. Dis 2016, 16 (1), 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Sengupta A; Ghosh S; Das BK; Panda A; Tripathy R; Pied S; Ravindran B; Pathak S; Sharma S; Sonawat HM Mol. BioSyst 2016, 12 (11), 3324–3332. [DOI] [PubMed] [Google Scholar]
- (132).Palama TL; Canard I; Rautureau GJ; Mirande C; Chatellier S; Elena-Herrmann B Analyst 2016, 141 (15), 4558– 4561. [DOI] [PubMed] [Google Scholar]
- (133).Lam CW; Law CY; Sze KH; To KK Clin. Chim. Acta 2015, 438, 24–8. [DOI] [PubMed] [Google Scholar]
- (134).Chiu CY; Lin G; Cheng ML; Chiang MH; Tsai MH; Lai SH; Wong KS; Hsieh SY Sci. Rep 2016, 6, 24930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Phetcharaburanin J; Lees H; Marchesi JR; Nicholson JK; Holmes E; Seyfried F; Li JV J. Proteome Res 2016, 15 (6), 1897–1906. [DOI] [PubMed] [Google Scholar]
- (136).Gralka E; Luchinat C; Tenori L; Ernst B; Thurnheer M; Schultes B Am. J. Clin. Nutr 2015, 102 (6), 1313–1322. [DOI] [PubMed] [Google Scholar]
- (137).Elliott P; Posma JM; Chan Q; Garcia-Perez I; Wijeyesekera A; Bictash M; Ebbels TM; Ueshima H; Zhao L; van Horn L; Daviglus M; Stamler J; Holmes E; Nicholson JK Sci. Transl. Med 2015, 7 (285), 285ra62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Lee SH; Wang TY; Hong JH; Cheng TJ; Lin CY Nanotoxicology 2016, 10 (7), 924–934. [DOI] [PubMed] [Google Scholar]
- (139).Wagner ND; Simpson AJ; Simpson MJ Environ. Toxicol. Chem 2016, DOI: 10.1002/etc.3604. [Google Scholar]
- (140).Kovacevic V; Simpson AJ; Simpson MJ Comp. Biochem. Physiol., Part D: Genomics Proteomics 2016, 19, 199–210. [DOI] [PubMed] [Google Scholar]
- (141).Jung YS; Lee J; Seo J; Hwang GS Environ. Toxicol 2016. DOI: 10.1002/tox.22322. [DOI] [PubMed] [Google Scholar]
- (142).Deng Y; Zhang Y; Lu Y; Zhao Y; Ren H Sci. Total Environ 2016, 544, 507–514. [DOI] [PubMed] [Google Scholar]
- (143).Yuan TH; Chung MK; Lin CY; Chen ST; Wu KY; Chan CC Sci. Total Environ 2016, 548–549, 260–269. [DOI] [PubMed] [Google Scholar]
- (144).Dudka I; Kossowska B; Senhadri H; Latajka R; Hajek J; Andrzejak R; Antonowicz-Juchniewicz J; Gancarz R Environ. Int 2014, 68, 71–81. [DOI] [PubMed] [Google Scholar]
- (145).Wei F; Furihata K; Zhang M; Miyakawa T; Tanokura MJ Agric. Food Chem 2016, 64, 6459–6465. [DOI] [PubMed] [Google Scholar]
- (146).Wei F; Furihata K; Miyakawa T; Tanokura M Food Chem 2014, 152, 363–369. [DOI] [PubMed] [Google Scholar]
- (147).Tomassini A; Sciubba F; Di Cocco ME; Capuani G; Delfini M; Aureli W; Miccheli AJ Agric. Food Chem 2016, 64 (25), 5284–5291. [DOI] [PubMed] [Google Scholar]
- (148).Schievano E; Finotello C; Uddin J; Mammi S; Piana LJ Agric. Food Chem 2016, 64 (18), 3645–3652. [DOI] [PubMed] [Google Scholar]
- (149).Song EH; Kim HJ; Jeong J; Chung HJ; Kim HY; Bang E; Hong YS J. Agric. Food Chem 2016, 64 (15), 3009–3016. [DOI] [PubMed] [Google Scholar]
- (150).Masetti O; Ciampa A; Nisini L; Valentini M; Sequi P; Dell’Abate MT Food Chem 2014, 162, 215–222. [DOI] [PubMed] [Google Scholar]
- (151).Wu J; Yang L; Li S; Huang P; Liu Y; Wang Y; Tang H; J. Proteome Res 2016, 15 (7), 2299–2308. [DOI] [PubMed] [Google Scholar]
- (152).Schmedes M; Aadland EK; Sundekilde UK; Jacques H; Lavigne C; Graff IE; Eng Ø; Holthe A; Mellgren G; Young JF; Bertram HC; Liaset B; Clausen MR Mol. Nutr. Food Res 2016, 60 (7), 1661–1672. [DOI] [PubMed] [Google Scholar]
- (153).Elmsjö A; Rosqvist F; Engskog MK; Haglöf J; Kullberg J; Iggman D; Johansson L; Ahlström H; Arvidsson T; Risérus U; Pettersson C Nutr. Diabetes 2015, 5, e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Heinzmann SS; Holmes E; Kochhar S; Nicholson JK; Schmitt-Kopplin PJ Agric. Food Chem 2015, 63 (38), 8615–8621. [DOI] [PubMed] [Google Scholar]
- (155).Maisch J; Kreppenhofer K; Büchler S; Merle C; Sobich S; Görling B; Luy B; Ahrens R; Guber AE; Nick PJ J. Plant Physiol 2016, 200, 28–34. [DOI] [PubMed] [Google Scholar]
- (156).Yun DY; Kang YG; Yun B; Kim EH; Kim M; Park JS; Lee JH; Hong YS J. Agric. Food Chem 2016, 64 (29), 5773–5783. [DOI] [PubMed] [Google Scholar]
- (157).Hagel JM; Mandal R; Han B; Han J; Dinsmore DR; Borchers CH; Wishart DS; Facchini PJ BMC Plant Biol 2015. 15, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).Pereira SI; Figueiredo PI; Barros AS; Dias MC; Santos C; Duarte IF; Gil AM Food Chem 2014, 154, 291–298. [DOI] [PubMed] [Google Scholar]
- (159).Clausen MR; Edelenbos M; Bertram HC J. Agric. Food Chem 2014, 62 (19), 4392–4398. [DOI] [PubMed] [Google Scholar]
- (160).Misra BB; van der Hooft JJ Electrophoresis 2016, 37 (1), 86–110. [DOI] [PubMed] [Google Scholar]
- (161).Ravanbakhsh S; Liu P; Bjorndahl TC; Mandal R; Grant JR; Wilson M; Eisner R; Sinelnikov I; Hu X; Luchinat C; Greiner R; Wishart DS PLoS One 2015, 10 (5), e0124219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (162).Mohamed A; Nguyen CH; Mamitsuka H Bioinformatics 2016, 32 (13), 2067–2068. [DOI] [PubMed] [Google Scholar]
- (163).Gómez J; Brezmes J; Mallol R; Rodríguez MA; Vinaixa M; Salek RM; Correig X; Cañellas N Anal. Bioanal. Chem 2014, 406 (30), 7967–7976. [DOI] [PubMed] [Google Scholar]
- (164).Bingol K; Bruschweiler-Li L; Li DW; Brüschweiler R Anal. Chem 2014, 86 (11), 5494–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Bingol K; Li DW; Bruschweiler-Li L; Cabrera OA; Megraw T; Zhang F; Brüschweiler R ACS Chem. Biol 2015, 10 (2), 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Fitzpatrick MA; McGrath CM; Young SP BMC Bioinf 2014, 15, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Xia J; Wishart DS Curr. Protoc. Bioinformatics 2016, 55, 14.10.1–14.10.91. [DOI] [PubMed] [Google Scholar]
- (168).Worley B; Powers R ACS Chem. Biol 2014, 9 (5), 1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Sud M; Fahy E; Cotter D; Azam K; Vadivelu I; Burant C; Edison A; Fiehn O; Higashi R; Nair KS; Sumner S; Subramaniam S Nucleic Acids Res 2016, 44 (D1), D463–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Kale NS; Haug K; Conesa P; Jayseelan K; Moreno P; Rocca-Serra P; Nainala VC; Spicer RA; Williams M; Li X; Salek RM; Griffin JL; Steinbeck C Curr. Protoc. Bioinformatics 2016, 53 (14.13), 1–18. [DOI] [PubMed] [Google Scholar]
- (171).Markley JL; Brüschweiler R; Edison AS; Eghbalnia HR; Powers R; Raftery D; Wishart DS Curr. Opin. Biotechnol 2017, 43, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]