Abstract
Many areas of research suffer from poor reproducibility, particularly in computationally intensive domains where results rely on a series of complex methodological decisions that are not well captured by traditional publication approaches. Various guidelines have emerged for achieving reproducibility, but implementation of these practices remains difficult due to the challenge of assembling software tools plus associated libraries, connecting tools together into pipelines, and specifying parameters. Here, we discuss a suite of cutting-edge technologies that make computational reproducibility not just possible, but practical in both time and effort. This suite combines three well-tested components—a system for building highly portable packages of bioinformatics software, containerization and virtualization technologies for isolating reusable execution environments for these packages, and workflow systems that automatically orchestrate the composition of these packages for entire pipelines—to achieve an unprecedented level of computational reproducibility. We also provide a practical implementation and five recommendations to help set a typical researcher on the path to performing data analyses reproducibly.
Reproducible computational practices are critical to continuing progress within the life sciences. Reproducibility improves the quality of published research by facilitating the review process that involves replication and validation of results by independent investigators. Further, reproducibility speeds up research progress by promoting reuse and repurposing of published analyses to different data-sets or even to other disciplines. The importance of these benefits is clear, and vigorous discourse in the literature over the past several years (Nekrutenko and Taylor, 2012; Baker, 2016; McNutt, 2014; Begley, 2013; Begley et al., 2015; Leek and Peng, 2015) has led to reproducibility guidelines at the level of individual journals, as well as funding agencies.
However, achieving reproducibility on a practical, day-to-day level (and thus following these guidelines) still requires overcoming technical hurdles that are beyond the abilities of most life sciences researchers.
There have been successful efforts aimed at addressing some of these challenges: Galaxy (Afgan et al., 2016), GenePattern (Reich et al., 2006), Jupyter (Kluyver et al., 2016), R Markdown (Baumer et al., 2014), and VisTrails (Scheidegger et al., 2008). These environments automatically record details of analyses as they progress and therefore implicitly make them reproducible. Yet most still fall short from achieving full reproducibility because they fail to preserve the full computing environment in which analyses have been performed. For example, consider an analysis executed on Galaxy, a popular web-based scientific workbench. An analysis executed on a particular Galaxy server might include tools not found elsewhere and therefore cannot be reproduced outside that server. Another example is a Jupyter notebook that includes tools specific to a particular platform and a distinct set of software libraries. There is no guarantee that this notebook will produce the same results on a different computer. Here, we discuss a solution that greatly improves computational reproducibility by preserving the exact environment in which an analysis has been performed and enabling that environment to be recreated and used on other computing platforms.
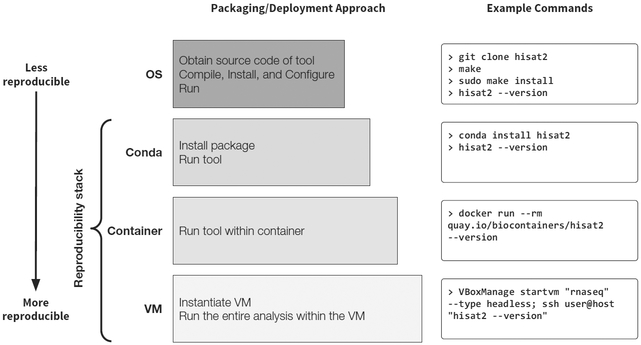
While the need for reproducibility is clear and initial guidelines are beginning to emerge, research practices will not change until ensuring reproducibility becomes straightforward and automated. Here, we focus on one important aspect of the reproducibility challenge: ensuring computational analysis can be run reproducibly, even in different environments. We describe a three-layer technology stack composed of open, well-tested, and community-supported components (Figure 1). This three-layer design reflects steps necessary to support fully reproducible analysis: (1) managing software dependencies, (2) isolating analyses from the idiosyncrasies of local computational environments, and (3) virtualizing entire analyses for complete portability and preservation against time. Fully reproducible analysis workflows require both ensuring reproducible computations and precise tracking of parameters and source data provenance. The solution presented here ensures that an encapsulated analysis tool can be run reproducibly and, when combined with a workflow system for capturing parameters and data provenance, can provide unprecedented reproducibility.
Figure 1.
Software Stack of Interconnected Technologies that Enables Computational Reproducibility It uses an example of the most basic RNA-seq analysis involving four tools. Our stack includes three components: (1) the cross-platform package manager Conda (https://conda.io) for installing analysis tools across operating systems, including virtualized environments that include all tools and dependencies at specified versions for performing a computational analysis, (2) lightweight software containers, such as Docker or Singularity, for using virtual environments and tool installations across different computing clusters, both local and in the cloud, and (3) hardware virtualization to achieve complete isolation and reproducibility. We have implemented this stack in the Galaxy scientific workbench (https://galaxyproject.org), enabling any Galaxy server to easily and automatically install all requirements for each Galaxy analysis workflow. This stack is also integrated into the CWL reference implementation. Integration of our reproducibility stack into Galaxy and CWL demonstrates, for the first time, how analysis workflows can be shared, rerun, and reproduced across platforms with no manual setup.
A Technology Stack for Reproducibility
The first step, managing software dependencies, ensures that one can obtain the exact versions of all software used in a given analysis. Because most software tools rely on external libraries and analysis workflows use multiple tools, it is necessary to record versions of numerous components. Given a multitude of operating systems and local configurations, ensuring the consistency of analysis software is a considerable challenge.
Conda (https://conda.io), a powerful and robust open source package and environment manager, has been developed to address this issue. It is operating system independent, does not require administrative privileges, and provides isolated virtual execution environments. These features make Conda exceptionally well-suited for use on existing high-performance computing (HPC) environments, as well as cloud infrastructure, because precise control over the execution environment does not depend on system-level configuration or access. Conda explicitly supports installation of specific tool versions, even old ones, and allows the creation of “environments” where specific tool versions are installed and run. While Conda is implemented in Python, it is able to package software written in any programming language. Creating and maintaining Conda package definitions (called recipes) requires no programming knowledge beyond basic shell scripting, and this feature has led to rapid uptake of Conda by the scientific community.
Leveraging Conda, Bioconda (https://bioconda.github.io) is a community project dedicated to data analysis in life sciences that contains over 4,000 tool packages with contributions by more than 400 authors (Grüning et al., 2017). Despite the fact that Bioconda is one of the most recent package managers dedicated to biomedical tools, it contains by far the largest number of software tools, underscoring its rapid uptake by the community (see Figure 2 in Grüning et al., 2017). Bioconda packages are well maintained and include a testing system to ensure their quality. They are built in a minimal environment to allow maximum portability and are provided as compiled binaries that are archived, ensuring the exact executables used for an analysis can always be obtained. In contrast to other solutions such as Debian-Med (Möller et al., 2010) or linuxbrew (http://linuxbrew.sh), unlike most OS-provided package managers, Conda allows multiple versions of any software tool at the same time, provides isolated environments, and runs on all major Linux distributions, macOS, and Windows. Thus, Bioconda is particularly enabling because it overcomes issues that, to some degree, plague other package managers and is championed by the community—an extremely important metric for ensuring future sustainability.
While Conda and Bioconda provide an excellent solution for packaging software components and their dependencies, archiving them, and recreating analysis environments, they are still dependent on and can be influenced by the host computer system (Beaulieu-Jones and Greene 2017). Moreover, since Conda packages are frequently updated, if a Conda virtual environment is created by specifying only the top-level tools and versions, recreating it at a later point in time using the same specifications may easily result in slightly different dependencies being installed. An additional level of isolation to solve this problem is provided by containerization platforms (or, simply, containers), such as Docker (https://www.docker.com), Singularity (Kurtzer et al., 2017), or rkt (http://coreos.com/rkt). Containers are run directly on the host operating system’s kernel but encapsulate every other aspect of the runtime environment, providing a level of isolation that is far beyond of what Conda environments can provide. Containers are easy to create, which is a great strength of this technology. Yet it is also its Achilles’ heel, because the ways in which containers are created need to be trusted and, again, reproducible.
This is why we generate containers automatically from Bioconda packages, and these automatically created containers form the second layer of our reproducibility stack (Figure 1). This has several advantages. First, container creation requires no user intervention; every container is created automatically and consistently using exactly the same process. A user of the container knows exactly what the container will include and how to use it. Second, this approach allows creation of large numbers of containers; in particular, we automatically generate and archive a container for every Bioconda package. Third, this approach can easily target multiple container types. We currently build containers for Docker, rkt, and Singularity. Our goal in supporting multiple container engines is for users to be able to adopt this solution regardless of the infrastructure they have access to. Docker is the most popular container environment currently and has broad platform support, including macOS (https://www.docker.com/docker-mac) and Windows (https://www.docker.com/docker-windows); however, it still has some security concerns and should only be run by trusted users on multi-user systems. Singularity provides better security guarantees for multi-user systems and works well in HPC environments (Kurtzer et al., 2017). Other container engines will likely be developed in the future; thus, our container creation approach does not rely on the specification format of any particular system, allowing additional container platforms to easily be added as they become available. Container registries provide an authoritative mapping from a container name (e.g., https://quay.io/repository/biocontainers/hisat2?tag=2.1.0-py27pl5.22.0_0&tab=tags) to a specific container image. We register containers with Quay (https://quay.io), and since we build standard open container initiative (OCI) containers (https://www.opencontainers.org), other registries (e.g., DockerHub, Cook, 2017; BioContainers, da Veiga Leprevost et al., 2017; or Dockstore, O’Connor et al., 2017) can also be used. Finally, in addition to creating containers for single Bioconda packages, it is possible to automatically create containers for combinations of packages. This is useful when a step in an analysis workflow has multiple dependencies. Given any combination of packages with version, we can generate a uniquely named container that contains all of the required dependencies. When the combinations of dependencies required are known in advance, these containers can be created automatically as well; for example, we can create containers for all tool dependencies used in the Galaxy ToolShed (https://galaxyproject.org/toolshed).
Containers provide isolated and reproducible compute environments, but still depend on the operating system kernel version and underlying hardware. An even greater isolation can be achieved through virtualization, which runs analysis within an emulated virtual machine (VM) with precisely defined hardware specifications. Virtualization, which provides the third layer of our reproducibility stack (Figure 1), can be achieved via commercial clouds, on public clouds, such as Jetstream (https://jetstream-cloud.org), or by using hypervisors or virtual machine applications on a local computer (such as VMware, KVM, Xen, and VirtualBox). Conda packages can be installed directly into VMs; however, using containers in VMs provides even greater portability and isolation when relocating analysis to another container execution environment. While introducing this layer adds complexity and overhead, it provides maximal isolation and security, as well as resistance to time, as emulated environments can be recreated in the future, regardless of whether the physical hard ware still exists. Although virtualization environments may change over time, older versions can be preserved and run as needed, using emulation within emulation if required.
Performance and scalability are important considerations in evaluating scientific workflow solutions. Because containers share a kernel with the host environment, the impact on execution performance is negligible. There are some fixed costs associated with downloading and assembling the container on first execution. Because VMs require emulation of the execution environment, the overhead is more significant; however, leveraging hardware virtualization features of modern CPUs this overhead is 5%–30% (Younge et al., 2011). Some workflows also need to be able to parallelize analysis and scale well in HPC environments. Support for scheduling and running analyses within containers already exists in popular resource managements systems, including Slurm, LSF, HTCondor, and Univa GridEngine. Additionally, Singularity is particularly well suited for running containers in HPC environments, including support for multi-node parallelization using the message passing interface (MPI).
Practical Implementation
To make it easy for analysis environments and workflow engines to adopt this solution, we have implemented our suite in a Python library called galaxy-lib. Given a set of required software packages and versions, galaxy-lib provides utilities to create a container with all dependencies for a given analysis. Thus, a workflow can be executed in which every step of the analysis runs either using a dedicated Conda environment or an isolated container. Support for this reproducibility stack has been integrated into the Galaxy platform, the Common Workflow Language (CWL) reference implementation, and the Snakemake workflow engine (Köster and Rahmann, 2012). Additional isolation and reproducibility can be achieved by running in a virtualized or cloud environment—for example, by using Galaxy CloudMan (Afgan et al., 2010) to run Galaxy on Amazon or Jetstream. Figure 1 shows examples of running a specific analysis tool, HISAT2 (Kim et al., 2015), using each level of isolation provided by the proposed stack: (1) installing as a package using Conda and running directly, (2) running in a container using “docker run,” and (3) launching in a local virtual machine—in this case using VirtualBox.
Recommendations
Reproducibility in computational life sciences is now truly possible. It is no longer a technological issue of “how do we achieve reproducibility?” Instead, it is now an educational (or even sociological) issue of “how to make sure that the community uses existing practices.” In other words, how do we set a typical researcher (e.g., a graduate student or a post-doc) performing data analyses on the path of performing them reproducibly? While there are now several platforms that enable reproducibility, the technologies we describe here are both very general and easy to use. Thus, we offer the following recommendations:
Carefully define a set of tools for a given analysis. In many cases, such as variant discovery, DNA/protein interactions assays, and transcriptome analyses, best-practice tool sets have been established by consortia such as 1000 Genomes, ENCODE, and modENCODE. In other, less common cases, selection of appropriate tools must be done by consulting published studies, Q&A sites, and trusted, community supported blogs. Because methodological decisions are vital to today’s biomedical research, it is essential to capture them so they can be shared with the scientific community. Recommendations 2–4 summarize our best practices for capturing and sharing these methodological decisions. To put this discussion on a practical footing, consider the simplest possible analysis of RNA sequencing (RNA-seq) data in which one needs to map reads against a genome, assemble transcripts, and estimate their abundance. The most basic set of tool for an analysis like this might include Trimmomatic (Bolger et al., 2014) to trim the reads, HISAT2 (Kim et al., 2015) to map the reads to a genome, StringTie (Pertea et al., 2015) to assemble and quantify transcripts, and Ballgown to perform differential expression testing.
Use tools from the Bioconda registry and help it grow. The registry at https://bioconda.github.ioprovides a list of available packages. The four tools from our example are all available in Bioconda and can be used directly (Figure 1). If a tool is not already available, one can either write a Bioconda recipe for the tool in question or request the tool to be packaged by the Bioconda community by opening an issue at the project’s GitHub page. Note that using Bioconda-enabled tools is not just “good behavior for the sake of reproducibility.” It is often the truly easiest way to use these tools. First, it makes installation easy. Conda automatically obtains and installs all necessary dependencies, so the only requirement for installing, say, StringTie, is opening a shell and typing “conda install stringtie.” Second, it makes analyses reproducible. Simply providing the output of “conda env export” with a manuscript allows anyone to easily obtain the exact version of the software used, as well as all its dependencies.
Adopt containers to guarantee consistency of results. Analysis tools installed with Bioconda can be used directly. However, the consistency of results (ability to guarantee that the same version of a tool gives exactly the same output every time it is run on a given input dataset) can be influenced by local computational environment. Because every Bioconda package is automatically packaged as a container, the tools can be run from within the container in isolation, providing a guarantee of result consistency. An example of this process for our RNA-seq example is shown in Figure 1. As container technologies become widely available, we see this as the preferred way to use analysis tools in the majority of research scenarios.
Use virtualization to make analyses “resistant to time.” Containers still depend on the host operating system, which will become outdated with time along with the hardware. To make an analysis “time proof” it is possible to use virtualization by encapsulating all tools, their dependencies, and the operating system with a VM image. Executing analysis within an archived VM will always afford maximum reproducibility, and this will become easier as an increasing number of compute resources providers move toward cloud-style operations.
Use a platform to orchestrate complete analysis reproducibility. Making Bioconda packages, software containers, and virtual machines all work together is critical for reproducibility, but it is also difficult. Furthermore, while this approach addresses reproducibility of running analysis tools, it does not handle tracking of parameters or provenance of data. Using a platform that helps connect and manage these layers of our reproducibility stack, as well as provides data and parameter tracking, is essential. For instance, Galaxy, Snakemake, and the CWL reference implementation can all automatically download, install, and manage both Bioconda packages and software containers while providing various levels of parameter and data tracking.
In conclusion, we are reaching the point where not performing data analyses reproducibly becomes unjustifiable and inexcusable. Aside from hardening the software, the main challenges ahead are in education and outreach that will be critical for fostering the next generation of researchers. There are also substantial cultural differences among research fields in the degree of software openness that will need to be tackled. We believe that this work is the first step toward making computational life sciences as robust as well-established quantitative and engineering disciplines. After all—our health depends on it!
ACKNOWLEDGMENTS
The authors are grateful to the Bioconda, BioContainers, and Galaxy communities, as without these resources, this work would not be possible. Nate Coraor provided critical advice on the project and edited the manuscript. This project was supported in part by NIH grants U41 HG006620 and R01 AI134384–01, as well as NSF grant 1661497 to J.T., A.N., and J.G. R.D. was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. N.S. was supported by the BBSRC Core Capability Grant BB/CCG1720/1 to the Earlham Institute. Additional funding was provided by German Federal Ministry of Education and Research (BMBF grants 031A538A & 031L0101C de.NBI-RBC & de.NBI-epi) to R.B. and B.G.
Footnotes
DECLARATION OF INTERESTS
Authors declare no competing financial interests.
REFERENCES
- Afgan E, Baker D, Coraor N, Chapman B, Nekrutenko A, and Taylor J (2010). Galaxy CloudMan: delivering cloud compute clusters. BMC Bioinformatics 11 (Suppl 12), S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44 (W1), W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M (2016). 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454. [DOI] [PubMed] [Google Scholar]
- Baumer B, Cetinkaya-Rundel M, Bray A, Loi L, and Horton NJ (2014). R Markdown: Inte grating A Reproducible Analysis Tool into Introductory Statistics. arXiv, arXiv:1402.1894. [Google Scholar]
- Beaulieu-Jones BK, and Greene CS (2017). Reproducibility of computational workflows is automated using continuous analysis. Nat. Biotechnol 35, 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley CG (2013). Six red flags for suspect work. Nature 497, 433–434. [DOI] [PubMed] [Google Scholar]
- Begley CG, Buchan AM, and Dirnagl U (2015). Robust research: Institutions must do their part for reproducibility. Nature 525, 25–27. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J (2017). Docker Hub. In Docker for Data Science (Apress), pp. 103–118.
- da Veiga Leprevost F, Grüning BA, Alves Aflitos S, Röst HL, Uszkoreit J, Barsnes H, Vaudel M, Moreno P, Gatto L, Weber J, et al. (2017). BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics 33, 2580–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüning B, Dale R, Sjödin A, Rowe J, Chapman BA, Tomkins-Tinch CH, Valieris R; The Bioconda Team, and Köster J (2017). Bioconda: A sustainable and comprehensive software distribution for the life sciences. bioRviv. https://doi.org/10.1101/207092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluyver T, Ragan-Kelley B, Pérez F, Granger B, Bussonnier M, Frederic J, Kelley K, Ham-rick J, Grout J, Corlay S, et al. (2016). Jupyter Notebooks-A Publishing Format for Reproducible Computational Workflows (ELPUB), pp. 87–90. [Google Scholar]
- Köster J, and Rahmann S (2012). Snakemake–a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Kurtzer GM, Sochat V, and Bauer MW (2017). Singularity: Scientific containers for mobility of compute. PLoS One 12, e0177459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, and Peng RD (2015). Statistics. What is the question? Science 347, 1314–1315. [DOI] [PubMed] [Google Scholar]
- McNutt M (2014). Reproducibility. Science 343, 229. [DOI] [PubMed] [Google Scholar]
- Möller S, Krabbenhöft HN, Tille A, Paleino D, Williams A, Wolstencroft K, Goble C, Holland R, Belhachemi D, and Plessy C (2010). Community-driven computational biology with Debian Linux. BMC Bioinformatics 11 (Suppl 12), S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko A, and Taylor J (2012). Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat. Rev. Genet 13, 667–672. [DOI] [PubMed] [Google Scholar]
- O’Connor BD, Yuen D, Chung V, Duncan AG, Liu XK, Patricia J, Paten B, Stein L, and Ferretti V (2017). The Dockstore: enabling modular, community-focused sharing of Docker-based genomics tools and workflows. F1000Res. 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, and Mesirov JP (2006). GenePattern 2.0. Nat. Genet 38, 500–501. [DOI] [PubMed] [Google Scholar]
- Scheidegger CE, Vo HT, Koop D, Freire J, and Silva CT (2008). Querying and Re-using Workflows with VsTrails. Proc. ACM SIGMOD Int. Conf. Manag. Data (ACM), pp. 1251–1254. [Google Scholar]
- Younge AJ, Henschel R, Brown JT, von Laszewski G, Qiu J, and Fox GC (2011). Analysis of Virtualization Technologies for High Performance Computing Environments. Proc. IEEE Int. Conf. Cloud Comput. 9–16. [Google Scholar]