Summary
Fundamental differences exist between males and females, encompassing anatomy, physiology, behaviour, and genetics. Such differences undoubtedly play a part in the well documented, yet poorly understood, disparity in disease susceptibility between the sexes. Although traditionally attributed to gonadal sex hormone effects, recent work has begun to shed more light on the contribution of genetics — and in particular the sex chromosomes — to these sexual dimorphisms. Here, we explore the accumulating evidence for a significant genetic component to mammalian sexual dimorphism through the paradigm of sex chromosome evolution. The differences between the extant X and Y chromosomes, at both a sequence and regulatory level, arose across 166 million years. A functional result of these differences is cell autonomous sexual dimorphism. By understanding the process that changed a pair of homologous ancestral autosomes into the extant mammalian X and Y, we believe it easier to consider the mechanisms that may contribute to hormone-independent male–female differences. We highlight key roles for genes with homologues present on both sex chromosomes, where the X-linked copy escapes X chromosome inactivation. Finally, we summarise current experimental paradigms and suggest areas for developments to further increase our understanding of cell autonomous sexual dimorphism in the context of health and disease.
In this review, Snell and Turner provide an overview of the genetic basis of sexual dimorphisms in mammals and discuss models to disentangle the effects of hormones versus genes in differences between the sexes.
Main Text
Introduction
Men and women differ in their physical appearance, indicative of an anatomical and physiological sexual dimorphism that is widespread in the natural world [1]. In primates, for example, males of Gorilla and Mandrillus taxa species are significantly larger than females; in contrast, females are generally larger than males in the Lorisid and Cheirogalid taxa [2]. Ultimately such differences must be attributed to male–female variation at the genetic level, which in turn drives the development of the gonads, and production of gonadal sex hormones in utero. Prior to this point of physiological differentiation, though, the sex chromosomes have already induced sex-specific aspects of organ development in the absence of gonadal sex hormones 3, 4. Subsequently, however, as mammals and their gonads do not each exist in isolation, these two variables must be separated in order to further understand their relative contributions to sexual dimorphism in human health and disease. In this review, we seek first to highlight some of the key evidence of human sexual dimorphism. Subsequently, we use the evolution of the sex chromosomes as a paradigm with which to understand the possible sources of genetic sexual dimorphism in mammals. Finally, we summarise a small number of the model systems available for investigating the mechanisms of mammalian genetic sexual dimorphism.
Evidence for Sexual Dimorphism from Human Health and Disease
The difference in disease prevalence rates between males and females has been recognised for many years, with examples from cradle to grave. Boys are more likely to be born with pyloric stenosis or malformations of the genitourinary tract, whereas girls are more likely to have developmental dysplasia of the hip or scoliosis 5, 6. In early childhood, boys have a higher incidence of bacterial and viral infections, including meningitis, septicaemia, influenza A and respiratory syncytial viruses 7, 8, 9, 10. In adult life, autoimmune disease, depression and dementia are more common in females, whereas cardiovascular disease, schizophrenia, and Parkinson’s disease are more prevalent in males 11, 12, 13, 14, 15, 16. Although these associations have been shown to be reproducible, the underlying mechanisms are yet to be definitively elucidated. The most prominent two hypotheses attribute these sexual dimorphisms to either the gonadal sex hormones or the sex chromosomes.
Gonadal Sex Hormones
Both male and female human fetuses are exposed to high levels of maternal oestrogens in utero, in addition to hormones produced by the placenta [17]. Furthermore, males start producing testosterone following testis determination at around eight weeks gestation [18]. At birth, sex hormone levels drop significantly in both sexes as the feto-placental unit is separated, before rising again transiently at around two to three months during ‘mini-puberty’ [19]. Subsequently, girls enter puberty slightly earlier than boys, and both sexes achieve maximum sex hormone levels during their mid-teens. In later life, from around the age of 50, testosterone levels in men drop gradually, whereas oestrogen levels in women fall precipitously during menopause [19].
A number of diseases have been associated with these sex-specific patterns of hormone secretion. For example, boys have a high prevalence of asthma in the pre-pubertal years [20]. Following puberty, when testosterone production is markedly increased, the burden of disease is significantly reduced. In contrast, the prevalence of asthma during childhood in girls is low, but this increases significantly during puberty, as does the risk of severe asthma [21]. Interestingly, there is a subsequent drop in asthma severity in women aged 50–65, correlating with the timing of menopause and reduced oestrogen production [21]. Post-menopausal women are also at increased risk of developing cardiovascular disease, which has similarly been attributed to reduced oestrogen levels [22]. The endocrine system therefore appears to play a significant role in mediating some sexual dimorphisms in disease. However, with our ever-increasing understanding of sex chromosomes, it has become clear that some of the differences between males and females are due to genetics.
Genetic Causes of Sexual Dimorphism: Understanding from Evolution
Genetic testis determination triggered the evolution of the mammalian sex chromosomes, producing a pair of chromosomes fundamentally different from the autosomes in terms of gene content, regulation of gene expression, and inheritance. The extant X and Y chromosomes, and the females and males in which they exist, also differ from each other as a result of this process. We can therefore use the evolution of the sex chromosomes as a paradigm for understanding possible genetic mechanisms underlying male–female differences.
The mammalian sex chromosomes have evolved from a pair of autosomes during the past 166 million years (Figure 1A) 23, 24. Between 148 and 166 million years ago, mutations on the proto-Y chromosome resulted in the creation of the testis-determining gene SRY: carriers of SRY develop with testes, while non-carriers develop with ovaries 25, 26. SRY-based genetic testis determination is conserved in most eutherian mammals, and the sequence is present in metatherians [27], though whether it retains a role in testis determination in this mammalian clade remains an open question.
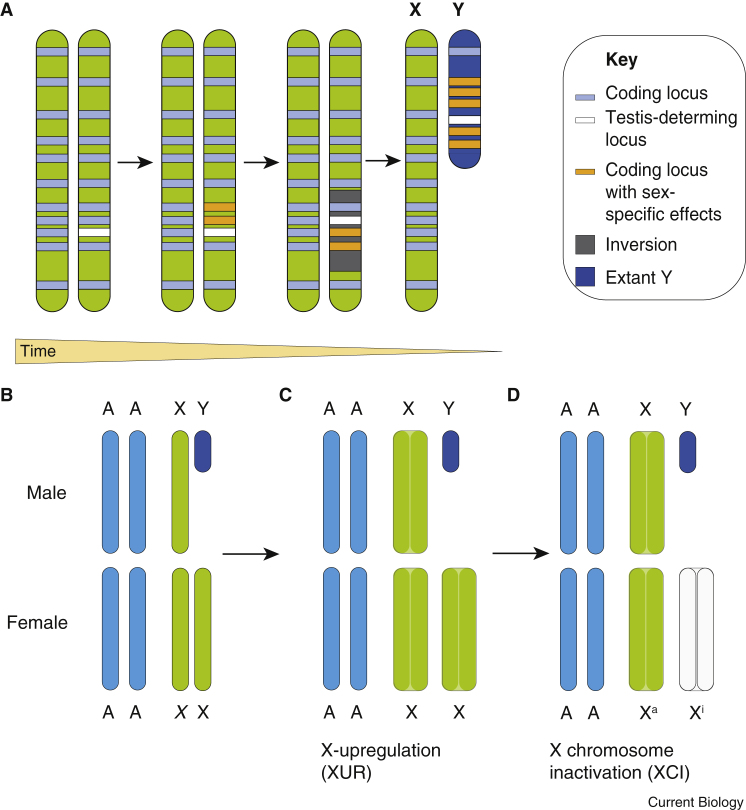
Figure 1.
The evolution of the mammalian sex chromosomes and dosage compensation mechanisms.
(A) A testis-determining locus (proto-SRY, white) was acquired on an autosome around 148–166 million years ago. Sexually antagonistic alleles (orange) then evolved at nearby loci, selected for in males due to their tight linkage to SRY. Recombination suppression between the proto-X and -Y chromosomes likely followed on from chromosomal inversions (grey), which were subsequently only carried by males. Over evolutionary time, the lack of sexual recombination led to the appearance of repetitive DNA sequences and short-term expansion. In the longer term, large deletions took place. The outcome of this process is the small, relatively gene poor Y chromosome observed in most eutherian mammals today. Concurrent with this process, X upregulation (XUR) evolved to balance X gene dosage between the single X chromosome and the autosomes in males: this is depicted as the doubled surface area of the X chromosomes in (C) compared to (B). However, XUR was passed on to XX offspring, resulting in X:autosome dosage disparity between males and females. X chromosome inactivation (XCI) then evolved to repress one of the two X chromosomes in XX cells (D). This is depicted as the loss of colour of the X chromosome. Abbreviations: Xa, active X chromosome; Xi, inactive X chromosome.
After the acquisition of SRY, the proto-Y chromosome picked up a number of male-beneficial mutations. As a result of linkage with the testis-determining locus, these mutations provided the selective force to suppress recombination between proto-X and proto-Y [28]. Mechanistically, the suppression and eventual elimination of recombination was possibly achieved by a series of local inversions [29]. Non-recombining regions also accumulated deleterious mutations that could not be repaired. Over evolutionary time, lack of recombination led to the accrual of repetitive DNA sequences and a short-term increase in the size of the chromosome, though this eventually resulted in large deletions and explains the relatively diminutive size of the Y chromosome in many mammals 28, 30. Most genes from the ancestral autosome pair were therefore lost from the Y chromosome, whereas the X chromosome largely maintained its gene content 23, 27, 31. Taking humans as an example, the extant Y chromosome encodes fewer than 78 proteins; in contrast, the X chromosome contains around 800 genes [28].
As a result of the evolution of XY testis determination, female mammals carry two copies of the relatively gene-rich X chromosome, whereas male mammals carry a single copy of the X chromosome and a gene-poor Y chromosome. The difference in X-linked gene dosage between males and females led to the appearance of compensation mechanisms aiming, firstly, to balance X expression with that of the autosomes and, secondly, to balance X expression between the homogametic (XX) and heterogametic (XY) sexes. Susumu Ohno hypothesised that X:autosome balance was achieved in males by X chromosome upregulation (XUR), the two-fold transcriptional upregulation of the X chromosome (Figure 1B,C). However, XUR alone would leave females expressing X genes at twice the level of autosomal genes. In order to correct this X:autosome imbalance, a further step is the inactivation of one X chromosome in the homogametic sex (two of the same sex chromosomes) — X chromosome inactivation (XCI, Figure 1D). In female human embryos, XCI is random, resulting in the silencing of either the maternally derived (Xm) or paternally derived X chromosome (Xp) in each cell 32, 33. The process of XCI is effected by the long non-coding RNAs XIST in eutherians 34, 35, 36 and RSX in metatherians [37]. XIST RNA is expressed from and coats the future inactive X chromosome. Subsequently, a number of other mechanisms lock-in the inactive state, including the histone modification H3K27 tri-methylation 38, 39, DNA methylation 40, 41, 42, and a shift in replication timing relative to the rest of the nucleus 43, 44.
The X chromosome has the potential to cause differences between males and females in a number of ways. Firstly, XCI can be skewed, resulting in preferential expression of either Xm or Xp. Secondly, a number of genes escape XCI and are thus expressed from both X chromosomes. These genes are therefore more highly expressed in XX females compared to XY males, resulting in further potential for cell autonomous sexual dimorphism. Thirdly, the parental origin of the X chromosome in males and females is not equivalent, and differential gene expression between the sexes could result from genomic imprinting.
X Chromosome Inactivation: Mosaicism and Skewing
As a result of XCI, XX females are mosaic, with each cell expressing either Xm- or Xp-genes. A well-known representation of this phenomenon is the tortoiseshell cat, which is a mosaic of black and orange X-linked coat colours [45]. X chromosome mosaicism has long been recognised as a way in which individuals with two X chromosomes differ from those with a single X chromosome, both in terms of normal physiology and disease [46]. Physiologically, XX females express paternal X alleles in 50% of cells, whereas XY males express maternal X alleles in 100% of cells. Any subtle difference in function between the two alleles could therefore manifest as sexual dimorphism (Figure 2). Significant differences in function present as X-linked disease. In males, the presence of a single X chromosome means that X-linked recessive mutations have a fully-penetrant phenotype, but in females this is usually mild or not clinically apparent. X-linked diseases present a range of phenotypes, from relatively benign colour blindness [47], through life-limiting Duchenne and Becker muscular dystrophies [48], to embryonic lethality, as in incontinentia pigmenti [49].
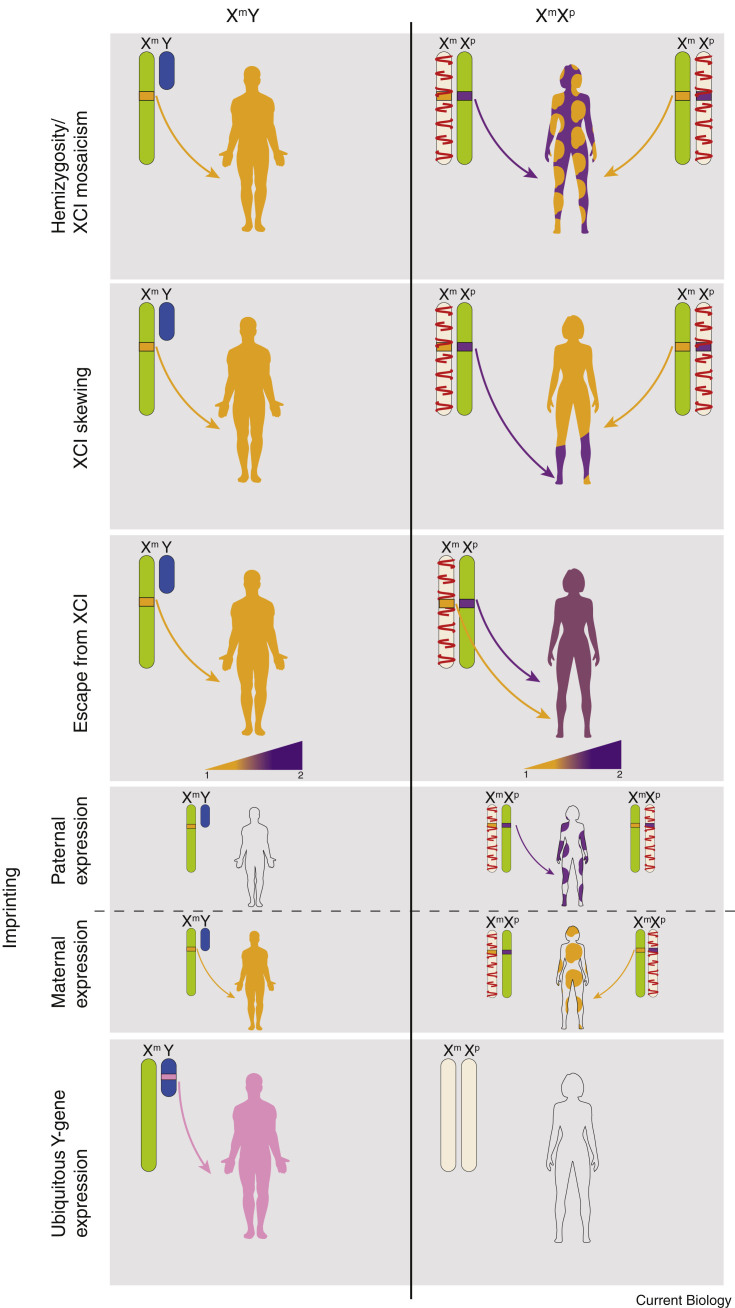
Figure 2.
Possible mechanisms underlying male–female genetic sexual dimorphism in eutherian mammals.
The organism-wide expression of an individual gene allele is represented by block colour, with XY males in the left-hand column and XX females in the right-hand column. (A) A single allele of an X-linked gene is expressed in all cells in the male, whereas due to X chromosome inactivation (XCI), the same allele is only expressed in 50% of cells in the female. (B) XCI skewing can result in a change to the percentage of cells expressing any given X allele in females. (C) As both alleles of XCI escapee genes are expressed in females, the relative expression is increased compared to males. (D) Imprinting resulting in Xp allelic expression would be absent in males due to the absence of Xp, and would be present in 50% of cells in females. Imprinting resulting in Xm allelic expression would be present in all cells in males and 50% of cells in females. (E) Ubiquitously expressed Y-linked genes are only present in males. Abbreviations: Xm, maternally derived X chromosome; Xp, paternally derived X chromosome. Gene expression is depicted in arbitrary units, taking 1 as normal expression for a single chromosome.
Occasionally females also have a typically male disease phenotype, denoted as manifesting heterozygosity. In these individuals, XCI is no longer random, and a skew is present. Such a skew can be classified as either primary, if it arose at the onset of XCI, or secondary if it arose later [50]. There is abundant evidence for the existence of primary skewing in mouse, resulting from the influence of a locus denoted the X controlling element (Xce). Cattanach observed that certain mouse strains have stronger Xces, such that the X chromosomes carrying these Xces are more likely to remain active in F1 hybrid crosses [51]. In humans, there is little evidence for either the presence of an Xce or primary skewing. Some studies have suggested there may be a genetic component to XCI choice, i.e. inferring the existence of a human XCE 52, 53; however, more work is required to definitively address this question.
Secondary skewing has been observed at a population level in humans [53]. In a situation where no primary skew is present, two populations of cells will exist in an XX female: each expressing either the Xm or Xp allele of any given X-linked gene (Figure 2). Each of these alleles may differentially affect cellular growth, such that the rate of proliferation varies between the two populations, and a competition ensues [54]. The cell population with the growth advantage will outgrow the other — usually, but not always, the normal and mutant alleles, respectively 46, 55. Based on evidence in the literature, it is likely that this secondary skewed XCI is largely tissue-specific, and is common in normal, healthy individuals 53, 56. Skewed XCI has also been proposed as one of the causes underlying the female sex bias in autoimmune disease, and in this context it is known as the loss of mosaicism hypothesis [57].
Escape from X Chromosome Inactivation
A number of X-linked genes are not silenced by XCI, and could therefore effect male–female differences in expression (Figure 2). Some of these genes are located within the pseudoautosomal region (PAR), and others have been found outside this region.
The PAR is an area of sequence homology between the X and Y chromosomes [58]. This homology enables X–Y pairing, synapsis, and recombination during meiosis 59, 60, 61. As males and females have PAR genes in equal copy number, it was expected that expression levels would be equivalent between the sexes. However, recent work indicates the existence of a male expression bias in humans [62]. This bias likely results from XCI spreading into the PAR on the inactive X (Xi) in females, but increased expression from the Y-linked genes in males could also contribute. The PAR is perhaps one of the most poorly studied genomic regions in sequenced eutherian mammals, as the assembly quality is not equivalent to the rest of the genome. Further work may build upon the recently reported male expression bias to reveal an unexpected role for the PAR in mammalian sexual dimorphism.
Outside of the human PAR, it has been estimated that around 12% of X genes show consistent escape, and a further 8% escape variably in different individuals and different tissues 62, 63, 64, whereas in mouse, the equivalent numbers are 3% and 4%, respectively 65, 66. The disparity in constitutive escape genes between human and mouse has been attributed to the arrangement of the genes on the chromosome. In mouse, escapees are situated in blocks of only one or two genes, whereas in human these blocks contain 10–15 genes [67]. Even within species, though, the process has inherent variability, creating the potential for sexually dimorphic effects. A recent study in humans showed that only 41% of XCI-escape genes are consistently expressed from both alleles across multiple tissues. For the remainder, inter-tissue variability in XCI-escape was observed. There was also significant variability in Xi gene expression between the two X chromosome haplotypes within an individual. Furthermore, escapee expression from the Xi was on average only one-third of the level of expression from the active X (Xa) [62]. Importantly, 52 of the 67 non-PAR escape genes showed female-biased expression. Taken together, these data suggest that the process of XCI-escape is tightly regulated for some genes and highly variable for others. This may reflect absolute limits on gene dosage for tightly regulated genes and flexibility of expression for those showing variability. The outcomes following XCI, i.e. silencing, variable escape and consistent escape, have the potential to give rise to male–female and female–female phenotypic variation, as evidenced by the female-biased expression of those consistent escapees. More work will be required to elucidate whether expression bias at the RNA level translates into phenotypic sexual dimorphism at the organism level.
Further evidence of the role of escape genes in sexual dimorphism has emerged from studies of human cancers. Many cancers show a sex bias, including those affecting the kidney and renal pelvis, blood, and brain 68, 69, 70, 71. Recent work has associated part of this bias with mutations in genes that escape XCI, so-called escape from X inactivation tumor suppressors (EXITS) [72]. By analysing 21 different tumor types, loss-of-function mutations in X genes ATRX, CNKSR2, DDX3X, KDM5C, KDM6A, and MAGEC3 were found more commonly in males than females [72]. These genes could be tumor suppressors that are required in at least one copy to prevent oncogenesis. The single X gene copy in males is therefore more vulnerable to a mutation event than the two copies present in females [73]. A number of these X genes have Y homologues, which may or may not carry out the same function as the X copy (also see next section). As loss-of-function mutations in X genes were found more commonly in males, this would argue against functional conservation between X and Y genes. The study further investigated whether the Y homologue could also act as a tumor suppressor by looking at chromosome loss instead of mutations. It was found that tumors from female patients with an EXITS gene mutation lose the second X chromosome more commonly than males with an EXITS gene mutation lose the Y chromosome 74, 75, 76. This result suggested that EXITS genes were more effective tumor suppressors than their Y-linked homologues, as male patients received a cancer diagnosis without the loss of Y-linked genes. Moreover, it supports the hypothesis that X–Y gene pairs have functionally diverged, and thus contribute to male–female sexual dimorphism.
X–Y Gene Pairs and the Y Chromosome
Males have a Y chromosome, whereas females do not: this is the most recognisable genetic difference between the mammalian sexes and, therefore, an obvious focus for the study of male–female genetic sexual dimorphism. However, 166 million years of evolution has led the Y chromosome to become specialised for reproduction, with primary roles in testis determination and spermatogenesis [30]. These roles are reflected in gene content and expression patterns, suggesting the Y may have less of an impact on male–female differences outside of the gonad than appearances might initially suggest. For example, SRY drives testis determination and testis specification, resulting in gonadal sex hormone production, and a number of ampliconic genes are expressed exclusively in the testis and contribute to sperm production 23, 30. Nevertheless, a small number of single copy, ubiquitously expressed Y genes have been maintained across the mammalian group and through evolutionary time (Figure 2) 23, 27. Based on Gene Ontology annotations, these genes are involved in the regulation of transcription and translation 23, 27, functions that are acutely sensitive to haploinsufficiency [77]. Furthermore, each of these genes has an X-linked homologue that escapes XCI. It is therefore likely that this group was initially maintained on the ancestral mammalian Y chromosome in order to balance dosage between XX females and XY males 23, 27. This newly ascribed role for the Y chromosome as a ‘balancer’ could mean that the sexes are more similar, if both X and Y genes retain ancestral homologous function [78]. However, emerging evidence suggests a degree of functional divergence in such X–Y gene pairs, as mentioned previously for EXITS genes [72]. This small group of widely expressed genes is therefore of considerable interest in the investigation of male–female sexual dimorphism and the development of new therapeutics. While more work is required to more comprehensively profile the functions of the Y-linked homologues, some of the X-linked genes have already been characterised. Among these, genes KDM5C, KDM6A and DDX3X are of specific relevance. All three genes are conserved across almost all eutherian species and are implicated in sexually dimorphic diseases (Figure 3), including the cancers described above, and X-linked intellectual disability (XLID).
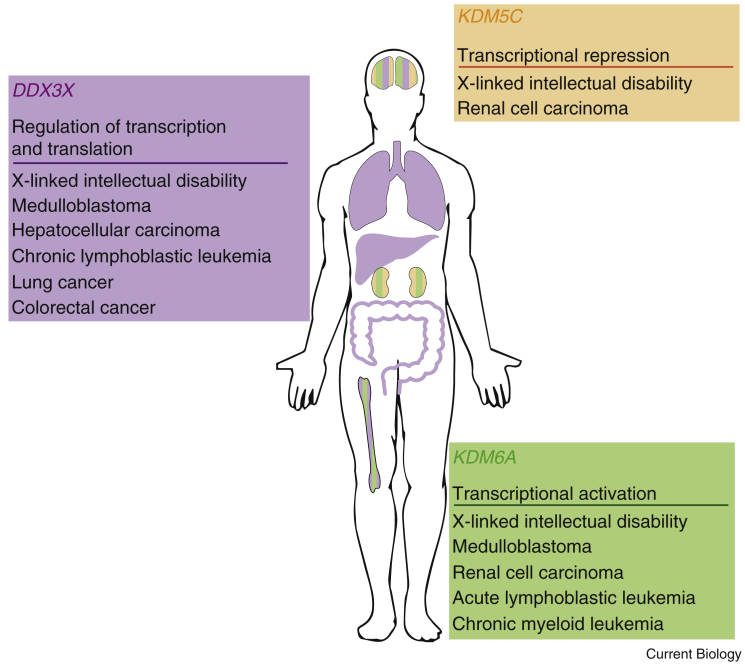
Figure 3.
Genes escaping XCI are implicated in a range of sexually dimorphic diseases.
A small number of X-linked genes show ubiquitous expression, have extant Y-linked homologues, and escape XCI. These genes have roles in the regulation of gene expression and are implicated in male–female sexual dimorphism in a wide range of diseases. Specific organs affected are indicated by the gene-related colours, i.e. KDM5C in orange.
KDM5C is a histone lysine demethylase active at both di- and tri-methylated histone H3 position K4, and has a key role in transcriptional repression 79, 80, 81. Although widely expressed, KDM5C has been clinically implicated in impaired neuronal function and XLID, suggesting a key role in brain development 82, 83, 84, 85, 86, 87, 88, 89. A high proportion of reported KDM5C mutations primarily affect males and, when females are affected, the phenotype is generally less severe. The presence of the XLID phenotype in males suggests that the Y homologue, KDM5D, cannot fully compensate for loss of KDM5C, possibly because of divergence in expression or function of these two genes. The intermediate phenotype observed in heterozygous females carrying a wildtype copy of KDM5C supports the case for dosage sensitivity in this gene, implying that a single wild-type copy is unable to fully compensate for loss of the second copy.
KDM6A (UTX) is a histone lysine demethylase that catalyses removal of H3K27me2 and me3, and possibly also functions as a transcriptional activator 90, 91, 92. Similar to mutations in KDM5C, mutations in KDM6A have been reported in the context of XLID and are linked to the developmental disorder Kabuki syndrome [93]. Males with mutations in KDM6A show a more severe phenotype than females, supporting the hypothesis that the X–Y gene pair KDM6A and KDM6D (UTY) have diverged 94, 95, 96. In mice, males with mutations in Kdm6a survive to birth, whereas females carrying homozygous mutant alleles are lost mid-way through gestation [97]. This result suggests that Uty is able to partly compensate for the loss of Kdm6a in embryonic development. However, mutant males are born at sub-Mendelian frequency and show life-long grow deficiency relative to wild-type littermates, and heterozygous female mutants have no detectable phenotype [97]. Collectively, these data further support the case for divergence between Kdm6a and Kdm6d and, moreover, implicate this gene pair in male–female sexual dimorphism.
DDX3X is a member of the DEAD box protein family, with diverse roles in RNA splicing and export, translation initiation, cell cycle regulation, and apoptosis 98, 99, 100, 101, 102, 103, 104, 105, 106, 107. DDX3X mutations have been described in the context of XLID, almost exclusively affecting females 108, 109, though two recent cases have been described in males with predicted hypomorphic gene variants [110]. The absence of live males carrying loss-of-function mutations in DDX3X can be explained by an absolute requirement for the DDX3X protein product, which is present in heterozygous females but not in mutant males [110]. Additionally, this observation implies that DDX3X has functionally diverged from its Y homologue DDX3Y, as presence of the Y homologue does not facilitate survival. A similar picture is noted in the mouse model, whereby males carrying a mutated Ddx3x allele are lost earlier in embryonic development than heterozygous mutant females [111]. DDX3X/Y functional divergence likely contributes to phenotypic differences between males and females.
X Imprinting
Eutherian embryos derived solely from paternal genomes (androgenotes) or from maternal genomes (gynogenotes) do not survive in utero development 112, 113, 114, 115. The requirement during embryogenesis for a paternal and maternal genome is explained by genomic imprinting, in which genes are expressed monoallelically in a parent-of-origin-specific manner [116]. As females inherit an X chromosome from both parents, whereas males inherit only a maternal X chromosome, imprinted expression of X-linked genes could result in phenotypic differences between the sexes regardless of XCI. For example, a gene expressed only from Xp would be expressed in half the cells in a female, but in no cells in a male. In contrast, a gene expressed only from Xm would be present in 100% of male cells but only half of female cells (Figure 2).
There is emerging evidence for X-linked imprinted genes in mammals. Any effects of X-linked imprinting may be shown more clearly in women with Turner syndrome, who have a single X chromosome (XO), than women with two X chromosomes. Women with Turner syndrome (XO) can inherit their single X chromosome paternally or maternally. Skuse and colleagues observed that XmO women had poorer social and verbal skills than XpO women [117], based on evidence from a set of neuropsychological tests. An imprinted X locus responsible for this effect has not yet been identified. These two populations also differ in their abdominal fat accumulation: XmO women show a typically male distribution of fat, whereas XpO women have a typically female fat distribution [118]. As all males carry a single Xm, whereas the Xp is inherited only by females, this result in XO females is consistent with X-linked imprinting.
In the mouse, a small number of X-linked imprinted genes have been reported, with the majority expressed in the placenta 119, 120, 121 and brain [122]. Similar to the result from humans, Davies and colleagues found that XmO female mice have poorer cognitive function than XpO female mice, as assessed by a reversal learning task. X-linked imprinted genes expressed in the placenta may underlie a growth phenotype long known to affect XpO female embryos. XpO embryos are growth retarded relative to XX littermates at preimplantation [123], egg cylinder (E7.25), and E10.5 stages of development 124, 125, 126. In contrast, XmO embryos, along with XY embryos, are developmentally advanced relative to XX littermates at E10.5 [126]. Interestingly, the ectoplacental cone (part of the early placenta) was found to have reduced volume in XpO conceptuses compared to XX controls [127]. Together, these data strongly suggest a role for parental origin of the X chromosome in mouse embryonic growth. However, despite the identification of a number of X imprinting candidates in other studies 119, 120, 121, no specific locus has yet been linked to the growth deficit phenotype.
X imprinting is still at the nascent stage of development as an area of study and, as such, accurate and appropriate models are not yet in place. Person-to-person variability clouds the picture in humans when looking at the phenotypic level, while at the molecular level, any effect size is likely to be subtle and difficult to detect. The inbred mouse model provides a greater degree of control over genetics and therefore phenotype, though with the caveat of evolutionary distance between mouse and human.
Paradigms for Investigating Cell-Autonomous Sexual Dimorphism
In order to identify the contribution of these mechanisms to male–female differences, relative contributions from genes and gonadal sex hormones must first be teased apart. Traditionally, linkage and association analyses have been used to look for sexual dimorphism in gene expression. Initial genome-wide association studies (GWAS) neglected sex chromosome data in their analyses: the sexually dimorphic expressed quantitative trait loci (eQTLs) they identified associated with polygenic traits such as waist–hip ratio 1, 128, and bone density [129], were autosomal [130]. More recently, X-linked sexually dimorphic eQTLs have been identified, associated with height and fasting insulin [131], and genomic regulatory variation [132]. While association studies are useful for unbiased, observational work at the population level in humans, there is little room for experimental manipulation, and no control for hormonal differences. Herein lies the strength of the mouse as a model organism, which is the most genetically tractable in vivo system available for recapitulating areas of human health and disease.
Mouse Models
In the study of genetic sex differences between males and females, the main confounding factor is gonadal sex hormones. An ideal experimental system would therefore facilitate the separation of these two key variables, in order to attribute phenotypic effects to either genetics or hormones. Such a system would also allow for the manipulation of sex chromosome copy number, enabling further investigation into dosage of escapees from XCI, and functional redundancy in X–Y gene pairs.
A model denoted ‘four core genotypes’ (FCG) achieves the most important aim of the ideal system. It can be used to detect XX versus XY differences that are independent of the gonad and its hormonal influence [133]. The FCG model utilises a Y chromosome with a mutated, inactive copy of Sry (denoted Y−) and an autosomal Sry transgene [25]. As a result, testis determination is separated from inheritance of the Y chromosome, and so XY females and XX males can be generated in addition to XX females and XY males (Figure 4).
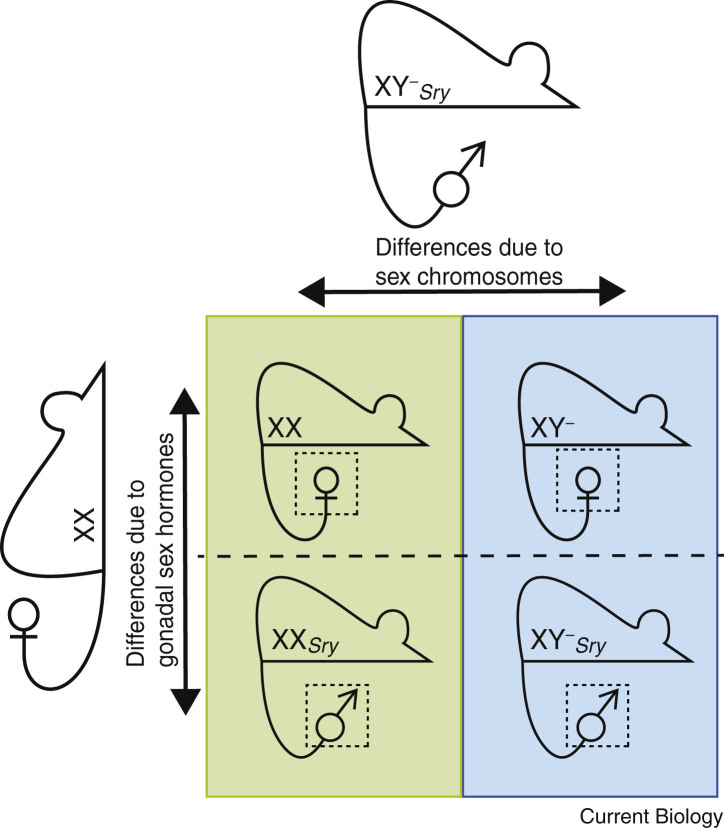
Figure 4.
The Four Core Genotypes (FCG) model.
In this Punnett square, the maternal genotype is depicted on the left-hand side and the paternal genotype is depicted at the top. A gamete from each parent carries a single sex chromosome, which come together to create the two possible offspring genotypes, XX (green column) and XY− (blue column). Furthermore, the father carries Sry as a transgene, the inheritance of which determines the gonadal sex of the offspring: female above the dashed line (XX, XY−), and male below the dashed line (XXSry, XY−Sry). The FCG model can therefore be used to separate sex chromosome effects (XX, green; XY, blue) from gonadal sex hormone effects (ovarian hormones above dotted line, and testicular hormones below dotted line) in mouse.
XX and XY mice with testes have been shown to produce similar levels of gonadal sex hormones to one another, as have XX and XY mice with ovaries [134]. Phenotypic differences are therefore more likely to be attributable to sex chromosome complement 135, 136. In a further modification, the gonads can be removed (gonadectomy), which serves to reduce gonadal sex hormone levels to zero. Following this procedure, further phenotypic differences between XX and XY mice have been identified.
Despite its utility, the FCG model does not control for X chromosome copy number or the presence of a Y chromosome. For example, the XX versus XY− comparison varies the number of X chromosomes, but the presence of a Y chromosome is a confounding factor. When looking to uncover the specific mechanism underlying a sex difference detected by the FCG model, a supplementary model can be used, in which a male with a variant Y chromosome (also known as XY∗ model) generates XX, XO, XY, and XXY offspring [137]. The effects of X chromosome copy number can be tested either in the presence of ovaries (XO versus XX) or testes (XY versus XXY), and the effects of Y chromosome presence can be tested with one (XO versus XY) or two X chromosomes (XX versus XXY). In the latter comparison, however, gonadal sex hormones are not controlled for.
Both the FCG and Y chromosome variant mouse models have been used to great effect in a number of experiments, facilitating the identification of sex differences in cardiovascular disease, metabolism and adiposity, and behaviour. For example, it is well known that men and women differ in their susceptibility to cardiovascular disease, though any mechanism has remained elusive. The FCG and Y chromosome variant models were combined with a mouse model of ischaemia reperfusion injury to mimic myocardial infarction, and removal of the gonads was performed one month prior in order to isolate sex chromosome specific effects [138]. Using both in vivo and ex vivo modelling to characterise infarct size and functional recovery, respectively, hearts from XX mice were found to perform significantly worse than hearts from XY mice, independent of gonadal sex [138]. The presence of a Y chromosome, assessed using gonadectomised variant Y chromosome model mice, seemed to have no effect on outcome. The study concluded that presence of two X chromosomes burdens the organism with greater disease risk than a single X chromosome, and therefore provides a potential focus for future human work.
The FCG model has also been used to identify sex differences in gene regulation. Euchromatic genes are silenced in a proportion of cells following translocation near to regions of heterochromatin, an effect known as position effect variegation (PEV) [139]. Using a heterochromatin-sensitive reporter transgene, Festenstein and colleagues demonstrated X chromosome dosage-dependent differences in PEV [140]. Mice with a single X chromosome showed a greater degree of silencing than those with two X chromosomes. Utilising the FCG and an XO mouse model, this effect was found to be independent of the hormonal milieu or presence of a Y chromosome. Mechanistically, it is possible that the inactive X chromosome acts as a heterochromatic sink, reducing availability of factors required for gene silencing at other heterochromatic loci [140].
Additional evidence for sexually dimorphic gene regulation has recently been observed during mammalian germ cell development. While female and male somatic cells show similar dosage compensation states in the form of XUR 141, 142, 143, the status of XUR in the germline was previously unknown. Sangrithi and colleagues (2017) found that both male and female germ cells initially exhibit upregulation of the active X chromosome. XX female germ cells then exhibited a period of X dosage excess, whereas XY male germ cells showed a period of X dosage decompensation. Interestingly, these patterns were conserved in human germ cells. In order to differentiate sex chromosome dosage effects from gonadal hormone effects, FCG and XO mouse models were used. These data revealed that like XX female germ cells, XX male germ cells had X dosage excess. Furthermore, XO female germ cells had X dosage decompensation, like XY male germ cells [144]. Mammalian germ cells therefore have an X dosage compensation state that is determined by the number of X chromosomes, and not the phenotypic sex or hormonal milieu. This contrast between X dosage compensation state and phenotypic sex could contribute to the infertility widely associated with XXY and XO genotypes and represents an interesting therapeutic angle for future work [144].
By maintaining the same autosomes and hormonal milieu, these mouse models can be used to isolate the phenotypic effects of different sex chromosome complements. However, to better understand cell autonomous differences between men and women, a human experimental system is necessary.
Human Isogenic Cell Lines
It has been known for many years that stem cells can lose sex chromosomes when cultured in vitro for long periods, even when the originating clone was karyotypically normal [145]. Recent work has shown that cellular reprogramming, which generates induced pluripotent stem cells (iPSCs), also results in sex chromosome loss [146]. This process can be utilised to generate iPSC lines that are autosomally isogenic but carry different sex chromosome complements. For example, reprogramming of XXY cells generates XX, XY, XXY, and XO iPSCs. These iPSCs could potentially be screened using a multi-omics approach for differences in transcription, translation and metabolism [147] to characterise cell autonomous sexual dimorphism in the stem cell population. Additionally, iPSCs can be differentiated to study the effects of sex chromosome complement on specific cell types (i.e. as in [148]). Furthermore, mutations of interest could be introduced via genome editing to explore sexually dimorphic gene expression in a given disease model (i.e. as in [149]). Such a system would be excellent for isolating the specific effects of sex chromosome complement on gene expression at a cellular level, fully removing hormonal effects. Models of increasing complexity could then be built up, utilising multiple cell types to form tissues and introducing hormones to further understand the relationship between the two variables.
Summary and Outlook
In 2001, the Institute of Medicine Committee on Understanding the Biology of Sex and Gender Differences reported on why sex matters in human health and disease [150], and a number of recommendations were made. Although seemingly aspirational at the time, significant progress has been made towards “determining the functions and effects of X-chromosome and Y-chromosome-linked genes in somatic cells as well as germ-line cells”. However, it was not until 2014 that the NIH mandated the consideration of sex as a biological variable in grant proposals [151], to partially address the seemingly simple recommendation “(to) determine and disclose the sex of origin of biological research materials”. In this review, we sought to build on this mandate and recommendation, providing an introduction to sex chromosome effects on male–female dimorphism, and describing a number of useful experimental models for hypothesis testing. Through fundamental sex chromosome biology, we have highlighted potential mechanisms underlying cell autonomous differences between the sexes, drawing particular attention to the role of X–Y pairs. The difference between human males and females, at the level of the DNA, is less than 80 genes on the Y chromosome. However, in order to unpick the effects of this genetic difference from that of the gonadal sex hormones and truly understand what it is to be XX versus XY, no single model provides enough data. We must use multiple models in combination. Human population work allows us to make associations between genotype and phenotype, and cell lines provide a faithful recapitulation of human physiology, but at the most basic cellular level. Animal models currently bridge the gap, allowing organism-level investigation and facilitating genetic manipulation. In order to more completely understand genetic sexual dimorphism in the context of human health and disease, we must invest in higher level models that bring together multiple cell types into tissues, tissues into organs and, eventually, organs into organisms.
Acknowledgements
The authors would like to thank Daria Beldzik and Valdone Maciulyte for useful discussions and critical reading of the manuscript.
References
- 1.Randall J.C., Winkler T.W., Kutalik Z., Berndt S.I., Jackson A.U., Monda K.L., Kilpeläinen T.O., Esko T., Mägi R., Li S. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9:e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plavcan J.M. Sexual dimorphism in primate evolution. Am. J. Phys. Anthropol. 2002;116:25–53. doi: 10.1002/ajpa.10011.abs. [DOI] [PubMed] [Google Scholar]
- 3.Arnold A.P. Sex chromosomes and brain gender. Nat. Rev. Neurosci. 2004;5:701–708. doi: 10.1038/nrn1494. [DOI] [PubMed] [Google Scholar]
- 4.Burgoyne P.S., Thornhill A.R., Boudrean S.K., Darling S.M., Bishop C.E., Evans E.P., Capel B., Mittwoch U. The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse [and Discussion] Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995;350:253–261. doi: 10.1098/rstb.1995.0159. [DOI] [PubMed] [Google Scholar]
- 5.Cui W., Ma C.-X., Tang Y., Chang V., Rao P.V., Ariet M., Resnick M.B., Roth J. Sex differences in birth defects: A study of opposite-sex twins. Birth Defect Res. A Clin. Mol. Teratol. 2005;73:876–880. doi: 10.1002/bdra.20196. [DOI] [PubMed] [Google Scholar]
- 6.Konieczny M.R., Senyurt H., Krauspe R. Epidemiology of adolescent idiopathic scoliosis. J. Child. Orthop. 2013;7:3–9. doi: 10.1007/s11832-012-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowell N.R., Burch P.R.J. Genetic origin of some sex differences among human beings. Pediatrics. 1965;36:658–659. [PubMed] [Google Scholar]
- 8.Cao B., Li X.-W., Mao Y., Wang J., Lu H.-Z., Chen Y.-S., Liang Z.-A., Liang L., Zhang S.-J., Zhang B. Clinical features of the initial cases of 2009 pandemic influenza A (H1N1) virus infection in China. N. Engl. J. Med. 2009;361:2507–2517. doi: 10.1056/NEJMoa0906612. [DOI] [PubMed] [Google Scholar]
- 9.Eshima N., Tokumaru O., Hara S., Bacal K., Korematsu S., Tabata M., Karukaya S., Yasui Y., Okabe N., Matsuishi T. Sex- and age-related differences in morbidity rates of 2009 pandemic influenza A H1N1 virus of swine origin in Japan. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019409. e19409–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glezen W.P., Loda F.A., Clyde W.A., Senior R.J., Sheaffer C.I., Conley W.G., Denny F.W. Epidemiologic patterns of acute lower respiratory disease of children in a pediatric group practice. J. Pediatr. 1971;78:397–406. doi: 10.1016/s0022-3476(71)80218-4. [DOI] [PubMed] [Google Scholar]
- 11.Rubtsova K., Marrack P., Rubtsov A.V. Sexual dimorphism in autoimmunity. J. Clin. Invest. 2015;125:2187–2193. doi: 10.1172/JCI78082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cyranowski J.M., Frank E., Young E., Shear M.K. Adolescent onset of the gender difference in lifetime rates of major depression: a theoretical model. Arch. Gen. Psychiatry. 2000;57:21–27. doi: 10.1001/archpsyc.57.1.21. [DOI] [PubMed] [Google Scholar]
- 13.Mielke M.M., Vemuri P., Rocca W.A. Clinical epidemiology of Alzheimer's disease: assessing sex and gender differences. Clin. Epidemiol. 2014;6:37–48. doi: 10.2147/CLEP.S37929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aleman A., Kahn R.S., Selten J.-P. Sex differences in the risk of schizophrenia: evidence from meta-analysis. Arch. Gen. Psychiatry. 2003;60:565–571. doi: 10.1001/archpsyc.60.6.565. [DOI] [PubMed] [Google Scholar]
- 15.Gillies G.E., Pienaar I.S., Vohra S., Qamhawi Z. Sex differences in Parkinson’s disease. Front. Neuroendocrinol. 2014;35:370–384. doi: 10.1016/j.yfrne.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lerner D.J., Kannel W.B. Patterns of coronary heart disease morbidity and mortality in the sexes: a 26-year follow-up of the Framingham population. Am. Heart J. 1986;111:383–390. doi: 10.1016/0002-8703(86)90155-9. [DOI] [PubMed] [Google Scholar]
- 17.Costa M.A. The endocrine function of human placenta: an overview. Reprod. Biomed. Online. 2016;32:14–43. doi: 10.1016/j.rbmo.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Scott H.M., Mason J.I., Sharpe R.M. Steroidogenesis in the fetal testis and its susceptibility to disruption by exogenous compounds. Endocr. Rev. 2009;30:883–925. doi: 10.1210/er.2009-0016. [DOI] [PubMed] [Google Scholar]
- 19.Alonso L.C., Rosenfield R.L. Oestrogens and puberty. Best Pract. Res. Clin. Endocrinol. Metab. 2002;16:13–30. doi: 10.1053/beem.2002.0177. [DOI] [PubMed] [Google Scholar]
- 20.Fuseini H., Newcomb D.C. Mechanisms driving gender differences in asthma. Curr. Allergy Asthma Rep. 2017;17:19. doi: 10.1007/s11882-017-0686-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zein J.G., Erzurum S.C. Asthma is different in women. Curr. Allergy Asthma Rep. 2015;15 doi: 10.1007/s11882-015-0528-y. 363–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi B.G., McLaughlin M.A. Why men's hearts break: cardiovascular effects of sex steroids. Endocrinol. Metab. Clin. North Am. 2007;36:365–377. doi: 10.1016/j.ecl.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Bellott D.W., Hughes J.F., Skaletsky H., Brown L.G., Pyntikova T., Cho T.-J., Koutseva N., Zaghlul S., Graves T., Rock S. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capel B. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat. Rev. Genet. 2017;18:675–689. doi: 10.1038/nrg.2017.60. [DOI] [PubMed] [Google Scholar]
- 25.Gubbay J., Collignon J., Koopman P., Capel B., Economou A., Münsterberg A., Vivian N., Goodfellow P., Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 26.Sinclair A., Berta P., Palmer M., Hawkins J., Griffiths B., Palmer M.S. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 27.Cortez D., Marin R., Toledo-Flores D., Froidevaux L., Liechti A., Waters P.D., Grützner F., Kaessmann H. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508:488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- 28.Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright A.E., Dean R., Zimmer F., Mank J.E. How to make a sex chromosome. Nat. Commun. 2016;7:1–8. doi: 10.1038/ncomms12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes J.F., Page D.C. The biology and evolution of mammalian Y chromosomes. Annu. Rev. Genet. 2015;49:507–527. doi: 10.1146/annurev-genet-112414-055311. [DOI] [PubMed] [Google Scholar]
- 31.Soh Y.Q.S., Alföldi J., Pyntikova T., Brown L.G., Graves T., Minx P.J., Fulton R.S., Kremitzki C., Koutseva N., Mueller J.L. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159:800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petropoulos S., Edsgärd D., Reinius B., Deng Q., Panula S.P., Codeluppi S., Reyes A.P., Linnarsson S., Sandberg R., Lanner F. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1–33. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto I., Patrat C., Thépot D., Peynot N., Fauque P., Daniel N., Diabangouaya P., Wolf J.-P., Renard J.-P., Duranthon V. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- 34.Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R., Willard H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 35.Borsani G., Tonlorenzi R., Simmler M.C., Dandolo L., Arnaud D., Capra V., Grompe M., Pizzuti A., Muzny D., Lawrence C. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 36.Brockdorff N., Ashworth A., Kay G.F., Cooper P., Smith S., McCabe V.M., Norris D.P., Penny G.D., Patel D., Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 37.Grant J., Mahadevaiah S.K., Khil P., Sangrithi M.N., Royo H., Duckworth J., McCarrey J.R., VandeBerg J.L., Renfree M.B., Taylor W. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature. 2012;487:254–258. doi: 10.1038/nature11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., de la Cruz C.C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto I., Otte A.P., Allis C.D., Reinberg D., Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 40.Norris D.P., Brockdorff N., Rastan S. Methylation status of CpG-rich islands on active and inactive mouse X chromosomes. Mamm. Genome. 1991;1:78–83. doi: 10.1007/BF02443782. [DOI] [PubMed] [Google Scholar]
- 41.Pfeifer G.P., Tanguay R.L., Steigerwald S.D., Riggs A.D. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990;4:1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- 42.Pfeifer G.P., Steigerwald S.D., Hansen R.S., Gartler S.M., Riggs A.D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc. Natl. Acad. Sci. USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takagi N., Sugawara O., Sasaki M. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- 44.Sugawara O., Takagi N., Sasaki M. Allocyclic early replicating X chromosome in mice: Genetic inactivity and shift into a late replicator in early embryogenesis. Chromosoma. 1983;88:133–138. doi: 10.1007/BF00327333. [DOI] [PubMed] [Google Scholar]
- 45.Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 46.Migeon B.R. The role of X inactivation and cellular mosaicism in women's health and sex-specific diseases. JAMA. 2006;295:1428–1433. doi: 10.1001/jama.295.12.1428. [DOI] [PubMed] [Google Scholar]
- 47.Deeb S.S. The molecular basis of variation in human color vision. Clin. Genet. 2005;67:369–377. doi: 10.1111/j.1399-0004.2004.00343.x. [DOI] [PubMed] [Google Scholar]
- 48.Guiraud S., Aartsma-Rus A., Vieira N.M., Davies K.E., van Ommen G.-J.B., Kunkel L.M. The pathogenesis and therapy of muscular dystrophies. Annu. Rev. Genom. Human Genet. 2015;16:281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- 49.Ardelean D., Pope E. Incontinentia pigmenti in boys: a series and review of the literature. Pediatr. Dermatol. 2006;23:523–527. doi: 10.1111/j.1525-1470.2006.00302.x. [DOI] [PubMed] [Google Scholar]
- 50.Peeters S.B., Yang C., Brown C.J. Have humans lost control: the elusive X-controlling element. Semin. Cell Dev. Biol. 2016;56:71–77. doi: 10.1016/j.semcdb.2016.01.044. [DOI] [PubMed] [Google Scholar]
- 51.Cattanach B.M., Isaacson J.H. Controlling elements in the mouse X chromosome. Genetics. 1967;57:331–346. doi: 10.1093/genetics/57.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renault N.K.E., Pritchett S.M., Howell R.E., Greer W.L., Sapienza C., rstavik K.H.O., Hamilton D.C. Human X-chromosome inactivation pattern distributions fit a model of genetically influenced choice better than models of completely random choice. Eur. J. Hum. Genet. 2013;21:1–7. doi: 10.1038/ejhg.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amos-Landgraf J.M., Cottle A., Plenge R.M., Friez M., Schwartz C.E., Longshore J., Willard H.F. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am. J. Hum. Genet. 2006;79:493–499. doi: 10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Migeon B.R. Non-random X chromosome inactivation in mammalian cells. Cytogenet. Cell Genet. 1998;80:142–148. doi: 10.1159/000014971. [DOI] [PubMed] [Google Scholar]
- 55.Drenckhahn J.-D., Schwarz Q.P., Gray S., Laskowski A., Kiriazis H., Ming Z., Harvey R.P., Du X.-J., Thorburn D.R., Cox T.C. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev. Cell. 2008;15:521–533. doi: 10.1016/j.devcel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 56.El Kassar N., Hetet G., Brière J., Grandchamp B. X-chromosome inactivation in healthy females: incidence of excessive lyonization with age and comparison of assays involving DNA methylation and transcript polymorphisms. Clin. Chem. 1998;44:61–67. [PubMed] [Google Scholar]
- 57.Libert C., Dejager L., Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat. Rev. Genet. 2010;10:594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 58.Burgoyne P.S. Genetic homology and crossing over in the X and Y chromosomes of Mammals. Hum. Genet. 1982;61:85–90. doi: 10.1007/BF00274192. [DOI] [PubMed] [Google Scholar]
- 59.Koller P.C., Darlington C.D. The genetical and mechanical properties of the sex-chromosomes. J. Genet. 1934;29:159–173. [Google Scholar]
- 60.Moses M.J., Counce S.J., Paulson D.F. Synaptonemal complex complement of man in spreads of spermatocytes, with details of the sex chromosome pair. Science. 1975;187:363–365. [PubMed] [Google Scholar]
- 61.Solari A.J. The behavior of the XY pair in mammals. Int. Rev. Cytol. 1974;38:273–317. doi: 10.1016/s0074-7696(08)60928-6. [DOI] [PubMed] [Google Scholar]
- 62.Tukiainen T., Villani A.-C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carrel L., Cottle A.A., Goglin K.C., Willard H.F. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl. Acad. Sci. USA. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 65.Berletch J.B., Ma W., Yang F., Shendure J., Noble W.S., Disteche C.M., Deng X. Escape from X Inactivation varies in mouse tissues. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005079. e1005079–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berletch J.B., Yang F., Disteche C.M. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Balaton B.P., Dixon-McDougall T., Peeters S.B., Brown C.J. The eXceptional nature of the X chromosome. Hum. Mol. Genet. 2018;27(R2):R242–R249. doi: 10.1093/hmg/ddy148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van der Meulen J., Sanghvi V., Mavrakis K., Durinck K., Fang F., Matthijssens F., Rondou P., Rosen M., Pieters T., Vandenberghe P. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13–21. doi: 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Haaften G., Dalgliesh G.L., Davies H., Chen L., Bignell G., Greenman C., Edkins S., Hardy C., O'Meara S., Teague J. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dalgliesh G.L., Furge K., Greenman C., Chen L., Bignell G., Butler A., Davies H., Edkins S., Hardy C., Latimer C. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson G., Parker M., Kranenburg T.A., Lu C., Chen X., Ding L., Phoenix T.N., Hedlund E., Wei L., Zhu X. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dunford A., Weinstock D.M., Savova V., Schumacher S.E., Cleary J.P., Yoda A., Sullivan T.J., Hess J.M., Gimelbrant A.A., Beroukhim R. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 2016;49:10–16. doi: 10.1038/ng.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knudson A.G. Antioncogenes and human cancer. Proc. Natl. Acad. Sci. USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dumanski J.P., Lambert J.-C., Rasi C., Giedraitis V., Davies H., Grenier-Boley B., Lindgren C.M., Campion D., Dufouil C., Alzheimer’s Disease Initiative Investigators European. Mosaic loss of chromosome Y in blood is associated with alzheimer disease. Am. J. Hum. Genet. 2016;98:1208–1219. doi: 10.1016/j.ajhg.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Forsberg L.A., Rasi C., Malmqvist N., Davies H., Pasupulati S., Pakalapati G., Sandgren J., de Ståhl T.D., Zaghlool A., Giedraitis V. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat. Genet. 2014;46:624–628. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Forsberg L.A. Loss of chromosome Y (LOY) in blood cells is associated with increased risk for disease and mortality in aging men. Hum. Genet. 2017;136:657–663. doi: 10.1007/s00439-017-1799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kondrashov F.A., Koonin E.V. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 2004;20:287–290. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 78.Arnold A.P. Y chromosome’s roles in sex differences in disease. Proc. Natl. Acad. Sci. USA. 2017;114:3787–3789. doi: 10.1073/pnas.1702161114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H.H., Whetstine J.R., Bonni A., Roberts T.M., Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 80.Christensen J., Agger K., Cloos P.A.C., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 81.Tahiliani M., Mei P., Fang R., Leonor T., Rutenberg M., Shimizu F., Li J., Rao A., Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 82.Jensen L.R., Amende M., Gurok U., Moser B., Gimmel V., Tzschach A., Janecke A.R., Tariverdian G., Chelly J., Fryns J.-P. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tzschach A., Lenzner S., Moser B., Reinhardt R., Chelly J., Fryns J.-P., Kleefstra T., Raynaud M., Turner G., Ropers H.-H. Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum. Mutat. 2006;27 doi: 10.1002/humu.9420. 389–389. [DOI] [PubMed] [Google Scholar]
- 84.Santos C., Rodriguez-Revenga L., Madrigal I., Badenas C., Pineda M., Milà M. A novel mutation in JARID1C gene associated with mental retardation. Eur. J. Hum. Genet. 2006;14:583–586. doi: 10.1038/sj.ejhg.5201608. [DOI] [PubMed] [Google Scholar]
- 85.Abidi F.E., Holloway L., Moore C.A., Weaver D.D., Simensen R.J., Stevenson R.E., Rogers R.C., Schwartz C.E. Mutations in JARID1C are associated with X-linked mental retardation, short stature and hyperreflexia. J. Med. Genet. 2008;45:787–793. doi: 10.1136/jmg.2008.058990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rujirabanjerd S., Nelson J., Tarpey P.S., Hackett A., Edkins S., Raymond F.L., Schwartz C.E., Turner G., Iwase S., Shi Y. Identification and characterization of two novel JARID1C mutations: suggestion of an emerging genotype-phenotype correlation. Eur. J. Hum. Genet. 2010;18:330–335. doi: 10.1038/ejhg.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Õunap K., Puusepp-Benazzouz H., Peters M., Vaher U., Rein R., Proos A., Field M., Reimand T. A novel c.2T > C mutation of the KDM5C/JARID1C gene in one large family with X-linked intellectual disability. Eur. J. Hum. Genet. 2012;55:178–184. doi: 10.1016/j.ejmg.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 88.Peng Y., Suryadi J., Yang Y., Kucukkal T.G., Cao W., Alexov E. Mutations in the KDM5C ARID domain and their plausible association with syndromic claes-jensen-type disease. Int. J. Mol. Sci. 2015;16:27270–27287. doi: 10.3390/ijms161126022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fieremans N., Van Esch H., de Ravel T., Van Driessche J., Belet S., Bauters M., Froyen G. Microdeletion of the escape genes KDM5C and IQSEC2 in a girl with severe intellectual disability and autistic features. Eur. J. Hum. Genet. 2015;58:324–327. doi: 10.1016/j.ejmg.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 90.Agger K., Cloos P.A.C., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 91.Lan F., Bayliss P.E., Rinn J.L., Whetstine J.R., Wang J.K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 92.Lee M.G., Villa R., Trojer P., Norman J., Yan K.P., Reinberg D., Croce L.D., Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 93.Adam M.P., Hudgins L. Kabuki syndrome: a review. Clin. Genet. 2005;67:209–219. doi: 10.1111/j.1399-0004.2004.00348.x. [DOI] [PubMed] [Google Scholar]
- 94.Miyake N., Mizuno S., Okamoto N., Ohashi H., Shiina M., Ogata K., Tsurusaki Y., Nakashima M., Saitsu H., Niikawa N. KDM6A point mutations cause Kabuki syndrome. Hum. Mut. 2012;34:108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- 95.Lederer D., Grisart B., Digilio M.C., Benoit V., Crespin M., Ghariani S.C., Maystadt I., Dallapiccola B., Verellen-Dumoulin C. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miyake N., Koshimizu E., Okamoto N., Mizuno S., Ogata T., Nagai T., Kosho T., Ohashi H., Kato M., Sasaki G. MLL2 and KDM6A mutations in patients with Kabuki syndrome. Am. J. Med. Genet. A. 2013;161A:2234–2243. doi: 10.1002/ajmg.a.36072. [DOI] [PubMed] [Google Scholar]
- 97.Shpargel K.B., Sengoku T., Yokoyama S., Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cordin O., Banroques J., Tanner N.K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 99.Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C.L., Kastner B., Stark H., Urlaub H., Lührmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yedavalli V.S.R.K., Neuveut C., Chi Y.-H., Kleiman L., Jeang K.-T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 101.Beckham C., Hilliker A., Cziko A.-M., Noueiry A., Ramaswami M., Parker R. The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol. Biol. Cell. 2008;19:984–993. doi: 10.1091/mbc.E07-09-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shih J.-W., Tsai T.-Y., Chao C.-H., Lee Wu, Y.-H Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene. 2008;27:700–714. doi: 10.1038/sj.onc.1210687. [DOI] [PubMed] [Google Scholar]
- 103.Chang P.-C., Chi C.-W., Chau G.-Y., Li F.-Y., Tsai Y.-H., Wu J.-C., Wu Lee Y.-H. DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control. Oncogene. 2005;25:1991–2003. doi: 10.1038/sj.onc.1209239. [DOI] [PubMed] [Google Scholar]
- 104.Chao C.-H., Chen C.-M., Cheng P.-L., Shih J.W., Tsou A.-P., Lee Y.-H.W. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006;66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- 105.Su C.-Y., Lin T.-C., Lin Y.-F., Chen M.-H., Lee C.-H., Wang H.-Y., Lee Y.-C., Liu Y.-P., Chen C.-L., Hsiao M. DDX3 as a strongest prognosis marker and its downregulation promotes metastasis in colorectal cancer. Oncotarget. 2015;6:18602–18612. doi: 10.18632/oncotarget.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sekiguchi T., Iida H., Fukumura J., Nishimoto T. Human DDX3Y, the Y-encoded isoform of RNA helicase DDX3, rescues a hamster temperature-sensitive ET24 mutant cell line with a DDX3X mutation. Exp. Cell Res. 2004;300:213–222. doi: 10.1016/j.yexcr.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 107.Sun M., Song L., Li Y., Zhou T., Jope R.S. Identification of an antiapoptotic protein complex at death receptors. Cell Death Differ. 2008;15:1887–1900. doi: 10.1038/cdd.2008.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Blok L.S., Madsen E., Juusola J., Gilissen C., Baralle D., Reijnders M.R.F., Venselaar H., Helsmoortel C., Cho M.T., Hoischen A. Mutations in DDX3X are a common cause of unexplained intellectual disability with gender-specific effects on Wnt signaling. Am. J. Hum. Genet. 2015;97:343–352. doi: 10.1016/j.ajhg.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dikow N., Granzow M., Graul-Neumann L.M., Karch S., Hinderhofer K., Paramasivam N., Behl L.-J., Kaufmann L., Fischer C., Evers C. DDX3Xmutations in two girls with a phenotype overlapping Toriello-Carey syndrome. Am. J. Med. Genet. 2017;173:1369–1373. doi: 10.1002/ajmg.a.38164. [DOI] [PubMed] [Google Scholar]
- 110.Kellaris G., Khan K., Baig S.M., Tsai I.-C., Zamora F.M., Ruggieri P., Natowicz M.R., Katsanis N. A hypomorphic inherited pathogenic variant in DDX3X causes male intellectual disability with additional neurodevelopmental and neurodegenerative features. Hum. Genomics. 2018;12:11. doi: 10.1186/s40246-018-0141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen C.-Y., Chan C.-H., Chen C.-M., Tsai Y.-S., Tsai T.Y., Wu Lee Y.H., You L.-R. Targeted inactivation of murine Ddx3x: essential roles of Ddx3x in placentation and embryogenesis. Hum. Mol. Genet. 2016;25:2905–2922. doi: 10.1093/hmg/ddw143. [DOI] [PubMed] [Google Scholar]
- 112.Kaufman M.H., Barton S.C., Surani M.A. Normal postimplantation development of mouse parthenogenetic embryos to the forelimb bud stage. Nature. 1977;265:53–55. doi: 10.1038/265053a0. [DOI] [PubMed] [Google Scholar]
- 113.Surani M.A., Barton S.C. Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos. Science. 1983;222:1034–1036. doi: 10.1126/science.6648518. [DOI] [PubMed] [Google Scholar]
- 114.Surani M.A., Barton S.C., Norris M.L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 115.McGrath J., Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 116.Cleaton M.A.M., Edwards C.A., Ferguson-Smith A.C. Phenotypic outcomes of imprinted gene models in mice: elucidation of pre- and postnatal functions of imprinted genes. Annu. Rev. Genomics. Hum. Genet. 2014;15:93–126. doi: 10.1146/annurev-genom-091212-153441. [DOI] [PubMed] [Google Scholar]
- 117.Skuse D.H., James R.S., Bishop D.V., Coppin B., Dalton P., Aamodt-Leeper G., Bacarese-Hamilton M., Creswell C., McGurk R., Jacobs P.A. Evidence from Turner's syndrome of an imprinted X-linked locus affecting cognitive function. Nature. 1997;387:705–708. doi: 10.1038/42706. [DOI] [PubMed] [Google Scholar]
- 118.Van P.L., Bakalov V.K., Zinn A.R., Bondy C.A. Maternal X chromosome, visceral adiposity, and lipid profile. JAMA. 2006;295:1373–1374. doi: 10.1001/jama.295.12.1373. [DOI] [PubMed] [Google Scholar]
- 119.Li Y., Behringer R.R. Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nat. Genet. 1998;20:309–311. doi: 10.1038/3129. [DOI] [PubMed] [Google Scholar]
- 120.Rodriguez T.A., Sparrow D.B., Scott A.N., Withington S.L., Preis J.I., Michalicek J., Clements M., Tsang T.E., Shioda T., Beddington R.S.P. Cited1 is required in trophoblasts for placental development and for embryo growth and survival. Mol. Cell Biol. 2004;24:228–244. doi: 10.1128/MCB.24.1.228-244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shi W., van den Hurk J.A., Alamo-Bethencourt V., Mayer W., Winkens H.J., Ropers H.-H., Cremers F.P.M., Fundele R. Choroideremia gene product affects trophoblast development and vascularization in mouse extra-embryonic tissues. Dev. Biol. 2004;272:53–65. doi: 10.1016/j.ydbio.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 122.Davies W., Isles A., Smith R., Karunadasa D., Burrmann D., Humby T., Ojarikre O., Biggin C., Skuse D., Burgoyne P. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat. Genet. 2005;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- 123.Banzai M., Omoe K., Ishikawa H., Endo A. Viability, development and incidence of chromosome anomalies of preimplantation embryos from XO mice. Cytogenet. Genome Res. 1995;70:273–277. doi: 10.1159/000134050. [DOI] [PubMed] [Google Scholar]
- 124.Burgoyne P.S., Tam P., Evans E.P. Retarded development of XO conceptuses during early pregnancy in the mouse. J. Reprod. Fertil. 1983;68:387–393. doi: 10.1530/jrf.0.0680387. [DOI] [PubMed] [Google Scholar]
- 125.Ishikawa H., Banzai M., Yamauchi T. Developmental retardation of XO mouse embryos at mid-gestation. J. Reprod. Fertil. 1999;115:263–267. doi: 10.1530/jrf.0.1150263. [DOI] [PubMed] [Google Scholar]
- 126.Thornhill A.R., Burgoyne P.S. A paternally imprinted X chromosome retards the development of the early mouse embryo. Development. 1993;118:171–174. doi: 10.1242/dev.118.1.171. [DOI] [PubMed] [Google Scholar]
- 127.Jamieson R.V., Tan S.-S., Tam P.P. Retarded postimplantation development of X0 mouse embryos: impact of the parental origin of the monosomic X chromosome. Dev. Biol. 1998;201:13–25. doi: 10.1006/dbio.1998.8972. [DOI] [PubMed] [Google Scholar]
- 128.MAGIC. Heid I.M., Jackson A.U., Randall J.C., Winkler T.W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M.C., Speliotes E.K. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu C.-T., Estrada K., Yerges-Armstrong L.M., Amin N., Evangelou E., Li G., Minster R.L., Carless M.A., Kammerer C.M., Oei L. Assessment of gene-by-sex interaction effect on bone mineral density. J. Bone Miner. Res. 2012;27:2051–2064. doi: 10.1002/jbmr.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chang D., Gao F., Slavney A., Ma L., Waldman Y.Y., Sams A.J., Billing-Ross P., Madar A., Spritz R., Keinan A. Accounting for eXentricities: analysis of the X chromosome in GWAS reveals X-linked genes implicated in autoimmune diseases. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113684. e113684–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tukiainen T., Pirinen M., Sarin A.-P., Ladenvall C., Kettunen J., Lehtimäki T., Lokki M.-L., Perola M., Sinisalo J., Vlachopoulou E. Chromosome X-wide association study identifies loci for fasting insulin and height and evidence for incomplete dosage compensation. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004127. e1004127–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kukurba K.R., Parsana P., Balliu B., Smith K.S., Zappala Z., Knowles D.A., Favé M.-J., Davis J.R., Li X., Zhu X. Impact of the X chromosome and sex on regulatory variation. Genome Res. 2016;26:768–777. doi: 10.1101/gr.197897.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.De Vries G.J., Rissman E.F., Simerly R.B., Yang L.-Y., Scordalakes E.M., Auger C.J., Swain A., Lovell-Badge R., Burgoyne P.S., Arnold A.P. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J. Neurosci. 2002;22:9005–9014. doi: 10.1523/JNEUROSCI.22-20-09005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen X., McClusky R., Chen J., Beaven S.W., Tontonoz P., Arnold A.P., Reue K. The number of X chromosomes causes sex differences in adiposity in mice. PLoS Genet. 2012;8:e1002709. doi: 10.1371/journal.pgen.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gatewood J.D. Sex chromosome complement and gonadal sex influence aggressive and parental behaviors in mice. J. Neurosci. 2006;26:2335–2342. doi: 10.1523/JNEUROSCI.3743-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Smith-Bouvier D.L., Divekar A.A., Sasidhar M., Du S., Tiwari-Woodruff S.K., King J.K., Arnold A.P., Singh R.R., Voskuhl R.R. A role for sex chromosome complement in the female bias in autoimmune disease. J. Exp. Med. 2008;205:1099–1108. doi: 10.1084/jem.20070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cox K.H., Bonthuis P.J., Rissman E.F. Mouse model systems to study sex chromosome genes and behavior: Relevance to humans. Front. Neuroendocrinol. 2014;35:405–419. doi: 10.1016/j.yfrne.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li J., Chen X., McClusky R., Ruiz-Sundstrom M., Itoh Y., Umar S., Arnold A.P., Eghbali M. The number of X chromosomes influences protection from cardiac ischaemia/reperfusion injury in mice: one X is better than two. Cardiovasc. Res. 2014;102:375–384. doi: 10.1093/cvr/cvu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weiler K.S., Wakimoto B.T. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 140.Wijchers P.J., Yandim C., Panousopoulou E., Ahmad M., Harker N., Saveliev A., Burgoyne P.S., Festenstein R. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Dev. Cell. 2010;19:477–484. doi: 10.1016/j.devcel.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 141.Deng X., Hiatt J.B., Nguyen D.K., Ercan S., Sturgill D., Hillier L.W., Schlesinger F., Davis C.A., Reinke V.J., Gingeras T.R. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 2011;43:1179–1185. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lin H., Halsall J.A., Antczak P., O'Neill L.P., Falciani F., Turner B.M. Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno's hypothesis. Nat. Genet. 2011;43:1169–1170. doi: 10.1038/ng.992. [DOI] [PubMed] [Google Scholar]
- 143.Yildirim E., Sadreyev R.I., Pinter S.F., Lee J.T. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat. Struct. Mol. Biol. 2011;19:56–61. doi: 10.1038/nsmb.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sangrithi M.N., Royo H., Mahadevaiah S.K., Ojarikre O., Bhaw L., Sesay A., Peters A.H.F.M., Stadler M., Turner J.M.A. Non-canonical and sexually dimorphic X dosage compensation states in the mouse and human germline. Dev. Cell. 2017;40:289–301.e3. doi: 10.1016/j.devcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Robertson E.J., Evans M.J., Kaufman M.H. X-chromosome instability in pluripotential stem cell lines derived from parthenogenetic embryos. J. Embryol. Exp. Morphol. 1983;74:297–309. [PubMed] [Google Scholar]
- 146.Hirota T., Ohta H., Powell B.E., Mahadevaiah S.K., Ojarikre O.A., Saitou M., Turner J.M.A. Fertile offspring from sterile sex chromosome trisomic mice. Science. 2017;357:932–935. doi: 10.1126/science.aam9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gnecchi M., Stefanello M., Mura M. Induced pluripotent stem cell technology: Toward the future of cardiac arrhythmias. Int. J. Cardiol. 2017;237:49–52. doi: 10.1016/j.ijcard.2017.03.085. [DOI] [PubMed] [Google Scholar]
- 149.Calatayud C., Carola G., Consiglio A., Raya A. Modeling the genetic complexity of Parkinson's disease by targeted genome edition in iPS cells. Curr. Opin. Genet. Dev. 2017;46:123–131. doi: 10.1016/j.gde.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 150.Institute of Medicine (US) Committee on Understanding the Biology of Sex and Gender Differences, Wizemann, T.M., and Pardue, M.-L. (2001). Exploring the Biological Contributions to Human Health: Does Sex Matter? (Washington DC: National Academies Press (US).) [PubMed]
- 151.Clayton J.A., Collins F.S. Policy: NIH to balance sex in cell and animal studies. Nature. 2014;509:282–283. doi: 10.1038/509282a. [DOI] [PMC free article] [PubMed] [Google Scholar]