Abstract
Despite breakthroughs in immune checkpoint inhibitor (ICI) immunotherapy, not all human cancers respond to ICI immunotherapy and a large fraction of patients with the responsive types of cancers do not respond to current ICI immunotherapy. This clinical conundrum suggests that additional immune checkpoints exist. We report here that interferon regulatory factor 8 (IRF8) deficiency led to impairment of cytotoxic T lymphocyte (CTL) activation and allograft tumor tolerance. However, analysis of chimera mice with competitive reconstitution of WT and IRF8-KO bone marrow cells as well as mice with IRF8 deficiency only in T cells indicated that IRF8 plays no intrinsic role in CTL activation. Instead, IRF8 functioned as a repressor of osteopontin (OPN), the physiological ligand for CD44 on T cells, in CD11b+Ly6CloLy6G+ myeloid cells and OPN acted as a potent T cell suppressor. IRF8 bound to the Spp1 promoter to repress OPN expression in colon epithelial cells, and colon carcinoma exhibited decreased IRF8 and increased OPN expression. The elevated expression of OPN in human colon carcinoma was correlated with decreased patient survival. Our data indicate that myeloid and tumor cell–expressed OPN acts as an immune checkpoint to suppress T cell activation and confer host tumor immune tolerance.
Keywords: Immunology
Keywords: Cancer immunotherapy, Immunotherapy, T cells
Introduction
CD8+ cytotoxic T lymphocytes (CTLs) are the central component of the host adaptive immune system. A typical CD8+ T cell immune response starts from T cell receptor (TCR) recognition of cognate antigenic peptides presented by the MHC class I molecule. The interaction between the TCR and the antigen–MHC class I complex, in coordination with costimulatory signals delivered by interactions between costimulatory receptor CD28 on T cells and costimulatory ligands B7.1 and/or B7.2 on antigen-presenting cells (APCs), induces CD8+ T cell activation (1–3). However, T cell–mediated cytotoxicity must spare destruction of normal cells and maintain self-tolerance, which is accomplished by several corepressive mechanisms and also through receptor-ligand interactions between CD8+ T cells and APCs. The major corepressive receptor, programmed cell death protein 1 (PD-1), interacts with programmed cell death protein 1 ligand 1 (PD-L1) and/or PD-L2 expressed by APCs (4–6), resulting in dephosphorylation of both CD28 and TCR and repression of T cell activation (4, 7–9). In addition, CTLA-4, LAG-3, and TIM-3 also act as corepressive receptors to keep chronically activated effector T cells in check (10). These corepressive receptors thus function as immune checkpoints to maintain the balance during the CD8+ T cell adaptive immune response.
CTLs are the primary immune cells that act to eradicate tumors (11, 12). On one hand, CTLs recognize tumor cells through tumor-specific antigens to mount an antitumor immune response and suppress tumor progression. However, tumor cells often mount a counterattack by multiple mechanisms including loss of antigen expression, and both primary and acquired resistance mechanisms, which in turn shut down the tumor-reactive CTLs in the tumor microenvironment (13–16). Therefore, tumor cells hijack the corepressive receptor-based immune checkpoint mechanism to suppress tumor-reactive CTLs to avoid immune rejection (17–20). Based on this mechanism, antibody-based inhibitors to block CTLA-4 and PD-1/PD-L1 immune checkpoints have been developed and shown durable efficacy in many types of human cancers (21–23). However, not all human cancers respond to these ICI immunotherapies and a large fraction of patients in the responsive cancer types also do not respond favorably. This clinical conundrum suggests that there may exist other immune checkpoints that suppress CTL function in tumor rejection. We aimed to test this hypothesis and observed that interferon regulatory factor 8 (IRF8) functions as a repressor of osteopontin (OPN) expression and loss of IRF8 expression in CD11b+Ly6CloLy6G+ myeloid cells and colon tumor cells leads to elevated expression of OPN that acts as a potent T cell suppressor. Our data demonstrate that IRF8 regulates T cell activation through repressing OPN expression, and myeloid and tumor cells use the silencing of IRF8 expression to upregulate the expression of OPN, which functions as an immune checkpoint to suppress CTL activation to promote host tumor immune tolerance and tumor immune evasion.
Results
IRF8 null mice tolerate allograft tumor.
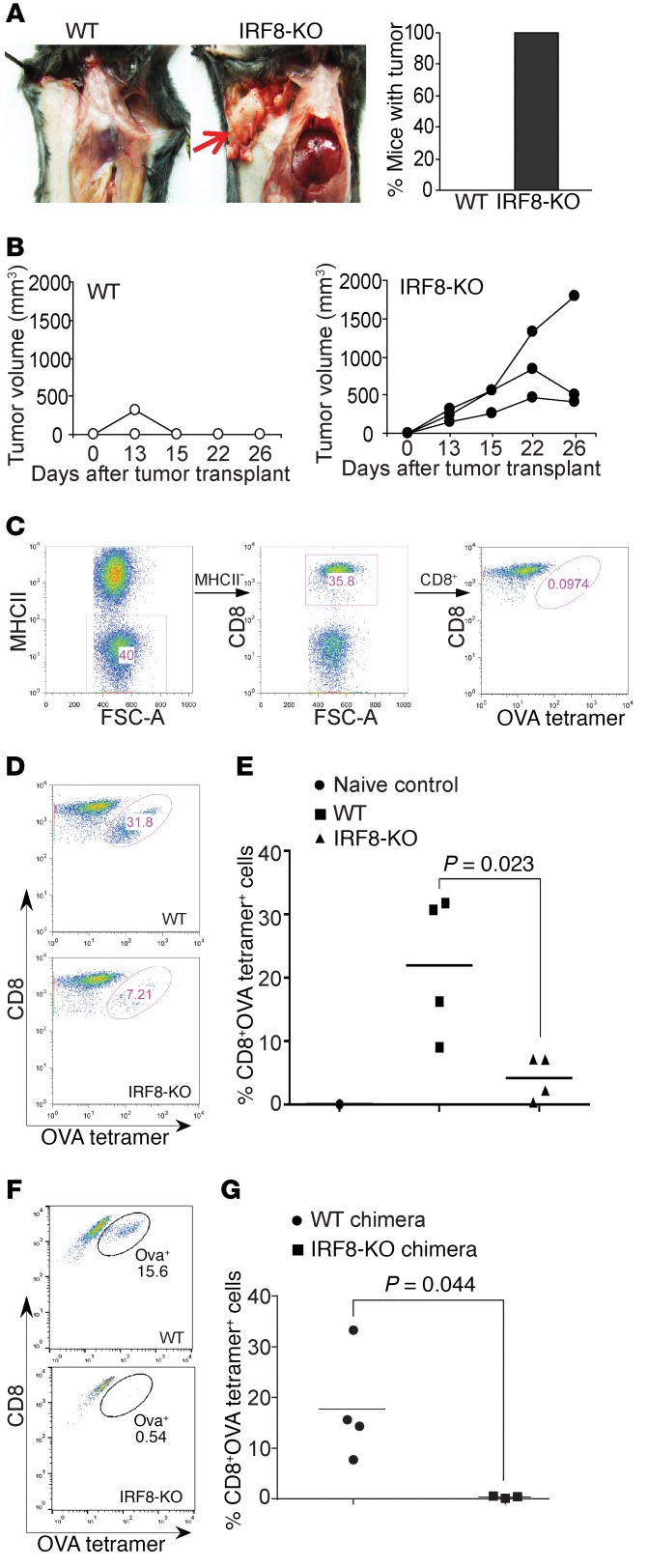
4T1 tumor cells, a mouse tumor cell line of BALB/c origin, were orthotopically injected into the mammary gland of WT C57BL/6 mice and IRF8 knockout (IRF8-KO) mice of C57BL/6 origin. 4T1 tumors grew initially in the WT C57BL/6 mice, but were quickly rejected within 2 weeks after tumor transplant (Figure 1, A and B). Surprisingly, 4T1 tumors continued growing and formed relatively large tumors in all IRF8-KO mice (Figure 1, A and B).
Figure 1. IRF8 is essential for tumor rejection and antigen-specific CD8+ T cell activation.
(A) The BALB/c mouse–derived mammary carcinoma 4T1 cells (1 × 104 cells/mouse) were injected into the mammary gland of WT (C57BL6/J, n = 4) and IRF8-KO (C57BL/6, n = 3) mice. Mice were sacrificed at day 26 and dissected for examination of tumor presence. The image is representative of WT and IRF8-KO mice. The red arrow indicates location of 4T1 tumor. The right panel shows percentage of mice with tumor. Shown are representative images of 1 of 3 independent experiments. (B) Tumor growth was monitored over time. Each line represents the tumor growth kinetics of an individual mouse. (C–E) WT (n = 4) and IRF8-KO (n = 4) mice were vaccinated with OVA peptide, followed by a boost with the same peptide regime 14 days later. Peripheral blood was collected 7 days after boost and stained with MHCII-, CD8-, and OVA tetramer–specific antibodies. MHCII–CD8+ cells were gated for OVA tetramer+ cells. Naive C57BL/6 mice were used as negative and gating controls (C). FSC-A, forward scatter–area. Shown are representative plots of one pair of WT and IRF8-KO mice from 1 of 2 independent experiments (D). The tetramer+ CD8+ T cells were quantified (E). (F) WT C57BL/6 and IRF8-KO BM cells were adoptively transferred into lethally irradiated C57BL/6 recipient mice to recreate chimera mice with IRF8 deficiency only in the hematopoietic cells. The chimera WT (n = 4) and IRF8-KO (n = 3) mice were vaccinated as in A–C and analyzed for OVA-specific CD8+ T cells. Shown are representative plots from one pair of mice. (G) Quantification of OVA-specific CD8+ T cells in WT and IRF8-KO chimera mice.
IRF8-deficient mice are deficient in generation of antigen-specific CD8+ T cells.
Allograft rejection is mediated by host T cells (24). The above observations thus suggest that IRF8 deficiency might lead to T cell functional deficiency in the IRF8-KO mice (25). To test this hypothesis, we made use of the ovalbumin (OVA) peptide vaccination system to determine IRF8 function in T cell response to antigen in vivo. WT and IRF8-KO mice were vaccinated with OVA peptide to activate CD8+ T cells. As expected, WT mice responded to the OVA peptide robustly to generate OVA-specific CD8+ T cells (Figure 1, C–E). In contrast, IRF8-KO mice exhibited a significantly decreased response to generate OVA-specific CD8+ T cells (Figure 1, D and E). A complementary approach was then taken to validate this finding. IRF8-KO chimera mice with IRF8 deficiency only in hematopoietic cells, and control WT chimera mice were vaccinated with the OVA vaccine. The WT chimera mice responded efficiently as determined by generation of OVA-specific CD8+ T cells (Figure 1F). Consistent with what was observed in IRF8-KO mice, the IRF8-KO chimera mice also generated significantly fewer OVA-specific CD8+ T cells (Figure 1, F and G). Our data thus indicate that global deletion of Irf8 in mice leads to deficiency in the generation of antigen-specific CD8+ T cells in vivo.
IRF8-deficient CD8+ T cells have a CD44hi memory T cell phenotype.
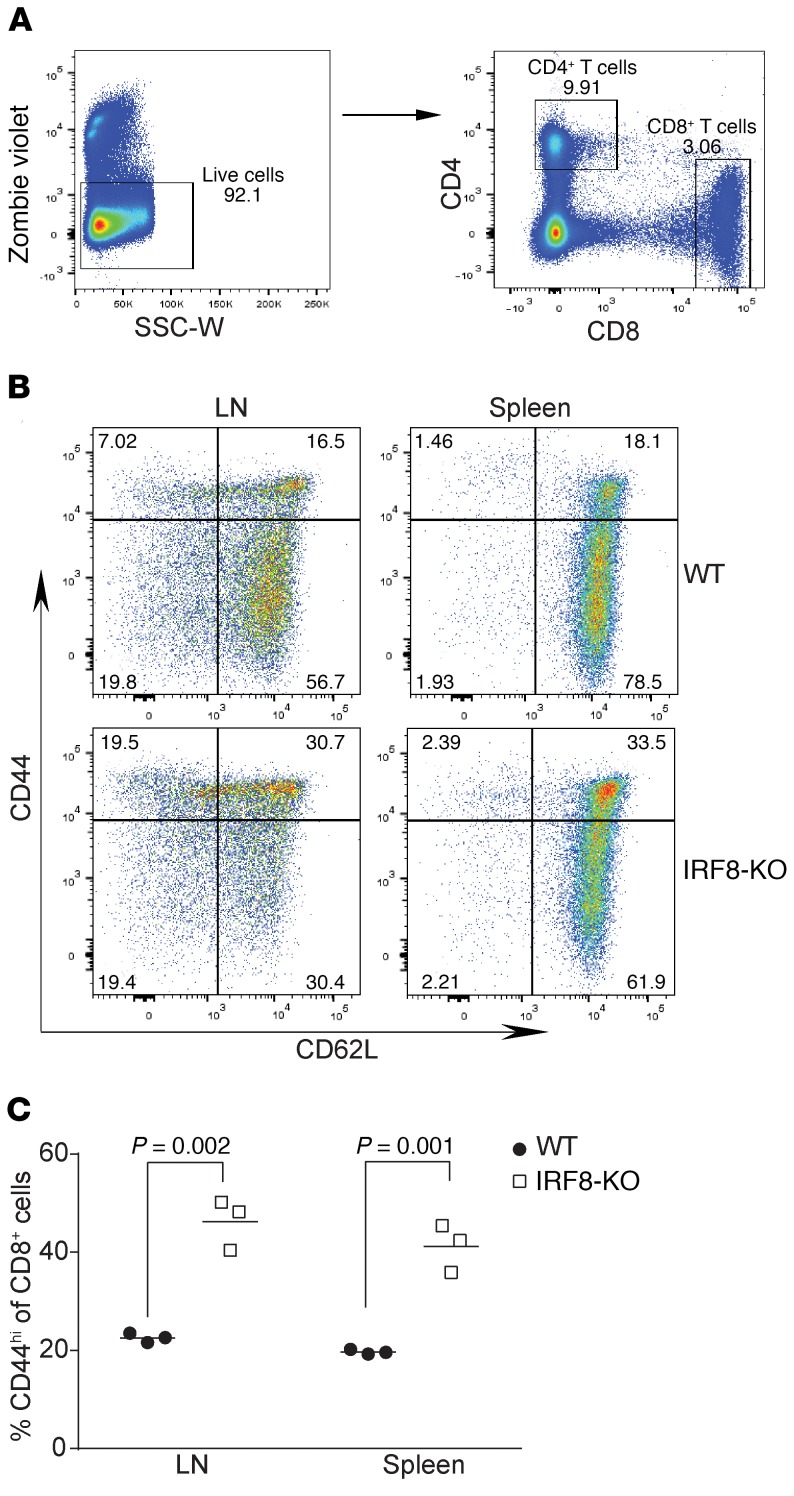
To identify the cellular mechanisms underlying why IRF8-deficient CD8+ T cells fail to be activated in response to antigen in vivo, we performed flow cytometric analysis of cell surface markers on CD8+ T cells comparing those from WT to IRF8-KO mice and identified that the CD44 level is markedly different between the 2 populations (Figure 2, A and B). The percentage of the subset of CD44hi cells is significantly higher on CD8+ T cells in lymphoid organs of IRF8-KO mice compared with WT mice (Figure 2C).
Figure 2. IRF8 deficiency increases CD44hiCD8+ memory T cells.
(A) Peripheral blood cells were stained with Zombie violet to exclude dead cells and the live cells were analyzed for CD4+ and CD8+ T cells. SSC-W, side scatter–width. (B) LN and spleen cells were collected from WT (n = 3) and IRF8-KO (n = 3) mice. The CD8+ cells gated out as in A were further analyzed for CD44hi cells with CD62L as reference. Shown are representative plots of 1 pair of the mice. (C) The percentage of CD8+CD44hi cells as shown in A were quantified.
IRF8 regulates OPN expression in myeloid cells.
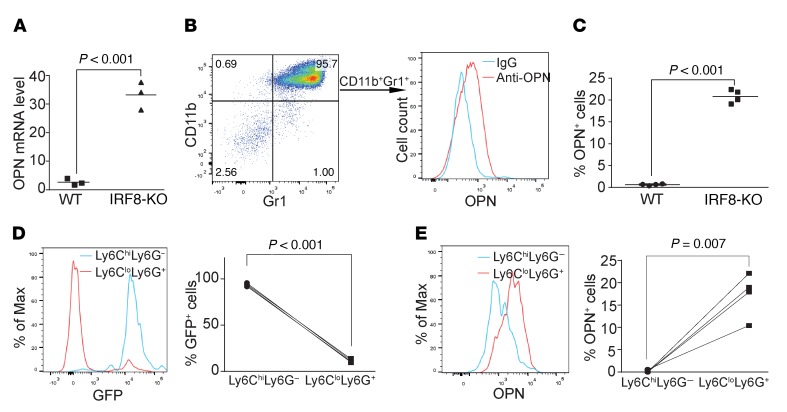
CD44 is known to interact with various ligands, which are crucial for its cellular function (26, 27). The above observation that IRF8 deficiency leads to significantly increased CD44hi CD8+ T cells in mice suggests that CD44 may contribute to the deficiency of CD8+ T cell activation. To test this hypothesis, we first analyzed the expression level of major CD44 ligands in spleen cells. Hyaluronic acid is considered the major ligand for CD44 (28). Quantitative polymerase chain reaction (qPCR) analysis of total spleen cells indicated that the expression levels of the major genes encoding enzymes of the hyaluronic acid metabolism pathways, including Has1, Has2, Has3, Hyal1, Hyal2, Hyal3, and Hyal5, are not significantly different between WT and IRF8-KO mice (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI123360DS1). OPN is a secreted matricellular protein that also acts as the physiological ligand for CD44 (29). qPCR analysis revealed that total spleen cells from IRF8-KO mice expressed a more than 10-fold higher level of OPN than WT spleen cells (Figure 3A). To determine what types of cells express OPN, spleen cells were intracellularly stained for OPN concomitantly with surface staining for B cells (CD19), T cells (CD3), and myeloid cells (CD11b and Gr1). Gating OPN+ cells revealed that about 95% of OPN+ cells are CD11b+Gr1+ in IRF8-KO mice. Therefore, we determined that these OPN+ cells are primarily CD11b+Gr1+ myeloid cells (Figure 3B). IRF8-KO mice have a significantly higher level of OPN+ myeloid cells than WT mice (Figure 3C).
Figure 3. IRF8 represses the expression of OPN expression in myeloid cells.
(A) RNA was prepared from total spleens of WT (n = 3) and IRF8-KO (n = 3) mice and analyzed by qPCR for OPN mRNA level. (B) Spleen cells of WT (n = 3) and IRF8-KO (n = 3) mice were stained with CD19-, CD3-, CD11b-, and Gr1-specific mAbs, followed by intracellular staining of OPN. The OPN+ cells were gated to show the CD11b+Gr1+ myeloid cells from IRF8-KO mice (left panel). OPN protein level in these CD11b+Gr1+ myeloid cells is shown in the right panel. (C) The OPN+ cells in total spleen cells of WT (n = 3) and IRF8-KO (n = 3) mice as shown in B were quantified. (D) Spleen cells from IRF8-GFP mice were stained with Ly6G- and Ly6C-specific mAbs. Ly6ChiLy6G+ and Ly6CloLy6G– cells were overlaid for GFP intensity. Shown are representative plots of 1 of 3 mice (left panel). The GFP+ cells of the Ly6ChiLy6G+ and Ly6CloLy6G– cells were quantified and are presented in the right panel. (E) The spleen cells were stained with Ly6C- and Ly6G-specific mAbs, followed by intracellular staining with OPN-specific antibody. The Ly6ChiLy6G+ and Ly6CloLy6G– cells were gated and analyzed for OPN expression level. Representative plot of 1 of 3 mice is shown. The OPN+ cells in Ly6ChiLy6G+ and Ly6CloLy6G– cells were quantified and are presented in the right panel.
A complementary approach was used to further determine the relationship between IRF8 and OPN. Myeloid cells in the IRF8-GFP reporter mice (30) were analyzed for GFP intensity (a surrogate marker for IRF8 protein level) and OPN expression level. CD11b+Ly6CloLy6G+ myeloid cells are GFP– myeloid cells (Figure 3D) that have a significantly higher percentage and level of OPN+ cells than the GFP+ CD11b+Ly6ChiLy6G– myeloid cells (Figure 3, D and E). Thus, IRF8 expression level is inversely correlated with OPN expression level under physiological conditions.
OPN inhibits T cell activation in vitro.
The observations that IRF8-KO mice have a significantly higher percentage of CD44hi CD8+ T cells than WT mice, and that the OPN expression level is significantly higher in the CD11b+Gr1+ myeloid cells in IRF8-KO mice than those in WT mice, suggest that the CD44-OPN axis may suppress CD8+ T cell activation. To test this hypothesis, we cultured T cells in the presence of recombinant OPN protein and analyzed T cell proliferation. Indeed, OPN protein reproducibly inhibited CD8+ T cell activation and proliferation in a dose-dependent manner (Figure 4, A and B). Consistent with the inhibited T cell proliferation, OPN inhibited IFN-γ production by T cells in vitro (Figure 4C). To determine whether OPN inhibits T cell activation, we analyzed T cell activation markers. OPN decreased CD69+CD8+ T cells as early as 2 hours after stimulation (Figure 4D). Similarly, the levels of CD25- and PD-1–expressing CD8+ T cells were also decreased by OPN (Figure 4D). Taken together, these data indicate that OPN is highly expressed in CD11b+Ly6CloLy6G+ myeloid cells and acts as a potent suppressor for CD8+ T cell activation.
Figure 4. OPN inhibits T cell activation in vitro.
(A) CD3+ T cells from WT mouse spleen were labeled with CFSE and cultured in plates coated with anti-CD3 (0.8 μg/ml) and anti-CD28 (10 μg/ml) mAbs and OPN at the indicated concentrations for 3 days. Cells were then stained with CD8-specific mAb and CD8+ T cells were analyzed for CFSE intensity. The CFSE labeled and unstimulated cells were used as control. Representative data of cells from 1 of the 3 mice are shown. (B) CFSE intensity as shown in A was quantified as division index. (C) CD3+ T cells were cultured in plates coated with anti-CD3 (0.8 μg/ml) and anti-CD28 (10 μg/ml) mAbs and OPN at the indicated concentrations in triplicate for 3 days. Culture supernatant was collected and measured for IFN-γ protein level by ELISA. Data from B and C were analyzed using a 1-way ANOVA, with Dunnett’s test for multiple comparisons. (D) CD3+ T cells were cultured in plates coated with anti-CD3 (0.8 μg/ml) and anti-CD28 (10 μg/ml) mAbs in the presence of IgG (5 μg/ml) or OPN (1 μg/ml and 5 μg/ml, respectively). Cells were collected at the indicated time points, stained with CD69-, CD25-, PD-1–, and CD8-specific mAbs, and analyzed by flow cytometry. Data are mean ± SD. Significance was calculated using a 2-way ANOVA with Tukey’s test.
IRF8 regulates antigen-specific CD8+ T cell activation by a cell-extrinsic mechanism.
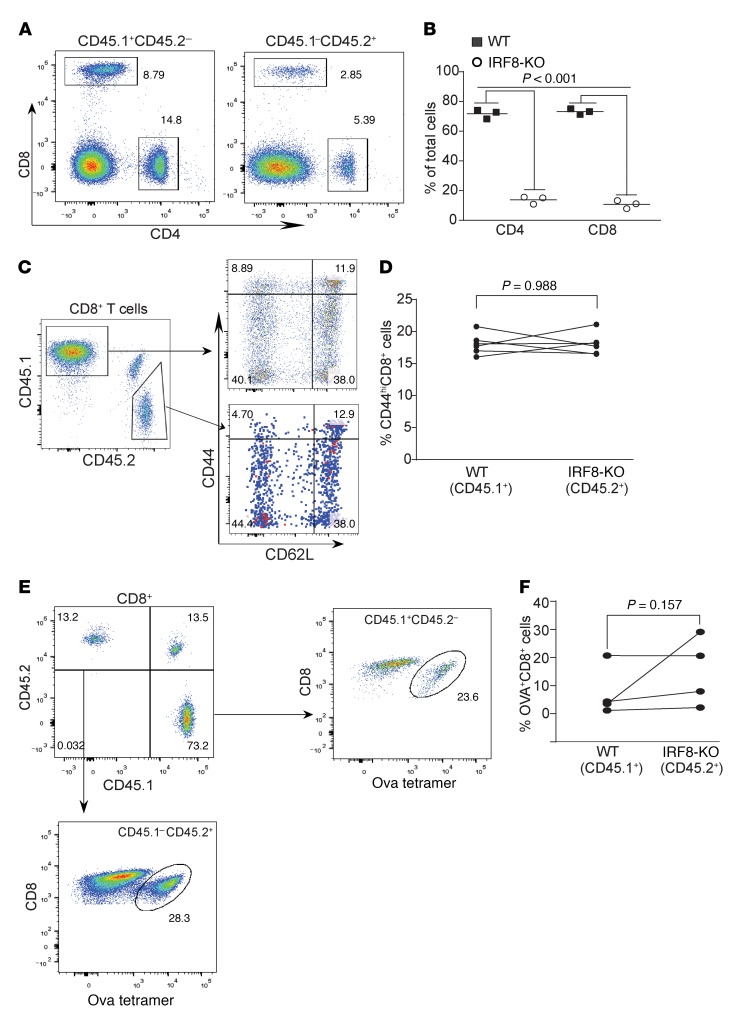
The above observations that IRF8-KO mice are deficient in response to vaccine to generate antigen-specific CD8+ T cells and that OPN expression level is elevated in CD11b+Ly6CloLy6G+ myeloid cells in IRF8-KO mice suggest that IRF8 might regulate CD8+ T cell response to antigen in a cell-extrinsic manner. To test this hypothesis, we analyzed antigen-specific CD8+ T cell response in competitive mixed bone marrow (BM) chimeras. Chimeric mice were generated by transplanting a mixture of Irf8–/– (CD45.2+) BM cells and BM cells from WT SJL (B6.SJL-Ptprca Pepcb/BoyJ) congenic donors (CD45.1+) into irradiated F1 congenic recipients (CD45.1+CD45.2+) (Supplemental Figure 2). Because BM in IRF8-KO BM contains a higher level of CD11b+Gr1+ myeloid cells than WT mice, the number of BM cells injected from Irf8–/– and WT animals was mixed at adjusted ratios (2:1 of Irf8–/–/WT) (31). We first analyzed the development of mature T cells 8 weeks after transplantation. Although it has been reported that the total numbers of CD4+ and CD8+ T cells are not markedly different between IRF8-KO and WT mice in the lymphoid organs (32), the Irf8–/– BM cells exhibit a competitive disadvantage over the WT BM cells in both CD4+ and CD8+ T cell maturation. The levels of Irf8–/– CD4+ and CD8+ T cells are significantly lower than the WT CD4+ and CD8+ T cells in the mixed BM chimeras (Figure 5, A and B). We repeated this competitive mixed BM chimera experiment at a ratio of 5:1 of Irf8–/–/WT, and still observed that the levels of Irf8–/– CD4+ and CD8+ T cells are significantly lower than the WT CD4+ and CD8+ T cells in the mixed BM chimeras. Furthermore, unlike the WT and IRF8-KO mice, there is no significant difference in CD44 expression level between WT and Irf8–/– CD8+ T cells in the mixed BM chimeras 8 weeks after BM cell transplantation (Figure 5, C and D).
Figure 5. IRF8 regulates antigen-specific CD8+ T cell differentiation and activation in a cell-extrinsic manner.
(A) Competitive mixed BM chimeras were created by adoptively transferring SJL (CD45.1+) WT whole BM cells with Irf8–/– BM cells into lethally irradiated C57BL/6×SJL) F1 recipients (CD45.1+CD45.2+). Peripheral blood cells were collected from WT and IRF8-KO mixed BM chimera mice, stained with CD45.1-, CD45.2-, CD4-, and CD8-specific mAbs, and analyzed by flow cytometry. Shown are representative plots of phenotypes of WT (CD45.1) and IRF8-KO (CD45.2) CD4+ and CD8+ T cells in the mixed BM chimeras. (B) The CD4+ and CD8+ cells from WT (CD45.1) and IRF8-KO (CD45.2) as shown in A were quantified. (C) Blood cells from WT and IRF8-KO mixed BM chimera mice were stained with CD45.1-, CD45.2-, CD8, CD44-, and CD62L-specific mAbs. CD8+ T cells were gated out for CD45.1 and CD45.2 cells. The WT and IRF8-KO CD8+ cells were then analyzed for CD44hi and CD62L+ cells. Representative plots of 1 of 3 mice are shown. (D) The percentage of CD44hi cells of the WT CD8+ and IRF8-KO CD8+ T cells was quantified. (E) WT (CD45.1) and IRF8-KO (CD45.2) mixed BM chimera mice were vaccinated with OVA peptide, followed by a boost with OVA peptide 14 days later. Peripheral blood was collected 7 days after boost and stained with MHCII-, CD8-, and OVA tetramer–specific antibodies. MHCII-CD8+ cells were gated for OVA tetramer+ cells. Shown are representative plots of OVA-specific WT and IRF8-KO CD8+ T cells. (F) The WT and IRF8-KO CD8+ OVA-specific T cells as shown in E were quantified.
Next, the mixed BM chimeras were vaccinated with the OVA peptide regimen. Using the same analysis strategy as in the WT and IRF8-KO mice (Figure 1), CD8+ T cells in the MHC class II– cell population were further gated into CD45.1+ (WT) and CD45.2+ (Irf8–/–) cells (Figure 5E). Analysis of OVA+ cells indicates that, although Irf8–/– CD8+ T cells are at a lower level, these Irf8–/– CD8+ T cells respond to the vaccine as efficiently as the WT CD8+ T cells in the same host (Figure 5E). There is no significant difference in the percentage of OVA-specific WT and Irf8–/– CD8+ T cells (Figure 5F).
Antigen-specific CD8+ T cell differentiation and allograft tumor tolerance is independent of intrinsic IRF8 function in T cells.
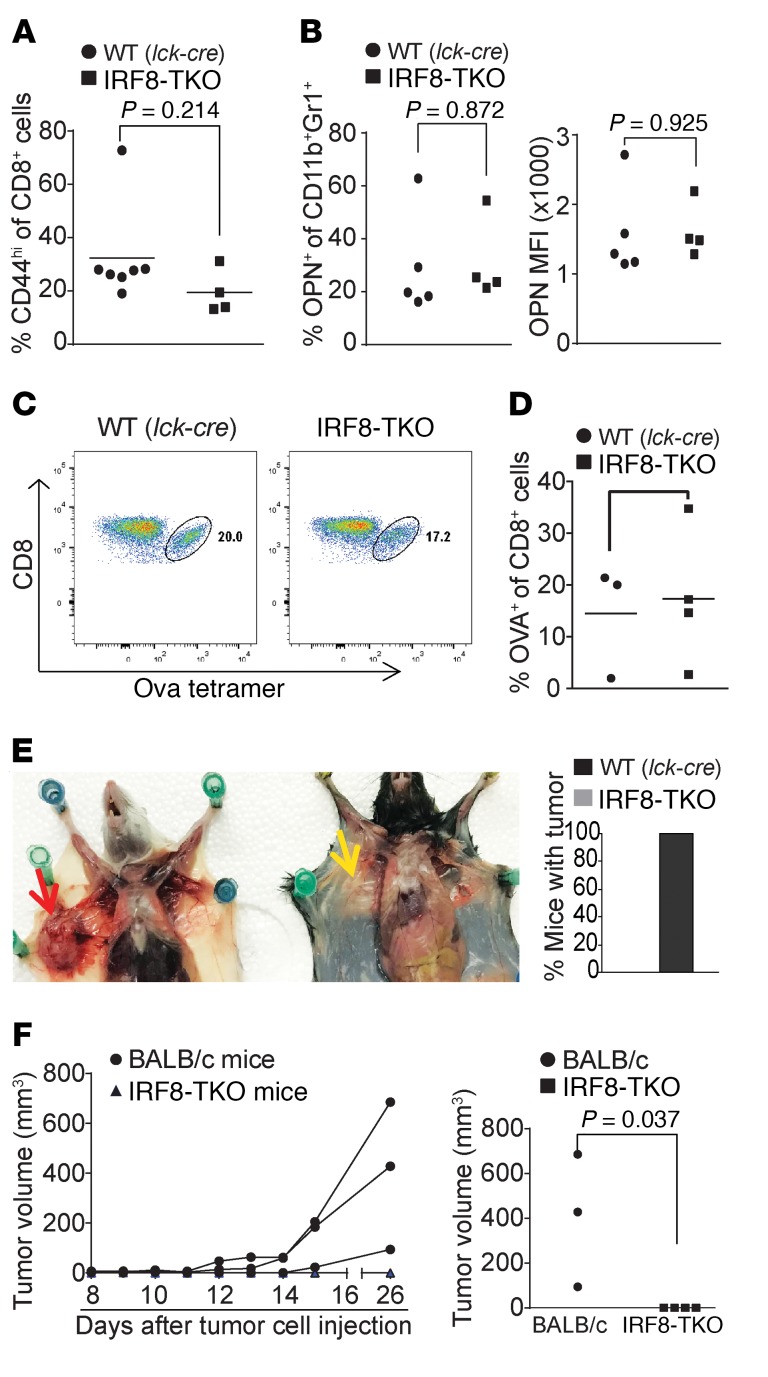
A complementary approach was then used to strengthen our above finding that the deficiency in generation of antigen-specific CD8+ T cells in IRF8-KO mice is not due to intrinsic IRF8 function. We developed a mouse model with IRF8 deficiency only in T cells (IRF8-TKO). Unlike the IRF8-KO mice, IRF8-TKO mice have a similar CD44hiCD8+ T cell phenotype as the WT (Lck-cre+/–Irf8+/+) mice (Figure 6A). Furthermore, there are no significant differences in the percentage of OPN+CD11b+Gr1+ myeloid cells. OPN protein levels of CD11b+Gr1+ myeloid cells are also not significantly different between IRF8-TKO and WT mice (Figure 6B). Consistent with the normal CD44 and OPN expression patterns, IRF8-TKO mice responded to OVA peptide vaccination in a similar degree as the WT mice in the generation of OVA-specific CD8+ T cells (Figure 6, C and D).
Figure 6. Mice with IRF8 deficiency only in T cells exhibit no deficiency in generation of antigen-specific CD8+ T cells and reject allograft tumor.
(A) Blood cells were collected from WT (Lck-cre+/–Irf8+/+, n = 7) and IRF8-TKO (n = 4) mice. Cells were stained with CD8- and CD44-specific mAbs and analyzed by flow cytometry. The CD8+ and CD44hi cells were quantified. Column: mean; bar: SD. (B) Spleen cells were collected from WT (Lck-cre+/–Irf8+/+, n = 7) and IRF8-TKO (n = 4) stained with CD11b- and Gr1-specific mAbs, followed by intracellular staining with OPN-specific mAb. The CD11b+Gr1+ cells were then gated and analyzed for percentage of OPN+ cells (left panel) and OPN MFI (right panel). (C) WT (Lck-cre+/–Irf8+/+, n = 4) and IRF8-TKO (n = 3) mice vaccinated with OVA peptide, followed by a boost with OVA peptide 14 days later. Peripheral blood was collected 7 days after boost and stained with MHCII-, CD8-, and OVA tetramer–specific antibodies. MHCII-CD8+ cells were gated for OVA tetramer+ cells. Shown are representative plots of OVA-specific CD8+ T cells in WT and IRF8-TKO mice. (D) WT and IRF8-KO CD8+ OVA-specific T cells as shown in C were quantified. (E) 4T1 cells (1 × 104 cells/mouse) were injected into the mammary gland of BALB/c (n = 3) and IRF8-TKO (C57BL/6, n = 4) mice. Mice were sacrificed at day 26 and dissected for examination of tumor presence. Shown is a representative image of 4T1 tumor-bearing BALB/c and 4T1 tumor-challenged IRF8-TKO mice. The red arrow indicates location of the 4T1 tumor. Yellow area indicates lack of tumor in injected area. The right panel shows percentage of mice with tumor. (F) Tumor growth was monitored over time and the tumor growth kinetics is presented in the left panel. Each line represents the tumor growth kinetics of an individual mouse. The tumor size at day 31 after tumor injection is presented in the right panel.
To determine whether IRF8-TKO mice tolerate an allograft tumor, 4T1 tumor cells were injected into IRF8-TKO mice. 4T1 tumor cells were also injected into WT BALB/c mice as an autograft tumor control. As expected, the 4T1 tumor grew aggressively in syngeneic BALB/c mice (Figure 6E). However, unlike what was observed in IRF8-KO mice that tolerate the allograft 4T1 tumor (Figure 1), IRF8-TKO mice rejected the allograft 4T1 tumor completely (Figure 6F). Taken together, these data demonstrate that IRF8 regulates antigen-specific CD8+ T cell activation and allograft tumor tolerance in a cell-extrinsic manner, and myeloid cell–expressed OPN may suppress CD8+ T cell activation in vivo.
IRF8 also represses OPN expression in colon epithelial cells.
The above findings determined that OPN is a potent suppressor of T cells and IRF8 functions as a repressor of OPN expression in CD11b+Ly6CloLy6G+ myeloid cells. In addition to being silenced in myeloid cells such as the CD11b+Ly6CloLy6G+ myeloid cells (Figure 3, D and E), IRF8 is often silenced in colon carcinoma cells by DNA methylation (33), which raises the possibility that tumor cells may also use silencing IRF8 expression as a mechanism to upregulate OPN to CTL activation in the tumor microenvironment. To test this hypothesis, we made use of a spontaneous azoxymethane-dextran sodium sulphate (AOM-DSS) colon cancer mouse model. Normal colon tissues and colon tumors were collected and analyzed by qPCR. As expected, IRF8 is significantly downregulated in colon tumor tissues as compared with normal colon (Figure 7A). Consistent with what was observed in the CD11b+Ly6CloLy6G+ myeloid cells, OPN expression is significantly upregulated in the tumor tissues as compared with the normal colon in vivo (Figure 7B). Normal colon epithelial cells have a high level of IRF8 protein, but IRF8 protein is undetectable in the colon tumor cells (Figure 7C). Consistent with the elevated OPN mRNA level in the colon tumor tissue (Figure 7B), the OPN protein level is significantly higher in serum of the AOM-DSS–induced colon tumor–bearing mice as compared with tumor-free mice (Figure 7D).
Figure 7. IRF8 functions as a transcriptional repressor of OPN in colon epithelial cells.
(A and B) Total RNA was isolated from mouse colon (n = 5) and AOM-DSS–induced colon carcinoma (n = 3) tissues and analyzed by qPCR for IRF8 (A) and OPN (B) expression levels. Each dot represents data from one mouse. Significance was determined using the nonparametric Mann-Whitney U test. (C) Colon tissues (c1 and c2) from tumor-free IRF8-GFP reporter mice (n = 3) and tumor tissues (c3) from AOM-DSS–induced colon tumor mice (n = 3) were collected analyzed for GFP intensity under a confocal microscope. Scale bars: 100 μM (c1 and c3) and 20 μM (c2). Shown are representative images of each group. (D) Serum was collected from tumor-free (n = 6) and AOM-DSS–induced colon tumor-bearing (n = 5) mice and analyzed for OPN protein level by ELISA. (E) The Spp1 promoter structure showing the 2 putative ISRE consensus sequence elements. The ChIP PCR-amplified regions are also indicated. +1 indicates Spp1 gene transcription initiation site. (F) Normal mouse colon tissues were analyzed by ChIP using IgG (negative control) and anti-IRF8 antibody. The Spp1 promoter–specific qPCR-amplified regions are indicated at the top panel. The ChIP qPCR were normalized to input DNA. (G) CD3+ T cells were stimulated on anti-CD3– and anti-CD28–coated plates for 3 days. Nuclear extracts were prepared and analyzed for IRF8 binding by using EMSA with the Pdcd1 promoter ISRE consensus sequence DNA probe (Supplemental Table 1). Anti-IRF8 antibody was used to identify the IRF8-DNA complexes. IgG was used as a negative control. Red arrows point to the IRF8-Pdcd1 ISRE DNA complexes. (H) Nuclear extract was prepared from normal mouse colon and incubated with the 2 ISRE DNA probes as shown in D. The unlabeled Pdcd1 ISRE DNA probe (cold probe) was used at the indicated amount (fold over the labeled Spp1 ISRE probes) to compete the Spp1 IRRE probes. Green arrow points IRF8-DNA complex.
IRF8 functions as either a transcriptional activator or repressor depending on its associated protein factors and the target gene promoter consensus sequence (34). We analyzed the mouse Spp1 gene promoter and identified 2 putative IRF8 consensus interferon-stimulated response elements (ISRE1 and ISRE2) (Figure 7E). Analysis of normal colon tissues by chromatin immunoprecipitation (ChIP) detected IRF8 association with the ISRE consensus sequence chromatin at the Spp1 promoter region (Figure 7F). As a complementary approach, electrophoretic mobility shift assay (EMSA) was then used to determine IRF8 binding to the 2 putative ISRE elements of the Spp1 promoter. IRF8-specific antibody did not supershift the IRF8-DNA complexes when nuclear extracts from the colon were used. To determine the specific colon epithelial cell IRF8 protein complex–DNA interaction, we used a cold DNA probe competition approach. A DNA probe containing the ISRE consequence sequence element of the mouse Pdcd1 promoter (Supplemental Table 1) was incubated with nuclear extracts from activated CD3+ T cells. Two protein-DNA complexes were detected and anti-IRF8 antibody displaced them (Figure 7G), indicating IRF8 binding to this DNA probe. We next used the Pdcd1 promoter ISRE-containing DNA probe to compete the Spp1 promoter ISRE probes. Two major protein-DNA complexes were detected when colon nuclear extract was incubated with Spp1 ISRE1 and ISRE2 DNA probes (Figure 7G) and cold Pdcd1 ISRE DNA probe competed away the 2 protein-DNA complexes (Figure 7H). These observations determined that IRF8 protein binds to the ISRE elements at the Spp1 promoter to repress OPN expression in colon epithelial cells.
OPN is elevated in human colon cancer patient periphery and is correlated with decreased disease-specific survival.
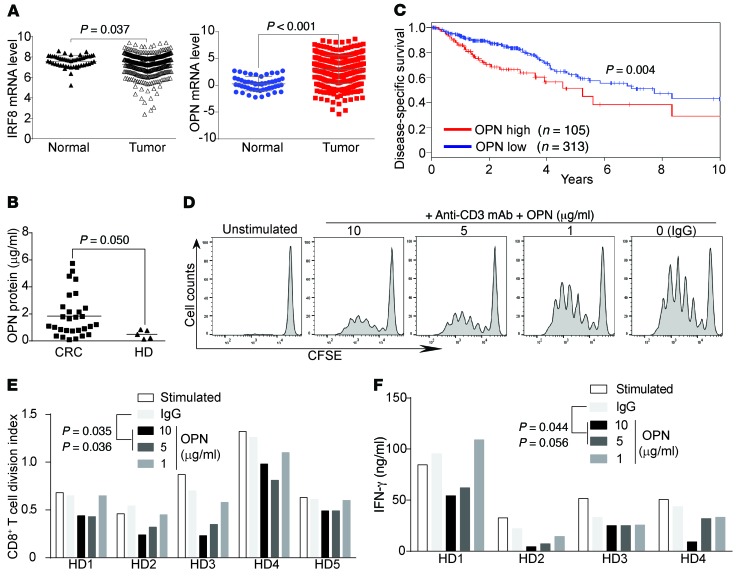
To determine whether the above findings can be translated to human colon cancer patients, mRNA expression data sets of colon cancer and matched normal tissues were extracted from The Cancer Genome Atlas (TCGA) data sets. IRF8 expression level is significantly downregulated in human colon carcinoma as compared with normal colon, whereas OPN expression is significantly upregulated in human colon carcinoma as compared with the normal colon (Figure 8A). As OPN is a secreted protein, it is therefore possible that OPN protein level may be elevated in patient periphery. To test this hypothesis, we analyzed serum specimens from healthy donors and patients with colon cancer. As expected, OPN protein level is significantly higher in serum from patients with colon cancer as compared with healthy donors (Figure 8B). We determined that OPN is a potent suppressor of CD8+ T cells (Figure 4, A and B) and CD8+ T cells are the primary adaptive immune cells of the host cancer immunosurveillance. It is thus likely that elevated OPN will decrease host cancer immunosurveillance to promote tumor progression. To test this hypothesis, we analyzed OPN expression level and colon cancer patient clinical outcomes. Indeed, Kaplan-Meier survival analysis revealed that OPN expression level is inversely correlated with survival time of patients with colon cancer (Figure 8C). To determine whether OPN inhibits human CD8+ T cell activation, human T cells were activated with anti-CD3 mAb in the absence or presence of recombinant human OPN protein. Analysis of cellular proliferation indicated that OPN significantly inhibited human CD8+ T cell proliferation at a concentration of 5 μg/ml (Figure 8, D and E). Consistent with the decreased proliferation, OPN also significantly inhibited IFN-γ secretion by human CD8+ T cells (Figure 8F). Taken together, our data indicate that OPN is an inhibitor of human CD8+ T cells and OPN expression is elevated in human colon cancer and secreted into the periphery. OPN promotes human colon cancer progression.
Figure 8. OPN is elevated in human colon carcinoma and inversely correlated with patient survival.
(A) IRF8 and OPN mRNA expression data sets in normal colon and colon carcinoma tissues were extracted from TCGA database and compared as indicated. (B) Serums were collected from healthy donors and patients with colon cancer, and analyzed for OPN protein level by ELISA. Each dot represents serum OPN protein level from 1 donor or patient. (C) OPN mRNA expression levels in human patients with colon cancer were extracted from TCGA database and analyzed by Kaplan-Meier survival analysis. (D) CD3+ human T cells were purified from healthy donors, labeled with CFSE, and cultured in plates coated with anti-CD3 (1 μg/ml) mAb and OPN at the indicated concentrations for 3 days. Cells were then stained with CD8-specific mAb and CD8+ T cells were analyzed for CFSE intensity. The CFSE-labeled and unstimulated cells were used as control. Representative data of cells from 1 of 5 donors are shown. (E) CFSE intensity as shown in D was quantified as division index. Data from 5 healthy donors (HD1-HD5) are shown. (F) Human CD3+ T cells were cultured in plates coated with anti-CD3 (1 μg/ml) mAb and OPN at the indicated concentrations for 3 days. Culture supernatant was collected and measured for IFN-γ protein level by ELISA. Data from 4 healthy donors are shown. Statistical significance for each treatment in E and F was determined by ANOVA, using Dunett’s test for multiple comparisons.
Discussion
T cell activation, particularly the transition from the resting stage of naive T cells to the functional effector phase of CD8+ cytotoxic T cells, requires marked changes in gene expression, which is regulated by T cell–specific transcription factors (35). In addition to the well-characterized T-box transcription factor (T-bet) and eomesodermin (Eomes) (36, 37), IRF8 has recently emerged as another key transcription factor that regulates CD8+ T cell activation and differentiation (38). IRF8 was previously identified as a lineage-specific transcription factor for myeloid cell differentiation. Mice with a null mutation of IRF8 exhibit massive accumulation of CD11b+Gr1+ immature myeloid cells, revealing an essential role of IRF8 in myelopoiesis (32). IRF8 regulates myeloid cell differentiation through repressing granulocyte development and promoting differentiation of monocytic cells such as dendritic cells and macrophages (39–43). IRF8 functions in hematopoietic cell differentiation and activation have since been extended to other hematopoietic cells, including B, NK, and T cells (25, 31, 41, 43–56). However, IRF8 functions in T cells are apparently tissue- and disease-specific (25, 30, 38, 46, 53, 57). The mechanism underlying IRF8 function in regulation of T cell activation is largely unknown. Here, we determined that the intrinsic IRF8 function is not required for T cell activation, and that IRF8 regulates T cell activation through repressing the expression of OPN in myeloid cells and tumor cells.
IRF8 is an essential lineage-specific transcription factor for myeloid cell differentiation (41), and IRF8 deficiency leads to impaired differentiation of myeloid cells, including plasmacytoid dendritic cells (pDCs), CD8α+ conventional DCs, and CD103+ DCs (39, 40, 49, 58). IRF8-KO mice are deficient in the generation of effector T cells in both the EAE and the experimental graft-versus-host disease (GVHD) mouse models (57, 59), but the underlying mechanism is unknown. In this study, we observed that IRF8-KO mice do not respond to antigenic stimulation to generate antigen-specific effector CD8+ T cells. Therefore, it is possible that IRF8 deficiency may diminish APCs to impair host immune responses to generate antigen-specific T cells in vivo. Analysis of APCs in IRF8-KO mice revealed that although the levels of several subsets of APCs are decreased, IRF8-KO mice still have substantial levels of conventional DCs, CD8α- DCs, and CD8a+ DCs (Supplemental Figure 3). Furthermore, IRF8-KO mice exhibited enhanced expansion of Th17 cells and developed more severe inflammation in the experimental colitis mice (46), suggesting that IRF8-KO mice do have functional APCs and are capable of generating effector T cells. Therefore, although IRF8 deficiency may decrease the levels of certain APC populations, a decreased level of APCs is unlikely the main cause of impairment of CD8+ T cell activation and immune tolerance against allograft tumor challenge in IRF8-KO mice.
IRF8 is not expressed in naive T cells, but is rapidly induced by T cell receptor (TCR) stimulation and γc-cytokine (30, 59), suggesting a role of IRF8 in T cell activation and differentiation. Indeed, knocking out IRF8 abrogated naive CD8+ T cell differentiation into effector cells in the experimental GVHD mouse model (59). Furthermore, in the EAE mouse model, while all WT mice responded to myelin oligodendrocyte glycoprotein (MOG) peptide vaccination and showed clear signs of EAE, IRF8-KO mice showed no signs of response to MOG vaccination and were completely resistant to EAE in the absence of effector T cells (57). In this study, we observed that IRF8-KO mice tolerated an allograft tumor challenge. Furthermore, we observed that naive CD8+ T cells do not respond to antigen stimulation to generate antigen-specific CD8+ effector T cells. Our observations thus support the notion that IRF8 is essential for CD8+ T cell activation and effector function (46, 57, 59) and determined a critical role of IRF8 in T cell–mediated immune surveillance. However, strikingly, competitive reconstitution of WT and Irf8–/– BM revealed that intrinsic IRF8 function is not required for naive CD8+ T cell response to antigen to differentiate into antigen-specific CD8+ effector cells, suggesting a CD8+ T cell–extrinsic mechanism that underlies IRF8 function in regulating naive CD8+ T cell activation and differentiation in vivo. This notion was further supported by our observation that IRF8-TKO mice respond to antigenic stimulation and reject allograft tumors as efficiently as WT mice. We believe we have therefore identified a previously uncharacterized mechanism that IRF8 regulates CD8+ T cell activation through a cell-extrinsic mechanism. More importantly, we have identified the molecular link between myeloid cells and T cells in the context of IRF8 function in T cell activation. We demonstrated that OPN is a potent suppressor of T cell activation and its expression is repressed by IRF8 in myeloid cells. In the absence of IRF8, OPN is highly expressed in CD11b+Ly6CloLy6G+ myeloid cells under physiological conditions. IRF8 therefore acts as a repressor of OPN expression in myeloid cells to facilitate T cell activation in a cell-extrinsic mechanism. Our data indicate that OPN functions as a repressive ligand that negatively regulates T cell activation.
However, OPN is known to be markedly elevated after irradiation and persistent throughout the course of GVHD (60). OPN apparently acts to enhance activation and survival of T cells during GVHD (60, 61). The mechanism underlying these contrasting functions of OPN in GVHD and cancer is unknown and thus requires further study.
Although IRF8 was originally identified and extensively studied in myeloid cells (62), IRF8 is also expressed and functional in nonhematopoietic cells (63, 64) and acts as a tumor suppressor (65, 66). Interestingly, we observed that the highly expressed IRF8 binds to the Spp1 promoter region to repress OPN expression in colon epithelial cells. However, IRF8 expression is silenced and OPN is elevated during colon epithelial cells transformation into colon tumor cells, suggesting that tumor cells may use downregulation of IRF8 to upregulate OPN as a mechanism to suppress host CTL antitumor immune response. We believe, therefore, that our data revealed a previously uncharacterized mechanism of action of IRF8 as a tumor suppressor and indicate that IRF8 acts to repress OPN to suppress OPN-mediated immune suppression to suppress tumor development. Elevated OPN expression is known to link to the progression of colon cancer and multiple other human cancers (67, 68). Our data indicate that OPN is highly expressed in CD11b+Ly6CloLy6G+ myeloid cells and tumor cells, 2 major components of the tumor microenvironment. Therefore, OPN may act as another immune checkpoint and contribute, at least in part, to CTLA-4/PD-1/PD-L1–independent immune suppression and cancer patient nonresponse to the current ICI immunotherapy.
Methods
Mice.
IRF8-KO mice were generated as previously described (32). Mice with the loxp-flanked Irf8 gene B6(Cg)-Irf8tm1.1Hm/J were generated as previously described (44). B6.Cg-Tg(Lck-cre)548Jxm/J, SJL (B6.SJL-Ptprca Pepcb/BoyJ), and C57BL/6 mice were obtained from the Jackson Laboratory. BALB/c mice were obtained from the Charles River Frederick Facility. IRF8-TKO mice were created by crossing the B6(Cg)-Irf8tm1.1Hm/J mouse with the B6.Cg-Tg(Lck-cre)548Jxm/J mouse. The IRF8-GFP reporter mice (B6(Cg)-Irf8tm2.1Hm/J) were generated as previously described (30). WT and IRF8-KO chimera mice were created by transferring 5 × 106 to 10 × 106 BM cells from C57BL/6 and IRF8-KO mice to lethally (8.5 Gy) radiated C57BL/6 recipient mice, respectively. To create mixed BM chimera mice, C57BL/6 and SJL mice were crossed to generate F1 hybrid mice. BM cells from SJL and IRF8-KO mice were then mixed (at 1:2 or 1:5 ratio of SJL/IRF8-KO) and adoptively transferred to the lethally radiated F1 hybrid mice to generate mixed chimera mice (Supplemental Figure 3).
Mouse tumor model.
Mice were injected with AOM (Sigma-Aldrich, 10 mg/kg body weight) intraperitoneally once and treated with 2.5% DSS (MP Biomedicals, 35,000–50,000 mol wt) 1 day after AOM injection for 1 week, followed by 2 weeks with sterile, untreated water. The AOM-DSS cycle was repeated 2 more times. Mice were maintained with regular drinking water after the third AOM-DSS cycle and sacrificed for analysis. 4T1 mammary carcinoma cells were obtained from American Type Culture collection (ATCC). 4T1 cells were tested for mycoplasma and were mycoplasma-free at the time of the study. 4T1 tumor cells (1 × 104 cells/mouse) were injected into the no. 3 mammary gland of female BALB/c mice.
Human colon carcinoma data set and peripheral blood specimens.
The gene expression data set was extracted from the TCGA Colon and Rectal Cancer (COADREAD) data set using the Xena Functional Genomics Explorer (UCSD). Human colon cancer patient serum was obtained from the Georgia Cancer Center Biorepository (Augusta, GA). Human blood specimens were obtained with written informed consent from healthy donors enrolled in the Shepeard Community Blood Bank (Augusta, GA).
GFP fluorescence visualization.
GFP fluorescence was visualized as previously described (30). The tissues were then examined under a LSM780 Meta confocal laser microscope (Carl Zeiss). The captured images were viewed and analyzed using Zeiss Zen Meta imaging 2012 software.
Activation of CD8+ T cells by vaccination in vivo.
Mice were given immunizations with the OVA peptide (SIINFEKL) using the reported procedures (69). The vaccine consists of a prime followed by a boost 14 days later, and is administered by injecting a mixture of the OVA peptide (100 μg, Genscript), CD40 mAb (prime 100 μg, boost 25 μg; BioXcell), and poly-IC (50 μg, Invivogen). Seven days after each vaccination, blood cells were collected and stained with MHCII-, CD45.1-, CD45.2-, and CD8-specific mAbs (BioLegend) and OVA tetramer (BML Intern Corp). An Fc receptor blocker (BioLegend) was used jointly with the OVA tetramer. Stained cells were analyzed on a LSR II flow cytometer (BD Biosciences).
Cell surface marker analysis.
Cells were stained with antibodies and analyzed by flow cytometry. The following antibodies and dye were obtained from BioLegend: CD4 (clone RM4-5), CD8 (clone 53-6.7), CD25 (clone PC61), CD11b (clone M1/70), Gr1 (clone RB6-8C5), Ly6G (clone 1A8), Ly6C (clone HK1.4), CD45.1 (clone A20), CD45.2 (clone 104), CD44 (clone IM7), CD62L (clone MEL-14), and Zombie Violet. Stained cells were analyzed on an Accuri C6, LSRII, or LSR Fortessa (BD Biosciences).
T cell activation in vitro.
CD3+ T cells were purified from mouse spleen and lymph nodes with the MojoSort mouse CD3 T cell isolation kit (BioLegend) according to the manufacturer’s instructions. Human CD3+ T cells were purified from peripheral blood cells of healthy donors using the MojoSort human CD3+ T cell isolation kit (BioLegend). For mouse T cell activation, a 96-well culture plate was coated with anti-mouse CD3 (clone 145-2C11) and anti-mouse CD28 (clone 37.51) mAbs (BioXcell) in PBS overnight in 4°C. For human T cell activation, a 96-well culture plate was coated with anti-human CD3 (clone OKT3) mAbs (BioLegend). Mouse OPN protein (catalog 763604, BioLegend) and human OPN protein (catalog 1433-OP, R&D Systems). The purified T cells (1.5 × 105 cells/well) were then seeded in the coated plate in RPMI-1640 medium plus 10% FBS. For cell proliferation assay, the purified CD3+ T cells were labeled with 0.1 μM CFSE according to the manufacturer’s instructions (Invitrogen). CFSE-labeled cells were analyzed using an Accuri Flow Cytometer (BD Biosciences).
EMSA of protein-DNA interactions.
CD3+ T cells were purified from spleen and lymph nodes using the MojoSort CD3+ T Cell Isolation Kits as described above. T cells were activated in anti-CD3 and anti-CD28 mAb-coated plates for 3 days and used to prepare nuclear extract as previously described (70). Colon tissues were collected from mice and homogenized in a glass homogenizer for nuclear extract preparation as described (70). Complementary oligonucleotides containing the ISRE consensus sequence of the mouse Spp1 promoter (Supplemental Table 1) were synthesized and annealed to make double-stranded DNA probes. Complementary oligonucleotides containing ISRE consequence sequence of the mouse Pdcd1 promoter (Supplemental Table 1) were also synthesized and annealed to make double-stranded DNA probes. The DNA probes were end-labeled with 32P and incubated with nuclear extracts in the presence of IgG, anti-IRF8 antibody (C-19, Santa Cruz Biotechnology), or cold probes as indicated. The DNA-protein complexes were analyzed by polyacrylamide gel electrophoresis and detected by a phosphoImager.
ChIP assay.
ChIP assays were carried out using the anti-IRF8 antibody (C-19, Santa Cruz Biotechnology) and protein A-agarose beads (Millipore). The mouse Spp1 promoter DNA was detected by qPCR and semiquantitative PCR using gene-specific primers (Supplemental Table 1).
Intracellular staining and flow cytometry.
Cells were stained with anti-CD11b and anti-Gr1 mAbs, fixed with IC Fixation Buffer (BD Biosciences), incubated with permeabilization buffer, and stained with PE–anti-mouse OPN (catalog IC808P, R&D Systems).
IFN-γ and OPN protein analysis by ELISA.
Serum and cell culture medium were analyzed for IFN-γ and OPN protein level using the mouse IFN-γ ELISA kit (catalog 430805, BioLegend), human IFN-γ ELISA kit (catalog 430105, BioLegend), mouse OPN ELISA kit (catalog MOST00, R&D Systems), and the human OPN ELISA kit (catalog DOST00, R&D Systems).
Gene expression analysis.
Total RNA was isolated from cells using GenElute Direct mRNA Miniprep Kits (Sigma-Aldrich) according to the manufacturer’s instructions. cDNA was synthesized using the MMLV reverse transcriptase (Promega), and used for qPCR using the StepOne Real-Time PCR system (Applied Biosystems). The PCR sequences are listed in Supplemental Table 1.
Statistics.
Except where indicated, all statistical analyses were performed using a 2-tailed Student’s t test. P value for survival analysis was calculated using a log-rank test on a Cox hazard-proportional model. Significance for activation marker kinetics was calculated using a 2-way ANOVA with Tukey’s test. Significance for IFN-γ production and division index was calculated using a 1-way ANOVA with Dunnett’s correction. Tumor volume was calculated as (length × width2)/2. P less than 0.05 was considered statistically significant. All data are mean ± SD.
Study approval.
Use of mice was performed according to protocol 2008-0162 approved by the Augusta University institutional animal use and care committee. All studies with human specimens were reviewed and determined as “not human subject research” by the Augusta University Institutional Review Board (approval 933148-1).
Author contributions
JDK, AVP, EC, SIA, KO, and KL developed the concept and study design; EC and KO provided key reagents; JDK, AVP, PSR, MLI, CL, and DY conducted the experiments; JDK and KL wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA133085 and CA182518 to KL and CA172105 to SA), Veterans Affairs award (BX001962 to KL), and in part by the Intramural Research Program of the National Institute of Child Health and Human Development (to KO). We thank Roni Bollag at the Georgia Cancer Center Biorepository for pathological advice on human specimens and for providing human colon cancer patent serum. We also thank Hussein Sultan for assistance in mouse vaccination and analysis of antigen-specific T cells.
Version 1. 11/05/2018
Electronic publication
Version 2. 12/03/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(12):5549–5560.https://doi.org/10.1172/JCI123360.
See the related Commentary at Osteopontin controls immunosuppression in the tumor microenvironment.
Contributor Information
Amy V. Paschall, Email: Apaschall@uga.edu.
Priscilla S. Redd, Email: Predd@augusta.edu.
Chunwan Lu, Email: Clu@augusta.edu.
Dafeng Yang, Email: Dyang@augusta.edu.
Esteban Celis, Email: Ecelis@augusta.edu.
Keiko Ozato, Email: ozatok@dir6.nichd.nih.gov.
Kebin Liu, Email: Kliu@augusta.edu.
References
- 1.Thaker YR, Schneider H, Rudd CE. TCR and CD28 activate the transcription factor NF-κB in T-cells via distinct adaptor signaling complexes. Immunol Lett. 2015;163(1):113–119. doi: 10.1016/j.imlet.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13(4):227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kägi D, et al. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science. 1994;265(5171):528–530. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- 4.Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med. 2016;375(18):1767–1778. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor A, Harker JA, Chanthong K, Stevenson PG, Zuniga EI, Rudd CE. Glycogen synthase kinase 3 inactivation drives T-bet-mediated downregulation of co-receptor PD-1 to enhance CD8(+) cytolytic T cell responses. Immunity. 2016;44(2):274–286. doi: 10.1016/j.immuni.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui E, et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017;355(6332):1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamphorst AO, et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science. 2017;355(6332):1423–1427. doi: 10.1126/science.aaf0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krueger J, Rudd CE. Two strings in one bow: PD-1 negatively regulates via co-receptor CD28 on T cells. Immunity. 2017;46(4):529–531. doi: 10.1016/j.immuni.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12(6):492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 11.Hanson HL, et al. Eradication of established tumors by CD8+ T cell adoptive immunotherapy. Immunity. 2000;13(2):265–276. doi: 10.1016/S1074-7613(00)00026-1. [DOI] [PubMed] [Google Scholar]
- 12.Golstein P, Griffiths GM. An early history of T cell-mediated cytotoxicity. Nat Rev Immunol. 2018;18(8):527–535. doi: 10.1038/s41577-018-0009-3. [DOI] [PubMed] [Google Scholar]
- 13.Ribas A. Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov. 2015;5(9):915–919. doi: 10.1158/2159-8290.CD-15-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marisa L, et al. The balance between cytotoxic T-cell lymphocytes and immune checkpoint expression in the prognosis of colon tumors. J Natl Cancer Inst. 2018;110(1):68–77. doi: 10.1093/jnci/djx136. [DOI] [PubMed] [Google Scholar]
- 15.Juneja VR, et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med. 2017;214(4):895–904. doi: 10.1084/jem.20160801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TK, Herbst RS, Chen L. Defining and understanding adaptive resistance in cancer immunotherapy. Trends Immunol. 2018;39(8):624–631. doi: 10.1016/j.it.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirano F, et al. Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005;65(3):1089–1096. [PubMed] [Google Scholar]
- 18.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271(5256):1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 19.Lin H, et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest. 2018;128(2):805–815. doi: 10.1172/JCI96113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang H, et al. PD-L1 on host cells is essential for PD-L1 blockade-mediated tumor regression. J Clin Invest. 2018;128(2):580–588. doi: 10.1172/JCI96061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ariyan CE, et al. Robust antitumor responses result from local chemotherapy and CTLA-4 blockade. Cancer Immunol Res. 2018;6(2):189–200. doi: 10.1158/2326-6066.CIR-17-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8(328):328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harper SJ, et al. CD8 T-cell recognition of acquired alloantigen promotes acute allograft rejection. Proc Natl Acad Sci U S A. 2015;112(41):12788–12793. doi: 10.1073/pnas.1513533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, et al. Interferon regulator factor 8 (IRF8) limits ocular pathology during HSV-1 infection by restraining the activation and expansion of CD8+ T cells. PLoS One. 2016;11(5):e0155420. doi: 10.1371/journal.pone.0155420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baaten BJ, Li CR, Deiro MF, Lin MM, Linton PJ, Bradley LM. CD44 regulates survival and memory development in Th1 cells. Immunity. 2010;32(1):104–115. doi: 10.1016/j.immuni.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtsinger JM, Lins DC, Mescher MF. CD8+ memory T cells (CD44high, Ly-6C+) are more sensitive than naive cells to (CD44low, Ly-6C-) to TCR/CD8 signaling in response to antigen. J Immunol. 1998;160(7):3236–3243. [PubMed] [Google Scholar]
- 28.Lesley J, He Q, Miyake K, Hamann A, Hyman R, Kincade PW. Requirements for hyaluronic acid binding by CD44: a role for the cytoplasmic domain and activation by antibody. J Exp Med. 1992;175(1):257–266. doi: 10.1084/jem.175.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin YH, Yang-Yen HF. The osteopontin-CD44 survival signal involves activation of the phosphatidylinositol 3-kinase/Akt signaling pathway. J Biol Chem. 2001;276(49):46024–46030. doi: 10.1074/jbc.M105132200. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, et al. A reporter mouse reveals lineage-specific and heterogeneous expression of IRF8 during lymphoid and myeloid cell differentiation. J Immunol. 2014;193(4):1766–1777. doi: 10.4049/jimmunol.1301939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker AM, Michael DG, Satpathy AT, Sciammas R, Singh H, Bhattacharya D. IRF-8 extinguishes neutrophil production and promotes dendritic cell lineage commitment in both myeloid and lymphoid mouse progenitors. Blood. 2012;119(9):2003–2012. doi: 10.1182/blood-2011-06-364976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holtschke T, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87(2):307–317. doi: 10.1016/S0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 33.McGough JM, et al. DNA methylation represses IFN-gamma-induced and signal transducer and activator of transcription 1-mediated IFN regulatory factor 8 activation in colon carcinoma cells. Mol Cancer Res. 2008;6(12):1841–1851. doi: 10.1158/1541-7786.MCR-08-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura T, Thotakura P, Tanaka TS, Ko MS, Ozato K. Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages. Blood. 2005;106(6):1938–1947. doi: 10.1182/blood-2005-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urso K, et al. NFATc3 regulates the transcription of genes involved in T-cell activation and angiogenesis. Blood. 2011;118(3):795–803. doi: 10.1182/blood-2010-12-322701. [DOI] [PubMed] [Google Scholar]
- 36.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc Natl Acad Sci U S A. 2003;100(26):15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kao C, et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat Immunol. 2011;12(7):663–671. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyagawa F, Zhang H, Terunuma A, Ozato K, Tagaya Y, Katz SI. Interferon regulatory factor 8 integrates T-cell receptor and cytokine-signaling pathways and drives effector differentiation of CD8 T cells. Proc Natl Acad Sci U S A. 2012;109(30):12123–12128. doi: 10.1073/pnas.1201453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grajales-Reyes GE, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat Immunol. 2015;16(7):708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sichien D, et al. IRF8 transcription factor controls survival and function of terminally differentiated conventional and plasmacytoid dendritic cells, respectively. Immunity. 2016;45(3):626–640. doi: 10.1016/j.immuni.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 41.Kurotaki D, et al. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. 2013;121(10):1839–1849. doi: 10.1182/blood-2012-06-437863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurotaki D, et al. IRF8 inhibits C/EBPα activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat Commun. 2014;5:4978. doi: 10.1038/ncomms5978. [DOI] [PubMed] [Google Scholar]
- 43.Netherby CS, et al. The granulocyte progenitor stage is a key target of IRF8-mediated regulation of myeloid-derived suppressor cell production. J Immunol. 2017;198(10):4129–4139. doi: 10.4049/jimmunol.1601722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng J, Wang H, Shin DM, Masiuk M, Qi CF, Morse HC. IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J Immunol. 2011;186(3):1458–1466. doi: 10.4049/jimmunol.1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365(2):127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouyang X, et al. Transcription factor IRF8 directs a silencing programme for TH17 cell differentiation. Nat Commun. 2011;2:314. doi: 10.1038/ncomms1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J, et al. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat Immunol. 2017;18(8):877–888. doi: 10.1038/ni.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taffurelli M, Pellegrini A, Ghigone F, Santini D, Zanotti S, Serra M. Positive predictive value of breast lesions of uncertain malignant potential (B3): can we identify high risk patients? The value of a multidisciplinary team and implications in the surgical treatment. Surg Oncol. 2016;25(2):119–122. doi: 10.1016/j.suronc.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Luda KM, et al. IRF8 transcription-factor-dependent classical dendritic cells are essential for intestinal T cell homeostasis. Immunity. 2016;44(4):860–874. doi: 10.1016/j.immuni.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Lee J, et al. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat Immunol. 2017;18(8):877–888. doi: 10.1038/ni.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Humblin E, et al. IRF8-dependent molecular complexes control the Th9 transcriptional program. Nat Commun. 2017;8(1):2085. doi: 10.1038/s41467-017-01070-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mace EM, et al. Biallelic mutations in IRF8 impair human NK cell maturation and function. J Clin Invest. 2017;127(1):306–320. doi: 10.1172/JCI86276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim SH, et al. Dual function of the IRF8 transcription factor in autoimmune uveitis: loss of IRF8 in T cells exacerbates uveitis, whereas Irf8 deletion in the retina confers protection. J Immunol. 2015;195(4):1480–1488. doi: 10.4049/jimmunol.1500653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newman DM, Leung PS, Putoczki TL, Nutt SL, Cretney E. Th17 cell differentiation proceeds independently of IRF8. Immunol Cell Biol. 2016;94(8):796–801. doi: 10.1038/icb.2016.33. [DOI] [PubMed] [Google Scholar]
- 55.Lee W, Kim HS, Baek SY, Lee GR. Transcription factor IRF8 controls Th1-like regulatory T-cell function. Cell Mol Immunol. 2016;13(6):785–794. doi: 10.1038/cmi.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams NM, et al. Transcription factor IRF8 orchestrates the adaptive natural killer cell response. Immunity. 2018;48(6):1172–1182.e6. doi: 10.1016/j.immuni.2018.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida Y, et al. The transcription factor IRF8 activates integrin-mediated TGF-β signaling and promotes neuroinflammation. Immunity. 2014;40(2):187–198. doi: 10.1016/j.immuni.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tailor P, Tamura T, Morse HC, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 2008;111(4):1942–1945. doi: 10.1182/blood-2007-07-100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyagawa F, et al. IL-15 serves as a costimulator in determining the activity of autoreactive CD8 T cells in an experimental mouse model of graft-versus-host-like disease. J Immunol. 2008;181(2):1109–1119. doi: 10.4049/jimmunol.181.2.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao F, et al. Blockade of osteopontin reduces alloreactive CD8+ T cell-mediated graft-versus-host disease. Blood. 2011;117(5):1723–1733. doi: 10.1182/blood-2010-04-281659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawakami K, et al. Osteopontin attenuates acute gastrointestinal graft-versus-host disease by preventing apoptosis of intestinal epithelial cells. Biochem Biophys Res Commun. 2017;485(2):468–475. doi: 10.1016/j.bbrc.2017.02.047. [DOI] [PubMed] [Google Scholar]
- 62.Karki R, et al. IRF8 regulates transcription of Naips for NLRC4 inflammasome activation. Cell. 2018;173(4):920–933.e13. doi: 10.1016/j.cell.2018.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W, Nagineni CN, Ge H, Efiok B, Chepelinsky AB, Egwuagu CE. Interferon consensus sequence-binding protein is constitutively expressed and differentially regulated in the ocular lens. J Biol Chem. 1999;274(14):9686–9691. doi: 10.1074/jbc.274.14.9686. [DOI] [PubMed] [Google Scholar]
- 64.Yan M, et al. Cutting edge: expression of IRF8 in gastric epithelial cells confers protective innate immunity against helicobacter pylori infection. J Immunol. 2016;196(5):1999–2003. doi: 10.4049/jimmunol.1500766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee KY, et al. Epigenetic disruption of interferon-gamma response through silencing the tumor suppressor interferon regulatory factor 8 in nasopharyngeal, esophageal and multiple other carcinomas. Oncogene. 2008;27(39):5267–5276. doi: 10.1038/onc.2008.147. [DOI] [PubMed] [Google Scholar]
- 66.Yang D, et al. Repression of IFN regulatory factor 8 by DNA methylation is a molecular determinant of apoptotic resistance and metastatic phenotype in metastatic tumor cells. Cancer Res. 2007;67(7):3301–3309. doi: 10.1158/0008-5472.CAN-06-4068. [DOI] [PubMed] [Google Scholar]
- 67.Agrawal D, et al. Osteopontin identified as lead marker of colon cancer progression, using pooled sample expression profiling. J Natl Cancer Inst. 2002;94(7):513–521. doi: 10.1093/jnci/94.7.513. [DOI] [PubMed] [Google Scholar]
- 68.Coppola D, et al. Correlation of osteopontin protein expression and pathological stage across a wide variety of tumor histologies. Clin Cancer Res. 2004;10(1 Pt 1):184–190. doi: 10.1158/1078-0432.ccr-1405-2. [DOI] [PubMed] [Google Scholar]
- 69.Nagato T, Lee YR, Harabuchi Y, Celis E. Combinatorial immunotherapy of polyinosinic-polycytidylic acid and blockade of programmed death-ligand 1 induce effective CD8 T-cell responses against established tumors. Clin Cancer Res. 2014;20(5):1223–1234. doi: 10.1158/1078-0432.CCR-13-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu C, Redd PS, Lee JR, Savage N, Liu K. The expression profiles and regulation of PD-L1 in tumor-induced myeloid-derived suppressor cells. Oncoimmunology. 2016;5(12):e1247135. doi: 10.1080/2162402X.2016.1247135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.