Abstract
ASXL1 is frequently mutated in myeloid malignancies and is known to co-occur with other gene mutations. However, the molecular mechanisms underlying the leukemogenesis associated with ASXL1 and cooperating mutations remain to be elucidated. Here, we report that Asxl1 loss cooperated with haploinsufficiency of Nf1, a negative regulator of the RAS signaling pathway, to accelerate the development of myeloid leukemia in mice. Loss of Asxl1 and Nf1 in hematopoietic stem and progenitor cells resulted in a gain-of-function transcriptional activation of multiple pathways such as MYC, NRAS, and BRD4 that are critical for leukemogenesis. The hyperactive MYC and BRD9 transcription programs were correlated with elevated H3K4 trimethylation at the promoter regions of genes involving these pathways. Furthermore, pharmacological inhibition of both the MAPK pathway and BET bromodomain prevented leukemia initiation and inhibited disease progression in Asxl1Δ/Δ Nf1Δ/Δ mice. Concomitant mutations of ASXL1 and RAS pathway genes were associated with aggressive progression of myeloid malignancies in patients. This study sheds light on the effect of cooperation between epigenetic alterations and signaling pathways on accelerating the progression of myeloid malignancies and provides a rational therapeutic strategy for the treatment of myeloid malignancies with ASXL1 and RAS pathway gene mutations.
Keywords: Hematology, Stem cells
Keywords: Epigenetics, Leukemias
Introduction
Alteration of epigenetic regulation provides a critical mechanism in the development of myeloid leukemia (1–3). Recent genomic studies have resulted in an emerging understanding of the significance of chromatin regulators in cancer (4–6). Additional sex combs-like 1 (ASXL1) is frequently mutated in a wide range of myeloid malignancies (7–11), including myelodysplastic syndrome (MDS), myeloproliferative neoplasm (MPN), chronic myelomonocytic leukemia (CMML), and acute myeloid leukemia (AML). Notably, ASXL1 mutations are generally associated with aggressiveness and poor clinical outcomes (12, 13). We and others have established several Asxl1-KO mouse models and demonstrated in mice that loss of Asxl1 leads to MDS-like disease, which can transform into myeloid leukemia with age (14, 15). These studies suggest that additional mutations may cooperate with Asxl1 loss to induce leukemia transformation.
Mutations of genes involved in the MAPK pathway, such as activating mutations of NRAS or KRAS and inactivating mutations of NF1, are common genetic events in AML (16, 17). Observations in juvenile myelomonocytic leukemia (JMML) and CMML, along with studies of genetically engineered mice, provide compelling evidence that NF1 and RAS mutations may function as either early/initiating or cooperating mutations for leukemia progression (6, 18, 19).
Integrated genomic approaches identified potential cooperating events in AML (20, 21), such as comutations of genes involved in chromatin modifiers (e.g., ASXL1) and activated signaling (e.g., NF1) (22). Therefore, elucidation of the molecular mechanisms underlying alterations of both ASXL1 and NF1 has translational significance for patients with myeloid malignancies. Malignancies in NF1 result from a combination of ubiquitous NF1 heterozygosity and somatic loss of the residual NF1 allele (i.e., loss of heterozygosity) (23, 24). Epigenetic dysregulation leads to altered transcriptional events that are key for cell fates and that may prime for oncogenesis when mutations of signaling pathways occur. Abdel-Wahab et al. have shown that viral transduction of NRasG12D with Asxl1 shRNA into bone marrow (BM) cells accelerates myeloproliferation (25). However, the cellular and molecular mechanism underlying the cooperative effect of ASXL1 and RAS pathway gene mutations in myeloid malignancies remains to be elucidated. Furthermore, an effective treatment for such patients with myeloid malignancies with comutations in ASXL1 and RAS pathway genes is desperately needed.
In the current study, we show that haploinsufficiency of both Asxl1 and Nf1 (Asxl1+/– Nf1+/–) cooperates to accelerate myeloid leukemia transformation in vivo. RNA-sequencing (RNA-Seq) analyses of Asxl1+/– Nf1+/– hematopoietic stem/progenitor cells (HSCs/HPCs) reveal aberrant transcriptional activation of multiple pathways, such as MYC, NRAS, and BRD9, that are critical for leukemogenesis, indicating a gain of function of the alterations of Asxl1 and Nf1 in epigenetic regulation. Importantly, pharmacological inhibition of both the BET bromodomain and the MAPK pathway prevents leukemia initiation and inhibits disease progression. Furthermore, concomitant mutations of ASXL1 and NF1 or other RAS pathway genes are associated with a more aggressive disease status in patients with myeloid malignancies. This study provides a therapeutic strategy for the treatment of patients with myeloid malignancies with ASXL1 and RAS pathway gene mutations.
Results
Haploinsufficiency of Asxl1 and Nf1 leads to myeloid leukemia in mice.
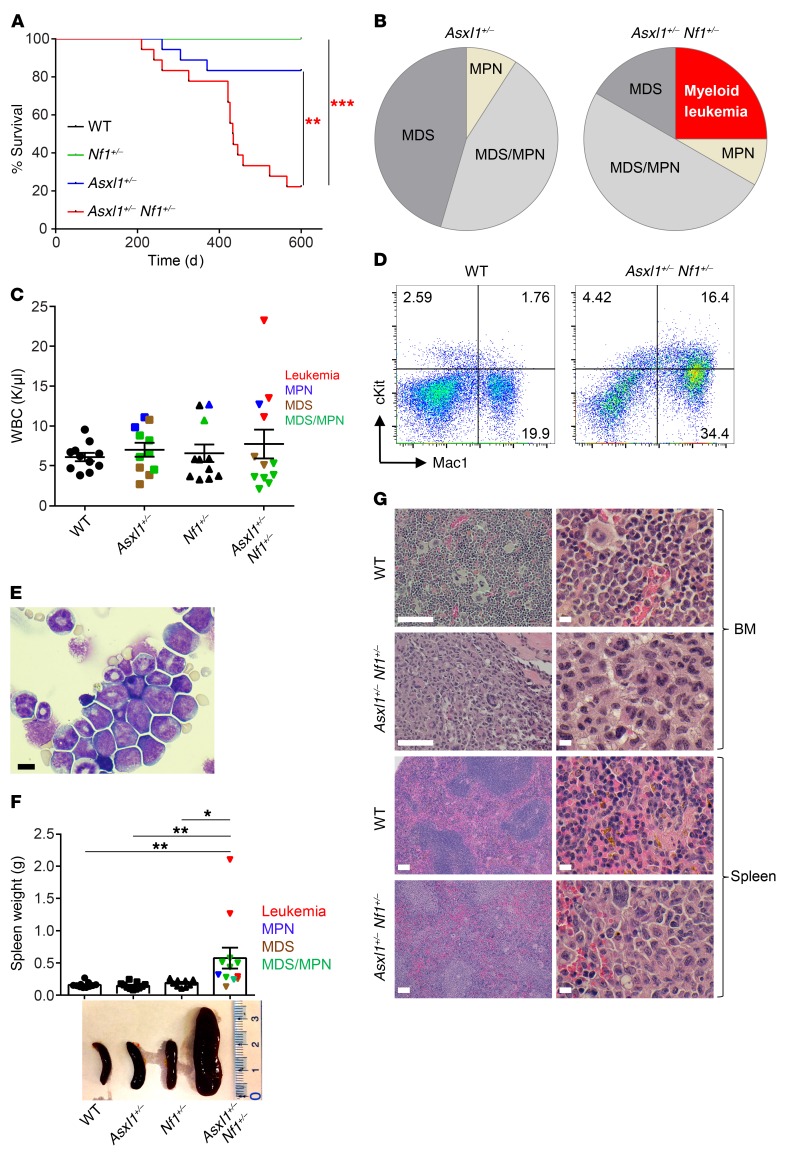
To determine the functional significance of comutations of ASXL1 and NF1 in the disease progression of myeloid malignancies, we intercrossed Asxl1 heterozygous (Asxl1+/–) mice with Nf1+/– mice and generated Asxl1+/– Nf1+/– mice. Quantitative reverse transcription PCR (RT-qPCR) confirmed a 40%–60% reduction in mRNA expression of Asxl1 and Nf1 in Asxl1+/– Nf1+/– cells compared with expression in WT cells (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI121366DS1). Of note, we observed no obvious difference in Asxl1 or Nf1 mRNA expression levels between young mice and diseased mice (Supplemental Figure 1A). Consistent with our previous work, the survival rate of Asxl1+/– mice was 83% up to 600 days of age, and the deceased Asxl1+/– mice died of myeloid malignancies such as MDS or MDS/MPN (Figure 1, A and B). In contrast, the survival rate of Asxl1+/– Nf1+/– mice was significantly lower (22%) than that for mice of the 3 other genotypes (Figure 1A and Supplemental Table 1).
Figure 1. Development of myeloid leukemia in Asxl1+/– Nf1+/– mice.
(A) Kaplan-Meier survival curve representing the survival percentage of WT (n = 16), Asxl1+/– (n = 17), Nf1+/– (n = 16), and Asxl1+/– Nf1+/– (n = 20) mice over time. No lethality was observed for WT or Nf1+/- mice during this period. A log-rank test was used to determine the survival statistics. (B) Pie charts illustrate the relative frequency of different hematopoietic diseases found in Asxl1+/- (n = 11) and Asxl1+/– Nf1+/– (n = 12) mice. (C) Peripheral WBC counts for WT, Asxl1+/-, Nf1+/-, and Asxl1+/– Nf1+/– mice (n = 11–12 per group). (D) Expansion of myeloid-lineage cells in Asxl1+/– Nf1+/– mice. Flow cytometric analysis of BM cells revealed increased Mac1+ cells as well as an expanded cKit+ cell population. (E) May-Grunwald-Giemsa–stained cytospin preparations of BM cells from WT and Asxl1+/– Nf1+/– mice. Scale bar: 10 μm. (F) Representative size and weight of spleens from mice of each genotype (n = 11–12 per group). (G) Representative H&E-stained sections from BM and spleens from WT and Asxl1+/– Nf1+/– mice. Scale bars: 100 μm (left); 10 μm (right). Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by 1-way ANOVA with Tukey’s multiple comparisons test.
Examination of peripheral blood (PB) parameters showed that a subset of Asxl1+/– Nf1+/– mice developed profound anemia and thrombocytopenia compared with age-matched WT mice (Supplemental Figure 1B). A subset of Asxl1+/– Nf1+/– mice (n = 3) developed myeloid leukemia (Figure 1B). These leukemic Asxl1+/– Nf1+/– mice had high WBC counts compared with those of mice of the other genotypes (Figure 1C and Supplemental Table 2). Phenotypic analyses by flow cytometry revealed a higher percentage of the cKit+Mac1+ cell population in the BM of Asxl1+/– Nf1+/– mice (Figure 1D). Consistently, morphologic analysis of cytospins prepared from the BM cells revealed an accumulation of blastic cells in Asxl1+/– Nf1+/– mice (Figure 1E). Necropsy of the deceased or moribund Asxl1+/– Nf1+/– mice demonstrated hepatosplenomegaly (Figure 1F and Supplemental Figure 1, C and D), lower body weights (Supplemental Figure 1E), and pale foot pads, indicating cachexia. Histological analyses of BM, splenic, and liver sections from Asxl1+/– Nf1+/– mice showed a disrupted architecture with significant myeloid cell infiltration (Figure 1G and Supplemental Figure 1F). These results suggest a cooperative effect of Nf1 and Asxl1 haploinsufficiency on accelerating the pathogenesis of myeloid leukemia.
To verify the malignant nature of the cells from the moribund Asxl1+/– Nf1+/– mice, we performed a tumor transfer assay by injecting splenic cells from a deceased mouse into sublethally irradiated WT recipient mice. Transfer of splenic cells from a moribund Asxl1+/– Nf1+/– mouse led to the early death of the WT recipients (at the age of 6 weeks, Supplemental Figure 1G). All the recipient mice had splenomegaly, and the femurs were pale with cKit+Mac1+ cell infiltration (Supplemental Figure 1, H–J), a phenocopy of the myeloid leukemia observed in the primary Asxl1+/– Nf1+/– mice.
To determine whether additional mutations could occur in the leukemic cells on the Asxl1 and Nf1 haploinsufficient background, we performed whole-exome sequencing using leukemic cells from 2 deceased Asxl1+/– Nf1+/– mice. The tail tissue from 1 mouse was used as a control. Surprisingly, we did not identify any gene mutations known to be critical for leukemogenesis in the Asxl1+/– Nf1+/– leukemic cells (Supplemental Table 3). Of note, we did not observe a loss of heterozygosity at the Asxl1 and Nf1 loci in the hematopoietic cells of these Asxl1+/– Nf1+/– mice (Supplemental Figure 1K). Collectively, these results suggest an intrinsic cooperative effect of Asxl1 and Nf1 haploinsufficiency in accelerating myeloid leukemia.
Asxl1 and Nf1 haploinsufficiency alters HSC/HPC cell fates.
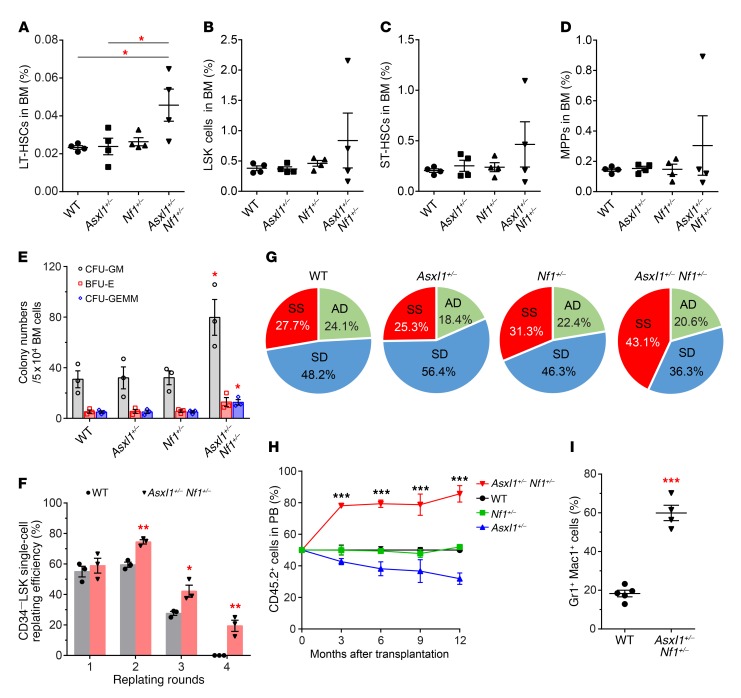
To determine whether haploinsufficient loss of Asxl1 and Nf1 affects stem cell self-renewal and differentiation, we next compared the composition of the HSC/HPC compartment in the BM of mice of the 4 different genotypes. The frequency of long-term HSCs (LT-HSCs) was significantly increased in the BM of Asxl1+/– Nf1+/– mice compared with that of mice from the other groups (Figure 2A), while the frequencies of Lin−Sca1+cKit+ (LSK) cells, short-term HSCs (ST-HSCs), and multipotent progenitors (MPPs) were comparable among the 4 groups of mice (Figure 2, B–D). To determine the effect of Asxl1 and Nf1 loss on the colony-forming capacity of BM cells, we next prepared methylcellulose cultures and compared the frequency of CFU cells (CFU-Cs) in BM from mice of the 4 different genotypes. We detected significantly higher frequencies of CFU-Cs, CFU-granulocytes/macrophages (CFU-GMs), and mixed colonies of GMs, erythrocytes, and megakaryocytes (GEMMs) in BM cells from Asxl1+/– Nf1+/– mice compared with BM cells from mice of the other genotypes (Figure 2E).
Figure 2. Altered HSC self-renewal and cell fates in Asxl1+/– Nf1+/– mice.
(A–D) Flow cytometric quantification of the percentages of LT-HSCs (A), LSK cells (B), ST-HSCs (C), and MPPs (D) in total BM cells from mice of each genotype (n = 4 per group). (E) CFU-Cs in BM cells from WT, Asxl1+/–, Nf1+/–, and Asxl1+/– Nf1+/– mice (n = 3 per group) were assessed in semisolid media in the presence of mSCF, mIL-3, mEPO, mTPO, mGM-CSF, and human IL-6. BFU-E, burst-forming unit–erythrocytes; GEMM, mixed colonies of GMs, erythrocytes, and megakaryocytes. (F) CD34−LSK single-cell replating assays show the increased self-renewal capacity of Asxl1+/– Nf1+/– cells compared with that of WT control cells (n = 3 mice per group). (G) Paired daughter cell assays were performed at the single-cell level on CD34−LSK cells isolated from WT, Asxl1+/–, Nf1+/–, and Asxl1+/– Nf1+/– mice (n = 67–102 paired cells from 3 mice per group), and each was analyzed for symmetric self-renewal (SS, red), asymmetric division (AD, green) or symmetric differentiation (SD, blue). (H) Competitive repopulation assay using BM cells from Asxl1+/– Nf1+/– mice. CD45.2+ BM cells from WT, Asxl1+/–, Nf1+/–, or Asxl1+/– Nf1+/– mice were mixed with equal numbers of CD45.1+ competitor cells and transplanted into lethally irradiated CD45.1+ recipient mice (n = 4–5 per group). The percentage of CD45.2+ cells was then monitored every 3 months. (I) Percentage of Gr1+Mac1+ cells within the CD45.2+ cell population in PB from WT and Asxl1+/– Nf1+/– recipient mice (n = 4–5 per group). Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by 1-way ANOVA with Tukey’s multiple comparisons test (A–E and H) or unpaired Student’s t test (F and I).
The preservation of a normal stem cell pool and the production of blood cells require a balance between self-renewal and differentiation of HSCs/HPCs. To determine the replating potential of CD34−LSK cells over time, we performed CFU-C replating assays. Although we observed a steady decline of replating potential in WT cultures, we detected significantly higher replating activity over 4 successive replating periods in Asxl1+/– Nf1+/– CD34−LSK cell cultures (Figure 2F), suggesting an increased self-renewal capacity of Asxl1+/– Nf1+/– CD34–LSK cells. To further determine whether haploinsufficient loss of Asxl1 and Nf1 affects HSC/HPC symmetric/asymmetric cell division, we next performed paired daughter cell assays (26) to assess the proportions of symmetric self-renewal, symmetric differentiation, and asymmetric division using sorted CD34−LSK cell populations from mice of the 4 genotypes. Haploinsufficiency of Asxl1 and Nf1 resulted in a higher proportion of cells with symmetric self-renewal capacity (43.1%) compared with WT cells (27.7%) (Figure 2G). In contrast, the symmetric differentiation division potential was significantly reduced in Asxl1+/– Nf1+/– CD34–LSK cells (36.3%) compared with that of WT cells (48.2%). The frequency of cells that underwent asymmetric division in Asxl1+/– Nf1+/– CD34−LSK cells was similar to that of cells from the other groups of mice. These data suggest that heterozygous deletion of both Asxl1 and Nf1 cooperatively affects the cell fates of HSCs/HPCs.
To evaluate whether Asxl1 and Nf1 haploinsufficiency–mediated alteration in HSC/HPC functions is cell autonomous, we next performed BM transplantation assays. Mice transplanted with Asxl1+/– Nf1+/– BM cells developed myeloid leukemia as evidenced by stomach sarcoma infiltrated with Gr1+Mac1+ myeloid cells (Supplemental Figure 2, A and B). To examine the repopulating capacity of Asxl1+/– Nf1+/– BM cells in vivo, we performed competitive transplantation assays by transplanting WT, Asxl1+/–, Nf1+/–, or Asxl1+/– Nf1+/– BM cells (CD45.2+) with competitor cells (CD45.1+) at a 1:1 ratio into lethally irradiated recipient mice (CD45.1+). Kinetic analysis of the donor cell chimerism in the PB of recipient mice revealed higher levels of CD45.2+ cell populations in mice receiving Asxl1+/– Nf1+/– BM cells, whereas a stable percentage of CD45.2+ cell populations was observed in the PB of recipient mice transplanted with WT or Nf1+/– BM cells (Figure 2H). Notably, the PB of chimeric mice receiving Asxl1+/– Nf1+/– BM cells was dominated by myeloid-lineage cells (Figure 2I). These results indicate that Asxl1+/– Nf1+/– cells possess a competitive advantage with a skewed differentiation toward the myeloid lineage.
Deletion of Asxl1 and Nf1 accelerates the development of myeloid leukemia in vivo.
Asxl1–/– mice exhibited severe MDS and died of BM failure (15), while mice with Nf1 loss in the hematopoietic system (Nf1Δ/Δ) were characterized by myeloproliferative features (27). To assess the cooperative effect of complete loss of Asxl1 and Nf1 on leukemogenesis in vivo, we generated Mx1Cre Asxl1fl/fl Nf1fl/fl mice by intercrossing MxCre Asxl1fl/fl mice with Nf1flfl mice. Following polyinosinic-polycytidylic acid (pIpC) injection, we analyzed the hematopoietic phenotypes. Asxl1Δ/Δ Nf1Δ/Δ double-KO (DKO) mice had a lower survival rate than did Asxl1+/–Nf1+/–, Asxl1Δ/Δ, or Nf1Δ/Δ mice (Supplemental Figure 2C). The DKO mice displayed a rapid, progressive leukocytosis with severe anemia and thrombocytopenia 3–6 months after pIpC injection (Supplemental Figure 2, D–G), which was indicative of aggressive myeloid leukemia with 100% penetrance.
Examination of the PB, BM, and spleen revealed that the DKO mice had a high frequency of myeloid blastic cells (Supplemental Figure 2H). Flow cytometric analysis verified a higher percentage of the cKit+ cell population in the BM cells of DKO mice (Supplemental Figure 2I). In addition, DKO mice had splenomegaly along with massive myeloid infiltration (Supplemental Figure 2, J and K). These data demonstrate that deletion of Asxl1 and Nf1 accelerates the progression of myeloid leukemia in vivo.
Cooperative effect of Asxl1 and Nf1 loss on the induction of a MYC-driven transcription signature.
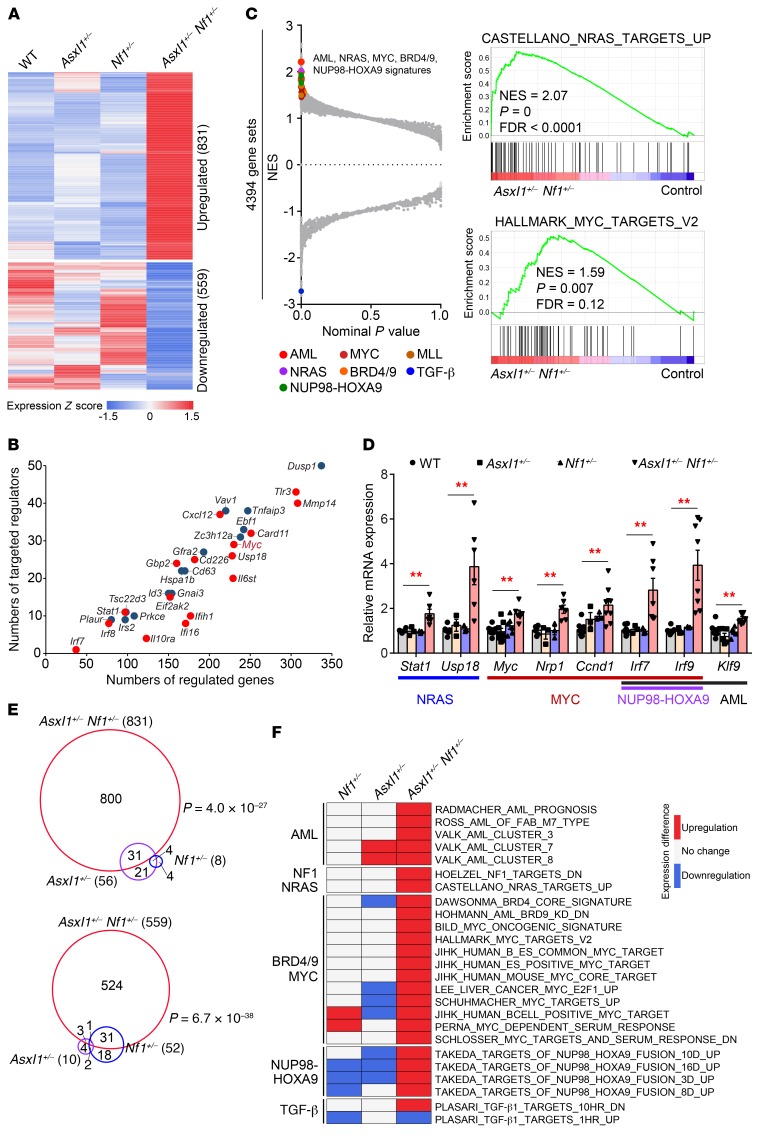
To understand the molecular mechanism underlying Asxl1 and Nf1 loss–associated myeloid transformation, we examined gene expression profiles of the WT, Asxl1+/–, Nf1+/–, and Asxl1+/– Nf1+/– cKit+ cells by RNA-Seq. Compared with WT and single mutant cells, haploinsufficiency of both Asxl1 and Nf1 resulted in a distinct gene expression signature in cKit+ cells consisting of 831 upregulated and 559 downregulated genes (fold change [FC] ≥1.2 and FDR <0.25), with elevated expression of known oncogenes such as Myc and Ccnd1 (Figure 3A, Supplemental Figure 3, A and B, and Supplemental Table 4). To identify key transcriptional hubs that regulate these expression changes, we used the Upstream Regulator analysis tool (www.nihlibrary.nih.gov/resources/tools/ingenuity-pathways-analysis-ipa) and identified 16 upregulated and 14 downregulated genes as key master regulators (overlap P < 1 × 10–20, network P < 0.01 and |Z score| ≥1.5) (Figure 3B). Notably, top-ranked regulators in Asxl1+/– Nf1+/– cKit+ cells include the oncogene Myc, the cell migration– and development–related gene Mmp14, as well as the immune response–related genes Tlr3, Cxcl12, and Gfra2. Consistently, gene set enrichment analysis (GSEA) identified multiple key transcription signatures, including AML, NRAS, NUP98-HOXA9, and MYC and MYC upregulators BRD4 and BRD9, in Asxl1+/– Nf1+/– cells compared with WT and single mutant cells (P < 0.01 and FDR <0.20) (Figure 3C and Supplemental Figure 3C). In support of this finding, several known self-renewal and differentiation genes that were activated by NRAS, MYC, and NUP98-HOXA9 were validated with RT-qPCR (Figure 3D). Immunofluorescence revealed an accumulation of the MYC protein in Asxl1+/– Nf1+/– cells (Supplemental Figure 3D). Furthermore, Western blotting showed that Asxl1+/– Nf1+/– cells had much higher levels of MYC and RAS-GTP compared with levels in WT control cells (Supplemental Figure 3, E and F). It is noteworthy that patients with AML who harbor ASXL1 and/or RAS pathway mutations also exhibit an elevation in RNA expression of MYC downstream targets (NRP1 and IRF7) (Supplemental Figure 3, G and H). Thus, the aberrant upregulation of gene expression in Asxl1+/– Nf1+/– HSCs/HPCs parallels that seen in AML patients, which could be a characteristic, if not driving, event in leukemogenesis.
Figure 3. Cooperative effect of Asxl1 and Nf1 loss on the induction of a MYC-driven transcription signature.
(A) Heatmap shows a unique expression signature in Asxl1+/– Nf1+/– cKit+ cells compared with that of control cells. Averaged expression values of DEGs (FC ≥1.2 and FDR <0.25) in 2 mice of each genotype were normalized and are shown. (B) Scatter plot displaying the number of regulated genes and targeted regulators for master regulators. Red and blue indicate up- and downregulated master regulators, respectively. (C) GSEA of the RNA-Seq data for Asxl1+/– Nf1+/– cKit+ cells compared with control cells. Selected dysregulated signatures are indicated in color and shown with the NES, P, and FDR values. (D) RT-qPCR verified the increase in mRNA levels of the indicated genes associated with NRAS, MYC, NUP98-HOXA9, and AML (n = 3–8 mice per group). Data represent the mean ± SEM. **P < 0.01, by 1-way ANOVA with Tukey’s multiple comparisons test. (E) Numbers and overlap of DEGs in Asxl1+/– Nf1+/– and single mutant cells. The significance of the overlaps was tested using GeneOverlap (a Fisher’s exact test–based method). (F) Heatmap displays the expression difference of selected dysregulated signatures from C in Asxl1+/– Nf1+/– and single mutant cells compared with controls (P < 0.05 and FDR <0.25).
Loss of Asxl1 and Nf1 cooperates to promote myeloid transformation. This led us to hypothesize a gain of function in gene expression. We identified a pool of differentially expressed genes (DEGs) in Asxl1+/– (66 genes) and Nf1+/– (60 genes) cKit+ cells compared with WT cells (FC ≥1.2 and FDR <0.25, Supplemental Table 4). More than half of the DEGs seen in the cKit+ cells with single-allele mutations (Asxl1+/– or Nf1+/–) had dramatic changes in Asxl1+/– Nf1+/– cells (P < 1 × 10–22, Figure 3E). In contrast, we found that approximately 95% of DEGs in Asxl1+/– Nf1+/– cKit+ cells were not significantly changed in single-mutant cells. Importantly, 84% (21 of 25) of the activated signatures related to NRAS/NF1, MYC, AML, and NUP98-HOXA9 in Asxl1+/– Nf1+/– cells did not display a significant rise in RNA expression in the single-mutant cells (Figure 3F and Supplemental Figure 3I). Furthermore, 93.3% (28 of 30) of the master regulators aberrantly expressed were specific to double-mutant cells (Supplemental Figure 3J). Therefore, the impact of the loss of both Asxl1 and Nf1 on the transcriptome was much greater than would be expected from the additive effects of individual mutant alleles, implying a gain of function in gene regulation by the combined loss of Asxl1 and Nf1.
Increased H3K4me3 enrichment in Asxl1+/– Nf1+/– cells contributes to hyperactivation of key transcription programs.
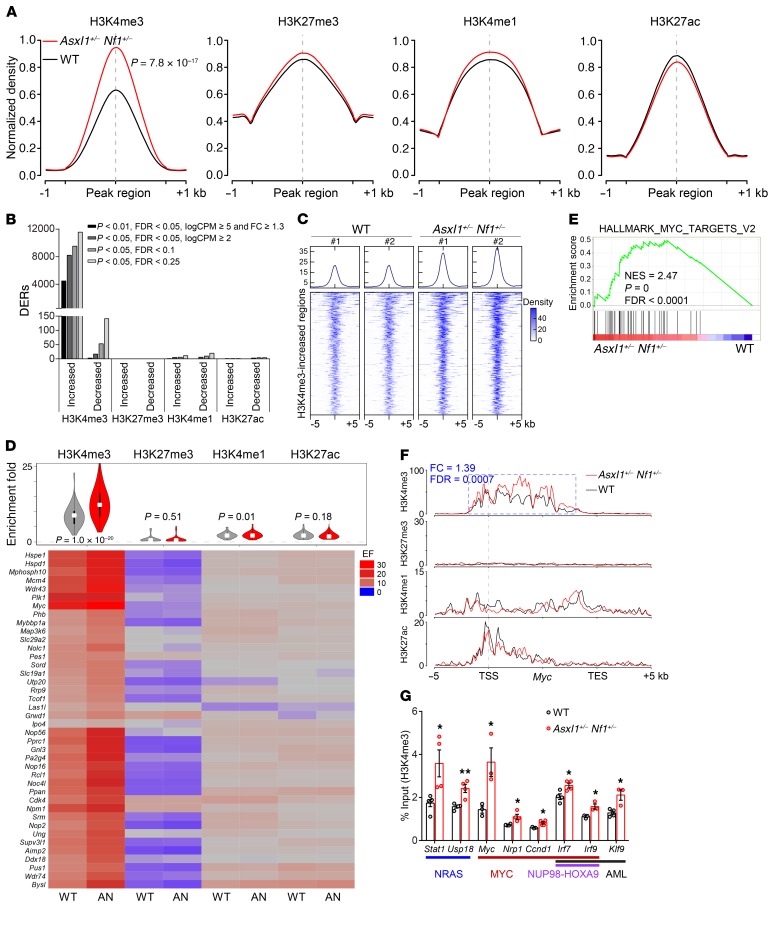
While Asxl1 loss promotes myeloid transformation through abrogation of PRC2-mediated H3K27me3 (25), RAS pathway mutations alter H3K27ac marking at active typical and super enhancers (28). To test whether haploinsufficiency of both Asxl1 and Nf1 affects gene-specific histone modifications, we performed ChIP assays followed by sequencing (ChIP-Seq) for H3K27me3, H3K4me3, H3K4me1, and H3K27ac in WT and Asxl1+/– Nf1+/– cKit+ cells. Normalized global read density and locus-level enrichment consistently revealed a significant increase (30% higher) in genome-wide H3K4me3 occupancy in Asxl1+/– Nf1+/– cells compared with WT cells (P = 7.8 × 10−17) (Figure 4A and Supplemental Figure 4, A–D). In support of this finding, thousands of differentially H3K4me3-enriched regions were identified regardless of cutoffs, in which more than 98% displayed an increase of H3K4me3 enrichment in Asxl1+/– Nf1+/– cKit+ cells (Figure 4B). We observed no significant effect on the other 3 histone marks in WT or Asxl1+/– Nf1+/– cKit+ cells. Western blotting verified the increased levels of global H3K4me3 in Asxl1+/– Nf1+/– cKit+ cells (Supplemental Figure 4E). Furthermore, we detected a significant increase of H3K4me3 enrichment in DKO cKit+ cells compared with WT control cells (Supplemental Figure 4, F and G). Using a more stringent cutoff, 610 H3K4me3-enriched regions had at least a 1.4-fold increase in H3K4me3, while only 2 regions had a 1.4-fold reduction in H3K4me3 in Asxl1+/– Nf1+/– (P < 0.001, FDR < 0.01 and logarithm of counts per million reads [logCPM] ≥5) (Figure 4C). These results demonstrate that alterations of both Asxl1 and Nf1 lead to a marked increase in genome-wide H3K4me3 occupancies in HSCs/HPCs.
Figure 4. Increased H3K4me3 enrichment contributes to aberrant activation of key signatures in Asxl1+/– Nf1+/– cKit+ cells.
(A) Global levels of histone modifications at peaks and flanking 1-kb regions. For ease of comparison, normalized coverages by sequencing depth were scaled to 100% and averaged in 2 biological replicates. (B) Number of DERs for each histone modification identified in Asxl1+/– Nf1+/– cells compared with WT cells according to different cutoffs. (C) Density plot of H3K4me3 ChIP-Seq data sets centered on the midpoints of 610 H3K4me3-increased regions. The plot depicts tag counts in 50-bp bins in the ± 5-kb region surrounding the center. Each row represents a single region. (D) Heatmap displays the densities of histone modification marks on 40 activated genes from the hallmark MYC signature. The ChIP signal density was calculated to be enrichment fold (EF) for the TSS ± 5-kb regions that were averaged in 2 biological replicates. AN, Asxl1+/– Nf1+/–. (E) GSEA plot showing enrichment of the hallmark MYC signature consisting of 40 activated genes in genes marked by promoter H3K4me3 occupancies in Asxl1+/– Nf1+/– versus WT cells. NES, P, and FDR values are shown. (F) Normalized ChIP-binding signals are displayed for 4 histone modifications on the Myc gene. The y axis represents the normalized read density that was averaged in 2 biological replicates. The identified DER and its statistical difference between Asxl1+/– Nf1+/– versus WT cells are indicated in a blue box. TES, transcription end site. (G) ChIP-qPCR verified the increase in H3K4me3 occupancies at the promoters of the indicated genes associated with NRAS, MYC, NUP98-HOXA9, and AML (n = 3–4 mice per group). Data represent the mean ± SEM. *P < 0.05 and **P < 0.01, by unpaired Student’s t test.
The consensus from genome-wide scale analysis reveals that H3K4me3 is characteristic of a transcriptionally active status (29, 30). Genes significantly upregulated in Asxl1+/– Nf1+/– cells showed a significant increase in H3K4me3 occupancies at their promoters, as shown by GSEA analysis (Supplemental Figure 4H). To determine whether the increased H3K4me3 enrichment contributes to the activation of key transcription programs, we assessed the occupancies of histone marks on the MYC hallmark signature as well as on Myc and Ccnd1, two genes associated with cell proliferation. A substantial elevation of H3K4me3 in Asxl1+/– Nf1+/– cKit+ cells versus WT cells affected almost all genes of the MYC-driven transcription program (40 core activated genes), as shown by ChIP-Seq tag densities (Figure 4D). GSEA confirmed that genes comprising the MYC-driven program were enriched for increased H3K4me3 occupancies in Asxl1+/– Nf1+/– cKit+ cells compared with WT cells (Figure 4E). Myc was marked by the active histone modifications H3K4me1/3 and H3K27ac, which were distributed throughout the promoter and partial gene body (Figure 4F). Ccnd1 was occupied by activation and repression histone marks, including H3K4me1/3 and H3K27me3, at its promoter region (Supplemental Figure 4I). Expression levels and upregulated transcription of Myc and Ccnd1 in Asxl1+/– Nf1+/– cKit+ cells were closely correlated with a higher intensity of H3K4me3 rather than of other histone marks. We also examined H3K4me3 occupancies for other dysregulated signatures shown in Figure 3F. Interestingly, we found that signatures driven by MYC, BRD9, AML, and NUP98-HOXA9 had higher H3K4me3 occupancies at their promoter regions in Asxl1+/– Nf1+/– cKit+ cells, while the NRAS-driven signature did not change (Supplemental Figure 4J). ChIP-qPCR confirmed significant increases in H3K4me3 at the promoters of genes that were activated by MYC and NUP98-HOXA9 in Asxl1+/– Nf1+/– cKit+ cells, in contrast to minimal changes in H3K27me3 occupancies (Figure 4G and Supplemental Figure 4K). Thus, Asxl1+/– Nf1+/– cKit+ cells are characterized by a global increase of H3K4me3 that contributes to the activation of key transcription programs such as MYC/BRD9 signatures, suggesting that the AML oncogenic program and myeloid transformation may be associated with global elevation of H3K4me3.
Combined PD0325901 and JQ1 treatment inhibits the development of myeloid leukemia in Asxl1/Nf1 double-mutant mice.
Recently, the bromodomain and extraterminal (BET) family proteins BRD4 and BRD9 were identified as therapeutic targets in AML (31, 32). The inhibition of BRD4 and BRD9 abolished the abnormal self-renewal program in leukemia stem cells through a Myc-dependent mechanism (31–33). We next investigated the efficacy of JQ1 and/or a MEK inhibitor (PD0325901, hereafter referred to as PD-901) on DKO CFU-Cs using colony-forming assays in the presence of PD-901, JQ1, or a combination of PD-901 and JQ1 (PD-901/JQ1). Strikingly, the DKO cells were highly sensitive to the combination of PD-901 and JQ1, as evidenced by a significantly lower frequency of CFU-Cs compared with those from PD-901 or JQ1 single treatment (Figure 5A). In addition, we prepared liquid cultures of BM cells with PD-901 and/or JQ1 in the presence of a cytokine cocktail for 3 days, followed by assessment of the myeloid cell population using flow cytometry. Addition of PD-901 to the liquid culture reduced the percentage of cKit+ cells, while JQ1 decreased the Gr1+Mac1+ cell population without affecting the cKit+ cells in the DKO cultures. Interestingly, the combination of PD-901 and JQ1 dramatically reduced both Gr1+Mac1+ and cKit+Mac1+ cell populations, indicating that the combination of PD-901 plus JQ1 is capable of diminishing both myeloid blast cells and mature Gr1+Mac1+ cells in the cultures of DKO BM cells (Supplemental Figure 5A).
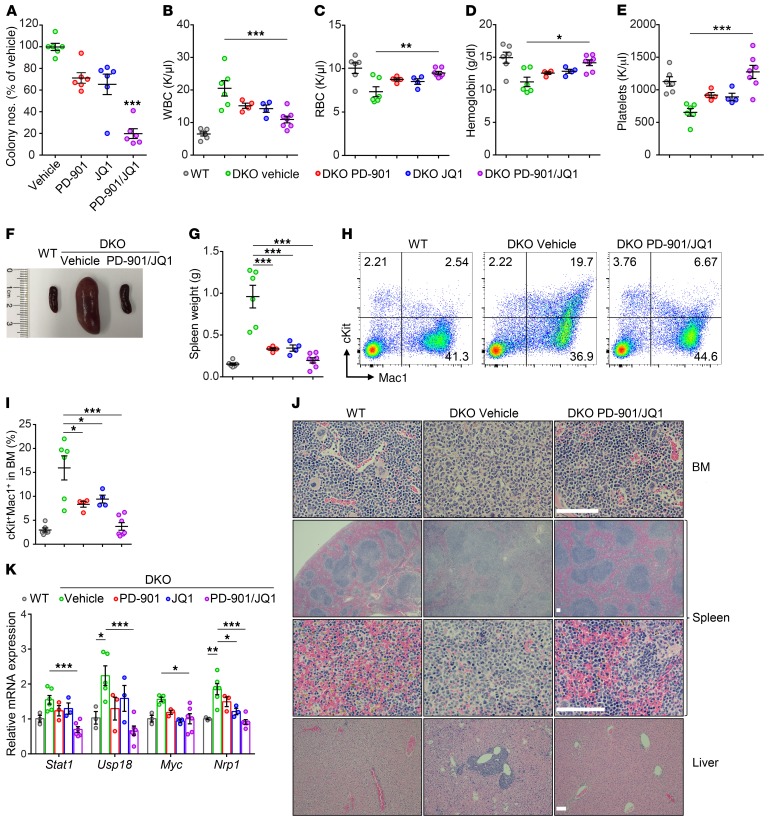
Figure 5. Inhibition of the MAPK pathway and MYC using PD-901 and JQ1 eradicates leukemogenesis in vivo.
(A) Colony-forming assays of splenic cells from DKO mice treated with vehicle, PD-901, JQ1, or PD-901/JQ1 (n = 6 mice per group from 4 independent experiments). (B–E) Peripheral WBC (B), RBC (C), hemoglobin (D), and platelet (E) counts for vehicle-treated (n = 6), PD-901–treated (n = 4), JQ1-treated (n = 4), and PD-901/JQ1–treated (n = 7) DKO mice. (F and G) Spleen size and weight significantly decreased in the PD-901–, JQ1-, and PD-901/JQ1–treated groups. (H and I) The frequency of cKit+Mac1+ cells significantly decreased in the treatment group. (J) Representative H&E-stained sections from BM, spleen, and liver are shown. Scale bar: 100 μm. (K) Expression levels of key genes were rescued in the PD-901/JQ1–treated group (n = 3–6 mice per group). Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by 1-way ANOVA with Tukey’s multiple comparisons test.
To determine whether targeting both MYC and MAPK pathways could prevent the myeloid leukemia initiation and/or slow down disease progression in vivo, we treated the DKO mice with PD-901 and JQ1 following pIpC injection (Supplemental Figure 5B). After 10 weeks of treatment, the mice were sacrificed, and the hematological phenotypes were characterized. Combined treatment with PD-901 and JQ1 significantly decreased WBC and monocyte counts, while RBC, hemoglobin, and platelet counts were significantly increased in the DKO mice (Figure 5, B–E, and Supplemental Figure 5C). The BM cellularity was significantly increased in PD-901/JQ1–treated mice compared with that seen in the vehicle-treated DKO mice (Supplemental Figure 5D). The spleen sizes of DKO mice were also significantly decreased following PD-901 plus JQ1 treatment compared with the spleens of vehicle-treated controls (Figure 5, F and G, and Supplemental Figure 5E). Flow cytometric analyses and cytospin preparation of BM cells revealed that PD-901/JQ1 treatment dramatically reduced the percentage of myeloid progenitor cells and blastic cells in DKO mice compared with the vehicle-treated DKO mice (Figure 5, H and I, and Supplemental Figure 5F), while PD-901/JQ1 treatment did not change the overall percentage of the Gr1+Mac1+ cell population (Supplemental Figure 5G). Interestingly, the spleen weights and BM cKit+Mac1+ cell populations in DKO mice treated with either PD-901 or JQ1 were significantly reduced compared with vehicle controls (Figure 5, G and I, and Supplemental Figure 5E). Furthermore, PD-901/JQ1 treatment significantly increased the CD71+Ter119+ cell population in the BM of DKO mice (Supplemental Figure 5H). Morphological analyses of the histological sections revealed that PD-901/JQ1–treated DKO BM and spleen had a comparable architecture of WT BM and spleen, and no myeloid infiltration was observed in the BM, spleens, or livers of PD-901/JQ1–treated DKO mice (Figure 5J).
Next, we asked whether pharmacological inhibition of the BET bromodomain and MAPK pathway could reverse the dysregulated gene profiling in DKO BM cells. RT-qPCR verified that expression levels of several selected upregulated genes key for hematopoiesis, including Stat1, Usp18, Myc, and Nrp1, were significantly reduced following PD-901/JQ1 treatment (Figure 5K). In contrast, expression of the erythroid transcription factors Tal1 and Klf1 were significantly elevated in BM cells following the treatment (Supplemental Figure 5I). Consistently, enrichment of H3K4me3 at the promoter regions of Myc and Nrp1 was also reduced in the BM cells treated with PD-901 plus JQ1, as determined by ChIP-qPCR (Supplemental Figure 5J). These data suggest that decreased gene expression is associated with the reduced enrichment of H3K4me3 at the promoter regions of these genes. Interestingly, PD-901/JQ1 treatment did not change the enrichment of H3K4me3 at the promoter regions of Stat1 and Usp18 (Supplemental Figure 5J), suggesting that the changes in these genes were H3K4me3 independent.
To validate the treatment effect by PD-901/JQ1 observed in the DKO mice, we further applied PD-901/JQ1 treatment to the WT recipient mice reconstituted with Asxl1+/– Nf1+/– leukemic cells. Compared with the vehicle control group, we found that PD-901/JQ1 treatment significantly extended the survival of Asxl1+/– Nf1+/– leukemic, reconstituted mice (Supplemental Figure 5, K and L). Furthermore, the human AML cell line NOMO1, harboring both ASXL1 and RAS mutations, was also sensitive to PD-901/JQ1 treatment, as assessed by viable cell counts in cultures (Supplemental Figure 5M). Consistently, PD-901/JQ1 dramatically reduced the expression of MYC, NRP1, and USP18 in NOMO1 cells (Supplemental Figure 5N). These results indicate that Asxl1 and Nf1 loss cooperates to promote leukemogenesis and that the combined targeting of the MYC and MAPK pathways may offer an optional therapeutic value for patients with myeloid malignancies harboring ASXL1 and RAS pathway gene mutations.
Concomitant mutations of ASXL1 and RAS pathway genes are more prevalent in AML and high-risk MDS.
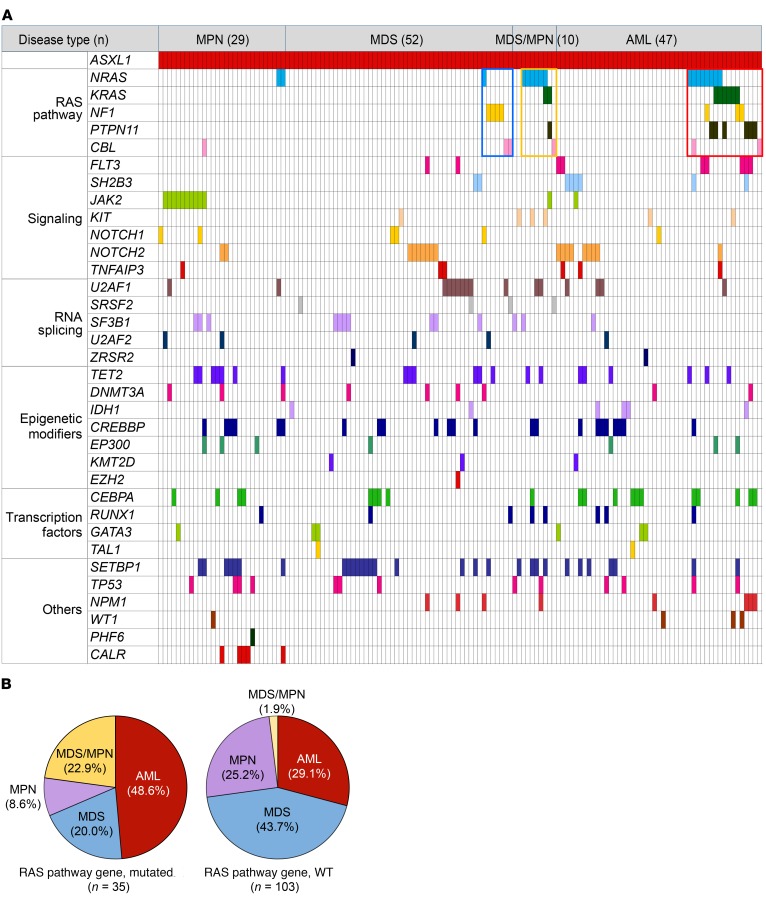
To determine whether the findings in our mouse models recapitulate human myeloid malignancies, we recruited a cohort of 138 patients with myeloid malignancies harboring ASXL1 mutations to the current study and analyzed the cooperating mutation genes. All the patients were diagnosed with myeloid malignancies, including AML, MDS, MPN, or MDS/MPN, according to the 2016 revised World Health Organization criteria (34, 35). We examined the gene mutational profiles of these patients on the basis of targeted sequencing of 67 frequently mutated genes in myeloid malignancies (Figure 6A and Supplemental Table 5). We identified 35 patients with gene mutations involving the RAS/MAPK signaling pathway, including NF1, NRAS, KRAS, PTPN11, or CBL, in the cohort of 138 patients with ASXL1 mutations (Supplemental Table 6). We did not observe loss of heterozygosity in either ASXL1 or RAS pathway genes in these patients (Supplemental Table 6). Notably, the incidence of AML was significantly higher in patients with RAS pathway gene mutations (48.6%) than in those without RAS pathway mutations (29.1%, Figure 6B and Supplemental Table 7). According to the revised International Prognostic Scoring System (36), the MDS patients with RAS pathway gene mutations had a higher frequency of very high-risk MDS (28.6%) than did patients without RAS pathway gene mutations (6.7%, Supplemental Table 8). These data reinforce the notion that concomitant mutations of ASXL1 and RAS pathway genes are probably associated with myeloid malignancies with worse prognosis.
Figure 6. Patterns of concomitantly mutated genes in patients with ASXL1-associated myeloid malignancies.
(A) Identified mutations of 34 genes in samples from the total cohort of 138 patients with ASXL1 mutations are shown by colored squares. Only cases with at least 1 mutation are shown. (B) The incidence of AML was higher in comutations of ASXL1 and RAS pathway genes.
Discussion
Cancers commonly harbor mutations in both signaling pathway components and chromatin regulators. Increasing evidence shows that mutations involving epigenetic regulation and signaling pathways cooperate to promote AML progression in mice. For example, loss of Tet2 or Dnmt3a cooperates with Flt3ITD to induce myeloid leukemia (37–39). Recent studies indicate that oncogenic RAS mutations or loss of Nf1 cooperate with chromatin regulators, such as Dnmt3a, Tet2, Mll3, and Suz12, to accelerate leukemogenesis (40–44). Given the high frequency of NF1/RAS mutations in myeloid malignancies and the need for an effective therapy targeting the oncogenic RAS pathway, we sought to determine whether ASXL1 and NF1 cooperate in leukemogenesis.
We found that RAS pathway gene mutations occurred in 25.4% of ASXL1-mutated myeloid malignancies with a poorer prognosis. Using mouse models of ASXL1 and NF1, we showed that loss of both Asxl1 and Nf1 in hematopoietic cells led to lethal myeloid leukemia, verifying the cooperative effect of mutations in ASXL1 and RAS pathway genes in aggressive forms of myeloid malignancies. Importantly, pharmacological inhibition of both MYC and MAPK pathways prevented disease initiation and inhibited the progression of myeloid leukemia. This study provides a therapeutic strategy for the treatment of myeloid malignancies with ASXL1 and RAS pathway gene mutations.
ASXL1 plays an important role in gene regulation (25, 45, 46). We and others have reported that loss of Asxl1 dysregulates the transcription profile in hematopoietic cells via modulation of repressive histone modifications, such as H3K27me3 (14, 15, 47, 48). However, it remains unclear whether concurrent ASXL1 and NF1 alterations result in a distinct epigenetic signature, triggering leukemogenesis. Our ChIP-Seq analyses revealed a substantial increase in global H3K4me3 occupancies in both Asxl1+/– Nf1+/– and DKO cKit+ cells compared with that observed in WT cells. Interestingly, using MLL-rearranged AML models, Wong et al. demonstrated that a hyper-H3K4me3 epigenetic state maintains a transcriptional program of leukemic stem cells and that its global decrease is associated with leukemic stem cell differentiation (49). Our data also suggest an association between H3K4me3 and the expression profile for key transcription programs, especially those for MYC-driven signature genes, that may contribute to leukemogenesis. MLL is one of the major methyltransferases responsible for H3K4me3 (50). We found that the MLL target gene signature was upregulated in Asxl1+/– Nf1+/– cKit+ cells and was accompanied by a significant increase in H3K4me3 occupancies at their promoter regions (Supplemental Figure 3C and Supplemental Figure 4J). Kim et al. reported that SON interacted with Menin to inhibit MLL complex assembly, leading to a reduction of H3K4me3 and transcription repression (51). Interestingly, our previous mass spectrometric data showed that SON was a candidate interacting partner of ASXL1 (47). It is conceivable that ASXL1 loss may cooperate with Nf1 loss to interrupt SON-Menin interaction, leading to increased H3K4me3-associated leukemogenesis. Future studies are warranted to determine whether the link between ASXL1/NF1 and H3K4me3 is direct.
We and others have shown that Asxl1 mutations can result in loss of function and lead to MDS-like disease in mice (14, 15). Studies from several groups have also shown that truncated mutations of Asxl1 generated gain of function and dominant-negative function, leading to myeloid leukemia (52–54). Future work to investigate the hematopoietic phenotypes in truncated Asxl1 and haploinsufficient Nf1-mutant mice is warranted.
PD-901 has been shown to rescue myeloproliferation and enhance erythropoiesis in Nf1-mutant cells in vivo (55). BET bromodomain inhibition by JQ1 downregulates MYC transcription as well as MYC-dependent target genes (33). Enrichment of MYC/BRD4/9 and NRAS signature genes in Asxl1 and Nf1-deficient mice led us to hypothesize that targeting both the MYC/BRD4/9 and MAPK pathways may be sufficient to rescue the disease phenotype. Indeed, we found that PD-901/JQ1 treatment was sufficient to eradicate the disease phenotype, including a restored WBC count and inhibition of myeloid differentiation skewing. Consistently, the expression levels of MYC and NRAS target genes were restored to WT levels. This study thus provides strong evidence that JQ1 and PD-901 combined therapy is of clinical significance for myeloid malignancies with compound mutations of ASXL1 and RAS pathway genes.
Collectively, our results demonstrate that alterations in both ASXL1 and NF1 cooperatively act to accelerate myeloid leukemia transformation with poorer outcomes. We determined that increased MYC and MAPK pathway activation contributes to the progression of myeloid malignancies driven by combined loss of Asxl1 and Nf1. Our studies establish a platform for testing the therapeutic strategy to treat myeloid malignancies harboring ASXL1 and RAS pathway gene mutations.
Methods
Mice and chemicals.
The generation of Asxl1+/–, Nf1+/–, Asxl1fl/fl, and Nf1fl/fl mice has been previously described (14, 15, 56, 57). Mx1-Cre–transgenic mice were purchased from The Jackson Laboratory. All mice were bred on a C57BL/6 genetic background. Chemicals were obtained from MilliporeSigma, unless otherwise indicated.
Phenotypic analyses of mice.
PB was collected and subjected to an automated blood count (Hemavet System 950FS, Drew Scientific). For morphological and lineage differential analysis, PB smears were subjected to May-Grünwald-Giemsa staining. Morphological analyses of BM and splenic cells were performed on cytospins followed by May-Grünwald-Giemsa staining. For histopathologic analyses, femurs and sternums were fixed for more than 24 hours in 10% neutral buffered formalin at room temperature and demineralized in a solution of 10% EDTA for 1 to 2 weeks. The specimens and other soft tissues (spleen and liver) were dehydrated using ethanol and cleared in xylenes. The specimens were then embedded in melted paraffin and allowed to harden. Thin sections (4–5 μm thickness) were cut and floated onto microscope slides. For routine assessment, slides were stained with H&E. The slides were visualized under a Nikon TE2000-S microscope.
Flow cytometry, cell sorting, and colony assays.
Total WBC were obtained after lysis of PB with red cell lysis buffer. Single-cell suspensions from BM and PB were stained with panels of fluorochrome-conjugated antibodies (Supplemental Table 9). Flow cytometric analysis of HSCs/HPCs was performed as previously described (26). The analyses were performed using a BD FACSCanto II or LSR Fortessa flow cytometer (BD Biosciences). All data were analyzed using FlowJo software, version 10. BM cKit+ cells were sorted using cKit (CD117) MicroBeads (Miltenyi Biotec). The purity of selected cKit+ cells was routinely greater than 95%. For CFU assays, BM or splenic cells were plated in triplicate in methylcellulose medium (MethoCult M3134, STEMCELL Technologies) supplemented with mouse stem cell factor (mSCF) (100 ng/ml), mouse IL-3 (mIL-3) (10 ng/ml), mouse erythropoietin (mEPO) (4 U/ml), mouse thrombopoietin (mTPO) (50 ng/ml), mouse granulocyte-macrophage CSF (mGM-CSF) (10 ng/ml), and human IL-6 (50 ng/ml, Peprotech). The cells were then scored on day 7 of the cultures at 37°C and 5% CO2.
Paired daughter cell assay.
Paired daughter cell assays were performed to examine the frequency of HSC self-renewal and differentiation capabilities (58). Single CD34−LSK cells from BM were sorted into 96-well plates at 1 cell per well containing RPMI 1640 medium supplemented with mSCF (100 ng/ml) and mTPO (50 ng/ml). After the first cell division, the 2 daughter cells were separated, 1 per well, for an additional 12 days in the medium supplemented with mSCF, mIL-3, mEPO, mTPO, and mGM-CSF. The self-renewal and differentiation capabilities of cultured CD34−LSK cells were determined by microscopic morphological analyses following H&E staining.
Single-cell colony assay.
To explore the proliferative potential of single LT-HSCs, we performed the single cell colony assay as previously described (59). Single CD34−LSK cells were sorted into 96-well plates with 10% FBS, 1% BSA, and 2 mM L-glutamine in α-MEM and supplemented with mSCF (100 ng/ml), mIL-3 (10 ng/ml), mEPO (4 U/ml), and mTPO (50 ng/ml). The colonies were scored after 7 days, and the progenitors of individual colonies were collected and replated onto a new 96-well plate.
Transplantation assays.
Total BM single-cell suspensions were harvested from 1-month-old WT and Asxl1+/– Nf1+/– mice (without obvious disease phenotype), and 1 × 106 cells were injected into lethally irradiated (9.5 Gy) WT recipients (CD45.1+) through the tail vein. For the tumor transfer assay, 1 × 106 splenic cells from a 13-month-old Asxl1+/– Nf1+/– mouse with myeloid leukemia were injected into sublethally irradiated (6.5 Gy) WT recipients (CD45.1+). The recipient mice were sacrificed when they became moribund. The mice were examined to determine their hematological phenotype and development of myeloid malignancies.
A competitive repopulation assay was performed by transplanting a mixture of 1 × 106 1-month-old WT CD45.1+ BM competitor cells and 1 × 106 of 1-month-old WT, Asxl1+/, Nf1+/–, or Asxl1+/– Nf1+/– CD45.2+ BM cells into lethally irradiated (9.5 Gy) WT recipients (CD45.1+). PB engraftment was analyzed every 3 months for the following 12 months after transplantation.
Immunofluorescence.
Cells were fixed in 4% paraformaldehyde (Alfa Aesar) in PBS for 15 minutes, permeabilized in 0.5% Triton X-100 in PBS for 5 minutes, blocked with 4% goat serum in PBS for 60 minutes, and incubated with rabbit monoclonal anti–c-MYC (5605S, 1:200, Cell Signaling Technology) for 2 hours at room temperature. DyLight 488–conjugated goat anti-rabbit was used as a secondary antibody (35552, 1:500, Thermo Fisher Scientific). Samples were mounted in VECTASHIELD Antifade Mounting Medium containing DAPI (H-1200, Vector Laboratories) on glass slides and photographed on a fluorescence microscope (Nikon ECLIPSE Ci).
Inhibitor treatment.
For CFU assays, splenic cells from Asxl1Δ/Δ Nf1Δ/Δ (DKO) mice were plated in methylcellulose medium in the presence of either DMSO (vehicle), 200 nM PD0325901 (PD-901, LC Laboratories), 200 nM JQ1 (APExBIO), or a combination of PD-901 and JQ1. Inhibitors were prepared in DMSO and plated in a total volume of 1 μl/ml. All colonies were scored after 7 days of culture.
The human leukemic cell lines K562 and NOMO-1 were obtained from American Type Culture Collection (ATCC) and cultured in RPMI1640 medium with 10% FBS and 1% penicillin-streptomycin. Cells were treated with PD-901, JQ1, or a combination of PD-901 and JQ1 at the indicated concentrations (Supplemental Figure 5M) for 3 days, after which the viable cells were counted.
In vivo treatment studies.
DKO mice were randomized to begin treatment with either the MEK inhibitor PD-901 (5 mg/kg, oral gavage), the MYC inhibitor JQ1 (50 mg/kg, i.p. injection), a combination of PD-901 and JQ1, or vehicle followed by injection of pIpC. The mice used for in vivo treatment studies were 1 month old, and both males and females were included. After 10 weeks of treatment, the mice were sacrificed and analyzed.
Asxl1+/–Nf1+/– leukemic cells (1 × 106 cells per recipient mouse) were transplanted into sublethally irradiated WT recipient mice (CD45.1+). The mice were then randomized into 2 groups that received either vehicle or a combination of PD-901 and JQ1 two weeks after the transplantation. The recipient mice were monitored for hematopoietic phenotypes and signs of drug toxicity and then euthanized when the disease was evident.
Whole-exome sequencing analysis.
DNA was prepared from 2 Asxl1+/– Nf1+/– leukemic cells, and 1 matched normal tail was used as a control. Exome libraries were captured with an Agilent SureSelectXT 50 mb Mouse All Exon Kit and followed by sequencing with a read length of 2 × 100 bp using the Illumina HiSeq system. Approximately 3.5 Gb data per sample were aligned to the mouse genome (GRCm38/mm10) with the Burrows-Wheeler Alignment (BWA) tool (60). The alignments were preprocessed and duplications removed with the Picard tool (http://broadinstitute.github.io/picard/) and GATK (61). Single nucleotide variants (SNVs) and small indels were called with both VarScan2 (62) and MuTect2 (63) (max_alt_allele_in_normal_fraction = 0.75) in leukemia cells compared with control cells (at least 8 and 6 reads that covered the mutation loci in normal and cancer cells, respectively, with at least 2 reads supporting the variant allele), and were related to protein changes with ANNOVAR (64). Mutations that were found in genomic repeat, intergenic, and intronic regions as well as known polymorphism sites were excluded. Finally, 67 potential functional or regulatory point mutations with a variant allele frequency of at least 15% were kept for further analysis. The genes with potential point mutations were assigned to pathways with ToppGene (65). The commonly mutated genes or pathways were searched for in the 2 leukemic mice.
RNA-Seq analyses.
Purified cKit+ cells from 8-month-old Asxl1+/– Nf1+/– preleukemic mice with splenomegaly and age-matched WT, Asxl1+/–, and Nf1+/– mice were used. The RNA libraries were prepared with the Illumina TruSeq Strand-Specific mRNA Sample Preparation System and followed by sequencing with a read length of 2 × 76 bp using the Illumina NextSeq 500. Approximately 50 million paired-end reads per sample were aligned to the mouse genome (GRCm38/mm10), and RefSeq annotation (released June 28, 2017) was obtained from UCSC using TopHat (version 2.1.1) (66) with a parameter of --max-multihits = 1. The rates of overall mapped reads reached approximately 90% to 97%. Cufflink software (version 2.2.1) (66) was used to quantify the gene expression values by calculating the fragments per kilobase per million mapped reads (FPKM) value. Cuffdiff (66) was used to detect DEGs, and DEGs with a cutoff of P < 0.05, FC ≥1.2, and a FDR <0.25 were selected for further analyses. The identified DEGs shown in Supplemental Table 4 were used for the master regulator prediction analyses with Ingenuity Pathway Analysis (IPA) (QIAGEN; www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/). Master regulators predicted (P value of overlap < 1 × 10–20, network P < 0.01 and |Z score| ≥1.5) were selected if differentially expressed in Asxl1+/– Nf1+/– cells compared with controls. GSEA analysis (67) was performed with C2 curated gene sets and hallmark signatures in the GSEA Molecular Signatures Database (MSigDB), version 6.0, as well as with published signatures. Enriched gene sets in Asxl1+/– Nf1+/– cells compared with cells of other genotypes were selected using a cutoff of P < 0.05 and a FDR < 0.25.
Four expression data sets for AML patients with ASXL1 and/or RAS pathway mutations (NF1, NRAS, KRAS, PTPN11, and CBL) were obtained from TCGA database (21) and the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE42064 [ref. 68] and GSE84359 [ref. 69]). DEGs in patients with ASXL1/RAS pathway mutations compared with the individuals without ASXL1/RAS pathway mutations were identified with edgeR (70) (P < 0.05 and FC ≥1.5 or 2).
RT-qPCR analysis.
Total RNA was extracted with TRIzol Reagent (Invitrogen, Thermo Fisher Scientific), and cDNA was synthesized using the QuantiTect Reverse Transcription Kit (QIAGEN) according to the manufacturer’s instructions. RT-qPCR was performed in triplicate using an ABI 7500 with the Fast SYBR Green Master Mix (both from Applied Biosystems). Expression of the gene of interest was normalized to the housekeeping gene Actb using the 2–ΔΔCt method. All RT-qPCR primers used are listed in Supplemental Table 10.
Western blot analysis.
BM cKit+ cells were harvested from 8- to 10-month-old preleukemic mice with splenomegaly. Lysates from cKit+ cells were prepared and then resolved on NuPAGE 4%–12% Bis-Tris Gels (Invitrogen, Thermo Fisher Scientific). Immunoblotting was performed with the following antibodies: rabbit polyclonal anti-H3K4me3 (C15410003, 1:1,000, Diagenode); rabbit polyclonal anti-H3 (ab1791, 1:2,000, Abcam); rabbit monoclonal anti–c-MYC (5605S, 1:500, Cell Signaling Technology); and mouse monoclonal anti–β-actin (A2228, 1:2,000, MilliporeSigma). After incubation with anti–rabbit IgG or anti–mouse IgG antibodies conjugated with HRP (GE Healthcare), signals were detected using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific).
Quantification of RAS-GTP levels was performed using a RAS Activation Assay Kit (MilliporeSigma) according to the manufacturer’s instructions. The product was detected using a mouse monoclonal anti-RAS antibody (05-516, 1:1,000, MilliporeSigma).
ChIP-Seq and data analysis.
ChIP assays were performed using cKit+ cells from 8-month-old Asxl1+/– Nf1+/– and WT mice as previously reported (26). Genomic DNA regions of interest were isolated using antibodies against H3K4me3 (C15410003, Diagenode), H3K4me1 (C15410194, Diagenode), H3K27ac (C15410196, Diagenode), and H3K27me3 (07-449, MilliporeSigma). Additional H3K4me3 ChIP assays were performed using cKit+ cells from 8-month-old Asxl1+/– and Nf1+/– mice and cKit+ cells from Asxl1Δ/Δ, Nf1Δ/Δ, and DKO mice (3 months after pIpC). The ChIP libraries were prepared using the MicroPlex Library Preparation Kit (Diagenode) and sequenced with a read length of 76 bp on an Illumina NextSeq 500 system.
Approximately 20 to 50 million single-end reads per sample were aligned to the mouse genome (GRCm38/mm10), and enrichment peaks of histone modifications were called as described previously (26). Using MULTOVL version 1.2 (71), with the parameters ‘-u -m 1’, a total of 31277, 35312, 87995, and 37637 peak regions were present in at least 1 biological replicate of H3K4me3, H3K27me3, H3K4me1, and H3K27ac ChIP-Seqs, respectively (the union of overlapping regions was reported; minimum overlap required 1 bp). A total of 109,419 union regions of 4 histone modifications were assigned to the nearest genes within 100 kb using RefSeq annotation (released June 28, 2017) from UCSC as described previously (26). Read counting and differential enrichment analysis for each histone modification were done with CisGenome (72) and edgeR (70), respectively. Significant differences in H3K27me3, H3K4me1, and H3K27ac enrichments were observed at only approximately 10 regions in Asxl1+/– Nf1+/– cells compared with WT cells that were filtered according to several sets of cutoffs. A total of 612 differentially enriched regions (DERs) of H3K4me3 were identified in Asxl1+/– Nf1+/– cells compared with WT cells (P ≤ 0.001, FC ≥1.4, FDR < 0.01, and logCPM ≥5), in which only 2 regions had a 1.4-fold reduction of H3K4me3 enrichment. Similarly, when comparing with matched WT cells, 20, 2,581, and 619 DERs of H3K4me3 were identified in Asxl1Δ/Δ, Nf1Δ/Δ, and Asxl1Δ/Δ Nf1Δ/Δ cells, respectively (P ≤ 0.001, FC ≥1.3, FDR <0.01, and logCPM ≥5). ChIP-Seq read counts were normalized to reads per million (RPM) and combined into 1 matrix with deepTools (73) for heatmap and average profile visualization. For each gene, an enrichment fold of H3K4me3 at the transcription start site (TSS) ± a 5-kb region was defined as log2-transformed RPM in ChIP divided by RPM in the input, log2(ChIP/input). This enrichment fold matrix was used to perform GSEA analysis. All figures were visualized in R and GraphPad Prism software.
Patients’ samples.
A cohort of 138 patients with myeloid malignancies harboring ASXL1 mutations was included in this study. The patients were diagnosed according to the 2016 revised World Health Organization criteria (35). The mutational status for 67 myeloid panel genes (Supplemental Table 5) was analyzed by targeted exome sequencing in BM mononuclear cells from 138 patients with MDS (n = 52), MPN (n = 29), MDS/MPN (n = 10), or AML (n = 47).
Targeted exome-sequencing and data analysis.
Ion Torrent AmpliSeq (Thermo Fisher Scientific) was used for detecting the mutational status of 67 myeloid panel genes. SNVs and small indels (≤50 bp) were identified with Ion Reporter directly from within Torrent Suite Software (Thermo Fisher Scientific) and annotated with the COSMIC cancer mutation database, human population variant databases including 1000 genomes, ExAC, and ESP6500, and functional tools and databases including SIFT, Polyphen2, and MutationTaster, as well as our collected mutation hotspots. SNVs and small indels that affect protein coding or splice sites were kept to determine the cooperativity of mutated genes if they had a minor allele frequency of less than 0.05 in human populations.
Statistics.
Differences between experimental groups were determined by the log-rank test, unpaired 2-tailed Student’s t test, or 1-way ANOVA followed by an appropriate post hoc correction. P values of less than 0.05 were considered statistically significant.
Study approval.
Animal care and experiments were conducted in accordance with IACUC regulatory guidelines of the University of Miami Miller School of Medicine. BM samples for research were obtained from patients according to a study protocol approved by the ethics committee of the Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences, with written informed consent provided by all patients prior to their inclusion in the study.
Data availability.
The raw sequencing data reported here were deposited in the Genome Sequence Archive database (74) at the Beijing Institute of Genomics (BIG) Data Center (75), Chinese Academy of Sciences, under accession numbers CRA000666 and CRA000668 (http://bigd.big.ac.cn/gsa).
Author contributions
FCY conceived the project. PZ and FCY designed the study. PZ, SY, SC, YG, NM, HY, and SW performed the experiments. PZ, FH, Lin Zhang, and QFW performed whole-exome sequencing, RNA-Seq, and ChIP-Seq. JB, MS, Lei Zhang, and YZ provided patients’ samples and analyzed the clinical data. PZ, FH, TC, SDN, MX, QFW, and FCY discussed and analyzed the data. PZ, FH, and FCY wrote the manuscript. All authors reviewed, edited, and approved the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (CA172408 and CA185751, to FCY and MX); the Leukemia and Lymphoma Society Specialized Center of Research grant (to FCY, MX, and SDN); the National Natural Science Foundation of China (91731302, to QFW; 81570113, to YZ; 81421002, to TC; 81470302, to LZ; and 81470340, to FH); the Social Development Project of Jiangsu Province (CXTDA2017014); and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (to FH). We thank the members of the Satellite Histological Core, Flow Cytometry Core, and Oncogenomics Core Facilities of the Sylvester Comprehensive Cancer Center at the University of Miami Miller School of Medicine.
Version 1. 09/18/2018
In-Press Preview
Version 2. 10/29/2018
Electronic publication
Version 3. 11/06/2018
Label corrections have been made in Figures 2 and 3, as well as minor changes in the text for clarity.
Version 4. 12/03/2018
Print issue publication
Footnotes
SY’s present address is: Showa University Fujigaoka Hospital, Yokohama, Japan.
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(12):5383–5398.https://doi.org/10.1172/JCI121366.
Contributor Information
Peng Zhang, Email: pxz36@miami.edu.
Fuhong He, Email: hefh@big.ac.cn.
Jie Bai, Email: janebai86@hotmail.com.
Shohei Yamamoto, Email: shohei-y@med.showa-u.ac.jp.
Shi Chen, Email: sxc1137@med.miami.edu.
Lin Zhang, Email: zhanglin17m@big.ac.cn.
Mengyao Sheng, Email: mengyao0714@163.com.
Lei Zhang, Email: zhanglei1@ihcams.ac.cn.
Ying Guo, Email: yxg365@med.miami.edu.
Na Man, Email: manna@med.miami.edu.
Hui Yang, Email: hxy211@med.miami.edu.
Suyun Wang, Email: suyunwcn@163.com.
Tao Cheng, Email: chengtao@ihcams.ac.cn.
Stephen D. Nimer, Email: snimer@med.miami.edu.
Yuan Zhou, Email: yuanzhou@ihcams.ac.cn.
Mingjiang Xu, Email: mxx51@med.miami.edu.
Qian-Fei Wang, Email: wangqf@big.ac.cn.
Feng-Chun Yang, Email: fxy37@med.miami.edu.
References
- 1.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12(9):599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 2.Ntziachristos P, Mullenders J, Trimarchi T, Aifantis I. Mechanisms of epigenetic regulation of leukemia onset and progression. Adv Immunol. 2013;117:1–38. doi: 10.1016/B978-0-12-410524-9.00001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenblatt SM, Nimer SD. Chromatin modifiers and the promise of epigenetic therapy in acute leukemia. Leukemia. 2014;28(7):1396–1406. doi: 10.1038/leu.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 5.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papaemmanuil E, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gelsi-Boyer V, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145(6):788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 8.Boultwood J, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24(5):1062–1065. doi: 10.1038/leu.2010.20. [DOI] [PubMed] [Google Scholar]
- 9.Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24(6):1128–1138. doi: 10.1038/leu.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdel-Wahab O, et al. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 2010;70(2):447–452. doi: 10.1158/0008-5472.CAN-09-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou WC, et al. Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations. Blood. 2010;116(20):4086–4094. doi: 10.1182/blood-2010-05-283291. [DOI] [PubMed] [Google Scholar]
- 12.Gelsi-Boyer V, et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol. 2010;151(4):365–375. doi: 10.1111/j.1365-2141.2010.08381.x. [DOI] [PubMed] [Google Scholar]
- 13.Gelsi-Boyer V, Brecqueville M, Devillier R, Murati A, Mozziconacci MJ, Birnbaum D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol. 2012;5:12. doi: 10.1186/1756-8722-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdel-Wahab O, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210(12):2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123(4):541–553. doi: 10.1182/blood-2013-05-500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowen DT, et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood. 2005;106(6):2113–2119. doi: 10.1182/blood-2005-03-0867. [DOI] [PubMed] [Google Scholar]
- 17.Parkin B, et al. NF1 inactivation in adult acute myelogenous leukemia. Clin Cancer Res. 2010;16(16):4135–4147. doi: 10.1158/1078-0432.CCR-09-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vries AC, Zwaan CM, van den Heuvel-Eibrink MM. Molecular basis of juvenile myelomonocytic leukemia. Haematologica. 2010;95(2):179–182. doi: 10.3324/haematol.2009.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward AF, Braun BS, Shannon KM. Targeting oncogenic Ras signaling in hematologic malignancies. Blood. 2012;120(17):3397–3406. doi: 10.1182/blood-2012-05-378596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatlen MA, et al. Integrative genetic analysis of mouse and human AML identifies cooperating disease alleles. J Exp Med. 2016;213(1):25–34. doi: 10.1084/jem.20150524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen SJ, Shen Y, Chen Z. A panoramic view of acute myeloid leukemia. Nat Genet. 2013;45(6):586–587. doi: 10.1038/ng.2651. [DOI] [PubMed] [Google Scholar]
- 23.Gottfried ON, Viskochil DH, Couldwell WT. Neurofibromatosis type 1 and tumorigenesis: molecular mechanisms and therapeutic implications. Neurosurg Focus. 2010;28(1):E8. doi: 10.3171/2009.11.FOCUS09221. [DOI] [PubMed] [Google Scholar]
- 24.Staser K, Yang FC, Clapp DW. Plexiform neurofibroma genesis: questions of Nf1 gene dose and hyperactive mast cells. Curr Opin Hematol. 2010;17(4):287–293. doi: 10.1097/MOH.0b013e328339511b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdel-Wahab O, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22(2):180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, et al. Loss of Asxl2 leads to myeloid malignancies in mice. Nat Commun. 2017;8:15456. doi: 10.1038/ncomms15456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le DT, et al. Somatic inactivation of Nf1 in hematopoietic cells results in a progressive myeloproliferative disorder. Blood. 2004;103(11):4243–4250. doi: 10.1182/blood-2003-08-2650. [DOI] [PubMed] [Google Scholar]
- 28.Nabet B, et al. Deregulation of the Ras-Erk signaling axis modulates the enhancer landscape. Cell Rep. 2015;12(8):1300–1313. doi: 10.1016/j.celrep.2015.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 30.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27(12):1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hohmann AF, et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat Chem Biol. 2016;12(9):672–679. doi: 10.1038/nchembio.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, et al. The usefulness of mutational data on prognosis of myelodysplastic syndromes: alone or incorporated into the IPSS-R? [published online ahead of print November 28, 2017] Br J Haematol. doi: 10.1111/bjh.15036. https://doi.org/10.1111/bjh.15036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 36.Greenberg PL, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120(12):2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shih AH, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27(4):502–515. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, et al. DNMT3A Loss Drives Enhancer Hypomethylation in FLT3-ITD-Associated Leukemias. Cancer Cell. 2016;30(2):363–365. doi: 10.1016/j.ccell.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 39.Meyer SE, et al. DNMT3A Haploinsufficiency Transforms FLT3ITD Myeloproliferative Disease into a Rapid, Spontaneous, and Fully Penetrant Acute Myeloid Leukemia. Cancer Discov. 2016;6(5):501–515. doi: 10.1158/2159-8290.CD-16-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25(5):652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Raedt T, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514(7521):247–251. doi: 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- 42.Chang YI, et al. Loss of Dnmt3a and endogenous Kras(G12D/+) cooperate to regulate hematopoietic stem and progenitor cell functions in leukemogenesis. Leukemia. 2015;29(9):1847–1856. doi: 10.1038/leu.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu R, et al. Epigenetic perturbations by Arg882-mutated DNMT3A potentiate aberrant stem cell gene-expression program and acute leukemia development. Cancer Cell. 2016;30(1):92–107. doi: 10.1016/j.ccell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunimoto H, et al. Cooperative epigenetic remodeling by TET2 loss and NRAS mutation drives myeloid transformation and MEK inhibitor sensitivity. Cancer Cell. 2018;33(1):44–59.e8. doi: 10.1016/j.ccell.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheuermann JC, et al. Histone H2A deubiquitinase activity of the polycomb repressive complex PR-DUB. Nature. 2010;465(7295):243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang P, et al. Loss of ASXL1 in the bone marrow niche dysregulates hematopoietic stem and progenitor cell fates. Cell Discov. 2018;4:4. doi: 10.1038/s41421-017-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, et al. ASXL1 interacts with the cohesin complex to maintain chromatid separation and gene expression for normal hematopoiesis. Sci Adv. 2017;3(1):e1601602. doi: 10.1126/sciadv.1601602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi H, et al. ASXL1 plays an important role in erythropoiesis. Sci Rep. 2016;6:28789. doi: 10.1038/srep28789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong SH, et al. The H3K4-methyl epigenome regulates leukemia stem cell oncogenic potential. Cancer Cell. 2015;28(2):198–209. doi: 10.1016/j.ccell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang P, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29(22):6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JH, et al. SON and its alternatively spliced isoforms control MLL complex-mediated H3K4me3 and transcription of leukemia-associated genes. Mol Cell. 2016;61(6):859–873. doi: 10.1016/j.molcel.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balasubramani A, et al. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat Commun. 2015;6:7307. doi: 10.1038/ncomms8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H, et al. Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood. 2018;131(3):328–341. doi: 10.1182/blood-2017-06-789669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagase R, et al. Expression of mutant Asxl1 perturbs hematopoiesis and promotes susceptibility to leukemic transformation. J Exp Med. 2018;215(6):1729–1747. doi: 10.1084/jem.20171151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang T, et al. Sustained MEK inhibition abrogates myeloproliferative disease in Nf1 mutant mice. J Clin Invest. 2013;123(1):335–339. doi: 10.1172/JCI63193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7(3):353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 57.Zhu Y, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15(7):859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takano H, Ema H, Sudo K, Nakauchi H. Asymmetric division and lineage commitment at the level of hematopoietic stem cells: inference from differentiation in daughter cell and granddaughter cell pairs. J Exp Med. 2004;199(3):295–302. doi: 10.1084/jem.20030929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ema H, et al. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nat Protoc. 2006;1(6):2979–2987. doi: 10.1038/nprot.2006.447. [DOI] [PubMed] [Google Scholar]
- 60.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grossmann V, et al. CEBPA double-mutated acute myeloid leukaemia harbours concomitant molecular mutations in 76·8% of cases with TET2 and GATA2 alterations impacting prognosis. Br J Haematol. 2013;161(5):649–658. doi: 10.1111/bjh.12297. [DOI] [PubMed] [Google Scholar]
- 69.Micol JB, et al. ASXL2 is essential for haematopoiesis and acts as a haploinsufficient tumour suppressor in leukemia. Nat Commun. 2017;8:15429. doi: 10.1038/ncomms15429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aszódi A. MULTOVL: fast multiple overlaps of genomic regions. Bioinformatics. 2012;28(24):3318–3319. doi: 10.1093/bioinformatics/bts607. [DOI] [PubMed] [Google Scholar]
- 72.Ji H, Jiang H, Ma W, Wong WH. Using CisGenome to analyze ChIP-chip and ChIP-seq data. Curr Protoc Bioinformatics. 2011;Chapter 2:Unit2.13. doi: 10.1002/0471250953.bi0213s33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42(Web Server issue):W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, et al. GSA: Genome Sequence Archive<sup/>. Genomics Proteomics Bioinformatics. 2017;15(1):14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.BIG Data Center Members The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 2017;45(D1):D18–D24. doi: 10.1093/nar/gkw1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data reported here were deposited in the Genome Sequence Archive database (74) at the Beijing Institute of Genomics (BIG) Data Center (75), Chinese Academy of Sciences, under accession numbers CRA000666 and CRA000668 (http://bigd.big.ac.cn/gsa).