Abstract
The ubiquitin-proteasome system (UPS) degrades a protein molecule via 2 main steps: ubiquitination and proteasomal degradation. Extraproteasomal ubiquitin receptors are thought to couple the 2 steps, but this proposition has not been tested in vivo with vertebrates. More importantly, impaired UPS performance plays a major role in cardiac pathogenesis, including myocardial ischemia-reperfusion injury (IRI), but the molecular basis of UPS impairment remains poorly understood. Ubiquilin1 is a bona fide extraproteasomal ubiquitin receptor. Here, we report that mice with a cardiomyocyte-restricted knockout of Ubiquilin1 (Ubqln1-CKO mice) accumulated a surrogate UPS substrate (GFPdgn) and increased myocardial ubiquitinated proteins without altering proteasome activities, resulting in late-onset cardiomyopathy and a markedly shortened life span. When subject to regional myocardial ischemia-reperfusion, young Ubqln1-CKO mice showed substantially exacerbated cardiac malfunction and enlarged infarct size, and conversely, mice with transgenic Ubqln1 overexpression displayed attenuated IRI. Furthermore, Ubqln1 overexpression facilitated proteasomal degradation of oxidized proteins and the degradation of a UPS surrogate substrate in cultured cardiomyocytes without increasing autophagic flux. These findings demonstrate that Ubiquilin1 is essential to cardiac ubiquitination-proteasome coupling and that an inadequacy in the coupling represents a major pathogenic factor for myocardial IRI; therefore, strategies to strengthen coupling have the potential to reduce IRI.
Keywords: Cardiology
Keywords: Cardiovascular disease, Protein traffic, Ubiquitin-proteosome system
Introduction
Targeted removal of individual abnormal protein molecules in the cell is primarily performed by the ubiquitin-proteasome system (UPS). Hence, the UPS is vital to cellular protein quality control (PQC), which functions to minimize the level and toxicity of misfolded proteins. By regulatory degradation of most cellular proteins that are normal but no longer needed, the UPS also participates in the regulation and execution of virtually all cellular processes (1, 2). Thus, the proper functioning of the UPS is pivotal to both the functioning and survival of the cell. UPS-mediated degradation of a protein molecule is generally known to take 2 main steps: step 1 is the covalent attachment of a ubiquitin (Ub) chain to a target protein molecule via a process known as ubiquitination, in which Ub ligases (E3) are rate limiting and determine substrate specificity; step 2 is proteasome-mediated degradation of the ubiquitinated protein molecule (1, 2). The pathophysiological significance of defects in either step is being rapidly elucidated, as exemplified by identification of genetic mutations of Ub and Ub ligases in human diseases (3–7), by revelation of the essential roles of Ub ligases in organ/system (patho)physiology (6–14), and by demonstration of the therapeutic effectiveness of measures priming the proteasome in some of the most common and debilitating diseases or pathological processes in animal models (15-18). These exciting recent advances in UPS research have illustrated that dysfunction in either the ubiquitination or the proteasomal steps can impair UPS functioning and play important pathogenic roles. Notably, studies using cell culture and lower species have suggested that the coupling between ubiquitination and proteasomal degradation also represents an important step in UPS-mediated proteolysis (19); however, the coupling factors have rarely been investigated using vertebrates. More importantly, the pathophysiological significance of impaired or inadequate coupling has, for the most part, not been explored. The present study was conducted to fill these critical gaps.
Polyubiquitination and proteasomal degradation are coupled by a family of proteins known as Ub receptors, which recruit polyubiquitinated proteins and dock them to the proteasome for degradation (20). The 19S proteasome subunits Rpn10/S5a and Rpn13 harbor Ub interacting motif (UIM) domains and a pleckstrin-like receptor for Ub (Pru) domain, respectively; hence, they can serve as intraproteasomal Ub receptors to directly bind polyubiquitinated proteins for degradation (21). However, the recruitment and docking of polyubiquitinated proteins to the 26S proteasome seem to also employ shuttling factors or extraproteasomal Ub receptors. The latter must be capable of both binding ubiquitinated substrates and interacting with the 19S proteasome (21). The UBL-UBA family proteins, including Ubiquilin1 (Ubqln1) and the mammalian homologs of Rad23 and Ddi1, are ideally suited to do so and are regarded as the shuttling Ub receptors. These include the Ub-like (UBL) domain at the N terminus and the Ub-associated (UBA) domain or domains at the C terminus. The UBA binds polyubiquitinated proteins, while the UBL can interact with the 19S proteasome via Rpn10 or Rpn1 (20).
Among UBL-UBA proteins, Rad23 is extensively studied for its role in DNA repair (22); however, very few reported studies have examined the physiological and pathophysiological significance of the remote recruitment of ubiquitinated proteins to the proteasome in a mammalian organ (23, 24). Defective or inadequate ubiquitination-proteasome coupling has been implicated by the frequent coexistence of increased ubiquitinated proteins with normal or even increased proteasome peptidase activities in the heart under many pathological conditions, such as proteinopathy (25, 26), pressure overload (27, 28), and myocardial ischemia (29). Therefore, unraveling the pathophysiological significance of this inadequate coupling in the heart is expected to improve understanding of cardiac UPS dysfunction and its role in cardiac pathogenesis.
Members of the Ubiquilin (Ubqln) family of Ub receptors are mammalian orthologues of yeast Dsk2, which is the primary Ub receptor for shuttling ubiquitinated misfolded proteins to the proteasome for degradation in yeast (19). Ubqln genes are highly conserved among mammals. Ubqln1 is ubiquitously expressed and Ubqln2 and Ubqln4 are expressed in most tissues, including the heart, but Ubqln3 is only expressed in testis (30). Ubqln proteins share a high degree of sequence and domain structural homology. Like other UBL-UBA proteins, Ubqln1 harbors a UBL domain at the N terminus and a UBA domain at the C terminus. Compared with 7 other Ub-binding domains, the UBA domain of Ubqln1 was found to bind the broadest range of Ub moieties, capturing most types of Ub moieties (31), with binding affinity similar to that of K48- and K63-linked poly-Ub chains (32). Located between UBL and UBA domains is a central region containing multiple STI1 motifs that may confer Ubqln1 with a chaperone-like function. Hence, Ubqln1 is purported to deliver polyubiquitinated proteins to the 26S proteasome for degradation (33). Endoplasmic reticulum–associated (ER-associated) PQC is responsible for the quality control of proteins targeted to the secretary pathway, which is pivotal to cell functioning and survival (34). Terminally misfolded ER proteins are retrotranslocated to the cytosolic side of the ER membrane, where they are degraded via the ER-associated degradation (ERAD) pathway, which is activated by the UPS. ER stress is induced by an accumulation of misfolded proteins in the ER and triggers the classical unfolded protein response (UPR), which attempts to resolve the ER stress through suppressing nonessential protein synthesis and increasing ER chaperoning and ERAD capacity via various transcriptional and posttranscriptional mechanisms (34, 35). Work using mammalian model cell lines (e.g., HEK293 cells) has shown that Ubqln1 is upregulated by ER stress and is recruited to the ERAD complex, where it interacts with UBXD2 (also known as erasin) along with p97/VCP (36), indicating that the shuttling role of Ubqln1 is likely important for ERAD, in addition to ER-independent PQC (37). To date, the role of Ubqln in cardiomyocytes and in the heart remains undocumented.
In the present study, we demonstrate that Ubqln1 plays an important role in cardiac ubiquitination-proteasome coupling and that mice with perinatal cardiomyocyte-restricted knockout of the Ubqln1 gene (Ubqln1-CKO) develop late-onset cardiomyopathy. Moreover, we provide proof-of-principle evidence that inadequacy in ubiquitination-proteasome coupling represents a major pathogenic factor for myocardial ischemia-reperfusion injury (IRI), suggesting that facilitating the coupling may be a novel therapeutic strategy to reduce IRI.
Results
Ubqln1 colocalizes with the proteasome and is recruited to the ERAD upon ER stress in cardiomyocytes.
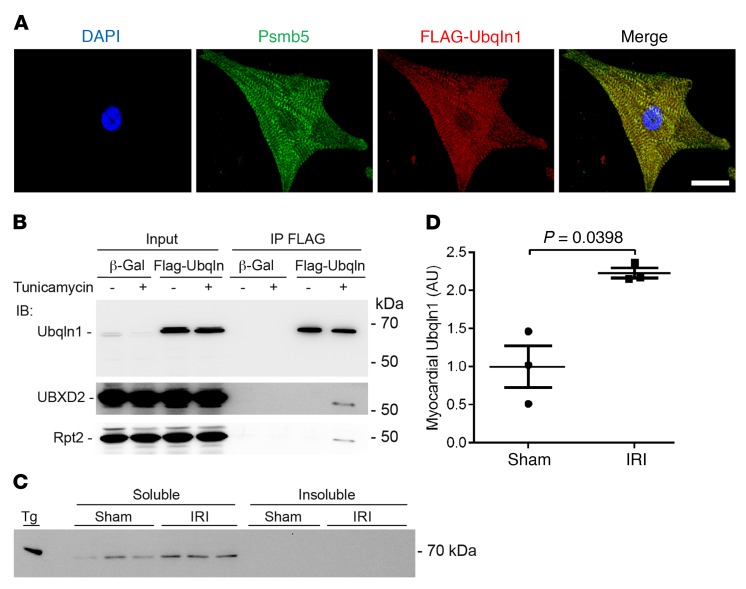
To investigate the distribution of Ubqln1 proteins in cardiomyocytes, we performed immunofluorescence confocal microscopy of cultured neonatal rat ventricular myocytes (NRVMs) overexpressing a FLAG-tagged full-length Ubqln1. Double-immunostaining revealed that Ubqln1 and Psmb5 (a stoichiometric subunit of the 20S proteasome) are colocalized, showing a striated pattern of enrichment (Figure 1A). The Psmb5-marked proteasome distribution in the cultured cardiomyocytes was in agreement with prior reports showing the enrichment of cardiac proteasomes in the intermyofibrillar space at the z-line level (38, 39). The colocalization between Ubqln1 and proteasomes is consistent with the purported role of Ubqln1 in coupling ubiquitination to proteasomal degradation. The physical interaction of Ubqln1 with the proteasome is further confirmed by our coimmunoprecipitation (co-IP) studies, which revealed that Rpt2, a stoichiometric subunit of the 19S proteasome, was detected in the Ubqln1 IP (Figure 1B). In HEK293 cells, Ubqln1 was shown to bind UBXD2 (also known as Erasin) (36), a known component of the ERAD machinery (40); our co-IP experiments confirmed that this also occurs in cardiomyocytes when ER stress is induced with tunicamycin, a prototype inducer of ER stress that achieves that induction through inhibition of protein glycosylation (Figure 1B), suggesting that Ubqln1 is likely involved in ERAD in cardiomyocytes during ER stress.
Figure 1. Ubqln1 colocalizes with the proteasome and is recruited to the ERAD pathway by ER stress in NRVMs.
(A) Representative confocal micrographs of double-immunofluorescence staining for Ubqln1 (red) and Psmb5 (green). FLAG-tagged Ubqln1 was overexpressed in NRVMs grown on chamber slides through adenovirus-mediated gene delivery. The cells were fixed and processed for indirect immunofluorescence staining for FLAG and Psmb5. Scale bar: 5 μm. (B) Immunoblot analyses for the indicated proteins in the FLAG-Ubqln1 immunoprecipitates. Cultured NRVMs were infected with adenoviruses expressing FLAG-Ubqln1 or Ad–β-Gal 24 hours before treatment with tunicamycin (60 μg/ml) for 6 hours. The cell lysates were subject to co-IP with a mouse monoclonal anti-FLAG Ab. The IP products were fractionated with 10% SDS-PAGE and transferred to PVDF membrane for immunoblot. Loading control used in-gel stain-free total protein imaging (Supplemental Figure 1). (C and D) Western blot image (C) and densitometry data (D) of Western blot analysis for myocardial Ubqln1 in WT mice subjected to myocardial IRI (30 minutes/24 hours) or sham surgery. Both 1% Triton X-100 PBS-soluble and -insoluble fractions were examined. Tg, Ubqln1 Tg mouse myocardial tissue used as a positive control to validate the Ubqln1 Ab. Loading controls used in-gel stain-free total protein imaging (Supplemental Figure 2). Mean ± SD are superimposed. Two-tailed t test with Welch’s correction. For each group of all mouse experiments presented here and after, equal or nearly equal numbers of male and female mice were used. Each dot and each lane in Western blot image represents an individual animal.
We performed Western blot analysis for myocardial Ubqln1 levels of WT mice subject to sham surgery or myocardial IRI induced via left anterior descending coronary artery (LAD) ligation for 30 minutes, followed by release for 24 hours. The result revealed that Ubqln1 was only detected in the soluble fraction of myocardial proteins in both IRI and sham surgery control groups and that Ubqln1 protein levels in the region remote to the IRI were considerably higher in the IRI group than in the sham group (Figure 1, C and D).
Mice with perinatal Ubqln1-CKO develop late-onset cardiomyopathy.
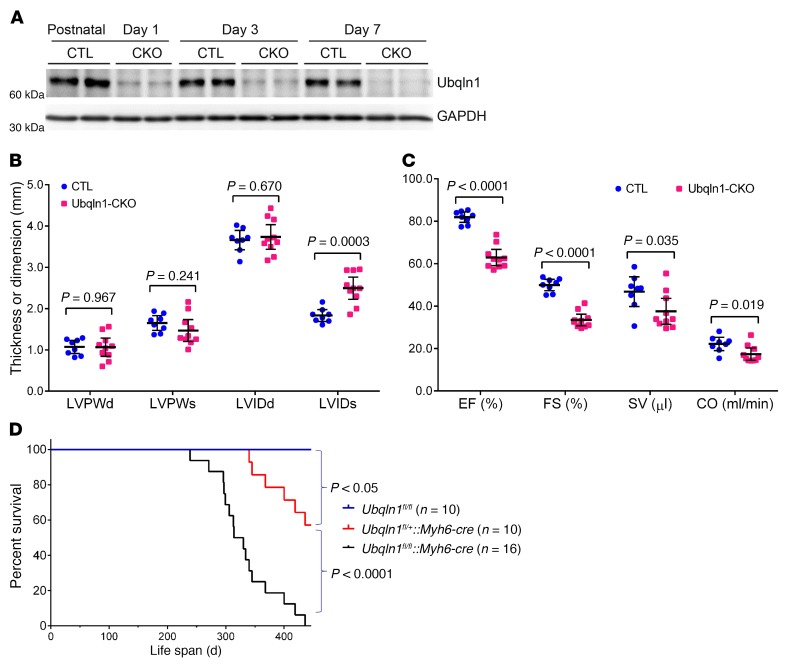
By coupling a Ubqln1 floxed allele (Ubqln1fl/fl) with the cre transgene driven by the Myh6 promoter (Myh6-cre) (41), we successfully generated perinatal Ubqln1-CKO mice. In homozygous Ubqln1-CKO (Ubqln1fl/fl::Myh6-cre) mice, myocardial Ubqln1 protein levels were markedly reduced at postnatal day 1 and further reduced at day 3 and day 7. By day 7, the remaining myocardial Ubqln1 proteins were less than 10% of those of the Ubqln1fl/fl and Myh6-cre control mice (Figure 2A). The residual Ubqln1 proteins were likely from the noncardiomyocyte compartment. Offspring from breeding pairs between Ubqln1fl/+ and Ubqln1fl/+::Myh6-cre or between Ubqln1fl/fl and Ubqln1fl/+::Myh6-cre displayed the expected genotypes of the Mendelian ratio at birth, suggesting no embryonic lethality. No gross abnormality was discernible during the first 7 months of age in any of these genotypes, including homozygous Ubqln1-CKO mice. However, by 5 months of age, echocardiography revealed cardiac malfunction, as reflected by moderate but statistically significant increases in left ventricle (LV) end-systolic internal diameter (LVIDs) and decreases in ejection fraction (EF), fractional shortening (FS), stroke volume (SV), and cardiac output (CO) in the homozygous Ubqln1-CKO mice compared with the control group (Figure 2, B and C). Strikingly, homozygous Ubqln1-CKO mice died prematurely, with a median life span of 322 days, which is significantly shorter than that of the heterozygous Ubqln1-CKO (Ubqln1fl/+::Myh6-cre) mice (Figure 2D). These observations show that perinatal Ubqln1-CKO causes a late-onset cardiomyopathy in mice, demonstrating that Ubqln1 is dispensable for perinatal and postnatal cardiac development, but that it inevitably plays an important role in maintaining normal cardiac function. Notably, the heterozygous Ubqln1-CKO mice lived longer than the homozygotes, but still showed a significantly shortened life span compared with the littermate Ubqln1fl/fl mice (Figure 2D), which suggests a gene-dosage effect. However, a gene-dosage effect on life span is uncertain, as the potential contribution of Myh6-cre to this phenotype observed in the heterozygous Ubqln1-CKO could not be ruled out.
Figure 2. Perinatal ablation of Ubqln1 in cardiomyocytes causes late-onset cardiomyopathy in mice.
Equal numbers of male and female mice were analyzed for each group. (A) Western blot analyses of myocardial Ubqln1 in Ubqln1-CKO (CKO) and littermate control (CTL) mice. (B and C) Echocardiographic data of mice at 5 months of age. Scatter dot plots are superimposed by mean ± SD. Two-tailed Student’s t test. LVPW, left ventricular posterior wall thickness; LVPWd, LVPW at end of diastole; LVPWs, LVPW at end of systole; LVIP, LV internal dimension; LVIDd, end-diastolic LVID; LVIDs, end-systolic LVID. Each lane (A) or dot (B and C) represents a mouse. (D) Kaplan-Meier survival curves, log-rank test.
Ubqln1-CKO compromises ubiquitination-proteasome coupling and impairs UPS performance in the heart.
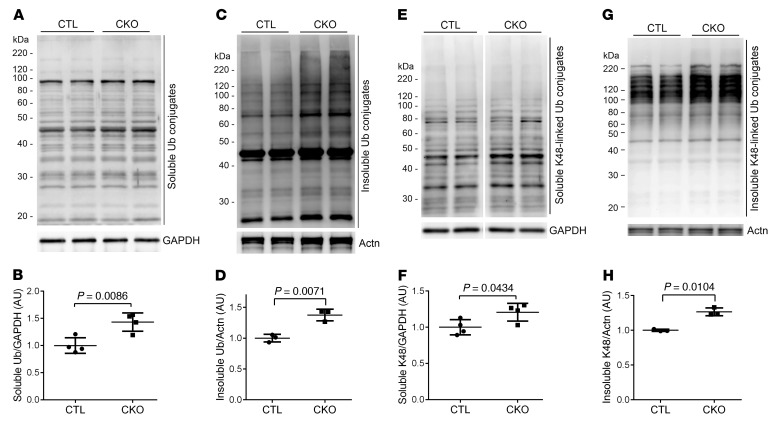
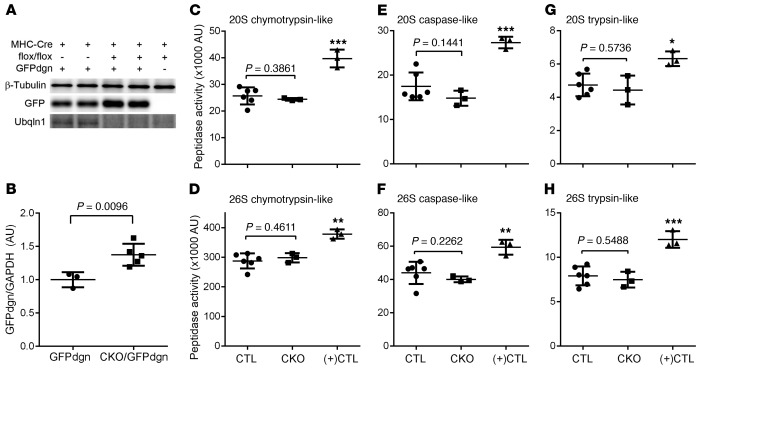
As a mammalian orthologue of yeast Dsk2, Ubqln1 is purported to function as a Ub receptor that shuttles ubiquitinated proteins to the proteasome for degradation, which is critical to UPS-mediated protein degradation (19); however, this proposition has not been formally tested in animals. Hence, we assessed cardiac UPS performance and the coupling between ubiquitination and proteasomal degradation in Ubqln1-CKO mice. First, we examined the myocardial levels of steady-state Ub conjugates and found that total ubiquitinated proteins and K48-linked poly-Ub chains (Figure 3) as well as K63-linked poly-Ub chains (Supplemental Figure 3; supplemental material available online with this article; https://doi.org/10.1172/JCI98287DS1) were moderately but statistically significantly increased in the homozygous Ubqln1-CKO mice. This suggests that the degradation of ubiquitinated proteins in cardiomyocytes is impaired by Ubqln1 deficiency. To further probe myocardial UPS proteolytic performance, we introduced transgenic (Tg) GFPdgn, a well-proven surrogate substrate of the UPS created via carboxyl fusion of degron CL1 to an enhanced green fluorescence protein (42), into the Ubqln1-CKO and littermate control mice via mouse crossbreeding. At 3 weeks of age, when gross and cardiac abnormalities had not become discernible yet, myocardial GFPdgn protein levels were significantly higher in the Ubqln1-CKO mice than in littermate controls (Figure 4, A and B), indicative of impairment of cardiac UPS performance by Ubqln1-CKO. The increased ubiquitinated proteins and impaired UPS performance in Ubqln1-CKO hearts are not caused by a decrease in the intrinsic function of the proteasome because all 3 types of myocardial proteasomal peptidase activities from both the 20S and the 26S proteasomes were comparable between Ubqln1-CKO and control mice (Figure 4, C–H). Taken together, these findings demonstrate that Ubqln1 plays an important role in UPS-mediated protein degradation through coupling ubiquitination with proteasomal degradation and that the Ubqln1-CKO mice represent a mouse model of cardiomyocyte-restricted impaired coupling between ubiquitination and the proteasome.
Figure 3. Western blot analyses for myocardial total and K48-linked Ub conjugates in Ubqln1-CKO and littermate control mice at 3 weeks of age.
The 1% Triton X-100 soluble and insoluble fractions of protein extracts from ventricular tissues were subject to SDS-PAGE and immunoblotting. GAPDH and sarcomeric α-actinin (Actn) were reprobed as the loading controls for the soluble and insoluble fractions, respectively. Representative images (A, C, E, G) and pooled densitometry data (B, D, F, H) are presented. A–D, Changes in total ubiquitinated proteins in the soluble (A and B) and insoluble (C and D) fractions. E–H, Changes in K48-linked ubiquitinated proteins in the soluble (E and F) and insoluble (G and H) fractions. P values were derived from the 2-tailed t test with Welch’s correction. Each dot and each lane in Western blot image represents an independent animal.
Figure 4. Ubqln1-CKO impairs myocardial UPS performance without affecting proteasome peptidase activities at 3 weeks of age.
(A and B) Representative image (A) and pooled densitometry data (B) of Western blot analyses for the indicated proteins in ventricular myocardial samples. Tg GFPdgn was introduced into Ubqlin1-CKO and control background through mouse crossbreeding. (C–H) Myocardial proteasomal peptidase activity assays. Crude protein extracts from the ventricular myocardium of homozygous Ubqln1-CKO mice and littermate control (combining Ubqln1fl/fl and heterozygous Ubqln1-CKO) mice as well as from a Tg mouse model of desmin-related cardiomyopathy serving as a positive control [(+)CTL] for the assays were assayed for their 20S and 26S proteasomal chymotrypsin-like (C and D), caspase-like (E and F), and trypsin-like (G and H) activities using specific fluorogenic substrates. The P values between control and Ubqln1-CKO groups were derived from 2-tailed unpaired t test with Welch’s correction. *P < 0.05; **P < 0.01; ***P < 0.005 vs. control and Ubqln1-CKO, Welch’s ANOVA followed by Tukey’s test for pair-wise comparison. Each dot and each lane in Western blot represents an independent mouse.
Ubqln1-CKO exacerbates myocardial IRI.
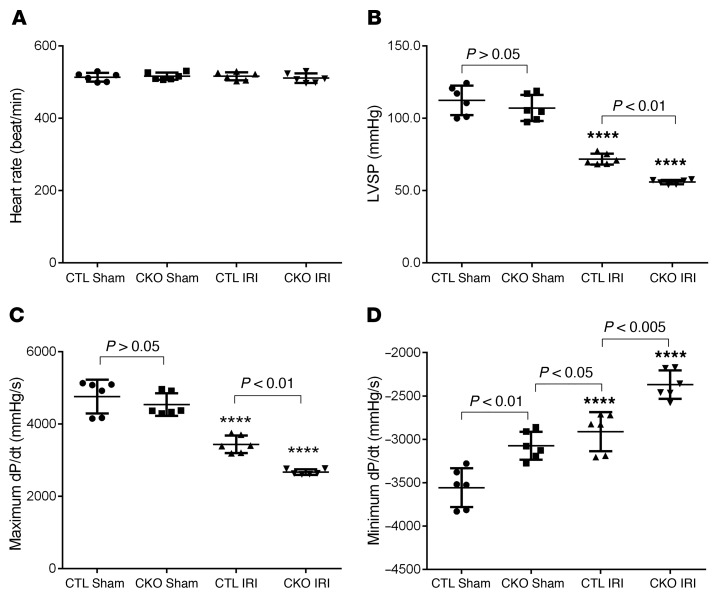
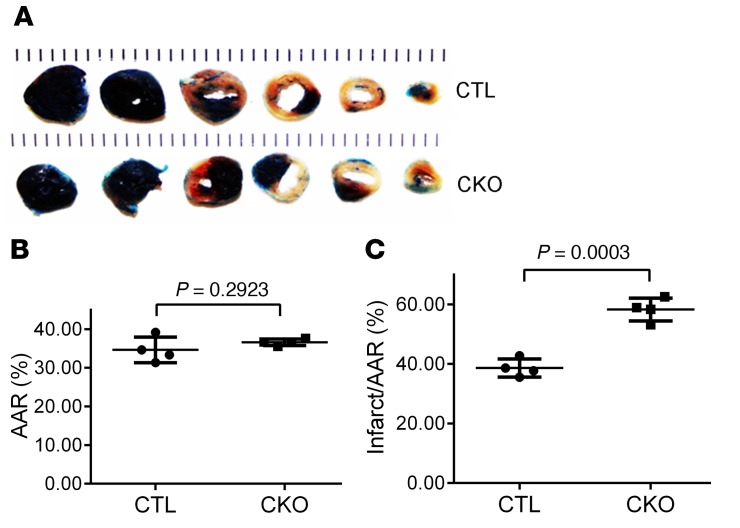
Perinatal Ubqln1-CKO mice do not show apparent abnormality until 5 months of age, and we observed a significant increase in myocardial Ubqln1 protein levels in WT mice subjected to myocardial IRI (Figure 1, C and D); hence, we hypothesized that Ubqln1 might play a more critical role in UPS-mediated protein degradation under a stress condition than at baseline. To test this hypothesis, we subjected 10-week-old Ubqln1-CKO and control mice to myocardial IRI, which was induced as described in Figure 5. Hemodynamic measurements revealed that the comparable IRI procedure induced more severe impairment of LV systolic and diastolic function in Ubqln1-CKO mice than in control mice, as reflected by significantly lower LV peak pressure (LVSP), diminished maximum rate of rise of left ventricular pressure (dP/dt), and elevated minimum dP/dt (Figure 5). Notably, compared with the control sham group, the Ubqln1-CKO sham group displayed a significantly elevated minimum dP/dt (P < 0.01), comparable to that induced by IRI in the control mice, although the parameters of systolic function (LVSP, maximum dP/dt) were not discernibly different between the 2 sham groups. This suggests 2 possibilities: (a) mild diastolic malfunction of Ubqln1-CKO mice at baseline that was not detected with echocardiography at this age became detectable with the more sensitive catheterization-based LV mechanical assessment; (b) the stress from the sham operation, which involves a major open chest surgery, unmasked the compromised LV diastolic function in Ubqln1-CKO mice. Furthermore, comparable initial ischemic insult (i.e., comparable area at risk [AAR]) resulted in a significantly larger infarct size in Ubqln1-CKO mice than in control mice, as determined with triphenyltetrazolium chloride (TTC) staining (Figure 6). These results demonstrate that Ubqln1-mediated ubiquitination-proteasome coupling is essential to cardiac function and cardiomyocyte survival under stress conditions.
Figure 5. Cardiac Ubqln1 deficiency exacerbates LV malfunction induced by acute myocardial IRI.
(A) Young adult homozygous Ubqln1-CKO mice and littermate control mice were subject to LAD ligation for 30 minutes, and then the ligation was released for reperfusion. At 23.5 hours after reperfusion, LV catheterization was performed to record LV pressure (B) and dP/dt (C, D) in vivo. ****P < 0.0001 vs. the respective sham group. Two-way ANOVA followed by Tukey’s test for pair-wise comparison. Each dot represents an individual animal.
Figure 6. Cardiac Ubqln1 ablation increases infarct size induced by acute myocardial IRI in mice.
Myocardial IRI was produced as described in Figure 5. At 24 hours of reperfusion, the animal was sacrificed, the original coronary ligature was retied, and the heart was subject to retrograde perfusion with 5% phthalocyanine blue to demarcate the original ischemic area or AAR before undergoing TTC staining to reveal the infarct area. (A) Representative pair of TTC staining images. (B) AAR presented is the percentage of LV that was subject to ischemia during the initial LAD ligation phase. (C) Changes in infarct size. Two-tailed Student’s t test. Each dot represents an individual animal.
Overexpression of Ubqln1 attenuates myocardial IRI.
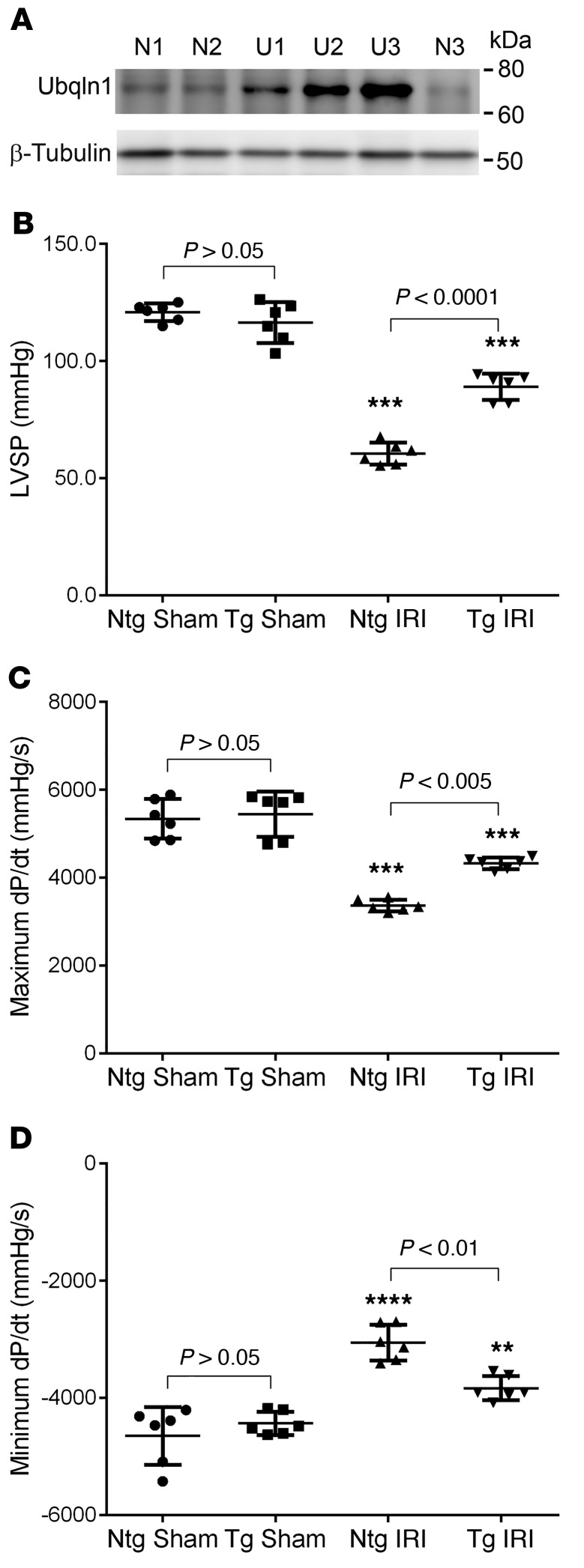
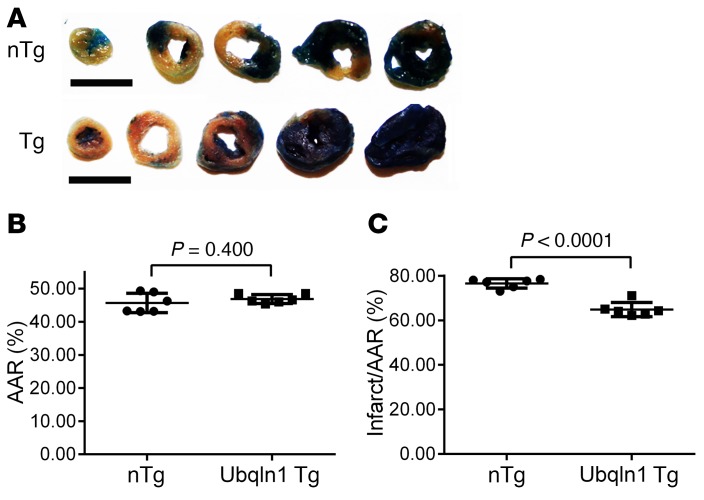
To test the impact of Ubqln1 gain of function, we have created a Tg mouse model of Ubqln1 overexpression (43), in which Ubqln1 overexpression is driven by the CAG promoter consisting of the CMV immediate early enhancer element and the chicken β-actin promoter (44). Three stable Tg lines were obtained with myocardial Ubqln1 overexpression of 3-, 5-, and 9-fold compared with the endogenous Ubqln1 protein level (Figure 7A). In these stable mouse lines, Ubqln1 overexpression did not lead to discernible abnormal phenotypes at baseline conditions. Mixed-sex adult littermate Tg and non-Tg (nTg) mice from the intermediate overexpression line were subject to IRI in exactly the same way as described earlier for the Ubqln1-CKO mice. LV catheterization and pressure measurement were performed at 24 hours after the initiation of reperfusion to assess changes in LV function. No statistically significant difference in any of the parameters was discerned between the nTg sham and the Tg sham groups. As reflected by marked decreases in LVSP and maximum dP/dt as well as by substantial elevation of minimum dP/dt, LV systolic and diastolic function were substantially impaired by IRI in both nTg and Tg mice. The IRI-induced LV functional impairment was remarkably less severe in the Tg mice than in the nTg mice (Figure 7). Infarct size assessment using TTC staining also revealed that the same initial ischemia caused significantly smaller infarct size in Ubqln1 Tg mice than in nTg mice (Figure 8). These experiments demonstrate that Ubqln1 gain of function protects myocardium against acute IRI, which indicates that inadequate coupling between ubiquitination and proteasomal degradation is a major pathogenic factor for IRI.
Figure 7. Tg Ubqln1 overexpression attenuates LV malfunction induced by acute myocardial IRI in mice.
(A) Representative image of Western blot analysis for myocardial Ubqln1 protein levels in Ubqln1 Tg mouse lines. Ventricular myocardial samples were collected from an adult mouse from each of the 3 Ubqln1 stable Tg mouse lines (U1, U2, and U3) and an nTg littermate mouse of each (N1, N2, and N3) and processed for analysis. β-Tubulin was probed for loading control. (B–D) Comparisons of hemodynamic parameters between Ubqln1 Tg and nTg mice at 24 hours after IRI or sham surgery. Mixed sex Tg and nTg littermate mice from the U2 line were subject to the IRI or sham surgical procedures; 24 hours after the initiation of reperfusion, LV was catheterized and changes in LV pressure were measured with the heart rate comparable among the 4 groups (data not shown). **P < 0.01; ***P < 0.005; ****P < 0.0001 vs. the respective sham group. Two-way ANOVA followed by Tukey’s test for pair-wise comparison. Each dot represents an individual animal.
Figure 8. Tg Ubqln1 overexpression reduces infarct size during acute myocardial IRI in mice.
(A–C) The induction of myocardial IRI in Ubqln1-Tg and nTg littermate mice and the subsequent processing and measurements at the terminal experiment were performed in the same way as described for Figures 5 and 6. Two-tailed Student’s t test. Each dot represents an individual animal. Scale bars = 5 mm.
Ubqln1 overexpression promotes proteasomal degradation of oxidized proteins and a surrogate misfolded protein without altering autophagic flux in cardiomyocytes.
In Ubqln1-CKO mice, the loss of Ubqln1 is restricted to cardiomyocytes, but Tg Ubqln1 overexpression is not; thus, it remains an important question whether the protective effects of Ubqln1 gain of function on IRI observed in the Ubqln1 Tg mice were necessarily derived from the Ubqln1 increase in the cardiomyocyte compartment. To answer this question, we performed cardiomyocyte culture studies.
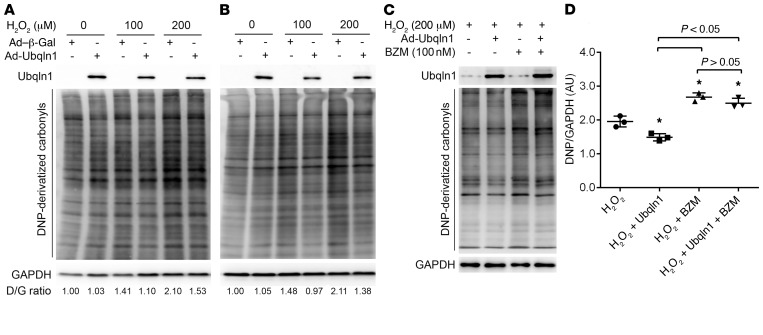
A rapid increase in oxidative stress in ischemic myocardium when reperfusion is initiated and resultant oxidative modifications to macromolecules, such as proteins and lipids, are well established causes of IRI. The oxidized proteins, for example, not only lose their normal function, but also gain toxicities and thereby further adversely affect cell function and viability. Therefore, once formed, a timely removal of these oxidized proteins should help reduce their toxicities to the cell. Given the supposed role of Ubqln1 in UPS-mediated protein degradation, we hypothesized that the primary mechanism underlying protection by Ubqln1 against myocardial IRI works to facilitate UPS-mediated degradation of oxidized proteins in cardiomyocytes. To examine this hypothesis, we subjected cultured NRVMs to increased oxidative stress produced by hydrogen peroxide and examined the impact of forced Ubqln1 overexpression on the dynamic changes in the levels of steady-state protein carbonyls. The latter is the signature change of oxidative modifications on proteins and can be derivatized to 2,4-dinitrophenyl–hydrazone (DNP-hydrazone) by reaction with 2,4 dinitrophenylhydrazine (DNPH) and then detected with anti-DNP Abs (45). We found that hydrogen peroxide treatment induced dose-dependent increases in DNP-derivatized protein carbonyls and that the increases were substantially less in NRVMs overexpressing Ubqln1 compared with cells without Ubqln1 overexpression. Moreover, proteasome inhibition significantly increased protein carbonyls in H2O2-treated NRVMs, and this increase could not be attenuated by Ubqln1 overexpression. In other words, the decreasing protein carbonyl levels by Ubqln1 overexpression were nearly completely prevented by proteasome inhibition with bortezomib (Figure 9). These findings demonstrate that, under acute oxidative stress, there is an inadequate coupling between ubiquitination and proteasomal degradation and that overexpression of Ubqln1 is sufficient to promote UPS-mediated removal of oxidized proteins.
Figure 9. Ubqln1 overexpression promotes proteasomal degradation of oxidized proteins in cultured NRVMs.
Twenty-four hours after plating, NRVMs were infected with Ad-Ubqln1 or Ad–β-Gal. Further experiments were initiated at 48 hours after the adenoviral infection. (A and B) Representative images of Western blot analyses for DNP-derivatized protein carbonyls. The Ubqln1 overexpressing and control NRVMs were treated with H2O2 at the indicated doses. The cells were harvested at either 15 minutes (A) or 3 hours (B) after the H2O2 treatment for DNPH derivatization to measure the level of protein carbonyls. D/G, DNP/GAPDH. Representatives of 3 biological repeats are shown. (C and D) Proteasome inhibition attenuates the Ubqln1 overexpression-induced reduction of oxidized proteins. The cultured NRVMs were treated with proteasome inhibitor bortezomib (BZM, +) or vehicle control (saline, –) at 10 minutes before H2O2 treatment was initiated. Three hours later, cells were harvested for DNPH derivatization. Representative images (C) and pooled densitometry data from 3 biological repeats (D) are shown. *P < 0.05 vs. the H2O2 group; 1-way ANOVA followed by Tukey’s test.
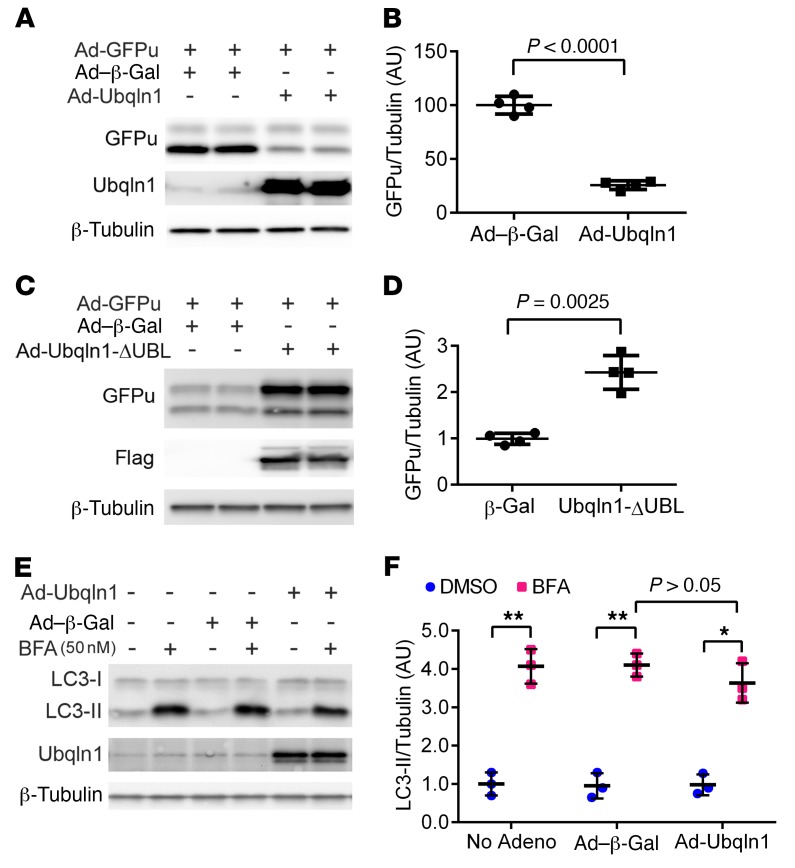
To determine whether Ubqln1 overexpression enhances UPS performance in cardiomyocytes, we tested the effect of Ubqln1 overexpression on the protein level of a known UPS substrate, GFPu, in cultured NRVMs (46), a slightly different version of GFPdgn, as expressed in the GFPdgn Tg mice used in the Ubqln1-CKO in vivo study (Figure 4, A and B). It should be pointed out that GFPu/GFPdgn is also a surrogate for misfolded proteins (15, 16, 47). Consistently with what was observed in Ubqln1-CKO hearts, overexpression of full-length Ubqln1 significantly decreased, while overexpression of a UBL-deleted form of Ubqln1 (Ubqln1-ΔUBL) markedly increased, the steady-state GFPu protein level (Figure 10, A–D), indicating that Ubqln1 gain of function promotes UPS-mediated degradation of misfolded proteins and that the Ubqln1-ΔUBL, which is able to bind ubiquitinated proteins but unable to interact with the proteasome, acts as a decoy Ub receptor and thereby prevents ubiquitinated substrates from proteasomal degradation in cardiomyocytes. Furthermore, we tested to determine whether autophagy is involved in the Ubqln1 promotion of oxidized protein removal and cardioprotection, and we found that Ubqln1 overexpression did not alter LC3-II flux, a commonly used indicator of autophagic activity (48), in cardiomyocytes (Figure 10, E and F).
Figure 10. The effect of Ubqln1 overexpression on the degradation of a UPS surrogate substrate (GFPu) and autophagic flux in cultured cardiomyocytes.
(A–D) Cultured NRVMs were infected with Ad-Ubqln1 or Ad-Ubqln1-ΔUBL (or Ad–β-Gal as control) to overexpress Flag-tagged full-length or UBL domain–deleted Ubqln1, respectively. Representative images of Western blot analyses for the indicated proteins (A and C) and densitometry data pooled from 4 repeats (B and D). Ad-GFPu infection to express GFPu, a surrogate substrate for the UPS, was initiated at 24 hours after Ad-Ubqln1 infection; 48 hours later, total cell lysates were collected for Western blot analyses. (E and F) Autophagic flux assay. At 46 hours after Ad-Ubqln1 or Ad–β-Gal infection, bafilomycin A1 (BFA) or vehicle control (DMSO) was administered to inhibit lysosomes. Two hours later, cells were collected for Western blot analyses for the indicated proteins. β-Tubulin was probed for loading control. No Adeno, cells not subject to adenoviral infection. *P < 0.005; **P < 0.001, 2-tailed t test with Welch’s correction. Each dot represents a biological repeat.
Discussion
Failure of cardiac UPS to meet the increased demand for timely removal of unwanted and no longer needed proteins in cardiomyocytes contributes to acute myocardial IRI (16, 49). Prior to the present study, proteasome functional insufficiency, or proteasomal impairment, was the only factor established as the cause for UPS inadequacy in IRI myocardium (16, 38, 49). Here, we demonstrate for the first time, to our knowledge, that Ubqln1 exerts an essential role in coupling ubiquitination to proteasomal degradation in heart muscle cells, and moreover, we have identified inadequate coupling between ubiquitination and proteasomal degradation as another major cause for the UPS functional deficit that thereby contributes to myocardial IRI. This provides compelling evidence that, not only priming the proteasome, but also enhancing the coupling between ubiquitination and the proteasome, should be explored as therapeutic strategies to reduce myocardial IRI, a major and common pathological process that occurs in both the natural progression and therapeutic intervention of ischemic heart disease as well as during heart transplantation.
Ubqln1 contributes to cardiac ubiquitination-proteasome coupling.
Since Dsk2 plays an essential role in shuttling K48-linked ubiquitinated proteins to the proteasome for degradation in yeast (50), Ubqln1 as an orthologue of yeast Dsk2 was naturally purported to serve a similar role in mammals. However, this supposed role of Ubqln1 has not been formally tested in vivo. Our creation and characterization of the Ubqln1-CKO mice allowed this critical gap to be filled. Taking advantage of a well-established reverse reporter (GFPdgn) of UPS performance, we detected impairment of cardiac UPS performance in Ubqln1-CKO mice, as reflected by significantly higher myocardial GFPdgn protein levels in Ubqln1-CKO::GFPdgn mice compared with control::GFPdgn (CTL::GFPdgn) mice (Figure 4, A and B). This myocardial UPS impairment by Ubqln1-CKO was detected at 3 weeks of age when no abnormal phenotypes were discerned, indicating that this is a primary effect of Ubqln1 deficiency. Similarly, we previously found that conditional ablation of Ubqln1 in neurons also caused accumulation of GFPdgn in mouse brains (43). Tests from both mouse hearts and brains consistently demonstrate a crucial role for Ubqln1 in UPS performance in mammals. Similarly to what we had found in mouse brains with neuronal ablation of Ubqln1 (43), here we found Ubqln1-CKO caused myocardial accumulation of total ubiquitinated proteins (Figure 3, A–D). Moreover, we unveiled for what we believe is the first time that in both the soluble and insoluble fractions of Ubqln1-CKO hearts, the level of proteins conjugated with K48-linked poly-Ub chains and the canonical signal targeting proteins for proteasomal degradation (51) were significantly increased (Figure 3, E–H), indicating that Ubqln1 deficiency impairs the degradation of K48-linked polyubiquitinated proteins by the proteasome. This impairment conceivably can result from either impaired intrinsic activity of the proteasome or reduced delivery of the polyubiquitinated proteins to the proteasome. Our data support the latter. This is because proteasomal activities were examined for the first time, to our knowledge, in a mammalian organ deficient of Ubqln1 by the present study, and it was revealed that all 3 types of proteasomal peptidase activities in either the 20S or the 26S proteasomes were not discernibly altered by Ubqln1-CKO (Figure 4, C–H). In general, proteins conjugated with polyubiquitin chains, especially those with K48-linked Ub chains, are efficiently degraded by the proteasome (51); hence, this coexistence of increased K48-linked Ub conjugates with unaltered proteasome proteolytic activities in Ubqln1-CKO mouse hearts indicates an apparent uncoupling between ubiquitination and proteasomal degradation by Ubqln1 deficiency. The uncoupling and the resultant UPS impairment are not without consequence, as Ubqln1-CKO mice developed late-onset cardiomyopathy. The latter is evidenced by echocardiography that detected in 5-month-old (Figure 2), but not 10-week-old (Supplemental Table 1), mice with perinatal Ubqln1-CKO significant morphometric and functional changes, as reflected by statistically significant decreases in EF, FS, SV, and CO and an increase in LVIDs in the absence of significant alterations in LV posterior wall thickness and end-diastolic LVID. The initial phenotype was relatively mild, but significantly, a shortened life span was observed in homozygous Ubqln1-CKO mice (Figure 2). These in vivo results provide compelling evidence that Ubqln1 acts as a coupling factor to shuttle ubiquitinated proteins to the proteasome for degradation in mammalian heart muscle cells and is indispensable to normal cardiac proteostasis at baseline. The feature of the cardiomyopathy being late onset rather than early onset suggests that Ubqln1 is likely responsible for shuttling only a subset of proteins in cardiomyocytes under basal physiological conditions. Supporting this proposition, a recent study suggests that ubiquilins act to triage mitochondrial membrane proteins for proteasomal degradation (37). Although there are at least 4 ubqln genes encoding Ubqln1, -2, -3, and -4, respectively, in mice and humans and they all are orthologous to yeast Dsk2 and share the same domain structure, the development of abnormal phenotypes in the Ubqln1-CKO mice indicates that Ubqln1 is not redundant to other Ubqlns in cardiomyocytes. This observation is also supported by the clinical discovery that a loss-of-function mutation of the Ubqln2 gene causes impairment of protein degradation and neurodegeneration in humans (52).
That mice with perinatal Ubqln1-CKO do not display discernible abnormal phenotypes at baseline when they are young (e.g., <10 weeks of age) prompted us to test the impact of cardiac Ubqln1 deficiency on the heart under a condition of increased demand for UPS-mediated protein degradation. Compared with the littermate control mice, young Ubqln1-CKO mice showed more severe cardiac malfunction and myocardial injury when subject to IRI, as reflected by greater reduction of LV-maximum dP/dt, more elevation of LV-minimum dP/dt, and larger infarct size after myocardial IRI (Figures 5 and 6). It is well known that IRI increases oxidative stress, which mediates IRI via increasing oxidative modifications of macromolecules, including producing highly toxic oxidized proteins (53). Once produced, a timely removal of the oxidized proteins by the UPS becomes a determining factor for the longevity of their toxicities (54, 55). Hence, these results indicate that Ubqln1 is also important for maintaining cardiac proteostasis under stress conditions with increased production of misfolded/damaged proteins.
Inadequate or impaired ubiquitination-proteasome coupling contributes to IRI.
Prior studies have detected a moderate decrease in myocardial proteasomal peptidase activities as well as accumulation of ubiquitinated proteins in IRI hearts (38, 56). Moreover, cardiomyocyte-restricted inhibition of proteasomal function exacerbates, and conversely, enhancement of proteasome function via overexpression of PA28α protects against acute IRI (16, 38), identifying reduced proteasomal function as an important contributor to UPS impairment in IRI hearts, which thereby plays a major pathogenic role in IRI. In the present study, we were able to demonstrate that improving the coupling between ubiquitination and proteasomal degradation protects against IRI despite a significant increase in Ubqln1 proteins in IRI myocardium (Figure 1A). This is because the same myocardial IRI insult caused significantly less cardiac malfunction (Figure 7) and smaller infarct size (Figure 8) in mice with Tg overexpression of Ubqln1 compared with the nTg littermate mice. Complemented by the Ubqln1 loss-of-function study described earlier, the protection of Ubqln1 overexpression against myocardial IRI provides compelling evidence that inadequate coupling between ubiquitination and proteasomal degradation represents a major cause for UPS functional insufficiency observed in IRI hearts, identifying enhancement of ubiquitination-proteasome coupling as a strategy for reducing myocardial IRI.
It should be pointed out that diastolic malfunction undetected by echocardiography at baseline was detected in the Ubqln1-CKO sham control group (Figure 5D); hence, it is possible that the diastolic malfunction induced by Ubqln1-CKO might have contributed to the IRI exacerbation observed in the Ubqln1-CKO mice. Nevertheless, this inevitable ambiguity in the interpretation of the in vivo studies of this nature is compellingly compensated by the findings from our in vivo and in vitro gain-of-function studies.
Mechanisms underlying Ubqln1 protection against IRI.
Ubqln1 overexpression had been found to facilitate the degradation of overexpressed neural degenerative diseases causing polyglutamine-expanded — or polyalanine-expanded — proteins and protect against their cytotoxicity in cultured noncardiac cells (57–59). More recently, it was shown that the protection of Ubqln1 overexpression against brain damage in an ischemic stroke model was associated with marked attenuation of the accumulation of insoluble ubiquitinated proteins in the affected brain tissues (60). However, the exact pathway taken by Ubqln1 to enhance degradation of misfolded proteins remains unclear, although promoting the UPS-mediated proteolysis has been implicated (33, 43, 60). In the present study, the results from our cell-culture experiments demonstrate that Ubqln1 overexpression promotes the degradation of oxidized proteins in cardiomyocytes and, more importantly, that this promotion can be completely abolished by proteasome inhibition (Figure 9), thereby establishing the UPS as the primary pathway by Ubqln1 to facilitate the degradation of oxidized/damaged proteins.
The UPR triggered by ER stress attempts to resolve ER stress, but the UPR can also, by sustained ER stress, cause cell death. ER stress/UPR can occur in myocardial IRI and has proven to be a major factor affecting IRI. For example, the activating transcription factor 6α (ATF6) branch of the UPR mediates the transcription of ER stress-response genes (34); overexpression of an activated form of ATF6 protects, and conversely, ATF6 knockout exacerbates IRI in mouse hearts and cultured cardiomyocytes (61, 62). Since our data confirm that Ubqln1 is recruited to the ERAD machinery by ER stress in cardiomyocytes (Figure 1), it is very likely that promoting ERAD and thereby alleviating ER stress is an underlying mechanism for Ubqln1 to protect against myocardial IRI.
By removal of protein aggregates and damaged organelles, macroautophagy (commonly known as autophagy) is known to play a role in quality control in the cell (63, 64). Although opposing reports do exist (65), some recently reported studies support a protective role of increasing macroautophagy against IRI (66). This raises a question of whether enhancing macroautophagy plays a role in the protection of Ubqln1 against IRI. Addressing this question is important because a prior report showed that Ubqln1 could be found in autophagosomes and LC3 complexes and that siRNA-mediated Ubqln1/Ubqln2 knockdown reduced LC3-II levels in cultured HeLa cells, leading to a conclusion that Ubqln1 may function in macroautophagy (67). However, this conclusion was made without a rigorous assessment of autophagic flux, and more importantly, the effect of Ubqln1 gain of function on macroautophagy was not examined in the study (67). In the present study, we have tested the impact of Ubqln1 overexpression on autophagic flux in cultured NRVMs and the results do not support the notion that Ubqln1 overexpression significantly increases autophagic activity (Figure 10). Thus, it is very unlikely that modulating autophagy plays a significant role in the protection of Ubqln1 overexpression against myocardial IRI. Nevertheless, it is possible that Ubqln1 may exert cardiac protection through additional mechanisms, beyond enhancing removal of damaged and toxic proteins. For example, it may inhibit cell death by acting directly on specific proteins that regulate cell-death pathways. Beverly et al. reported that BCL2L10/BCLb, an antiapoptotic BCL2-like protein, was stabilized by Ubqln1 in its monoubiquitinated form (68). It remains unclear how Ubqln1 stabilizes monoubiquitinated BCLb while it promotes the degradation of polyubiquitinated proteins.
In summary, this study provides compelling evidence that the coupling of ubiquitination to proteasomal degradation becomes inadequate during IRI and that such inadequacy contributes to myocardial IRI, identifying improving the coupling via, for example, upregulation of Ublqn1 as a therapeutic strategy for preventing or better treating acute myocardial IRI.
Methods
Genetically modified mouse models.
The creation of mice harboring a floxed Ubqln1 allele in a C57BL/6 inbred background and the creation of Ubqln1 Tg overexpression mouse lines were reported (43). The Ubqln1-floxed mice were crossbred with the Mhc6-cre–Tg mice (Tg[Myh6-cre]2182Mds; The Jackson Laboratory, catalog 011038) (41) to achieve Ubqln1-CKO. The Myh6-cre–Tg mice were used as the control, unless indicated otherwise. The Ubqln1-Tg overexpression mice had been backcrossed into the C57BL/6 inbred background through more than 9 generations of backcross. The creation and validation of the GFPdgn-Tg mice were previously described (42).
Echocardiography.
2D echocardiogram-guided M-mode echocardiography was performed on mice as previously described (69). In brief, mice were anesthetized by inhalation of isoflurane (4.5% for induction and 1.5% for maintenance) via a nose cone. Transthoracic echocardiograph was recorded using the Vevo2100 echocardiography system with a 40 MHz transducer. The LV morphometric and functional parameters were analyzed offline (69).
Myocardial IRI.
Under anesthesia with isoflurane, mice of both sexes (male/female = roughly 1:1) at approximately 10 weeks of age were subject to surgical ligation (30 minutes) and subsequent release (24 hours) of the LAD.
Cardiac function assessment.
At 23.5 hours of reperfusion, a microtipped pressure transducer catheter (1.4F, model SPR-835, Millar Instruments Inc.) was placed into the LV via the apex, and the LV pressure and dP/dt were recorded and analyzed as we previously described (16).
Ischemic area and infarct size determination.
At the end of 24 hours of reperfusion, the suture was retied and 250 μl of 5% phthalocyanine blue was injected into the LV chamber. The heart was quickly excised, immediately frozen in a–20°C freezer for 30 minutes, and sliced into 5 short-axis sections, which were incubated in 1% TTC solution for 15 minutes. Each section was weighed and digitally photographed. The area not at risk (phthalocyanine blue stain), the AAR (including viable myocardium and infarcted area), and the total LV areas from both sides of each section were measured using Image-Pro Plus software (Media Cybernetics). The AAR was expressed as percentage of total LV, and infarct size was expressed as percentage of the AAR and LV (16).
Western blot analysis.
Frozen ventricular myocardial tissues were homogenized in 1× SDS sampling buffer (50 mM Tris-HCl at pH 6.8, 2% SDS, and 10% glycerol). The supernatant was obtained following a 12,000 g centrifugation for 10 minutes at 4°C. The protein concentration was measured using the bicinchoninic acid (BCA) method. Equal amounts of protein sample were resolved by SDS-PAGE, transferred to PVDF membrane, and probed with appropriate primary Ab (see Supplemental Table 2 for sources of primary Abs) and horseradish peroxidase–conjugated secondary Ab, followed by detection with enhanced chemiluminescence (ECL-Plus) reagents (GE Healthcare) and a VersaDoc 4000 imaging system (Bio-Rad). The signal was quantified with Quantity One software (Bio-Rad) (16).
NRVM culture and adenoviral gene delivery.
The isolation and culture of NRVMs as well as the infection of replication-deficient recombinant adenoviruses expressing Flag-tagged murine full-length Ubqln1 (Ad-Ubqln1), UBL domain–deleted Ubqln1 (Ad-Ubqln1-ΔUBL), GFPu (Ad-GFPu), or β-galactosidase (Ad–β-Gal) as control were performed as we previously reported (47).
Immunofluorescence confocal microscopy.
This was conducted as we previously described (70). In brief, NRVMs cultured in fibronectin-coated chamber slides were fixed with 4% paraformaldehyde at 48 hours after infection of Ad-Ubqln1. The fixed cells were then subject to double-immunostaining with the mouse mAb against the FLAG-tag (catalog 8146, Cell Signaling Technology) and the custom-made and previously validated rabbit anti-rodent Psmb5 (i.e., the β5 subunit of the 20S proteasome) primary Ab (38). The bound primary Abs were stained with Alexa Fluor 568 donkey anti-mouse IgG H&L (ab175700, Abcam) and Alexa Fluor 488 donkey anti-rabbit IgG H&L (ab150073, Abcam) secondary Ab (Molecular Probes). The nuclei were stained with DAPI. The triple fluorescence staining was imaged with a triple-laser confocal microscope (Olympus Fluoview 500, Olympus).
Co-IP.
Co-IP was done with the Pierce Co-Immunoprecipitation Kit (catalog 26149, Thermo Fisher Scientific), which provides covalent Ab immobilization so that potential interference of the IP Abs is completely avoided. All the components required for a co-IP experiment, including cell-lysis buffer, were from the kit. IP flag-Ubqln1 used the mouse mAb against FLAG tag (catalogv8146, Cell Signaling Technology). Immunoblot for Ubqln1 and UBXD2 (also known as Erasin) used, respectively, the rabbit polyclonal anti-Ubqln1 Ab (ab3341, Abcam) and a rabbit polyclonal Ab against UBXD2 (catalog 21052-1-AP, Proteintech Group).
Detection of oxidized proteins.
The Oxidized Protein Western Blot Detection Kit (ab178020, Abcam) was used for detection and quantification of carbonyl groups, a hallmark of proteins with oxidative modifications. In brief, the protein carbonyl groups were derivatized to DNP-hydrazone by reaction with DNPH. The DNP-derived protein samples were then fractionated with SDS-PAGE, followed by transferring to a PVDF membrane and immunoblotting with a rabbit anti-DNP primary Ab. The bound anti-DNP primary Abs on the membrane were detected with an HRP-conjugated goat anti-rabbit secondary Ab and chemiluminescent detection reagents (47).
Statistics.
Unless indicated otherwise, data of continuous variables are presented as scatter dot plots with mean ± SD superimposed, where differences between 2 groups or among multiple groups were evaluated in SPSS for statistical significance using, respectively, unequal variance 2-tailed t test or Welch’s ANOVA followed by Tukey’s test. Statistical tests for other results are specified in figure legends. P < 0.05 was considered statistically significant.
Study approval.
The animal care and use protocol of this study was approved by the IACUC of the University of South Dakota. The protocol for animal use and care in this study conforms to the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85-23, revised 1996).
Author contributions
XW, YT, CH, and JL designed research studies. CH, YT, BP, and PW conducted experiments. CH, YT, HX, EMT, and PW acquired data. XW, CH, YT, HX, FL, and JL analyzed data. HW and FL provided reagents. XW, YT, and CH wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants HL072166, HL085629, and HL131667. We wish to thank Andrea Jahn and Megan Lewno for their assistance in maintaining mouse colonies and genotype determination.
Version 1. 09/11/2018
In-Press Preview
Version 2. 10/22/2018
Electronic publication
Version 3. 12/03/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(12):5294–5306.https://doi.org/10.1172/JCI98287.
See the related Commentary at Ushering in the cardiac role of Ubiquilin1.
Contributor Information
Chengjun Hu, Email: chengjunhu@whu.edu.cn.
Yihao Tian, Email: yihaotian@whu.edu.cn.
Hongxin Xu, Email: 303876340@qq.com.
Bo Pan, Email: bo.pan@usd.edu.
Erin M. Terpstra, Email: Erin.J.Terpstra@coyotes.usd.edu.
Penglong Wu, Email: penglong.wu@usd.edu.
Hongmin Wang, Email: hongmin.wang@usd.edu.
Faqian Li, Email: lif@umn.edu.
Jinbao Liu, Email: jliu@gzhmu.edu.cn.
Xuejun Wang, Email: xuejun.wang@usd.edu.
References
- 1.Collins GA, Goldberg AL. The logic of the 26S proteasome. Cell. 2017;169(5):792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99(12):1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 3.van Leeuwen FW, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer’s and Down patients. Science. 1998;279(5348):242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 4.Chen SN, et al. Human molecular genetic and functional studies identify TRIM63, encoding muscle RING finger protein 1, as a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2012;111(7):907–919. doi: 10.1161/CIRCRESAHA.112.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Yacoub N, et al. FBXO32, encoding a member of the SCF complex, is mutated in dilated cardiomyopathy. Genome Biol. 2016;17:2. doi: 10.1186/s13059-015-0861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaglia T, et al. Atrogin-1 deficiency promotes cardiomyopathy and premature death via impaired autophagy. J Clin Invest. 2014;124(6):2410–2424. doi: 10.1172/JCI66339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garg A, et al. KLHL40 deficiency destabilizes thin filament proteins and promotes nemaline myopathy. J Clin Invest. 2014;124(8):3529–3539. doi: 10.1172/JCI74994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moresi V, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143(1):35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fielitz J, et al. Loss of muscle-specific RING-finger 3 predisposes the heart to cardiac rupture after myocardial infarction. Proc Natl Acad Sci U S A. 2007;104(11):4377–4382. doi: 10.1073/pnas.0611726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fielitz J, et al. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Invest. 2007;117(9):2486–2495. doi: 10.1172/JCI32827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willis MS, et al. Cardiac muscle ring finger-1 increases susceptibility to heart failure in vivo. Circ Res. 2009;105(1):80–88. doi: 10.1161/CIRCRESAHA.109.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100(4):456–459. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li HH, et al. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J Clin Invest. 2007;117(11):3211–3223. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papizan JB, et al. Deficiency in Kelch protein Klhl31 causes congenital myopathy in mice. J Clin Invest. 2017;127(10):3730–3740. doi: 10.1172/JCI93445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranek MJ, Terpstra EJ, Li J, Kass DA, Wang X. Protein kinase g positively regulates proteasome-mediated degradation of misfolded proteins. Circulation. 2013;128(4):365–376. doi: 10.1161/CIRCULATIONAHA.113.001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121(9):3689–3700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myeku N, et al. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat Med. 2016;22(1):46–53. doi: 10.1038/nm.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lokireddy S, Kukushkin NV, Goldberg AL. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc Natl Acad Sci U S A. 2015;112(52):E7176–E7185. doi: 10.1073/pnas.1522332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu K, den Brave F, Jentsch S. Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat Cell Biol. 2017;19(6):732–739. doi: 10.1038/ncb3531. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Terpstra EJ. Ubiquitin receptors and protein quality control. J Mol Cell Cardiol. 2013;55:73–84. doi: 10.1016/j.yjmcc.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clague MJ, Urbé S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143(5):682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Dantuma NP, Heinen C, Hoogstraten D. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) 2009;8(4):449–460. doi: 10.1016/j.dnarep.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Ng JM, et al. Developmental defects and male sterility in mice lacking the ubiquitin-like DNA repair gene mHR23B. Mol Cell Biol. 2002;22(4):1233–1245. doi: 10.1128/MCB.22.4.1233-1245.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Safren N, et al. Ubiquilin-1 overexpression increases the lifespan and delays accumulation of Huntingtin aggregates in the R6/2 mouse model of Huntington’s disease. PLoS One. 2014;9(1):e87513. doi: 10.1371/journal.pone.0087513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, et al. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J. 2006;20(2):362–364. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 26.Chen Q, et al. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97(10):1018–1026. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 27.Depre C, et al. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation. 2006;114(17):1821–1828. doi: 10.1161/CIRCULATIONAHA.106.637827. [DOI] [PubMed] [Google Scholar]
- 28.Ranek MJ, et al. Genetically induced moderate inhibition of 20S proteasomes in cardiomyocytes facilitates heart failure in mice during systolic overload. J Mol Cell Cardiol. 2015;85:273–281. doi: 10.1016/j.yjmcc.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu X, Kem DC. Proteasome inhibition during myocardial infarction. Cardiovasc Res. 2010;85(2):312–320. doi: 10.1093/cvr/cvp309. [DOI] [PubMed] [Google Scholar]
- 30.Conklin D, Holderman S, Whitmore TE, Maurer M, Feldhaus AL. Molecular cloning, chromosome mapping and characterization of UBQLN3 a testis-specific gene that contains an ubiquitin-like domain. Gene. 2000;249(1-2):91–98. doi: 10.1016/S0378-1119(00)00122-0. [DOI] [PubMed] [Google Scholar]
- 31.Nakayasu ES, et al. Evaluation of selected binding domains for the analysis of ubiquitinated proteomes. J Am Soc Mass Spectrom. 2013;24(8):1214–1223. doi: 10.1007/s13361-013-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377(1):162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko HS, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566(1-3):110–114. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Glembotski CC. Roles for ATF6 and the sarco/endoplasmic reticulum protein quality control system in the heart. J Mol Cell Cardiol. 2014;71:11–15. doi: 10.1016/j.yjmcc.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glembotski CC. Roles for the sarco-/endoplasmic reticulum in cardiac myocyte contraction, protein synthesis, and protein quality control. Physiology (Bethesda) 2012;27(6):343–350. doi: 10.1152/physiol.00034.2012. [DOI] [PubMed] [Google Scholar]
- 36.Lim PJ, et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol. 2009;187(2):201–217. doi: 10.1083/jcb.200903024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, Hegde RS. Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol Cell. 2016;63(1):21–33. doi: 10.1016/j.molcel.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian Z, Zheng H, Li J, Li Y, Su H, Wang X. Genetically induced moderate inhibition of the proteasome in cardiomyocytes exacerbates myocardial ischemia-reperfusion injury in mice. Circ Res. 2012;111(5):532–542. doi: 10.1161/CIRCRESAHA.112.270983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomes AV, et al. Mapping the murine cardiac 26S proteasome complexes. Circ Res. 2006;99(4):362–371. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- 40.Liang J, et al. Characterization of erasin (UBXD2): a new ER protein that promotes ER-associated protein degradation. J Cell Sci. 2006;119(Pt 19):4011–4024. doi: 10.1242/jcs.03163. [DOI] [PubMed] [Google Scholar]
- 41.Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100(1):169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumarapeli AR, et al. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. FASEB J. 2005;19(14):2051–2053. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, et al. Ubiquilin-1 protects cells from oxidative stress and ischemic stroke caused tissue injury in mice. J Neurosci. 2014;34(8):2813–2821. doi: 10.1523/JNEUROSCI.3541-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marber MS, Mestril R, Chi SH, Sayen MR, Yellon DM, Dillmann WH. Overexpression of the rat inducible 70-kD heat stress protein in a transgenic mouse increases the resistance of the heart to ischemic injury. J Clin Invest. 1995;95(4):1446–1456. doi: 10.1172/JCI117815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griendling KK, et al. Measurement of reactive oxygen species, reactive nitrogen species, and redox-dependent signaling in the cardiovascular system: a scientific statement from the American Heart Association. Circ Res. 2016;119(5):e39–e75. doi: 10.1161/RES.0000000000000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong X, et al. In situ dynamically monitoring the proteolytic function of the ubiquitin-proteasome system in cultured cardiac myocytes. Am J Physiol Heart Circ Physiol. 2004;287(3):H1417–H1425. doi: 10.1152/ajpheart.01233.2003. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Powell SR, Wang X. Enhancement of proteasome function by PA28α overexpression protects against oxidative stress. FASEB J. 2011;25(3):883–893. doi: 10.1096/fj.10-160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gottlieb RA, Andres AM, Sin J, Taylor DP. Untangling autophagy measurements: all fluxed up. Circ Res. 2015;116(3):504–514. doi: 10.1161/CIRCRESAHA.116.303787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Divald A, et al. Myocardial ischemic preconditioning preserves postischemic function of the 26S proteasome through diminished oxidative damage to 19S regulatory particle subunits. Circ Res. 2010;106(12):1829–1838. doi: 10.1161/CIRCRESAHA.110.219485. [DOI] [PubMed] [Google Scholar]
- 50.Tsuchiya H, et al. In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 axis contributes to K48-linked chain specificity of the proteasome. Mol Cell. 2017;66(4):488–502.e7. doi: 10.1016/j.molcel.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 51.Grice GL, Nathan JA. The recognition of ubiquitinated proteins by the proteasome. Cell Mol Life Sci. 2016;73(18):3497–3506. doi: 10.1007/s00018-016-2255-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477(7363):211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Granger DN, Kvietys PR. Reperfusion injury and reactive oxygen species: The evolution of a concept. Redox Biol. 2015;6:524–551. doi: 10.1016/j.redox.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Divald A, Powell SR. Proteasome mediates removal of proteins oxidized during myocardial ischemia. Free Radic Biol Med. 2006;40(1):156–164. doi: 10.1016/j.freeradbiomed.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 55.Powell SR, et al. Oxidized and ubiquitinated proteins may predict recovery of postischemic cardiac function: essential role of the proteasome. Antioxid Redox Signal. 2005;7(5-6):538–546. doi: 10.1089/ars.2005.7.538. [DOI] [PubMed] [Google Scholar]
- 56.Calise J, Powell SR. The ubiquitin proteasome system and myocardial ischemia. Am J Physiol Heart Circ Physiol. 2013;304(3):H337–H349. doi: 10.1152/ajpheart.00604.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H, Monteiro MJ. Ubiquilin overexpression reduces GFP-polyalanine-induced protein aggregates and toxicity. Exp Cell Res. 2007;313(13):2810–2820. doi: 10.1016/j.yexcr.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, Monteiro MJ. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun. 2007;360(2):423–427. doi: 10.1016/j.bbrc.2007.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington’s disease by ubiquilin. Hum Mol Genet. 2006;15(6):1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- 60.Liu Y, Qiao F, Wang H. Enhanced proteostasis in post-ischemic stroke mouse brains by ubiquilin-1 promotes functional recovery. Cell Mol Neurobiol. 2017;37(7):1325–1329. doi: 10.1007/s10571-016-0451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin JK, et al. ATF6 decreases myocardial ischemia/reperfusion damage and links ER stress and oxidative stress signaling pathways in the heart. Circ Res. 2017;120(5):862–875. doi: 10.1161/CIRCRESAHA.116.310266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martindale JJ, et al. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98(9):1186–1193. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- 63.Bhuiyan MS, et al. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest. 2013;123(12):5284–5297. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tannous P, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105(28):9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsui Y, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100(6):914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 66.Ma X, et al. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125(25):3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rothenberg C, et al. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19(16):3219–3232. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beverly LJ, Lockwood WW, Shah PP, Erdjument-Bromage H, Varmus H. Ubiquitination, localization, and stability of an anti-apoptotic BCL2-like protein, BCL2L10/BCLb, are regulated by Ubiquilin1. Proc Natl Acad Sci U S A. 2012;109(3):E119–E126. doi: 10.1073/pnas.1119167109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su H, et al. Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res. 2011;108(1):40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su H, Li F, Ranek MJ, Wei N, Wang X. COP9 signalosome regulates autophagosome maturation. Circulation. 2011;124(19):2117–2128. doi: 10.1161/CIRCULATIONAHA.111.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.