Abstract
After the initial responsiveness of triple-negative breast cancers (TNBCs) to chemotherapy, they often recur as chemotherapy-resistant tumors, and this has been associated with upregulated homology-directed repair (HDR). Thus, inhibitors of HDR could be a useful adjunct to chemotherapy treatment of these cancers. We performed a high-throughput chemical screen for inhibitors of HDR from which we obtained a number of hits that disrupted microtubule dynamics. We postulated that high levels of the target molecules of our screen in tumors would correlate with poor chemotherapy response. We found that inhibition or knockdown of dynamin 2 (DNM2), known for its role in endocytic cell trafficking and microtubule dynamics, impaired HDR and improved response to chemotherapy of cells and of tumors in mice. In a retrospective analysis, levels of DNM2 at the time of treatment strongly predicted chemotherapy outcome for estrogen receptor–negative and especially for TNBC patients. We propose that DNM2-associated DNA repair enzyme trafficking is important for HDR efficiency and is a powerful predictor of sensitivity to breast cancer chemotherapy and an important target for therapy.
Keywords: Oncology
Keywords: Breast cancer, DNA repair
Introduction
DNA double-strand breaks (DSBs) are believed to be the most critical lesions induced by many cancer therapies, including chemotherapy and ionizing radiation (IR). Homology-directed repair (HDR), or homologous recombination repair, plays an important role in tumor resistance to treatment because it repairs these breaks. Hence, the clinical ability to inhibit HDR may lead to improved outcome for patients undergoing DSB-inducing cancer therapy. On the other hand, defects in HDR result in genomic instability and increase carcinogenesis, explaining the fact that tumor cells in many aggressive cancers, including some forms of breast, ovarian, and pancreatic cancers, are defective in HDR (1). HDR defects are thought to be involved in the etiology of these cancers in such cases. As an example, women with germline mutations in BRCA1, a tumor suppressor with crucial roles in HDR and maintenance of genome stability, have a high risk of developing breast and ovarian tumors. Since HDR is involved in repair of DNA DSBs and interstrand cross-links (ICLs), cells with defective BRCA1 are unable to repair these types of damage and display sensitivity to DSB- and ICL-inducing agents, such as IR and nitrogen mustards (e.g., chlorambucil and cyclophosphamide).
Mutations in BRCA1 are particularly prevalent in triple-negative breast cancers (TNBCs), i.e., those that do not express estrogen receptor and progesterone receptor and lack overexpression or amplification of human epidermal growth factor receptor 2 (HER2/NEU, or erbB2). TNBCs have a significant overlap with basal-like breast cancers (BLBCs), and the majority of BRCA1-related tumors are both triple-negative and basal-like (2, 3). These cancers are characterized by high genomic instability, fast growth, and early metastasis, and have the worst prognosis among breast cancer types. Sporadic TNBCs also display a genome instability phenotype and sensitivity to chemotherapy similar to those of the BRCA1-related TNBCs, suggesting that deficiency in BRCA1 or other DNA repair defects may also be involved in their etiology. In fact, promoter methylation and transcriptional inactivation of BRCA1, defects in DNA excision repair, and low expression of DNA repair genes from HDR, nucleotide excision repair, and Fanconi anemia pathways (4, 5) are observed in TNBCs.
Since TNBCs lack the expression of hormone receptors and do not overexpress HER2, no targeted therapy is available for these breast cancers, and standard DNA-damaging chemotherapy remains the main treatment of choice for TNBC/BLBC patients (2, 3). On a positive note, as mentioned, early-stage TNBCs/BLBCs display high sensitivity to chemotherapy, likely due to DNA repair defects, and if they do not relapse within the first 3–5 years after chemotherapy, the risk of relapse at later times is significantly lower than for other types of breast cancer (6, 7). However, in an apparent paradox, tumors that present at late stage or have relapsed are very resistant to treatment and often display upregulation of HDR (8–12). Mechanistically, this HDR upregulation may represent selective adjustment to DNA repair deficiencies characteristic of TNBCs. For example, in cells with BRCA1 deficiency the HDR defect can be rescued by upregulation of RAD51, the protein with a central role in the homologous recombination reaction (13, 14). In fact, overexpression of RAD51 is a common feature of tumors from TNBC patients (13), but the mechanisms of HDR reactivation in TNBCs are largely unknown.
HDR inhibitors may be particularly useful for targeting upregulated HDR in TNBCs. In addition, HDR inhibitors may benefit tumor therapy in general, as they target S/G2 populations and spare normal tissues that are mostly in G1/G0 (15). S/G2 populations are often enriched within tumors as a result of chemotherapy or fractionated IR therapy (16).
In a search for new and more effective HDR inhibitors, we conducted a small-molecule screen based on potentiation of the cytotoxicity of the ICL-inducing agent chlorambucil. Cells defective in HDR cannot perform a recombination step of ICL repair and are therefore hypersensitive to DNA cross-linking agents (17, 18). Unexpectedly, we identified a group of tubulin binders as potent inhibitors of HDR. We also show that inhibition of dynamin 2 (DNM2) — mostly known for its role in endocytosis and interaction with microtubules (19–21) — has a rapid and profound inhibitory effect on HDR capability. We considered the possibility that the DNM2-mediated role in HDR involved trafficking of DNA repair enzymes and studied the distribution of RAD51 within different cellular compartments. We found that, under unperturbed conditions, a considerable fraction of RAD51 in cytoplasm is found in vesicles tightly associated with the microtubules, and that DNM2 inhibition leads to reduced nuclear accumulation of RAD51 at the sites of DSBs.
We reasoned that if dynamin levels were important in enabling HDR in cells, then high HDR activity might correlate with high dynamin expression, which then potentially could predict the response to therapy of patients with TNBC/BLBC. In a retrospective analysis, we find that low DNM2 expression in tumors at the time of chemotherapy is associated with significantly increased survival of patients with TNBC/BLBC. Taken together, our results suggest that DNM2 and associated cell trafficking of the RAD51 recombinase are important determinants of HDR efficiency and treatment outcome for patients with TNBC/BLBC.
Results
High-throughput screen to identify HDR inhibitors.
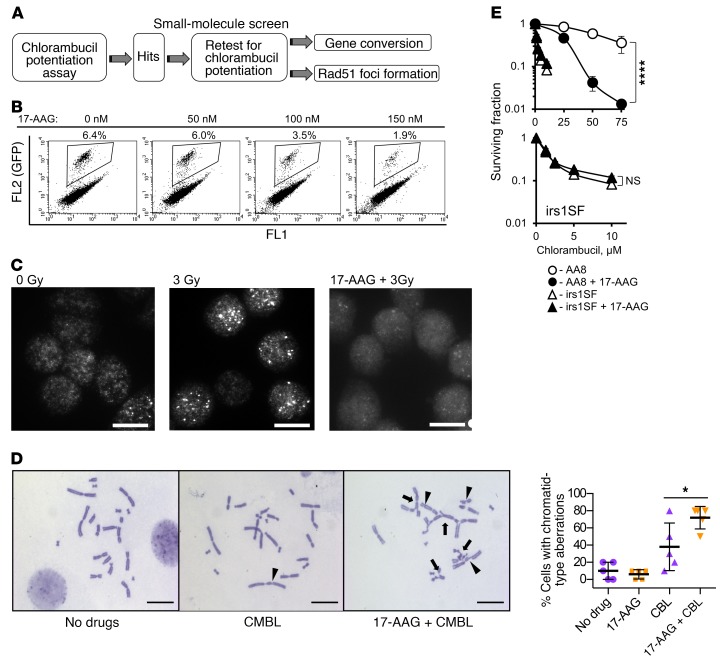
HDR deficiency is associated with extreme sensitivity to agents that induce DNA ICLs (17, 18). Therefore, inhibitors of HDR are expected to further increase the sensitivity of cells to these agents. As nitrogen mustards (e.g., chlorambucil) are potent inducers of ICLs, we monitored the increase in cellular sensitivity to chlorambucil as a readout to identify HDR inhibitors (Figure 1A). In secondary screens, 2 HDR-specific assays — for the capabilities for RAD51 foci formation and for gene conversion — were tested. The RAD51 foci formation assay relies on quantitation of subnuclear RAD51 foci formed in response to DNA damage. The gene conversion assay uses a direct-repeat DR-GFP recombination substrate that quantitates HDR efficiency by detecting GFP signal in cells that used HDR to restore broken GFP sequence (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI87191DS1) (22). As a positive control, we chose to treat cells with 17-AAG (17-allylamino-17-demethoxygeldanamycin; see structure in Supplemental Figure 1B), an agent belonging to the geldanamycin family (23) that binds to the ATP-binding region of HSP90 and inhibits HDR by blocking the HSP90-dependent maturation of RAD51 and BRCA1 (24). Treatment of cells with 17-AAG inhibited both gene conversion (Figure 1B and Supplemental Figure 1C) and IR-induced Rad51 foci formation (Figure 1C and ref. 24) in CHO AA8 cells, as well as in CHO irs1SF cells, which are defective in HDR because of loss of the Xrcc3 gene (Supplemental Figure 1D). Exposure to 17-AAG also significantly elevated chromatid-type aberrations after chlorambucil (Figure 1D and Supplemental Figure 1E). Notably, 17-AAG increased chlorambucil sensitivity of repair-proficient CHO AA8 cells, but had no effect on the chlorambucil sensitivity of HDR-defective CHO irs1SF cells (Figure 1E), suggesting that 17-AAG potentiates chlorambucil cytotoxicity through inactivation of HDR. This conclusion is further supported by the knockdown of the HDR mediator Rad51C in AA8 cells (Supplemental Figure 1F): both knockdown of Rad51C and pretreatment with 17-AAG separately increase the sensitivity of AA8 cells to chlorambucil, while 17-AAG does not further increase chlorambucil sensitivity in cells with shRad51C knockdown. Combined, our data suggest that 17-AAG can be used as a positive control in the screen to identify agents compromising HDR. As expected, in our library screen of known compounds for HDR inhibition (see Methods), 17-AAG (and other geldanamycins) came up among the positive hits. Interestingly and unexpectedly, our screen also identified agents that disrupt tubulin dynamics and endocytosis (Figure 2A).
Figure 1. Overview of the small-molecule screen performed to identify inhibitors of homology-directed repair (HDR).
(A) Diagram of the screen. (B–E) 17-Allylamino-17-demethoxygeldanamycin (17-AAG) is used as a positive control for the screen. (B) 17-AAG inhibits gene conversion in the U2OS-DR-GFP cells. Details on gene conversion assay and quantification are provided in Supplemental Figure 1, A and C. (C) 17-AAG (100 nM) inhibits formation of Rad51 foci in the CHO AA8 cells after 3 Gy. Images were taken at 2 hours after irradiation. Representative images from n ≥ 3 experiments are shown. Scale bars: 10 μm. Quantification of signals is provided in Figure 2D. (D) Chlorambucil (CMBL; 5 μM) induces chromatid-type aberrations in CHO AA8 cells, and 17-AAG (150 nM) potentiates this effect. Arrowheads point to chromatid gaps and breaks, and arrows to complex chromatid exchanges. Scale bars: 20 μm. Graph on the right shows quantitation for data exemplified on the left. Significance analysis: 2-way ANOVA (P = 0.0343). Distribution of chromatid-type aberrations for each treatment is shown in Supplemental Figure 1E. (E) 17-AAG (50 nM) increases sensitivity of CHO AA8 cells to chlorambucil, but does not affect sensitivity of HDR-deficient CHO irs1SF cells, as measured by MTS assay. Bottom: The same data as in the top panel for the irs1SF cells at lower concentrations of chlorambucil. Shown are means ± SDs from n ≥ 3 experiments. Significance analysis: 2-way ANOVA (P < 0.0001). *P < 0.05, ****P < 0.0001.
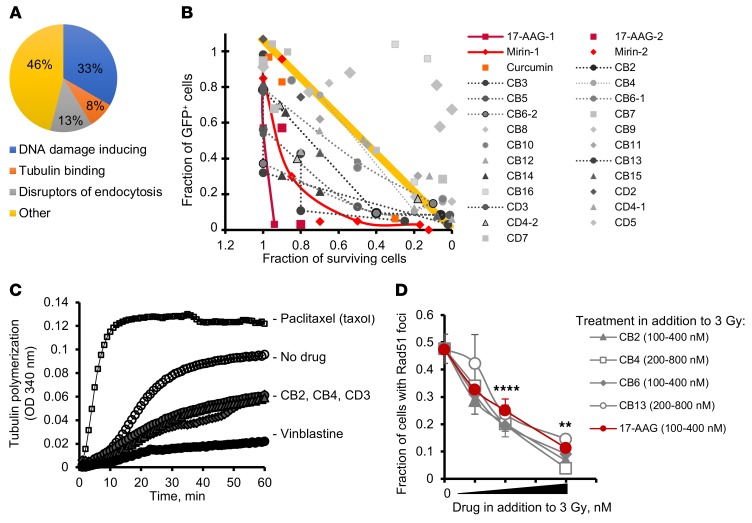
Figure 2. High-throughput chemical screen identifies tubulin binders as inhibitors of HDR.
(A) A pie chart of the prescreen using the libraries of known compounds shows that 21% of compounds potentiating the chlorambucil effect classify as disruptors of cell trafficking. (B) Fraction of cells undergoing gene conversion after DSBs induced by I-SceI (GFP-positive cells) versus fraction of surviving cells. The plot identifies compounds that are able to inhibit gene conversion without affecting cell viability. The yellow line and the area above it show compounds (light gray symbols) for which inhibition of gene conversion (low fraction of GFP-positive cells) could have resulted from increased cell death associated with compound toxicity. True inhibitors of gene conversion are clustering in the left lower quadrant (below the yellow line), where compounds inhibit gene conversion at concentrations not affecting cell viability. (C) CB2, CB4, and CD3 (see Supplemental Figure 2B) inhibit tubulin polymerization similarly to vinblastine. The results for taxol are shown for comparison purposes. Taxol, vinblastine, CB2, CB4, and CD3 were used at 10 μM. (D) CB2, CB4, CB6, and CB13 inhibit radiation-induced Rad51 formation in CHO AA8 cells. CB2 and CB6 were used at 100, 200, and 400 nM each. CB4 and CB13 were used at 200, 400, and 800 nM each. 17-AAG (control) was used at 100, 200, and 400 nM. Shown are means ± SDs from n > 2 experiments. Significance analysis: ANOVA. **P < 0.01, ****P < 0.0001.
A high-throughput screen reveals that microtubule-binding agents impair HDR.
We screened chemical libraries of more than 130,000 diverse compounds. We found 640 hits in the primary screen using the chlorambucil sensitivity assay, of which 46 were confirmed in a dose-response assay to indeed increase cellular sensitivity to chlorambucil. These 46 compounds were further tested in the gene conversion assay. To separate inhibitors of HDR from compounds that reduce GFP expression because of their cytotoxicity or cytostatic effects, we plotted gene conversion levels versus cell growth (Figure 2B). Inhibition of HDR at concentrations compatible with cell survival should significantly reduce the number of GFP-positive cells. Therefore, HDR inhibitors should cluster in the lower left (low gene conversion and high survival) quadrant of the chart (dark gray symbols). As expected, this quadrant contained 17-AAG, and also mirin and curcumin, 2 other compounds previously reported to inhibit HDR (25, 26). All 3 compounds reduced I-SceI–induced HDR at concentrations compatible with cell survival (Figure 2B). In agreement with the results of our screen using the known compound libraries (Figure 2A), the “low gene conversion and high survival” quadrant contained several compounds from a family related to a naphthopyran, LY290181 (Supplemental Figure 2, A and B), which binds microtubules (27, 28). These hits were tested for inhibition of microtubule polymerization. We found that they acted similarly to the Vinca alkaloid vinblastine, a well-characterized and clinically used inhibitor of microtubule dynamics (Figure 2C). In addition to inhibiting gene conversion (Figure 2B and Supplemental Figure 2C), all tested compounds from the LY290181 family also impaired IR-induced Rad51 foci formation (Figure 2D). Importantly, these compounds inhibited gene conversion and Rad51 foci at low concentrations known to affect tubulin dynamics but not tubulin polymerization, and at these concentrations they did not affect cell growth and cell cycle distribution (Supplemental Figure 2D and Supplemental Figure 3).
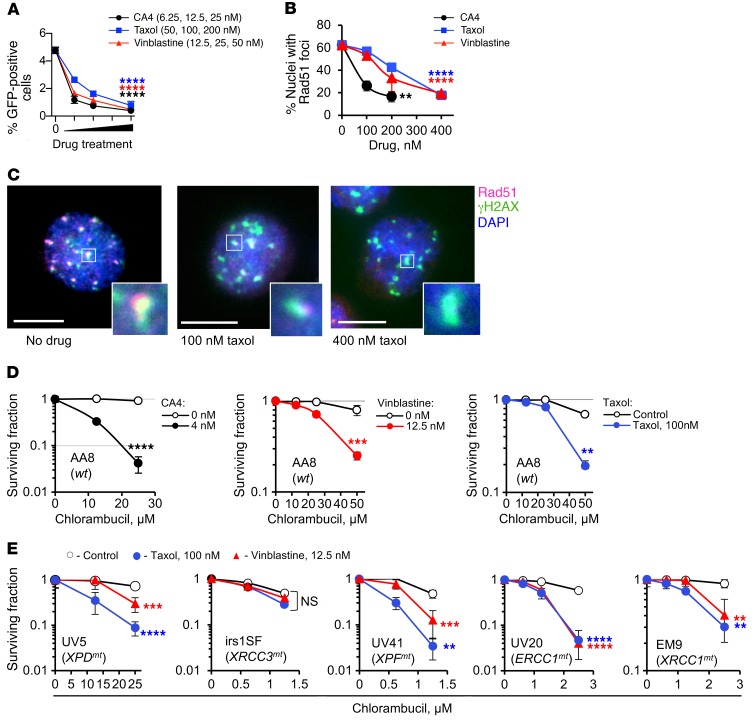
To test whether other tubulin binders affected HDR in a similar manner, we chose to investigate the effects of 3 drugs that bind to different microtubule domains, namely vinblastine, combretastatin A4 (CA4), and taxol. All 3 inhibited both gene conversion and the formation of IR-induced Rad51 foci (Figure 3, A and B). In AA8 cells costained with γH2AX, IR-induced Rad51 foci overlapped with γH2AX foci (Figure 3C). Treatment with tubulin binders led to a reduction in the number of cells with Rad51-positive γH2AX foci (Figure 3C and Supplemental Figure 4A). Importantly, tubulin binders did not inhibit the nonhomologous end-joining (NHEJ) pathway of DSB repair (Supplemental Figure 4B). We note that γH2AX foci in cells treated with tubulin binders were noticeably larger in size (Figure 3C). To the highest dose used, they still contained some Rad51, suggesting that Rad51 recruitment to DSBs was reduced rather than abolished. In a popular view of Rad51 foci assembly, foci are formed from the available nuclear pool of Rad51 (29). Cell fractionation studies further show that the full response to DNA damage involves a cytoplasmic-to-nuclear transport of Rad51 (30) and that this process may be tubulin-dependent (31). Our data obtained by quantification of total Rad51 nuclear fluorescence after treatment with tubulin binders support these cell fractionation studies (Supplemental Figure 4C).
Figure 3. Tubulin binders inhibit HDR.
(A) The tubulin binders combretastatin A4 (CA4), taxol, and vinblastine inhibit gene conversion in the U2OS-DR-GFP cells. (B) CA4, taxol, and vinblastine inhibit formation of Rad51 foci in CHO AA8 cells at 2 hours after 3 Gy. (C) Colocalization of Rad51 and γH2AX foci shows that Rad51 foci are induced at DSBs and that taxol reduces Rad51 accumulation at DSBs. Scale bars: 10 μm. Insets show higher magnification of selected γH2AX foci. (D) CA4, taxol, and vinblastine increase the sensitivity of CHO AA8 cells to chlorambucil. (E) Taxol and vinblastine increase chlorambucil sensitivity of CHO mutant cells defective in nucleotide excision repair (UV5, UV20, and UV41) and base excision/single-strand break repair (EM9), but do not increase chlorambucil sensitivity of irs1SF cells defective in HDR. UV5, irs1SF, UV41, UV20, and EM9 cells have mutations in the XPD, XRCC3, XPF, ERCC1, and XRCC1 genes, respectively. Shown are means ± SDs from n ≥ 3 experiments (A, B, and D) and means ± SDs from n > 2 experiments (E; ranges are shown for points with 2 replicates). Significance analysis: 1-way ANOVA (A and B) and 2-way ANOVA (D and E). Drug-treated samples were compared with the solvent controls. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Tubulin binders have pleiotropic effects (32). Therefore, some of our results might have been impacted by the known effects of tubulin binders on cell cycle progression (32). Importantly, our data showed that the compounds from the LY290181 family did not affect the cell cycle progression at concentrations used in our study, but we did observe cell cycle changes at the highest concentrations of CA4 and vinblastine (Supplemental Figure 3A). To minimize cell cycle perturbations induced by CA4 and vinblastine, we reduced the treatment time to 3 hours (which did not affect cell cycle distribution; Supplemental Figure 3B) and still observed the impaired formation of Rad51 foci (Supplemental Figure 4A).
As expected, vinblastine, CA4, and taxol sensitized cells to chlorambucil (Figure 3D). To address the possibility that tubulin binders may increase sensitivity to chlorambucil by interfering with repair of lesions other than DSBs, we used a panel of CHO mutant cell lines with mutations in DNA repair pathways that may be involved in processing chlorambucil-induced DNA damage. These include mutants in nucleotide excision repair (NER) (with defects in XPD, XPG, XPF, and ERCC1 genes) and single-strand break repair (with a defect in XRCC1). Repair of DNA ICLs induced by chlorambucil involves excision and removal of DNA ICLs by NER and repair of DSB intermediates by HDR (17). Among NER proteins only XPF and ERCC1 are involved in repair of ICLs (33). Cells with defects in these proteins (UV41 and UV20, respectively) cannot repair ICLs and therefore, like HDR mutants irs1SF and irs1, are extremely sensitive to chlorambucil (note the scale differences between panels, Figure 3, D and E). Cells defective in other NER proteins, such as XPD and XPG (UV5 and UV135 cells, respectively), are proficient in ICL excision, but have defects in repair of other (less critical) types of DNA damage induced by chlorambucil and are therefore mildly sensitive to this drug. We show that tubulin binders increased sensitivity to chlorambucil in all mutants tested, with the exception of HDR-deficient irs1SF cells (Figure 3E). Importantly, the lack of effect of tubulin binders on sensitivity of irs1SF cells cannot be explained by the extreme sensitivity of these cells to chlorambucil, as tubulin binders sensitized the equally hypersensitive UV41, UV20, and EM9 cells. Since, in contrast to all other mutants tested, there was no further sensitization to chlorambucil in HDR-defective cells, we conclude that tubulin binders potentiate chlorambucil sensitivity largely through impairment of HDR.
Response to DNA damage initiates reorganization of tubulin and involves dynamin 2.
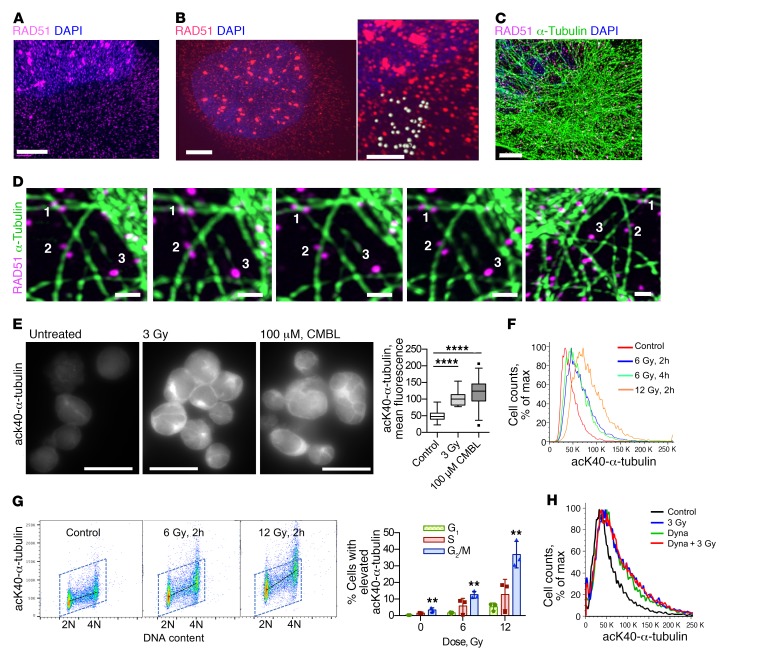
Our data (Figures 2 and 3) suggest that microtubule-dependent trafficking of Rad51 may be required for efficient HDR. Super-resolution microscopy (34) showed that Rad51 was present as 2 populations: as Rad51 foci in the nucleus and as a separate punctate population in both the nucleus and the cytoplasm (Figure 4, A and B). These puncta were much smaller than the nuclear Rad51 foci, with an estimated maximum size of 280 ± 50 nm (Figure 4B). The size of these Rad51 puncta suggests that they might be associated with vesicles formed during clathrin-mediated endocytosis (35), which mediates vesicular transport between different compartments within the cell. 3D super-resolution microscopy analysis showed that Rad51 is associated with tubulin in a manner suggestive of its transport along microtubules (Figure 4, C and D). We observed a significant reduction of those tubulin-associated vesicles upon knockdown of Rad51, thus ruling out the possibility of a staining artifact (Supplemental Figure 5, A and B). We also observed the direct physical association of Rad51 with microtubules in a coimmunoprecipitation assay (Supplemental Figure 5C).
Figure 4. Response to DNA damage changes tubulin dynamics and involves DNM 2.
(A and B) Super-resolution microscopy analysis reveals a discrete punctate pattern of Rad51 in cytoplasm and nucleus. Human MDA-MB-231-BR3 (A) and CHO AA8 (B) cells fixed at 2 hours after 3 Gy show Rad51 foci in the nucleus. Note the much smaller Rad51 dots in both the cytoplasm and the nucleus. Scale bars: 3 μm. B, right: Higher-magnification image taken from the same cell as in the left panel; scale bar: 1 μm. Estimation of Rad51 dot size: light gray spheres drawn around Rad51 dots have an average diameter of 280 ± 50 nm. (C) Super-resolution microscopy images of MDA-MB-231-BR3 cells show that Rad51 associates with microtubules in unperturbed cells. Scale bar: 3 μm. (D) 3D super-resolution analysis: images of the same Rad51 vesicles (white numbers) viewed from different angles. Numbers correspond to the same objects. Scale bars: 1 μm. (E) Lysine 40 (K40) α-tubulin acetylation is increased after DNA damage in human B lymphoma cell line PW. Left: Representative images. Scale bars: 20 μm. CMBL, chlorambucil. Right: Quantification of the fluorescence intensity per cell. (F) K40 α-tubulin acetylation increases after DNA damage. FACS of CHO AA8 cells fixed 2 and 4 hours after treatment with x-rays. (G) FACS analysis of K40 α-tubulin acetylation in CHO AA8 cells shows that G2/M-phase cells compared with G1 have the highest increase in tubulin acetylation after radiation. Left: Representative dot plots. Right: Quantification. Shown are means ± SDs from n = 3 experiments. (H) FACS analysis of CHO AA8 cells irradiated with 3 Gy in the absence and presence of the dynamin 2 (DNM2) inhibitor dynasore. (E and G) Significance analysis: ANOVA. **P < 0.01, ****P < 0.0001.
Regulation of vesicular trafficking is strongly associated with tubulin acetylation, which facilitates the recruitment of molecular motors and the microtubule-associated transport of cargo proteins (reviewed in ref. 36). Tubulin acetylation is required for efficient protein translocation to the nucleus in response to stress (37). We found that tubulin acetylation was significantly elevated after exposure to IR or chlorambucil in a dose-dependent manner (Figure 4, E–H). Consistent with HDR taking place later in the cell cycle, cells in G2/M phase had the highest increase in tubulin acetylation signal per cell in response to DNA-damaging treatment (Figure 4G). To explore this further, we searched for a target associated with vesicular trafficking and tubulin acetylation. Dynamin 2 (DNM2) was an obvious choice, because this GTPase plays key roles in receptor-mediated endocytosis and regulates tubulin acetylation and inhibitors of DNM2 are now readily available. DNM2 inhibitors were absent in the library that we used for our screen. Treatment of cells with dynasore (38), an inhibitor of DNM2, increased tubulin acetylation to the same level as treatment with moderate doses of IR or chlorambucil (Figure 4H). Dynasore pretreatment did not further increase tubulin acetylation after 3 Gy (Figure 4H), suggesting that DNM2 might be acting downstream of tubulin acetylation in response to DNA damage. Overall, these results suggest that tubulin and DNM2 may increase HDR efficiency through improved trafficking of Rad51 and possibly other HDR proteins.
Inhibition of DNM2 impairs HDR.
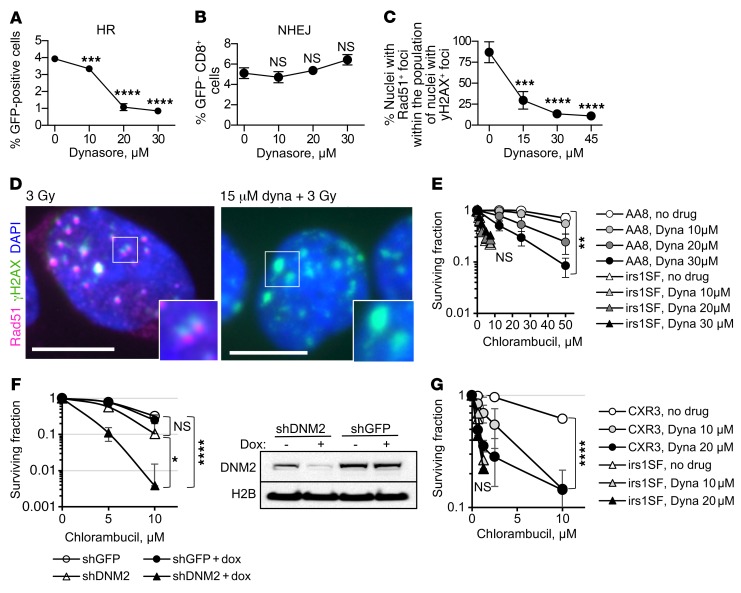
The dynamin inhibitor dynasore effectively reduced I-SceI–induced gene conversion without impairing NHEJ (Figure 5, A and B) and reduced the number of cells with Rad51-positive γH2AX foci (Figure 5, C and D), suggesting that it interfered with the HDR of DSBs. Dynasore also increased the sensitivity of HDR-proficient cells to chlorambucil (Figure 5E and Supplemental Figure 6, A and B). Other inhibitors of DNM2 (39, 40) acted similarly to dynasore (Supplemental Figure 6C). Dynamin inhibition had no significant effect on the cell cycle distribution (Supplemental Figure 3B).
Figure 5. Inhibition of DNM2 impairs HDR.
(A) Dynasore reduces gene conversion in the U2OS-DR-GFP cells. (B) Dynasore does not inhibit NHEJ, as measured by the appearance of CD8+GFP– cells after induction of DSB by the I-SceI enzyme in 293-1040 cells. (C) Dynasore reduces numbers of cells with Rad51-positive/γH2AX-positive foci after IR. CHO AA8 cells were fixed 2 hours after 3 Gy. (D) Left: Rad51 foci in the CHO AA8 cells at 2 hours after 3 Gy are positive for γH2AX, showing that HR repair occurs at the sites of DSBs. Right: Treatment with dynasore reduces the accumulation of Rad51 at the γH2AX foci. Insets show higher magnification of selected areas. Scale bars: 10 μm. (E) Treatment with dynasore increases sensitivity of HDR-proficient CHO AA8 cells to chlorambucil, while it does not alter the sensitivity to chlorambucil in HDR-deficient XRCC3-mutant irs1SF cells. See Supplemental Figure 6A for the survival of irs1SF cells at lower concentrations of chlorambucil. (F) DNM2 knockdown results in increased sensitivity to chlorambucil in the human TNBC cell line MDA-MB-231-BR3. DNM2 knockdown was induced by the addition of doxycycline, as shown in the Western blot (right). Cells were treated with chlorambucil on day 5 after the addition of doxycycline. Shown are means ± SDs from n ≥ 5 experiments. (G) Correction of HDR defect in the irs1SF cells by introduction of XRCC3 cDNA restores both the resistance to chlorambucil and the potentiation of chlorambucil effect by inhibition of DNM2. (A–C, E, and G) Shown are means ± SDs (ranges) from n ≥ 2 MTS experiments. (A–C and E–G) Significance analysis: ANOVA. Dynasore-treated groups were compared with no-drug group (A–C, E, and G). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Dynasore, as other DNM2 inhibitors, also inhibits dynamin 1 (DNM1) and mitochondrial dynamin (DRP1) (38–40). To rule out possible off-target effects of DNM2 inhibitors, we created a doxycycline-inducible knockdown of DNM2 in human MDA-MB-231-BR3 breast cancer cells. DNM2 knockdown impaired IR-induced Rad51 foci formation and sensitized cells to chlorambucil and cisplatin (Figure 5F and Supplemental Figure 7), in accord with our data obtained for DNM2 inhibitors.
Importantly, while dynasore effectively sensitized HDR-proficient AA8 and V79 cells to chlorambucil, it had no effect on the corresponding isogenic HDR-deficient irs1SF and irs1 cell lines with mutations in XRCC3 and XRCC2, respectively (Figure 5E and Supplemental Figure 6, A and B). These observations suggest that potentiation of chlorambucil sensitivity by the dynamin inhibitor occurred because of the impairment of HDR and not because of the effects on other targets of dynamin inhibition. Indeed, potentiation of the chlorambucil effect by dynasore is restored in the CXR3 cell line (41) obtained by expression of human XRCC3 in XRCC3-deficient irs1SF cells (Figure 5G).
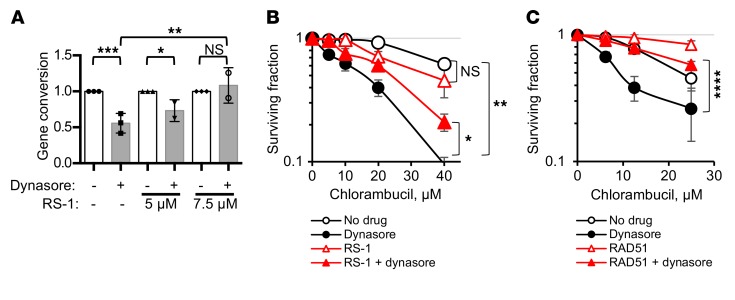
We reasoned that if dynasore acts through impairment of HDR, then selective upregulation of the HDR function should rescue the compound sensitivities. A moderate increase in HDR activity by either overexpression or stimulation of RAD51 has been reported to improve cell survival and to partially complement (i.e., suppress) defects in a number of different HDR genes that encode recombination mediator and comediator proteins (13, 14, 42). Hence, overexpression or stimulation of Rad51 may overcome impairment of HDR by dynasore. To stimulate HDR, we used a sulfonamido-benzamide compound, RS-1, that increases DNA binding and recombination activities of RAD51 by stabilizing RAD51 filaments and promotes resistance to chemotherapy (42, 43). We show that treatment with RS-1 reverses both the inhibition of gene conversion and potentiation of chlorambucil sensitivity by dynasore (Figure 6, A and B). Similarly, overexpression of RAD51 also reverses dynasore-induced sensitization to chlorambucil (Figure 6C and Supplemental Figure 8).
Figure 6. Upregulation of HDR by direct stimulation of RAD51 activity or overexpression of RAD51 overcomes dynasore-induced sensitization to chlorambucil.
(A) Stimulation of RAD51 activity by RS-1 overcomes dynasore-induced inhibition of gene conversion in U2OS-DR-GFP cells. Shown are means and SDs from n ≥ 2 experiments. Ranges are shown for points with 2 replicates. (B) CHO AA8 cells are effectively sensitized to chlorambucil by dynasore, and RS-1 (7.5 μM) reverses this dynasore effect. MTS assay: Shown are means ± SDs from n ≥ 2 experiments. Ranges are shown for points with 2 replicates. (C) U2OS cells overexpressing the RAD51-HA construct show increased resistance to chlorambucil. While U2OS cells are effectively sensitized to chlorambucil by dynasore, the effect of dynasore is lost or reduced in cells overexpressing RAD51. MTS assay: Shown are means and SDs from n ≥ 3 experiments. See Supplemental Figure 8A for Western blots showing the overexpression of RAD51 and for the significance analysis. (A–C) Significance analysis: ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Taken together, our results show that DNM2 function impacts HDR. First, low or impaired DNM2 is associated with the major phenotypic manifestations of HDR deficiency. Second, DNM2 inhibition does not further increase the sensitivity of HDR-defective hamster cell lines. Last, HDR defects stemming from DNM2 impairment can be rescued by direct chemical stimulation of RAD51 activity or by overexpression of RAD51.
RAD51 upregulation does not rescue dynamin inhibition effects in TNBC cells.
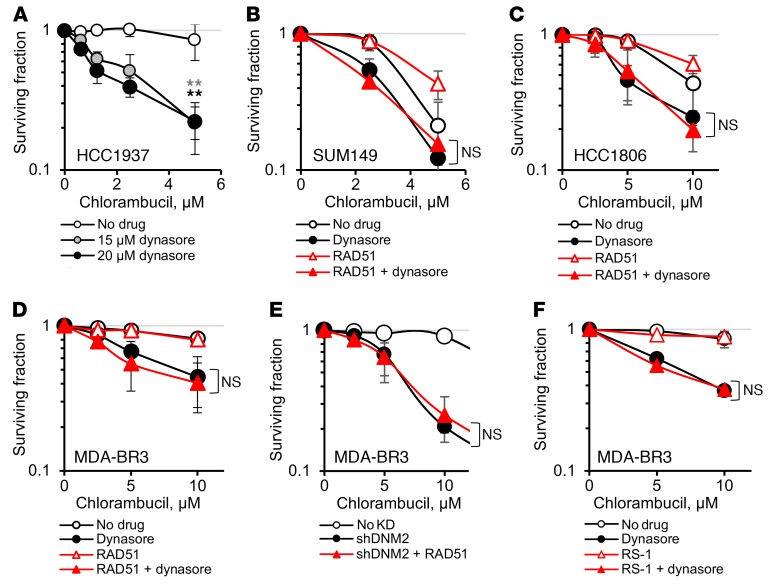
BRCA1-mutant cells are deficient in HDR. Hence, we expected that, similarly to HDR-deficient XRCC2- or XRCC3-mutant hamster cells (Figure 5, E and G, and Supplemental Figure 6, A and B), BRCA1-defective human cells would not be further sensitized to chlorambucil by inhibition of dynamin. In contrast to our expectations, we observed that the sensitivity to chlorambucil was enhanced by dynasore treatment in BRCA1-mutant TNBC cell lines (Figure 7, A and B, and Supplemental Figure 9). Overall, dynasore effectively increased the sensitivity to chlorambucil in cells representing different types of TNBC (as shown for BRCA1-mutant inflammatory breast cancer SUM149, basaloid HCC1806, and metastatic mesenchymal-like MDA-MB-231-BR3 cells) (Figure 7, B–F).
Figure 7. Upregulation of HDR by direct stimulation of RAD51 activity or overexpression of RAD51 fails to rescue dynasore-induced sensitization to chlorambucil in TNBC cells.
(A and B) Dynasore increases chlorambucil sensitivity of the human BRCA1-mutant TNBC cells HCC1937 (A) and SUM149 (B). (B–D) Sensitization to chlorambucil by dynasore is not reversed by overexpression of RAD51-HA in TNBC cell lines SUM149 (B), HCC1806 (C), and MDA-MB-231-BR3 (MDA-BR3) (D). (E) Overexpression of RAD51-HA does not reduce sensitivity to chlorambucil in MDA-MB-231-BR3 cells with DNM2 knockdown. (F) RS-1 does not reduce the sensitizing effect of dynasore in MDA-MB-231-BR3 cells. (A–F) Shown are means ± SDs (ranges) from n ≥ 3 (A) and n ≥ 2 (B–F) MTS experiments. (B–F) See Supplemental Figures 10 and 11 for Western blots showing RAD51 overexpression and for significance analyses. **P < 0.01 by ANOVA.
Martin et al. (13) showed that cells from BRCA1-deficient tumors compensate for the HDR defects by elevated RAD51 expression; they predicted that any additional RAD51 may have little or no effect on tumor-derived BRCA1-deficient cells, as optimum levels of RAD51 may already be reached in TNBC tumors. In agreement with those predictions, we show that RAD51 overexpression in SUM149 cells has only a small effect on resistance to chlorambucil (dose-modifying factor [DMF] = 1.3; Figure 7B), compared with a much stronger effect in the U2OS cells (DMF = 2.5; Figure 6C). Similarly, overexpression of RAD51 in other TNBC cell lines had no effect or only minor effects on chlorambucil sensitivity (DMF = 1.2 and 1, respectively; Figure 7, C and D, and Supplemental Figure 10).
Importantly, while RAD51 overexpression in U2OS rescued dynasore-induced sensitization to chlorambucil (DMF = 2.5; Figure 6C), it failed to do so in all TNBC cell lines tested (Figure 7, B–D, and Supplemental Figure 10). Also, both overexpression of RAD51 in MDA-MB-231-BR3 cells with DNM2 knockdown and chemical stimulation of RAD51 in MDA-MB-231-BR3 cells treated with dynasore failed to increase their resistance to chlorambucil (Figure 7, E and F, and Supplemental Figure 11). The fact that all our attempts to rescue dynasore-induced sensitization to chlorambucil did not work in TNBC cells (while they did in other cell types) argues that the upregulated recombination that occurs in TNBC may be dependent on DNM2 to a greater extent or in additional ways compared with that in other cell types, and therefore, DNM2 inhibition may constitute a new way to enhance the therapy of TNBCs.
In vivo tests and implications for DNM2 inhibition in cancer therapy.
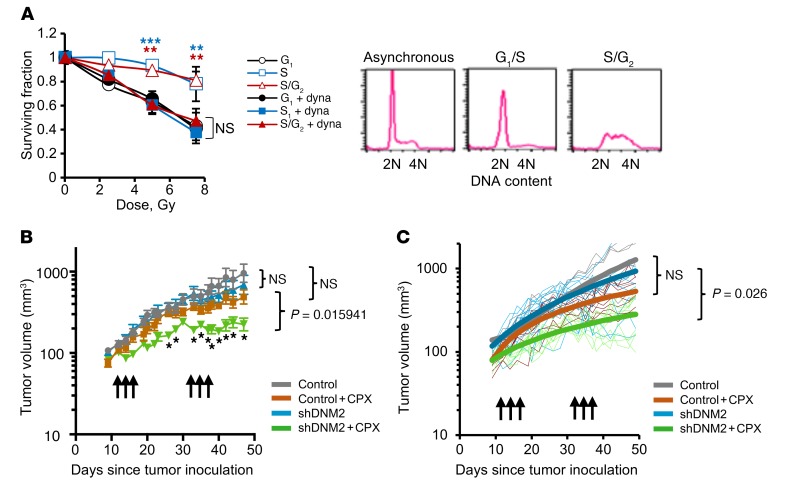
Since HDR takes place predominantly in the S/G2 phase, an inhibitor of HDR is expected to increase the IR sensitivity of cells in the S and G2 phases, largely without affecting cells in G0/G1 (15). This could be useful in cancer therapy, as an HDR inhibitor would specifically sensitize rapidly dividing tumor cells, which at any given time include a significant fraction of cells in the more IR-resistant S and G2 phases, without sensitizing the cells of the normal tissues surrounding the tumor, mostly found in G0/G1. To test this, we collected cells in G1, early S, and late S/G2 phases using mitotic shake-off as previously described (44). As predicted, dynasore increased the IR sensitivity of cells in the S and late S/G2 phases, without affecting the radiation sensitivity of cells in the G1 phase (Figure 8A). We conclude that dynasore acts as a true inhibitor of DSB repair pathways specific to S/G2 phase.
Figure 8. Inhibition of DNM2 sensitizes cells in S/G2 phase and improves tumor response to cytotoxic therapy in an orthotopic mouse model.
(A) Dynasore increases radiation sensitivity of AA8 cells in S and S/G2 phases, but does not affect radiation sensitivity of G1-phase cells. Shown are means ± SDs (ranges) from n ≥ 2 MTS experiments. ANOVA: all groups were compared with the G1. **P < 0.01, ***P < 0.001. Cell cycle distribution is shown on the right. (B) DNM2 knockdown increases tumor sensitivity to cyclophosphamide (CPX) in an orthotopic model of TNBC. MDA-MB-231-BR3 cells with an inducible DNM2 knockdown (described in Figure 5F) were injected into mammary fat pads of female Nu/Nu mice and treated with 2 cycles of 100 mg/kg CPX on days 12, 14, and 16. After a 2-week break, mice were treated with CPX again (days 33, 35, and 37). DNM2 knockdown was induced by the addition of doxycycline to drinking water 3 days before and during CPX treatment. Symbols and error bars are means ± SEMs. Significance analysis: ANOVA, *P < 0.05. (C) Same experiment as in B analyzed using repeated-measures method. Post hoc pairwise comparisons were performed using a Tukey adjustment for multiple comparisons. P values are shown at day 49. See Supplemental Table 1 for details of the significance analysis.
We then tested whether lowering DNM2 levels would increase chemosensitivity in vivo. For this purpose, MDA-MB-231-BR3 cells with doxycycline-inducible expression of shRNA against DNM2 (shDNM2) were implanted into mammary fat pads in Nu/Nu mice. The tumors were subjected to 2 rounds of treatment with cyclophosphamide, a nitrogen mustard widely used in chemotherapy of breast cancers. We show that reduced expression of DNM2 significantly improved tumor response to cyclophosphamide (Figure 8, B and C, and Supplemental Table 1). Lowering DNM2 levels also marginally increased tumor sensitivity to cisplatin (Supplemental Figure 7, B and C). This effect, however, was not as pronounced as for cyclophosphamide, likely because DNA ICLs represent only a minor portion of all DNA cross-links induced by cisplatin (45, 46).
In summary, lowering or inhibiting DNM2 sensitized cells to IR and improved response to chemotherapy in the orthotopic breast cancer model. This suggests that high DNM2 likely interferes with the response to chemotherapy of patients with late-stage estrogen receptor–negative (ER–) breast cancers and TNBCs.
DNM2 predicts the outcome of chemotherapy of hormone receptor–negative breast cancers.
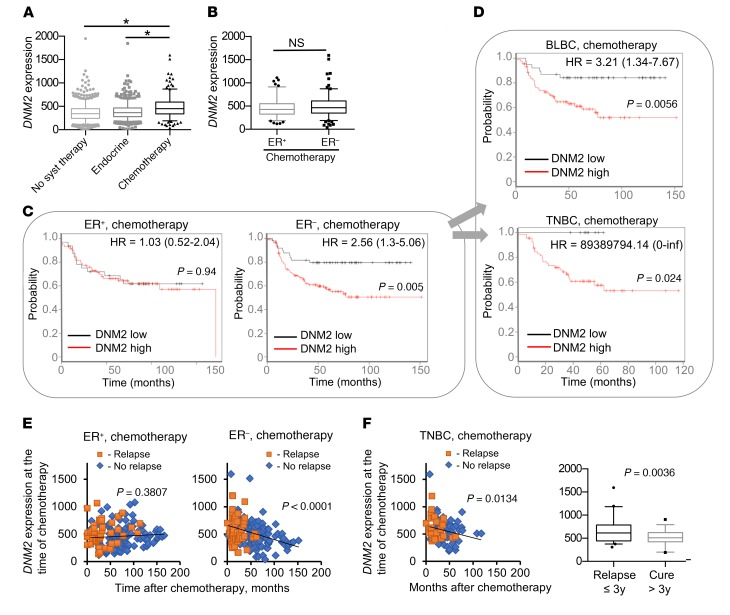
To evaluate the role of DNM2 in resistance to chemotherapy, we performed a survival analysis on an integrated breast cancer data set derived from publicly available gene expression studies (see Methods). We classified each breast cancer patient into 1 of 3 groups based on treatment: “chemotherapy-treated,” “endocrine therapy–treated,” and “systemically untreated” (47). We then compared DNM2 expression in tumors between each group at the time of chemotherapy. Compared with the systemically untreated patients and those on endocrine therapy, chemotherapy-treated breast cancer patients had significantly higher DNM2 expression, likely correlating with the high aggressiveness of cancers for which chemotherapy is prescribed (Figure 9A). While further separation of chemotherapy-treated patients into ER+ and ER– groups did not show significant differences in average DNM2 expression between the groups (Figure 9B), a striking difference was observed in the effects of DNM2 levels on treatment outcome. In the ER– group, DNM2 expression levels below the median of the total breast cancer population were associated with highly improved post-chemotherapy outcome (P = 0.005; Figure 9C). Low DNM2 expression remained significantly correlated with good treatment outcome after adjustment for lymph node status, grade, and size in the Cox proportional hazards multivariate analysis (P = 0.0134). Of the clinical variables, only lymph node status (P = 0.0131) reached significance, while grade and size were not significant (P = 0.525 and P = 0.721, respectively).
Figure 9. DNM2 expression predicts chemotherapy outcome for ER– breast cancer patients.
(A) DNM2 expression in 1,000 systemically untreated patients, 752 patients treated with endocrine therapy, and 274 breast cancer patients treated with chemotherapy. *P < 0.05, ANOVA. (B) DNM2 expression levels are not significantly different between chemotherapy-treated ER+ and ER− tumors. P = 0.167, Mann-Whitney test. (C and D) Decreased DNM2 is associated with better chemotherapy outcome in ER− patients. Patients were split by median expression of DNM2 over the entire breast cancer data set (3,455 patients). Patients with higher than median expression of DNM2 are denoted in red, and those with lower than median expression of DNM2 are in black. Note the hazard ratio increase in the ER−, but not in the ER+, group. (C) Kaplan-Meier analysis of 105 patients with ER+ breast tumors treated with chemotherapy and 169 patients with ER− breast tumors treated with chemotherapy. (D) Kaplan-Meier analysis of 125 patients with BLBC and 76 patients with TNBC. (E) Longer disease-free survival after chemotherapy is associated with lower tumor DNM2 levels at the time of chemotherapy in ER− patients, but not in ER+ patients. Patients diagnosed with disease recurrence at the time of follow-up are designated as “Relapse,” and those remaining disease-free at the time of follow-up as “No relapse.” (F) Longer disease-free survival after chemotherapy is associated with lower tumor DNM2 levels at the time of chemotherapy in TNBC patients. Box plot: TNBCs that relapse during the first 3 years after treatment have significantly higher DNM2 than those that do not relapse or relapse later. (E and F) Linear regression analysis: P < 0.05 indicates that the slope is significantly non-zero. (A, B, and F) The boxes and whiskers contain points within the 25th to 75th and the 5th to 95th percentiles, respectively.
The association of DNM2 expression with chemotherapy outcome was especially pronounced in basal-like and/or triple-negative breast cancers (TNBCs/BLBCs) (Figure 9D), which constitute a significant fraction of ER– cancers and are associated with the worst prognosis. Remarkably, patients in this group with DNM2 levels below the median showed no relapse in a 5-year follow-up period, in contrast to a greater than 40% relapse rate for those with above-median DNM2 levels (Figure 9D). In contrast, the ER+ group showed no difference in survival outcome after chemotherapy when classified by DNM2 expression (Figure 9C). Even in the ER+ luminal B subgroup, which may be more suitable for a comparison with highly proliferative ER– cancers (48), DNM2 expression was no predictor of treatment outcome (Supplemental Figure 12). Furthermore, ER– and TNBC patients with longer disease-free post-chemotherapy survival showed lower DNM2 expression in tumors at the time of chemotherapy (Figure 9, E and F). No such association was found for the ER+ cancers (Figure 9E). The data in Figure 9, E and F, are in agreement with the reported relapse rates for ER– breast cancers and especially for TNBCs, which peak between the first and third year after chemotherapy (6, 7). Our analysis shows that DNM2 expression at the time of chemotherapy in TNBCs that relapsed within the first 3 years after chemotherapy was significantly higher compared with DNM2 expression in TNBCs that did not relapse until later or did not relapse at all (Figure 9F).
Taxanes (e.g., paclitaxel and docetaxel) have been increasingly used as an addition to standard chemotherapy since their approval as chemotherapy agents in 1995. Exposure to taxanes has been shown to be associated with alterations in tubulin expression (49). Although most of the regimen for the cohorts in our analysis did not include taxanes, some studies did not provide the specifics on the kinds of chemotherapy used. Therefore, it is possible that the increased patient survival we observed for the ER– breast cancers in Figure 9, C and D, might have resulted from the exposure to taxanes. To rule out any possible exposure to taxanes as a confounding factor in the integrated breast cancer data set, we next analyzed breast cancer patient data from the highly annotated Stanford Cancer Institute Research Database. In this group of patients there was no difference in taxane exposure between the chemotherapy-treated ER+ and ER– breast cancers (Supplemental Figure 13A), making it unlikely that taxane treatment affected our findings for the integrated breast cancer data set. Moreover, no difference was observed in high-grade/low-grade tumor composition between the ER+ and ER– groups (Supplemental Figure 13B). In addition, we show that while the addition of taxanes to chemotherapy was clearly beneficial for the patients with ER+ breast cancers, no statistical significance for taxane treatment was observed in patients with TNBCs (Supplemental Figure 13C). Together our retrospective analyses show that DNM2 expression predicts the response to chemotherapy in patients with hormone receptor–negative cancers, particularly TNBCs/BLBCs.
Discussion
In this study, we propose a new mechanism for the regulation of HDR involving microtubule-dependent molecular trafficking. We identified dynamin 2 (DNM2), involved in endocytic transport of molecular cargo between cell compartments, regulation of microtubule dynamics (19), and interaction between microtubules (20, 21), as a critical factor in regulation of HDR activity. We propose that changes in tubulin- and DNM2-associated cell trafficking in response to DNA damage are critical for HDR and are of specific relevance to the treatment of ER– breast cancers and TNBCs/BLBCs (Figure 10). Targeting DNM2 may therefore provide a new way of enhancing the effects of chemotherapy, which at present is the only option available for these difficult-to-treat cancers.
Figure 10. Proposed model: DNM2 expression differentiates between good and nonfavorable chemotherapy outcomes in TNBC patients.
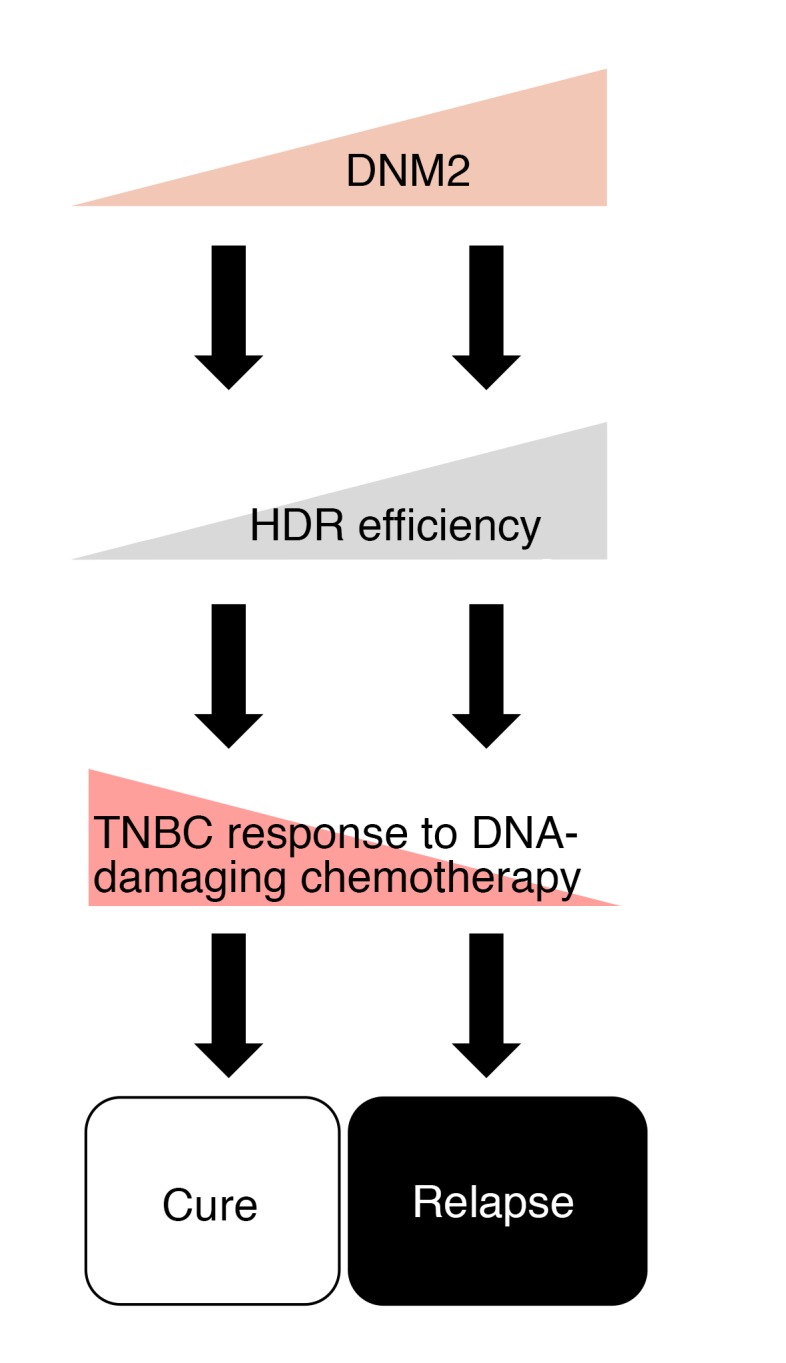
Cells with high DNM2 may have more efficient HDR and may be selected for during DNA-damaging chemotherapy. High HDR proficiency, as well as higher migratory and metastatic potential in these cells, may constitute a mechanism for resistance to chemotherapy in late-stage and recurrent TNBCs.
The unifying feature of TNBCs is that they trace their origin to genomic instability associated with a DNA repair (or HDR) defect, but regain HDR capability in late-stage, aggressive tumors (8–10, 13). We propose that upregulated DNM2/tubulin dynamics play a role in the HDR reactivation in these cancers. Notably, we show that elevated expression of DNM2 is associated with worse chemotherapy outcome in TNBC/BLBC patients (Figure 9, D–F). Importantly, DNM2 expression does not appear to affect the survival of patients treated with endocrine therapy, but is an important determinant of patient survival following cytotoxic therapy, particularly in the ER– group (Figure 9C and Supplemental Figure 14).
RAD51 overexpression is typically observed in late-stage TNBCs, likely as a defense mechanism to counter high genomic instability (13, 14). RAD51 overexpression has been demonstrated to rescue proliferation defects and sensitivity to genotoxic agents in BRCA1- and BRCA2-defective cells (13, 50). Also, in the absence of BRCA/Fanconi anemia proteins, RAD51 overexpression has been shown to improve the stability of stalled replication forks (51, 52). We hypothesize that in the absence of BRCA and other proteins that control recruitment of RAD51 to the sites of DNA damage and/or stalled replication forks (30, 51), RAD51 trafficking largely depends on DNM2. This may explain why overexpression of RAD51 failed to reverse DNM2 inhibition effects in TNBC cells (Figure 7), suggesting that the upregulated recombination that occurs in TNBC may be dependent on DNM2 to a greater extent than in non-TNBC cells.
DNM2-associated trafficking of RAD51 provides a rational explanation for the recent observation that ties tumor aggressiveness, and specifically the basal and triple-negative status, to high cytoplasmic expression of RAD51 (53, 54). We note that overexpression of RAD51 may be toxic to the cells as a result of the formation of RAD51 complexes on undamaged DNA. In this regard, DNM2-mediated trafficking of RAD51 may alleviate the toxic effects of RAD51 overexpression and contribute to survival of TNBC cells, by maintaining low nuclear/high cytoplasmic levels of RAD51.
Our data may have immediate implications for improved treatment options for TNBC patients. Our finding of the role of disturbed microtubule dynamics and its negative impacts on HDR capacity may be useful in fine-tuning current chemo-/radiotherapy regimens to achieve higher cure rates. In fact, greater efficacy of tubulin binders when used in combination with DNA-damaging agents has been reported (reviewed in refs. 3, 55–57), although the interplay of the molecular mechanisms and associated benefits has not been understood.
Using dynamin inhibitors to improve chemotherapy outcome is an attractive possibility that might reduce systemic toxicity associated with the use of tubulin binders. Also, since we demonstrated that a DNM2 inhibitor acted as a true inhibitor of HDR, by specifically sensitizing S- and G2-phase cells to IR without affecting cells in G0/G1, combining DNM2 inhibitors with localized radiotherapy might offer further advantage to reducing systemic toxicity.
The involvement of DNM2 in HDR may provide a long-awaited explanation for 2 observations. First, DNM2 was isolated as a genomic fragment that corrected mitomycin C hypersensitivity of cells from a Fanconi anemia group D [FA(D)] patient (58, 59). Importantly, the DNM2 message was present and not mutated in the FA(D) patient studied, and therefore the correction of mitomycin C hypersensitivity by DNM2 could not be explained by a complementation of a possible defect in the DNM2 gene. The newly identified function of DNM2 in HDR suggests that in that case the correction of mitomycin C hypersensitivity could be due to upregulation of HDR by additional copies of DNM2. Separately, mutation in DNM2 has been shown to be associated with the neuropathic disorder Charcot-Marie-Tooth syndrome (CMT) (19). CMT patients display unusual sensitivity to both tubulin binding and ICL agents (60). Sensitivity to each of these classes of drugs is consistent with and can be explained by our findings that show that DNM2- and tubulin-associated trafficking of RAD51 is important in HDR.
While we favor the hypothesis that high resistance to chemotherapy in TNBCs results from the upregulation of HDR mediated by DNM2 in late-stage TNBC, we are aware of the fact that elevated DNM2 also is associated with increased cell motility and metastasis (61), which could provide an alternative explanation for the increased relapse rates observed in TNBC patients with high DNM2. The 2 mechanisms are by no means mutually exclusive, as it is plausible that a tumor cell population that survives primary tumor chemotherapy is selected for high DNM2 expression, and is, therefore, more resistant to chemotherapy and more metastatic.
In summary, we found that microtubule-associated trafficking affects the efficiency of HDR. Agents that disrupt microtubule dynamics, including tubulin binders and DNM2 inhibitors, impaired all assays conducted to monitor HDR capacity. Similarly to tubulin-binding agents, DNM2 inhibitors sensitized cells to chemotherapy and IR, supporting the targeted clinical application of these compounds as radio- and chemosensitizers. In addition, the fact that reduced expression of DNM2 is associated with improved outcome of cytotoxic therapy in patients with hormone receptor–negative breast cancers, and specifically TNBCs/BLBCs, strongly suggests that DNM2 expression status could be used to stratify patient groups in the future.
Methods
Cell culture and transfection.
All cell lines used in this study were cultured for 2 passages to expand the cell population and frozen for future use. The CHO AA8, UV41, UV5, UV135, EM9, UV20, and HCC1806 cells were obtained from the American Type Culture Collection and the SUM149 cells from Asterand Bioscience. CHO irs1SF and hamster V79 and irs1 cells were provided by L.H. Thompson (Lawrence Livermore National Laboratory, Livermore, California, USA), human U2OS-DR-GFP cells by M. Jasin (Memorial Sloan Kettering Cancer Center, New York, New York, USA), PW human B lymphoma cells by S.J. Knox (Stanford University, Palo Alto, California, USA), MDA-MB-231-BR3 cells by J.E. Price (M.D. Anderson Cancer Center, Houston, Texas, USA), the BRCA1-deficient HCC1937 cell line and its derivatives bearing an empty pcDNA3.1 or a plasmid with BRCA1 cDNA (62, 63) by S.N. Powell (Memorial Sloan Kettering Cancer Center), and the human 293/1040 and HT1080 cells with an inducible knockdown of RAD51 by A. Aroumougame (University of Texas Southwestern Medical Center, Dallas, Texas, USA). RAD51-HA expression construct was a gift from Z. Shen (Rutgers Cancer Institute, New Brunswick, New Jersey, USA). Cells were cultured in DMEM supplemented with 10% FBS and maintained in a humidified incubator with 5% CO2 at 37°C. Plasmid DNA transfections were done using LipofectAMINE-2000 (Life Technologies) following the manufacturer’s instructions.
High-throughput chemical screen.
Small-molecule chemical screening was performed at the High-Throughput Bioscience Center at Stanford University Medical School. Screening was performed using libraries from ChemDiv, Specs, ChemBridge, Sigma LOPAC1280, the NIH Clinical Center, the National Cancer Institute Developmental Therapeutics Program, Microsource Spectrum, Biomol ICCB Known Bioactives, and Biomol FDA-approved drug library. More details are provided in Supplemental Methods.
Gene conversion assay.
Gene conversion assay was performed as described in ref. 64. Briefly, U2OS-DR-GFP cells were transfected with I-SceI enzyme and, 6 hours after transfection, treated with chemical compounds of interest. Seventy-two hours after transfection, cells were analyzed for the presence of GFP-expressing cells by FACS.
Nonhomologous end-joining assay.
Nonhomologous end-joining (NHEJ) assay was performed as described in ref. 65. Briefly, we used a 293/1040 cell line, which stably expresses an end-joining reporter that contains a GFP gene flanked by I-SceI recognition sites. Downstream of the GFP gene the reporter contains a CD8a gene, which is not constitutively expressed because of the lack of an internal ribosome entry site. To induce DSBs, 293/1040 cells were transfected with the I-SceI expression plasmid. If NHEJ occurs, cells lose expression of GFP and gain expression of CD8, which is measured by PE-conjugated anti-CD8 mAb (anti-CD8–PE, BD Biosciences, 555635). Cells were analyzed for the loss of GFP expression and gain of CD68 expression by FACS 4 days after transfection with I-SceI–expressing plasmid. The end-joining rate was determined by counting of the percentage of GFP-CD8+ cells and normalizing for the transfection efficiency.
Immunofluorescence.
Rad51 and γ-H2AX staining were performed as previously described (66). Briefly, cells were plated at 70%–80% confluence. Cells were pretreated with desired compounds for 30 minutes or 1, 3, or 16 hours and irradiated with 3 Gy. Cells were fixed in 2% formaldehyde for 15 minutes on ice, permeabilized in 0.2% Triton X-100 for 15 minutes on ice, and blocked in 5% goat serum for 30 minutes. Cells were incubated with rabbit anti-Rad51 (clone H-92, sc-8349, Santa Cruz Biotechnology, or ab63801, Abcam) and/or with anti-γH2AX (pS139, 16-202A, Millipore) antibody. Secondary antibodies were from Molecular Probes, Invitrogen. Tubulin staining was performed as previously described (67) with anti–α-tubulin (05-829, DM1A, Millipore), anti–acK40–α-tubulin (ab24610, Abcam), and anti–β-tubulin (F2043, Sigma-Aldrich) antibodies.
Tubulin polymerization assay.
Tubulin polymerization was conducted in the 96-well format using HTS-tubulin polymerization assay kit BK004P (Cytoskeleton Inc.). Optical density at 340 nm was measured using a Tecan Infinite M1000 PRO microplate reader (Tecan Group Ltd.).
Super-resolution microscopy.
Structured illumination microscopy (34) images were acquired with an OMX V4 microscope (GE Healthcare) equipped with an Olympus U-Plan Apo 60×1.42 NA oil immersion objective. Image reconstruction and channel alignment processing were performed with softWoRx software (GE Healthcare).
Immunoprecipitation.
Cell pellets from triple-negative breast cancer MDA-MB-231-BR3 cells were washed twice with PBS and resuspended in hypotonic lysis buffer (10 mM HEPES-KOH, pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and protease inhibitors). Cells were broken down in Dounce homogenizer and centrifuged 5 minutes at 200 x g. Supernatants were collected as cytoplasmic fraction and precleared with rabbit IgG followed by Protein A/G PLUS agarose beads (sc-2003, Santa Cruz Biotechnology; 1 μg of IgG and 50 μl of Dynabeads per each milligram of lysate). Precleared lysates were divided for incubation with IgG and anti-Rad51 antibody (1 μg of antibody and 10 μl of beads per each 1 mg of protein) and incubated for 4 hours, followed by 1-hour incubation with Dynabeads. Beads were washed 3 times with lysis buffer and resuspended in 50 μl NuPAGE loading buffer. All procedures were performed at room temperature to preserve the structure of microtubules, which depolymerize on ice.
Western blotting.
Western blotting was performed with anti-Rad51 (H-92, sc-8349, Santa Cruz Biotechnology, or ab63801, Abcam), anti–α-tubulin (05-829, DM1A, Millipore), anti-H2B (07-371, Upstate), anti-DNM2 (ab3457, Abcam), and anti-HA tag (clone 6E2, 2367S, Cell Signaling Technology). See complete unedited blots in the supplemental material.
Cell cycle synchronization.
CHO AA8 cells were synchronized in mitosis using mitotic shake-off procedure, as previously described (44). Cell cycle distribution was confirmed by FACS analysis, and cells were considered to be in G1 phase at 3 hours, S phase at 8 hours, and S/G2 at 11 hours after shake-off. Synchronized cells were pretreated for a half hour with 30 μM dynasore and irradiated using an X-rad 320 irradiator (Precision X-Ray).
RNA interference.
For downregulation of DNM2 by RNAi, we used a lentiviral RNAi system based on the BLOCK-iT system (Invitrogen) modified by Campeau et al. (68). The following sequences were used to create shDNM2 constructs: 5′-GGACTTACGACGGGAGATC-3′ and 5′-GACATGATCCTGCAGTTCA-3′. A doxycycline-inducible GFP shRNA was used as a control.
Survival analysis in integrated breast cancer data set.
Gene chip data sets for breast cancer were identified in PubMed GEO (http://www.ncbi.nlm.nih.gov/gds) using the keywords “breast,” “cancer,” and “survival.” The database was constructed as described previously (47). Only data sets with available raw data and clinical survival information and including at least 30 patients were considered. Altogether, 4,142 breast cancer patients were processed; of these, 3,458 had relapse-free survival data. Of these, 1,000 had not received a systemic treatment, 274 had received chemotherapy only, and 752 had received endocrine therapy only. For the remaining patients, either the applied therapy was not documented or they received both endocrine and chemotherapy. Chemotherapy for most of the cohorts (where described) included cyclophosphamide as part of the CMF regimen (cyclophosphamide, methotrexate, 5-fluorouracil) or FEC regimen (5-flourouracil, epirubicin, cyclophosphamide). As many genes are represented by multiple probe sets on the Affymetrix gene chips, we selected the most reliable probe sets using JetSet (69). For DNM2 the probe set 202253_s_at was used. The median expression of the genes was calculated over the entire data set, and this was used as cutoff in each analysis. Kaplan-Meier survival plot, and the hazard ratio with 95% confidence intervals and log-rank P value, were calculated and plotted in R using Bioconductor packages (47). For the multivariate Cox proportional hazards regression analysis, the clinical variables (lymph node status, tumor grade and size) were included.
Chemotherapy in the orthotopic xenograft model of TNBC.
The MDA-MB-231-BR3 cells expressing doxycycline-inducible shDNM2 or shGFP were used to create an orthotopic model of TNBC in Nu/Nu mice. The details of tumor inoculation and monitoring are provided in Supplemental Methods. Treatment began when tumors reached 60–90 mm3 and consisted of 2 cycles of cyclophosphamide or 1 cycle of cisplatin (300 mg/kg/cycle or 16 mg/kg/cycle, respectively). To induce DNM2 knockdown, doxycycline was given 3 days before and during cyclophosphamide or cisplatin administration. Cisplatin was given by i.p. injections on days 11, 13, 15, and 17 after tumor cell inoculations. Cyclophosphamide cycle was composed of 3 i.p. injections given every other day followed by a 2-week rest period.
Statistics.
The specific details of statistical tests and number of samples and experimental repeats are included in the figure legends. A Student’s t test was used for comparing 2 groups. ANOVA models were used for comparing 3 or more groups with post hoc testing for pairwise comparisons. A Tukey adjustment was applied in testing all possible pairwise comparisons, and a Dunnett’s adjustment was applied in comparing treatment groups with a control. To find interactions between compounds or to compare dose-response curves, we used 2-way ANOVA. The in vivo experiments were randomized and analyzed in a generalized linear model to account for the within-mouse correlations. The treatment groups were compared across time, and at each time point and post hoc, pairwise comparisons were done with a Tukey adjustment. All tests were 2-sided with an α level of 0.05; and all tests were performed in Prism (GraphPad Software) or SAS version 9.4 (SAS Institute Inc.). A P value less than 0.05 was considered significant.
Study approval.
All animal procedures were approved by Stanford University’s Administrative Panel on Laboratory Animal Care.
Additional methods are provided in Supplemental Methods.
Author contributions
SBC, JCG, and JMB designed the studies. SBC, RBN, JTT, SSM, BG, JHS, AO, LD, DESC, DJW, SH, AA, and CW performed various parts of the study. SBC, JCG, RBN, JTT, SSM, BG, MPH, DESC, DJW, RVE, MHG, and JMB analyzed the data. AA, CW, and MHG provided reagents. SBC wrote the manuscript. SBC, JCG, SSM, CW, BG, and JMB edited the manuscript.
Supplementary Material
Acknowledgments
We thank M. Jasin, S.N. Powell, L.H. Thompson, S.J. Knox, J.E. Price, and Z. Shen for the gifts of cell lines and constructs used in the study. We thank O.V. Razorenova (University of California, Irvine, California, USA) for valuable manuscript critiques, and Jason Wu from the Stanford High-Throughput Bioscience Center (Stanford University, Stanford, California, USA) for technical assistance with the chemical screen. This work was supported by NIH grants P01-CA067166 to JMB, ES021454 to CW, K08-NS901527 to MHG, and R01-AG053341 to AA. High-resolution microscopy images were obtained with Stanford Neuroscience Microscopy Service, supported by NIH NS069375. Stanford Cancer Institute Research Database was supported by National Cancer Institute Cancer Center Support Grant 5P30CA124435 and Stanford NIH/National Center for Research Resources Clinical and Translational Science Award UL1 RR025744. BG was supported by grant NVKP_16-1-2016-0037 of the National Research, Development and Innovation Office, Hungary. LD was supported by the China Scholarship Council.
Version 1. 10/29/2018
Electronic publication
Version 2. 12/03/2018
Print issue publication
Funding Statement
These are listed in the acknowledgments
Footnotes
SBC’s present address is: Department of Neurosurgery, Stanford University, Stanford, California, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(12):5307–5321.https://doi.org/10.1172/JCI87191.
Contributor Information
Sophia B. Chernikova, Email: sophiac1@stanford.edu.
Stephano S. Mello, Email: ssmello@stanford.edu.
Michael P. Hay, Email: m.hay@auckland.ac.nz.
Andrew Olson, Email: aolson6@stanford.edu.
Solomon Henry, Email: shenry@stanford.edu.
Rie von Eyben, Email: rieve@stanford.edu.
Lei Deng, Email: cbhdddf@gmail.com.
Melanie Hayden Gephart, Email: mghayden@gmail.com.
Claudia Wiese, Email: Claudia.Wiese@colostate.edu.
Balázs Győrffy, Email: zsalab2@yahoo.com.
J. Martin Brown, Email: mbrown@stanford.edu.
References
- 1.Riaz N, et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun. 2017;8(1):857. doi: 10.1038/s41467-017-00921-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaspers JE, Rottenberg S, Jonkers J. Therapeutic options for triple-negative breast cancers with defective homologous recombination. Biochim Biophys Acta. 2009;1796(2):266–280. doi: 10.1016/j.bbcan.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Crown J, O’Shaughnessy J, Gullo G. Emerging targeted therapies in triple-negative breast cancer. Ann Oncol. 2012;23(suppl 6):vi56–vi65. doi: 10.1093/annonc/mds196. [DOI] [PubMed] [Google Scholar]
- 4.Ribeiro E, et al. Triple negative breast cancers have a reduced expression of DNA repair genes. PLoS One. 2013;8(6):e66243. doi: 10.1371/journal.pone.0066243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alli E, Sharma VB, Sunderesakumar P, Ford JM. Defective repair of oxidative dna damage in triple-negative breast cancer confers sensitivity to inhibition of poly(ADP-ribose) polymerase. Cancer Res. 2009;69(8):3589–3596. doi: 10.1158/0008-5472.CAN-08-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dent R, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15 pt 1):4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 7.Tischkowitz M, et al. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer. 2007;7:134. doi: 10.1186/1471-2407-7-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maacke H, et al. Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer. Int J Cancer. 2000;88(6):907–913. doi: 10.1002/1097-0215(20001215)88:6<907::AID-IJC11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 9.Wiegmans AP, et al. Rad51 supports triple negative breast cancer metastasis. Oncotarget. 2014;5(10):3261–3272. doi: 10.18632/oncotarget.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woditschka S, et al. DNA double-strand break repair genes and oxidative damage in brain metastasis of breast cancer. J Natl Cancer Inst. 2014;106(7):dju145. doi: 10.1093/jnci/dju145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao Z, Jiang Y, Liu X, Seluanov A, Gorbunova V. DNA repair by homologous recombination, but not by nonhomologous end joining, is elevated in breast cancer cells. Neoplasia. 2009;11(7):683–691. doi: 10.1593/neo.09312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ossovskaya V, et al. Exploring molecular pathways of triple-negative breast cancer. Genes Cancer. 2011;2(9):870–879. doi: 10.1177/1947601911432496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007;67(20):9658–9665. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
- 14.Schild D, Wiese C. Overexpression of RAD51 suppresses recombination defects: a possible mechanism to reverse genomic instability. Nucleic Acids Res. 2010;38(4):1061–1070. doi: 10.1093/nar/gkp1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chernikova SB, Game JC, Brown JM. Inhibiting homologous recombination for cancer therapy. Cancer Biol Ther. 2012;13(2):61–68. doi: 10.4161/cbt.13.2.18872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Somaiah N, Yarnold J, Lagerqvist A, Rothkamm K, Helleday T. Homologous recombination mediates cellular resistance and fraction size sensitivity to radiation therapy. Radiother Oncol. 2013;108(1):155–161. doi: 10.1016/j.radonc.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 17.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20(21):7980–7990. doi: 10.1128/MCB.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinz JM. Role of homologous recombination in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51(6):582–603. doi: 10.1002/em.20577. [DOI] [PubMed] [Google Scholar]
- 19.Tanabe K, Takei K. Dynamic instability of microtubules requires dynamin 2 and is impaired in a Charcot-Marie-Tooth mutant. J Cell Biol. 2009;185(6):939–948. doi: 10.1083/jcb.200803153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda K, Nakata T, Noda Y, Sato-Yoshitake R, Hirokawa N. Interaction of dynamin with microtubules: its structure and GTPase activity investigated by using highly purified dynamin. Mol Biol Cell. 1992;3(10):1181–1194. doi: 10.1091/mbc.3.10.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shpetner HS, Vallee RB. Identification of dynamin, a novel mechanochemical enzyme that mediates interactions between microtubules. Cell. 1989;59(3):421–432. doi: 10.1016/0092-8674(89)90027-5. [DOI] [PubMed] [Google Scholar]
- 22.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulte TW, Neckers LM. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother Pharmacol. 1998;42(4):273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 24.Noguchi M, et al. Inhibition of homologous recombination repair in irradiated tumor cells pretreated with Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Biochem Biophys Res Commun. 2006;351(3):658–663. doi: 10.1016/j.bbrc.2006.10.094. [DOI] [PubMed] [Google Scholar]
- 25.Dupré A, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4(2):119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chirnomas D, et al. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol Cancer Ther. 2006;5(4):952–961. doi: 10.1158/1535-7163.MCT-05-0493. [DOI] [PubMed] [Google Scholar]
- 27.Panda D, Singh JP, Wilson L. Suppression of microtubule dynamics by LY290181. A potential mechanism for its antiproliferative action. J Biol Chem. 1997;272(12):7681–7687. doi: 10.1074/jbc.272.12.7681. [DOI] [PubMed] [Google Scholar]
- 28.Kemnitzer W, et al. Discovery of 4-aryl-4H-chromenes as a new series of apoptosis inducers using a cell- and caspase-based high throughput screening assay. 4. Structure-activity relationships of N-alkyl substituted pyrrole fused at the 7,8-positions. J Med Chem. 2008;51(3):417–423. doi: 10.1021/jm7010657. [DOI] [PubMed] [Google Scholar]
- 29.Tarsounas M, Davies AA, West SC. RAD51 localization and activation following DNA damage. Philos Trans R Soc Lond B Biol Sci. 2004;359(1441):87–93. doi: 10.1098/rstb.2003.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gildemeister OS, Sage JM, Knight KL. Cellular redistribution of Rad51 in response to DNA damage: novel role for Rad51C. J Biol Chem. 2009;284(46):31945–31952. doi: 10.1074/jbc.M109.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poruchynsky MS, et al. Microtubule-targeting agents augment the toxicity of DNA-damaging agents by disrupting intracellular trafficking of DNA repair proteins. Proc Natl Acad Sci U S A. 2015;112(5):1571–1576. doi: 10.1073/pnas.1416418112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4(4):253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 33.Niedernhofer LJ, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24(13):5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gustafsson MG, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94(12):4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12(8):517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 36.Perdiz D, Mackeh R, Poüs C, Baillet A. The ins and outs of tubulin acetylation: more than just a post-translational modification? Cell Signal. 2011;23(5):763–771. doi: 10.1016/j.cellsig.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 37.Giustiniani J, et al. Tubulin acetylation favors Hsp90 recruitment to microtubules and stimulates the signaling function of the Hsp90 clients Akt/PKB and p53. Cell Signal. 2009;21(4):529–539. doi: 10.1016/j.cellsig.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10(6):839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Hill TA, et al. Long chain amines and long chain ammonium salts as novel inhibitors of dynamin GTPase activity. Bioorg Med Chem Lett. 2004;14(12):3275–3278. doi: 10.1016/j.bmcl.2004.03.096. [DOI] [PubMed] [Google Scholar]
- 40.McCluskey A, et al. Building a better dynasore: the dyngo compounds potently inhibit dynamin and endocytosis. Traffic. 2013;14(12):1272–1289. doi: 10.1111/tra.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tebbs RS, et al. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc Natl Acad Sci U S A. 1995;92(14):6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mason JM, et al. The RAD51-stimulatory compound RS-1 can exploit the RAD51 overexpression that exists in cancer cells and tumors. Cancer Res. 2014;74(13):3546–3555. doi: 10.1158/0008-5472.CAN-13-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jayathilaka K, et al. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc Natl Acad Sci U S A. 2008;105(41):15848–15853. doi: 10.1073/pnas.0808046105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chernikova SB, Lindquist KL, Elkind MM. Cell cycle-dependent effects of wortmannin on radiation survival and mutation. Radiat Res. 2001;155(6):826–831. doi: 10.1667/0033-7587(2001)155[0826:CCDEOW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 45.Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc Natl Acad Sci U S A. 1994;91(22):10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zamble DB, Mu D, Reardon JT, Sancar A, Lippard SJ. Repair of cisplatin — DNA adducts by the mammalian excision nuclease. Biochemistry. 1996;35(31):10004–10013. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- 47.Györffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 48.Creighton CJ. The molecular profile of luminal B breast cancer. Biologics. 2012;6:289–297. doi: 10.2147/BTT.S29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tegze B, et al. Parallel evolution under chemotherapy pressure in 29 breast cancer cell lines results in dissimilar mechanisms of resistance. PLoS One. 2012;7(2):e30804. doi: 10.1371/journal.pone.0030804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown ET, Holt JT. Rad51 overexpression rescues radiation resistance in BRCA2-defective cancer cells. Mol Carcinog. 2009;48(2):105–109. doi: 10.1002/mc.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22(1):106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolinjivadi AM, Sannino V, de Antoni A, Técher H, Baldi G, Costanzo V. Moonlighting at replication forks — a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett. 2017;591(8):1083–1100. doi: 10.1002/1873-3468.12556. [DOI] [PubMed] [Google Scholar]
- 53.Alshareeda AT, et al. Clinical and biological significance of RAD51 expression in breast cancer: a key DNA damage response protein. Breast Cancer Res Treat. 2016;159(1):41–53. doi: 10.1007/s10549-016-3915-8. [DOI] [PubMed] [Google Scholar]
- 54.Sosińska-Mielcarek K, et al. Immunohistochemical prediction of brain metastases in patients with advanced breast cancer: the role of Rad51. Breast. 2013;22(6):1178–1183. doi: 10.1016/j.breast.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 55.Isakoff SJ. Triple-negative breast cancer: role of specific chemotherapy agents. Cancer J. 2010;16(1):53–61. doi: 10.1097/PPO.0b013e3181d24ff7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arnedos M, Bihan C, Delaloge S, Andre F. Triple-negative breast cancer: are we making headway at least? Ther Adv Med Oncol. 2012;4(4):195–210. doi: 10.1177/1758834012444711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.André F, Zielinski CC. Optimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agents. Ann Oncol. 2012;23(suppl 6):vi46–vi51. doi: 10.1093/annonc/mds195. [DOI] [PubMed] [Google Scholar]
- 58.Diatloff-Zito C, Papadopoulo D, Averbeck D, Moustacchi E. Abnormal response to DNA crosslinking agents of Fanconi anemia fibroblasts can be corrected by transfection with normal human DNA. Proc Natl Acad Sci U S A. 1986;83(18):7034–7038. doi: 10.1073/pnas.83.18.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diatloff-Zito C, Gordon AJ, Duchaud E, Merlin G. Isolation of an ubiquitously expressed cDNA encoding human dynamin II, a member of the large GTP-binding protein family. Gene. 1995;163(2):301–306. doi: 10.1016/0378-1119(95)00275-B. [DOI] [PubMed] [Google Scholar]
- 60.Weimer LH, Podwall D. Medication-induced exacerbation of neuropathy in Charcot Marie Tooth disease. J Neurol Sci. 2006;242(1–2):47–54. doi: 10.1016/j.jns.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 61.Eppinga RD, Krueger EW, Weller SG, Zhang L, Cao H, McNiven MA. Increased expression of the large GTPase dynamin 2 potentiates metastatic migration and invasion of pancreatic ductal carcinoma. Oncogene. 2012;31(10):1228–1241. doi: 10.1038/onc.2011.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evans JW, et al. Homologous recombination is the principal pathway for the repair of DNA damage induced by tirapazamine in mammalian cells. Cancer Res. 2008;68(1):257–265. doi: 10.1158/0008-5472.CAN-06-4497. [DOI] [PubMed] [Google Scholar]
- 63.Treszezamsky AD, Kachnic LA, Feng Z, Zhang J, Tokadjian C, Powell SN. BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res. 2007;67(15):7078–7081. doi: 10.1158/0008-5472.CAN-07-0601. [DOI] [PubMed] [Google Scholar]
- 64.Nakanishi K, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102(4):1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25(14):3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chernikova SB, et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 2012;72(8):2111–2119. doi: 10.1158/0008-5472.CAN-11-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Azarenko O, Okouneva T, Singletary KW, Jordan MA, Wilson L. Suppression of microtubule dynamic instability and turnover in MCF7 breast cancer cells by sulforaphane. Carcinogenesis. 2008;29(12):2360–2368. doi: 10.1093/carcin/bgn241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campeau E, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4(8):e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics. 2011;12:474. doi: 10.1186/1471-2105-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.