Abstract
DNA-RNA hybrids associated with R-loops promote DNA damage and genomic instability. The capacity of hybrids at different genomic sites to cause DNA damage was not known, and the mechanisms leading from hybrid to damage were poorly understood. Here, we adopt a new strategy to map and characterize the sites of hybrid-induced damage genome-wide in budding yeast. We show that hybrid removal is essential for life because persistent hybrids cause irreparable DNA damage and cell death. We identify that a subset of hybrids is prone to cause damage, and that the chromosomal context of hybrids dramatically impacts their ability to induce damage. Furthermore, persistent hybrids affect the repair pathway generating large regions of ssDNA by two distinct mechanisms, likely resection and re-replication. These damaged regions may act as potential precursors to gross chromosomal rearrangements like deletions and duplications that are associated with R-loops and cancers.
Summary
One Sentence Summary: Persistence of DNA-RNA hybrids leads to the formation of different aberrant DNA damage structures.
ETOC paragraph
RNA can hybridize to genomic loci creating DNA-RNA hybrids and ssDNA filaments called R-loops, which can cause DNA damage. Constantino and Koshland discovered that only a subset of persistent R-loops causes irreparable damage. When the hybrid persists in the damaged region, it blocks correct repair, generating potential precursors of genome instability.
Genomic instability is crucial for evolution, but is also a key driver of diseases like cancer and neurodegeneration (Tubbs and Nussenzweig, 2017). Genomic instability is often caused by errors in endogenous processes like DNA replication and transcription. One type of endogenous error occurs when RNA inappropriately anneals to a homologous genomic locus, generating a DNA-RNA hybrid and a displaced single-stranded DNA (ssDNA) segment. These unusual structures, called R-loops, are a potent source of DNA damage and gross chromosomal rearrangements (GCRs) (Aguilera and Gomez-Gonzalez, 2017; Costantino and Koshland, 2015; Sollier and Cimprich, 2015).
A major indication that R-loops cause DNA damage and genomic instability came from studies of enzymes that prevent the formation of R-loops or remove R-loops from the genome (Huertas and Aguilera, 2003; Li and Manley, 2005; Wahba et al., 2011). One class of enzymes that removes hybrids include two isoforms of RNase-H. These enzymes degrade the RNA specifically in hybrids (Figure 1A) (Cerritelli and Crouch, 2009; Reijns et al., 2012; Wahba et al., 2011). Budding yeast mutants that lack RNases-H exhibit an increase in nuclear foci of DNA-repair proteins, a readout of DNA damage, and an increase in the frequency of genomic rearrangements, the output of the DNA damage. Furthermore, RNase-H overexpression suppresses genome instability that result from mutants that promote hybrid formation. Thus, by preventing R-loop persistence, RNases-H prevent DNA damage and genome instability.
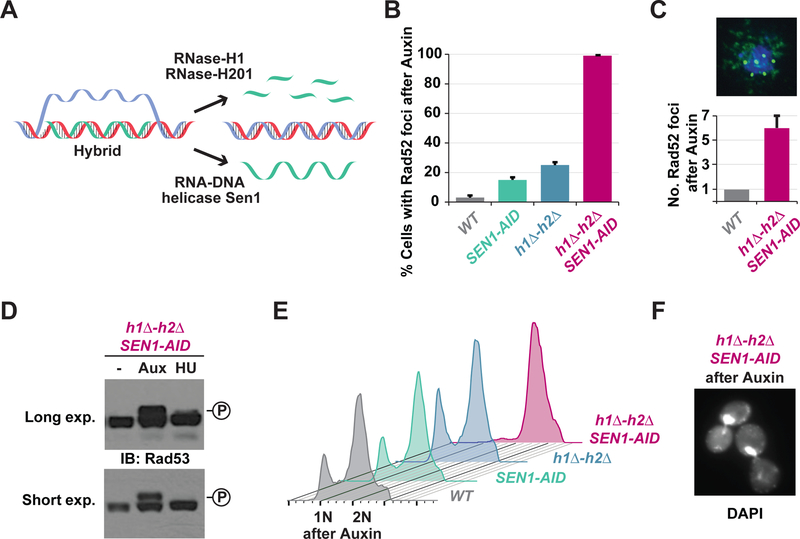
Fig. 1. Blocking the R-loop removing enzymes causes high levels of irreparable DNA damage and cell-cycle arrest.
(A) Cartoon of hybrid removal pathways. RNase-H1 and RNase-H2 degrade the RNA while Sen1 helicase unwinds hybrids. (B) DNA damage in cells with persistent hybrids. Asynchronous cultures of wild-type (WT), auxin-inducible degron allele of Sen1 (SEN1-AID), deletion alleles of RNase-H1 and RNase-H2 (h1Δ-h2Δ), or both (h1Δ-h2Δ SEN1-AID) were treated with auxin for 4h and then scored for Rad52-GFP foci. (C) Asynchronous cultures of WT and h1Δ-h2Δ SEN1-AID were treated with auxin for 4h and then prepared for chromosome spreads using an antibody against endogenous Rad52 (green), the DNA was stained with DAPI (blue). Quantification of 100 cells is shown with bar graph. (D) Rad53-phosphorylation in cells with persistent hybrids. Asynchronous cultures of h1Δ-h2Δ SEN1-AID cells were untreated (-), treated for 4h with auxin (Aux) or 2h with hydroxyurea (HU). Western-blot was performed using Rad53 antibody. The levels of phosphorylated-Rad53 (upper band) were significantly greater when cells were treated with auxin compared to HU. (E) DNA content in cells with persistent hybrids. Cultures in (B) were analyzed by FACS to assess DNA content. Only the h1Δ-h2Δ SEN1-AIDculture arrested in G2/M (2N). (F) Cell morphology of h1Δ-h2Δ SEN1-AIDtreated with auxin for 4h. DNA was stained with DAPI. Cells presented a large budded morphology typical of G2/M arrested cells.
Cells have an additional class of enzymes to remove hybrids, the RNA-DNA helicases that unwind the RNA. Senataxin (Sen1) is the DNA-RNA helicase in budding yeast (Figure 1A) (Becherel et al., 2013; Groh et al., 2017; Mischo et al., 2011; Skourti-Stathaki et al., 2011). Understanding the impact of Sen1 on hybrid induced-damage has been complicated by the fact that Sen1 forms a complex with Nrd1-Nab3 and together they perform an essential function in transcription termination (Porrua and Libri, 2015).
Cells that lack the RNases-H exhibit a uniformly distributed nuclear staining with an antibody specific to DNA-RNA hybrids (Wahba et al., 2011). This staining suggests that R-loops must form at many sites in every cell, but are quickly removed by the RNases-H. The potential to form hybrids at many sites is further confirmed by genome-wide sequencing of hybrid-prone regions (Chan et al., 2014; El Hage et al., 2014; Wahba et al., 2016). While every cell accumulates R-loops at multiple loci when the RNases-H are inactivated, DNA damage occurs only in a fraction of cells (Amon and Koshland, 2016). This observation suggests that R-loops that accumulate when RNases H are depleted are inefficiently converted to damage. This inefficiency could reflect that every R-loop region has a common low probability of causing DNA damage. Alternatively, only a specific subset of genomic R-loops might be prone to induce damage, generating a subset of fragile sites. The question of whether all hybrids are prone to induce damage has become of increasing interest with the realization that a subset of R-loops perform important functions in gene regulation (Castellano-Pozo et al., 2013; Skourti-Stathaki et al., 2011; Sun et al., 2013). Assessing the damage potential of a given hybrid has been hampered by an inability to map the sites of DNA damage relative to the sites of R-loop formation.
In addition to causing DNA damage, recent evidence suggests that hybrids influence the repair of damaged DNA. Hybrid formation is induced at the site of double strand break (DSB) (Ohle et al., 2016). Blocking the removal of the DSB-induced hybrid impairs repair by homologous recombination, implying that hybrid removal is required for recombination-based repair. Furthermore, yeast cells lacking Topoisomerase-I and the RNases-H resulted in damage in the repetitive ribosomal-DNA locus. This damage appears to activate repair by break-induced replication (BIR) (Amon and Koshland, 2016) and origin-independent replication (Stuckey et al., 2015). These observations lead to many questions. What are the structures of DNA associated with hybrid induced damage that are modulating DNA repair? Are they the same at all damage loci? Do pre-existing hybrids influence repair of neighboring DNA damage?
To address these questions about hybrid-induced damage and hybrid modulation of repair, we exploited our genome-wide map of approximately 800 hybrid-prone loci (Wahba et al., 2016). Using this hybrid map as a foundation, we employed a two-steps strategy to identify hybrid sites with DNA damage. First, we increased the signal for hybrid-induced damage through mutations that elevated the fraction of cells experiencing damage. Second, we developed a genome-wide assay to detect and characterize this signal. The success of our strategy allowed us to reveal an essential function for hybrid removal in cell survival, to identify the subset of hybrid sites prone to damage, and to uncover properties of the DNA associated with hybrid-induced damage that inform on how hybrids modulate DNA repair.
Results
Persistent DNA-RNA hybrids cause multiple DNA damage events
We reasoned that we could increase the levels of hybrid-induced damage by prolonging R-loops persistence. In budding yeast S. cerevisiae, the two RNases-H isoforms are RNase-H1 and RNase-H2, and the essential DNA-RNA helicase is Sen l. We eliminated these activities by deleting the non-essential RNases-H genes, RNH1 and RNH201 (h1Δ-h2Δ) or by generating an auxin-inducible degron allele of SEN1 (SEN1-AID), where addition of auxin to the media drove protein degradation (Figure S1A–C).
To assess the levels of hybrid-induced damage, we measured the number of cells displaying nuclear foci of green fluorescent tagged Rad52 (Rad52-GFP), a DNA repair protein that binds to ssDNA at the sites of damage (Lisby and Rothstein, 2009). We observed that 15–25% of cells had Rad52-GFP foci when either RNases-H (h1Δ-h2Δ) or Sen1 activity (SEN1-AID) was eliminated, a 5-fold increase over wild type (WT) (Figure 1B). However, when Sen1-AID was depleted from cells lacking both RNase-H enzymes (h1Δ-h2Δ SEN1-AIDhenceforth called Sen1-AID triple), virtually all cells became positive for Rad52-GFP foci (Figure 1B). These results suggested that elevating hybrid persistence causes DNA damage in every cell.
Rad52-GFP tends to aggregate and form a single focus in cells, so to estimate the number of DNA damage events per cell, we prepared chromosome spreads and probed them with antibody specific for the endogenous Rad52 (Grubb et al., 2015). The few cells positive for Rad52 foci in WT strain possessed a single focus suggesting the presence of a single DNA damage event; while Sen1-AID triple cells presented an average of 6 Rad52 foci per cell (Figure 3C), suggesting multiple DNA damage sites in every cell experiencing elevated hybrid persistence. We conclude that blocking the hybrid-removal machineries results in DNA damage at multiple loci in virtually every cell.
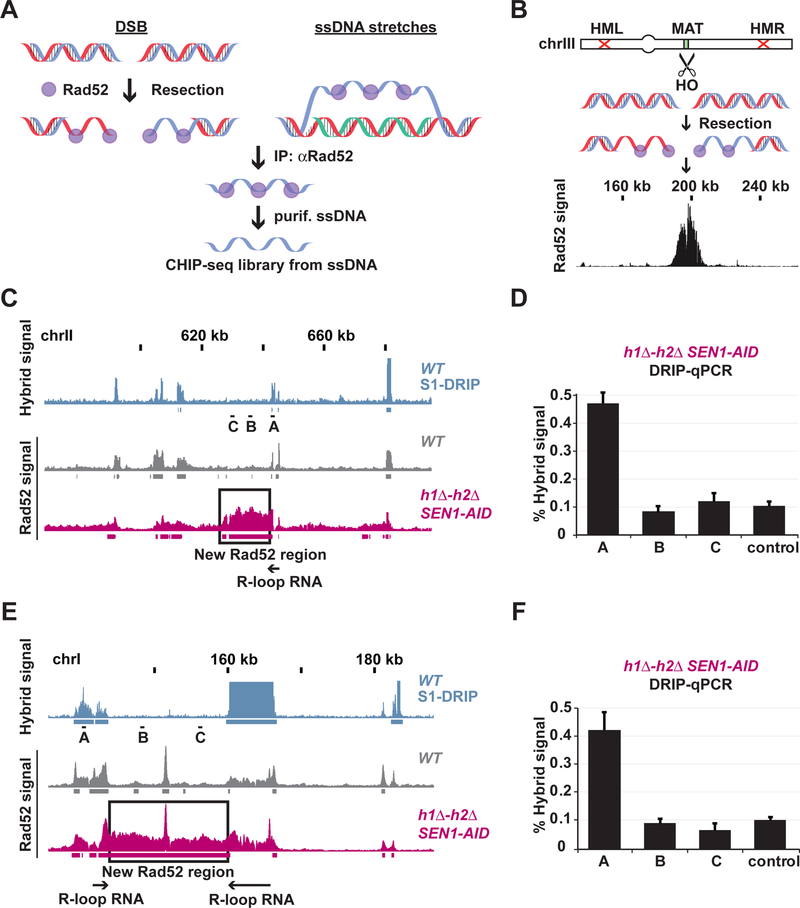
Fig. 3. Rad52-ChIPseq as method to map sites of hybrid-induced DNA damage.
(A) Schematic for Rad52 chromatin immuno-precipitation followed by sequencing (Rad52- ChIPseq) method. (B) Expression of the endonuclease-HO makes a single DSB at the MAT-locus. The DSB is not repaired by recombination since the homologous sequences (HML and HMR) have been deleted. The DSB is resected and bound by Rad52. The black bar-chart shows Rad52-ChIPseq signal that peaks around the break at the MAT locus on chromosome III. (C and E) Rad52-ChIPseq signal from a representative portion of chromosome I (C) and II (E) are presented with bar-charts, and identified regions of enrichment are shown with underlining boxes. Signal coming from hybrids (S1-DRIPseq) in a WT strain is in blue, signal from Rad52 (Rad52-ChIPseq) from WT is in grey and h1Δ-h2Δ SEN1-AID treated with auxin in purple. The new Rad52 regions identified in h1Δ-h2Δ SEN1-AID strain are boxed in black, and the flanking hybrid-RNAs are depicted with black arrows. (D and F) Hybrid signal in the h1Δ-h2Δ SEN1- AID treated with auxin was measured with DRIP-qPCR at the chromosomal locations A, B and C (black dashes in Fig. 2C and E). The new Rad52 regions present in h1Δ-h2Δ SEN1-AID strain are devoid of new hybrids.
Persistent DNA-RNA hybrids result in DNA damage checkpoint activation and cell cycle arrest.
DNA damage can activate the DNA damage checkpoint, resulting in cell cycle arrest to allow the repair of the damage. To assess the DNA damage checkpoint activation, we monitored the phosphorylation of the checkpoint protein, Rad53 (Harrison and Haber, 2006) When we blocked the hybrid removal in Sen1-AID triple cells with auxin, we observed a robust Rad53 phosphorylation (Figure 1D and Figure S1D). The Rad53 phosphorylation in cells treated with auxin was significantly higher than cells treated with hydroxyurea (HU), a common positive control for robust DNA checkpoint activation. Furthermore, the presence of an active DNA damage checkpoint was corroborated by phosphorylation of histone H2A (Figure S1E). These results suggest that hybrid persistency results in high levels of DNA damage and a robust DNA damage checkpoint activation.
Consistent with the activation of the DNA damage checkpoint cells were arrested in G2/M as showed by the fluorescence-activated cell sorting (FACS) profile of Sen1-AID triple cells with a G2/M DNA content (2N) (Figure 1E). Cell morphology of Sen1-AID triple cells stained with DAPI showed a large-budded morphology with the DNA mass trapped in the neck of the dividing cells, typical of G2/M arrested cells (Figure 1F and Figure S1F–G). The SEN1-AID or h1Δ-h2Δ strains did not exhibit cell cycle arrest or Rad53 phosphorylation as expected since they did not exhibit high levels of DNA damage. These results demonstrate that the multiple DNA damage events that are induced by hybrid persistence led to checkpoint activated cell-cycle arrest.
Blocking the DNA-RNA hybrid removal machineries result in irreparable damage and cell death.
The Sen1-AID triple cells with high levels of damage were inviable after transient Sen1-AID depletion (Figure 2A and Figure S2A–B). The cell death was likely due to Sen1 activity in hybrid-removal and not in transcription-termination for two reasons. First, transient depletion of Sen1-AID in the SEN1-AID strain did not impair viability. This strain was defective for termination but could remove hybrids because of the presence of the RNases-H. Second, h1Δ-h2Δ cells transiently depleted of the other two components of the termination complex, Nrd1 (h1Δ-h2Δ NRD1-AID) and Nab3 (h1Δ-h2Δ NAB3 AID), were viable (Figure S2C–D). Thus, transient inactivation of this alternative transcription termination complex in cells lacking RNases-H did not cause cell death. These results show that hybrid removal is essential for cell survival, since a transient block of the three enzymes that remove hybrids generates multiple irreparable damage events leading to cell death.
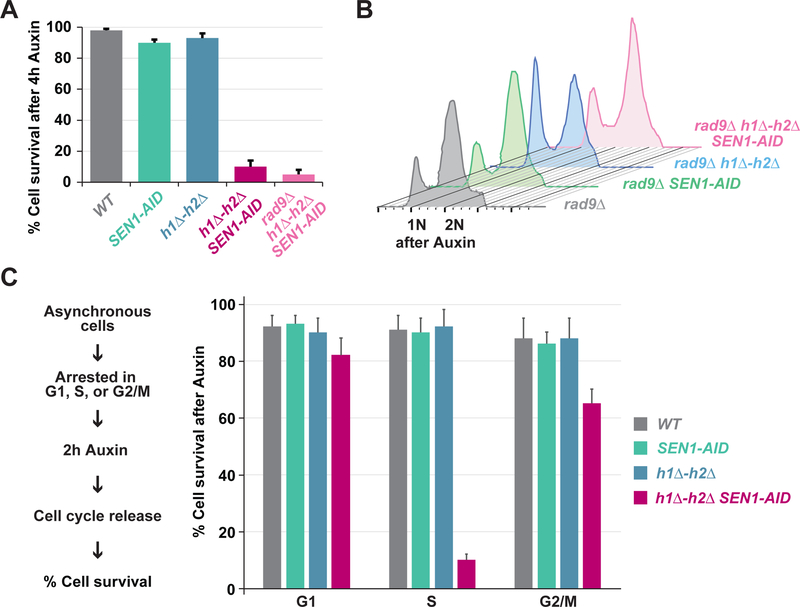
Fig. 2. Blocking R-loop removing enzymes leads to irreparable DNA damage and cell death.
(A) Cell death in cells with persistent hybrids. Asynchronous culture of WT, SEN1-AID, h1Δ-h2Δ, h1Δ-h2Δ SEN1-AID and Rad9Δ h1Δ-h2Δ SEN1-AID were treated for 4h with auxin and then placed on plates lacking auxin to stabilize Sen1-AID expression. Cell survival was measured as colony forming units of cells transiently-depleted versus untreated. Cells lacking the hybrid removing machinery are inviable independent of the DNA damage checkpoint. (B) DNA content in cells with persistent hybrids. Asynchronous culture of Rad9 deletion strain (Rad9Δ), Sen1 with auxin-inducible degron in Rad9 deletion strain (Rad9Δ SEN1-AID), RNase-H1 and RNase-H2 in Rad9 deletion strain (Rad9A h1Δ-h2Δ) or both (Rad9Δ h1Δ-h2Δ SEN1-AID) were treated for 4h with auxin and analyzed by FACS to assess DNA content. Deletion of the DNA damage checkpoint protein Rad9 prevents the cell-cycle arrest observed in cell lacking the hybrid-removing machineries. (C) Asynchronous culture of WT, SEN1-AID, h1Δ-h2Δ, and h1Δ-h2Δ SEN1-AID were synchronized in G1 using alpha-factor, S using HU, or G2/M using nocodazole for 2h. Cultures were then treated with auxin for 2h and then released from the cell-cycle arrest in fresh media without auxin. Cell survival was measured as colony forming units. Cells are inviable when the hybrid-removing machineries are blocked during S-phase.
Cell death could have resulted from an inability to repair hybrid-induced damage or from a constitutive activation of cell-cycle arrest (Shaltiel et al., 2015). To distinguish between the two possibilities, we eliminated the cell cycle arrest by deleting the DNA damage checkpoint gene RAD9 (Harrison and Haber, 2006) in the Sen1-AID triple strain (Rad9Δ h1Δ-h2Δ SEN1-AID). This strain no longer activated Rad53 phosphorylation upon auxin treatment (Figure S2E–F), and no longer arrested in G2/M (Figure 2B and Figure S1F–G). Blocking the DNA damage checkpoint activation by Rad9 deletion did not restore viability in Rad9Δ h1Δ-h2Δ SEN1-AID cells after a transient depletion of Sen1-AID (Figure 2A and Figure S2A–B), suggesting that cell death likely stemmed from the inability to repair the damage. Thus, persistent R-loops induce robust amounts of unrepairable damage leading to cell death.
To determine whether the lethal damage occurred at a particular cell-cycle phase, we depleted Sen1-AID transiently during G1 S or G2/M. First, we arrested cells in G1, S or G2/M with the appropriate reagents and then depleted Sen1-AID with auxin. We washed the cells to remove the arresting reagent and auxin, thereby allowing the cells to reenter the cell cycle and to recover Sen1-AID expression. Cells were plated to monitor cell survival (Figure 2C). Cell death occurred only in Sen1-AID triple cells after transient Sen1-AID depletion during S-phase but not G1 or G2/M (Figure 2C). No lethality was observed during transient cell cycle arrest of the SEN1-AID or h1Δ-h2Δ strains with reduced DNA damage. Thus, the lethality of the Sen1-AID triple strain correlated with the induction of DNA damage during S-phase. This result is consistent with the formation of lethal damage by collision of the S-phase replication-machinery with persistent hybrids (Alzu et al., 2012; Gan et al., 2011; Hamperl et al., 2017; Lang et al., 2017; Mischo et al., 2011).
Rad52-ChIPseq as genome-wide method to map sites of irreparable DNA damage.
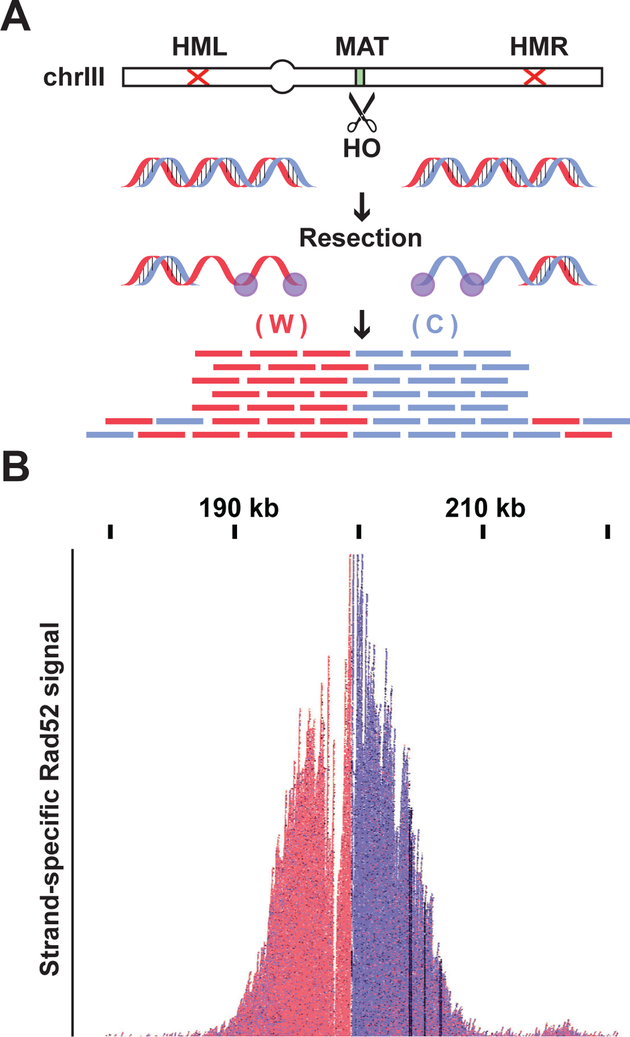
The abundance of irreparable DNA damage in every Sen1-AID triple cell, provided a robust signal to map the loci of hybrid-induced damage. We took advantage of the fact that these damaged sites would recruit DNA-repair proteins. Rad52 was a good candidate (Zhou et al., 2013), since it formed foci in Sen1-AID triple strain, presumably due to its binding to ssDNA associated with hybrid-induced DNA damage. Therefore, we developed a method for chromatin immuno-precipitation (ChIP) of Rad52-bound to ssDNA that was compatible with next- generation sequencing (Rad52-ChIPseq, see STAR Methods and Figure 3A). Briefly, we cross-linked the proteins bound to DNA, fragmented and isolated the Rad52-ssDNA using an antibody specific for the endogenous Rad52. We used the purified ssDNA to build libraries compatible with next generation sequencing technology in place of the usual dsDNA substrate used in ChIP. To validate this method we used a strain where we induced the expression of the endonuclease HO to make a single double-strand break (DSB) at the MAT locus on chromosome III (Figure 3B) (Lee et al., 1998). The DSB is not repairable since the homologous sequences HML and HMR have been deleted. Upon DSB induction, we observed a robust Rad52 signal centered around the MAT locus (Figure 3B and Figure S3A), confirming that Rad52-ChIPseq could be used to identify sites of irreparable damage.
Rad52-ChIPseq to map sites of hybrid-induced DNA damage.
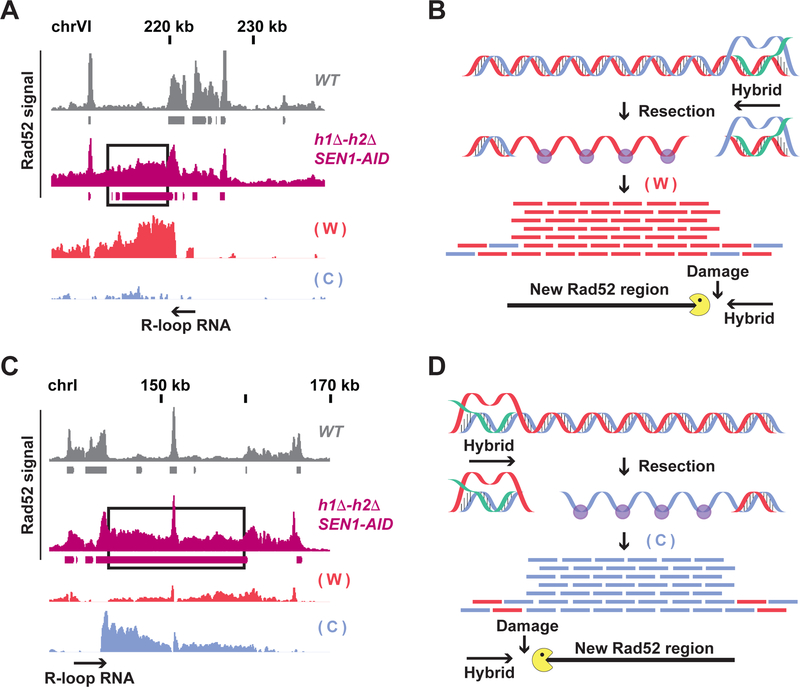
We performed Rad52-ChIPseq in the strains defective for hybrid-removal to identify the genome-wide location of hybrid-induced damage. We expected that the irreparable damage in the Sen1-AID triple strain that was absent from WT, SEN1-AID and h1Δ-h2Δ strains would generate a unique Rad52-ChIPseq signature. Indeed, we observed about 100 new Rad52 regions in the Sen1-AID triple strain, absent from the other strains. Each new region was immediately adjacent to a previously mapped hybrid (Figure 3C and Figure S3B) or sandwiched between two hybrids (Figure 3E and Figure S3B) consistent with induction of an irreparable damage by a flanking R-loop. Since we observed a baseline of Rad52 signal at hybrid sites in all strains, we had to test whether the new Rad52 signal in the Sen1-AID triple strain was not due to new regions of hybrid formation. The new Rad52 regions were devoid of hybrids by using DNA- RNA immuno-precipitation followed by qPCR (DRIP-qPCR) (Figure 3D and Figure 3F), indicating that they were indeed caused by hybrid-induced damage. The new 100 regions of Rad52 damage in the Sen1-AID triple were around 10–20 kilobases long and each region is proximal to at least one of the 800 regions prone to hybrid formation. These results suggest that only a subset of hybrids is prone to induce damage, and therefore, proximal chromosomal features must play a key role in hybrid-induced damage.
Rad52-ChIPseq identifies rDNA as region of DNA damage in h1Δ-h2Δ TOP1-AID.
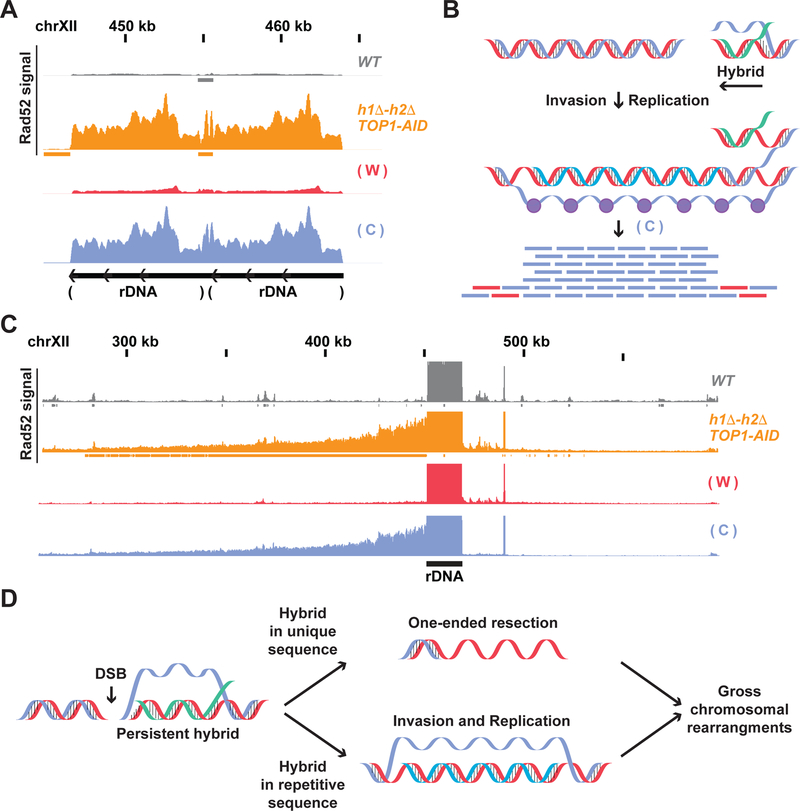
The success of our Rad52-ChIPseq analysis with the Sen1-AID triple prompted us to examine another strain that accumulates lethal damage induced by R-loops. We and others had shown that cells depleted of Topoisomerase-I in combination with deletion of the RNases-H (h1Δ-h2Δ TOP1-AID, henceforth Top1-AID triple), also experienced high levels of irreparable damage leading to cell death (Amon and Koshland, 2016; El Hage et al., 2010; French et al., 2011; Stuckey et al., 2015). Rad52-repair foci were localized mainly in the nucleolus, the compartment containing the ribosomal DNA (rDNA), a hybrid-prone locus constituted of around 150 rDNA tandem-repeats. Indeed, Rad52-ChIPseq of Top1-AID triple upon auxin treatment revealed one major Rad52 region located at the rDNA, whose signal was around 20-fold higher compared to wild type, Sen1-AID or h1Δ-h2Δ strains (Figure 4A and Figure S4). These results corroborated hybrid-induced damage in the rDNA in the Top1 triple strain seen previously.
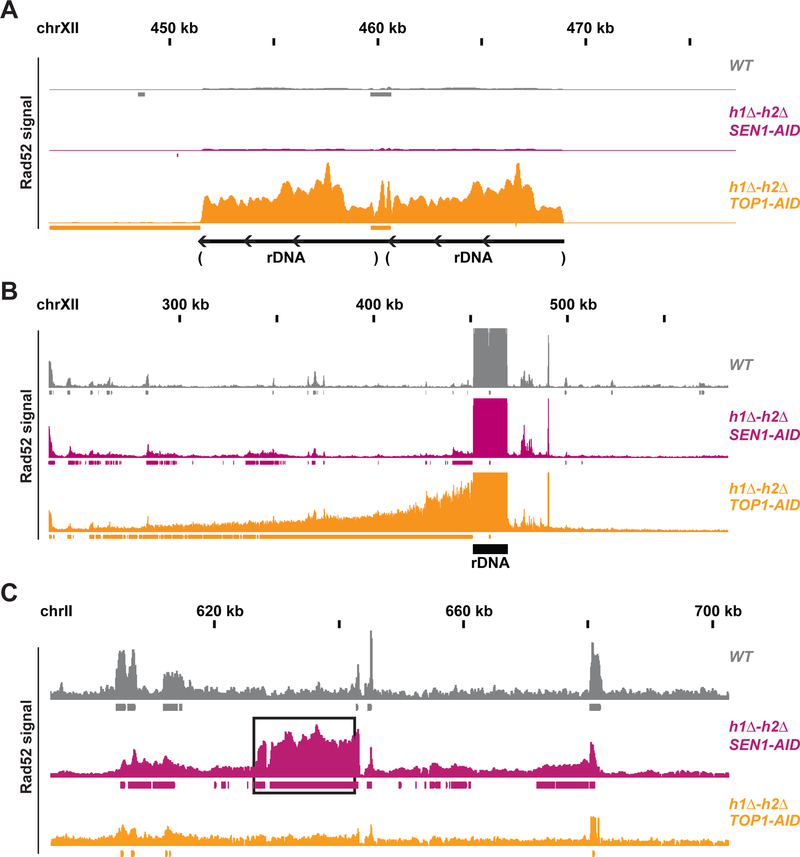
Fig. 4. Rad52-ChIPseq identifies rDNA as region of DNA damage in h1Δ-h2Δ TOP1-AID.
(A) Rad52-ChIPseq signal from WT(grey), h1Δ-h2Δ SEN1-AID (purple) and h1Δ-h2Δ TOP1-AID (orange) on the ribosomal-DNA locus (rDNA), after auxin treatment to deplete Sen1-AID and Top1-AID. Just two repeats of the rDNA are present in the annotated genome, while cells possess around 150 copies of the rDNA. Upon Top1 depletion, h1Δ-h2Δ TOP1-AID cells present a 20-fold increase in Rad52 signal on the rDNA region compared to other strains. (B) Rad52-ChIPseq signal from a larger portion of chromosome XII containing the rDNA locus. Rad52 signal from strains as in (A). The signal on the repetitive rDNA locus is around 75-fold higher than the one from unique regions; because the reference genome used in the alignment has just 2 rDNA copies while cells possess around 150 copies. By lowering the maximum signal by around 75 folds and showing a larger portion of the chromosome XII with the unique sequence flanking the rDNA, we detected a new Rad52 region large 150kb on the centromere-side of the rDNA, but not in the telomere side. H1Δ-h2Δ TOP1-AID presents a new Rad52 region in the rDNA and in the 150kb on the centromere-flanking unique region. (C) Rad52 signal from strains as in (A). A representative new Rad52 region identified in h1Δ-h2Δ SEN1-AID (black box) is not present in h1Δ-h2Δ TOP1-AID cells.
The annotated genome used in the sequencing alignment contains just two copies of the rDNA, while cells possess around 150 copies. Therefore, the signal registered at the rDNA locus was 75-fold higher than any unique sequence in the genome. After lowering the signal 75-fold and zooming out from the rDNA locus we observed one additional Rad52 region in the genome, starting at the centromere proximal side of the rDNA and extending around 150kb into the unique flanking sequence (Figure 4B and Figure S4B). We conclude that Top1-AID triple strain accumulated DNA damage encompassing the rDNA locus and around 150 kb of unique flanking sequence on the centromere proximal side.
A comparison of the Rad52-ChIPseq of the Sen1-AID triple and Top1-AID triple strains was informative. The new Rad52 regions found in Sen1-AID triple were not present in the Top1-AID triple strain. Conversely, the new Rad52 signal coming from the rDNA and flanking region in the Top1-AID triple was not present in the Sen1-AID triple strain (Figure 4C). From these results, we draw two conclusions. Rad52-ChIPseq can successfully identify different regions of hybrid-induced damage in different strains, and Sen1 and Top1 regulate R-loops homeostasis at distinct loci to prevent genomic instability.
Rad52-ChIPseq preserves the information on Rad52 strand-specific binding on irreparable DSB.
We then investigated the nature of the hybrid-induced damage and the possible reasons leading to block of repair (Ohle et al., 2016) and cell death. The Rad52 regions in both Sen1-AID triple and Top1-AID triple strains had several common features: these regions were particularly large for the yeast genome, 10’s of kb in Sen1-AID triple and 100’s of kb in Top1-AID triple, and the new Rad52 region was always adjacent to the 3’-end of the hybrid-RNA. We reasoned that identifying the strand(s) bound by Rad52 in the new Rad52 regions might provide key information on the initial damage event and/or its repair pathway.
To confirm that strand-specific information was correctly preserved by Rad52-ChIPseq, we examined the Rad52 binding adjacent to the HO-induced DSB. After the induction of a break, the 5’ strand is progressively resected on both ends leading to Rad52 binding to ssDNA Watson (in red) on one side, and Crick (in blue) on the other (Figure 5A). Indeed, we detected a striking separation of the Watson-Crick signal precisely around the DSB site. Thus, our method successfully preserves the information on the strand-specificity of Rad52 binding.
Fig. 5. Rad52-ChIPseq preserves strand-specific Rad52 signal from an irreparable DSB.
(A) In the DBS model system strain (Fig. 3B) induction of HO nuclease makes an irreparable DSB at the MAT locus, both ends are then resected 5′-3′ leaving the ssDNA Watson (W) strand on one side and the Crick (C) on the other side to be coated by Rad52. (B) Rad52-ChIPseq signal from the MAT locus after HO induction. Signal coming from the W strand is shown in red while C strand signal is in blue. Rad52-ChIPseq correctly preserve the strand-specific information on Rad52 binding around a DSB.
The strand-specific signal of Rad52-ChIPseq identifies the nature of hybrid-induced DNA damage.
We expected the hybrid to induce a “classical” DSB at a precise location within the new Rad52 region, resulting in a Watson-Crick signal separated around the DSB (Figure S5A). The DSB might occur at different locations within the new Rad52 region, resulting in a mixed Watson- Crick signal (Figure S5B). Surprisingly, we observed a striking asymmetry in the strand signal coming from the new Rad52 regions in both the Sen1-AID triple and Top1-AID triple strains.
In Sen1-AID triple strain, the signal from the new Rad52 regions had two striking features. First, it originated almost exclusively from one strand (Figure 6A, 6C and Figure S6A–B). Furthermore, the peak of new Rad52 signal was located next to the 3’ end of the hybrid-RNA. When the hybrid-RNA annealed to the Watson strand, the Rad52 signal from the damaged DNA was located left to the hybrid and originated from the Watson strand (Figure 6B). Conversely when the hybrid-RNA annealed to the Crick strand, the signal was located on the right side and is Crick biased (Figure 6D) These two features were compatible with a model where hybrids induced a DSB right next to the 3’ of the hybrid-RNA, due to head-to-head collision between the replication machinery with the 3’ of the hybrid-RNA. The DSB was progressively resected on one broken end, generating the Rad52 signal from one strand. The resection of the other broken end was blocked by the persistent hybrid preventing a signal from the opposite strand on the other broken end (Figure 6B and D)
Fig. 6. The strand-specific signal of Rad52-ChIPseq identifies signatures of hybrid-induced DNA damage.
(A and C) Rad52-ChIPseq signal from WT (gray) and h1Δ-h2Δ SEN1-AID (purple) strains treated with auxin to deplete Sen1-AID on two representatives new Rad52 regions (black boxes). The strand-specific signal of the new Rad52 region is showed with a red bar-chart for the Watson strand, and blue for the Crick strand. The hybrid-RNA is indicated with a black arrow. The signal from the new Rad52 region is originates mainly from only the Watson strand (A) or Crick strand (C). (B and D) Model for hybrid-induced damage in h1Δ-h2Δ SEN1-AID. When Sen1-AID is depleted with auxin in h1Δ-h2Δ SEN1-AID strain, R-loop persistency induces a DSB near the 3’of the hybrid-RNA. The DSB-end with the hybrid is protected from resection, while the other is progressively resected. The direction of the hybrid-RNA determines which strand is coated by Rad52. Rad52 coats the Watson strand (forming a new Rad52 regions) when the damage is occurring left of the hybrid (Fig. 6B); or the Crick strand if the damage is on the right side (Fig. 6D).
This model made several predictions. For a head-on collision between the replication fork and the 3’ end of the hybrid to generate damage, the origin of replication 3’ to the hybrid (head-on, Figure S7A) should be closer to the hybrid than the origin of replication 5’ to the hybrid (co- directional Figure S7A). This prediction was fulfilled for 75% of new Rad52 regions. In fact, analyzing the distances between the hybrids flanking the new Rad52 regions and the neighboring replication origins showed that the head-on origins are significantly closer than the co-directional ones (Figure S7B, first and second bar). There was no statistical difference in the distance distribution between head-on and co-directional forks for all hybrid-prone regions (Figure S7B, third and fourth bar). Therefore, the bias was characteristic of hybrids with new Rad52 regions. We assessed whether having a head-on replication origin significantly closer than a co-directional one was sufficient to result in damage. The distance distribution in the hybrids that did not form damage and had a head-on fork closer than the co-directional one (around 50% of the no-damage hybrids) (Figure S7B, fifth and sixth bar) was similar to the distribution in hybrids causing damage (Figure S7B, first and second bar). This result suggests that having a head-on replication fork closer to the hybrid than a co-directional one is not sufficient to result in DNA damage. Therefore, other unidentified features must contribute to the fragility of the hybrid.
If the Rad52 region was generated by resection, the size of the region would be determined by the rate of resection. Indeed, the resection speed at an irreparable DSB (~4.5kb/h (Vaze et al., 2002) and Figure 5B), can generate the observed 10–20 kb Rad52 regions after 4 hours of Sen1-AID inactivation. Together these results are consistent with a model of collision induced DSB followed by one-ended resection.
In the Top1-AID triple strain the Rad52 signal in the rDNA was bound to the Crick strand (Figure 7A), and this bias persisted for around 150kb in the centromere proximal unique sequence (Figure 7C). This rDNA strand bias was not present in any other strain analyzed (Figure S7A–B). The Crick bias in Top1-AID triple is not compatible with the uni-directional resection-model we proposed for Sen1-AID triple. The resection model would predict a Watson bias since the hybrid RNA from ribosomal RNA should anneal to the Watson strand. Furthermore, the 100’s of kb of Crick Rad52 signal was incompatible with the speed of resection. Thus Top1-AID triple strain must generate the Crick strand bias by another mechanism.
Fig. 7. Hybrids persistence induces different types of aberrant DNA structures.
(A) Rad52-ChIPseq signal from WT (gray) and h1Δ-h2Δ TOP1-AID (orange) strains treated with auxin to deplete Top1-AID on the rDNA locus. The strand signal in the h1Δ-h2Δ TOP1-AID strain is showed with a red bar-chart for the Watson strand, and blue for the Crick strand. The signal from the rDNA is originates mainly from the Crick strand. (B) Model for hybrid-induced damage in h1Δ-h2Δ TOP1-AID. When Top1 is depleted with auxin in the hΔ-h2Δ TOP1-AID strain, a DSB is induced at the 3’of the hybrid-RNA. The ssDNA from the R-loop (or the hybrid-RNA) triggers origin-independent DNA replication. The aberrant replication fork replicates only the Watson strand leaving a single stranded Crick coated by Rad52. (C) Rad52 signal from strains as in (A) from the unique region next to the rDNA. The signal from the unique sequence on the centromere-side of the rDNA locus is originated mainly from the Crick strand. (D) Model for hybrid-induced DNA damage. Hybrid persistence drives different aberrant DNA structure formation: persistent hybrids at unique sequences result in tens of kilobases of one-ended resected DNA, while hybrids at highly repetitive sequence drive origin-independent replication that proceeds for hundreds of kilobases. These aberrant ssDNA structures can be precursors of gross chromosomal rearrangements like deletions and duplications.
One possible mechanism is that origin-independent replication (Stuckey et al., 2015) of the Watson strand would produce a D-loop with a long Crick strand coated by Rad52 (Fig. 7B). This aberrant replication fork could be triggered by the hybrid-RNA functioning as RNA-primer, or by invasion of the ssDNA present in the R-loop after a DSB of an adjacent rDNA repeat. Hybrids from one rDNA repeat would replicate neighboring repeats generating the rDNA Crick- specific signal, and then proceed into the adjacent unique sequence. Consistent with this model, the known replication rate (Anand et al., 2013) could generate within 4 hours the observed large (150kb) Rad52 region in the unique DNA. We propose that hybrids can also modulate repair to induce uni-directional DNA replication of one DNA strand.
Discussion
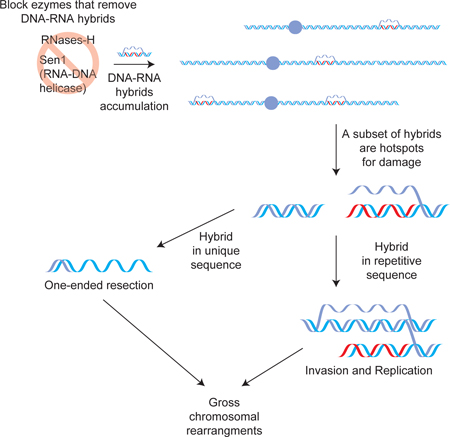
Here we engineered a yeast strain that lacks the RNA-DNA helicase Sen1, and the two RNases- H (Sen1-AID triple), thereby inhibiting two major pathways of hybrid removal. We provide multiple lines of evidence that Sen1-AID triple strain accumulates irreparable DNA damage at multiple independent loci during S phase in every cell. We had previously observed hybrid- induced irreparable damage in a strain lacking Topoisomearse-1 and the two RNases-H (Top1- AID triple). We exploit Top1-AID triple and Sen1-AID triple strains and a modified Rad52-ChIPseq to identify and characterize the sites of hybrid-induced damage genome wide.
We previously demonstrated that hybrid formation occurs frequently at distinct classes of loci characterized by high level of transcription including highly transcribed ORF, tRNA, ribosomal- DNA, transposons elements, and LTRs (Wahba et al., 2016). In the Sen1-AID triple strain a subset of hybrids in each of these classes give rise to the 100 sites of DNA damage. This partial correlation between hybrid formation and damage suggests that the damage is not driven solely by hybrid features. Rather, the chromosomal features proximal to some hybrid must contribute to their propensity to cause damage.
Another potential feature for hybrid-induced damage is the head-to-head collision of the replication fork with hybrids (Alzu et al., 2012; Brambati et al., 2015; Castellano-Pozo et al., 2012; Gan et al., 2011; Hamperl et al., 2017; Helmrich et al., 2011; Lang et al., 2017; Tuduri et al., 2009). Indeed, we observed that most hybrid induced damage occurred at sites where head- on replication origins were closer than co-directional ones. However, this feature is not sufficient to cause damage, as many of the 700 hybrids without irreparable damage also had closer head-on replication origin. Furthermore, this feature was not present in 25% of the hybrids that induced damage. In these cases, the head-on collision might still occur because of delayed firing of proximal co-directional origin. Alternatively, mechanisms other than head-on collision might contribute to hybrid-induced damage. Our genome-wide map of damaged loci provides a critical foundation for future studies to identify the additional features and possible mechanisms that make some hybrids “fragile”.
The fragility of the 700 sites may be underestimated as our method does not detect hybrid induced damage that is repaired by non-homologous end joining (NHEJ). However, NHEJ is not particularly efficient in yeast and we could not detect any NHEJ hotspots by NHEJ specific ChIP-seq (data not shown). Thus, most hybrids are likely benign for DNA damage, providing an explanation for why many organisms can use hybrids as positive regulators of gene expression without affecting their genome stability.
Features of Rad52-ChIPseq signal also provide important insights into the mechanism of hybrid-induced damage. One prominent feature in the Sen1-AID triple strain is that the peak of Rad52 signal in each damaged region is usually proximal to the 3’ end of a hybrid (this study). This pattern is consistent with a DNA break occurring at the 3’ end of the persistent hybrid-RNA, supporting a mechanism of DNA break formation by head-to head collision between the replication machinery and the hybrid. Our genome-wide data strengthen the model by providing many in vivo examples of hybrid-induced damage at the 3’ end of hybrids at endogenous loci. We further support the model by showing that transient persistence of hybrids in S-phase greatly increases the formation of irreparable damage, as expected if the damage were occurring by collisions with replication machinery.
A second feature of the Rad52-ChIPseq signal in the damage regions of Sen1-AID triple is that it results from Rad52 mainly binding to one of the two DNA strands, dictated by the direction of the hybrid RNA. We show that this pattern is different from a classical DSB, where 5’ to 3’ resection of both sides of the break lead to Rad52 binding to the Watson strand on one side and the Crick on the other. A simple explanation for the pattern in the Sen1-AID triple strain is that resection from one end occurs normally, but resection from the other side is blocked by the hybrid. We support this model by showing that the size of the damaged regions is consistent with the known rate of resection. Our results provide molecular evidence to support the notion that hybrids can block resection (Ohle et al., 2016). The fact that we observe no detectable ssDNA from one side of the break even after 4 hours suggests that persistent hybrids are potent blocks of resection. We note that even under more physiological condition where hybrids do not persist long enough to completely inhibit resection, they could alter it and affect the final DNA repair outcome.
A third feature of Rad52-ChIPseq signal from hybrid-induced damage comes from the differences in damaged regions in the Sen1-AID triple and Top1-AID triple strains. In the Top1-AID triple strain, Rad52 signal in the damaged region also results from Rad52 binding to only one the two DNA strands. However, the bound strand was consistent with unidirectional replication of one strand rather than resection. Furthermore, the damaged regions in the Top1-AID triple strain were at the rDNA and rDNA-proximal unique sequences, rather than the 100 unique sites in the Sen1-AID triple strain. This result opens up possibility that defects in other enzymes acting on DNA homeostasis may synergize with R-loops to alter the position of DNA damage and its repair.
The damage in the rDNA of the Top1-AID triple strain likely arises by depletion of Top1 function rather than persistent hybrids for two reasons. First, the absence of damage in the rDNA in the Sen1-AID triple strain suggests that persistent rDNA hybrids are like the 700 other hybrid loci; they lack the chromosomal features to induce hybrid damage. Second, Top1 mutants alone have been shown to cause DNA damage in the rDNA and to induce rDNA rearrangements (Andersen et al., 2015; El Hage et al., 2010; French et al., 2011). Since the damage in the Top1-AID triple is irreparable, the repair of Top1 induced damage in the rDNA must be impaired by the partial stabilization of rDNA-hybrids. These results suggest that when DNA damage occurs next to a site prone to R-loop formation, persistency of the hybrid can impair the DNA repair of the damage. Furthermore, these results open the possibility that the repair of DNA damage by other endogenous or exogenous mutagens may also be influenced by R-loops.
One explanation for the differences in the repair pathway activated by hybrid-induced damage in the two strains, resection versus re-replication, is the location of the damage. We favor a model in which when the damage is located next to unique sequence, the lack of a close homologous sequence triggers extensive resection on one side of the break, while resection is blocked on the other side by the hybrid (Figure 7D top). But if the damage occurs in highly repetitive sequence, like the rDNA locus, the broken end can invade another rDNA repeat activating a unidirectional replication of one strand (Figure 7D bottom). These large regions of resected DNA could be a powerful precursor of genomic deletions, while re-replicated DNA could lead to genomic duplications, both gross chromosomal rearrangements associated with R-loops and key signatures of genomic instability observed in cancers.
The hybrid removal machineries are thought to be conserved from bacteria to human as an essential barrier to the threat hybrids pose to genomic instability. By pushing the biological system to an extreme situation when all the enzymes that remove hybrids are blocked, we discovered that persistent hybrids efficiently generate irreparable DNA damage in every cell, leading to cell death. Our results reveal that these machineries not only prevent genomic instability, but play an essential function for cell survival, providing an even stronger selective pressure for their conservation.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal Rad52 | D. Bishop lab | N/A |
| Rabbit polyclonal Rad53 | Abeam | ab104232 |
| Rabbit polyclonal H2A phospho serl29 | Abeam | abl5083 |
| Mouse monoclonal V5-tag | Thermo fisher | R960–25 |
| HRP goat anti-mouse | Biorad | 1706516 |
| HRP goat anti-rabbit | Biorad | 1706515 |
| Mouse monoclonal S9.6 DNA/RNA hybrid | S9.6 (ATCC HB-8730) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Hydroxyurea | Sigma | H8627 |
| Nocodazole | Sigma | Ml 404 |
| Alpha-factor | Sigma | T6901 |
| Auxin (3-Indoleacetic acid) | Sigma | 13750 |
| SYBR Green DNA I | Thermo fisher | S7563 |
| RNase-A | Thermo fisher | EN0531 |
| Proteinase K | Roche | 03115828001 |
| AccI | NEB | R0161S |
| EcoRV | NEB | R3195S |
| Ncol | NEB | R3193S |
| Haelll | NEB | R0108S |
| BsrGI | NEB | R0108S |
| Critical Commercial Assays | ||
| Accel-NGS IS Plus DNA Library Kit (ssDNA ChIP library) | SWIFT Biosciences | 10024 |
| Deposited Data | ||
| Rad52-ChIPseq | This paper | GSE110575 |
| Experimental Models: Organisms/Strains | ||
| Strains list | This work | See Table S1 |
| Oligonucleotides | ||
| qPCR oligos | This work | See Table S2 |
| Software and Algorithms | ||
| Bowtie2 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | N/A |
| MACS | http://liulab.dfci.harvard.edu/MACS/ | N/A |
| SamTools | http://samtools.sourceforge.net/ | N/A |
| BedTools | http://bedtools.readthedocs.io/en/latest/ | N/A |
| FlowJo | https://www.flowjo.com/ | N/A |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to Lead Contact Douglas Koshland: koshland@berkeley.edu
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Growth condition
Yeast strains used in this study are the 288c background, and their genotypes are listed in Table S1. Yeast strains were grown using yeast extract/peptone/dextrose (YPD) media at 23°C. For experiments using the AID system, a 1 M stock of 3-indoleacetic acid (Sigma-Aldrich, St. Louis, MO) was made in dimethyl sulfoxide and added to plates or liquid cultures at a final concentration of 500 μM (unless different concentrations are indicated), with cooling agar used in plates to ~55°C before addition of auxin to each batch.
METHOD DETAILS
Dilution plating assays
Cells were grown to saturation in YPD medium at 23°C, diluted to OD66O 1.0 using YPD, and then plated in 10-fold serial dilutions. Cells were incubated on plates at 23 °C.
Microscopy
Images were acquired with an Axioplan2 microscope (100× objective, numerical aperture [NA] 1.40; Zeiss, Thornwood, NY) equipped with a Quantix charge-coupled device camera (Photometrics, Tucson, AZ).
Chromosome spreads
Chromosome spreads were performed as previously published (Grubb et al., 2015).
Western blotting
Primary antibodies used were a mouse monoclonal anti-V5 used at a 1:5000 dilution (Invitrogen, Carlsbad, CA), mouse monoclonal anti-Tub1 used at 1:20,000 dilution, Anti-Rad53 antibody (ab104232, Abcam) at 1:2000 dilution, Anti-Histone H2A (phospho S129) antibody - ChIP Grade (ab15083, Abcam) at 1:1000 dilution. Secondary antibody used were an HRP-conjugated goat anti-mouse or rabbit at 1:25,000 (BioRad, Hercules, CA).
FACS
Fixed cells were washed twice in 50 mM sodium citrate (pH 7.2), then treated with RNase A (50 mM sodium citrate [pH 7.2]; 0.25 mg/ml RNase A; 1% Tween-20 [v/v]) overnight at 37°C. Proteinase K was then added to a final concentration of 0.2 mg/ml and samples were incubated at 50°C for 2 hr. Samples were sonicated for 30s or until cells were adequately disaggregated. SYBR Green DNA I dye (Life Technologies, Carlsbad, CA) was then added at 1:20,000 dilution and samples were run on a Guava easyCyte flow cytometer (Millipore, Billerica, MA). 20,000 events were captured for each time point. Quantification was performed using FlowJo analysis software.
Growth curve
0.1 OD of the indicated strain culture were seeded in a 96well plate with the indicated amount of Auxin. Growth was measured for 3 days using a TECAN Spark.
Survival rate
For survival rate after transient AID depletion experiments, cells of the indicated genotype were treated with 500 μM Auxin for the indicated time, washed counted and plated on plate with fresh media. Colony-forming units were counted after 3 days. For colony formation rates on Auxin plates, cells from exponentially growing culture were counted and plated on agar-plates containing 500 μM auxin for the indicated time and the scored for colony-forming units.
Survival rate of arrested cells
Cells of the indicated genotype from exponentially growing culture were treated with hydroxyurea (HU) 50mM, nocodazole 15 μl/ml or alpha-factor 10−8 M for 2h at 23°C. Auxin was added at 500 μM to deplete Senl-AID for 2h. Cells were collected, washed and plated on fresh plates without drugs. Colony-forming units were scored and normalized to no-Auxin culture.
RAD52-ChIPseq protocol
1×1010 cells of exponentially growing cells (in 0.5 L of YEP+ 2% glucose) were fixed with paraformaldehyde (final 1%) for 1h. Glycine was added to stop the reaction at 0.125M for 10’. Cells were collected by centrifugation and washed once with PBS 1X buffer, once with TE 1X buffer and stored at -80°C. After cell lysis, chromatin was sheared 20 times for 45 s each (settings at duty cycle, 20%; intensity, 10; cycles/burst, 200; 30 s of rest between cycles) using a Covaris S2 (Covaris, Woburn, MA). Immunoprecipitation of DNA-protein was performed using anti-Rad52 (gift from Doug Bishop). 20ug of anti-Rad52 antibody was pre-bound to 160ul of magnetic Protein A Dynabeads (Thermo Fisher) for 1 hr at 4°C. Sonicated samples in final 1X FA (1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 50 mM HEPES, 150 mM NaCl, 1 mM EDTA) was incubated with the anti-Rad52—protein A magnetic beads for 2 hrs at 4°C. Beads were then washed and the DNA eluted according to standard ChIP protocols: 5 successive washes with 1X FA, 1X FA with 0.5M NaCl, Lithium chloride detergent (0.25M LiCl, 0.5% NP- 40, 0.5% Sodium deoxycholate, 1mM EDTA, and 10mM Tris-HCl), and finally twice with TE buffer. DNA was then eluted from the beads with 300ul of Elution buffer (1% SDS, 0.1M NAHCO3). To ensure antibody is fully removed samples are treated with 1ul proteinase K (20mg/ml) for 1 hour at 42° while shaking at 1000 rpm, and the ssDNA was then isolated using Qiagen PCR purification kit (28104 Qiagen) and eluted in 20ul low EDTA TE. The library was prepared using the Accel-NGS 1S Plus DNA Library Kit (Swift Bioscience) following the manufacturer protocol. Libraries were sequenced using Illumina Hiseq 4000. The sequencing files were aligned to the SacCer3 yeast genome using Bowtie2 tool and identified regions of enrichment were called using MACS, with default settings.
DRIP-qPCR
DRIP experiments were performed as described previously (Wahba et al., 2016). Briefly, 3×l010 cells of exponentially growing cells (in 1.2 L of YEP+ 2% glucose) were pelleted at 5500 rpm in a Sorvall RC-5B centrifuge, and pellets were either frozen at -80°C or used directly for genomic DNA extraction. DNA extractions were done with Qiagen tip 500/G following standard protocol, with three exceptions: 30ul of 100T 20mg/ml zymolyase was used for spheroplasting, RNase A was not added to the G2 buffer, and only 200ul of Proteinase K was used in the deproteinization step. DNA is resuspended in 1.5ml nuclease-free TE buffer and nutated overnight at 4°C. 3×1010 cells should yield ~200– 300ug of DNA, all of which is used for an IP. Note that despite the omission of RNase A treatment, no RNA contamination is detectable in the genomic preps as assayed by ethidium gel and 260/280 ratio. Then 100 μg of genomic DNA was digested overnight at 37 °C with AccI, EcoRV, Ncol, Haelll and BsrGI. For the IP, 35ug of S9.6 antibody was pre-bound to 160ul of magnetic Protein A Dynabeads (Thermo Fisher) for 1 hr at 4°C. Digested DNA in final IX FA (1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 50 mM HEPES, 150 mM NaCl, 1 mM EDTA) was incubated with the S9.6—protein A magnetic beads for 2 hrs at 4°C. Beads were then washed and the DNA eluted according to standard ChIP protocols: 5 successive washes with 1X FA, 1X FA with 0.5M NaCl, Lithium chloride detergent (0.25M LiCl, 0.5% NP-40, 0.5% Sodium deoxycholate, 1mM EDTA, and 10mM Tris-HCl), and finally twice with TE buffer. DNA was then eluted from the beads with 300ul of Elution buffer (1% SDS, 0.1M NAHCO3). To ensure antibody is fully removed samples are treated with 1ul proteinase K (20mg/ml) for 1 hour at 42° while shaking at 1000 rpm, extracted with phenol:chloroform, and ethanol precipitated. Final DNA pellet was resuspended in 50ul TE buffer. The percentage of the hybrid signal was quantified using qPCR on DNA from immunoprecipitation and total input with the DyNAmo HS SYBR Green qPCR kit (Thermo Scientific). Oligos used are listed in the table S2.
Supplementary Material
Highlights.
R-loop removal is essential since persistent R-loops cause lethal DNA damage
Only a subset of persistent R-loops induces unrepairable DNA damage
Persistent hybrids block proper DNA repair generating large regions of ssDNA
These damaged regions are potential precursors to gross chromosomal rearrangements
Acknowledgments:
We thank Barbara Meyer, James Haber, Don Rio, Elcin Unal, Silvia Ramundo, Lamia Wahba and Rebecca Lamothe for comments and proofreading of the manuscript. We thank Douglas Bishop for the Rad52 antibody, James Haber for Gal-HO strain and Frederick J. Tan for helping in the bioinformatics analysis. This work was supported by a grant from the NIH (1R35GM118189–01 to D.K.) and Swiss National Science Foundation Fellowships (P300P3_154624 and P300PA_167654 to L.C.). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. The data generated are available at NCBI’s Sequence Read Archive (SRA) under accession number GSE110575.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
References and Notes:
- Aguilera A, and Gomez-Gonzalez B (2017). DNA-RNA hybrids: the risks of DNA breakage during transcription. Nature structural & molecular biology 24, 439–443. [DOI] [PubMed] [Google Scholar]
- Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, et al. (2012). Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 151, 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon JD, and Koshland D (2016). RNase H enables efficient repair of R-loop induced DNA damage. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand RP, Lovett ST, and Haber JE (2013). Break-induced DNA replication. Cold Spring Harbor perspectives in biology 5, a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Sloan RS, Petes TD, and Jinks-Robertson S (2015). Genome-destabilizing effects associated with top1 loss or accumulation of top1 cleavage complexes in yeast. PLoS genetics 11, e1005098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherel OJ, Yeo AJ, Stellati A, Heng EY, Luff J, Suraweera AM, Woods R, Fleming J, Carrie D, McKinney K, et al. (2013). Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS genetics 9, e1003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambati A, Colosio A, Zardoni L, Galanti L, and Liberi G (2015). Replication and transcription on a collision course: eukaryotic regulation mechanisms and implications for DNA stability. Frontiers in genetics 6, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Garcia-Muse T, and Aguilera A (2012). R-loops cause replication impairment and genome instability during meiosis. EMBO reports 13, 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Santos-Pereira JM, Rondon AG, Barroso S, Andujar E, Perez- Alegre M, Garcia-Muse T, and Aguilera A (2013). R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Molecular cell 52, 583–590. [DOI] [PubMed] [Google Scholar]
- Cerritelli SM, and Crouch RJ (2009). Ribonuclease H: the enzymes in eukaryotes. The FEBS journal 276, 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, and Hieter P (2014). Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP- chip. PLoS genetics 10, e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L, and Koshland D (2015). The Yin and Yang of R-loop biology. Current opinion in cell biology 34, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, and Tollervey D (2010). Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes & development 24, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Webb S, Kerr A, and Tollervey D (2014). Genome-wide distribution of RNA- DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS genetics 10, e1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Sikes ML, Hontz RD, Osheim YN, Lambert TE, El Hage A, Smith MM, Tollervey D, Smith JS, and Beyer AL (2011). Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Molecular and cellular biology 31, 482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, and Li X (2011). R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes & development 25, 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh M, Albulescu LO, Cristini A, and Gromak N (2017). Senataxin: Genome Guardian at the Interface of Transcription and Neurodegeneration. Journal of molecular biology 429, 3181–3195. [DOI] [PubMed] [Google Scholar]
- Grubb J, Brown MS, and Bishop DK (2015). Surface Spreading and Immunostaining of Yeast Chromosomes. J Vis Exp, e53081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Bocek MJ, Saldivar JC, Swigut T, and Cimprich KA (2017). Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 170, 774–786 e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JC, and Haber JE (2006). Surviving the breakup: the DNA damage checkpoint. Annual review of genetics 40, 209–235. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, and Tora L (2011). Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Molecular cell 44, 966–977. [DOI] [PubMed] [Google Scholar]
- Huertas P, and Aguilera A (2003). Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Molecular cell 12, 711–721. [DOI] [PubMed] [Google Scholar]
- Lang KS, Hall AN, Merrikh CN, Ragheb M, Tabakh H, Pollock AJ, Woodward JJ, Dreifus JE, and Merrikh H (2017). Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell 170, 787–799 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, and Haber JE (1998). Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94, 399–409. [DOI] [PubMed] [Google Scholar]
- Li X, and Manley JL (2005). Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378. [DOI] [PubMed] [Google Scholar]
- Lisby M, and Rothstein R (2009). Choreography of recombination proteins during the DNA damage response. DNA repair 8, 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, and Proudfoot NJ (2011). Yeast Sen1 helicase protects the genome from transcription-associated instability. Molecular cell 41, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, and Fischer T (2016). Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell 167, 1001–1013 e1007. [DOI] [PubMed] [Google Scholar]
- Porrua O, and Libri D (2015). Transcription termination and the control of the transcriptome: why, where and how to stop. Nature reviews. Molecular cell biology 16, 190–202. [DOI] [PubMed] [Google Scholar]
- Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, et al. (2012). Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149, 1008–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaltiel IA, Krenning L, Bruinsma W, and Medema RH (2015). The same, only different - DNA damage checkpoints and their reversal throughout the cell cycle. Journal of cell science 128, 607–620. [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, and Gromak N (2011). Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Molecular cell 42, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, and Cimprich KA (2015). Breaking bad: R-loops and genome integrity. Trends in cell biology 25, 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey R, Garcia-Rodriguez N, Aguilera A, and Wellinger RE (2015). Role for RNA:DNA hybrids in origin-independent replication priming in a eukaryotic system. Proceedings of the National Academy of Sciences of the United States of America 112, 5779–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, and Dean C (2013). R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340, 619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs A, and Nussenzweig A (2017). Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 168, 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. (2009). Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nature cell biology 11, 1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, and Haber JE (2002). Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Molecular cell 10, 373–385. [DOI] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, and Vuica-Ross M (2011). RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Molecular cell 44, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Costantino L, Tan FJ, Zimmer A, and Koshland D (2016). S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes & development 30, 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZX, Zhang MJ, Peng X, Takayama Y, Xu XY, Huang LZ, and Du LL (2013). Mapping genomic hotspots of DNA damage by a single-strand-DNA-compatible and strand- specific ChIP-seq method. Genome research 23, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.