Abstract
Background
Selection of resistance mutations may play a major role in the development of endocrine resistance. ESR1 mutations are rare in primary breast cancer but have high prevalence in patients treated with aromatase inhibitors (AI) for advanced breast cancer. We investigated the evolution of genetic resistance to first line AI therapy using sequential ctDNA sampling in patients with advanced breast cancer.
Patients and Methods
83 patients on first line AI therapy for metastatic breast cancer were enrolled in a prospective study. Plasma samples were collected every 3 months to disease progression and ctDNA analysed by digital droplet PCR and enhanced tagged-amplicon sequencing (eTAm-Seq). Mutations were tracked back through samples prior to progression to study the evolution of mutations on therapy. The frequency and impact of KRAS mutations were validated in an independent cohort of available baseline plasma samples in the SoFEA study, which enrolled patients with prior sensitivity to AI.
Results
Of the 39 patients who progressed on first line AI, 56.4%(22/39) had ESR1 mutations detectable at progression, which were polyclonal in 40.9%(9/22) patients. In serial tracking, ESR1 mutations were detectable median 6.7 months (95%CI 3.7-NA) prior to clinical progression. Utilising eTAm-Seq ctDNA sequencing of progression plasma, ESR1 mutations were demonstrated to be sub-clonal in 72.2%(13/18) patients. Mutations in RAS genes were identified in 15.4%(6/39) of progressing patients (4 KRAS, 1 HRAS, 1 NRAS). In SoFEA, KRAS mutations were detected in 21.2%(24/113) of patients, although there was no impact of KRAS mutations on progression free or overall survival.
Conclusions
Cancers progressing on first line AI show high levels of genetic heterogeneity, with frequent sub-clonal mutations. Sub-clonal KRAS mutations are found at a high frequency, although the detection had no impact on progression on subsequent endocrine therapy in the SoFEA study. The genetic diversity of AI resistant cancers may limit subsequent targeted therapy approaches.
Keywords: Breast cancer, ESR1, KRAS, ctDNA
Introduction
Selection of resistance mutations may play a major role in the development of resistance to therapy. Many examples are described, such as KRAS mutations emerging in colorectal cancer treated with anti-epidermal growth factor receptor (EGFR) therapy (1)(2) and the development of EGFR T790M mutations in patients with non small cell lung cancer treated with EGFR inhibitors (3)(4). In breast cancer, ESR1 mutations are rare in primary disease (5) but have a high prevalence in patients treated with aromatase inhibitor (AI) therapy in the advanced setting. ESR1 mutations mainly occur within the ligand binding domain and result in ligand independent activation of the estrogen receptor (ER) (6). They are an acquired mechanism of resistance and mutations in ctDNA predict resistance to AI (7)(8). In a retrospective study (9), circulating ESR1 mutations were found in 30.6% of patients at progression on an AI and were detectable in 75% of those patients prior to progression.
We investigated the development and evolution of genetic resistance to first line AI therapy in a prospective study using sequential ctDNA sampling in patients with advanced breast cancer. Samples were analysed with ESR1 multiplex droplet digital PCR (ddPCR) assays and by enhanced tagged-amplicon sequencing (eTAm-Seq, InVision™) to investigate for mutations that may contribute to AI resistance. Sub-clonal KRAS mutations were found relatively frequently in ctDNA of patients progressing on first line AI therapy, suggesting that KRAS mutations could be selected as a potential mechanism of resistance. We validated the frequency of KRAS mutations in baseline plasma of the SoFEA study, a large phase III trial of patients who had progressed on prior AI therapy.
Materials and Methods
Study Design
Eighty-three patients on first line AI therapy for metastatic breast cancer were enrolled in the prospective plasmaDNA AI study (CCR3297, REC 10/H0805/50) to collect plasma samples for ctDNA analysis every three months on therapy, and at disease progression. All plasma samples were analysed with ESR1 multiplex ddPCR assays, and samples at disease progression were analysed by eTAm-Seq. Mutations identified by eTAm-Seq were tracked back through samples prior to disease progression, to study the evolution of mutations on therapy. Written informed consent was obtained from all patients.
ER, progesterone receptor (PgR), and human epidermal growth factor receptor 2 (HER2) were assessed in a single laboratory at the Royal Marsden Histopathology department (or reviewed by the RMH when reported from a referring hospital) using standard criteria.
Plasma collection and processing
In the plasmaDNA AI study, plasma samples were collected every 3 months and at end of treatment in EDTA Blood Collection Tubes (BCT). Samples were processed within 2 hours of collection by centrifugation at 1600g for 20 minutes at room temperature. Plasma was separated from buffy coat and red blood cells, aliquoted and stored at -80°C until DNA extraction.
In the SoFEA trial, baseline blood was collected in EDTA BCT and processed within 0 to 9 days of sample collection. Plasma was separated by centrifugation 1600g for 20 minutes. We have previously demonstrated that archival EDTA plasma samples can be used for ctDNA analysis with ddPCR (8).
DNA extraction
Following thawing, ctDNA was extracted from 2 or 4ml of plasma using the MagMax Cell-Free DNA Isolation kit (Thermo A29319) on a Kingfisher Flex Purification System (Thermo) according to manufacturer instructions. The DNA was quantified and stored at -20°C until analysis.
Droplet digital PCR
DNA concentration was estimated in each sample as previously described (7).
For ESR1 mutation analysis we used two commercially available ddPCR multiplexes from Bio-Rad, multiplex 1 (dHsaMDXE91450042) and multiplex 2 (dHsaMDXE65719815). Multiplex 1 contained FAM-labelled probes for E380Q (c.1138G>C), L536R (c.1607T>G), Y537C (c.1610A>G), D538G (c.1613A>G). Multiplex 2 contained FAM-labelled probes for S463P (c.1387T>C), Y537N (c.1609T>A) and Y537S (c.1610A>C).
For KRAS mutation analysis we used a commercially available ddPCR multiplex from Bio-Rad (Cat Number 1863506). The multiplex assay contains FAM-labelled probes to 7 commonly occurring hotspot mutations on codons 12 and 13 of KRAS.
Samples were analyzed using DNA extracted from 1 ml plasma (1ml plasma equivalent). Multiplex reaction volumes were made up to 20μl with 10μl of Bio-Rad ddPCR Supermix for probes, 1μl of assay and 9μl nuclease-free water, then partitioned to a mean of 15,000 droplets using a ddPCR Auto Droplet Generator (Bio-Rad). For ESR1 mutation analysis the following conditions were used: 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds then 52°C for 60 seconds, ramp rate 2.5°C/second, and final incubation 98°C for 10 minutes. For KRAS mutation analysis the following conditions were used: 95°C for 10minutes followed by 40 cycles of 94°C for 30 seconds then 55°C for 60 seconds, ramp rate 2.5°C/second, and final incubation 98°C for 10 minutes. The subsequent analysis was done on a Bio-Rad QX200 droplet reader, and analyzed using QuantaSoft software v1.7.4.0917 (Bio-Rad). A multiplex assay was called as mutation positive if there were at least 2 FAM-positive droplets. Samples were only called negative if there were at least 300 wild type alleles detected and <2 FAM-positive droplets. If this criterion was not met the sample was repeated or failed if there was insufficient material to repeat.
InVision™ / eTAm-Seq analysis
The InVision liquid biopsy platform combines efficient next-generation sequencing library preparation and statistical algorithms to identify and quantify low frequency tumor-derived single nucleotide variants (SNVs), insertions/deletion (Indels) and CNVs in cell-free DNA, based on methods previously described (10)(3). Next generation sequencing libraries were prepared using a two-step amplification process, with primers targeting 36 cancer-related genes (Supplementary figure 1) designed to hotspot and entire coding regions of interest. The panels (v1.4/v1.5) are optimised for amplification of highly fragmented DNA with amplicon sizes ranging 72bp −154bp.
Pooled libraries were quantified using Kapa Library Quantification Kit, and 1.8pM libraries analysed on an Illumina NextSeq 500 (300 cycle PE). Sequencing files were analysed using the Inivata Somatic Mutation Analysis (ISoMA) analytical pipeline (V1.15-1.17), and sequencing reads clipped, merged and aligned. Coding and splice-site mutations in SNVs and Indels were annotated using Variant Effect Predictor (VEP) using the canonical transcript for each gene. Sub-clonal mutations were defined as mutations with an aggregate allele frequency <0.25 of driver mutation allele frequency identified in the analyzed samples.
Orthogonal validation of the detected mutations by eTAm-Seq was performed using ddPCR as described above. KRAS, PIK3CA and TP53 mutations were validated using either commercially available assays or in-house designed assays as previously described (11).
Statistical analysis
All statistical analysis was performed with GraphPad Prism version 6.0, Stata or R. Lead time was calculated using the Turnbull estimator. For analyses of progression free survival (PFS), Kaplan-Meier curves were plotted and groups compared using the log-rank test.
Results
ESR1 mutations are frequently subclonal and polyclonal at progression on AI
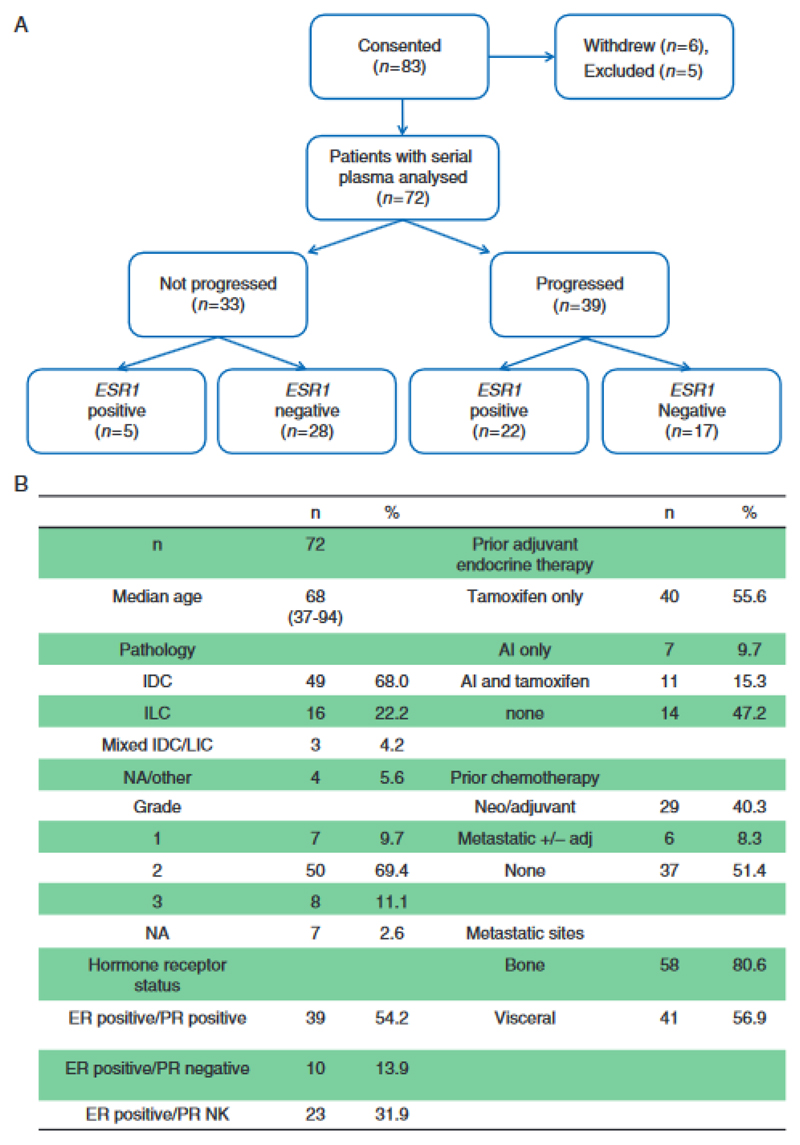
83 patients with ER positive metastatic breast cancer on 1st line AI therapy were enrolled into a prospective study to collect plasma samples for ctDNA analysis every 3 months and at disease progression (Figure 1A). The clinical and pathological characteristics of the study cohort are described in Figure 1B.
Figure 1.
PlasmaDNA AI study of sequential plasma DNA sampling during first line aromatase inhibitor therapy for advanced breast cancer. (A) Consort diagram of plasma samples analysed for ESR1 mutations on the plasmaDNA AI study. (B) Baseline characteristics of patients in the PlasmaDNA AI study.
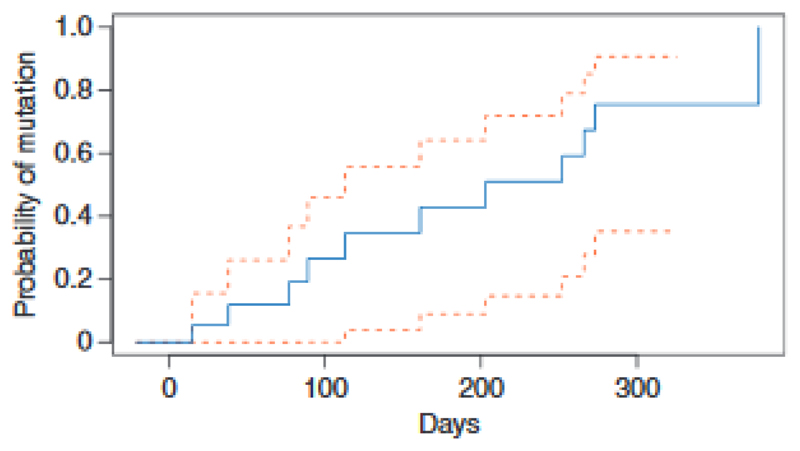
We initially studied the evolution of ESR1 mutations on AI therapy, using ultra high sensitivity multiplex ddPCR assays for 7 commonly occurring ESR1 mutations to track these mutations in plasma until clinical progression. Of the 39 patients who progressed on first line AI, 56.4% (22/39) had ESR1 mutations detectable at progression. In the patients with ESR1 mutations detected, the mutations were polyclonal in 40.9% (9/22) of patients (Figure 2). In serial tracking prior to progression, ESR1 mutations were detectable in plasma prior to progression in 86.4% (19/22) of patients, with ESR1 mutations detectable a median of 6.7 months (95% CI 3.7-NA) prior to clinical progression (Figure 4). In patients who progressed on AI, all patients who had ESR1 mutations detected prior to progression also had ESR1 mutations detected at progression, suggesting the early detection of ESR1 mutations robustly predicted the presence of the mutation at progression. ESR1 mutations were detectable in 15.2% (5/33) patients who had not yet clinically progressed (Figure 1B).
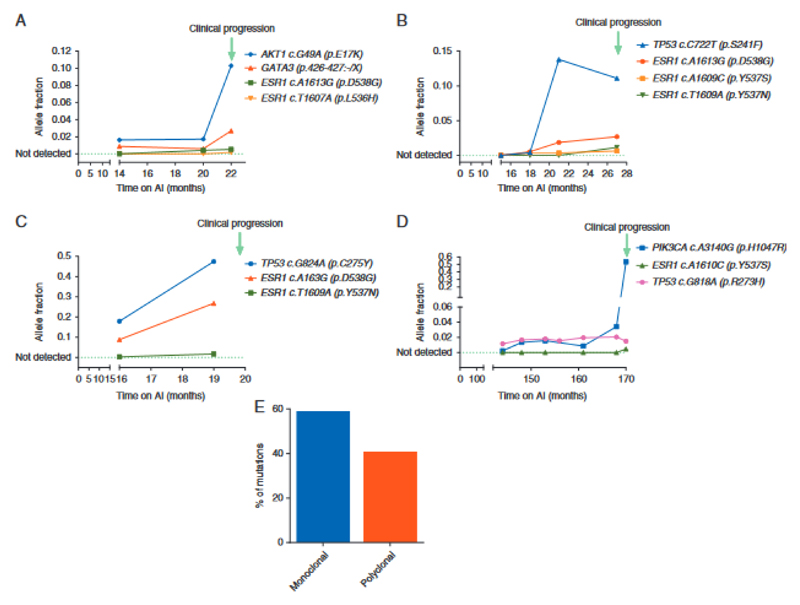
Figure 2.
Evolution of ESR1 mutations during aromatase inhibitor (AI) therapy. (A–D) Mutation tracking in ctDNA collected during first line AI therapy. Data from four patients with ESR1 subclonal mutations detectable in ctDNA tracked until clinical progression. Allele fractions are shown as determined by sequencing. TP53 mutation in grey likely to have arisen from Clonal Haematopoesis of Indeterminate Potential (CHIP). (E) Percentage of cases with monoclonal (59.1%) or polyclonal ESR1 mutations (40.9%).
Figure 4.
Lead time to development of ESR1 mutations. Serial tracking before progression, ESR1 mutations were detectable in plasma median 6.7 months [95% confidence interval (CI) 3.7–NA] before clinical progression.
AI resistant breast cancers are genomically diverse
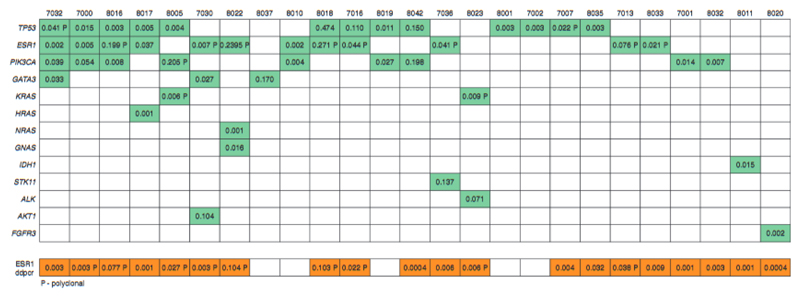
We investigated the genetics of breast cancers progressing on first line AI, with eTAm-Seq deep sequencing of ctDNA from progression plasma samples. Consistent with other studies (5) (12) (13), TP53 (36.1% (13/36)), ESR1 (33.3% (12/36)) and PIK3CA (25.0% (9/36)) mutations were the most frequent mutations detected. ESR1 mutations were identified in more samples by ddPCR than by eTAm-Seq. Of the 10 discordant cases, one ESR1 mutation detected by eTAm-Seq but not ddPCR occurred at an allele fraction (AF) of 0.002, whereas 9 ESR1 mutations detected only by ddPCR occurred at AF’s ranging from 0.0004 to 0.032. For 1 case, there was weak evidence of an ESR1 mutation but this was below the eTAm-Seq calling threshold. These cases had lower mutant copies per ml in ddPCR compared to concordant cases (median 14.3 vs 51.5 respectively, p=0.048 Mann Whitney U Test), suggesting that ddPCR was detecting low levels of ESR1 mutation in ctDNA. In patients with additional driver mutations detected in ctDNA, ESR1 mutations were sub-clonal in 72.2% (13/18) of the patients (Figure 2), found at aggregate relative allele frequency <0.25, with ESR1 mutation diversity increasingly detectable at the point of progression compared to samples taken prior to progression (Figure 2). In patients with polyclonal mutations, individual mutations were observed to be on different DNA strands in eTAm-Seq, further supporting the sub-clonality of the observed ESR1 mutations.
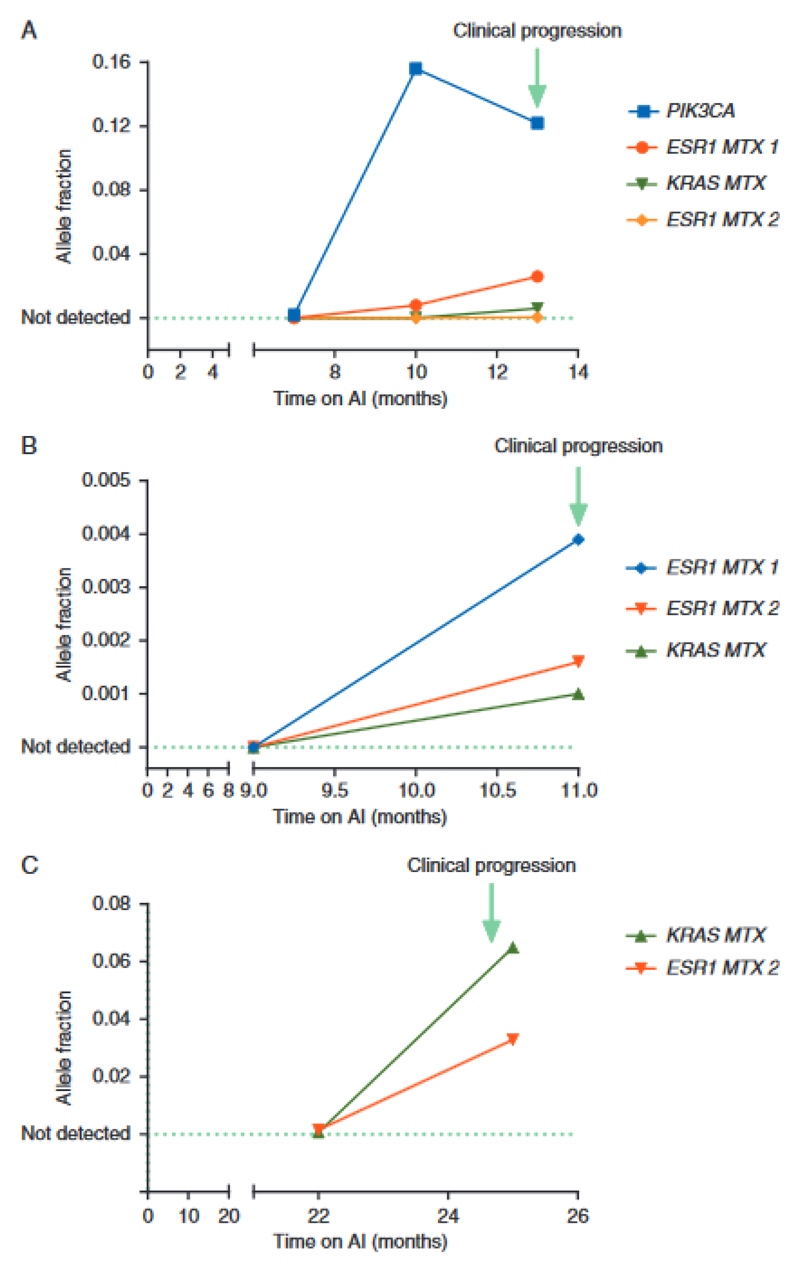
Deep ctDNA sequencing of progression samples identified previously unrecognized genetic diversity. Polyclonal KRAS mutations were identified in two patients, 8005 (p.G12V, p.G12S) and 8023 (p.G12V, p.G12C, p.G12R), a monoclonal HRAS mutation (p.G12V) in one patient and a monoclonal NRAS mutation (p.G12D) in another one. An activating p.R248C FGFR3 mutation was identified in a further patient ctDNA sample. Deep sequencing or ddPCR of ctDNA obtained from plasma identified RAS mutations in 15.4% (6/39) of progressing patients (4 KRAS (2 of which were polyclonal), 1 monoclonal HRAS and 1 monoclonal NRAS) (Figure 5). In patients where an additional driver mutation was detected in ctDNA, all identified RAS mutations were sub-clonal. In two patients with KRAS mutations detected at progression primary tumour was available, with the KRAS mutation being undetectable in both patients.
Figure 5.
Evolution of KRAS mutations during the first-line aromatase inhibitor (AI) therapy. (A–C) Mutation tracking in ctDNA collected during first line AI therapy. Data from three patients with KRAS subclonal mutations detectable in ctDNA tracked until clinical progression. Allele fractions are shown as determined by ddPCR. Patient B had an ALK mutation detected on sequencing at progression with an allele faction of 0.07. In two patients with KRAS mutations detected at progression primary tumour was available, with the KRAS mutation being undetectable in both patients.
Clonal Haematopoesis of Indeterminate Potential (CHIP) is an age related clonal expansion that is detectable in a high proportion of ageing people (14) (15). Mutations arising from CHIP may be detected in ctDNA analysis and present a potential confounder to discovery of resistance mutations in ctDNA. Although KRAS mutations are not a classic CHIP mutation, they are reported at low level. To ascertain whether detected KRAS mutations were arising from ctDNA or CHIP, we tracked KRAS mutations back through serial samples prior to progression (Figure 5). KRAS mutations arose in line with driver and ESR1 mutations at disease progression, demonstrating that the KRAS mutations were detected in ctDNA. In contrast, a TP53 mutation detectable at progression was shown to arise from CHIP with high-likelihood, as the AF of the mutation stayed constant through serial tracking, whilst mutations arising from ctDNA rose to the point of progression (Figure 2).
Identified RAS mutations are selected on AI therapy
To validate our novel discovery of KRAS mutations in AI resistant cancer, and to assess clinical significance of KRAS mutations in patients who progressed on endocrine therapy, we analysed baseline plasma samples from the phase III SoFEA study by ddPCR. The SoFEA study was a multicentre, randomized phase III trial in postmenopausal women with advanced, hormone receptor positive breast cancer who had progressed on a non-steroidal AI. All patients had demonstrated prior sensitivity to AIs, and were randomized to fulvestrant plus anastrozole, fulvestrant plus placebo, or exemestane.
We retrospectively analysed KRAS mutational status on 117 available baseline plasma samples of the 723 patients enrolled on the study. We investigated the association of KRAS mutations detected in ctDNA and clinical outcome. These samples had previously been analysed for ESR1 mutation status (8). KRAS mutational status was successfully interrogated in 96.6% (113/117) of available plasma samples, with KRAS mutations detected in 21.2% (24/113) of patients, with no KRAS mutations detected in controls (Supplementary table 2). 19.0% (8/42) of ESR1 mutant cancers also had KRAS mutations. KRAS mutations were detected at low levels in the majority of patients. There were no significant differences in baseline characteristics between patients with and without KRAS mutations (Supplementary table 3).
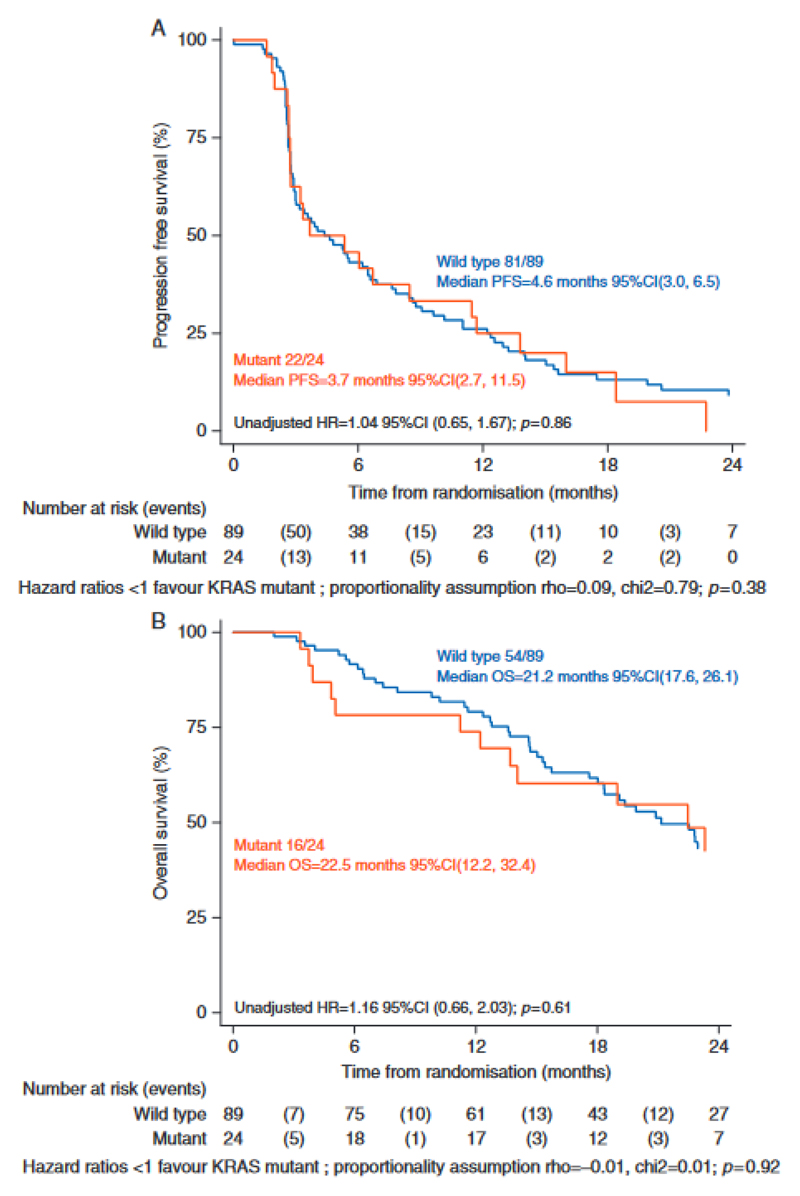
We assessed the impact of KRAS mutations on progression free and overall survival (Figure 6). For patients with KRAS mutations the median PFS was 3.7 months (95% CI, 2.7, 11.5) and for patients with wild type KRAS a median PFS of 4.6 months (95% CI, 3.0, 6.5) (HR=1.04 95% CI (0.65, 1.67) p=0.86). There was no significant difference in overall survival in those with and without KRAS mutations.
Figure 6.
Independent validation of KRAS mutations in baseline plasma from the SoFEA study. (A) Progression-free survival (PFS) in SoFEA by KRAS mutation status. HR, hazard ratio. (B) Overall survival (OS) in SoFEA by KRAS mutation status.
Discussion
In the prospective plasmaDNA AI study we demonstrate that ER positive advanced breast cancer progressing on AI shows substantial genetic diversity, with a high rate of ESR1 mutations and previ mutations in KRAS and a classical activating mutation in FGFR3. Many selected mutations are demonstrated to be sub-clonal, although our findings identify a potential major role for selected KRAS mutations in resistance to AI therapy in the treatment of advanced breast cancer.
In this cohort of patients progressing on first line AI, ESR1 mutations are found at high prevalence in plasma, detectable in over half of patients. Resistance to therapy can be anticipated with a long lead-time over clinical progression, with ESR1 mutations detectable prior to progression in 86.4% of patients. These results are consistent with a prior retrospective study that reported ESR1 mutations were detectable in 75% of patients prior to progression (9). This prior study reported a lower frequency of ESR1 mutations at progression on an AI but only 4 ESR1 mutations were analysed using 4ng preamplified DNA which likely explains the higher frequency reported here. The incidence of ESR1 mutations we report is in line with the rate we previously reported in the SoFEA study, with ESR1 mutations detected in 39.1% of baseline samples (8).
We show that many ESR1 mutations detected in plasma are likely sub-clonal in the cancer, with the aggregate allele fraction of ESR1 mutations frequently substantially lower than that of other identified driver mutations. This suggests that in an individual patient, ESR1 mutations may not be the sole driver of resistance in the cancer. Multiple drugs that degrade the mutant ER are in early clinical, and pre-clinical development, and this finding emphasises the importance of assessing clonal dominance of ESR1 mutations in clinical development. Also, due to the sub-clonal nature of these mutations, the amount of plasma DNA analysed may have a major impact on frequencies of ESR1 mutations identified.
In this study, we identify selection of KRAS activating mutations as a potential novel mechanism of resistance to AI, with a substantial prevalence of 21.2% (24/113) in the SoFEA validation series.
KRAS mutations are identified in approximately 2% of primary ER positive breast cancer (5) (12), and KRAS mutations are undetectable in the primary of two patients with selected KRAS mutations, suggesting selection by therapy. The KRAS mutations identified are frequently sub-clonal, possibly due to geographic development of KRAS mutations in individual metastases. Multiple prior studies have linked activation of MAP kinase pathway signaling to resistance to endocrine therapy (16, 17), suggesting the KRAS mutations may drive resistance to endocrine therapy in individual sub-clones. In SoFEA the presence of a KRAS mutation detected in ctDNA had no impact on PFS or OS, although this analysis used a relatively small number of samples and would need confirmation in a larger set. This finding suggests the importance of determining whether sub-clonal KRAS mutations continue to expand through subsequent therapy, or whether the mutations become undetectable once endocrine therapy is ceased, which will be an important area of future research.
This study has limitations. Some patients joined the study mid-AI therapy and had ESR1 mutations detected at the first sample. Although, this was taken into account when calculating lead time to progression, this adds imprecision to the median estimate of lead-time. There were a relatively small number of progression samples in the plasma AI study and it was not possible to perform sequencing on all progression samples due to amounts of DNA available. Although the ctDNA sequencing strategy we employ substantially expanded our ability of investigate the genetics of AI resistant cancer, leading to the discovery of KRAS mutations, the panel covered a limited number of genes. There may be other relevant selected mutations present at progression on AI that were not detected in this panel. Most of the KRAS mutations detected were present at low levels, and although some mutations were present at relatively high level it was not possible due to limited number of high level mutations to assess whether there is a different impact on outcome for those with high levels of KRAS mutation.
Our study demonstrates that selected genetic mechanisms of resistance are frequent in first line AI therapy. ESR1 mutations are found at high prevalence in this setting, along with high frequency sub-clonal KRAS mutations. AI resistant cancers are genetically heterogeneous and may consist of several clones that may limit the effectiveness of subsequent targeted therapies that target only one of the clones.
Supplementary Material
Key message.
Breast cancers progressing on first line aromatase inhibitors show high levels of genetic heterogeneity, with frequent sub-clonal mutations that may limit subsequent targeted therapy approaches.
Figure 3.
Error corrected ctDNA sequencing of plasma samples taken after progression on the first-line aromatase inhibitor (AI). Mutations identified in plasma DNA by eTAm-Seq error corrected sequencing, with ESR1 mutation analysis by ddPCR. Discordant cases for ESR1 between ddPCR ard ctDNA sequencing had lower mutant copies per ml in ddPCR compared with concordant cases (median 14.3 versus 51.5, respectively, P = 0.048 Mann–Whitney U test) and likely represent very low levels of mutant copies and random sampling. 8037 also had FGFR1 and ERBB2 amplification Identified. Of 36 progression plasma samples sequenced, 25 with mutations are displayed, 11 plasma samples with no mutations detected are not displayed. Numbers in boxes represent allele fraction for indicated gene. Where there are multiple mutations detected in the same gene, indicating polyclonality (P), aggregate allele fractions are given.
Acknowledgments
We thank the patients who participated in the plasmaDNA AI and SoFEA studies, along with the investigators, study nurses, and staff who supported the trials.
Funding:
This research was funded by Le Cure, Breast Cancer Now and Inivata. The SoFEA trial was funded by Cancer Research UK (grant C1491/A10962, reference numbers CRUKE/03/021 and CRUK/09/007). ICR-CTSU receives programme grant funding from Cancer Research UK (grant C1491/A15955). We acknowledge National Institute for Health Research funding to the Royal Marsden and Institute of Cancer Research Biomedical Research Centre.
Footnotes
Disclosures: KH, ME and NR and EG are employees, consultants or shareholders of Inivata Ltd., and/or inventors of patent applications related to sequencing strategies described in this manuscript. AR has an advisory role for Novartis, Roche and Genomic health.
References
- 1.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4(11):1269–80. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 3.Remon J, Caramella C, Jovelet C, Lacroix L, Lawson A, Smalley S, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol. 2017;28(4):784–90. doi: 10.1093/annonc/mdx017. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–45. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7(313):313ra182. doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fribbens C, O'Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, et al. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J Clin Oncol. 2016;34(25):2961–8. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 9.Clatot F, Perdrix A, Augusto L, Beaussire L, Delacour J, Calbrix C, et al. Kinetics, prognostic and predictive values of ESR1 circulating mutations in metastatic breast cancer patients progressing on aromatase inhibitor. Oncotarget. 2016;7(46):74448–59. doi: 10.18632/oncotarget.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 12.Fumagalli D, Wilson TR, Salgado R, Lu X, Yu J, O'Brien C, et al. Somatic mutation, copy number and transcriptomic profiles of primary and matched metastatic estrogen receptor-positive breast cancers. Ann Oncol. 2016;27(10):1860–6. doi: 10.1093/annonc/mdw286. [DOI] [PubMed] [Google Scholar]
- 13.Lajos Pusztai JC, Young Lauren, Schrock Alexa Betzig, Hartmaier Ryan, Frampton Garrett Michael, Gay Laurie M, Stephens Phil, Miller Vincent A, Ali Siraj Mahamed, Ross Jeffrey S, Vahdat Linda T, et al. Genomic profiling of circulating tumor DNA (ctDNA) from patients (pts) with metastatic breast cancer (mBC). 2017 ASCO Annual Meeting; Abstract 1016. [Google Scholar]
- 14.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez MC, Detre S, Johnston S, Mohsin SK, Shou J, Allred DC, et al. Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. J Clin Oncol. 2005;23(11):2469–76. doi: 10.1200/JCO.2005.01.172. [DOI] [PubMed] [Google Scholar]
- 17.Cui Y, Parra I, Zhang M, Hilsenbeck SG, Tsimelzon A, Furukawa T, et al. Elevated expression of mitogen-activated protein kinase phosphatase 3 in breast tumors: a mechanism of tamoxifen resistance. Cancer Res. 2006;66(11):5950–9. doi: 10.1158/0008-5472.CAN-05-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.