Abstract
Resource limitation is a fundamental factor governing the composition and function of ecological communities. However, the role of resource supply in structuring the intestinal microbiome has not been established and represents a challenge for mammals that rely on microbial symbionts for digestion: too little supply might starve the microbiome while too much supply might starve the host. Here, we present evidence that microbiota occupy a habitat limited in total nitrogen supply within the large intestines of 30 mammal species. Furthermore, lowering dietary protein levels in mice reduced bacterial fecal concentrations. A gradient of stoichiometry along the length of the gut was consistent with the hypothesis that intestinal nitrogen limitation results from host absorption of dietary nutrients. Nitrogen availability though is also likely shaped by host-microbe interactions: levels of host-secreted nitrogen were altered in germfree mice and when bacterial loads were reduced via experimental antibiotic treatment. Single-cell spectrometry revealed that members of the phylum Bacteroidetes consumed nitrogen in the large intestine more readily than other commensal taxa. Collectively, our findings support a model where nitrogen limitation arises from preferential host utilization of dietary nutrients, and we speculate that this resource limitation could enable hosts to regulate microbial communities in the large intestine. Furthermore, commensal microbiota may have adapted to nitrogen-limited settings, suggesting why excess dietary protein has been associated with degraded gut microbial ecosystems.
The mammalian large intestine (i.e., the colon) is typically considered a hospitable environment for microbes. Microbial communities there are among the densest on the planet (~1013 cells total from hundreds of species in humans)1,2 and are full of active cells3 that aid in host nutrient acquisition, waste processing, pathogen resistance, and immune regulation4–6. Relative to elsewhere in the gut, the large intestine has a more neutral pH, larger volume, and longer retention time, leading to a greater proliferation of microbes7. By contrast, the host’s primary uptake of dietary proteins, fatty acids, and simple carbohydrates occurs in the small intestine8,9. We therefore hypothesized that concentrations of essential elements are diminished in the large intestine, especially relative to demand, and thus become increasingly limiting for growth and replication of microbial cells along the length of the gut.
Nitrogen is likely to be among the limiting nutrients for bacteria in the large intestine as it is for myriad organisms in diverse environments worldwide10. Animals are frequently nitrogen limited and have evolved multiple physiological strategies to capture sufficient nitrogen from food6. These include peptide transport systems11, precise regulation of amino acid transporters12, and even the cultivation of obligate symbionts that fix nitrogen gas13,14. Yet, aside from the small subset of microbial taxa that can increase nitrogen supply to their hosts through fixation, studies using germ-free mammals have demonstrated that intestinal microbes, collectively, are more often net consumers of nitrogen and increase the protein requirement of their hosts15. Additionally, bacterial cells often have even higher nitrogen requirements than do eukaryotic cells16. Bacteria may consequently compete with their animal hosts for nitrogen. However, hosts have the opportunity to access the nitrogen in their food before many of their associated microbes do. Hosts therefore may reduce resource supply to the large intestine and thus influence the composition and functioning of resident microbial assemblages.
If nitrogen is limiting for the bacteria in the large intestine, a mismatch should exist between the carbon:nitrogen (C:N) ratio of gut microbes and that of digesta or feces. In particular, elevated C:N in resources relative to consumers, a phenomenon known as stoichiometric limitation, indicates that large amounts of food must be processed to gain sufficient nitrogen for organismal maintenance and growth16. Excess digested carbon must then be excreted to maintain stoichiometric balance, which in turn is expected to diminish growth efficiency17–19. Because the range of percent carbon content in organisms is generally narrow, C:N of digestible material is an indicator of overall nitrogen availability16. In addition, total nitrogen can be considered in an absolute sense, independent of carbon, such as in the Geometric Framework model20. Long term effects of reduced nitrogen diets have been shown to improve health outcomes in animals models when considered through the lens of the Geometric Framework21,22 and this is hypothesized to be due to nitrogen limiting the utilization of carbohydrates and thus altering microbial composition23. Still the relative importance of removal of nitrogen to increased carbon intake as well as the importance of microbial responses to these diets is not currently well-established. If the modulation of nitrogen levels in the gut results in changes in abundance of total microbes, nitrogen would be termed absolutely limiting for microbial growth (i.e., cellular replication) in the gut. In other words, such a result would imply that total gut bacterial load is constrained by an overall lack of nitrogen rather than by an imbalance between nitrogen and other resources.
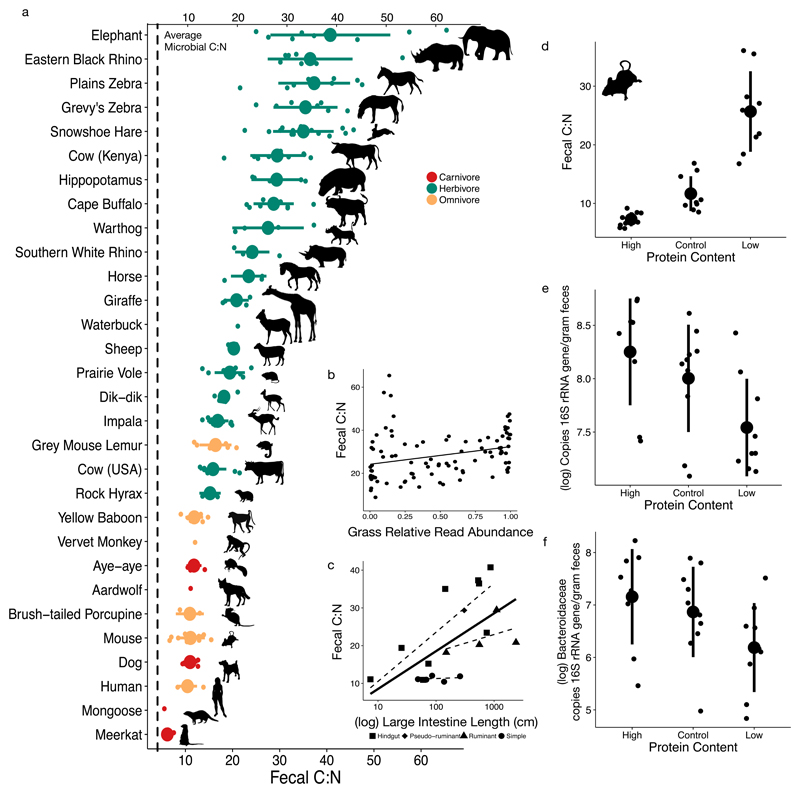
Fecal C:N (22.91±11.22) of all 30 mammal species tested was 36%-1000% higher than gut bacterial C:N (Fig. 1a), suggesting a widespread signature across the mammals of limited total nitrogen supply for gut bacteria. We found C:N to be low (4.07±0.24) in bacterial strains isolated from the human gut and grown in vitro (Supplementary Information Table 1), which is similar to C:N of non-host associated bacteria in both aquatic and terrestrial environments (4.67±1.38)24,25. Feces, of course, includes microbial cells, host cells, undigested food, and other waste. We did not separate these components, but the relatively low C:N of microbial cells and their high abundance in fecal material26 implies that the non-microbial material in feces has an even higher C:N than the feces as a whole. In the mammals sampled, fecal C:N values were similarly high for both wild (24.27±12.38, n=20) and domestic/captive species (21.14±9.26, n=10; Supplementary Information Table 2). Overall, values varied more than sevenfold among species, with approximately 300% greater C:N in herbivores than carnivores (Fig. 1a).
Fig. 1. Fecal C:N varied between mammals, was linked to diet and physiology, and controls microbial abundance in vivo.
a-c, Fecal C:N varied between mammals based on taxonomy, diet, and physiology. Fecal C:N from 30 mammal species (n=1-15 individuals per species, mean=7, see Supplementary Information Table 2) is higher than the average bacterial C:N (4.07, vertical dashed line) from gut isolates grown in vitro (n=35 strains, see Supplementary Information Table 1; a). Fecal C:N from East African herbivores and omnivores (n=16 species, see Methods for list) was positively correlated (linear regression fit shown) with proportional representation of grasses in the diet based on DNA metabarcoding (ρ=0.42, P<0.0001, Spearman correlation; n=95 fecal samples; b). Mammalian fecal C:N was also associated with the large intestine length (log transformed; P=0.04, ANCOVA) and gut architecture (P=0.03, ANCOVA; n=4-9 species per physiological group except pseudoruminant which has n=1; c). The solid line shows linear regression fit for large intestine length overall, while dashed lines show linear regression fits for each gut architecture group. d-f, Altering dietary protein (Supplementary Table 5) for two weeks impacted murine gut nitrogen and microbiota. Murine fecal C:N differed under altered-protein diets (P=5.87x10-6, Kruskal-Wallis test; n=9-10 mice per diet group; d). Microbial load, estimated by 16S rRNA gene copy number via qPCR, also changed under altered protein diets (P=0.017, Kruskal-Wallis test; n=9-10 mice per diet group; e). Bacteroidaceae abundance, calculated as 16S rRNA gene copy number multiplied by their relative abundance, changed under altered protein diets (P=0.05, Kruskal-Wallis test; n=9-10 mice per diet group; f). Large circles are means; bars show standard deviations.
In addition to the difference arising from broad diet type, we expected differences in nitrogen supply related to subtler variation within feeding guilds. Animals that eat plants with low C:N, such as woody plants and forbs27, should have intestines in which total nitrogen is less limiting than animals that feed primarily on comparatively high-C:N grasses. Indeed, within a group of African large mammals, we found a significant positive correlation between grass consumption (as assessed using both DNA metabarcoding28) and C:N (ρ=0.42, P<0.001, Spearman correlation; Fig. 1b). Physiology, including body length, intestine length, and gut type (i.e. simple, hindgut fermenter, or ruminant), were also associated with C:N (P=0.002, P=0.04 P=0.03, respectively, ANCOVA; Fig 1c). These associations may reflect the well-known ability of hindgut fermenters and larger animals to subsist on lower-quality (i.e., higher C:N) food than ruminants29 and smaller animals30, respectively. Therefore, the total nitrogen can differ dramatically with fine-scale variation in dietary composition, with lowest nitrogen availability in species that consume diets rich in carbon relative to nitrogen. Additional variation may be introduced by other physiological idiosyncrasies, e.g. in snowshoe hare (and other Leporidae), which engage in coprophagy to allow a second round of digestion of their high C:N diet31.
Having observed evidence of stoichiometric limitation in the mammalian gut, we next tested if nitrogen was absolutely limiting for microbial growth (i.e., an overall lack of nitrogen constrains total gut bacterial load). Experimental modulation of total dietary nitrogen input (via protein concentration) in mice led to shifts in fecal gut bacterial load consistent with the absolute nitrogen-limitation hypothesis. We fed mice three isocaloric diets (Supplementary Information Table 6) which varied in their casein protein levels: low (6%), control (20%), and high (40%). Higher-protein diets decreased fecal C:N (P<0.001, Kruskal-Wallis test; Fig. 1d, Supplementary Information Fig. 1a), which was in turn associated with changes in microbial load: bacterial concentration was greater in high-protein (low C:N) conditions and reduced in low-protein conditions (P=0.017, Kruskal-Wallis test; Fig. 1e, Supplementary Information Fig. 1b). Differences in diet did not significantly affect host weight gain or loss (P=0.9, Kruskal-Wallis test; Supplementary Information Fig. 1d), suggesting that changes in the microbiota did not simply reflect poor host condition. However, microbiota changes may also have been responses to mouse diet intake23 or varying fecal transit time from altered cellulose levels32, neither of which were tracked here.
We next examined the extent to which mammalian hosts might induce nitrogen limitation via selective nitrogen uptake and delivery. Because 80-90% of host absorption of dietary nitrogen takes place in the small intestine9, we predicted to exist a longitudinal gradient of total nitrogen along the length of the gut. Indeed, we found in laboratory mice that C:N increased from the proximal small intestine to the large intestine in laboratory mice (Supplementary Information Fig. 2a), with the distal small intestine and the large intestine exhibiting significantly higher C:N than the proximal small intestine (P<0.05, Tukey’s Honest Significant Difference test). This variation was independent of microbial load in gut contents (P=0.14, Spearman correlation; Supplementary Information Fig. 2b). These results are consistent with a model in which colonic microbes could experience total nitrogen limitation as a consequence of upstream host absorption of dietary nutrients.
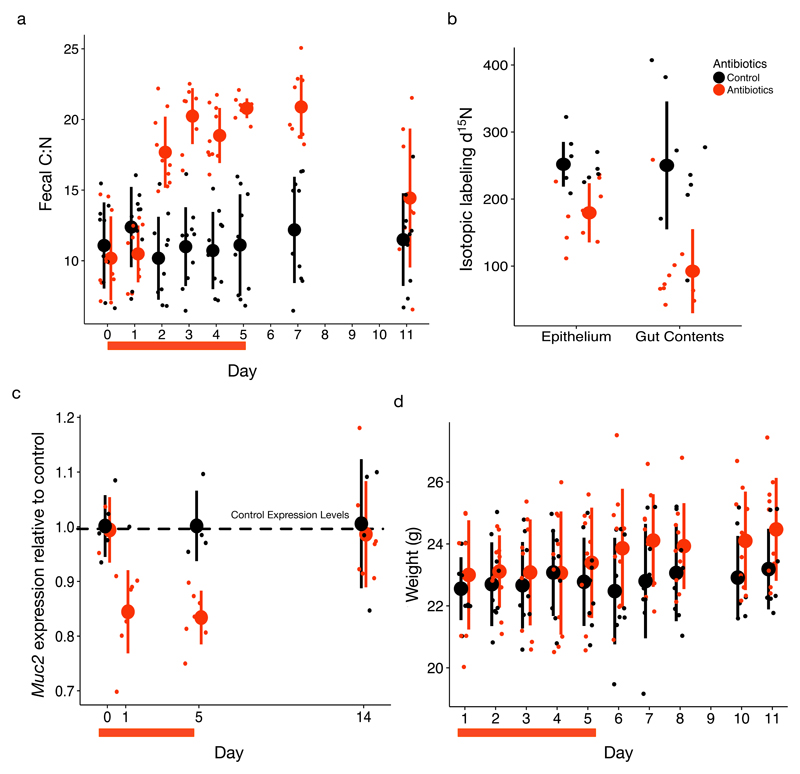
Although hosts may alter nitrogen availability through dietary absorption, we hypothesized that gut bacterial loads could also shape the nitrogen landscape. We therefore treated mice with a broad spectrum antibiotic cocktail (ampicillin, vancomycin, metronidazole, and neomycin33). Fecal C:N increased significantly relative to untreated animals after two days (P<0.001, linear mixed effects model likelihood test; Fig. 2a). This change may have been due to load reductions altering nitrogen available in bacterial cells or through gut bacterial metabolism and subsequent host absorption. But, we found evidence that this increase reflected, at least in part, changes in host provisioning of nitrogen. During antibiotic treatment, host delivery of 15N from an injected isotopically labeled amino acid, threonine, into the gut decreased more than two-fold (P=0.001, Mann-Whitney U test; Fig. 2b). We observed that expression of murine Muc2, a mucus protein coding gene, was also significantly reduced (P<0.05, Wilcoxon signed rank tests; Fig. 2c); moreover mucus thickness (reflecting overall mucus quantity) in the proximal colon tended to be lower in treated mice (P=0.1, Mann-Whitney U test; Supplementary Information Fig. 3b), as has been seen elsewhere34. Consistent with the finding that bacterial load influences nitrogen levels, we also observed that levels of delivered 15N isotopically labeled threonine across the gut were lower in germfree mice than conventional mice (P<0.001, mixed-design ANOVA; Supplementary Information Fig. 5b). This finding agrees with prior work demonstrating germfree mice have a thinner colonic firm mucus layer35,36 or thinner colonic mucus layers overall37. Together, our data support a model where host secretion of nitrogen is responsive to changes in microbial load.
Fig. 2. Antibiotics change gut nitrogen and host secretions.
a, Antibiotic cocktail (ampicillin, vancomycin, metronidazole, and neomycin) treatment induced a significant increase in fecal C:N (P=2.647x10-11, linear mixed effects model likelihood test; n=9-10 mice per treatment group)), followed by re-convergence within 6 day post-treatment. b-c, This increase is concomitant with decreases in nitrogen secretion as measured by (b) isotopic label delivery to epithelial tissue and gut contents (P=0.001, Mann-Whitney U tests; n=10 mice per treatment group) and (c) mucin production (measured as Muc2 expression) relative to control levels (dotted line) during treatment (P=1.0, 0.016, 0.016, 0.58 Wilcoxon signed rank tests for treated mice relative to control average on days 0, 1, 5, and 14, respectively; n=6 treated mice). d, Mouse weight increased more over the 11 day experiment in antibiotic-treated mice than in control mice (P=0.0002, linear mixed effects model likelihood test). Red bars under the x-axis indicate the 5 day course of antibiotics. Large circles are means; bars show standard deviations.
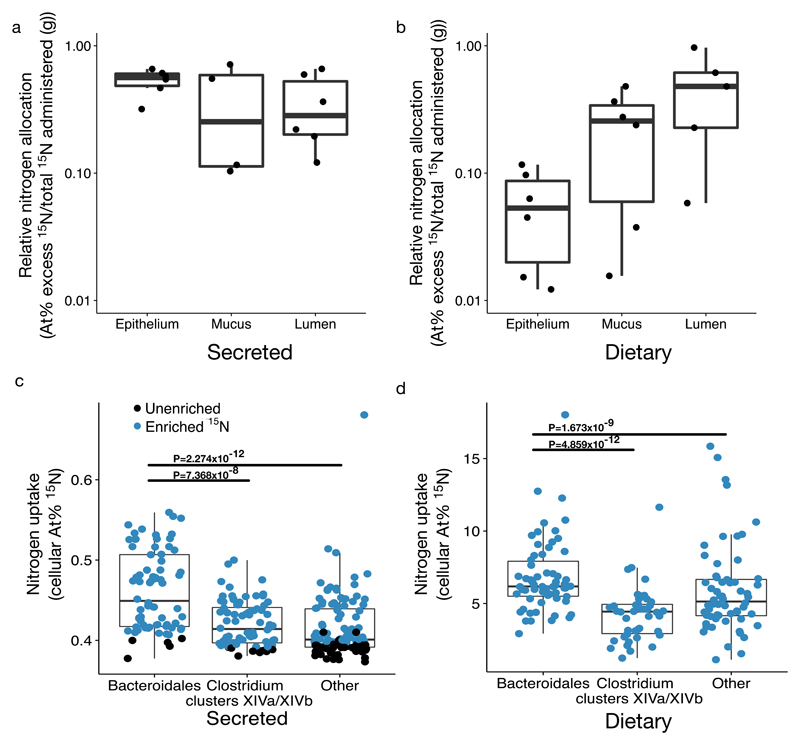
To obtain insights into the sources of nitrogen available to bacteria in the large intestine, we performed stable-isotope tracer experiments in mice using two complementary approaches. First, we tracked host allocation and microbial use of dietary nitrogen by feeding mice chow with typical protein levels in which all nitrogen was delivered via Spirulina in the form of 15N (the low natural abundance heavy isotope). While digestion of this diet may differ overall from standard mouse chow as Spirulina cells are not a typical component of mouse diet, consumption of it has not been found to negatively impact host health and this method ensured complete labeling of dietary nitrogen. Second, we considered the effect of host-secreted nitrogen by varying our source of labeled nitrogen in a separate cohort of mice, which received 15N/13C labeled threonine via tail-vein injection38 as in the antibiotic and germfree mouse experiments. This method represents a conservative estimate of host nitrogen secretion as it only includes labeled threonine; host secreted compounds including non-threonine amino acids or amino sugars are found39 but are not counted here. In concert, these experiments revealed significant isotopic enrichment of gut tissue and gut contents from both secretions (P<0.05, one-sample Wilcoxon tests; Fig. 3a, Supplementary Information Fig. 4a) and diet (P<0.05, one-sample Wilcoxon tests; Fig. 3b, Supplementary Information Fig. 4a). Moreover, the efficiency with which labeled nitrogen accumulated in large intestine gut contents (lumen and mucosa) did not differ significantly between the host-secreted and dietary delivery pathways (P>0.05, Mann-Whitney U tests Fig. 3a-b). Our results fit with prior findings23,38, and thus support an overall hypothesis that host amino acid secretions account for an appreciable fraction of nitrogen available in the colon.
Fig. 3. Microbes use nitrogen from host diet and secretions.
(a-b) Nitrogen in the gut is sourced both from host diet and host secretions. 15N isotope enrichment (adjusted for total 15N administered) was significantly different from zero for both (a) injected threonine secretions and (b) dietary nitrogen in large intestine epithelium, mucosa, and lumen layers (null hypothesis: μ=0; P=0.03, one-sample Wilcoxon tests; n=6 mice per treatment). (c-d) For large intestine gut microbiota from mice treated with labeled nitrogen, single cell isotopic enrichment was quantified on a NanoSIMS following FISH to distinguish between microbial taxonomic groups (n=2 mice per treatment group). Bacteroidales were disproportionate nitrogen consumers relative to other bacterial taxa: cells targeted by the Bacteroidales probe (Bac303) were more highly enriched for 15N from host secreted labeled threonine (n=72-110 cells per target; c) and also from host diet (n=42-62 cells per target; d) than the Clostridium cluster XIVa and XIVb-specific (Erec482) or other DAPI stained cells (P<0.05, Bonferroni-corrected Mann-Whitney U tests). Bars indicate groups which differed significantly from Bacteroidales. Isotope enrichment is reported as atom percent (i.e., the proportional representation of the heavy isotope times 100; c-d) or as atom percent excess (i.e., the difference between atom percent of the treated sample and the average control; a-b; see Methods). Blue points refer to cells significantly enriched in 15N. Boxplots summarize all cells (enriched and unenriched) and show median and quartiles; whiskers show the 1.5*interquartile range.
We next tested the hypothesis that nitrogen has distinct effects on different bacterial taxa in the gut. We employed high-resolution secondary ion mass spectrometry (NanoSIMS) to measure uptake of both host-secreted and diet-sourced 15N in the large intestine bacterial community with single cell resolution. We confirmed gut bacteria used nitrogen derived from both host diet and host secretions (P<0.001, Kruskal-Wallis test; Supplementary Information Fig. 6, 7). Combining NanoSIMS measurements with fluorescence in situ hybridization (FISH), we then tested for nitrogen uptake by gut bacteria, including members of the Bacteroidales and Clostridium clusters XIVa/XIVb compared to other bacteria (see Supplementary Information Table 3, 4, Methods). Select Bacteroidales contribute to intestinal health and function40, produce short chain fatty acids for host uptake41, and digest glycans into products upon which other commensal microbiota can feed42. Consistent with previous observations38, single cell measurements in mice treated with 15N/13C labeled threonine injections showed that the Bacteroidales consumed host-secreted nitrogen more readily than the targeted Clostridia (P<0.05, Bonferroni-corrected Mann-Whitney U tests; Fig. 3c). These patterns are congruous with a model where members of the order Bacteroidales are initially sensitive, more so than typical gut microbiota species, and likely promoted by increases in host-secreted nitrogen. In support of this model, Bacteroidales abundance is known to be heightened in hibernating43 and fasting44 animals, where secretions are the primary source of all nutrients to the microbiota.
We also found evidence that members of the Bacteroidales were sensitive to dietary nitrogen. Single cell measurements in mice treated with 15N labeled Spirulina indicated the Bacteroidales were more likely to consume dietary nitrogen (P<0.05, Bonferroni-corrected Mann-Whitney U tests; Fig. 3d). Notably, we observed in our dietary manipulation experiments that Bacteroidaceae abundance (the most abundant family in the Bacteroidales in laboratory mice) increased with greater nitrogen supply (P=0.05, Kruskal-Wallis test; Fig. 1f, Supplementary Information Fig. 1d. These findings contrast with a recent model suggesting that the Bacteroidetes should decrease with higher nitrogen intake23—a difference potentially due to that model's use of relative abundance data as compared to absolute bacterial abundances here. Their findings do, however, corroborate our observation that there is genus-level variation in response to greater dietary nitrogen supply, including increases in the Firmicutes family Lachnospiraceae (Supplementary Information Table 5). This observation may be explained by ecological phenomena like cross-feeding or variation in nitrogen utilization strategies at finer taxonomic levels, but will ultimately require further work to clarify.
Collectively, our findings suggest ecological and evolutionary mechanisms by which mammals and gut microbiota co-evolved45. By secreting nutrients into the large intestine or altering digestion, mammals could attenuate nitrogen limitation to upregulate preferred bacterial taxa like members of the Bacteroidales (or, conversely, downregulate these same taxa by withholding secretions), thereby adjusting the aggregate digestive and metabolic functions of the gut-microbial community. This regulatory mechanism would entail a cost; indeed a reduced need for nitrogen secretions may be one cause of antibiotic-associated weight gain, which is a phenomenon46 observed in our mice (P<0.001, linear mixed effects model likelihood test; Fig. 2d). Evolutionary theory, however, predicts such a cost worthwhile when animals interact with a diverse but beneficial microbiota, providing the host with a ‘dial’ to fine-tune gut microbial communities in response to dietary, physiological, and environmental variation47. Future work should elucidate the molecular and physiological mechanisms underpinning secretion dynamics, explore the magnitude of costs to the host, and probe the conditions associated with changes in nitrogen supply to the gut within hosts over time and between host species. In particular, co-limitation with other nutrients48 like phosphorous is likely in all animals, and we predict carnivores with high nitrogen diets, short guts, and low fecal C:N may be especially likely to modulate co-limiting nutrient levels to regulate their microbiota.
Our model would also predict that competitive advantages for secretion-consuming bacteria might be eliminated (and mutualistic benefits reduced) if nitrogen supply to the large intestine exceeded some critical threshold. Indeed, when the human gut experiences nitrogen excess (e.g. with high-protein diet interventions), the microbiota quickly shift composition49, and harmful metabolic products including ammonia, nitrosamines, and sulfide are expected to accumulate50. High protein diets are also associated with reductions in longevity in animals22. Thus, loss of microbial nitrogen limitation in the gut may resemble the environmental phenomenon of eutrophication in which excess nutrient delivery alters community composition and degrades ecosystem services.
Methods
Conventional mouse experiments
All mouse experiments were conducted in accordance with National Institute of Health Guide for the Care and Use of Laboratory Animals using protocols approved by the Duke University Institutional Animal Care & Use Committee. Male C57BL/6 mice (Charles River Laboratories) 8-10 weeks of age with a native microbiota were used for all manipulative experiments. Mice were kept in a conventional laboratory animal facility at Duke University and fed PMI Labdiet 5001 chow (20% protein; 12.5±0.9 C:N).
Animal fecal samples and gut-length estimates
Yellow baboon (Papio cynocephalus, n=8, sex unspecified) feces were collected from free-living baboons living in the Amboseli basin of Kenya. Freshly-dropped samples were collected within minutes when a known animal was observed defecating. These samples were collected under protocols approved by the Duke University Institutional Animal Care & Use Committee. Each sample was homogenized and then stored in 95% ethanol at a 2.5:1 ratio of ethanol to feces for transportation to the University of Nairobi. There the ethanol was allowed to evaporate and then the samples were stored at -20°C until freeze-drying at 30mTorr to below -50°C. Samples were then sifted and stored at -80°C until processing for elemental analysis. Gut length was extracted from data on olive baboon (Papio anubis)52.
Fresh fecal samples from Gunther’s dik dik (Madoqua guentheri, n=10), impala (Aepyceros melampus, n=10), domestic Boran cattle (Bos indicus, n=10), cape buffalo (Syncerus caffer, n=10), plains zebra (Equus quagga, n=10), Grevy’s zebra (Equus grevyi, n=9), African elephant (Loxodonta africana, n=10), southern white rhinoceros (Ceratotherium simum, n=5), vervet monkey (Chlorocebus pygerythrus, n=1), eastern black rhinoceros (Diceros bicornis, n=7), reticulated giraffe (Giraffa caemlopardalis reticulata, n=5), hippopotamus (Hippopotamus amphibious, n=5), crested porcupine (Hystrix cristata, n=2), white-tailed mongoose (Ichneumia albicauda, n=1), waterbuck (Kobus ellipsiprymnus, n=1), warthog (Phacochoerus africanus, n=6), rock hyrax (Procavia capensis, n=2), and aardwolf (Proteles cristata, n=1) were collected at the Mpala Research Centre and Conservancy in central Kenya. These species minus aardwolf and white-tailed mongoose make up the East African herbivores and omnivores included in Fig. 1b and 1c. All individuals were adults but data on their sex was not uniformly available. All samples were collected under protocols approved by the Princeton University Institutional Animal Care & Use Committee. We obtained adult gut-length estimates from the literature52–54 for a phylogenetically diverse subset of these species: aaardwolf, vervet monkey, hyrax, dik-dik, plains zebra, black rhinoceros, hippopotamus, giraffe, elephant, and cattle, as well as for the brush-tailed porcupine Atherurus africanus (as a proxy H. cristata). We used only African Bos indicus data and not North American cattle (Bos taurus) for analyzing C:N relationships because the former were free ranging.
Domestic sheep (Ovis aries, n=10), horse (Equus ferus caballus, n=4), and domestic cattle (Bos taurus, n=10) samples were obtained from adult animals on farms in New Jersey, USA. The sex of each individual was not available. Samples were collected non-invasively and were not subject to an animal care protocol. Gut length was identified for sheep52 and horse54.
Meerkat (Suricata suricatta, n=9) samples were collected from a wild population in South Africa’s Kalahari desert. Adults of both sexes were included. All samples were collected under protocols approved by the Duke University Institutional Animal Care & Use Committee.
Healthy human subjects (n=5), who reported no use of antibiotics in the month prior to enrollment, provided stool samples. Informed consent was obtained from all subjects and the protocol, approved by the Duke Health Institutional Review Board, complied with relevant ethical obligations. Subjects collected samples by placing disposable commode specimen containers (Fisher Scientific, Waltham, MA) under their toilet seats before bowel movements. Intact stool samples (~10g) were briefly stored in personal −20°C freezers before transport to the laboratory for long-term storage at -80°C in sterile collection tubes. Human gut-length data were obtained from a published report52.
Adult snowshoe hares (Lepus americanus, n=15), collected from wild populations in Washington and Montana, were kept in a photoperiod- and temperature-controlled research facility at the NCSU College of Veterinary Medicine. The sex of each individual was not recorded at time of collection. All animals are kept under protocols approved by the North Carolina State University Institutional Animal Care & Use Committee. Samples were collected within 8 hours of defecation and frozen at -20°C. Gut length data were obtained for European rabbit (Oryctolagus cuniculus)52.
Grey mouse lemur (Microcebus murinus, n=8) and aye-aye (Daubentoia madagascariensis, n=4) adults were housed in a breeding colony at the Duke Lemur Center. Fecal samples from mouse lemurs were collected fresh during regular technician handling during the non-torpor season then stored at -80°C. Fecal samples from aye-ayes were collected after an individual was observed defecating then stored at -80°C. The sex of each individual was not recorded at time of collection. All animals are kept under protocols approved by the Duke University Institutional Animal Care & Use Committee.
Prairie vole (Microtus ochrogaster, n=10) adults (4-5 months old) were sampled from a breeding population housed at NCSU for genetic and behavioral studies. Voles of both sexes in either single or group housing had feces collected fresh during normal technician handling. The sex of each individual was not recorded at time of collection. All animals are kept under protocols approved by the North Carolina State University Institutional Animal Care & Use Committee. Gut-length data were obtained for meadow vole (Microtus pennsylvanicus)52.
Dog samples (Canis lupus familiaris, n=5) were collected from a genetic model population of glycogen storage disease. Samples were collected fresh following feeding and then were prepared immediately for analysis. All individuals were adults but data on their sex was not recorded at collection. All animals are kept under protocols approved by the Duke University Institutional Animal Care & Use Committee. Gut length data were extracted from a previous report54.
Wild-type mouse (Mus musculus) data were extracted from baseline, control animals (n=10) in the antibiotic experiments (see below). Gut-length data were extracted from the control animals in a published report55.
To determine the impact of physiology on mammal species fecal C:N, we performed an analysis of covariance (ANCOVA). Individual physiological data were not available, so species mean C:N was calculated. Log large intestine length, gut physiology (simple, hindgut fermenter, or ruminant56), and total body length were extracted from the literature and included as predictor variables as such: C:N~log10(total body length)+log10(large intestine length)+gut physiology. Interaction terms were not found to be significant (p>0.05) and so were not included in the model. This test and all other statistical tests were carried out in R (R core team, ver. 3.3). All statistical tests performed were non-parametric except where a Shapiro-Wilks test indicated that data were normally distributed, in which case parametric tests were used.
Mouse whole-gut samples
We humanely sacrificed untreated wild-type mice and immediately removed their complete gastrointestinal tract. For total gut content analyses (n=10), lumen contents and mucosa were scraped from the proximal small intestine, the distal small intestine, the cecum, and the large intestine, and then immediately dried.
DNA from total gut samples was extracted using the MoBio PowerSoil extraction kit. To estimate total bacterial abundance, quantitative PCR (qPCR) was performed on fecal DNA using the following primers: forward, 5’-ACTCCTACGGGAGGCAGCAGT-3’, reverse, 5’-GTATTACCGCGGCTGCTGGCAC-3’57. qPCR assays were run using SYBR FAST qPCR Master Mix (KAPA) on a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). Cycle-threshold values were standardized against a dilution curve of known concentration and then adjusted for the weight of fecal matter extracted.
C:N analyses
All samples were dried to constant weight at 72°C in a vacuum oven and then ground and homogenized. Samples were packed into aluminum cups and processed on a Carlo Erba (Lakewood, NJ) Elemental Analyzer with zero-blank autosampler except for Kenyan and New Jersey samples which were analyzed at the University of California Santa Cruz Stable Isotope Facility (Dumas combustion in a Carlo Erba 1108 elemental analyzer coupled to a ThermoFinnigan (San Jose, CA) Delta Plus XP isotope ratio mass spectrometer) or the Duke Environmental Stable Isotope Laboratory (ThermoFinnigan MAT Delta Plus paired with a Carlo Erba elemental analyzer equipped with a zero blank auto-sampler). Fecal C:N measurements were conducted on whole feces which includes microbial cells, dietary material, host secretions, and sloughed host cells.
Reconstructing wild mammal diets with DNA metabarcoding
Grass typically has a higher C:N than woody plants and other herbs27 and so a diet rich in grass is expected to generate higher fecal C:N. Herbivore dietary composition was reconstructed using DNA metabarcoding58 on DNA extracted from the same fecal samples used for elemental analysis (following methods described in Kartzinel (2015)28). In short, DNA was extracted using the Zymo Xpedition Soil/Fecal Mini Kit and the composition of dietary plant DNA was quantified using targeted amplicon sequencing of the chloroplast trnL-P6 marker58,59. Thermocycling included denaturing at 95°C for 10 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and ending with a 2 min extension at 72°C. Sequence demultiplexing, quality control, and identification were performed with obitools and ecoPCR (see 28 for detailed processing information). Grass consumption by each herbivore was estimated using as the relative read abundance (RRA) of the grass family, Poaceae, (i.e., the proportion of all trnL-P6 sequence reads that were identified as grasses relative to non-grasses). These data are strongly correlated with δ13C enrichment (P<0.001, ρ=0.88, Spearman correlation), a well-established proxy for the consumption of C4 grasses relative to C3 plants (i.e., trees, shrubs, and herbs) by herbivores in African savannas28,60.
The present analysis supplements data available from Kartzinel et al. (2015) in multiple ways. First, 4-5 additional fecal samples were collected from each of the 7 wild herbivores at the same study site in Kenya during October 2014, and these were analyzed for C:N and DNA. Second, an additional set of 1-7 samples from 11 new species were collected from this Kenya study system in July 2016; all were analyzed for C:N and 9 were analyzed for DNA (excluding white-tailed mongoose and aardwolf, which are carnivores). Third, all new DNA metabarcode data were combined with raw data used in the analyses of Kartzinel et al. (2015) and reanalyzed in conjunction with improvements in local plant DNA reference library used to identify trnL-P6 DNA sequences—now including 1,828 fertile plant vouchers (~442 species). For the subsets of samples included in both analyses estimates of grass RRA were strongly correlated (R2 = 0.99). Altogether, we included estimates of grass abundance and C:N for 125 mammals (N=1-10 per species, mean=6) with both measurements taken from a single fresh fecal sample collected from each individual.
Gut isolate C:N
Published average values of bacterial C:N ratios (4.67±1.38) include only measurements of bacteria isolated from ocean and soil environments24,25. Here, we collected clonal population C:N values for 35 strains of 26 species of bacteria (Supplementary InformationTable 1). These species represent the five most abundant phyla in the mammalian gut56.
Bacterial strains were isolated from a single human fecal donor or obtained from a commercial strain collection of gut isolates (ATCC; Supplementary Information Table 1). Human gut isolates were identified with almost complete 16S rRNA gene Sanger sequencing (using primers 27-f and 1492-r numbered according to E. coli reference (Frank et al. 2008)). Samples were cultured anaerobically at 37°C on blood agar plates and checked for growth at 24 and 48 hours. These were then used to inoculate 5mL of modified Gifu Anaerobic Broth (mGifu; Gifu Anaerobic Medium (HiMedia Laboratories, Mumbai, India), 2mL/L hemin, 5mL/L menadione)61. This media has a C:N ratio of 3.77±0.01. After 24 hours of anaerobic incubation at 37 °C, these liquid cultures were used to inoculate sterile 250 mL bottles containing 150mL of mGifu. After another 24 hours of anaerobic incubation at 37 °C, OD600 levels of the cultures were measured using an Eppendorf BioPhotometer (Hamburg, Germany). If the OD600 levels did not exceed 1, cultures were re-incubated anaerobically for 24 hours. Once the OD600 levels exceeded 1, the bottle was removed from the incubator and centrifuged at 5000-7000 rpm for at least 10 minutes in an ultracentrifuge. The resulting supernatant was decanted. The pelleted cells were dried at 72 °C for 48 hours to a constant weight. The dried cells were then measured with a Carlo Erba Elemental Analyzer with zero-blank autosampler.
Dietary Protein Manipulation
Mouse diets
Conventional mice fed standard chow (LabDiet Picolab 5053 irradiated diet; 20% protein) were weighed and fresh fecal samples were collected before the initiation of the study. Mice were then randomly assigned a treatment and separated into cages, housing pairs which received the same treatment (n=10mice/treatment, n=5 cages/treatment). The sample size was chosen following a power analysis to allow for β less than 0.1. Treatments consisted of isocaloric diets which varied casein protein levels: low (6%; Envigo TD.90016), control (20%; TD.91352), and high (40%; TD.90018). Mice were allowed to feed ad libitum on these diets for the rest of the experiment. Fecal samples and mouse weights were collected 1, 2, 7, and 14 days after initiation of the diet (see Supplementary Information Fig. 1d). All fecal samples were immediately frozen. These samples were then used for C:N analysis and 16S rRNA gene qPCR as above (see C:N Analyses and Whole Gut Samples, respectively).
Microbial amplicon sequencing
We also performed 16S rRNA gene amplicon sequencing on mouse fecal samples throughout the dietary intervention to determine compositional responses to changes in nitrogen input. We performed sequencing using custom barcoded primers62 and published protocols62–64. DNA was extracted from frozen samples using the MoBio (Carlsbad CA) PowerSoil DNA extraction kit. Sequencing was conducted on an Illumina MiniSeq with paired end 150bp reads. The absolute abundance of the Bacteroidaceae was estimated by multiplying the total 16S rRNA gene copy number for a sample by the relative abundance of Bacteroidaceae identified by amplicon sequencing.
Antibiotic Treatment to Alter Microbial Load
In vivo nitrogen dynamics under antibiotics
Baseline fecal samples were collected at least twenty-four hours before the first dose and then mice were placed in individual housing with Supplementary Information enrichment and continued on their standard diet (LabDiet Picolab 5053 irradiated diet; 20% protein). Mice were orally gavaged with either 0.25ml autoclaved deionized water (control, n=10) or 0.25ml of an antibiotic cocktail (treated, n=10) daily for five days then tracked for one week following the end of treatment. Mice were weighed daily. The mice were randomly assigned a group with an equal number of mice in each group, and researchers collecting data were blinded to the groupings until after the final dose was administered. The sample size was chosen following a power analysis to allow for β less than 0.1.
The antibiotic cocktail consisted of ampicillin (Gold Biotechnology, St. Louis, MO) 1mg/ml, vancomycin (Alfa Aeser, Haverhill, MA) 5mg/ml, neomycin (EMD Millipore, Billerica, MA) 10mg/ml, and metronidazole (Alfa Aeser, Haverhill, MA) 10mg/ml (after Reikvam et al. (2011)33). Fresh antibiotic cocktails were prepared every day. Throughout the experiment freshly voided fecal samples were collected and stored at -80°C for downstream analysis. Load, as measured by 16S rRNA gene qPCR, was significantly reduced within 24 hours and remained low throughout treatment (P<0.05, Bonferroni-corrected Mann-Whitney U tests; Supplementary Information Fig. 3a). Fecal C:N ratio was measured with an elemental analyzer as above (C:N analyses).
We performed linear mixed effects analysis to determine the effects of antibiotics on C:N, microbial load, and mouse weight. As fixed effects, we entered antibiotic treatment and time with an interaction term into the model. We included mouse identity as a random effect. P values were obtained by likelihood ratio tests comparing the full model against a model including only time and mouse identity and were performed with the ‘anova’ function in the ‘lme4’ package65.
Isotopic labeling
For an additional cohort of mice, a stable isotope tracer experiment was performed on the last day of antibiotic treatment using the above protocol. Four hours before euthanasia on the final day of treatment, mice treated with 1.8μmol 98at% 15N/13C threonine in autoclaved deionized water via a 50μl lateral tail vein injection38. This treatment allowed nitrogen secretion into the gut to be conservatively estimated by measuring heavy label delivery. Immediately following euthanasia, the whole large intestine was removed and sectioned into equal thirds, longitudinally, and the distal section was used for isotope measurements. Total gut contents (mucus and lumen) were scraped out and dried. Colon epithelial tissue was dried separately. Samples were homogenized and ground for δ 15N quantification at the Duke Environmental Isotope Laboratory.
RNA Isolation and RT-PCR
Host mucin production was quantified as Muc2 expression (Muc2 is the major murine intestinal mucin66) measured in feces. Total RNA was isolated from fecal pellets stored in RNALater (Thermo Fisher, Waltham, MA) using the MoBio PowerMicrobiome RNA Isolation kit with an added phenol chloroform extraction step. ≤15ng RNA was reverse transcribed using cDNA Prep Reverse Transcription Master Mix (Fluidigm, South San Francisco, CA) following manufacturer’s instructions. Target transcripts were preamplified for 18 cycles and then diluted 10x. RT-PCR was performed using a BioMark (Fluidigm) on a 48 x 48 chip with Taqman Fast Advanced Master Mix (Thermo Fisher). Three ERCC RNA Spike-in Mix (Thermo Fisher) positive controls and a nontarget negative control of nuclease-free water were also run on the chip. Muc2 expression levels were normalized to mouse Actb (ΔCt) expression for each time point for each mouse and then compared to the average of control mice for that time point (ΔΔCt). Fold change (2^ΔΔCt) is presented in Fig. 2c.
Mucus Thickness Measurements
The proximal third of the large intestine from mice sacrificed on the final day of antibiotic treatment was stored in a tissue cassette for mucus thickness measurements. Tissue samples were fixed in Carnoy’s solution (ethanol6:acetic acid3:chloroform1, v/v/v) for four hours before being moved to 70% ethanol solution. Paraffin sections were then prepared, sectioned transversely, and stained with Alcian blue stain to highlight mucus. Mucus thickness was measured on a light microscope with 8-10 fields measured and averaged per sample. Thickness measurements were carried out in a blinded fashion and only in regions where the mucus layer was flanked on the luminal edge by intestinal contents. Records of how many measurements were made for each sample are provided in Supplementary Information Table 7.
15N Tracer Experiments
Mass Balance Studies
Stable isotope tracer experiments were performed to quantify nitrogen delivery from the host via dietary and secreted pathways. The dietary group (n=6 per time point) received a 25ul gavage of a dilute heavy-labeled mouse chow solution (0.01g chow per 1ml PBS) where the chow had all nitrogen sourced from Spirulina cells (Cambridge Isotope Laboratories, Tewksbury, MA). This chow can be fed to rodents for extended periods of time without adverse health impacts67,68 and is expected to provide a similar nitrogen content as conventional chow. The secretion group (n=6 per time point) received 1.8umol 15N/13C threonine in autoclaved deionized water via a 50μl lateral tail vein injection38. The control group (n=6) received a 14N/12C L-threonine (Sigma-Aldrich) injection. 6 mice per experimental treatment group were euthanized at four hours after treatment and another six mice were euthanized per experimental treatment group at six hours after treatment. All six control mice were euthanized after four hours. Mice were kept in single housing with enrichment and all fecal pellets were collected from the time of treatment through euthanasia. The mice were randomly assigned a group, but researchers were not blinded to treatment during sample collection. The sample size was chosen following a power analysis to allow for β less than 0.1. The short time span of the experiment was chosen to allow for appearance of dietary material in the gut while still minimizing the likelihood of re-secretion of diet delivered 15N by limiting the time for host processing of dietary material. Nevertheless some 15N made available to the host through the Spirulina chow may have nonetheless appeared in the gut after uptake and then subsequent re-secretion. This phenomenon would only lessen differences between the two experimental groups, however; it would not result in overestimating the importance of secretions.
Immediately after euthanasia, the gut contents were dissected out. Lumen material and then mucosa material were scraped from the large intestine, cecum, and small intestine of each mouse. Total material from each of these compartments and the epithelium were dried to a constant weight at 72°C then weighed. These samples were then ground and homogenized to prepare as above for C:N analysis and sent to the Duke Environmental Isotope Laboratory for isotope enrichment analysis. Overall C:N was not affected by treatment (P=0.13, Bonferroni-corrected Mann-Whitney U tests; Supplementary Information Fig. 4c) but did vary between compartments and layers (P<0.001, Kruskal-Wallis tests; Supplementary Information Fig. 4d,e).
Atom percent excess 15N was calculated for each sample. We also calculated atom percent excess 15N as a function of total label administered to determine the relative nitrogen allocation to gut site or microbial cells. First, atom percent 15N was calculated wherein for an element X, with heavy isotope H, and light isotope: at %H X = H/(H+L) * 100; in percent (%). Atom percent excess was then calculated as the difference between the atom percent of a treated sample and the average atom percent of all control (unlabeled) samples. Atom percent excess was then divided by the total 15N label administered (atom percent label multiplied by %N and weight) to correct for differences in total 15N added by the dietary and injection routes. Patterns were not meaningfully different between the four and six hour time points and so only large intestine, four hour data is presented in the main text. All results can be found in Supplementary Information Fig. 4a.
Germfree studies
The same protocol as above was followed for quantification of nitrogen allocation in germfree mice with only minor deviations as outlined below. 15 male C57BL/6 mice over the age of 8 weeks, derived and maintained in the Duke Gnotobiotic facility, were allocated into three treatment groups (control, dietary, secreted; n=5/group). Sample size was determined by the availability of age and sex matched C57BL/6 mice. Mice received treatment as outlined in Mass Balance Studies above and were maintained in sterile containers with Supplementary Information hydration under a fume hood for the four hour study period. Feces was collected and quantified per group for each hour following treatment. Mice were sacrificed after four hours, and total guts were harvested and processed as above. Atom percent excess was calculated for each gut section (small intestine, cecum, large intestine) and tissue type (mucus, lumen, and epithelium). Mucus was the attached portion, generally of host derivation, whereas the lumen samplers were the loose contents that ultimately get integrated into fecal pellets. Epithelium was all host tissue. Germfree versus conventional mouse isotope data were analyzed with a mixed-design ANOVA in the ‘ez’ package in R. Mouse was the case identifier with gut layer (epithelium, mucus, and lumen) and compartment (small intestine, cecum, large intestine) as predictor variables which varied within cases. Germfree status was included as a predictor variable which varied between cases. Greenhouse-Geisser epsilon corrections were used for variables that did not pass Mauchly’s tests for sphericity.
For analysis of fecal C:N (Supplementary Information Fig. 5a), germfree C57BL/6 mouse samples were collected in a sterile manner during regular technician handling from the National Gnotobiotic Rodent Research Center at University of North Carolina at Chapel Hill and from the Duke Gnotobiotic facility. Samples were stored temporarily at -20°C before being moved to -80°C for long-term storage.
Single Cell Study Tracers
To quantify microbial utilization of host and dietary nitrogen, we performed a slightly different stable isotope tracer experiment than for the Mass Balance Studies above (Supplementary Information Fig. 7a). All mice were fasted overnight and then offered chow with all protein sourced from Spirulina cells (Cambridge Isotope Laboratories) for one hour before being returned to normal mouse chow for four hours before euthanasia. The dietary treatment group (n=10) received one hour of chow with 15N labeled Spirulina and no other treatment. This setup allowed for greater label delivery than could be achieved via gavage of chow in solution as performed above. The secretion group (n=10) were offered 14N Spirulina chow for one hour before receiving 1.8umol 15N/13C threonine in autoclaved deionized water via a 50μl lateral tail vein injection38. The control group (n=10) received 14N chow and a 14N/12C L-threonine (Sigma-Aldrich) injection. The mice were randomly assigned a group, but researchers were not blinded to treatment during sample collection. The sample size was chosen following a power analysis to allow for β less than 0.1.
Immediately after euthanasia, the gut contents were dissected out and all lumen and mucosal material was scraped from the large intestine and small intestine. Samples were homogenized for each compartment for each mouse. Of these gut contents, 2/3 were immediately frozen for downstream sequencing and elemental analysis; 1/3 were fixed in 2% paraformaldehyde overnight at 4°C and then washed in PBS before being stored at -20°C in 60% ethanol/40% PBS until preparation for FISH.
Bulk Analyses
We prepared 16S rRNA gene amplicons as above but the sequencing was performed on an Illumina MiSeq with 250 paired end reads and the V2 kit at the Duke Molecular Physiology Institute. The experimental procedures did not result in significant differences in microbial community composition (P>0.05, PERMANOVA computed with ‘adonis’ function in the ‘vegan’ package; Supplementary Information Fig. 7d). We did observe differences between the large intestine and small intestine composition across all treatment groups (R2=0.60 P=0.001, PERMANOVA).
Samples were prepared as above for C:N analysis and sent to the Cornell Isotope Laboratory for isotope enrichment analysis on a ThermoFinnigan MAT Delta Plus paired with a Carlo Erba NC2500 elemental analyzer equipped with a low blank AS200 auto-sampler. Overall, both treatments produced significant bulk enrichment relative to 14N controls (P<0.001, Kruskal-Wallis test; Supplementary Information Fig. 7c) as well as significant cellular enrichment (P<0.001, Kruskal-Wallis test; Supplementary Information Fig. 7b), indicating that they could be used to track microbial uptake. Similar to the compositional patterns, bulk C:N measurements were not affected by treatment (P=0.09, Kruskal-Wallis test) but did vary between compartments (P=0.02, Mann-Whitney U test; Supplementary Information Fig. 7e).
Single Cell Analyses
Duplicate samples, randomly selected from each treatment group, were chosen for single cell analysis. Flushed gut contents fixed with 4% formaldehyde stored in a 60% ethanol/40% PBS solution were used for fluorescence in situ hybridization (FISH) and nano-scale resolution secondary ion mass spectrometry (NanoSIMS) imaging. FISH was performed with fluorescently-labeled rRNA-targeted oligonucleotide probes38 Bac303 (S-*-Bacto-0303-a-A-17-Cy3, 5’-CCA ATG TGG GGG ACC TT -3’) and Erec482 (S-*-Erec-0482-a-A-179-Cy5; GCT TCT TAG TCA RGT ACC G) (see Supplementary Information Table 3) using a standard protocol69. To evaluate potential non-specific FISH probe binding, parallel samples were hybridized with the reverse complement of the bacterial probe EUB338 for all used dyes (NONEUB-5’-ACTCCTACGGGAGGCAGC-3’; 38). Samples were subsequently stained with DAPI (1 μg/mL; Sigma–Aldrich) for 5 min. Hybridized, DAPI-stained samples were imaged and marked on an epifluorescence laser microdissection microscope (LMD, Leica LMD 7000) as previously described38.
NanoSIMS measurements were performed on an NS50L (Cameca, Gennevilliers France) at the University of Vienna, Austria. Data were recorded as multilayer image stacks obtained by sequential scanning of a finely focused Cs+ primary ion beam (ca. 80 nm spot size with 2 pA beam current) and detection of negative secondary ions and secondary electrons. Recorded images had a 512 x 512 pixel resolution and a field-of-view ranging from 47 x 47 to 72 x 72 µm2. The mass spectrometer was tuned to achieve a mass resolving power (MRP) of > 10.000 (according to Cameca's definition) for detection of C2- and CN- secondary ions. Prior to data acquisition, which was performed as long-runs for sampling of entire cells, analysis areas were gently pre-sputtered by application of a Cs+ dose density in the range from 2,3E15 to 7,0E15 at/cm2. All images were recorded with a dwell time of 5-10 msec/pixel/cycle and accumulation of 23 to 30 cycles per image.
NanoSIMS images were processed using the WinImage software package (Cameca). Cells were identified in drift-corrected, stack-accumulated NanoSIMS images and manually verified with aligned FISH images (see Supplementary Information Fig. 6 for representative images). Cells which overlapped or were otherwise indistinguishable were not measured. Signal intensities were corrected for detector dead time on a per-pixel basis and quasi-simultaneous arrival (QSA) of C2- and CN- secondary ions on a per-ROI basis. The QSA correction was performed according to the formalism suggested by previous work70, applying sensitivity factors of 1,06 and 1,05 for C2- and CN- ions, respectively (experimentally determined on dried yeast cells). 15N/(14N + 15N) isotope fractions, designated as atom percent (at%) 15N throughout the text, were calculated from 15N/(14N + 15N) = 12C15N-/(12C14N- + 12C15N-). Summary statistics from each region of interest were calculated for single-cell analysis. A single field of view was collected for each treatment with technical replicates of 91–162 cells per field of view. Individual cells were considered significantly enriched in 15N if the mean cellular at% 15N was greater than 5 standard deviations above the mean at% 15N of the unlabeled control cells and if the measurement error (5σ, Poisson) was smaller than the difference between the at% of the labeled cell and the mean at% of unlabeled control cells. The Poisson error (random measurement error due to counting statistics) was calculated by where L- and H- refer to the signal intensity (in counts) associated with the light and heavy isotope, respectively, and n = 1 for detection of CN- and n = 2 for detection of C2- secondary ions. Significant enrichment of cells relative to controls was documented from both labeling delivery paths (P<0.05, Mann-Whitney U tests; Supplementary Information Fig. 7b).
Supplementary Material
Acknowledgments
C:N measurements were carried out by Will Cook in the Duke Environmental Isotope Laboratory. Samples were provided by Scott Mills and Diana Lafferty (snowshoe hare); Erin Ehmke (lemurs); Lisa McGraw, Andrea Vogel, and Caitlin Clement (prairie vole); Dwight Koeberl, Vanessa Sakach, and Lindsey Morgan (dog); Christine Drea (meerkat). Statistical advice was provided by Kingshuk Choudhury and Sayan Mukherjee. The manuscript was improved thanks to comments from Jim Heffernan, John Rawls, and Peter Turnbaugh.
This work was funded by a NSF Doctoral Dissertation Improvement grant to ATR, JPW and LAD (DEB-1501495) and grants from the Hartwell Foundation, Alfred P. Sloan Foundation, and Searle Scholars Program to LAD. ATR was supported by the NSF Graduate Research Fellowship Program under Grant No. DGE 1106401. FCP was supported by a European Research Council Marie Curie Individual Fellowship (grant No.658718). DB was supported in part by Austrian Science Fund (P26127-B20 and P27831-B28) and European Research Council (Starting Grant: FunKeyGut 741623). MW was supported by the European Research Council via the Advanced Grant project “NITRICARE 294343”. The contents are of this paper solely the responsibility of the authors and do not necessarily represent the views of the funding institutions.
Footnotes
Data availability
The 16S rRNA gene nucleotide sequences generated in this study can be downloaded from the European Nucleotide Archive under study accession numbers PRJEB26478 (protein manipulation and NanoSIMS experiments) and PRJEB26446 (antibiotics experiment). NanoSIMS and bulk isotopic data for the dietary and injected 15N study is included in Supplementary Information Table 4. Other data that support these findings are available from the corresponding author upon request.
Author Contributions: ATR, FP, AS, DB, and MW carried out FISH/NanoSIMS work. ATR, XZ, and RP performed diet manipulation experiments. LPH, SJ and HKD processed samples. TMO, SCA, TRK, and RMP contributed data. ATR performed all other experiments. AMD, RRD, and JPW were involved in study design. ATR and LAD designed the study, analyzed data, and wrote the paper. All authors discussed the results and commented on the manuscript.
Conflict of interest: The authors declare no conflict of interest.
References
- 1.McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;1145:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 2.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nature Reviews Microbiology. 2011;9:119–130. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 4.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nature Reviews Immunology. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 5.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 6.Fuller MF, Reeds PJ. Nitrogen cycling in the gut. Annu Rev Nutr. 1998;18:385–411. doi: 10.1146/annurev.nutr.18.1.385. [DOI] [PubMed] [Google Scholar]
- 7.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 8.Carmody RN, Turnbaugh PJ. Gut Microbes Make for Fattier Fish. Cell Host Microbe. 2012;12:259–261. doi: 10.1016/j.chom.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borgstrom B, Dahlqvist A, Lundh G, Sjovall J. Studies of intestinal digestion and absorption in the human. J Clin Invest. 1957;36:1521–1536. doi: 10.1172/JCI103549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elser JJ, et al. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett. 2007;10:1135–1142. doi: 10.1111/j.1461-0248.2007.01113.x. [DOI] [PubMed] [Google Scholar]
- 11.Gardner MLG. Gastrointestinal absorption of intact proteins. Annu Rev Nutr. 1988;8:329–350. doi: 10.1146/annurev.nu.08.070188.001553. [DOI] [PubMed] [Google Scholar]
- 12.Ferraris RP, Carey HV. Intestinal transport during fasting and malnutrition. Annu Rev Nutr. 2000;20:195–219. doi: 10.1146/annurev.nutr.20.1.195. [DOI] [PubMed] [Google Scholar]
- 13.Lilburn TG, et al. Nitrogen fixation by symbiotic and free-living spirochetes. Science. 2001;292:2495–2498. doi: 10.1126/science.1060281. [DOI] [PubMed] [Google Scholar]
- 14.Vecherskii MV, Naumova EI, Kostina NV, Umarov MM. Some specific features of nitrogen fixation in the digestive tract of the European beaver (Castor fiber) Dokl Biol Sci. 2006;411:452–454. doi: 10.1134/s001249660606007x. [DOI] [PubMed] [Google Scholar]
- 15.Wostman BS. The germfree animal in nutritional studies. Annu Rev Nutr. 1981;1:257–279. doi: 10.1146/annurev.nu.01.070181.001353. [DOI] [PubMed] [Google Scholar]
- 16.Sterner RW, Elser JJ. Ecological Stoichiometry: The Biology of Elements from Molecules to the Biosphere. Vol. 464 Princeton University Press; 2002. [Google Scholar]
- 17.Frost PC, Elser JJ. Growth response of littoral mayflies to the phosphorus content of their food. Ecol Lett. 2002;5:232–240. [Google Scholar]
- 18.Elser JJ, et al. Growth rate–stoichiometry couplings in diverse biota. Ecol Lett. 2003;6:936–943. [Google Scholar]
- 19.Fink P, Von Elert E. Physiological responses to stoichiometric constraints: nutrient limitation and compensatory feeding in a freshwater snail. Oikos. 2006;115:484–494. [Google Scholar]
- 20.Simpson SJ, Raubenheimer D. The Nature of Nutrition: A Unifying Framework from Animal Adaptation to Human Obesity. Princeton University Press; 2012. [Google Scholar]
- 21.Le Couteur D, et al. The influence of macronutrients on splanchnic and hepatic lymphocytes in aging mice. Journals of Gerontology Series A: Biomedical Sciences and Medical Sciences. 2014;70:1499–1507. doi: 10.1093/gerona/glu196. [DOI] [PubMed] [Google Scholar]
- 22.Solon-Biet SM, et al. Macronutrient balance, reproductive function, and lifespan in aging mice. PNAS. 2015;112:3481–3486. doi: 10.1073/pnas.1422041112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes AJ, et al. Diet-microbiome interactions in health are controlled by intestinal nitrogen source constraints. Cell Metabolism. 2017;25:140–151. doi: 10.1016/j.cmet.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Zimmerman AE, Allison SD, Martiny AC. Phylogenetic constraints on elemental stoichiometry and resource allocation in heterotrophic marine bacteria. Environ Microbiol. 2013;16:1398–1410. doi: 10.1111/1462-2920.12329. [DOI] [PubMed] [Google Scholar]
- 25.Mouginot C, et al. Elemental stoichiometry of Fungi and Bacteria strains from grassland leaf litter. Soil Biology & Biochemistry. 2014;76:278–285. [Google Scholar]
- 26.Stephen AM, Cummings JH. The microbial contribution to human faecal mass. J Med Microbiol. 1980;13:45–56. doi: 10.1099/00222615-13-1-45. [DOI] [PubMed] [Google Scholar]
- 27.Tjoelker MG, Craine JM, Wedin D, Reich PB, Tilman D. Linking leaf and root trait syndromes among 39 grassland and savannah species. New Phytol. 2005;167:493–508. doi: 10.1111/j.1469-8137.2005.01428.x. [DOI] [PubMed] [Google Scholar]
- 28.Kartzinel TR, et al. DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. PNAS. 2015;112:8019–8024. doi: 10.1073/pnas.1503283112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleynhans EJ, Jolles AE, Bos M, Olff H. Resource partitioning along multiple niche dimensions in differently sized African savanna grazers. Oikos. 2011;120:591–600. [Google Scholar]
- 30.Müller DWH, et al. Assessing the Jarman–Bell Principle: Scaling of intake, digestibility, retention time and gut fill with body mass in mammalian herbivores. Comparative biochemistry and physiology. Part A, Molecular & integrative physiology. 2013;164:129–140. doi: 10.1016/j.cbpa.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Hirakawa H. Coprophagy in leporids and other mammalian herbivores. Mamm Rev. 2001;31:61–80. [Google Scholar]
- 32.Kashyap PC, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013;144:967–977. doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reikvam DH, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wlodarska M, et al. Antibiotic Treatment Alters the Colonic Mucus Layer and Predisposes the Host to Exacerbated Citrobacter rodentium-Induced Colitis. Infect Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson ME, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. PNAS. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansson ME, et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015;18:582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. 2015;6 doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berry D, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. PNAS. 2013;110:4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Frontiers in Genetics. 2015;6 doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.den Besten G, et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carey HV, Walters WA, Knight R. Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. AJP: Regulatory, Integrative and Comparative Physiology. 2013;304:R33–R42. doi: 10.1152/ajpregu.00387.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costello EK, Gordon JI, Secor SM, Knight R. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J. 2010;4:1375–1385. doi: 10.1038/ismej.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. PNAS. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43–51. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaspari M, Powers JS. Biogeochemistry and geographical ecology: embracing all twenty-five elements required to build organisms. Am Nat. 2016;188:S62–S73. doi: 10.1086/687576. [DOI] [PubMed] [Google Scholar]
- 49.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res. 2013;69:52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 51.Smith VH, Tilman GD, Nekola JC. Eutrophication: impacts of excess nutrient inputs on freshwater, marine, and terrestrial ecosystems. Environ Pollut. 1999;100:179–196. doi: 10.1016/s0269-7491(99)00091-3. [DOI] [PubMed] [Google Scholar]
- 52.Stevens CE, Hume ID. Comparative Physiology of the Vertebrate Digestive System. 2 edn. Cambridge University Press; 1995. [Google Scholar]
- 53.Pérez W, Lima M, Clauss M. Gross anatomy of the intestine in the giraffe (Giraffa camelopardalis) Anatomia Histologia Embryologia. 2009;38:432–435. doi: 10.1111/j.1439-0264.2009.00965.x. [DOI] [PubMed] [Google Scholar]
- 54.Kararli TT. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharaceutics & Drug Disposition. 1995;16:351–380. doi: 10.1002/bdd.2510160502. [DOI] [PubMed] [Google Scholar]
- 55.Aust G, et al. Mice overexpressing CD97 in intestinal epithelial cells provide a unique model for mammalian postnatal intestinal cylindrical growth. Molecular Biology of the Cell. 2013;24:2256–2268. doi: 10.1091/mbc.E13-04-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bergström A, et al. Introducing GUt Low-Density Array (GULDA) - a validated approach for qPCR-based intestinal microbial community analysis. FEMS Microbiol Lett. 2012;337:38–47. doi: 10.1111/1574-6968.12004. [DOI] [PubMed] [Google Scholar]
- 58.Taberlet P, et al. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007;3:e14. doi: 10.1093/nar/gkl938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Barba M, et al. DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet. Molecular Ecology Resources. 2014;14:306–323. doi: 10.1111/1755-0998.12188. [DOI] [PubMed] [Google Scholar]
- 60.Cerling TE, Harris JM, Passey BH. Diets of East African Bovidae based on stable isotope analysis. J Mammal. 2003;84:456–470. [Google Scholar]
- 61.Rettedal EA, Gumpert H, Sommer MO. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat Commun. 2014;5:4714. doi: 10.1038/ncomms5714. [DOI] [PubMed] [Google Scholar]
- 62.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics Shape the Physiology and Gene Expression of the Active Human Gut Microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bates D, Maechler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software. 2015;67:1–48. [Google Scholar]
- 66.Park SW, et al. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. PNAS. 2009;106:6950–6955. doi: 10.1073/pnas.0808722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McClatchy DB, Dong M-Q, Wu CC, Venable JD, Yates JR., III 15N metabolic labeling of mammalian tissue with slow protein turnover. Journal of Proteome Research. 2007;6:2005–2010. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McClatchy DB, Liao L, Park SK, Venable JD, Yates JR., III Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 2007;17:1378–1388. doi: 10.1101/gr.6375007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daims H, Stoecker K, Wagner M. Fluorescence in situ hybridization for the detection of prokaryotes. Molecular Microbial Ecology. 2005;213 [Google Scholar]
- 70.Slodzian G, Hillion F, Stadermann FJ, Zinner E. QSA influences on isotopic ratio measurements. Applied Surface Science. 2004;231:874–877. [Google Scholar]
- 71.Franks AH, et al. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer KH. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga–Flavobacter–Bacteroides in the natural environment. Microbiology. 1996;142(Pt 5):1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.