Abstract
Harbored as relics of ancient germline infections, human endogenous retroviruses (HERVs) now constitute up to 8% of our genome. A proportion of this sequence has been co-opted for molecular and cellular processes, beneficial to human physiology, such as the fusogenic activity of the envelope protein, a vital component of placentogenesis. However, the discovery of high levels of HERV-K mRNA and protein and even virions in a wide array of cancers has revealed that HERV-K may be playing a more sinister role–a role as an etiological agent in cancer itself. Whether the presence of this retroviral material is simply an epiphenomenon, or an actual causative factor, is a hotly debated topic. This review will summarize the current state of knowledge regarding HERV-K and cancer and attempt to outline the potential mechanisms by which HERV-K could be involved in the onset and promotion of carcinogenesis.
Keywords: human endogenous retrovirus, HERV-K, carcinogenesis, melanoma, breast cancer, prostate cancer, HERV-K activation, oncogenesis, immunomodulation, Env, Gag, Np9, Rec
One of the most striking findings that arose from the publication of the human genome sequence was the enormous swathe of transposable elements (TEs) it harbored.1 Constituting ~45% of the entire sequence, they have co-evolved alongside the protein-coding component to contribute to modern-day phenotypes in ways which are still being deciphered. A subset of TEs, known as human endogenous retroviruses (HERVs), are ancestral relics of germline infections to which the genome succumbed over the course of evolution. The progenitors of these retroelements were exogenous retroviruses, which infected germline cells, subsequently became endogenized and subject to the laws of Mendelian inheritance.2 HERVs share the genomic structure universal to all retroviruses: 5′LTR-gag, pro, pol, env–3′LTR. In retroviruses, these open reading frames (ORFs) encode viral polyproteins, which, after post-translational modification, become the critical structural and functional proteins, such as the reverse transcriptase or the transmembrane envelope, while the long terminal repeats (LTRs) specify promoter, enhancer and polyadenylation signals.3 The vast majority of HERVs have acquired inactivating mutations such as stop codons or frameshifts, inhibiting the translation of functional proteins and thus making the production of a full, infectious retrovirus particle, from a single genetic locus, an impossibility.4
The HERV-K group is class II HERVs and exhibits closest homology to betaretroviruses, which cluster as class II retroelements. It consists of 11 subgroups (HML-1 to HML-11), each as the result of a separate germline infection.5 One of these subgroups, HML-2, has been subject to intensive research because it maintains an unrivalled coding competence with many of its proviruses maintaining complete, or near-complete, ORFs for all viral polyproteins (Fig. 1).3 Finally, it represents the most recently integrated HERV group into the human genome. Some HML-2 proviruses are both human specific and/or polymorphic indicating integration events subsequent to the human–chimpanzee split and within modern humans. This likely contributes to the fact that HML-2 is the least defective and most active retroviral family. In this regard, HML-2 is considered the most interesting HERV group to study in terms of potential oncogenic activity.
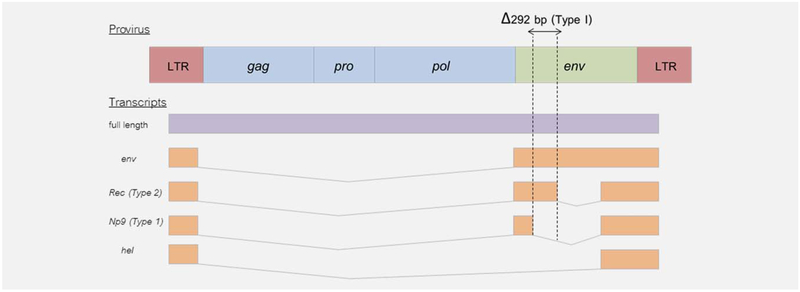
Figure 1.
Structure of HERV-K provirus. The full length (gag) HML-2 transcript encodes the gag, pro and pol polyproteins. A singly spliced transcript encodes the env polyprotein, while a doubly spliced transcript encodes either the Rec or Np9 accessory proteins depending on the presence or absence of a 292-bp deletion at the pol/env boundary–a characteristic that defines a HML-2 provirus as either Type 1 (deleted) or Type 2 (intact). HML-2 also expresses a 1.5-kb transcript of unknown function known as the hel transcript.
Overall, HML-2 is represented in the genome by 91 proviruses and 944 solitary (solo) LTRs. Solo LTRs are the result of unequal crossing over due to highly homologous sequences.5 Two main types of HERV-K (HML-2) are found in humans: type I is characterized by a 292 base pair deletion at the boundary of the pol and env (envelope) genes (Fig. 1), whereas type II lacks it. The deletion in type I proviruses leads to an alternative splicing event culminating in a protein known as Np9, while type II proviruses express a complete accessory protein known as Rec.3
HERVs play an important role in normal physiological function. For example, the protein syncytin 1 mediates cellular fusion of the placental trophoblast and is encoded by an env gene from the HERV-W group.6 Another syncytin— known as syncytin 2—plays a similar physiological role and is encoded by an env gene from the HERV-FRD group.7 Finally, the presence of HERVs, in particular their LTR elements, has added an additional layer of complexity to our genome, in that many of these LTRs have been co-opted by protein-coding genes and serve as regulatory elements directing tissue-specific expression.8
The association of HERVs with disease has garnered the most attention from researchers. HERVs have been implicated in autoimmune disorders,9,10 but with conflicting reports particularly involving multiple sclerosis (MS).11–13 Recent research refutes a role for deregulated HERV-W env in MS lesions, including the high-level-transcribed ERVWE1 locus encoding Syncytin-1.14 In this review, we discuss the most recent developments in the field of HERV-K and human tumor biology, in particular emerging evidence of a role for HERV-K in immunomodulation and the presence of HERV-K in tumor-derived exosomes, further indicating the potentially important role of HERV-K in human carcinogenesis.
HERV-K and Solid Tumors
To date there is evidence for HML-2 activation in ovarian cancer,15,16 melanoma,17–19 breast,20–24 prostate,25–28 lymphomas,29 leukemias30 and sarcomas.31 In the 1980s, Ono et al. successfully cloned HML-2, thanks to its similarity to mouse mammary tumor virus (MMTV).32 They also found that stimulation of human breast cancer cell lines with female steroid hormones led to an upregulation of HML-2 mRNA.33 Several groups followed with reports of HML-2 mRNA and viral particle expression in breast cancer.29,34–36 Wang-Johanning et al. refined this work to produce data that accurately quantified HML-2 env transcripts and spliced transcripts in breast tumors demonstrating elevated levels compared to unaffected controls.20,21 They also demonstrated an association between HML-2 Env protein expression in breast tumors and increased risk of lymph node metastasis and poor outcome in two separate US cohorts and a Chinese cohort of breast cancer patients,37,38 corroborating the findings of Golan et al.23 Most recently, Wang-Johanning et al. demonstrated that HML-2 serum mRNA and anti-Rec antibody titers are predictive of early-stage breast cancer. Additionally, HERV-K-gag copy number tended to be higher in breast cancer patients with a primary tumor who later on developed the metastatic.39
High levels of expression of HML-2 env, rec and np9 mRNA, and Env protein have been reported in ovarian cancer cell lines and tumors,16 whereas in another study Np9 mRNA was not detectible in two ovarian tumors tested.40 One possible mechanism of altered HML-2 expression in ovarian cancer may be due to alterations in its methylation status.15
Retrovirus-like particles and the expression of HML-2 mRNA and proteins are detectable in prostate cancer tissues. Ishida et al. isolated a HML-2 Gag protein in the serum of a prostate cancer patient using serological recombinant cDNA expression cloning (SEREX) technology.25 They subsequently detected HML-2 gag mRNA in the serum of six of nine prostate cancer patients, but failed to detect HML-2 gag mRNA in LnCAP, DU145 or PC3 prostate cancer cells.25 Gene fusions are a frequent occurrence in prostate cancer, the majority of which involve the fusion of the transcription factors ETS translocation variant (ETV1) or ETS-related gene (ERG1), to the transmembrane protease, serine 2 (TMPRSS2). In these fusions, the androgen-responsive TMPRSS2 drives expression of the ETV1 or ERG1 oncogenes. Recently, ETV1-HERV-K fusions have been described, corresponding to the 5′-untranslated region (UTR) of HERV-K-22q11.2326 and HERV-K17.41 Additionally, the ETV1-HERV-K-22q11.23 fusion is also inducible in LNCaP in response to androgen,26 similar to HML-2 induction by estrogen and progesterone in breast cancer cell lines.33
Goering et al. detected significant expression of HERV-K-22q11.23 and HERV-K17 in the androgen-responsive prostate cancer cell lines 22Rv1, LNCaP and MDA-PCa-2b.27 Normal prostate cells and androgen-insensitive prostate cancer cells (PC-3, DU-145 and BPH-1) exhibited expression near the limit of detection.27 Expression of two other proviruses HERV-K-11q23.3 and HERV-K-22q.11.21 was not detectable in prostate cancer cell lines. Assessing HERV-K-22q11.23 5′UTR-gag, env and Np9 gene expression in prostate tumors (n = 45) versus benign tissue (n = 11), the expression of the 5′UTR-gag and env region was significantly elevated in tumors compared to benign tissues. Np9 was detectable only in a subset of carcinomas (18/45). In contrast, HERV-K17 was reduced in prostate tumors compared to benign. Where HERV-K-22q11.23 and HERV-K17 were expressed, they correlated with PSA levels, suggesting that HERV-K-22q11.23 and HERV-K17 retroelements are under androgen-inducible control, whereas HERV-K-11q23.3 and HERV-K-22q.11.21 are not.27 Wallace et al. demonstrated that the HERV-K gag mRNA in peripheral blood mononuclear cells (PBMCs) is predictive of diagnosis with prostate cancer and correlates with elevated plasma interferon-γ and IP10.42
HERV-K and Hematological Malignancies
Brodsky et al. discovered a potential role for HERV-K in leukemia. They showed that HML-2 pol mRNA was expressed in the blood of patients suffering from chronic myeloid leukemia (CML) and acute myeloid leukemia (AML).43,44 Others also reported that HML-2 gag mRNA is present at higher levels in PBMCs of leukemia patients compared to healthy controls.30 Similar studies have reported HML-2 viral particles in lymphomas29 and HML-2 env expression in the H9 human T-cell lymphoma cell line.45 Additionally, the human lymphotropic herpesvirus Epstein–Barr virus (EBV), which has been implicated in the development of lymphoma, was shown to induce HERV-K18 env gene expression. The HERV-K18 env has been reported to have superantigen (SAg) activity by several groups,46,47 whereas others have found no evidence of SAg activity.48,49 Indeed, multiple HERV-K env proteins elicit antibody responses.22,50 A direct association between HERV-K18 env SAg and carcinogenesis has yet to be shown. HML-2 expression has also been seen to decrease after lymphoma therapy, indicating that it may be useful for monitoring therapeutic response.29
HERV-K and Melanoma
The prevalence of HML-2 pol, gag and env mRNA, and Gag and Env proteins in melanoma is well established.17–19,51–54 In 2002, a sequence homologous to HERV-K (HML-6) was identified in melanoma patients (HERV-K-MEL).31 HERV-K-MEL, which produces an antigen spliced from a defective noncoding env-like ORF, was reported in cutaneous and ocular melanomas, and nevi. Antibodies raised against the HERV-K-MEL antigen were detectable in melanoma patients.31 Melanoma cell lines (SKMel-28, SKMel-1, 518A2, MelJuso, HS-Mel2 and JH-Mel6 and HV-Mel7), but not cultured melanocytes (NHEM neo 5935, NHEM neo 4528 and NHEM neo 6083), produce retrovirus-like particles that exhibit reverse transcriptase activity,52 which contain mature Gag and Env proteins. HML-2 Pol, Gag and Env,52 and accessory proteins Rec and Np9 have also been detected in melanoma.18,51 Further studies sought to predict the prognostic value of HERV-K in melanoma and found that HERV-K was a statistically significant marker of acrolentiginous, mucosal and uveal melanoma. Patients with serological response against HERV-K had a significantly decreased disease-specific overall survival.55 Additionally, HML-2 rec mRNA is expressed in melanoma cells but not in benign nevi or normal skin, indicating aberrant activation in melanoma. Furthermore, rec mRNA positivity correlated with the vertical growth phase of melanoma, a step that increases the risk of metastatic melanoma.56 A recent study by Schmitt et al. defined the HML-2 transcriptome in melanoma, identifying 23 different HML-2 loci as transcribed to varying degrees in different patient specimens and melanoma cell lines.57
Polymorphic HML-2 Group Members
Of the 91 known HML-2 proviruses, 11 are polymorphic.5 The most recent insertions (~1 million years ago) include HERV-K-19p12 (K113) (29% of individuals) and HERV-K-8p23.1 (K115) (16% of individuals) as measured using a pool of mixed backgrounds.58 Other polymorphic HML-2 proviruses include: HERV-K-11q22.1 (K118), HERV-K-6q14.1 (K109), HERV-K-7p22.1a (K108R), HERV-K-8p23.1 (K115) and HERV-K-1p31.1(K116),59,60 in addition to HERV-K-3q13.2 (K106), HERV-K-7p22.1b (K108L), HERV-K-10p12.1 (K103), HERV-K-12q13.2 and finally HERV-K-U219 (K105) located in the unassembled centromeric region (Un_g1000219).5
It is currently not known whether inheriting specific HML-2 polymorphisms increases the risk of cancer. Burmeister et al. investigated the frequency of the polymorphic full-length HERV-K115 and HERV-K113 in 102 female breast cancer cases and 102 controls, but did not find a significant association with breast cancer (HERV-K-K113, 16.7 vs. 12.7%; HERV-K-K115, 4.9 vs. 9.8%). (Note the lower prevalence than reported above58 for both. This suggests ethnic differences in frequency of inheritance).24
Mechanisms of HERV Activation and Regulation
The abundance of inactive HERVs present in our genome suggests that active, integrating proviruses are largely deleterious to the host. Novel intrinsic restriction factors exist which impede retroviral infection and some of these have the ability to target both exogenous and endogenous infections. APOBEC proteins can inhibit viral RNA, thus blocking their translation.61 Additionally, APOBEC3G can hypermutate and inactivate HERV DNA.62 Activation of these retroelements can therefore be an indication that cellular programs, crucial to a healthy phenotype, have gone awry.
A crucial question that needs to be addressed is whether activation of HERVs is simply an epiphenomenon or is necessary for disease progression? A large proportion of HERV loci have become silenced via DNA hypermethylation, an epigenetic phenomenon.63 Many cancers display a globally hypomethylated state64; thus, activation of HERVs during tumorigenesis may simply be a bystander effect of this epigenetic state. It has become increasingly clear that genomic instability, including deregulated transcription and genome plasticity, is enabled as a result of epigenetic changes that take place within tumors. Demethylation of specific HERV families, including HERV-W, HERV-K and HERV-H, has been reported in various cancers.65 Moreover, demethylation of TEs correlates with their transcriptional activation in prostate cancer.27 This indicates that where HERV transcription is increased in cancer cells, it is likely due in part to hypomethylation of their LTRs. HML-2 DNA hypomethylation has been reported in melanoma cell lines,66 prostate tumors27 and ovarian tumor.15 Interestingly, age was negatively associated with HML-2 methylation in PBMCs from healthy donors aged 20–88 years. The average onset of HML-2 methylation in PBMCs occurred at 40–63 years, implicating HML-2 DNA hypomethylation in aging.67 Another important epigenetic mechanism that influences transcriptional activity is histone modification, but the influence of histone methylation, acetylation or other modifications on HERV expression in malignancy is still unknown.
Known inducers of HML-2 in vitro include ultraviolet radiation in melanoma,17,68 hormones, including progesterone, estrogen and androgen in breast20,33 and prostate26 cancer cell lines and bone morphogenetic proteins and retinoic acid in testicular germ cell tumor cell lines.69 Urine from smokers has also been shown to lead to an increase in HERV expression in normal human dermal fibroblasts and urothelium in vitro.70 Other factors that may activate or be activated by HERV-K include infectious viruses such as EBV71 and human immunodeficiency virus (HIV-1),72 and transcription factors including NF-κB, NF-AT,73 MITF-M,74 Sp1,Sp375 and YY1.76
Possible Mechanisms of HERV-K-Induced Oncogenesis
Insertional mutagenesis
HERVs may be oncogenic via insertional mutagenesis. However, to date, no fully intact and infectious HERV-derived retrovirus has been demonstrated in vivo. Retrovirus-like particles observed using electron microscopy in human placental trophoblasts,77 and teratocarcinoma78 and melanoma52 were identified as HERV-K derived. Efforts to identify an infectious HERV-K are compounded by the fact that the large majority are partially defective and also that a somatic integration event would be a relatively rare occurrence.3 Two independent groups have succeeded in resurrecting full retroviral particles after constructing consensus sequences representing ancestors of now defunct proviruses.79,80 Although these viruses were found to be only weakly infectious, these studies will prove invaluable in formulating hypotheses regarding the potential oncogenic mechanisms of an infectious HERV-K (Fig. 2).83,85,88,122,123
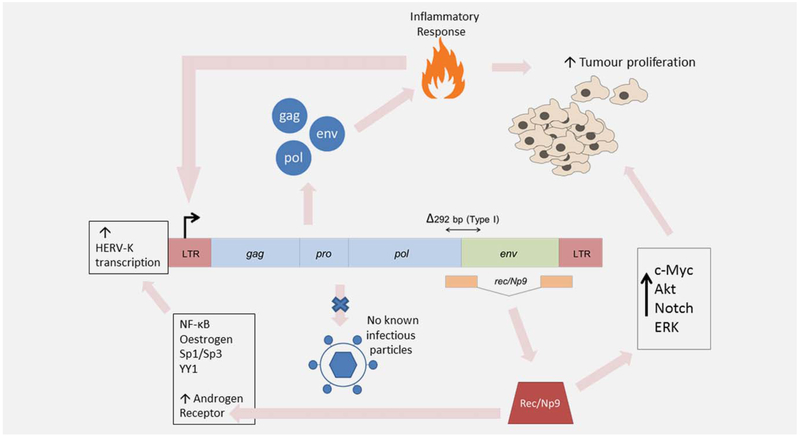
Figure 2.
Proposed model of HERV-K (HML-2)-driven cancer progression. Global DNA hypomethylation during early-stage cancer leads to activation of otherwise silenced TEs, including HERVs. A humoral response to HERV-K gag has been observed in some cancers.81 Such a response to high levels of HERV-K protein expression may culminate in chronic inflammation. Conversely, it has been hypothesized that HERV-K LTRs are responsive to inflammatory transcription factors–a phenomenon that may explain the high levels of HERV-K mRNA and protein seen in some inflammatory diseases.82 HERV-K (HML-2) accessory proteins Rec and Np9 have been shown to lead to the derepression of the c-myc protooncogene,83 while Np9 has been shown to co-activate Akt, Notch and ERK pathways in leukemia.84 Rec has also been observed to lead to the derepression of the androgen receptor, which directly or undirectly causes a further increase in HERV-K transcription.85 Overall, the synergistic effects of chronic inflammation and dysregulated signaling/protooncogene activation caused by HERV-K protein expression may help to create a protumorigenic microenvironment culminating in further proliferation and metastasis. Finally, it is important to note that an active, infectious HML-2 provirus has not been isolated to date, but the existence of such a particle cannot be ruled out. It would potentially be oncogenic via mechanisms such as insertional mutagenesis.
HERV-K113 and HERV-K115 are some of the most recently integrated HERVs in the human genome and represent obvious candidates for infectious proviruses. Boller et al. investigated this possibility and observed that HERV-K113 is able to produce fully intact retroviral particles in vitro.86 However, the authors concluded that an infectious HERV-K113 virus would be unlikely due to a lack of a functional reverse transcriptase.
HERV-K Rec and Np9 as putative oncogenes
Rec exhibits functional homology to the Rev protein of HIV-1, a nucleocytoplasmic shuttle protein.3 Np9 is spliced from an alternative splice donor site to Rec, and shares only 14aa with Rec and Env, with no homology to Rev.87 Functional studies found that both proteins bind the promyelocytic leukemia zinc finger (PLZF) protein, a transcriptional repressor of the C-MYC proto-oncogene,83 leading to the derepression of C-MYC. Rec also binds a related protein known as testicular zinc-finger protein (TZFP), a transcriptional repressor of the AR. Rec inhibits the ability of TZFP to repress AR transcription.88 Hanke et al. identified an additional binding partner of Rec known as human small glutamine-rich tetratricopeptide repeat protein (hSGT), which also acts as a co-repressor of the AR.85 Moreover, they proposed a “vicious cycle” model, whereby increased cellular AR led to increased transcription at HERV-K loci and thus increased levels of Rec leading to further AR derepression. The involvement of such hormonal regulators will be interesting to study in castration-resistant prostate cancer, in which disruption of the AR signaling axis is a key factor in development of resistance.
Reinforcing the possible importance of these proteins in tumorigenesis was the finding that mice transgenic for the Rec gene are prone to seminomas.89 Np9 has been shown to interact with LNX—an E3 ubiquitin ligase that targets members of the NUMB/NOTCH pathway.40 This pathway has been implicated in the regulation of proliferation of cancers of the breast and prostate.90 Finally, a recent study has shown that Np9 acts as a critical molecular switch for co-activating β-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells (Fig. 2).84
HERV-K-induced immunomodulation
In a Darwinian sense, cancerous tissue uses the inflammatory-associated milieu of the tumor microenvironment to confer a selective advantage.91 The apparent immunogenicity of HERV proteins therefore represents a potential contributor to, or initiator of a chronic inflammatory state, beneficial to tumor survival (Fig. 2). HML-2 antibodies have been observed in patients with melanoma,18 breast39 and ovarian cancers.16 In breast cancer, studies have found that both humoral and cell-mediated immune responses to HERVs were enhanced in patients when compared to controls.22 HERV-K18 Env protein has been shown to elicit T-cell responses and can be upregulated in response to EBV infection,46,92 and may be a prerequisite of B-cell lymphomas.93
Similar to discoveries in HIV-1, HERV-K may encode env proteins with immunosuppressive transmembrane domains. A recent study by Morozov et al. identified an immunosuppressive HERV-K env protein, which altered cytokine expression and suppressed immune cell proliferation in vitro.94
Nitric oxide (NO) is an endogenous free radical signaling molecule that has been intimately linked with inflammation, wound healing responses and cancer.95,96 A significant association between nitric oxide synthase 2 (NOS2) and HML-2 Env expression has been demonstrated in breast cancer.37 NOS2 is an independent predictor of poor outcome in estrogen receptor-negative breast cancer, associated with macrophage infiltration, deregulated p53 signaling, increased proliferation and resistance to apoptosis.95,97,98 Can HML-2 Env proteins mediate downstream inflammatory effects via their activation of NO signaling? Intriguingly, β-catenin, ERK and Akt, which are activated by Np9,84 are also activated by NO signaling.98,99
Exosomes
An evolving hypothesis in cancer research over the last few years has been the involvement of tumor exosomes in metastasis.100,101 Exosomes are nanoscale membrane vesicles that are secreted from cells and are thought to be important intercellular communicators, or, in a cancer setting, drivers of metastatic spread.102 A recent study has now implicated HERVs in this process, with the finding that HML-2 mRNA is selectively packaged into tumor exosomes and that this genetic material can be transferred to normal cells.103 The authors also demonstrated that these exosomes were enriched for the C-MYC protooncogene, which has been shown to be regulated by PLZF, a target of Rec and Np9.83 Therefore, it is possible that there is a link between the high levels of HERV-K mRNA and C-MYC in these exosomes, but further investigation will have to be done in this regard. Another important point is that HML-2-driven metastasis via exosomes would not require an envelope gene, as exosomes gain entry to target cells via a plasma membrane fusion event. In essence, exosomes could potentially empower the abundance of defective HERVs with a new-found infectivity.104
HERV-K viral proteins as potential vaccines
Although the direct oncogenic effects of HERVs in cancer remain to be fully elucidated, there is potential for their use as diagnostic or prognostic biomarkers and for immunotherapeutic purposes including vaccines. Independent groups have demonstrated a strong association between HERV-K antibodies and clinical manifestation of disease and therapeutic response.23,29 Antibodies recognizing synthetic HERV-K proteins were detected at a very low frequency in the sera of healthy donors.16,22 Humoral anti-HERV-K immune response may provide additional prognostic information to that of established melanoma markers.31,55 Data from these studies reveal a significant inverse correlation between serological anti-HERV-K reactivity and patient survival probability in melanoma patients. Among the different classes of tumor antigens recognizable by the immune system, mutated self-antigens and viral antigens are unique because they are foreign to the host and not subjected to preexisting antigen-specific tolerance.105–107 HML-2 exons coding for mature proteins are spread out over the genome and are a repository of immunogenic retroviral gene products that can be “reawakened” when genetic damage occurs through chromosome breaks, frameshifts and mutations, removing sequences normally silencing protein expression.
HERV-K MEL is an antigenic peptide that is encoded by a short ORF from a processed HERV-K (HML-6) pseudogene and has been shown to be recognized by cytotoxic T cells in human melanoma.31 BCG, vaccinia and yellow fever vaccinations are associated with a reduced risk of developing melanoma,108–110 although conflicting data exist for yellow fever vaccines.111 It is suggested that this effect is due to antigen sequence homology between these vaccines and HERV-K-MEL leading to cross-reaction between vaccine-elicited cytotoxic T cells and melanoma cells.112 Reintroduction of these vaccines has been suggested as a novel method of melanoma immunoprevention; otherwise, HERV-K MEL represents a legitimate target for cellular immunotherapy.112,113
Future Perspectives
Over the course of evolution, our genome has been locked in a molecular “war” with exogenous infectious agents. Ultimately, it is this very battleground, together with viral endogenization, which has bestowed upon us the diverse genetic repertoire we possess today. Constituting 8% of our genome, these HERVs have supplied us with an additional layer of plasticity and physiological functionality, yet scientists now believe that hidden detrimental processes fueled by HERVs may be present, which are inducing chronic diseases such as cancer and autoimmunity. As of yet, no truly infectious HERVs have been observed. However, as outlined in this review, a range of potential molecular mechanisms involving the retroviral proteins may be aiding and abetting both tumor formation and metastasis. Ultimately, it is likely that many of these mechanisms are working synergistically to produce these effects, and the heralding of a single molecular event induced by a HERV protein is improbable.
Ascribing a causative role for a particular agent to a disease has long been a challenging task. Criteria such as Hills criteria114 and Koch’s postulates115 have been formulated to address this problem. These criteria have recently been refined and built upon in light of HERVs-postulated role in human disease.113,116,117 However, even if a direct link between HERVs and carcinogenesis is never established, their presence may be highly advantageous in terms of the implementation of novel biomarkers for cancer. Further work will need to effectively correlate their presence with various disease stages and also make the necessary comparisons against “gold standard” biomarkers. Equally promising is the potential to take advantage of tumor-specific HERV expression for the use of targeted immunotherapies. Wang-Johanning et al. have demonstrated the potential of anti-HML-2-Env antibodies in inhibiting tumor growth and inducing apoptosis, both in vitro and in in vivo mouse xenograft models.37 This work represents a major milestone in research into HERVs and cancer and it is likely that targeting Env in a similar fashion in other cancers will be equally effective. However, it remains imperative that these studies are evaluated in a clinical setting. Additionally, it may also be possible to conjugate these antibodies to cytotoxic drugs for increased effect.118 Similarly, Kraus et al. demonstrated that HML-2-Env-targeted vaccine reduced renal tumor metastasis in a murine model.119 Novel therapies, such as these, are key to making inroads toward a future cure for the increasingly complex and multistep disease that is cancer. However, their safety must be assessed given the newly established role of HERV-K in embryonic stem cells and iPS cells,120 which may have implications for pregnancy. Their role in adult stem cells is not currently known.
Several limitations exist in the field of cancer-related HERV-K research, including a lack of adequately powered patient population studies to determine the role of HERV-K in the etiology of cancer, and/or its association with metastasis, therapeutic response and overall patient survival. A gap exists in our knowledge as to which HERV-K loci are specifically activated in cancer. A recent study by Schmitt et al. has defined the HML-2 transcriptome in melanoma, identifying 23 loci as transcribed,57 and it is an imperative that similar studies be initiated in other cancers. A causal role for HML-2 has yet to be established. Generally, retroviruses induce tumours by insertional mutagenesis targeting specific oncogenes, as is the case with HBV.121 This is an unlikely mechanism though in the case of HML-2. Evidence does suggest that Rec and Np9 may be putative oncogenes, but whether Gag or Env are also oncogenic is not known. In exceptional cases such as Jagsiekte sheep retrovirus (JSRV) the Env protein has been found to be causal (ovine pulmonary adenocarcinoma).122 However, it is unlikely that HML-2 Gag or Env have a similar causal effect; potentially they may influence carcinogenesis by activating or perturbing inflammation responses against cancer.
It is our belief that within the next decade these genetic “squatters” will have firmly established themselves within the modern multistep model of cancer progression and their expression will be viewed as an “enabling characteristic” of cancer, giving new meaning to the famous words of Nobel laureate J. Michael Bishop when he stated that “the seeds of cancer are within us”.123
Acknowledgements
This work was supported by the Galway University Foundation (RNR1008 to F.J.S. and S.A.G.); the Health Research Board of Ireland Clinical Research Facility, Galway (RSU004 to R.F.D. and F.J.G.); the Department of Defense Breast Cancer Research Program (BC113114 to F.J.W.); Breast Cancer Campaign UK (2013MayPR019 to S.A.G. and F.J.S.) and the Irish Cancer Society (PCT13MCD to S.A.G. and F.J.S.).
Grant sponsor: Galway University Foundation; Grant number: RNR1008; Grant sponsor: Health Research Board of Ireland Clinical Research Facility, Galway; Grant number: RSU004; Grant sponsor: Department of Defense Breast Cancer Research Program; Grant number: BC113114; Grant sponsor: Breast Cancer Campaign UK; Grant number: 2013MayPR019; Grant sponsor: Irish Cancer Society; Grant number: PCT13MCD
Abbreviations:
- AML
acute myeloid leukemia
- APOBEC3G
apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G
- AR
androgen receptor
- CML
chronic myeloid leukemia
- EBV
Epstein-Barr virus
- eMLV
ectropic murine leukemia virus
- ERG1
ETS-related gene
- ETV1
ETS translocation variant
- HERV
human endogenous retrovirus
- HIV-1
human immunodeficiency virus
- HPV
human papillomavirus
- hSGT
human small glutamine-rich tetratricopeptide repeat protein
- HTLV-1
human T-lymphotropic virus
- LTR
long terminal repeats
- MHC
major histocompatability complex
- MMTV
mouse mammary tumor virus
- NO
nitric oxide
- NOS2
nitric oxide synthase 2
- ORF
open reading frames
- PBMC
peripheral blood mononuclear cell
- PLZF
promyelocytic leukemia zinc finger
- Sags
superantigens
- SEREX
serological recombinant cDNA expression cloning
- TE
transposable element
- TLR
toll-like receptor
- TMPRSS2
transmembrane protease, serine 2
- TZFP
testicular zinc-finger protein
Footnotes
Conflicts of interest: Nothing to report
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860–921. [DOI] [PubMed] [Google Scholar]
- 2.Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA 1996;93:5177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet 2006;7:149–73. [DOI] [PubMed] [Google Scholar]
- 4.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA 2004;101 (Suppl 2):14572–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian RP, Wildschutte JH, Russo C, et al. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 2011;8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mi S, Lee X, Li X, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000;403:785–9. [DOI] [PubMed] [Google Scholar]
- 7.Blaise S, Ruggieri A, Dewannieux M, et al. Identification of an envelope protein from the FRD family of human endogenous retroviruses (HERV-FRD) conferring infectivity and functional conservation among simians. J Virol 2004; 78:1050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 2009;448: 105–14. [DOI] [PubMed] [Google Scholar]
- 9.Balada E, Ordi-Ros J, Vilardell-Tarres M. Molecular mechanisms mediated by human endogenous retroviruses (HERVs) in autoimmunity. Rev Med Virol 2009;19:273–86. [DOI] [PubMed] [Google Scholar]
- 10.Urnovitz HB, Murphy WH. Human endogenous retroviruses: nature, occurrence, and clinical implications in human disease. Clin Microbiol Rev 1996;9:72–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perron H, Lang A. The human endogenous retrovirus link between genes and environment in multiple sclerosis and in multifactorial diseases associating neuroinflammation. Clin Rev Allergy Immunol 2010;39:51–61. [DOI] [PubMed] [Google Scholar]
- 12.Curtin F, Lang AB, Perron H, et al. GNbAC1, a humanized monoclonal antibody against the envelope protein of multiple sclerosis-associated endogenous retrovirus: a first-in-humans randomized clinical study. Clin Ther 2012;34: 2268–78. [DOI] [PubMed] [Google Scholar]
- 13.Mameli G, Poddighe L, Mei A, et al. Expression and activation by Epstein Barr virus of human endogenous retroviruses-W in blood cells and astrocytes: inference for multiple sclerosis. PLoS One 2012;7:e44991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt K, Richter C, Backes C, et al. Comprehensive analysis of human endogenous retrovirus group HERV-W locus transcription in multiple sclerosis brain lesions by high-throughput amplicon sequencing. J Virol 2013; 87:13837–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iramaneerat K, Rattanatunyong P, Khemapech N, et al. HERV-K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance. Int J Gynecol Cancer 2011;21:51–7. [DOI] [PubMed] [Google Scholar]
- 16.Wang-Johanning F, Liu J, Rycaj K, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer 2007;120:81–90. [DOI] [PubMed] [Google Scholar]
- 17.Reiche J, Pauli G, Ellerbrok H. Differential expression of human endogenous retrovirus K transcripts in primary human melanocytes and melanoma cell lines after UV irradiation. Melanoma Res 2010;20:435–40. [DOI] [PubMed] [Google Scholar]
- 18.Buscher K, Trefzer U, Hofmann M, et al. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res 2005;65:4172–80. [DOI] [PubMed] [Google Scholar]
- 19.Serafino A, Balestrieri E, Pierimarchi P, et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res 2009; 315:849–62. [DOI] [PubMed] [Google Scholar]
- 20.Wang-Johanning F, Frost AR, Jian B, et al. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene 2003;22:1528–35. [DOI] [PubMed] [Google Scholar]
- 21.Wang-Johanning F, Frost AR, Johanning GL, et al. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin Cancer Res 2001;7:1553–60. [PubMed] [Google Scholar]
- 22.Wang-Johanning F, Radvanyi L, Rycaj K, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res 2008;68:5869–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golan M, Hizi A, Resau JH, et al. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia 2008;10:521–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burmeister T, Ebert AD, Pritze W, et al. Insertional polymorphisms of endogenous HERV-K113 and HERV-K115 retroviruses in breast cancer patients and age-matched controls. AIDS Res Hum Retroviruses 2004;20:1223–9. [DOI] [PubMed] [Google Scholar]
- 25.Ishida T, Obata Y, Ohara N, et al. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun 2008;8:15. [PMC free article] [PubMed] [Google Scholar]
- 26.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 2007;448:595–9. [DOI] [PubMed] [Google Scholar]
- 27.Goering W, Ribarska T, Schulz WA. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis 2011;32:1484–92. [DOI] [PubMed] [Google Scholar]
- 28.Agoni L, Guha C, Lenz J. Detection of human endogenous retrovirus K (HERV-K) transcripts in human prostate cancer cell lines. Front Oncol 2013;3:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Contreras-Galindo R, Kaplan MH, Leissner P, et al. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J Virol 2008;82:9329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Depil S, Roche C, Dussart P, et al. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia 2002;16:254–9. [DOI] [PubMed] [Google Scholar]
- 31.Schiavetti F, Thonnard J, Colau D, et al. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res 2002;62:5510–16. [PubMed] [Google Scholar]
- 32.Ono M, Yasunaga T, Miyata T, et al. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol 1986;60:589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol 1987;61:2059–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seifarth W, Skladny H, Krieg-Schneider F, et al. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J Virol 1995;69:6408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willer A, Saussele S, Gimbel W, et al. Two groups of endogenous MMTV related retroviral env transcripts expressed in human tissues. Virus Genes 1997;15:123–33. [DOI] [PubMed] [Google Scholar]
- 36.Etkind PR, Lumb K, Du J, et al. Type 1 HERV-K genome is spliced into subgenomic transcripts in the human breast tumor cell line T47D. Virology 1997;234:304–8. [DOI] [PubMed] [Google Scholar]
- 37.Wang-Johanning F, Rycaj K, Plummer JB, et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J Natl Cancer Inst 2012;104:189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Rycaj K, Geng S, et al. Expression of human endogenous retrovirus type K envelope protein is a novel candidate prognostic marker for human breast cancer. Genes Cancer 2011;2: 914–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang-Johanning F, Li M, Esteva FJ, et al. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int J Cancer 2014;134:587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armbruester V, Sauter M, Krautkraemer E, et al. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin Cancer Res 2002;8:1800–7. [PubMed] [Google Scholar]
- 41.Hermans KG, van der Korput HA, van Marion R, et al. Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer Res 2008;68:7541–9. [DOI] [PubMed] [Google Scholar]
- 42.Wallace TA, Downey RF, Seufert CJ, et al. Elevated HERV-K mRNA expression in PBMC is associated with a prostate cancer diagnosis particularly in older men and smokers. Carcinogenesis 2014, May 23, pii: bgu114 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brodsky I, Foley B, Gillespie D. Expression of human endogenous retrovirus (HERV-K) in chronic myeloid leukemia. Leuk Lymphoma 1993;11 (Suppl 1):119–23. [DOI] [PubMed] [Google Scholar]
- 44.Brodsky I, Foley B, Haines D, et al. Expression of HERV-K proviruses in human leukocytes. Blood 1993;81:2369–74. [PubMed] [Google Scholar]
- 45.Vogetseder W, Feng J, Dierich MP. Reactivity of monoclonal antibodies established against a recombinant human endogenous retrovirus-K (HERV-K) envelope protein. Immunol Lett 1995;46:129–34. [DOI] [PubMed] [Google Scholar]
- 46.Sutkowski N, Conrad B, Thorley-Lawson DA, et al. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 2001;15:579–89. [DOI] [PubMed] [Google Scholar]
- 47.Hsiao FC, Tai AK, Deglon A, et al. EBV LMP-2A employs a novel mechanism to transactivate the HERV-K18 superantigen through its ITAM. Virology 2009;385:261–6. [DOI] [PubMed] [Google Scholar]
- 48.Azar GA, Thibodeau J. Human endogenous retrovirus IDDMK(1,2)22 and mouse mammary tumor virus superantigens differ in their ability to stimulate murine T cell hybridomas. Immunol Lett 2002;81:87–91. [DOI] [PubMed] [Google Scholar]
- 49.Badenhoop K, Donner H, Neumann J, et al. IDDM patients neither show humoral reactivities against endogenous retroviral envelope protein nor do they differ in retroviral mRNA expression from healthy relatives or normal individuals. Diabetes 1999;48:215–18. [DOI] [PubMed] [Google Scholar]
- 50.Herve CA, Lugli EB, Brand A, et al. Autoantibodies to human endogenous retrovirus-K are frequently detected in health and disease and react with multiple epitopes. Clin Exp Immunol 2002;128:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buscher K, Hahn S, Hofmann M, et al. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res 2006;16:223–34. [DOI] [PubMed] [Google Scholar]
- 52.Muster T, Waltenberger A, Grassauer A, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res 2003;63:8735–41. [PubMed] [Google Scholar]
- 53.Humer J, Waltenberger A, Grassauer A, et al. Identification of a melanoma marker derived from melanoma-associated endogenous retroviruses. Cancer Res 2006;66:1658–63. [DOI] [PubMed] [Google Scholar]
- 54.Singh S, Kaye S, Gore ME, et al. The role of human endogenous retroviruses in melanoma. Br J Dermatol 2009;161:1225–31. [DOI] [PubMed] [Google Scholar]
- 55.Hahn S, Ugurel S, Hanschmann KM, et al. Serological response to human endogenous retrovirus K in melanoma patients correlates with survival probability. AIDS Res Hum Retroviruses 2008;24:717–23. [DOI] [PubMed] [Google Scholar]
- 56.Singh S, Kaye S, Francis N, et al. Human endogenous retrovirus K (HERV-K) rec mRNA is expressed in primary melanoma but not in benign naevi or normal skin. Pigment Cell Melanoma Res 2013;26:426–8. [DOI] [PubMed] [Google Scholar]
- 57.Schmitt K, Reichrath J, Roesch A, et al. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol Evol 2013;5:307–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner G, Barbulescu M, Su M, et al. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol 2001;11:1531–5. [DOI] [PubMed] [Google Scholar]
- 59.Hughes JF, Coffin JM. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc Natl Acad Sci USA 2004; 101:1668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belshaw R, Dawson AL, Woolven-Allen J, et al. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J Virol 2005;79: 12507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mullins CS, Linnebacher M. Human endogenous retroviruses and cancer: causality and therapeutic possibilities. World J Gastroenterol 2012; 18:6027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee YN, Malim MH, Bieniasz PD. Hypermutation of an ancient human retrovirus by APOBEC3G. J Virol 2008;82:8762–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol 2006; 310:211–50. [DOI] [PubMed] [Google Scholar]
- 64.Ehrlich M DNA methylation in cancer: too much, but also too little. Oncogene 2002;21: 5400–13. [DOI] [PubMed] [Google Scholar]
- 65.Gimenez J, Montgiraud C, Pichon JP, et al. Custom human endogenous retroviruses dedicated microarray identifies self-induced HERV-W family elements reactivated in testicular cancer upon methylation control. Nucleic Acids Res 2010;38:2229–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stengel S, Fiebig U, Kurth R, et al. Regulation of human endogenous retrovirus-K expression in melanomas by CpG methylation. Genes Chromosomes Cancer 2010;49:401–11. [DOI] [PubMed] [Google Scholar]
- 67.Jintaridth P, Mutirangura A. Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiol Genomics 2010;41:194–200. [DOI] [PubMed] [Google Scholar]
- 68.Schanab O, Humer J, Gleiss A, et al. Expression of human endogenous retrovirus K is stimulated by ultraviolet radiation in melanoma. Pigment Cell Melanoma Res 2011;24:656–65. [DOI] [PubMed] [Google Scholar]
- 69.Caricasole A, Ward-van Oostwaard D, Mummery C, et al. Bone morphogenetic proteins and retinoic acid induce human endogenous retrovirus HERV-K expression in NT2D1 human embryonal carcinoma cells. Dev Growth Differ 2000;42:407–11. [DOI] [PubMed] [Google Scholar]
- 70.Gabriel U, Steidler A, Trojan L, et al. Smoking increases transcription of human endogenous retroviruses in a newly established in vitro cell model and in normal urothelium. AIDS Res Hum Retroviruses 2010;26:883–8. [DOI] [PubMed] [Google Scholar]
- 71.Sutkowski N, Chen G, Calderon G, et al. Epstein-Barr virus latent membrane protein LMP-2A is sufficient for transactivation of the human endogenous retrovirus HERV-K18 superantigen. J Virol 2004;78: 7852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones RB, Garrison KE, Mujib S, et al. HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J Clin Invest 2012;122: 4473–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonzalez-Hernandez MJ, Swanson MD, Contreras-Galindo R, et al. Expression of human endogenous retrovirus type K (HML-2) is activated by the Tat protein of HIV-1. J Virol 2012; 86:7790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katoh I, Mirova A, Kurata S, et al. Activation of the long terminal repeat of human endogenous retrovirus K by melanoma-specific transcription factor MITF-M. Neoplasia 2011;13:1081–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuchs NV, Kraft M, Tondera C, et al. Expression of the human endogenous retrovirus (HERV) group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3. J Virol 2011;85:3436–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Knossl M, Lower R, Lower J. Expression of the human endogenous retrovirus HTDV/HERV-K is enhanced by cellular transcription factor YY1. J Virol 1999;73:1254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kalter SS, Helmke RJ, Heberling RL, et al. Brief communication: C-type particles in normal human placentas. J Natl Cancer Inst 1973;50: 1081–4. [DOI] [PubMed] [Google Scholar]
- 78.Boller K, Konig H, Sauter M, et al. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 1993;196:349–53. [DOI] [PubMed] [Google Scholar]
- 79.Dewannieux M, Harper F, Richaud A, et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res 2006;16:1548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog 2007;3:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sauter M, Schommer S, Kremmer E, et al. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J Virol 1995;69:414–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manghera M, Douville RN. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology 2013;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Denne M, Sauter M, Armbruester V, et al. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J Virol 2007;81:5607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen T, Meng Z, Gan Y, et al. The viral oncogene Np9 acts as a critical molecular switch for co-activating beta-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia 2013;27:1469–78. [DOI] [PubMed] [Google Scholar]
- 85.Hanke K, Chudak C, Kurth R, et al. The Rec protein of HERV-K(HML-2) upregulates androgen receptor activity by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT). Int J Cancer 2013;132: 556–67. [DOI] [PubMed] [Google Scholar]
- 86.Boller K, Schonfeld K, Lischer S, et al. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J Gen Virol 2008;89:567–72. [DOI] [PubMed] [Google Scholar]
- 87.Ruggieri A, Maldener E, Sauter M, et al. Human endogenous retrovirus HERV-K(HML-2) encodes a stable signal peptide with biological properties distinct from Rec. Retrovirology 2009;6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaufmann S, Sauter M, Schmitt M, et al. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J Gen Virol 2010;91: 1494–502. [DOI] [PubMed] [Google Scholar]
- 89.Galli UM, Sauter M, Lecher B, et al. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 2005;24:3223–8. [DOI] [PubMed] [Google Scholar]
- 90.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev 2007;17:52–9. [DOI] [PubMed] [Google Scholar]
- 91.Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420:860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stauffer Y, Marguerat S, Meylan F, et al. Interferon-alpha-induced endogenous superantigen. a model linking environment and autoimmunity. Immunity 2001;15:591–601. [DOI] [PubMed] [Google Scholar]
- 93.Zur Hausen H The search for infectious causes of human cancers: where and why. Virology 2009;392:1–10. [DOI] [PubMed] [Google Scholar]
- 94.Morozov VA, Dao Thi VL, Denner J. The transmembrane protein of the human endogenous retrovirus-K (HERV-K) modulates cytokine release and gene expression. PLoS One 2013;8: e70399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burke AJ, Sullivan FJ, Giles FJ, et al. The yin and yang of nitric oxide in cancer progression. Carcinogenesis 2013;34:503–12. [DOI] [PubMed] [Google Scholar]
- 96.Rapozzi V, Della Pietra E, Zorzet S, et al. Nitric oxide-mediated activity in anti-cancer photodynamic therapy. Nitric Oxide 2013;30:26–35. [DOI] [PubMed] [Google Scholar]
- 97.Ambs S, Glynn SA. Candidate pathways linking inducible nitric oxide synthase to a basal-like transcription pattern and tumor progression in human breast cancer. Cell Cycle 2011;10:619–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Glynn SA, Boersma BJ, Dorsey TH, et al. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J Clin Invest 2010;120:3843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Switzer CH, Glynn SA, Cheng RY, et al. S-Nitrosylation of EGFR and Src activates an oncogenic signaling network in human basal-like breast cancer. Mol Cancer Res 2012;10: 1203–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang C, Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol 2011;2011:842849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duijvesz D, Luider T, Bangma CH, et al. Exosomes as biomarker treasure chests for prostate cancer. Eur Urol 2011;59:823–31. [DOI] [PubMed] [Google Scholar]
- 102.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569–79. [DOI] [PubMed] [Google Scholar]
- 103.Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2011;2:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gould SJ, Booth AM, Hildreth JE. The Trojan exosome hypothesis. Proc Natl Acad Sci USA 2003;100:10592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smith C, Tsang J, Beagley L, et al. Effective treatment of metastatic forms of Epstein-Barr virus-associated nasopharyngeal carcinoma with a novel adenovirus-based adoptive immunotherapy. Cancer Res 2012;72:1116–25. [DOI] [PubMed] [Google Scholar]
- 106.Foster AE, Rooney CM. Improving T cell therapy for cancer. Expert Opin Biol Ther 2006;6: 215–29. [DOI] [PubMed] [Google Scholar]
- 107.Gottschalk S, Heslop HE, Rooney CM. Adoptive immunotherapy for EBV-associated malignancies. Leuk Lymphoma 2005;46:1–10. [DOI] [PubMed] [Google Scholar]
- 108.Pfahlberg A, Kolmel KF, Grange JM, et al. Inverse association between melanoma and previous vaccinations against tuberculosis and smallpox: results of the FEBIM study. J Invest Dermatol 2002;119:570–5. [DOI] [PubMed] [Google Scholar]
- 109.Krone B, Kolmel KF, Grange JM, et al. Impact of vaccinations and infectious diseases on the risk of melanoma—evaluation of an EORTC case-control study. Eur J Cancer 2003;39:2372–8. [DOI] [PubMed] [Google Scholar]
- 110.Mastrangelo G, Krone B, Fadda E, et al. Does yellow fever 17D vaccine protect against melanoma? Vaccine 2009;27:588–91. [DOI] [PubMed] [Google Scholar]
- 111.Hodges-Vazquez M, Wilson JP, Hughes H, et al. The yellow fever 17D vaccine and risk of malignant melanoma in the United States military. Vaccine 2012;30:4476–9. [DOI] [PubMed] [Google Scholar]
- 112.Krone B, Kolmel KF, Henz BM, et al. Protection against melanoma by vaccination with Bacille Calmette-Guerin (BCG) and/or vaccinia: an epidemiology-based hypothesis on the nature of a melanoma risk factor and its immunological control. Eur J Cancer 2005;41:104–17. [DOI] [PubMed] [Google Scholar]
- 113.Krone B, Grange JM. Melanoma, Darwinian medicine and the inner world. J Cancer Res Clin Oncol 2010;136:1787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hill AB. The environment and disease: association or causation? Proc R Soc Med 1965;58:295–300. [PMC free article] [PubMed] [Google Scholar]
- 115.Evans AS. Causation and disease: the Henle-Koch postulates revisited. Yale J Biol Med 1976; 49:175–95. [PMC free article] [PubMed] [Google Scholar]
- 116.Cegolon L, Salata C, Weiderpass E, et al. Human endogenous retroviruses and cancer prevention: evidence and prospects. BMC Cancer 2013;13:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sarid R, Gao SJ. Viruses and human cancer: from detection to causality. Cancer Lett 2011; 305:218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol 2006;6:343–57. [DOI] [PubMed] [Google Scholar]
- 119.Kraus B, Fischer K, Buchner SM, et al. Vaccination directed against the human endogenous retrovirus-K envelope protein inhibits tumor growth in a murine model system. PLoS One 2013;8:e72756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fuchs NV, Loewer S, Daley GQ, et al. Human endogenous retrovirus K (HML-2) RNA and protein expression is a marker for human embryonic and induced pluripotent stem cells. Retrovirology 2013;10:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guerrieri F, Belloni L, Pediconi N, et al. Molecular mechanisms of HBV-associated hepatocar-cinogenesis. Semin Liver Dis 2013;33: 147–56. [DOI] [PubMed] [Google Scholar]
- 122.Hofacre A, Fan H. Jaagsiekte sheep retrovirus biology and oncogenesis. Viruses 2010;2:2618–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Conti M. The selfish cell: an evolutionary defeat. Dordrecht, London: Springer, 2008. [Google Scholar]