Abstract
Diverse mechanisms and functions of post-transcriptional regulation by small regulatory RNAs (sRNAs) and RNA binding proteins (RBPs) have been described in bacteria. In contrast, little is known about the spatial organization of RNAs in bacterial cells. In eukaryotes, subcellular localization and transport of RNAs play important roles in diverse physiological processes, such as embryonic patterning, asymmetric cell division, epithelial polarity, and neuronal plasticity. It is now clear that bacterial RNAs also can accumulate at distinct sites in the cell. However, due the small size of bacterial cells, localized RNA and translation are more challenging to study in bacterial cells, and the underlying molecular mechanisms of transcript localization are less understood. Here we review the emerging examples of RNAs localized to specific subcellular locations in bacteria, with indications that subcellular localization of transcripts might be important for regulatory processes. Diverse mechanisms for bacterial RNA localization have been suggested, including close association to their genomic site of transcription, or to the localizations of their protein products in translation-dependent or independent processes. We also provide an overview of the state-of-the-art of technologies to visualize and track bacterial RNAs, ranging from hybridization-based approaches in fixed cells to in vivo imaging approaches using fluorescent protein reporters and/or RNA aptamers in single living bacterial cells. We conclude with a discussion of open questions in the field and ongoing technological developments regarding RNA imaging in eukaryotic systems that might likewise provide novel insights into RNA localization in bacteria.
Keywords: RNA localization, Regulatory RNA, Fluorescence labeling, Fluorescence microscopy, RNA binding protein, ribonuclease
INTRODUCTION
Spatial and temporal localization of macromolecules, including RNAs, reflects the compartmentalization of living cells and plays important roles in gene expression and regulation. In eukaryotic cells, physical separation between the transcription and translation machineries in the nucleus and cytoplasm, respectively, naturally results in the synthesis, processing, and translation of mRNA to be spatially disconnected. Both mRNA localization and localized-translation can be important regulatory mechanisms underlying embryonic patterning, asymmetric cell division, epithelial polarity, cell migration, and neuronal morphogenesis (1, 2). RNAs can be transported in the eukaryotic cell in several ways, such as (i) vectorial movement of mRNA by direct coupling to motor proteins, (ii) transport of mRNA by hitchhiking on another cargo, (iii) random transport of mRNA-motor complexes and local enrichment of mRNAs at target sites, or (iv) diffusion and motor-driven cytoplasmic flows with subsequent localized anchorage of the mRNA (3). Moreover, localized translation induction by phosphorylation and activation of translation initiation factors and their regulators in response to localized signals has been reported to impact gene regulation in eukaryotes (4).
In contrast, due to a lack of canonical membrane-bound organelles and a nuclear compartment, prokaryotic cells were long assumed to lack complex subcellular localization of macromolecules, and spatial localization has not been considered to play a significant role in expression and post-transcriptional regulation of bacterial mRNAs. This is also reflected by the classical picture of co-transcriptional translation of bacterial mRNAs, where transcription and protein synthesis are not spatially or temporally separated. Moreover, due to the much smaller size of bacterial cells compared to their eukaryotic counterparts, it has been more challenging to determine the subcellular organization of bacteria, to observe the subcellular distribution of biomolecules in bacterial cells, and to relate such organization and distribution to biological functions.
With the development of numerous labeling and imaging techniques as well as advanced microscopy approaches that can break the diffraction limit, it is now clear that the bacterial cells are also compartmentalized (5–7). Emerging evidence for differential localization of bacterial mRNAs indicates that the spatial organization in the cell can also impact gene expression and post-transcriptional regulation in prokaryotes, and that mRNAs can be differentially localized and/or translated at the site where their gene products are required (8–10). Commonly, localization patterns of mRNAs in bacteria include the nucleoid region, the cytoplasm, cell poles, and inner membrane. Frequently-observed organizations of bacterial biomolecules include uniform distribution, distinct foci, and a putative helical structure, often in the vicinity of the cell envelope. Along with the visualization of transcript localization, it has also been shown that many enzymes and complexes involved in RNA metabolism, such as RNA polymerase, ribosomes, and the degradosome, show distinct subcellular distributions, providing further support for the role of spatial organization in genetic information flow. Certain observations and conclusions in the study of bacterial RNA localization are still controversial and the mechanisms underlying observed examples of subcellular localized transcripts remain to be further explored. However, it has nonetheless become clear that spatial control of RNA and related cellular machineries are likely important for gene expression and regulation in prokaryotes, similar as it has been a well established concept in higher organisms. These preliminary observations of distinct RNA localization patterns have brought attention to new phenomena and questions in bacterial post-transcriptional control, such as transcription-coupled vs transcription-uncoupled translation, translation-dependent and translation-independent mRNA localization, as well as localized degradation, stabilization, or regulation by small regulatory RNAs (sRNAs) and RNA-metabolizing complexes.
Post-transcriptional regulation by regulatory RNAs, RNA-binding proteins (RBPs), and ribonucleases (RNases) is a central layer of gene expression control in all kingdoms of life. Bacterial sRNAs (typically 50–300 nucleotides in length) can control specific genes and/or coordinate expression of distinct regulons with clear physiological outcomes (11). While most sRNAs act as antisense RNAs by short and imperfect base-pairing, several can also directly bind to proteins and control their activity. Small RNA-mediated regulation requires numerous and dynamic interplay with various cellular machineries, including RNA polymerase, ribosomes, and degradosomes, and perturbs these machineries in the pathways of mRNA metabolism to broadly affect gene expression. The RNA chaperone Hfq serves as a key player in the sRNA regulatory pathways, where it functions in two main aspects: stabilization of sRNAs from degradation and promotion of the annealing between sRNAs and their target mRNAs (12). Base-pairing of sRNAs to their target mRNAs with the help of Hfq often leads to changes in translation and/or mRNA stability (positive or negative). Translation inhibition is often associated with RNase-mediated co-degradation of the sRNA-mRNA pair. While post-transcriptional regulation in bacteria has mainly been studied at the population level in batch cultures, little is known about sRNA-mediated regulation at the single cell level and even less about the extent and impact of subcellular localization of RNAs on regulatory processes in these organisms. Due to their important biological function, the subcellular localization of sRNAs and their interactions with target mRNAs, Hfq, and RNase E have become an intriguing research topic.
In this chapter, we describe recent advances in methods that allow for the investigation of RNA localization in bacterial systems, as well as findings regarding mRNA and sRNA localization in these organisms. We discuss the models and mechanisms revealed by these examples of spatial control of RNA. In addition, we introduce new labeling and imaging methods recently developed in eukaryotic cells, which have not yet been applied to bacteria, but have the potential to reveal new insights about prokaryotic transcript localization. Finally, we conclude by laying out open questions and future challenges in the field.
APPROACHES TO STUDY RNA LOCALIZATION
Biochemically, cell fractionation methods have been routinely used to study protein localization to the outer membrane, periplasm, inner membrane, and cytoplasm (13, 14). Similar approaches have also been used to investigate RNA localization. Particularly, when cell fractionation methods are combined with high-throughput RNA sequencing, the relative distribution of transcripts between the membrane and the cytoplasm can be estimated at the whole transcriptome level (15). Compared to fractionation-based approaches, visualization of RNAs by light microscopy techniques provides the most direct information on subcellular localization of individual RNA species. It is worth mentioning that 3D cryo-electron tomography provides another remarkable imaging category with an enhanced spatial resolution compared to conventional light microscopy, and has been applied to imaging subcellular organization of bacteria (reviewed in (16)). However, compared to light microscopy, as in general no specific tags are introduced to the specific biomolecules of interest, cryo-electron tomography usually cannot provide selectivity of the specific biomolecules of interest during imaging.
The majority of studies on bacterial RNA localization discussed in this book chapter are fluorescence imaging-based. Here, we first introduce labeling and imaging methods used in bacterial systems (see also several recent reviews on RNA imaging methods (2, 17, 18)).
FISH:
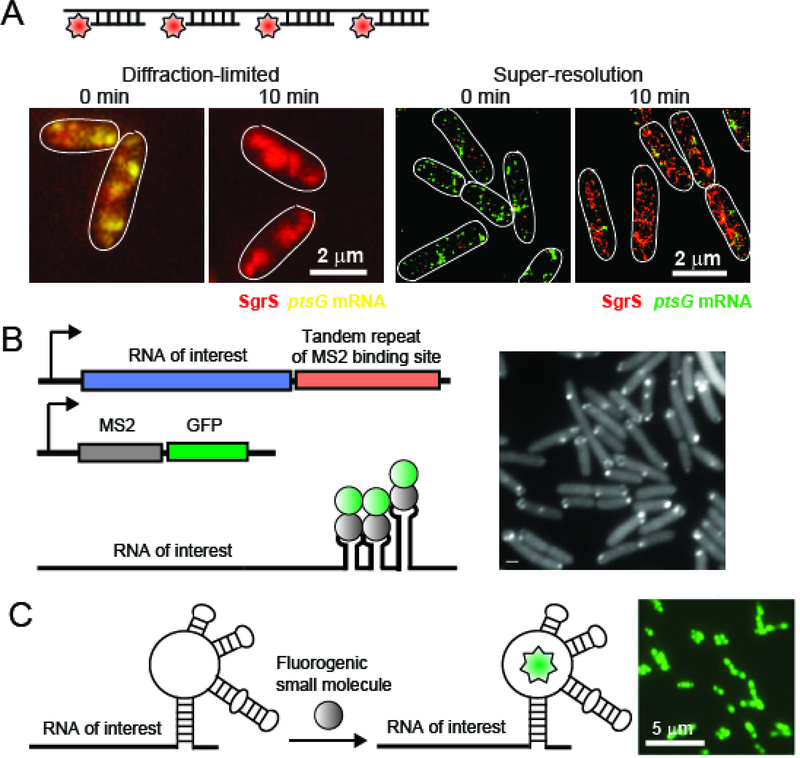
Fluorescence in situ hybridization (smFISH) is one of the most widely used RNA labeling strategies in both eukaryotic and bacterial systems (19–21). By fixing and permeabilizing the cells, DNA oligonucleotides covalently linked with fluorophores can access the interior and hybridize to the RNAs of interest, thereby labeling them (Figure 1A). To enhance the signal-to-noise and target specificity, a few to tens of labeled oligos are used for each RNA, tiling along the nucleotide sequence. Enhanced fluorescent signals from multiple oligos on the same RNA appear as a single spot under the diffraction-limited fluorescence microscope, providing single molecule sensitivity. Despite the limitation of FISH to “dead” cells, many important details can be gleaned from this approach, including the expression levels and localization of RNAs, which can be further used to derive the kinetic mechanisms of transcription or degradation (20, 22–24). FISH has been applied to both mRNA and sRNAs in bacteria (for references see sections on mRNA and sRNA localizations below). However, due to the relatively short length of sRNAs, the application of FISH to sRNAs may be case-dependent, and be more applicable to sRNAs existing in high copy number.
Figure 1: Methods to visualize bacterial RNAs.
(A) smFISH and its application to image SgrS and its target mRNA ptsG (adapted from (105)). Images from diffraction-limited and super-resolution microscopes are shown for comparison. (B) Illustration of the fluorescent protein RBP-RNA aptamer approach, using the MS2 system as an example, and its application to track mRNAs at the single-molecule level in live E.coli cells (image adapted from (67)). The scale bar in the image represents 2 μm. (C) The Spinach aptamer and its application to image mRNAs in live E.coli cells. The image is adapted from (58) (licensed under a Creative Commons Attribution 4.0 International License: http://creativecommons.org/licenses/by/4.0/), in which a tandem array of Spinach aptamers is fused to the RNA of interest to enhance the signal, and fluorescent detection upon addition of the organic ligand DFHBI.
FISH does not require genetic manipulation of the RNAs of interest, and normally does not perturb their function or certain features, such as lifetime. However, for fixed-cell imaging, fixation and permeabilization conditions can potentially affect the native localization of biomolecules and/or the accessibility of the labeling reagents (25). While the probe accessibility issue is less of a concern for short FISH probes (usually ~20 nucleotides) compared to sizable antibodies in immunostaining protocol, it is still recommended that imaging results are validated by using multiple fixation and permeabilization methods. In addition, negative controls (such as a knockout strain of the RNA of interest) should always be used to examine the level of non-specific binding of the FISH probes.
Fluorescent protein reporters:
Live-cell imaging of RNAs allows for a direct observation of transcript motion, as well as the kinetics of RNA-associated activities. While such approaches are more challenging and require genetic manipulation, several methodologies have been developed for live-cell RNA imaging. One category of approaches relies on orthogonal protein-RNA interactions, consisting of a RNA binding motif engineered into the transcript of interest, together with the cognate RNA-binding protein fused to a fluorescent protein (FP) (2). The FP-fused binding protein recognizes and binds to the RNA motif, thereby labeling the RNA. Repetitive RNA motifs of the same kind are often inserted into the RNA of interest to recruit multiple FP-fused binding proteins, thereby enhancing the signal, even to single transcript sensitivity (Figure 1B). Commonly-used RNA-protein pairs include MS2 and PP7 phage coat proteins with their respective RNA motifs, as well as λ-phage N-protein-boxB hairpin pair (26–28). There are several derivatives of this approach. To increase the signal brightness and photostability, FPs can be replaced by other visualizable tags on the RNA binding proteins, such as SNAP-tag or eDHFR-tag (29–32). In these approaches, the tags can be labeled by the addition of organic dyes introduced into the cell. Such methods rely on the ability to introduce the dyes into cells either by their natural membrane permeability or manual delivery via transfection or microinjection. Since FP-fused RNA binding proteins are constitutively expressed and fluorescent in the cell, even if not bound to the target RNAs, they can result in significant background signals. In order to lower the background, another derivative of the FP live-cell approach employs complementation of the fluorescent reporter upon RNA binding. In this method, FPs are expressed as two independent halves, fused to either two RNA binding proteins or halves of the same RNA binding protein, respectively. Only when the two FP halves are brought into close proximity through binding to the same RNA molecule, a complete, fluorescent FP is reconstituted. For example, the RNA-binding eukaryotic translation initiation factor eIF4A can be expressed as two independent halves, each fused to a half of a split enhanced GFP (EGFP) (33, 34). Upon binding of an eIF4A target encoded in a transcript of interest, fluorescent eGFP is reconstituted. It should be noted that the split eIF4A reporter system can only be applied to bacterial cells as they natively lack eIF4A. Similarly, split EGFP has been fused to two different Pumilio homology domains (PUM-HD) of human PUMILIO1 (35), and the FP Venus has been split into two domains, fused to either the PP7 or MS2 coat proteins, and used to detect RNA expressing adjacent PP7 and MS2 aptamers (36).
The fluorescent protein reporter systems use indirect labeling methods, in which significant modifications have to be introduced to the RNAs of interest. Therefore it is important to know the potential pitfalls of these methods. First, fluorescent proteins have a propensity to oligomerize (37, 38) also has such a propensity (39). Therefore, a careful choice of the FP-fused RNA binding protein is necessary to avoid the formation of artificial, RNA-independent foci. An independent labeling method, such as FISH, is recommended to be used to verify whether the fluorescence protein foci indeed also contain RNA of interest. Second, the fluorescent protein reporter systems have potential risks of changing the mRNA processing, lifetime and localization, due to the protection by the bound proteins or due to the tandem array of inserted RNA motif itself (40–44). Therefore, while this approach is very useful in revealing transcriptional activity, results have to be carefully interpreted when applied to studies of RNA degradation and localization. Recently, a re-engineered MS2 system has been developed that has minimal effect on mRNA half-life (45). In addition, an mRNA degradation assay using rifampicin can provide a good test on potential effects of the labeling method on mRNA turnover. Hereby, the signal of the fluorescent reporter on the mRNA should decay with the same kinetics as the native mRNA upon rifampicin treatment, e.g., to rule out accumulation of a reconstituted or aggregated reporter with the mRNA part already being degraded. A comparatively long-lived fluorescent signal is therefore unlikely to reflect the correct localization of the mRNA. A strong overexpression, e.g., from an artificial promoter, might also impact on the transcript’s properties, such as stability, function or localization. Therefore, preference should be given to expression from a native promoter.
Fluorescent RNA aptamers:
RNA-protein interaction-based methods often raise the concern of changes of molecular and functional properties of the tagged RNA, due to the binding of multiple bulky FPs to introduced repetitive, often structured, RNA motifs. This. may be especially problematic for short or structured transcripts such as sRNAs. The development of RNA aptamer based imaging methods provides another possibility of live-cell RNA imaging. This approach utilizes RNA aptamer sequences added to the transcript of interest and fluorogenic small molecules that can freely diffuse into the cell, and become fluorescent upon binding to the RNA aptamer (see also several recent reviews (17, 46–48)). While not quite suitable for live-cell imaging, earlier developments of malachite green (MG), thiazole orange (TO), and dimethyl indole red (DIR) aptamers, etc. provided proof-of-concept and illustrated the feasibility of such approaches (49–51). The Spinach system (Figure 1C) is an example of the first generation of RNA aptamers for live-cell imaging (52). Since then, new aptamer systems have been developed, including Spinach 2 (53), Mango (54), Broccoli (55, 56), and Corn (57). Repetitive Spinach aptamers have been shown to increase the brightness by a maximum of 17-fold (with 64 repeats) compared to a single Spinach monomer at a cost of a significant lengthening of the aptamer sequence (58). Alternatively, the fluorogenic small molecules have been designed into a covalently linked fluorophore-quencher pair, and the aptamers are selected to either bind the quencher or the fluorophore. In this design, the fluorophore is quenched by the linked quencher in the absence of the aptamer. Once either the quencher or the fluorophore binds to the aptamer, they are more physically separated and the fluorophore is dequenched (59, 60).
Despite their relatively broad applications in the area of metabolite sensing through engineering to various riboswitches (also see reviews (17, 46–48)), the use of RNA aptamers in single-molecule RNA imaging is still considered to be limited. This limited application of the aptamer-based method for single-molecule RNA imaging might be related to, e.g., the limited brightness of the tag, and the requirement for correct folding of the aptamer in vivo. Further engineering of smaller and more stable aptamers, brighter fluorophores, and tandem arrays of the aptamer could collectively help to achieve single-RNA sensitivity. Similar to the fluorescent protein reporter systems, the fusion of an aptamer sequence to a transcript of interest might also affect its properties, such as stability or function. Therefore, certain functional validation of the tagged RNA will still be necessary.
Super-resolution microscopy:
With the various labeling methods described above, RNA can be directly visualized under the fluorescence microscope. However, due to the diffraction limit of visible light, conventional optical microscopy has a limited resolution of around 200–300 nm in the lateral direction and 500–700 nm in the axial direction. While FISH can provide single-molecule sensitivity on RNA imaging, because of the small size of a bacterium (a few hundred nanometers to a few microns), only individual RNAs with very low abundance can be resolved in bacteria. Several super-resolution techniques that can break the diffraction limit have been developed. Among these techniques, single-molecule localization microscopies (SMLM) are most frequently applied in bacterial systems, in which photoactivatable FPs and photoswitchable dyes are used to label biomolecules, and can push the spatial resolution into the range of 10–20 nm (61–63). Therefore, the utilization of such super-resolution microscopy techniques can provide finer details on the RNA localization.
LOCALIZATION OF MESSENGER RNAS
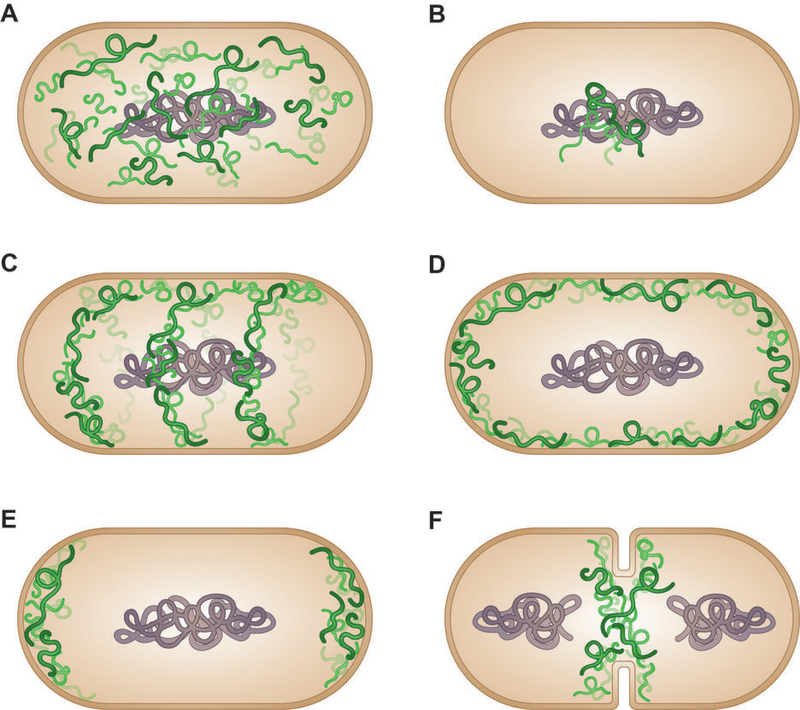
Besides distinct subcellular localizations of proteins and the nucleoid, diverse subcellular localizations of mRNAs have also been reported in various bacteria. Commonly-observed patterns of distribution of bacterial mRNAs include uniform expression throughout the cytoplasm, localization into distinct foci close to the nucleoid, formation of helical patterns along the cell axis, enrichment at the inner membrane, or concentration at the cell poles or the septum during cell division (Figure 2). For example, using the split eIF4A approach in live cells, Broude and co-workers observed that lacZ mRNA was evenly distributed along the E. coli cell (Figure 2A) (33). In contrast, another study observed limited dispersion of lacZ mRNA in E. coli as well as of several mRNAs in Caulobacter crescentus from their site of transcription using FISH in fixed cells (Figure 2B) (20, 64). The cat mRNA, encoding a cytoplasmic chloramphenicol acetyltransferase, demonstrates a helix-like pattern in the cytoplasm in E. coli (Figure 2C) (65). The lacY mRNA, encoding a membrane-bound lactose permease, localizes at or near the inner membrane (Figure 2D) (65). In the Gram-positive bacterium Bacillus subtilis, MS2-GFP tagging revealed that comE mRNA, encoding the late competence operon, localizes to the cell poles (Figure 2E) and the nascent septum that separates the daughter cells (Figure 2F) (66). The sometimes disparate results of observed subcellular localizations of the same transcript might be due to the different RNA visualization technologies and expression systems used.
Figure 2: Diverse patterns of subcellular mRNA localization in bacteria.
Schematic drawings of diverse mRNA localization patterns commonly reported in different bacteria. RNA molecules are shown in green, and the nucleoid in grey. (A) Distribution throughout the cytoplasm. (B) Localization at the site of transcription in the nucleoid. (C) Helical localization. (D) Enrichment at the inner membrane. (E) Localization at the cell poles and (F) septum.
mRNA accumulation near site of transcription:
The observed nucleoid-associated transcript foci led to the proposal that bacterial mRNAs may remain localized close to their genomic site of transcription (64). Using a combination of MS2-tagging and validation by RNA-FISH, one study reported that the mRNAs of groESL, creS, divJ, ompA and fljK of C. crescentus and the lacZ transcript in E. coli show limited dispersion from their site of transcription and are visible as distinct foci in the vicinity of the genomic DNA locus from where they are transcribed (64). Similarly, visualization of a transcript containing repeated MS2-binding aptamer sequences revealed that most transcripts moved randomly in a restricted spot near mid-point or quarter-point of the cells, which corresponds to localization of the F-plasmid which was used for expression of the RNAs, whereas a minority diffused throughout the cell or moved as chains (67). In this study, the observed distinct spots were suggested to be mRNAs that are still tethered to DNA by RNAP, while freely diffusing transcripts are completed transcripts.
mRNA accumulation at sites of protein requirement:
The observations that mRNAs accumulate at various other subcellular places in addition to close association with their genomic site of transcription (Figure 2), indicate that other cellular processes can affect the localization of the mRNA. For example, using a split aptamer approach for RNA labeling in live E. coli cells as well as independent validation by FISH, distinct spots of the endogenous ptsC mRNA, encoding an integral transmembrane transporter, could be detected, which do not colocalize with bulk DNA (68). Similarly, FISH analysis revealed distinct, uneven patterns of localization and specific foci at one or both poles for the dinitrogenase reductase-encoding nifH transcripts of Klebsiella oxytoca and Azotobacter vinelandii with specific foci at one or both poles in K. oxytoca (69). One intriguing hypothesis is that mRNAs can be targeted to the subcellular domains where their encoded protein products are required (cytoplasm, poles or inner-membrane) (65, 70, 71). For example, while the cat mRNA, encoding the cytoplasmic chloramphenicol acetyltransferase, was observed in a helix-like pattern in the cytoplasm of E. coli, the mRNA of lacY, encoding the membrane-bound lactose permease, was preferentially detected near the cytoplasmic membrane (65). In line with these microscopy observations, upon fractionation of E. coli, the authors found that the cat and lacY mRNAs are enriched in cytosolic and membrane samples, respectively (65). Moreover, the same study reported the polycistronic bglGFB mRNA, which encodes both membrane-bound (BglF sugar permease) and soluble (BglG transcription factor and BglB phospho-β-glucosidase) components, was enriched at the cell membrane, indicating that envelope-targeting signals may be dominant. Furthermore, inhibition of translation with kasugamycin or chloramphenicol revealed that this membrane localization occurs in a translation-independent manner (65). This observation was further supported by introducing mutations that abolish bglF translation, which did not affect localization, indicating that cis-acting signals in the RNA itself dictate membrane localization (65). The bglF membrane targeting signal, which is present in the sequence encoding the first two transmembrane helices of BglF, was found to be dominant over other operon components, because bglG mRNA expressed alone localizes to cell poles and bglB mRNA expressed alone is cytoplasmic (65). It has also been reported that cis-encoded RNA localization elements in the early coding region of mRNAs encoding Yop effector proteins (e.g. YopE and YopN) are required for the secretion of these effector proteins by type III secretion systems in Yersinia (72, 73). However, it is unknown if these transcripts are localized to the membrane or in proximity to the secretion apparatus. Similarly, N-terminal domains are required for secretion of flagellar proteins in diverse bacteria, and both mRNA and peptide signals are recognized by the secretion apparatus and contribute to secretion efficiency (74). For example, secretion of heterologous proteins was facilitated by fusing the 173-bp 5’UTR of the fliC gene (encoding flagellin) as well as a fliC transcriptional terminator (75).
While most studies have mainly looked at localization of only a single or small set of mRNAs, recently Zhuang and co-workers investigated the spatial organization of mRNAs in E. coli on the transcriptome scale (70). They designed complex FISH probe sets to visualize defined sets of mRNAs, categorized by the subcellular localizations of their encoded proteins, allowing examination of about 27% of all E. coli mRNAs. This revealed that mRNAs encoding inner-membrane proteins tended to be preferentially located at the inner membrane, whereas transcripts encoding cytoplasmic, periplasmic, and outer membrane proteins were generally detected more uniformly throughout the cytoplasm. Moreover, labeling of specific subgroups of mRNAs as a function of genomic localization of their genes revealed that spatial genome organization does not play a major role in the shaping of the cellular distribution of the transcriptome. Because inhibition of translation initiation using kasugamycin treatment disrupted envelope localization of membrane protein-encoding transcripts, Moffitt et al. concluded that membrane localization of these transcripts is translation-dependent. Most inner membrane proteins are secreted via the signal recognition particle (SRP) pathway, which co-translationally inserts proteins into the membrane, while outer membrane proteins are post-translationally secreted via the SecB pathway (76). Examination of localization by FISH of fluorescent reporter fusions to SRP or SecB signal peptides confirmed that mRNAs with SRP-signal sequences were found enriched at membranes, suggesting that these transcripts are recruited to the envelope during co-translational secretion of the ribosome-bound nascent signal peptide (70). Furthermore, introduction of stop codon mutations into the bglF mRNA abolished its localization at the membrane, indicating that translation of its SRP-signal sequence is required for membrane localization (70). Whether the protein signal sequence or solely the act of translation of its mRNA is required is so far unknown. Another study also reported, based on cell-fractionation, RNA-seq, and qPCR, that mRNAs of inner membrane proteins are enriched at the membrane and depletion of the SRP receptor FtsY reduced the amounts of all mRNAs at the membrane (15). However, mRNAs encoding inner membrane proteins were also found in the soluble ribosome-free fraction, which might represent a stage prior to their translation and targeting to the membrane. Mathematical modeling has suggested that translation initiation rates, the availability of secretory apparatuses, and the composition of the coding region defines mRNA abundance and residence time near the membrane (77). In addition, modeling suggested that formation of membrane protein clusters might be facilitated by a bursts of proteins translated from a single mRNA anchored to the membrane, and therefore spatiotemporal dynamics of mRNAs might strongly influence the organization of membrane protein complexes.
Recently, the mRNA encoding the major flagellin FlaA of the food-borne pathogen C. jejuni was also observed, using RNA-FISH, to localize to the cell poles in a translation-dependent manner (71). C. jejuni has two polar flagella, and polar flaA mRNA localization was primarily detected in short cells, which likely correspond to cells that have just divided and are building a new flagellum at the nascent cell pole. While polar localization was abolished for a translation-incompetent flaA mRNA with stop codon mutations before the N-terminal peptide, translation of the first 100 codons partially restored polar localization, suggesting that an N-terminal amino acid signal, or its translation, is sufficient for mRNA localization. Similarly, translation of the N-terminal peptide of type III section effector protein mRNAs has been shown to be required for secretion (72, 78).
mRNA localization by specific trans-acting factors:
Emerging examples have also indicated that there are specific trans-acting factors that post-transcriptionally regulate mRNA localization. For example, overexpression of the cold-shock protein CspE increased the fraction of membrane protein mRNAs in the ribosome-free fraction and their amount on the membrane and positively affected their translation, indicating potential regulation of subcellular RNA localization (15). Moreover, the 98-aa protein ComN, a post-transcriptional regulator of competence gene expression, localizes to the division site and cell poles via direct interaction with DivIVA, a key protein involved in cell pole differentiation in B. subtilis (66). ComN-DivIVA interaction promotes accumulation of comE mRNA to septal and polar sites, indicating that localized regulators can also impact mRNA localization in bacteria. Furthermore, localized mRNA translation of ComE proteins might be required for efficient competence development. During a global analysis of the direct RNA regulon of the translational regulator CsrA by RIP-seq, flaA mRNA was revealed as the major translationally repressed target of CsrA in C. jejuni and the above-mentioned polar flaA mRNA localization was detected to be post-transcriptionally regulated by the CsrA-FliW regulatory system (71). Deletion of the CsrA protein antagonist FliW releases CsrA and in turn allows translational repression of flaA mRNA, abrogating its polar localization. This observation suggests both that mRNA localization is translation-dependent and that spatial control of bacterial transcripts can be regulated post-transcriptionally. It remains to be seen how many other flagellar mRNAs localize to the flagella apparatus and whether polar localization of the flaA mRNA or membrane localization of inner membrane protein mRNAs has any effects on the efficiency of secretion and/or assembly of larger complexes, or is just a by-product of co-translational secretion.
LOCALIZED MRNA TRANSLATION AND DEGRADATION
While transcription and translation occur at distinct places in the nucleus and cytoplasm in eukaryotes, translation can already initiate co-transcriptionally in the cytoplasm of bacteria and translation can continue after transcription completion and release of the mRNA. Furthermore, the above-mentioned examples show that mRNAs can migrate outside the nucleoid and translation can take place at distinct locations in the cell, such as at the membrane during co-translational secretion of inner membrane proteins. Moreover, it has been reported that the RNA polymerase (RNAP) transcription machinery and ribosomes occupy partially different subcellular regions within different bacterial cells (79–81). For example, in B. subtilis, RNAP has been primarily detected inside and ribosomes outside the nucleoid, respectively (82), indicating that transcription and translation are spatially separated. Cryoelectron tomography indicated that 70S ribosomes of Spiroplasma melliferum are distributed throughout the cytoplasm, with 15% in close proximity to the membrane (83). Similar electron cryotomography analysis of the cellular ultrastructure of logarithmically growing cultures of C. jejuni revealed ribosome exclusion zones at cell poles (80). Also consistent with extra-nucleoid distribution of ribosomes, 5S rRNA was detected as an array of fluorescent particles distributed along the cell or the cell poles in E. coli (33), indicating specific localization of ribosomes outside the nucleoid in E. coli, which is in agreement with enrichment of ribosomal proteins at the cell periphery and cell poles in both E. coli (84) and B. subtilis (82, 85). While several ribosomal proteins (L1, S2, L7/L12) are enriched at either of the cell poles and the translation factor EF-Tu colocalizes with the bacterial cytoskeleton protein MreB, it remains unclear how many of these localized translation factors are incorporated into actively translated ribosomes (reviewed in (86)). It should be noted, that the cellular localization can differ for unbound subunits versus actively translating ribosomes or transcribing RNAP. For example, it has been reported that mRNA-free ribosome subunits are not fully excluded from the nucleoid, thereby allowing for translation initiation on nascent mRNAs throughout the nucleoid and co-transcriptional translation (81). Moreover, a large fraction of RNAP, presumably the transcribing population, has been reported to be primarily located at the periphery of the nucleoid and thus is close to the pool of ribosomes excluded from the nucleoid (87).
The specific organization of ribosomes has also been reported to change during different growth conditions in some bacteria (88). Overall, the distinct subcellular localization of ribosomes, mRNAs and RNAP indicate that transcription and translation are not necessarily coupled in bacteria and localized translation by specific ribosomes at subcellular locations might also play a role in bacterial gene regulation. For example, inner-membrane bound ribosomes in E. coli that are actively engaged in translation (89) might play a role in specific translation of inner membrane proteins. In C. crescentus and another alpha-proteobacterium Sinorhizobium meliloti, ribosomes are detected throughout the cell including the nucleoid region (64, 90), which is different from the ribosome/nucleoid segregation observed in E. coli and B. subtilis (82, 84). Hereby, it was suggested that, mRNA-bound ribosomal subunits show limited mobility in C. crescentus due to the observed limited dispersion of their mRNA targets from their site of transcription (64, 90, 91).
Similar to the specific and dynamic localization of ribosomes and transcripts in bacterial cells, distinct subcellular distribution of RNA degrading proteins and complexes has also been reported in prokaryotes (92–94). The E. coli degradosome initiates most of the RNA decay in bacteria and contains RNase E, the RNA helicase RhlB, polynucleotide phosphorylase (PNPase), and enolase (95). The B. subtilis degradosome consists of PNPase, RNases J1, J2, and Y, as well as the DEAD-box RNA helicase CshA, enolase, and phosphofructokinase (96). Components of the E. coli degradosome have been reported to associate with the membrane or assemble into helical filaments (97–100). Using super-resolution imaging of 24 fluorescent protein fusions to RNA degradation and processing enzymes in E. coli, Moffitt et al. detected that only the four proteins (RNase E, PNPase, RhlB, and the polyadenylation enzyme PAPI) were enriched at the membrane, whereas the other tested fusions were mainly uniformly distributed throughout the cytoplasm (70). The membrane localization of the degradosome is mediated by membrane anchoring of segment A of RNase E (98). In line with a co-localization of inner membrane protein-coding mRNAs and degradosome components at the cell envelope, it has been observed that these transcripts had shorter half-lives than the mRNAs of cytoplasmic, periplasmic, or outer membrane proteins (70). Moreover, artificial targeting of mRNAs to the membrane via fusion to SRP-signal sequences destabilizes these mRNAs. Thus, co-colocalization of certain mRNAs with RNA degradation components can lead to their specific degradation. Interestingly, the degradosome localization can be impacted by growth conditions: a redistribution of RNase E/enolase from membrane-associated patterns under aerobic to diffuse patterns under anaerobic conditions results in stabilization of DicF sRNA and filamentation of the bacteria (101). For more information on the E. coli degradosome see also the chapter by Bandyra & Luisi in this book (Bandyra & Luisi, 2018, RNase E and the High-Fidelity Orchestration of RNA Metabolism. Microbiology Spectrum).
Interestingly, RNase E shows a patchy localization pattern in C. crescentus and the clustered localization of this enzyme is determined by the location of DNA, independent of its mRNA substrates (64, 90). Hereby, the localization of RNase E clusters was found to correlate with two subcellular chromosomal positions that encode the highly expressed rRNA genes, indicating that RNase E mediated rRNA processing occurs at the site of rRNA synthesis (90). Although mediated by apparently different mechanisms compared to E. coli, the association of RNase E with DNA in C. crescentus also indicates that there is spatially organized mRNA decay in this organism. Such a subcellular organization of the RNA degradation machinery likely also applies to other prokaryotes. For example, the membrane-bound RNase Y of Gram-positive bacteria (e.g. B. subtilis and Staphylococcus aureus) appears to be assembled at similar cellular locations as other RNA degradation enzymes (see (93, 102)). Recently, it has been reported that RNase Y is recruited to lipid rafts via flotillin in S. aureus and that flotillin increases RNase Y function (103), indicating localized RNA degradation in bacterial membrane microdomains. How many and which mRNAs are mainly targeted by such localized degradation remains to be seen.
LOCALIZATION OF SMALL RNAS
Localization of bacterial small regulatory RNAs (sRNAs) has been predominately investigated with FISH. The housekeeping transcript transfer-messenger RNA (tmRNA, also known as SsrA), involved in stalled ribosome rescue, was one of the first sRNA investigated regarding its subcellular location (104). tmRNA contains both tRNA-like and mRNA properties. It forms a ribonucleoprotein (RNP) complex (tmRNP) with small protein B (SmpB) to function in trans-translation. During trans-translation, the tmRNP binds to stalled ribosomes and adds a proteolysis signaling tag to the stalled peptide, thereby recycling the ribosome and facilitating the degradation of the aberrant protein product. By FISH labeling of tmRNA and immunostaining of several relevant proteins, including SmpB and RNase R, in C. crescentus, Russell et al. demonstrated a helix-like pattern of tmRNA, SmpB and RNase R in the swarmer cells (G1 phase), whereas in S phase after initiation of DNA replication, tmRNA is largely degraded by RNase R with the remaining transcript becomings homogeneously distributed in the cytoplasm (104). Both tmRNA and SmpB show a high degree of colocalization, consistent with the formation of a tmRNP, whereas the helical structures formed by the tmRNP and RNase R are mostly distinct from each other. The helical organization of tmRNA relies on SmpB; however, the underlying molecular basis of this distinct organization remains unclear. In addition, such helical organization is not disrupted upon translation inhibition, suggesting that it is not related to trans-translation activity. These results indicate the most likely biological relevance of such a helical organization is to protect tmRNA from RNase R-mediated degradation during G1, as they are localized “out of phase”, whereas in the S phase, proteolysis of SmpB would release tmRNA from its helical location away from RNase R allowing it to be degraded in order to regulate the abundance and function of tmRNA in a cell-cycle dependent manner.
Compared to mRNAs, studies of several sRNAs suggest that the distribution of these transcripts is less compartmentalized compared to coding transcripts. With smFISH and super-resolution imaging Fei et al. showed that SgrS, an sRNA involved in the glucose-phosphate stress response, is roughly homogeneously distributed in the cytoplasm when its copy number is high, whereas when its expression is lower, it appears to be specifically absent from the central nucleoid region (105). A more comprehensive study of the localization of several several sRNAs, including GlmZ, OxyS, RyhB and SgrS, found equal preference for localization within cytoplasm and nucleoid region and no preferential membrane localization, or cell poles, as often observed for mRNAs by quantifying the signal overlap between RNA FISH and DAPI staining of DNA (106). Interestingly, the ability of a particular transcript to freely diffuse into the nucleoid region was correlated with the length and the translation activity of the RNA, because shortening and reducing translation of gfp mRNA led to the same localization as sRNAs and reduced nucleoid exclusion. Therefore, in general, longer RNAs, including coding RNAs, have a lower propensity towards nucleoid localization compared to shorter, often non-coding transcripts. So far, the available studies on a handful of examples of sRNAs suggest an unbiased cellular localization (105, 106). It should be noted that investigation of sRNA localizations are predominately focused on non-coding regulatory RNAs. Even though SgrS is a dual functional sRNA, which also encodes the small protein SgrT (107) under the conditions of previous investigations, SgrT was not translated actively. Therefore, SgrS is still considered as regulatory RNA only under the investigated conditions, and demonstrates the same localization behavior as other tested regulatory sRNAs (108).
The unbiased localization of regulatory sRNAs investigated so far seems to be consistent with the fact that typically each sRNA species can regulate multiple mRNA targets (11, 109), which themselves might each adopt different cellular localizations. Therefore, the availability of sRNAs throughout the cell will ensure that all targets can be regulated. On the other hand, the localization of the target mRNA might kinetically affect regulation by the local availability of its sRNA regulator or other protein factors. Computational simulations have shown that a biased localization of a sRNA (such as membrane vs. cytoplasmic localization) can lead to a distinct regulation of different target mRNAs, providing an opportunity for regulatory hierarchy among different targets (110). For example, a membrane-localized sRNA would regulate a membrane-localized target mRNA more efficiently compared to a cytoplasmic counterpart. However, the observation that sRNAs tested so far all exhibit an unbiased localization suggests that mRNA targets, regardless of their localization, would be equally sampled by the same sRNA species. Therefore, any regulatory hierarchy would be attributed to the other factors, such as localization of Hfq and RNase E, and the strength of base-pairing interactions, etc. As an example, the SgrS-mediated degradation of the mRNA encoding a fusion of ptsG to crp, for example, has been shown to be significantly reduced upon elimination of the transmembrane domains of PtsG (111). This observation is reminiscent of the observed faster endogenous turnover of membrane localized mRNAs compared to mRNAs with other cellular localizations (70) and of the membrane-localization of RNase E (see also the chapter by Bandyra & Luisi in this book: Bandyra & Luisi, 2018, RNase E and the High-Fidelity Orchestration of RNA Metabolism. Microbiology Spectrum).
Hfq is an important chaperone for small RNAs in bacteria and also facilitates sRNA-mediated gene regulation via base-pairing (12, 112). Therefore, understanding the localization of Hfq can provide insight into spatiotemporal themes of sRNA-based regulation. However, results from various studies using different labeling and imaging approaches to study Hfq localization are conflicting, and therefore its localization in the cell remains controversial. Hfq has been observed to adopt a diffuse cytoplasmic localization outside of the nucleoid region by immunofluorescence staining (79), as well as preferential membrane localization by electron microscopy (113), which was recently recapitulated in an in vitro system using artificial vesicles (113, 114). A helical organization of Hfq along the longitudinal direction of the cell has also been observed by immunofluorescence staining (99, 113, 115). Different from all the fixed-cell experiment, single-molecule tracking of fluorescent protein-tagged Hfq in live-cell reveals that Hfq is essentially freely diffusing inside the cell, with the diffusion rate slowed down when Hfq binds to the newly transcribed RNA attached to the nucleoid (116). Future experiments are needed to resolve this discrepancy in observation of Hfq localization. Despite absence of Hfq significantly affecting the stability and abundance of sRNAs (105), it has minimal effect on the localization of tested sRNAs (106). As mentioned above, localization of the ptsG mRNA to the inner membrane has been reported to strongly contribute to its efficient repression by the sRNA SgrS, together with Hfq, during phosphosugar stress (111). So far, it remains unclear if Hfq can actively localize SgrS to the membrane to facilitate sRNA-ptsG mRNA interactions, or if in contrast, SgrS-ptsG mRNA complexes might instead lead to Hfq localization. Moreover, membrane localization of Hfq might also be due to interactions with the degradosome.
EMERGING STRATEGIES AND APPROACHES
Recently, new developments in chemical biology and molecular engineering have expanded the toolkit for imaging RNA localization and measuring their activities in vivo. These tools have been developed mostly for RNA imaging in eukaryotic systems, but some of them might have the great potential to be applied to bacterial systems as well.
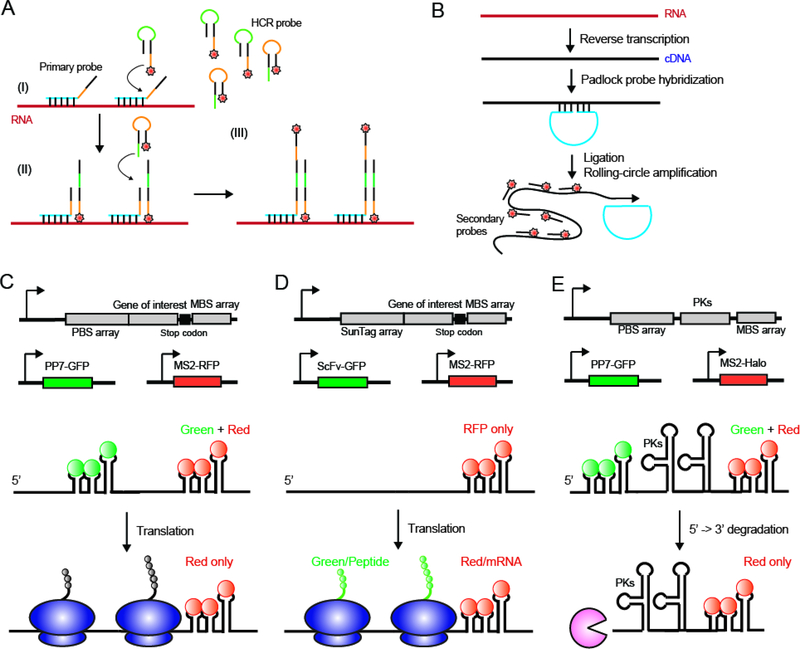
First, various new developments in FISH methods have allowed signal amplification and high-throughput imaging. For example, in the in situ hybridization chain reaction (HCR) (117, 118), two fluorescently-labeled oligonucleotides that alone exist as metastable hairpin structures are designed (Figure 3A). A third initiator probe is then introduced to bind to the target RNA of interest. This initiator probe has an overhang region that can open one of the two fluorescently labeled hairpins by branch-migration, making the first hairpin-containing oligo capable of hybridizing to the second hairpin oligo. The two fluorescent oligos bind to each other in an alternating fashion in the hybridization chain reaction, thereby linking multiple fluorescently labeled probes to the RNA of interest and amplifying the signal. In situ polymerase chain reaction (PCR) has also been used for FISH signal amplification (119–122). In this method, padlock probes (long oligonucleotides whose ends are complementary to the target RNA, where hybridization results in circularization of the probe) are hybridized to cDNAs generated from endogenous RNAs of interest in situ, which generates nicked ssDNAs that are further ligated to be circular DNAs (Figure 3B). Rolling circle amplification from these circular DNA templates by DNA polymerase can generate products carrying repetitive sequences for the hybridization of the fluorophore labeled secondary probes. Such FISH strategies with signal amplification can be adapted to bacterial cells, and could be beneficial for imaging sRNAs, to which it is difficult to attach multiple conventional FISH probes due to their short length. To increase the throughput of RNA imaging, various multiplexed FISH imaging methods have also been developed in eukaryotic cells. Multiplexed imaging can be achieved by fluorescent barcoding, in which different combinations of fluorophore-labeled oligos hybridize to separate RNA species to generate pseudo-colors (123), or repetitive hybridization and imaging cycles (124–127). However, such multiplexed imaging methods rely on the ability to spatially resolve individual transcripts. With diffraction-limited microscopy, only RNAs with very low abundance can be resolved individually in bacterial cells due to their small size, making the application of the multiplexed imaging method practically difficult in bacterial systems.
Figure 3: Emerging mRNA imaging methods in eukaryotic systems.
(A) In the in situ hybridization chain reaction (HCR), binding of the primary probe initiates the alternating binding of two HCR probes thereby amplifying the signal. (B) In the in situ polymerase chain reaction (PCR), a cDNA is first generated from the RNA of interest. Padlock probes are hybridized to the cDNA and ligated to be circular DNAs. Fluorophore labeled secondary probes are then hybridized to the products generated from rolling circle amplification of these circular DNA templates. (C) Schematic representation of the TRICK reporter construct. (D) Schematic representation of the SunTag construct. (E) Schematic representation of the TREAT reporter construct.
Recently, CRISPR-Cas systems have been engineered to label both DNA (128–130) and RNA (131, 132) in live cells. An early example of RNA labeling with CRISPR-Cas systems originated from the finding that Cas9, an RNA-guided DNase, can also target single-stranded RNA by providing the PAM (protospacer adjacent motif) as part of an oligonucleotide (PAMmer) (133). Thereby, a catalytically inactive Cas9 (dCas9), fused to a fluorescent protein and in complex with the PAMmer and single guide RNA (sgRNA), can target and label the RNA of interest in a programmable fashion (131). Recently, Cas13a, an RNA-guided RNA-targeting CRISPR–Cas effector, has been applied for RNA tracking in mammalian cells (132). Compared to the MS2/PP7 systems mentioned above, one advantage of CRISPR-based imaging is that no additional tagging sequence must be fused to the RNA of interest, as the sgRNA can be flexibly programmed to target any RNA sequence of interest. Nevertheless, similar to other fluorescent protein reporter systems, CRISPR-based labeling might affect mRNA processing, degradation or localization for a specific RNA of interest. Moreover, to distinguish target RNA-bound fusion proteins from those that are unbound, which could contribute to high levels of background fluorescence, additional modifications are necessary. One such strategy could be to utilize multiple sgRNAs to tile along the RNA sequence of interest to enhance the signal on the RNA compared to background levels, in a similar fashion as is employed in FISH labeling. Another strategy would be to reduce the unbound fraction of the fluorescent Cas proteins. For example, in a recent approach, a zinc-finger (ZF) binding domain and transcription repressor KRAB have been fused to Cas13a-FP, and the ZF binding site was inserted to the promoter controlling the expression of the fusion protein (132, 134). In this way, a negative-feedback loop is created, and the unbound Cas13a-FP therefore auto-represses its expression to control the background fluorescence.
Single-molecule RNA tracking has also been combined with additional reporters to correlate their translation or degradation activities in eukaryotic cells. A biosensor called TRICK (translating RNA imaging by coat protein knockoff) has been developed that reports the first round of translation (135). In this imaging system, cassettes of the PP7 binding site and the MS2 binding sites are integrated into the coding region and the 3’UTR, respectively (Figure 3C). Newly-synthesized mRNAs carry both GFP-fused PP7 proteins and RFP-fused MS2 proteins, generating colocalized green and red under the microscope. During the first round of translation after export of the mRNA to the cytoplasm, ribosomes displace the GFP-PP7 fusion from transcripts, leaving only RFP signals. The SunTag system allows real-time tracking of multiple cycles of translation on individual mRNAs (136–140). In this technique, a tandem array of sequence coding for a short peptide is inserted into the open reading frame of interest. Once translated, the resulting polypeptide, containing multiple such short peptide epitopes, are recognized and bound by a specific single-chain variable fragment (scFv) antibody fused with GFP co-expressed in the cell (Figure 3D). In parallel, the mRNA of interest is also labeled by either the MS2 or PP7 system with a different fluorescent protein (e.g. RFP) at the 3’UTR. Therefore, actively-translating mRNAs generate two fluorescence signals, whereas untranslated mRNAs only generate an RFP signal. In addition to reporters of translation, a biosensor called TREAT (3’-RNA end accumulation during turnover) has been developed to track mRNA turnover in eukaryotes ((126, 141)). In this approach, the 3’UTR is engineered to include two viral pseudoknots (PKs) flanked by PP7 and MS2 binding sites (Figure 3E). Since the PK structures block 5’-to-3’ degradation of the transcript by the 5’−3’ exoribonuclease Xrn1 (142), TREAT allows one to distinguish between intact and partially degraded mRNAs. The feasibility of applying such functional reporters to bacterial systems remains to be tested. For example, fluorescent protein reporters in these systems are generally tagged with a nuclear localization signal (NLS) sequence. The NLS can effectively reduce the amount of unbound fluorescent proteins in the cytoplasm and thereby reduce the imaging background. Such a segregation of unbound fluorescent reporter from the mRNA-bound signal cannot be applied in bacteria because they lack a separation of nucleus vs. cytoplasm. In addition, given that there are fundamental differences in mRNA metabolism between pro- and eukaryotes, such as transcription coupled translation in bacteria vs. spatially separated transcription and translation in eukaryotes, and the much shorter lifetimes of mRNA in bacteria compared to the ones of eukaryotic mRNAs, these functional reporters may need to be significantly re-engineered for their applications in bacterial cells.
In addition to imaging of mRNAs, a new method, FASTmiR, has been developed for the imaging of eukaryotic microRNAs (miRNAs) in live cells (143). FASTmiR was designed based on the Spinach system, in which a sensory domain that can base pair with the miRNA of interest is fused to a modified Spinach aptamer. Binding of a miRNA to the FASTmiR helps fold the Spinach aptamer and form the DHFBI binding pocket, therefore generating a fluorescence signal. The concept of FASTmiR may be further developed into platform for imaging bacterial sRNAs. However, given that sRNAs are in general longer than miRNAs, and most contain secondary structures, the structural design of FASTmiR might need to be significantly modified to allow efficient sRNA detection in bacteria.
Finally, in addition to optical approaches that enhance the spatial resolution, expansion microscopy utilizes swellable polymer networks to physically expand the specimen. Such physical magnification leads to an enhanced effective spatial resolution (126, 141). Recently, expansion microscopy has been combined with FISH imaging of RNA (144) in addition to immunofluorescence, and iterative expansion microscopy further enhances resolution from ~70 nm to ~25 nm (145). Expansion microscopy has already been applied to bacteria (146), and should allow for studies of biomolecular location with finer resolution.
OPEN QUESTIONS AND FUTURE CHALLENGES
Despite the emerging evidence of subcellular RNA localization in bacteria, we are still only at the beginning of understanding the underlying mechanisms and potential functions of this process. One of the major questions is what are the deterministic factors or the driving forces for mRNA localization in bacteria. So far, several cis- and trans-acting factors have been suggested to affect transcript localization. For example, many mRNAs encoding inner membrane proteins are preferentially localized at the membrane and this localization can be mediated in a translation-dependent manner during synthesis and secretion of N-terminal signal peptides. However, for certain mRNAs encoding inner membrane proteins or secreted effectors, it has been reported that signals within the mRNA itself are instead required for localization/secretion or that the mRNAs have multiple localization signals, including RNA sequence-based and those encoded in the N-terminal peptide amino acid sequence (10, 72, 74, 147). It remains to be seen how many bacterial mRNAs have a eukaryotic-like RNA zip-code, and what the sequence- and/or structural features of these elements are. Likewise, the features embedded in N-terminal signal sequences of the encoded proteins that might mediate localization of their cognate mRNA are unknown. While eukaryotic mRNAs can carry their zip codes at either the 3’UTR, 5’UTR or coding region (1), the few examples of RNA elements directing localization of bacterial mRNAs have been reported to be located at 5’ ends of mRNAs or within ORFs (10). It has been suggested that an enrichment for uracils in mRNAs encoding integral membrane proteins might be a physiologically relevant signature of this group of mRNAs (148). Furthermore, it will also be interesting to see which (protein) factors bind to bacterial RNA zip codes and how they transport RNAs in the cell. In eukaryotes, many mRNAs are actively transported by RNA-motor complexes along polarized cytoskeleton structures and localize at local anchor signals such as the dynein-1 motor (3). Similar mechanisms involving binding to cytoskeleton proteins, or even cell division factors, might apply in bacteria. In addition, localized protection from RNases, as observed in eukaryotes, might mediate subcellular localization of bacterial transcripts.
It also remains unclear what the functions and physiological roles of localized RNAs and/or localized translation are in the cell, and how perturbation of such localization affects phenotypes related to the encoded protein. The organization and co-expression of functionally-related genes in operons in bacteria might be in line with the observation that RNAs stay close to site of transcription and are also translated close to the nucleoid so that protein complexes can be assembled. On the other hand, localized translation, e.g. at the membrane, might increase the efficiency of assembly of larger protein complexes at the future site of action, especially for those which might be hydrophobic. Such a localized translation at the membrane especially would make sense for secreted factors such as T3SS effectors or flagellins that might be translationally repressed until the secretion machinery is completed and their translation might only be activated upon completion of the secretion machinery. Nevertheless, it is also possible that the membrane localization of mRNAs is just a by-product of secretion of the nascent N-terminal peptide of the translated protein. Moreover, it is still unclear whether the localization of certain mRNAs is regulated during changing growth or stress conditions, or whether this is connected to cell division. In eukaryotes, alternative splicing and polyadenylation can control incorporation of localization signals into mature transcripts and after nuclear export, diverse RNA binding proteins, adaptors and cytoskeleton motors are recruited to localizing mRNAs (1, 2). It has been observed that post-transcriptional regulators, such as the competence regulator ComN in B. subtilis or the CsrA-FliW regulatory network of C. jejuni, can impact RNA localization in bacteria (66, 71). Although this can be an indirect effect via regulation of translation, these first examples show that post-transcriptional regulatory networks can impact RNA localization.
Compared to mRNA localization, even less is known about sRNA localization in bacteria. Several sRNAs also encode small proteins (for a review, see (149)). It still remains to be studied whether these small protein encoding sRNAs might show a more defined cellular localization compared to solely non-coding RNAs. And if yes, it remains to be seen whether the final cellular locations of the encoded small protein products affect the localization of their encoding dual function sRNAs. Localization of sRNAs and their cognate mRNAs to certain sites in the cell might impact the efficiency and hierarchy of target gene regulation and in turn affect the functionality/outcome of genetic circuits. Therefore, considerations about subcellular localization might also be relevant for the design of synthetic gene regulatory circuits, since differential localization of the encoded protein and/or mRNA (membrane vs. cytoplasmic) might impact efficiency of regulation if not all components are expressed in close vicinity.
The ongoing technological developments listed above, together with additional new approaches that emerge, will help to further study RNA localization in bacteria. So far, it is more challenging to applying existing mRNA imaging methods to sRNAs. For example, single-molecule FISH has a significantly compromised signal-to-noise ratio for sRNA imaging because of the much shorter length of sRNAs compared to mRNAs and resulting lower number of probes that can be applied. Similarly, live-cell imaging of sRNAs remains challenging, because tagging with fluorescent proteins or RNA aptamers could impact their regulatory properties and/or localization. Beyond simple detection of transcripts, ideally one would also simultaneously image RNA and translation/protein localization in-vivo to decipher whether localization and localized translation impact translation, protein abundance, and physiology. Considering the sometimes controversial observations that have been reported so far regarding bacterial RNA localization, it is also recommended that findings are validated using different approaches, and experimental design maintains transcript expression and characteristics as close to native as possible. Certainly, continued technological advancements for studying RNA localization in these small organisms will reveal the previously-unappreciated extent of bacterial compartmentalization and its contribution to physiology.
Acknowledgements
We thank Dr. Sarah Svensson, Eric McLean and Dr. Seongjin Park for critical comments on this book chapter and Dr. Sandy Pernitzsch (www.scigraphix.com) for help with preparing Figure 2. Work in the lab of C.M.S. is supported by DFG (Sh580/4-1, GRK2157, SPP2002: Sh580/7-1 and Sh580/8-1), the BMBF (Infect-ERA (ERA-Net), 2nd call, CampyRNA), and a HIRI (Helmholtz Institute of RNA-Based Infection Research, Würzburg, Germany) seed grant (Project-No 6) through funds from the Bavarian Ministry of Economic Affairs and Media, Energy, and Technology (grant allocation numbers 0703/68674/5/2017 and 0703/89374/3/2017). J.F. acknowledges the support from the Searle Scholars Program and the NIH Director’s New Innovator Award (1DP2GM128185-01).
REFERENCES
- 1.Chin A and Lécuyer E (2017) RNA localization: Making its way to the center stage. Biochim. Biophys. Acta, 10.1016/j.bbagen.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Buxbaum AR, Haimovich G and Singer RH (2015) In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol, 16, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mofatteh M and Bullock SL (2017) SnapShot: Subcellular mRNA Localization. Cell, 169, 178–178.e1. [DOI] [PubMed] [Google Scholar]
- 4.Jung H, Gkogkas CG, Sonenberg N and Holt CE (2014) Remote control of gene function by local translation. Cell, 157, 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govindarajan S, Nevo-Dinur K and Amster-Choder O (2012) Compartmentalization and spatiotemporal organization of macromolecules in bacteria. FEMS Microbiol. Rev, 36, 1005–1022. [DOI] [PubMed] [Google Scholar]
- 6.Nevo-Dinur K, Govindarajan S and Amster-Choder O (2012) Subcellular localization of RNA and proteins in prokaryotes. Trends Genet, 28, 314–322. [DOI] [PubMed] [Google Scholar]
- 7.Campos M and Jacobs-Wagner C (2013) Cellular organization of the transfer of genetic information. Curr. Opin. Microbiol, 16, 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keiler KC (2011) RNA localization in bacteria. Curr. Opin. Microbiol, 14, 155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buskila AA, Kannaiah S and Amster-Choder O (2014) RNA localization in bacteria. RNA Biol, 11, 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kannaiah S and Amster-Choder O (2014) Protein targeting via mRNA in bacteria. Biochim. Biophys. Acta, 1843, 1457–1465. [DOI] [PubMed] [Google Scholar]
- 11.Storz G, Vogel J and Wassarman KM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell, 43, 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogel J and Luisi BF (2011) Hfq and its constellation of RNA. Nat. Rev. Microbiol, 9, 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLean R, Inglis GD, Mosimann SC, Uwiera RRE and Abbott DW (2017) Determining the Localization of Carbohydrate Active Enzymes Within Gram-Negative Bacteria. Methods Mol. Biol, 1588, 199–208. [DOI] [PubMed] [Google Scholar]
- 14.Fontaine F, Fuchs RT and Storz G (2011) Membrane localization of small proteins in Escherichia coli. J. Biol. Chem, 286, 32464–32474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benhalevy D, Biran I, Bochkareva ES, Sorek R and Bibi E (2017) Evidence for a cytoplasmic pool of ribosome-free mRNAs encoding inner membrane proteins in Escherichia coli. PLoS ONE, 12, e0183862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milne JLS and Subramaniam S (2009) Cryo-electron tomography of bacteria: progress, challenges and future prospects. Nat. Rev. Microbiol, 7, 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty K, Veetil AT, Jaffrey SR and Krishnan Y (2016) Nucleic Acid-Based Nanodevices in Biological Imaging. Annu. Rev. Biochem, 85, 349–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Gijtenbeek LA and Kok J (2017) Illuminating messengers: an update and outlook on RNA visualization in bacteria. Front. Microbiol, 8, 1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Femino AM, Fay FS, Fogarty K and Singer RH (1998) Visualization of single RNA transcripts in situ. Science, 280, 585–590. [DOI] [PubMed] [Google Scholar]
- 20.So L-H, Ghosh A, Zong C, Sepúlveda LA, Segev R and Golding I (2011) General properties of transcriptional time series in Escherichia coli. Nat. Genet, 43, 554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A and Tyagi S (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods, 5, 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raj A, Peskin CS, Tranchina D, Vargas DY and Tyagi S (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biol, 4, e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuert G, Munsky B, Tan RZ, Teytelman L, Khammash M and van Oudenaarden A (2013) Systematic identification of signal-activated stochastic gene regulation. Science, 339, 584–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones DL, Brewster RC and Phillips R (2014) Promoter architecture dictates cell-to-cell variability in gene expression. Science, 346, 1533–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnell U, Dijk F, Sjollema KA and Giepmans BNG (2012) Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods, 9, 152–158. [DOI] [PubMed] [Google Scholar]
- 26.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH and Long RM (1998) Localization of ASH1 mRNA particles in living yeast. Mol. Cell, 2, 437–445. [DOI] [PubMed] [Google Scholar]
- 27.Lange S, Katayama Y, Schmid M, Burkacky O, Bräuchle C, Lamb DC and Jansen R-P (2008) Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic, 9, 1256–1267. [DOI] [PubMed] [Google Scholar]
- 28.Hocine S, Raymond P, Zenklusen D, Chao JA and Singer RH (2013) Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat. Methods, 10, 119–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller LW, Cai Y, Sheetz MP and Cornish VW (2005) In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat. Methods, 2, 255–257. [DOI] [PubMed] [Google Scholar]
- 30.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H and Johnsson K (2003) A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol, 21, 86–89. [DOI] [PubMed] [Google Scholar]
- 31.Sun X, Zhang A, Baker B, Sun L, Howard A, Buswell J, Maurel D, Masharina A, Johnsson K, Noren CJ, et al. (2011) Development of SNAP-tag fluorogenic probes for wash-free fluorescence imaging. Chembiochem, 12, 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrocci TJ and Hoskins AA (2014) Imaging of RNAs in live cells with spectrally diverse small molecule fluorophores. Analyst, 139, 44–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valencia-Burton M, McCullough RM, Cantor CR and Broude NE (2007) RNA visualization in live bacterial cells using fluorescent protein complementation. Nat. Methods, 4, 421–427. [DOI] [PubMed] [Google Scholar]
- 34.Valencia-Burton M, Shah A, Sutin J, Borogovac A, McCullough RM, Cantor CR, Meller A and Broude NE (2009) Spatiotemporal patterns and transcription kinetics of induced RNA in single bacterial cells. Proc Natl Acad Sci USA, 106, 16399–16404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozawa T, Natori Y, Sato M and Umezawa Y (2007) Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat. Methods, 4, 413–419. [DOI] [PubMed] [Google Scholar]
- 36.Wu B, Chen J and Singer RH (2014) Background free imaging of single mRNAs in live cells using split fluorescent proteins. Sci. Rep, 4, 3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Moffitt JR, Dempsey GT, Xie XS and Zhuang X (2014) Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc Natl Acad Sci USA, 111, 8452–8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landgraf D, Okumus B, Chien P, Baker TA and Paulsson J (2012) Segregation of molecules at cell division reveals native protein localization. Nat. Methods, 9, 480–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeCuyer KA, Behlen LS and Uhlenbeck OC (1995) Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry, 34, 10600–10606. [DOI] [PubMed] [Google Scholar]
- 40.Golding I, Paulsson J, Zawilski SM and Cox EC (2005) Real-time kinetics of gene activity in individual bacteria. Cell, 123, 1025–1036. [DOI] [PubMed] [Google Scholar]
- 41.Haimovich G, Zabezhinsky D, Haas B, Slobodin B, Purushothaman P, Fan L, Levin JZ, Nusbaum C and Gerst JE (2016) Use of the MS2 aptamer and coat protein for RNA localization in yeast: A response to “MS2 coat proteins bound to yeast mRNAs block 5” to 3’ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system”.” RNA, 22, 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia JF and Parker R (2015) MS2 coat proteins bound to yeast mRNAs block 5’ to 3’ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system. RNA, 21, 1393–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinrich S, Sidler CL, Azzalin CM and Weis K (2017) Stem-loop RNA labeling can affect nuclear and cytoplasmic mRNA processing. RNA, 23, 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia JF and Parker R (2016) Ubiquitous accumulation of 3’ mRNA decay fragments in Saccharomyces cerevisiae mRNAs with chromosomally integrated MS2 arrays. RNA, 22, 657–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tutucci E, Vera M, Biswas J, Garcia J, Parker R and Singer RH (2018) An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods, 15, 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You M and Jaffrey SR (2015) Structure and mechanism of RNA mimics of green fluorescent protein. Annu. Rev. Biophys, 44, 187–206. [DOI] [PubMed] [Google Scholar]
- 47.Ouellet J (2016) RNA Fluorescence with Light-Up Aptamers. Front. Chem, 4, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolgosheina EV and Unrau PJ (2016) Fluorophore-binding RNA aptamers and their applications. Wiley Interdiscip. Rev. RNA, 7, 843–851. [DOI] [PubMed] [Google Scholar]
- 49.Babendure JR, Adams SR and Tsien RY (2003) Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc, 125, 14716–14717. [DOI] [PubMed] [Google Scholar]
- 50.Constantin TP, Silva GL, Robertson KL, Hamilton TP, Fague K, Waggoner AS and Armitage BA (2008) Synthesis of new fluorogenic cyanine dyes and incorporation into RNA fluoromodules. Org. Lett, 10, 1561–1564. [DOI] [PubMed] [Google Scholar]
- 51.Sando S, Narita A, Hayami M and Aoyama Y (2008) Transcription monitoring using fused RNA with a dye-binding light-up aptamer as a tag: a blue fluorescent RNA. Chem Commun (Camb), 10.1039/b808449a. [DOI] [PubMed] [Google Scholar]
- 52.Paige JS, Wu KY and Jaffrey SR (2011) RNA mimics of green fluorescent protein. Science, 333, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strack RL, Disney MD and Jaffrey SR (2013) A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods, 10, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dolgosheina EV, Jeng SCY, Panchapakesan SSS, Cojocaru R, Chen PSK, Wilson PD, Hawkins N, Wiggins PA and Unrau PJ (2014) RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol, 9, 2412–2420. [DOI] [PubMed] [Google Scholar]
- 55.Filonov GS, Moon JD, Svensen N and Jaffrey SR (2014) Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc, 136, 16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Filonov GS and Jaffrey SR (2016) RNA Imaging with Dimeric Broccoli in Live Bacterial and Mammalian Cells. Curr. Protoc. Chem. Biol, 8, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warner KD, Sjekloća L, Song W, Filonov GS, Jaffrey SR and Ferré-D’Amaré AR (2017) A homodimer interface without base pairs in an RNA mimic of red fluorescent protein. Nat. Chem. Biol, 13, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Fei J, Leslie BJ, Han KY, Kuhlman TE and Ha T (2015) Tandem Spinach Array for mRNA Imaging in Living Bacterial Cells. Sci. Rep, 5, 17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sunbul M and Jäschke A (2013) Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer. Angew Chem Int Ed Engl, 52, 13401–13404. [DOI] [PubMed] [Google Scholar]
- 60.Arora A, Sunbul M and Jäschke A (2015) Dual-colour imaging of RNAs using quencher- and fluorophore-binding aptamers. Nucleic Acids Res, 43, e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J and Hess HF (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science, 313, 1642–1645. [DOI] [PubMed] [Google Scholar]
- 62.Rust MJ, Bates M and Zhuang X (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods, 3, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang B, Wang W, Bates M and Zhuang X (2008) Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science, 319, 810–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T and Jacobs-Wagner C (2010) Spatial organization of the flow of genetic information in bacteria. Nature, 466, 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S and Amster-Choder O (2011) Translation-independent localization of mRNA in E. coli. Science, 331, 1081–1084. [DOI] [PubMed] [Google Scholar]
- 66.dos Santos VT, Bisson-Filho AW and Gueiros-Filho FJ (2012) DivIVA-mediated polar localization of ComN, a posttranscriptional regulator of Bacillus subtilis. J. Bacteriol, 194, 3661–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Golding I and Cox EC (2004) RNA dynamics in live Escherichia coli cells. Proc Natl Acad Sci USA, 101, 11310–11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toran P, Smolina I, Driscoll H, Ding F, Sun Y, Cantor CR and Broude NE (2014) Labeling native bacterial RNA in live cells. Cell Res, 24, 894–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pilhofer M, Pavlekovic M, Lee NM, Ludwig W and Schleifer K-H (2009) Fluorescence in situ hybridization for intracellular localization of nifH mRNA. Syst. Appl. Microbiol, 32, 186–192. [DOI] [PubMed] [Google Scholar]
- 70.Moffitt JR, Pandey S, Boettiger AN, Wang S and Zhuang X (2016) Spatial organization shapes the turnover of a bacterial transcriptome. elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dugar G, Svensson SL, Bischler T, Wäldchen S, Reinhardt R, Sauer M and Sharma CM (2016) The CsrA-FliW network controls polar localization of the dual-function flagellin mRNA in Campylobacter jejuni. Nat. Commun, 7, 11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorg JA, Miller NC and Schneewind O (2005) Substrate recognition of type III secretion machines--testing the RNA signal hypothesis. Cell. Microbiol, 7, 1217–1225. [DOI] [PubMed] [Google Scholar]
- 73.Anderson DM and Schneewind O (1997) A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science, 278, 1140–1143. [DOI] [PubMed] [Google Scholar]
- 74.Singer HM, Erhardt M and Hughes KT (2014) Comparative analysis of the secretion capability of early and late flagellar type III secretion substrates. Mol. Microbiol, 93, 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Majander K, Anton L, Antikainen J, Lång H, Brummer M, Korhonen TK and Westerlund-Wikström B (2005) Extracellular secretion of polypeptides using a modified Escherichia coli flagellar secretion apparatus. Nat. Biotechnol, 23, 475–481. [DOI] [PubMed] [Google Scholar]
- 76.Driessen AJM and Nouwen N (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem, 77, 643–667. [DOI] [PubMed] [Google Scholar]
- 77.Korkmazhan E, Teimouri H, Peterman N and Levine E (2017) Dynamics of translation can determine the spatial organization of membrane-bound proteins and their mRNA. Proc Natl Acad Sci USA, 114, 13424–13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anderson DM and Schneewind O (1999) Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol. Microbiol, 31, 1139–1148. [DOI] [PubMed] [Google Scholar]
- 79.Azam TA, Hiraga S and Ishihama A (2000) Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells, 5, 613–626. [DOI] [PubMed] [Google Scholar]
- 80.Müller A, Beeby M, McDowall AW, Chow J, Jensen GJ and Clemons WM (2014) Ultrastructure and complex polar architecture of the human pathogen Campylobacter jejuni. Microbiologyopen, 3, 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanamrad A, Persson F, Lundius EG, Fange D, Gynnå AH and Elf J (2014) Single-particle tracking reveals that free ribosomal subunits are not excluded from the Escherichia coli nucleoid. Proc Natl Acad Sci USA, 111, 11413–11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewis PJ, Thaker SD and Errington J (2000) Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J, 19, 710–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ortiz JO, Förster F, Kürner J, Linaroudis AA and Baumeister W (2006) Mapping 70S ribosomes in intact cells by cryoelectron tomography and pattern recognition. J. Struct. Biol, 156, 334–341. [DOI] [PubMed] [Google Scholar]
- 84.Bakshi S, Choi H and Weisshaar JC (2015) The spatial biology of transcription and translation in rapidly growing Escherichia coli. Front. Microbiol, 6, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mascarenhas J, Weber MH and Graumann PL (2001) Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep, 2, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keiler KC (2011) Localization of the bacterial RNA infrastructure. Adv. Exp. Med. Biol, 722, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stracy M, Lesterlin C, Garza de Leon F, Uphoff S, Zawadzki P and Kapanidis AN (2015) Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc Natl Acad Sci USA, 112, E4390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chai Q, Singh B, Peisker K, Metzendorf N, Ge X, Dasgupta S and Sanyal S (2014) Organization of ribosomes and nucleoids in Escherichia coli cells during growth and in quiescence. J. Biol. Chem, 289, 11342–11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Herskovits AA and Bibi E (2000) Association of Escherichia coli ribosomes with the inner membrane requires the signal recognition particle receptor but is independent of the signal recognition particle. Proc Natl Acad Sci USA, 97, 4621–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bayas CA, Wang J, Lee MK, Schrader JM, Shapiro L and Moerner WE (2018) Spatial organization and dynamics of RNase E and ribosomes in Caulobacter crescentus. Proc Natl Acad Sci USA, 115, E3712–E3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Montero Llopis P, Sliusarenko O, Heinritz J and Jacobs-Wagner C (2012) In vivo biochemistry in bacterial cells using FRAP: insight into the translation cycle. Biophys. J, 103, 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Evguenieva-Hackenberg E, Roppelt V, Lassek C and Klug G (2011) Subcellular localization of RNA degrading proteins and protein complexes in prokaryotes. RNA Biol, 8, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Redder P (2016) How does sub-cellular localization affect the fate of bacterial mRNA? Curr. Genet, 62, 687–690. [DOI] [PubMed] [Google Scholar]
- 94.Mackie GA (2013) RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol, 11, 45–57. [DOI] [PubMed] [Google Scholar]
- 95.Carpousis AJ (2007) The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol, 61, 71–87. [DOI] [PubMed] [Google Scholar]
- 96.Lehnik-Habrink M, Lewis RJ, Mäder U and Stülke J (2012) RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol. Microbiol, 84, 1005–1017. [DOI] [PubMed] [Google Scholar]
- 97.Taghbalout A and Rothfield L (2007) RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc Natl Acad Sci USA, 104, 1667–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khemici V, Poljak L, Luisi BF and Carpousis AJ (2008) The RNase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol, 70, 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taghbalout A, Yang Q and Arluison V (2014) The Escherichia coli RNA processing and degradation machinery is compartmentalized within an organized cellular network. Biochem. J, 458, 11–22. [DOI] [PubMed] [Google Scholar]
- 100.Strahl H, Turlan C, Khalid S, Bond PJ, Kebalo J-M, Peyron P, Poljak L, Bouvier M, Hamoen L, Luisi BF, et al. (2015) Membrane recognition and dynamics of the RNA degradosome. PLoS Genet, 11, e1004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Murashko ON and Lin-Chao S (2017) Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. Proc Natl Acad Sci USA, 114, E8025–E8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cascante-Estepa N, Gunka K and Stülke J (2016) Localization of Components of the RNA-Degrading Machine in Bacillus subtilis. Front. Microbiol, 7, 1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Koch G, Wermser C, Acosta IC, Kricks L, Stengel ST, Yepes A and Lopez D (2017) Attenuating Staphylococcus aureus Virulence by Targeting Flotillin Protein Scaffold Activity. Cell Chem. Biol, 24, 845–857.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Russell JH and Keiler KC (2009) Subcellular localization of a bacterial regulatory RNA. Proc Natl Acad Sci USA, 106, 16405–16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fei J, Singh D, Zhang Q, Park S, Balasubramanian D, Golding I, Vanderpool CK and Ha T (2015) RNA biochemistry. Determination of in vivo target search kinetics of regulatory noncoding RNA. Science, 347, 1371–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sheng H, Stauffer WT, Hussein R, Lin C and Lim HN (2017) Nucleoid and cytoplasmic localization of small RNAs in Escherichia coli. Nucleic Acids Res, 45, 2919–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wadler CS and Vanderpool CK (2007) A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci USA, 104, 20454–20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wadler CS and Vanderpool CK (2009) Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res, 37, 5477–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wagner EGH and Romby P (2015) Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet, 90, 133–208. [DOI] [PubMed] [Google Scholar]
- 110.Teimouri H, Korkmazhan E, Stavans J and Levine E (2017) Sub-cellular mRNA localization modulates the regulation of gene expression by small RNAs in bacteria. Phys. Biol, 14. [DOI] [PubMed] [Google Scholar]
- 111.Kawamoto H, Morita T, Shimizu A, Inada T and Aiba H (2005) Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev, 19, 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Updegrove TB, Zhang A and Storz G (2016) Hfq: the flexible RNA matchmaker. Curr. Opin. Microbiol, 30, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Diestra E, Cayrol B, Arluison V and Risco C (2009) Cellular electron microscopy imaging reveals the localization of the Hfq protein close to the bacterial membrane. PLoS ONE, 4, e8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Malabirade A, Jiang K, Kubiak K, Diaz-Mendoza A, Liu F, van Kan JA, Berret J-F, Arluison V and van der Maarel JRC (2017) Compaction and condensation of DNA mediated by the C-terminal domain of Hfq. Nucleic Acids Res, 45, 7299–7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Malabirade A, Morgado-Brajones J, Trépout S, Wien F, Marquez I, Seguin J, Marco S, Velez M and Arluison V (2017) Membrane association of the bacterial riboregulator Hfq and functional perspectives. Sci. Rep, 7, 10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Persson F, Lindén M, Unoson C and Elf J (2013) Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat. Methods, 10, 265–269. [DOI] [PubMed] [Google Scholar]
- 117.Choi HMT, Beck VA and Pierce NA (2014) Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano, 8, 4284–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shah S, Lubeck E, Zhou W and Cai L (2017) Editorial note to: in situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron, 94, 745–746. [DOI] [PubMed] [Google Scholar]
- 119.Qian X and Lloyd RV (2003) Recent developments in signal amplification methods for in situ hybridization. Diagn. Mol. Pathol, 12, 1–13. [DOI] [PubMed] [Google Scholar]
- 120.Bagasra O (2007) Protocols for the in situ PCR-amplification and detection of mRNA and DNA sequences. Nat. Protoc, 2, 2782–2795. [DOI] [PubMed] [Google Scholar]
- 121.Larsson C, Koch J, Nygren A, Janssen G, Raap AK, Landegren U and Nilsson M (2004) In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat. Methods, 1, 227–232. [DOI] [PubMed] [Google Scholar]
- 122.Larsson C, Grundberg I, Söderberg O and Nilsson M (2010) In situ detection and genotyping of individual mRNA molecules. Nat. Methods, 7, 395–397. [DOI] [PubMed] [Google Scholar]
- 123.Lubeck E and Cai L (2012) Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods, 9, 743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M and Cai L (2014) Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods, 11, 360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jungmann R, Avendaño MS, Woehrstein JB, Dai M, Shih WM and Yin P (2014) Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods, 11, 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen KH, Boettiger AN, Moffitt JR, Wang S and Zhuang X (2015) RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science, 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP and Zhuang X (2016) High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci USA, 113, 11046–11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Takei Y, Shah S, Harvey S, Qi LS and Cai L (2017) Multiplexed dynamic imaging of genomic loci by combined CRISPR imaging and DNA sequential FISH. Biophys. J, 112, 1773–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guan J, Liu H, Shi X, Feng S and Huang B (2017) Tracking multiple genomic elements using correlative CRISPR imaging and sequential DNA FISH. Biophys. J, 112, 1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, et al. (2013) Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell, 155, 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA and Yeo GW (2016) Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell, 165, 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. (2017) RNA targeting with CRISPR-Cas13. Nature, 550, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M and Doudna JA (2014) Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature, 516, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gross GG, Junge JA, Mora RJ, Kwon H-B, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GCR, McGee AW, Sabatini BL, et al. (2013) Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron, 78, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Halstead JM, Lionnet T, Wilbertz JH, Wippich F, Ephrussi A, Singer RH and Chao JA (2015) Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science, 347, 1367–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS and Vale RD (2014) A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell, 159, 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yan X, Hoek TA, Vale RD and Tanenbaum ME (2016) Dynamics of Translation of Single mRNA Molecules In Vivo. Cell, 165, 976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu B, Eliscovich C, Yoon YJ and Singer RH (2016) Translation dynamics of single mRNAs in live cells and neurons. Science, 352, 1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang C, Han B, Zhou R and Zhuang X (2016) Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells. Cell, 165, 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Morisaki T, Lyon K, DeLuca KF, DeLuca JG, English BP, Zhang Z, Lavis LD, Grimm JB, Viswanathan S, Looger LL, et al. (2016) Real-time quantification of single RNA translation dynamics in living cells. Science, 352, 1425–1429. [DOI] [PubMed] [Google Scholar]
- 141.Horvathova I, Voigt F, Kotrys AV, Zhan Y, Artus-Revel CG, Eglinger J, Stadler MB, Giorgetti L and Chao JA (2017) The Dynamics of mRNA Turnover Revealed by Single-Molecule Imaging in Single Cells. Mol. Cell, 68, 615–625.e9. [DOI] [PubMed] [Google Scholar]
- 142.Kieft JS, Rabe JL and Chapman EG (2015) New hypotheses derived from the structure of a flaviviral Xrn1-resistant RNA: Conservation, folding, and host adaptation. RNA Biol., 12, 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang K, Doyle F, Wurz ZE, Tenenbaum SA, Hammond RK, Caplan JL and Meyers BC (2017) FASTmiR: an RNA-based sensor for in vitro quantification and live-cell localization of small RNAs. Nucleic Acids Res, 45, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang J-B, Marblestone A, Church GM, et al. (2016) Nanoscale imaging of RNA with expansion microscopy. Nat. Methods, 13, 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chang J-B, Chen F, Yoon Y-G, Jung EE, Babcock H, Kang JS, Asano S, Suk H-J, Pak N, Tillberg PW, et al. (2017) Iterative expansion microscopy. Nat. Methods, 14, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang YS, Chang J-B, Alvarez MM, Trujillo-de Santiago G, Aleman J, Batzaya B, Krishnadoss V, Ramanujam AA, Kazemzadeh-Narbat M, Chen F, et al. (2016) Hybrid Microscopy: Enabling Inexpensive High-Performance Imaging through Combined Physical and Optical Magnifications. Sci. Rep, 6, 22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ramamurthi KS and Schneewind O (2005) A synonymous mutation in Yersinia enterocolitica yopE affects the function of the YopE type III secretion signal. J. Bacteriol, 187, 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Prilusky J and Bibi E (2009) Studying membrane proteins through the eyes of the genetic code revealed a strong uracil bias in their coding mRNAs. Proc Natl Acad Sci USA, 106, 6662–6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Storz G, Wolf YI and Ramamurthi KS (2014) Small proteins can no longer be ignored. Annu. Rev. Biochem, 83, 753–777. [DOI] [PMC free article] [PubMed] [Google Scholar]